ABSTRACT
Objective
The aim of the study was to assess the evidence on miRNAs as biomarkers for the diagnosis of endometriosis, as well as to provide insights into the challenges and strategies associated with the use of these molecules as accessible tools in clinical practice.
Methods
Systematic review conducted on PubMed®, Latin American and Caribbean Health Sciences Literature (LILACS), MEDLINE® and Web of Science databases using the search terms endometriosis (all fields) AND miRNA (all fields), evaluating all publication up to May 2019.
Results
Most miRNAs found to be dysregulated in this study were harvested from tissue samples, which precludes their use as a non-invasive diagnostic test. However, differential expression of 62 miRNAs was reported in samples that may be used for non-invasive diagnosis of endometriosis, such as blood, serum and plasma.
Conclusion
Despite the identification of several candidates, studies are investigatory in nature and have been conducted with small number of samples. Also, no particular miRNA has been validated for diagnostic purposes so far. Studies based primarily on biological samples and applicable to translational research are warranted. Large databases comprising information on sample type and the use of saliva and vaginal fluid for miRNAs identification may prove essential to overcome current barriers to diagnosis of endometriosis.
Keywords: Biomarkers, Saliva, Serum, Vaginal fluid, Body fluids banks, MicroRNAs, Endometriosis/diagnosis
INTRODUCTION
Endometriosis is a common disease that affects up to 10% of women of reproductive age(1,2) and is characterized by the presence of endometrial cells outside the uterine cavity. The disease has been the focus of many studies, however, the diagnosis is still very difficult. Clinical presentation varies widely, ranging from asymptomatic to severe, and no diagnostic biomarkers have been approved for routine clinical diagnosis of endometriosis to date.(1,3)
Diagnostic imaging tests such as pelvic ultrasonography and magnetic resonance have been used, especially in deep endometriosis. However, examiner expertise has a strong impact on imaging findings,(4-7) which ultimately makes the diagnosis difficult. In cases with no positive imaging findings, a final diagnosis of superficial endometriosis can only be made through histological analysis of the lesion, usually in samples obtained by laparoscopic surgery.(8,9) However, this procedure is invasive and requires general anesthesia.
The complexity of the disease, combined with the lack of precise and less invasive diagnostic methods, contributes to delayed diagnosis, which can take up to 11 years.(5,10,11) Therefore, there is great demand for accurate and less invasive diagnostic tests for endometriosis.(12-16)
Different research groups have investigated the role of miRNAs (microRNAs or miR) in the regulation of known genes, given their association with processes involved in disease pathogenesis and progression. miRNAs are a class of small endogenous, non-coding RNA molecules involved in post-transcriptional regulation of gene expression.(17) These small molecules have also been found in peripheral blood and may therefore be potential diagnostic biomarkers for endometriosis.(18,19)
This literature search was conducted to determine how close miRNAs are to being used as biomarkers for endometriosis. Findings of this review are expected to guide the next steps towards overcoming challenges associated with the use of miRNAs in clinical practice.
OBJECTIVE
To determine which miRNAs are applicable to the diagnosis of endometriosis and to outline the challenges and strategies involved in the use of these molecules as accessible diagnostic tools in clinical settings.
METHODS
To identify research articles addressing associations between endometriosis and miRNA, a search was conducted in PubMed®, Latin American and Caribbean Health Sciences Literature (LILACS), MEDLINE® and Web of Science databases using the search terms endometriosis (all fields) AND miRNA (all fields).
All publications listed up to May 2019 (automatically selected) were manually curated, and only those involving miRNA expression patterns, validated by polymerase chain reaction (PCR) in clinical samples of endometriosis, were discussed in this review. Articles published in languages other than English or based on cell culture, retracted articles and articles published in conference proceedings or inaccessible were excluded. Reports listed in more than one database were included only once in the pool of publications.
This study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement for systematic reviews adopted by Hospital Israelita Albert Einstein (HIAE), located in São Paulo (SP), Brazil. Data were extracted in duplicate and independently by two different investigators, then compared for confirmation. miRNAs and their respective expression levels in different types of samples and patient populations were examined. Studies were also analyzed according to year and country of publication.
RESULTS
Overview of publications on miRNA and endometriosis
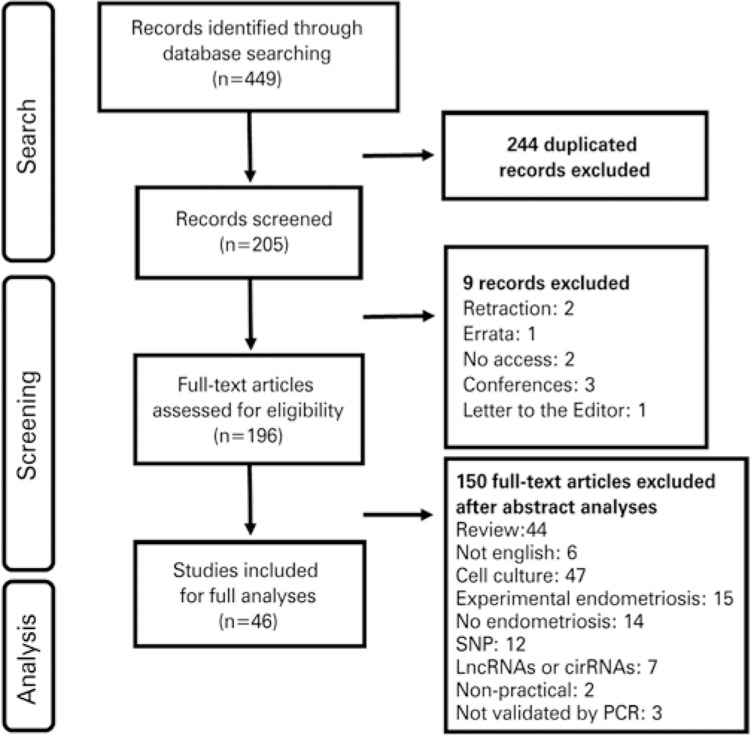
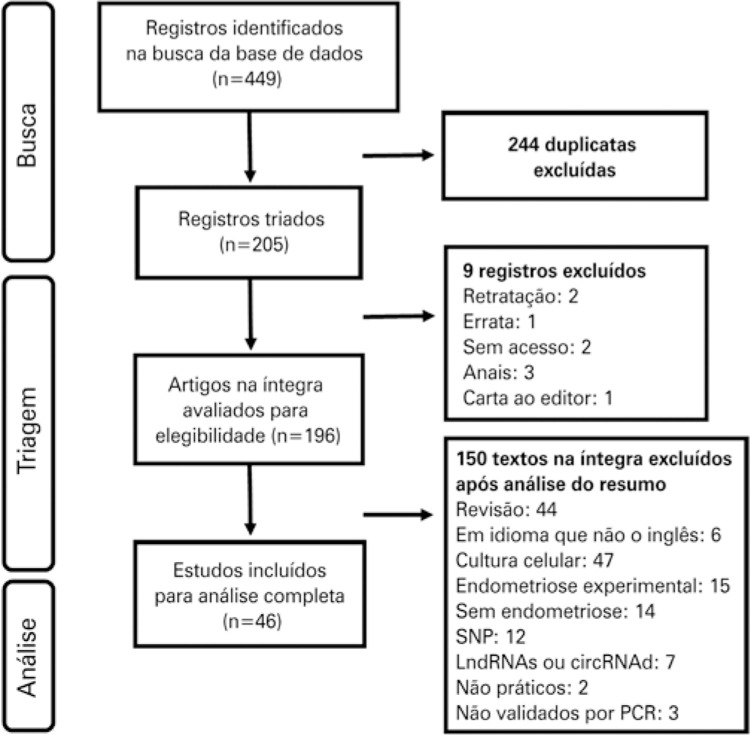
A total of 449 research articles addressing associations between endometriosis and miRNA were found in databases selected for this review. Most (185) were retrieved from PubMed®, followed by LILACS and MEDLINE® (158) and Web of Science (106). Out of this publication pool, 46 matched final selection criteria and were selected for further discussion in this review (Figure 1).
Figure 1. Summarized results of database screening for publications addressing associations between endometriosis and miRNAs.
SNP: single-nucleotide polymorphisms; LncRNAs: long non-coding ribonucleic acids; circRNAs: circular ribonucleic acids; PCR: polymerase chain reaction.
The number of publications investigating dysregulated miRNAs in women with endometriosis increased sharply since 2009, with approximately half of selected articles (23) published in the last 3 years. China and the United States were the countries with the largest number of publications (21 and 9 articles, respectively).
Within this pool of 46 studies, 43 investigated miRNAs found to be dysregulated in the ectopic (EC) relative to the eutopic endometrium of control patients (EN), 25 were detected in the EC relative to the eutopic endometrium (EU) of women with endometriosis and 23 were detected in the EU compared to the EN group. Furthermore, 27 were detected in the serum, 18 in the plasma, 30 in the blood and six in the peritoneal fluid of women with endometriosis compared to the Control Group. Blood seems to be the most widely investigated type of sample regarding potential applicability to non-invasive diagnosis. The summary of dysregulated miRNA found in selected published articles are listed in table 1A-1G .
Table 1A. miRNAs differentially expressed in eutopic endometrium of endometriosis patients compared with eutopic endometrium of control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| miR-126 | DR | 31 | 27 | Liu et al.(20) |
| miR-1281 | UR | 38 | 38 | Yang et al.(21) |
| miR-142-5p | UR | 38 | 38 | Yang et al.(21) |
| miR-145 | UR | 11 | 22 | Zheng et al.(22) |
| miR-146a-5p | UR | 38 | 38 | Yang et al.(21) |
| miR-183-5p | DR | N/A | N/A | Shi et al.(23) |
| miR-199a | DR | 12 | 12 | Dai et al.(24) |
| miR-202-3p | DR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-204 | DR | 38 | 9 | Haikalis et al.(26) |
| miR-29c | DR | 20 | 10 | Long et al.(27) |
| miR-30d-5p | UR | 21 | 25 | Laudanski et al.(28) |
| miR-3152-5p | UR | 21 | 25 | Laudanski et al.(28) |
| miR-34b | DR | 4 | 3 | Burney et al.(29) |
| miR-34c-5p | DR | 4 | 3 | Burney et al.(29) |
| miR-424-5p | DR | 51 | 32 | Braza-Boils et al.(25) |
| miR-4634 | UR | 38 | 38 | Yang et al.(21) |
| miR-483-5p | DR | 21 | 25 | Laudanski et al.(30) |
| miR-5187-3p | UR | 21 | 25 | Laudansk et al.(28) |
| miR-543 | DR | 38 | 38 | Yang et al.(21) |
| miR-556-3p | DR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-629* | DR | 21 | 25 | Laudanski et al.(30) |
| miR-9 | DR | 4 | 3 | Burney et al.(29) |
| miR-940 | UR | 38 | 38 | Yang et al.(21) |
miRNA: microRNA; DR: downregulated; UR: upregulated; N/A: not available.
Table 1B. miRNAs differentially expressed in ectopic endometrium of endometriosis patients and eutopic endometrium of control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| let-7g | UR | N/A | N/A | Wright et al.(31) |
| miR-100 | UR | N/A | N/A | Wright et al.(31) |
| miR-1304-3p | UR | 14 | 10 | Xu et al.(32) |
| miR-133a-3p | UR | 33 | 17 | Braicu et al.(33) |
| miR-138 | UR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-141 | UR | 22 | 24 | Saare et al.(34) |
| miR-143 | UR | 11 | 22 | Zheng et al.(22) |
| miR-145 | UR | 11 | 22 | Zheng et al.(22) |
| miR-148a | UR | N/A | N/A | Wright et al.(31) |
| miR-183-5p | DR | N/A | N/A | Shi et al.(23) |
| miR-191 | UR | 12 | 12 | Dong et al.(35) |
| miR-199a | DR | 12 | 12 | Dai et al.(24) |
| miR-200a | UR | 22 | 24 | Saare et al.(34) |
| miR-200b | UR | 22 | 24 | Saare et al.(34) |
| miR-200c | DR | 27 | 12 | Liang et al.(36) |
| miR-202-3p | UR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-205-5p | DR | 14 | 10 | Xu et al.(32) |
| miR-20a | UR | 40 | 20 | Zhao et al.(37) |
| miR-21-3p | UR | 7 | 7 | Qi et al.(38) |
| miR-223-3p | UR | 7 | 7 | Qi et al.(38) |
| miR-29a | UR | N/A | N/A | Wright et al.(31) |
| miR-29c | DR | 20 | 10 | Long et al.(27) |
| miR-29c | UR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-29c | UR | 15 | 11 | Joshi et al.(39) |
| miR-325 | UR | 33 | 17 | Braicu et al.(33) |
| miR-33b | DR | 20 | 15 | Yang et al.(40) |
| miR-34c | UR | 22 | 24 | Saare et al.(34) |
| miR-3663-3p | UR | 7 | 7 | Qi et al.(38) |
| miR-3684 | UR | 14 | 10 | Xu et al.(32) |
| miR-373-3p | UR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-3935 | DR | 14 | 10 | Xu et al.(32) |
| miR-411-5p | UR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-4427 | DR | 14 | 10 | Xu et al.(32) |
| miR-449a | UR | 22 | 24 | Saare et al.(34) |
| miR-450a-5p | DR | 7 | 7 | Qi et al.(38) |
| miR-451 | UR | 30 | 0 | Graham et al.(41) |
| miR-4683 | UR | 14 | 10 | Xu et al.(32) |
| miR-492 | UR | 33 | 17 | Braicu et al.(33) |
| miR-494-5p | UR | 14 | 10 | Xu et al.(32) |
| miR-503-5p | DR | 7 | 7 | Qi et al.(38) |
| miR-520e | UR | 33 | 17 | Braicu et al.(33) |
| miR-544b | UR | 14 | 10 | Xu et al.(32) |
| miR-5481 | DR | N/A | N/A | Wright et al.(31) |
| miR-652-5p | DR | 14 | 10 | Xu et al.(32) |
| miR-6747-3p | UR | 14 | 10 | Xu et al.(32) |
miRNA: microRNA; DR: downregulated; N/A: not available; UR: upregulated.
Table 1C. miRNAs differentially expressed in the ectopic and eutopic endometrium of endometriosis patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| miR-106a-5p | DR | 22 | 0 | Zhao et al.(42) |
| miR-106b-5p | UR | 32 | 19 | Yang et al.(43) |
| miR-10a | DR | 38 | 38 | Haikalis et al.(26) |
| miR-125a | UR | 58 | 38 | Ramón et al.(44) |
| miR-126 | DR | 31 | 27 | Liu et al.(20) |
| miR-126 | UR | 8 | N/A | Ohlsson Teague et al.(45) |
| miR-141 | DR | 8 | N/A | Ohlsson Teague et al.(45) |
| miR-145 | UR | 8 | N/A | Ohlsson Teague et al.(45) |
| miR-145-5p | UR | 32 | 19 | Yang et al.(43) |
| miR-146a-5p | DR | 32 | 19 | Yang et al.(43) |
| miR-15a-5p | DR | 32 | 19 | Yang et al.(43) |
| miR-16-5p | UR | 32 | 19 | Yang et al.(43) |
| miR-182 | DR | 16 | N/A | Filigheddu et al.(46) |
| miR-182-5p | DR | 22 | 0 | Zhao et al.(42) |
| miR-19b-1-5p | DR | 32 | 19 | Yang et al.(43) |
| miR-200a | DR | 16 | N/A | Filigheddu et al.(46) |
| miR-200a-3p | DR | 22 | 0 | Zhao et al.(42) |
| miR-200b | DR | 16 | N/A | Filigheddu et al.(46) |
| miR-200b | DR | 8 | N/A | Ohlsson Teague et al.(45) |
| miR-200b | DR | 32 | 19 | Yang et al.(43) |
| miR-200c | DR | 16 | N/A | Filigheddu et al.(46) |
| miR-200c | DR | 32 | 19 | Yang et al.(43) |
| miR-202 | UR | 16 | N/A | Filigheddu et al.(46) |
| miR-21 | DR | 38 | 38 | Haikalis et al.(26) |
| miR-222 | UR | 58 | 38 | Ramón et al.(44) |
| miR-34c | DR | 22 | 0 | Zhao et al.(42) |
| miR-424 | DR | 8 | N/A | Ohlsson Teague et al.(45) |
| miR-424 | DR | 38 | 38 | Haikalis et al.(26) |
| miR-449b | DR | 51 | 32 | Braza-Boïls et al.(25) |
| miR-449b | DR | 22 | 0 | Zhao et al.(42) |
| miR-451a | UR | 41 | 40 | Nothnick et al.(47) |
| miR-615 | UR | 22 | 0 | Zhao et al.(42) |
| miR-9 | DR | 38 | 38 | Haikalis et al.(26) |
| miR-99a | UR | 8 | N/A | Ohlsson Teague et al.(45) |
miRNA: microRNA; DR: downregulated; UR: upregulated; N/A: not available.
Table 1D. miRNAs differentially expressed in the serum of endometriosis and control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| let-7b | DR | 24 | 24 | Cho et al.(48) |
| let-7b-5p | DR | 20 | 26 | Nematian et al.(49) |
| miR-122 | UR | 60 | 25 | Wang et al.(19) |
| miR-122 | UR | 45 | 35 | Maged et al.(50) |
| miR-125b-5p | UR | 24 | 24 | Cosar et al.(51) |
| miR-125b | UR | 20 | 26 | Nematian et al.(49) |
| miR-127-3p | DR | 30 | 20 | Wang et al.(52) |
| miR-135a | DR | 24 | 24 | Cosar et al.(51) |
| miR-141 | DR | 60 | 25 | Wang et al.(19) |
| miR-143-3p | UR | 24 | 24 | Cosar et al.(51) |
| miR-145 | DR | 60 | 25 | Wang et al.(19) |
| miR-145-5p | UR | 24 | 24 | Cosar et al.(51) |
| miR-150-5p | UR | 24 | 24 | Cosar et al.(51) |
| miR-15b-5p | DR | 30 | 20 | Wang et al.(52) |
| miR-17 | DR | 80 | 60 | Wang et al.(53) |
| miR-185-5p | UR | 30 | 20 | Wang et al.(52) |
| miR-18a-5p | UR | 24 | 24 | Cosar et al.(51) |
| miR-191 | UR | 12 | 12 | Dong et al.(35) |
| miR-199a | UR | 60 | 25 | Wang et al.(19) |
| miR-199a | UR | 45 | 35 | Maged et al.(50) |
| miR-199a-5p | DR | 40 | 25 | Hsu et al.(54) |
| miR-20a-5p | DR | 30 | 20 | Wang et al.(52) |
| miR-30c-5p | DR | 30 | 20 | Wang et al.(52) |
| miR-342-3p | UR | 24 | 24 | Cosar et al.(51) |
| miR-3613-5p | DR | 24 | 24 | Cosar et al.(51) |
| miR-370 | DR | 20 | 26 | Hu et al.(55) |
| miR-424-3p | UR | 30 | 20 | Wang et al.(52) |
| miR-451a | UR | 41 | 40 | Nothnick et al.(47) |
| miR-451a | UR | 24 | 24 | Cosar et al.(51) |
| miR-500a-3p | UR | 24 | 24 | Cosar et al.(51) |
| miR-542-3p | DR | 60 | 25 | Wang et al.(19) |
| miR-6755-3p | DR | 24 | 24 | Cosar et al.(51) |
| miR-9 | DR | 60 | 25 | Wang et al.(19) |
| miR-99b-5p | DR | 30 | 20 | Wang et al.(52) |
miRNA: microRNA; DR: downregulated; UR: upregulated.
Table 1E. miRNAs differentially expressed in the plasma of endometriosis and control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| miR-139 | DR | 80 | 39 | Nisenblat et al.(56) |
| miR-141 | DR | 61 | 65 | Rekker et al.(57) |
| miR-145 | UR | 55 | 23 | Bashti et al.(58) |
| miR-154-5p | DR | 51 | 41 | Pateisky et al.(59) |
| miR-155 | DR | 80 | 39 | Nisenblat et al.(56) |
| miR-16 | UR | 33 | 20 | Suryawanshi et al.(60) |
| miR-17-5p | DR | 23 | 23 | Jia et al.(61) |
| miR-191 | UR | 33 | 20 | Suryawanshi et al.(60) |
| miR-195 | UR | 33 | 20 | Suryawanshi et al.(60) |
| miR-196b | DR | 51 | 41 | Pateisky et al.(59) |
| miR-200a | DR | 61 | 65 | Rekker et al.(57) |
| miR-200b | DR | 61 | 65 | Rekker et al.(57) |
| miR-20a | DR | 23 | 23 | Jia et al.(61) |
| miR-22 | DR | 23 | 23 | Jia et al.(61) |
| miR-31 | DR | 55 | 23 | Bashti et al.(58) |
| miR-33a | UR | 51 | 41 | Pateisky et al.(59) |
| miR-378a | DR | 51 | 41 | Pateisky et al.(59) |
| miR-574 | DR | 80 | 39 | Nisenblat et al.(56) |
miRNA: microRNA; DR: downregulated; UR: upregulated.
Table 1F. miRNAs differentially expressed in the blood of endometriosis and control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| let-3c | DR | 4 | 3 | Azmy et al.(62) |
| let-7e | DR | 4 | 3 | Azmy et al.(62) |
| let-7f | DR | 5 | 3 | Azmy et al.(62) |
| let-7g | DR | 4 | 3 | Azmy et al.(62) |
| miR-103 | DR | 4 | 3 | Azmy et al.(62) |
| miR-106b | DR | 4 | 3 | Azmy et al.(62) |
| miR-125a-5p | DR | 4 | 3 | Azmy et al.(62) |
| miR-126 | DR | 4 | 3 | Azmy et al.(62) |
| miR-15b | DR | 4 | 3 | Azmy et al.(62) |
| miR-16 | DR | 4 | 3 | Azmy et al.(62) |
| miR-17 | DR | 4 | 3 | Azmy et al.(62) |
| miR-181b | DR | 4 | 3 | Azmy et al.(62) |
| miR-18a | DR | 4 | 3 | Azmy et al.(62) |
| miR-194 | DR | 4 | 3 | Azmy et al.(62) |
| miR-195 | DR | 4 | 3 | Azmy et al.(62) |
| miR-19a | DR | 4 | 3 | Azmy et al.(62) |
| miR-19b | DR | 4 | 3 | Azmy et al.(62) |
| miR-20a | DR | 4 | 3 | Azmy et al.(62) |
| miR-21 | DR | 4 | 3 | Azmy et al.(62) |
| miR-22 | DR | 4 | 3 | Azmy et al.(62) |
| miR-26a | DR | 4 | 3 | Azmy et al.(62) |
| miR-26b | DR | 4 | 3 | Azmy et al.(62) |
| miR-27a | DR | 4 | 3 | Azmy et al.(62) |
| miR-27b | DR | 4 | 3 | Azmy et al.(62) |
| miR-30a | DR | 4 | 3 | Azmy et al.(62) |
| miR-374a | DR | 4 | 3 | Azmy et al.(62) |
| miR-374b | DR | 4 | 3 | Azmy et al.(62) |
| miR-424 | DR | 4 | 3 | Azmy et al.(62) |
| miR-7 | DR | 4 | 3 | Azmy et al.(62) |
| miR-93 | DR | 4 | 3 | Azmy et al.(62) |
miRNA: microRNA; DR: downregulated.
Table 1G. miRNAs differentially expressed in the peritoneal fluid of endometriosis and control patients.
| miRNA | Regulation | Endometriosis n | Control n | References |
|---|---|---|---|---|
| miR-106b-3p | UR | 126 | 45 | Marí-Alexandre et al.(63) |
| miR-122 | UR | 45 | 35 | Maged et al.(50) |
| miR-130b | UR | 6 | 3 | Chen et al.(64) |
| miR-199a | UR | 45 | 35 | Maged et al.(50) |
| miR-451a | UR | 126 | 45 | Marí-Alexandre et al.(63) |
| miR-486-5p | UR | 126 | 45 | Marí-Alexandre et al.(63) |
miRNA: microRNA; UR: upregulated.
A total of 33 miRNAs were examined in more than one study. Of these, 13 miRNAs were analyzed in the same types of samples. miRNAs identified in more than one study and body fluid are described in table 2.
Table 2. Summary of miRNA dysregulated identified in more than one study in different samples.
| Total | EU versus EN | EC versus EN | EC versus EU | Plasma | Serum | Blood | PF | References |
|---|---|---|---|---|---|---|---|---|
| 6 | miR-145 | miR-145 | miR-145 | miR-145 | miR-145 | Wang et al.,(19) Zheng et al.,(22) Yang et al.,(43) Ohlsson Teague et al.,(45) Cosar et al.(51) and Bashti et al.(58) | ||
| 5 | miR-200b | miR-200b | miR-200b | Saare et al.,(34) Yang et al.,(43) Ohlsson Teague et al.,(45) Filigheddu et al.(46) and Rekker et al.(57) | ||||
| 5 | miR-424 | miR-424 | miR-424 | miR-424 | Braza-Boils et al.,(25) Haikalis et al.,(26) Ohlsson Teague et al.,(45) Wang et al.(52) and Azmy et al.(62) | |||
| 4 | miR-199a | miR-199a | miR-199a | miR-199a | Wang et al.,(19) Dai et al.,(24) Maged et al.(50) and Hsu et al.(54) | |||
| 4 | miR-141 | miR-141 | miR-141 | miR-141 | Wang et al.,(19) Saare et al.,(34) Ohlsson Teague et al.(45) and Rekker et al.(57) | |||
| 4 | miR-20a | miR-20a | miR-20a | miR-20a | Zhao et al.,(37) Wang et al.,(52) Jia et al.(61) and Azmy et al.(62) | |||
| 4 | miR-200a | miR-200a | miR-200a | Saare et al.,(34) Zhao et al.,(42) Filigheddu et al.(46) and Rekker et al.(57) | ||||
| 3 | miR-29c | miR-29c | miR-29c | Braza-Boils et al.,(25) Long et al.(27) and Joshi et al.(39) | ||||
| 3 | miR-34c | miR-34c | miR-34c | Braza-Boïls et al.,(25) Saare et al.(34) and Joshi et al.(39) | ||||
| 3 | miR-200c | miR-200c | Liang et al.,(36) Yang et al.(43) and Filigheddu et al.(46) | |||||
| 3 | miR-21 | miR-21 | miR-21 | Haikalis et al.,(26) Qi et al.(38) and Azmy et al.(62) | ||||
| 3 | miR-126 | miR-126 | miR-126 | Liu et al.,(20) Ohlsson Teague et al.(45) and Azmy et al.(62) | ||||
| 3 | miR-16 | miR-16 | miR-16 | Yang et al.,(43) Suryawanshi et al.(60) and Azmy et al.(62) | ||||
| 3 | miR-451a | miR-451a | miR-451a | Nothnick et al.,(47) Cosar et al.(51) and Marí-Alexandre et al.(63) | ||||
| 3 | miR-9 | miR-9 | miR-9 | Wang et al.,(19) Haikalis et al.(26) and Burney et al.(29) | ||||
| 3 | miR-106b | miR-106b | miR-106b | Yang et al.,(43) Azmy et al.(62) and Marí-Alexandre et al.(63) | ||||
| 3 | miR-17 | miR-17 | miR-17 | Wang et al.,(53) Jia et al.(61) and Azmy et al.(62) | ||||
| 2 | miR-122 | miR-122 | Wang et al.(19) and Maged et al.(50) | |||||
| 2 | miR-449b | Braza-Boïls et al.(25) and Zhao et al.(42) | ||||||
| 2 | miR-191 | miR-191 | miR-191 | Dong et al.(35) and Suryawanshi et al.(60) | ||||
| 2 | miR-202 | miR-202 | miR-202 | Braza-Boïls et al.(25) and Filigheddu et al.(46) | ||||
| 2 | miR-143 | miR-143 | Zheng et al.(22) and Cosar et al.(51) | |||||
| 2 | miR-22 | miR-22 | Jia et al.(61) and Azmy et al.(62) | |||||
| 2 | miR let-7g | miR let-7g | Wright et al.(31) and Azmy et al.(62) | |||||
| 2 | miR-15b | miR-15b | Wanget al.(52) and Azmy et al.(62) | |||||
| 2 | miR-125a | miR-125a | Ramón et al.(44) and Azmy et al.(62) | |||||
| 2 | miR-195 | miR-195 | Suryawanshi et al.(60) and Azmy et al.(62) | |||||
| 2 | miR-18a | miR-18a | Cosar et al.(51) and Azmy et al.(62) | |||||
| 2 | miR-19b | miR-19b | Yanget al.(43) and Azmy et al.(62) | |||||
| 2 | miR-146a | miR-146a | Yang et al.(21) and Yang et al.(43) | |||||
| 2 | miR-182 | Zhao et al.(42) and Filigheddu et al.(46) | ||||||
| 2 | miR-125b | Nematian et al.(49) and Cosar et al.(51) | ||||||
| 2 | miR-let-7b | Cho et al.(48) and Nematian et al.(49) |
EU: eutopic endometrium of women with endometriosis; EN: eutopic endometrium of control patients; EC: ectopic endometrium; PF: peritoneal fluid.
Twenty out of 62 miRNAs identified in samples with potential applicability to minimally invasive diagnosis of endometriosis, such as blood, serum, and plasma, were also found to be dysregulated in other types of tissue, such as EC and eutopic endometrium, and in the peritoneal fluid. Of these, 35% were detected in the same type of tissue in more than one study, including miR-200b, miR-145, miR-199a, miR-424, miR-200a, miR-126, and miR-451a. Thirteen miRNAs were found to be up or downregulated, as follows: miR-125b, miR-let-7b, miR-122, miR-451a and miR-199a in serum; miR-29c in the EC relative to the EN Group; and miR-145, miR-200b, miR-424, miR-200a, miR-200c, miR-449b and miR-182 in the EC relative to the EC of women with endometriosis (Table 3).
Table 3. Characterization of miRNAs expression for upregulation and downregulation in different samples.
| miRNA | n | EU versus EN | EC versus EN | EC versus EU | Plasma | Serum | Blood | PF | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EU | EN | EC | EN | EC | EU | ||||||
| miR-145 | 6 | ↑ (1) | ↓ (1) | ↑ (1) | ↓ (1) | ↑ (2) | ↓ (2) | ↑ (1) | ↑ (1) /↓ (1) | ||
| miR-200b | 5 | ↑ (1) | ↓ (1) | ↓ (3) | ↑ (3) | ↓ (1) | |||||
| miR-424 | 5 | ↓ (1) | ↑ (1) | ↓ (2) | ↑ (2) | ↑ (1) | ↓ (1) | ||||
| miR-199a | 4 | ↓ (1) | ↑ (1) | ↓ (1) | ↑ (1) | ↑ (1) | ↑ (2)/↓ (1) | ↑ (1) | |||
| miR-141 | 4 | ↑ (1) | ↓ (1) | ↓ (1) | ↑ (1) | ↓ (1) | ↓ (1) | ||||
| miR-20a | 4 | ↑ (1) | ↓ (1) | ↓ (1) | ↓ (1) | ↓ (1) | |||||
| miR-200a | 4 | ↑ (1) | ↓ (1) | ↓ (2) | ↑ (2) | ↓ (1) | |||||
| miR-29c | 3 | ↓ (1) | ↑ (1) | ↑ (2) /↓ (1) | ↓ (2) / ↑(1) | ↓ (1) | ↑ (1) | ||||
| miR-34c | 3 | ↓ (1) | ↑ (1) | ↑ (1) | ↓ (1) | ↓ (1) | ↑ (1) | ||||
| miR-200c | 3 | ↓ (1) | ↑ (1) | ↓ (2) | ↑ (2) | ||||||
| miR-21 | 3 | ↑ (1) | ↓ (1) | ↓ (1) | ↑ (1) | ↓ (1) | |||||
| miR-126 | 3 | ↓ (1) | ↑ (1) | ↑ (1)/↓ (1) | ↓ (1)/↑ (1) | ↓ (1) | |||||
| miR-16 | 3 | ↑ (1) | ↓ (1) | ↑ (1) | ↓ (1) | ||||||
| miR-451a | 3 | ↑ (1) | ↓ (1) | ↑ (2) | ↑ (1) | ||||||
| miR-9 | 3 | ↓ (1) | ↑ (1) | ↓ (1) | ↑ (1) | ↓ (1) | |||||
| miR-106b | 3 | ↑ (1) | ↓ (1) | ↓ (1) | ↑ (1) | ||||||
| miR-17 | 3 | ↓ (1) | ↓ (1) | ↓ (1) | |||||||
| miR-122 | 2 | ↑ (2) | ↑ (1) | ||||||||
| miR-449b | 2 | ↓ (2) | ↑ (2) | ||||||||
| miR-191 | 2 | ↑ (1) | ↓ (1) | ↑ (1) | ↑ (1) | ||||||
| miR-202 | 2 | ↓ (1) | ↑ (1) | ↑ (1) | ↓ (1) | ↑ (1) | ↓ (1) | ||||
| miR-143 | 2 | ↑ (1) | ↓ (1) | ↑ (1) | |||||||
| miR-22 | 2 | ↓ (1) | ↓ (1) | ||||||||
| miR let-7g | 2 | ↑ (1) | ↓ (1) | ↓ (1) | |||||||
| miR-15b | 2 | ↓ (1) | ↓ (1) | ||||||||
| miR-125a | 2 | ↑ (1) | ↓ (1) | ↓ (1) | |||||||
| miR-195 | 2 | ↑ (1) | ↓ (1) | ||||||||
| miR-18a | 2 | ↑ (1) | ↓ (1) | ||||||||
| miR-19b | 2 | ↓ (1) | ↑ (1) | ↓ (1) | |||||||
| miR-146a | 2 | ↑ (1) | ↓ (1) | ↓ (1) | ↑ (1) | ||||||
| miR-182 | 2 | ↓ (2) | ↑ (2) | ||||||||
| miR-125b | 2 | ↑ (2) | |||||||||
| miR-let-7b | 2 | ↓ (2) | |||||||||
↑ upregulation; ↓ downregulation.
miRNA: microRNA; EU: eutopic endometrium of women with endometriosis; EN: eutopic endometrium of control patients; EC: ectopic endometrium; PF: peritoneal fluid.
DISCUSSION
Endometriosis can be a debilitating disease and may lead to poor quality of life.(65) The disease is associated with dysmenorrhea, deep dyspareunia, chronic pelvic pain and infertility(66,67) and is considered a public health concern, given the impact on patient physical and psychological health, and the socioeconomic impact of diagnosis, treatment and clinical control costs.(68)
The final diagnosis of endometriosis is currently based on histological analysis of the lesion, usually in samples obtained by laparoscopic surgery.(69) However, imaging modalities are important non-invasive diagnostic alternatives for ovarian and deep endometriosis. Both surgical and non-surgical approaches require considerable professional skill and availability of specific data, which may represent a huge economic and health burden in developing countries.(4-9)
In the last three decades, researchers worldwide have tried to identify a non-invasive test that could shorten the turnaround time for diagnosis of endometriosis. CA-125 can be detected in blood or peritoneal fluid and is one of the best studied biomarkers. In some case studies, measurement of CA-125 levels was deemed promising, especially for diagnosis of more invasive endometriosis, provided measurements are made in the beginning of the menstrual cycle.(70-72)
In spite of conflicting results regarding the value of CA-125 as a final and important biomarker reported in recent reviews, according to Socolov et al., CA-125 is still the most recommended biomarker for endometriosis diagnosis and monitoring.(73) In a more recent Cochrane review published in 2016, Nisenblat et al. compared the accuracy of any combination of non-invasive diagnostic tests to surgical diagnosis of pelvic endometriosis, using randomized controlled trials or cross-sectional studies published until early 2015 as a reference standard. Authors concluded that none of the biomarkers investigated (including CA-125) could be duly evaluated due to insufficient or poor-quality evidence, given the high heterogeneity and risk of bias in selected studies.(15)
CA-125 is most elevated in advanced stages of endometriosis. Therefore, the sensitivity of this marker is limited. Its specificity is also thought to be poor, since it is upregulated in other gynecological conditions.(74) In this context, the search for novel and effective noninvasive biomarkers capable of improving endometriosis diagnosis, management and monitoring remains high on the priority list.
Circulating miRNAs, first identified as non-invasive serological markers of tumors in 2008,(75-77) are promising alternative candidates. The high stability of circulating miRNAs in human plasma and their resistance to multiple sample handling procedures has been emphasized in these pioneer studies.
These same studies also established the concept of disease diagnosis based on specific cell-free miRNA signatures. Since then, miRNAs have been validated as noninvasive diagnostic markers for several diseases, including oncologic, inflammatory, cardiovascular, metabolic and reproductive disorders. miRNAs proved to be ideal diagnostic markers in oncology, as shown by differential circulating miRNA expression patterns in lung, ovarian, colorectal, prostate and breast cancer patients relative to healthy controls.(78)
In the female reproductive system in particular, dysregulated miRNA expression has been studied in uterine leiomyomata, in several gynecologic cancers (including adenocarcinomas), and in pregnancy disorders, such as preeclampsia and preterm birth.(79-83) These small noncoding molecules associated with several diseases have been proposed as useful diagnostic candidates for endometriosis.(84)
In this review, miR-145 was the miRNA found to be differentially expressed in the largest number of studies (six articles). In the 46 studies analyzed, most miRNAs found to be dysregulated in endometriosis were harvested from tissue samples. Bodily fluids were seldom investigated, even though they may be used as non- or minimally invasive diagnostic tools. Also, most studies compared miRNA expression differences between the eutopic and EC of patients with endometriosis and only a few compared the endometrium of patients with endometriosis, suggesting that examinations based endometrial biopsies are difficult.
As regards dysregulated miRNAs in endometriosis patients compared in this review, 30 were found in the blood, 27 in the serum and 18 in the plasma of women with endometriosis relative to control populations. Differences in the molecular composition of serum and plasma have been well-documented.(85,86)
When comparing the miRNA spectrum between serum and plasma, Wang et al.,(19) detected several differences in RNA levels driven by the release of certain miRNAs and other RNAs during the coagulation process, and suggested that use of plasma as the sample of choice for studying circulating miRNAs, since RNAs released during coagulation may alter the true repertoire of circulating miRNAs.
Differential expression of six miRNAs was detected in the peritoneal fluid of endometriosis patients relative to non-affected women. Hence, some miRNAs found in peritoneal fluid may play a role in the pathogenesis of endometriosis. However, given the nature of this fluid, its use is limited by the need for surgical (i.e., invasive) collection.
Some points are worthy of note and should be emphasized in these studies: conflicting results. They have been reported in studies investigating miR-145, -424, -199a, -29c, -126, -16, -195 and -18a expression in the same type of sample. Major characteristics of these studies are described below.
Upregulation of miR-145 was found in the serum, in a study with 24 stages III and IV endometriosis and 24 control patients,(51) and in plasma, in a study with 55 stages I and II endometriosis and 23 control patients.(58) In contrast, the same miRNA was found to be downregulated in the serum in a study including 60 cases and 25 controls,(19) in which disease stage was not reported.
miR-424 was downregulated in blood in a study with four patients with mild endometriosis and three controls.(62) However, it was also found to be upregulated in the serum of 30 patients with minimal-mild endometriosis relative to 20 control individuals.(52)
miR-199a was upregulated in the serum of patients with endometriosis in two studies, one with 60 stages III and IV endometriosis and 25 control patients,(19) and another with 45 endometriosis and 35 control patients.(50) However, the same miRNA was found to be downregulated in the serum in a different study with 40 endometriosis and 25 control patients.(54)
A study with 15 clinical cases and 11 controls revealed miR-29c upregulation in the EC of women with endometriosis relative to the eutopic endometrium in the Control Group.(39) This finding was further confirmed in a study including 51 women with endometriosis and 32 control women,(25) in the proliferative and secretory phases of the menstrual cycle. However, conflicting results suggesting miR-29c downregulation in the EC of 20 women with endometriosis relative to the eutopic endometrium of ten control patients,(27) all of them in the proliferative phase of the cycle, have been reported by a different researcher.
miR-126 was found to be upregulated in the ectopic compared to the eutopic endometrium of eight women with stages III to IV endometriosis(45) in the proliferative and secretory phases of the menstrual cycle. However, miR-126 downregulation was reported in the ectopic compared to the eutopic endometrium in 31 women with stages III to IV endometriosis,(20) all of them in the secretory phase of the menstrual cycle.
miR-16 and miR-195 were found to be upregulated in plasma of 33 women with endometriosis relative to 20 control patients.(60) However, another study identified both downregulated in the blood of four patients with mild endometriosis relative to three controls.(62)
miR-18a was upregulated in serum of 24 women with stage III and IV endometriosis compared to 24 control patients.(51) However, it was found to be downregulated in the blood of four patients with mild endometriosis compared to three controls.(62)
Conflicting results emphasize the relevance of criteria such as menstrual cycle phase, disease stage, type of sample and type of test procedure, and the need for studies with larger sample size to develop novel diagnostic tests for endometriosis.
The second objective of this review was to provide new directions for future studies aimed to identify a miRNA which may be used as a reliable biomarker and an accurate diagnostic tool for endometriosis. Sadly, according to this critical literature review no particular miRNA or miRNA combination has been validated for improved diagnosis of endometriosis to date. This may reflect the heterogeneity of the disease and resultant differences in tissue composition.(87) Thus, we support the World Endometriosis Research Foundation (WERF) and Endometriosis Phenome and Biobanking Harmonization Project (EPHect) initiatives. Endometriosis research teams worldwide must join forces in order to develop large databases comprising data derived from samples obtained from patients with well-characterized endometriosis.
This is an important tool for identification and validation of biomarkers and may play a key role in biomarker investigation in future endometriosis studies.(88) The inclusion of a large global pool of clinical samples collected from endometriosis patients is vital for the advancement of medical knowledge, and could be a key factor in the implementation of targeted therapies, which may enhance treatment effectiveness and improve the quality of life of endometriosis patients.
No studies investigating miRNA expression profile in the vaginal fluid were found in this literature review. This body fluid can be easily collected during gynecological examinations and, in spite of high rates of bacterial colonization, appears to be a promising source of diagnostic material.(89,90) The value of differential miRNA expression in vaginal fluid as potential screening test for HPV has been examined, with interesting results.(91-95)
Likewise, none of the papers examined investigated miRNAs in saliva. To date, there are no scientifically proven salivary biomarkers for endometriosis. Saliva is a suitable and desirable medium for biomarker detection(96,97) and its applicability to the diagnosis of endometriosis has been explored previously.(98,99) Saliva is widely available and can be easily collected in a non-invasive manner, at low cost and with minimal discomfort. Therefore, it is an ideal fluid for biomarker investigation and is attracting great interest in the public health sector. The use of saliva for miRNA identification could be a potential non-invasive solution to overcome current barriers to the diagnosis of endometriosis.
This study has some limitations. When evaluating papers with contrasting results, it was not possible to tease out the factors underlying such different outcomes. Reasons explaining miRNA heterogeneity were also not found.
In this review, different studies investigating miRNA expression in endometriosis patients were discussed. Most of these studies were based on pooled or small samples. Large, well-designed clinical trials aimed to validate endometriosis-related miRNAs are needed in order to develop accurate, low-invasiveness diagnostic methods for endometriosis. The clinical impact of scientifically proven miRNA biomarkers for endometriosis will translate into better access to care and less health disparities, with potential impacts on global health. The diagnosis of endometriosis at earlier stages of the disease may lead to dramatic reduction in health costs and provide significant benefits for patients through improved health and quality of life.
CONCLUSION
Differential miR-145 expression was reported in the largest number of studies (six articles). Most dysregulated miRNAs were harvested from tissue samples.
No particular miRNA or miRNA combination has been validated for improved diagnosis of endometriosis to date. This may have reflected the heterogeneity of the disease and resultant differences in tissue composition. Large databases comprising data derived from samples collected from patients with well-characterized endometriosis may play a key role in biomarker investigation in future studies. The use of saliva and vaginal fluid samples for miRNA identification could be a potential non-invasive solution to overcome current barriers to the diagnosis of endometriosis.
ACKNOWLEDGEMENTS
To Carlos Augusto Cardim de Oliveira for his instructions about the best practices for a good systematic review.
REFERENCES
- 1.1. Parasar P, Ozcan P, Terry KL. Endometriosis: epidemiology, diagnosis and clinical management. Curr Obstet Gynecol Rep. 2017;6(1):34-41. [DOI] [PMC free article] [PubMed]
- 2.2. Viganò P, Parazzini F, Somigliana E, Vercellini P. Endometriosis: epidemiology and aetiological factors. Best Pract Res Clin Obstet Gynaecol. 2004; 18(2):177-200. Review. [DOI] [PubMed]
- 3.3. Murphy AA. Clinical aspects of endometriosis. Ann N Y Acad Sci. 2002;955:1-10; discussion 34-36, 396-406. [DOI] [PubMed]
- 4.4. Goncalves MO, Podgaec S, Dias JA Jr, Gonzalez M, Abrao MS. Transvaginal ultrasonography with bowel preparation is able to predict the number of lesions and rectosigmoid layers affected in cases of deep endometriosis, defining surgical strategy. Hum Reprod. 2010;25(3):665-71. [DOI] [PubMed]
- 5.5. Carneiro MM, Filogônio ID, Costa LM, de Ávila I, Ferreira MC. Clinical prediction of deeply infiltrating endometriosis before surgery: is it feasible? A review of the literature. Biomed Res Int. 2013;2013:564153. Review. [DOI] [PMC free article] [PubMed]
- 6.6. Benacerraf BR, Groszmann Y. Sonography should be the first imaging examination done to evaluate patients with suspected endometriosis. J Ultrasound Med. 2012;31(4):651-3. [DOI] [PubMed]
- 7.7. Nisenblat V, Bossuyt PM, Farquhar C, Johnson N, Hull ML. Imaging modalities for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;2:CD009591. Review. [DOI] [PMC free article] [PubMed]
- 8.8. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67(5):817-21. [DOI] [PubMed]
- 9.9. Duffy JM, Arambage K, Correa FJ, Olive D, Farquhar C, Garry R, et al. Laparoscopic surgery for endometriosis. Cochrane Database Syst Rev. 2014;(4):CD011031. Review. [DOI] [PubMed]
- 10.10. D’Hooghe TM, Debrock S, Meuleman C, Hill JA, Mwenda JM. Future directions in endometriosis research. Obstet Gynecol Clin North Am. 2003; 30(1):221-44. Review. [DOI] [PubMed]
- 11.11. Ballard K, Lowton K, Wright J. What’s the delay? A qualitative study of women’s experiences of reaching a diagnosis of endometriosis. Fertil Steril. 2006;86(5):1296-301. [DOI] [PubMed]
- 12.12. Liu E, Nisenblat V, Farquhar C, Fraser I, Bossuyt PM, Johnson N, et al. Urinary biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2015;(12):CD012019. Review. [DOI] [PMC free article] [PubMed]
- 13.13. Gupta D, Hull ML, Fraser I, Miller L, Bossuyt PM, Johnson N, et al. Endometrial biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;4:CD012165. Review. [DOI] [PMC free article] [PubMed]
- 14.14. Nisenblat V, Bossuyt PM, Shaikh R, Farquhar C, Jordan V, Scheffers CS, et al. Blood biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;(5):CD012179. Review. [DOI] [PMC free article] [PubMed]
- 15.15. Nisenblat V, Prentice L, Bossuyt PM, Farquhar C, Hull ML, Johnson N. Combination of the non-invasive tests for the diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;7:CD012281. Review. [DOI] [PMC free article] [PubMed]
- 16.16. Somigliana E, Vercellini P, Viganò P, Benaglia L, Crosignani PG, Fedele L. Non-invasive diagnosis of endometriosis: the goal or own goal? Hum Reprod. 2010;25(8):1863-8. [DOI] [PubMed]
- 17.17. Marí-Alexandre J, Sánchez-Izquierdo D, Gilabert-Estellés J, Barceló-Molina M, Braza-Boïls A, Sandoval J. miRNAs regulation and its role as biomarkers in endometriosis. Int J Mol Sci. 2016;17(1):93. Review. [DOI] [PMC free article] [PubMed]
- 18.18. Lin PY, Yang PC. Circulating miRNA signature for early diagnosis of lung cancer. EMBO Mol Med. 2011;3(8):436-7. [DOI] [PMC free article] [PubMed]
- 19.19. Wang WT, Zhao YN, Han BW, Hong SJ, Chen YQ. Circulating microRNAs identified in a genome-wide serum microRNA expression analysis as noninvasive biomarkers for endometriosis. J Clin Endocrinol Metab. 2013;98(1):281-9. [DOI] [PubMed]
- 20.20. Liu S, Gao S, Wang XY, Wang DB. Expression of miR-126 and Crk in endometriosis: miR-126 may affect the progression of endometriosis by regulating Crk expression. Arch Gynecol Obstet. 2012;285(4):1065-72. [DOI] [PubMed]
- 21.21. Yang P, Wu Z, Ma C, Pan N, Wang Y, Yan L. Endometrial miR-543 is downregulated during the implantation window in women with endometriosis-related infertility. Reprod Sci. 2019;26(7):900-8. [DOI] [PubMed]
- 22.22. Zheng B, Xue X, Zhao Y, Chen J, Xu CY, Duan P. The differential expression of microRNA-143,145 in endometriosis. Iran J Reprod Med. 2014;12(8):555-60. [PMC free article] [PubMed]
- 23.23. Shi XY, Gu L, Chen J, Guo XR, Shi YL. Downregulation of miR-183 inhibits apoptosis and enhances the invasive potential of endometrial stromal cells in endometriosis. Int J Mol Med. 2014;33(1):59-67. [DOI] [PMC free article] [PubMed]
- 24.24. Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol Hum Reprod. 2012;18(3):136-45. [DOI] [PMC free article] [PubMed]
- 25.25. Braza-Boïls A, Marí-Alexandre J, Gilabert J, Sánchez-Izquierdo D, España F, Estellés A, et al. MicroRNA expression profile in endometriosis: its relation to angiogenesis and fibrinolytic factors. Hum Reprod. 2014;29(5):978-88. [DOI] [PubMed]
- 26.26. Haikalis ME, Wessels JM, Leyland NA, Agarwal SK, Foster WG. MicroRNA expression pattern differs depending on endometriosis lesion type. Biol Reprod. 2018;98(5):623-33. [DOI] [PubMed]
- 27.27. Long M, Wan X, La X, Gong X, Cai X. miR-29c is downregulated in the ectopic endometrium and exerts its effects on endometrial cell proliferation, apoptosis and invasion by targeting c-Jun. Int J Mol Med. 2015;35(4):1119-25. [DOI] [PubMed]
- 28.28. Laudanski P, Charkiewicz R, Tolwinska A, Szamatowicz J, Charkiewicz A, Niklinski J. Profiling of selected microRNAs in proliferative eutopic endometrium of women with ovarian endometriosis. Biomed Res Int. 2015;2015:760698. [DOI] [PMC free article] [PubMed]
- 29.29. Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, et al. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15(10):625-31. [DOI] [PMC free article] [PubMed]
- 30.30. Laudanski P, Charkiewicz R, Kuzmicki M, Szamatowicz J, Charkiewicz A, Niklinski J. MicroRNAs expression profiling of eutopic proliferative endometrium in women with ovarian endometriosis. Reprod Biol Endocrinol. 2013;11:78. Erratum in: Reprod Biol Endocrinol. 2015;13:50. [DOI] [PMC free article] [PubMed]
- 31.31. Wright KR, Mitchell B, Santanam N. Redox regulation of microRNAs in endometriosis-associated pain. Redox Biol. 2017;12:956-66. [DOI] [PMC free article] [PubMed]
- 32.32. Xu X, Li Z, Liu J, Yu S, Wei Z. MicroRNA expression profiling in endometriosis-associated infertility and its relationship with endometrial receptivity evaluated by ultrasound. J Xray Sci Technol. 2017;25(3):523-32. [DOI] [PubMed]
- 33.33. Braicu OL, Budisan L, Buiga R, Jurj A, Achimas-Cadariu P, Pop LA, et al. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225-38. [DOI] [PMC free article] [PubMed]
- 34.34. Saare M, Rekker K, Laisk-Podar T, Sõritsa D, Roost AM, Simm J, et al. High-throughput sequencing approach uncovers the miRNome of peritoneal endometriotic lesions and adjacent healthy tissues. PLoS One. 2014;9(11):e112630. [DOI] [PMC free article] [PubMed]
- 35.35. Dong M, Yang P, Hua F. MiR-191 modulates malignant transformation of endometriosis through regulating TIMP3. Med Sci Monit. 2015;21:915-20. [DOI] [PMC free article] [PubMed]
- 36.36. Liang Z, Chen Y, Zhao Y, Xu C, Zhang A, Zhang Q, et al. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell Res Ther. 2017;8(1):251. [DOI] [PMC free article] [PubMed]
- 37.37. Zhao M, Tang Q, Wu W, Xia Y, Chen D, Wang X. miR-20a contributes to endometriosis by regulating NTN4 expression. Mol Biol Rep. 2014;41(9):5793-7. [DOI] [PubMed]
- 38.38. Qi H, Liang G, Yu J, Wang X, Liang Y, He X, et al. Genome-wide profiling of miRNA expression patterns in tubal endometriosis. Reproduction. 2019;157(6):525-34. [DOI] [PubMed]
- 39.39. Joshi NR, Miyadahira EH, Afshar Y, Jeong JW, Young SL, Lessey BA, et al. Progesterone resistance in endometriosis is modulated by the altered expression of microRNA-29c and FKBP4. J Clin Endocrinol Metab. 2017;102(1):141-9. [DOI] [PMC free article] [PubMed]
- 40.40. Yang WW, Hong L, Xu XX, Wang Q, Huang JL, Jing L. Regulation of miR-33b on endometriosis and expression of related factors. Eur Rev Med Pharmacol Sci. 2017;21(9):2027-33. [PubMed]
- 41.41. Graham A, Falcone T, Nothnick WB. The expression of microRNA-451 in human endometriotic lesions is inversely related to that of macrophage migration inhibitory factor (MIF) and regulates MIF expression and modulation of epithelial cell survival. Hum Reprod. 2015;30(3):642-52. [DOI] [PMC free article] [PubMed]
- 42.42. Zhao L, Gu C, Ye M, Zhang Z, Li L, Fan W, et al. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod Biol Endocrinol. 2018;16(1):4. [DOI] [PMC free article] [PubMed]
- 43.43. Yang RQ, Teng H, Xu XH, Liu SY, Wang YH, Guo FJ, et al. Microarray analysis of microRNA deregulation and angiogenesis-related proteins in endometriosis. Genet Mol Res. 2016;15(2):gmr.15027826. [DOI] [PubMed]
- 44.44. Ramón LA, Braza-Boïls A, Gilabert-Estellés J, Gilabert J, España F, Chirivella M, et al. microRNAs expression in endometriosis and their relation to angiogenic factors. Hum Reprod. 2011;26(5):1082-90. [DOI] [PubMed]
- 45.45. Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23(2):265-75. [DOI] [PMC free article] [PubMed]
- 46.46. Filigheddu N, Gregnanin I, Porporato PE, Surico D, Perego B, Galli L, et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010;2010:369549. [DOI] [PMC free article] [PubMed]
- 47.47. Nothnick WB, Falcone T, Joshi N, Fazleabas AT, Graham A. Serum miR-451a levels are significantly elevated in women with endometriosis and recapitulated in baboons (Papio anubis) with experimentally-induced disease. Reprod Sci. 2017;24(8):1195-202. [DOI] [PMC free article] [PubMed]
- 48.48. Cho S, Mutlu L, Grechukhina O, Taylor HS. Circulating microRNAs as potential biomarkers for endometriosis. Fertil Steril. 2015;103(5)1252-60.e1. [DOI] [PMC free article] [PubMed]
- 49.49. Nematian SE, Mamillapalli R, Kadakia TS, Majidi Zolbin M, Moustafa S, Taylor HS. Systemic inflammation induced by microRNAs: endometriosis derived alterations in circulating microRNA 125b-5p and Let-7b-5p regulate macrophage cytokine production. J Clin Endocrinol Metab. 2018;103(1):64-74. [DOI] [PubMed]
- 50.50. Maged AM, Deeb WS, El Amir A, Zaki SS, El Sawah H, Al Mohamady M, et al. Diagnostic accuracy of serum miR-122 and miR-199a in women with endometriosis. Int J Gynaecol Obstet. 2017;141(1):14-9. [DOI] [PubMed]
- 51.51. Cosar E, Mamillapalli R, Ersoy GS, Cho S, Seifer B, Taylor HS. Serum microRNAs as diagnostic markers of endometriosis: a comprehensive array-based analysis. Fertil Steril. 2016;106(2):402-9. [DOI] [PubMed]
- 52.52. Wang L, Huang W, Ren C, Zhao M, Jiang X, Fang X, et al. Analysis of Serum microRNA Profile by Solexa Sequencing in Women With Endometriosis. Reprod Sci. 2016;23(10):1359-70. [DOI] [PubMed]
- 53.53. Wang F, Wang H, Jin D, Zhang Y. Serum miR-17, IL-4, and IL-6 levels for diagnosis of endometriosis. Medicine (Baltimore). 2018;97(24):e10853. [DOI] [PMC free article] [PubMed]
- 54.54. Hsu CY, Hsieh TH, Tsai CF, Tsai HP, Chen HS, Chang Y, et al. miRNA-199a-5p regulates VEGFA in endometrial mesenchymal stem cells and contributes to the pathogenesis of endometriosis. J Pathol. 2014;232(3):330-43. [DOI] [PubMed]
- 55.55. Hu Z, Mamillapalli R, Taylor HS. Increased circulating miR-370-3p regulates steroidogenic factor 1 in endometriosis. Am J Physiol Endocrinol Metab. 2019;316(3):E373-82. [DOI] [PMC free article] [PubMed]
- 56.56. Nisenblat V, Sharkey DJ, Wang Z, Evans SF, Healey M, Ohlsson Teague EM, et al. Plasma miRNAs display limited potential as diagnostic tools for endometriosis. J Clin Endocrinol Metab. 2019;104(6):1999-2022. [DOI] [PubMed]
- 57.57. Rekker K, Saare M, Roost AM, Kaart T, Sõritsa D, Karro H, et al. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil Steril. 2015;104(4):938-46.e2. [DOI] [PubMed]
- 58.58. Bashti O, Noruzinia M, Garshasbi M, Abtahi M. miR-31 and miR-145 as potential non-invasive regulatory biomarkers in patients with endometriosis. Cell J. 2018;20(1):84-9. Erratum in: Cell J. 2018;20(2):293. [DOI] [PMC free article] [PubMed]
- 59.59. Pateisky P, Pils D, Szabo L, Kuessel L, Husslein H, Schmitz A, et al. hsa-miRNA-154-5p expression in plasma of endometriosis patients is a potential diagnostic marker for the disease. Reprod Biomed Online. 2018;37(4):449-66. [DOI] [PubMed]
- 60.60. Suryawanshi S, Vlad AM, Lin HM, Mantia-Smaldone G, Laskey R, Lee M, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19(5):1213-24. [DOI] [PMC free article] [PubMed]
- 61.61. Jia SZ, Yang Y, Lang J, Sun P, Leng J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum Reprod. 2013;28(2):322-30. [DOI] [PMC free article] [PubMed]
- 62.62. Azmy OM, Elgarf WT. MiRNA-130a, a potential endometriosis inducing factor. Med Res J. 2012;11(2):40-7.
- 63.63. Marí-Alexandre J, Barceló-Molina M, Belmonte-López E, García-Oms J, Estellés A, Braza-Boïls A, et al. Micro-RNA profile and proteins in peritoneal fluid from women with endometriosis: their relationship with sterility. Fertil Steril. 2018;109(4):675-84.e2. [DOI] [PubMed]
- 64.64. Chen Y, Wang K, Xu Y, Guo P, Hong B, Cao Y, et al. Alteration of myeloid-derived suppressor cells, chronic inflammatory cytokines, and exosomal miRNA contribute to the peritoneal immune disorder of patients with endometriosis.. Reprod Sci. 2019;26(8):1130-8. [DOI] [PubMed]
- 65.65. Nogueira Neto J, Coelho TM, Aguiar GC, Carvalho LR, de Araújo AG, Girão MJ, et al. Experimental endometriosis reduction in rats treated with Uncaria tomentosa (cat’s claw) extract. Eur J Obstet Gynecol Reprod Biol. 2011;154(2):205-8. [DOI] [PubMed]
- 66.66. Berlanda N, Vercellini P, Somigliana E, Frattaruolo MP, Buggio L. Role of surgery in endometriosis-associated subfertility. Semin Reprod Med. 2013; 31(2):133-43. Review. [DOI] [PubMed]
- 67.67. Agostinis C, Zorzet S, De Leo R, Zauli G, De Seta F, Bulla R. The combination of N-acetyl cysteine, alpha-lipoic acid, and bromelain shows high anti-inflammatory properties in novel in vivo and in vitro models of endometriosis. Mediators Inflamm. 2015;2015:918089. [DOI] [PMC free article] [PubMed]
- 68.68. Rosa-E-Silva JC, Fortunato GG, Zanardi J, Meola J, Nogueira AA. Effect of cabergoline in experimental endometriosis in rats. J Minim Invasive Gynecol. 2015;22(6S):S168. [DOI] [PubMed]
- 69.69. Johnson NP, Hummelshoj L; World Endometriosis Society Montpellier Consortium. Consensus on current management of endometriosis. Hum Reprod. 2013;28(6):1552-68. [DOI] [PubMed]
- 70.70. Amaral VF, Ferriani RA, Sá MF, Nogueira AA, Rosa e Silva JC, Rossa e Silva AC, et al. Positive correlation between serum and peritoneal fluid CA-125 levels in women with pelvic endometriosis. Sao Paulo Med J. 2006;124(4):223-7. [DOI] [PMC free article] [PubMed]
- 71.71. Abrao MS, Podgaec S, Pinotti JA, de Oliveira RM. Tumor markers in endometriosis. Int J Gynaecol Obstet. 1999;66(1):19-22. [DOI] [PubMed]
- 72.72. Abrão MS, Podgaec S, Filho BM, Ramos LO, Pinotti JA, de Oliveira RM. The use of biochemical markers in the diagnosis of pelvic endometriosis. Hum Reprod. 1997;12(11):2523-7. [DOI] [PubMed]
- 73.73. Socolov R, Socolov D, Sindilar A, Pavaleanu I. An update on the biological markers of endometriosis. Minerva Ginecol. 2017;69(5):462-7. Review. [DOI] [PubMed]
- 74.74. O DF, Flores I, Waelkens E, D’Hooghe T. Noninvasive diagnosis of endometriosis: review of current peripheral blood and endometrial biomarkers. Best Pract Res Clin Obstet Gynaecol. 2018;50:72-83. Review. [DOI] [PubMed]
- 75.75. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513-8. [DOI] [PMC free article] [PubMed]
- 76.76. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997-1006. [DOI] [PubMed]
- 77.77. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672-5. [DOI] [PubMed]
- 78.78. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590-610. Review. [DOI] [PMC free article] [PubMed]
- 79.79. Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46(4):336-47. [DOI] [PubMed]
- 80.80. Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105(19):7004-9. [DOI] [PMC free article] [PubMed]
- 81.81. Traver S, Assou S, Scalici E, Haouzi D, Al-Edani T, Belloc S, et al. Cell-free nucleic acids as non-invasive biomarkers of gynecological cancers, ovarian, endometrial and obstetric disorders and fetal aneuploidy. Hum Reprod Update. 2014;20(6):905-23. [DOI] [PubMed]
- 82.82. Santamaria X, Taylor H. MicroRNA and gynecological reproductive diseases. Fertil Steril. 2014;101(6):1545-51. Review. [DOI] [PubMed]
- 83.83. Srivastava SK, Ahmad A, Zubair H, Miree O, Singh S, Rocconi RP, et al. MicroRNAs in gynecological cancers: Small molecules with big implications. Cancer Lett. 2017;407:123-38. Review. [DOI] [PMC free article] [PubMed]
- 84.84. Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidates†. Biol Reprod. 2019;100(5):1135-46. Erratum in: Biol Reprod. 2019;101(6):1179. Review. [DOI] [PMC free article] [PubMed]
- 85.85. Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5(13):3262-77. [DOI] [PubMed]
- 86.86. Hsieh SY, Chen RK, Pan YH, Lee HL. Systematical evaluation of the effects of sample collection procedures on low-molecular-weight serum/plasma proteome profiling. Proteomics. 2006;6(10):3189-98. [DOI] [PubMed]
- 87.87. Saare M, Rekker K, Laisk-Podar T, Rahmioglu N, Zondervan K, Salumets A, et al. Challenges in endometriosis miRNA studies - From tissue heterogeneity to disease specific miRNAs. Biochim Biophys Acta Mol Basis Dis. 2017;1863(9):2282-92. Review. [DOI] [PubMed]
- 88.88. Watson PH, Wilson-McManus JE, Barnes RO, Giesz SC, Png A, Hegele RG, et al. Evolutionary concepts in biobanking - the BC BioLibrary. J Transl Med. 2009;7:95. [DOI] [PMC free article] [PubMed]
- 89.89. Hanson EK, Ballantyne J. Circulating microRNA for the identification of forensically relevant body fluids. Methods Mol Biol. 2013;1024:221-34. [DOI] [PubMed]
- 90.90. Hanson EK, Lubenow H, Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem. 2009;387(2):303-14. [DOI] [PubMed]
- 91.91. Tian Q, Li Y, Wang F, Li Y, Xu J, Shen Y, et al. MicroRNA detection in cervical exfoliated cells as a triage for human papillomavirus-positive women. J Natl Cancer Inst. 2014;106(9):dju241. [DOI] [PMC free article] [PubMed]
- 92.92. Hesselink AT, Heideman DA, Steenbergen RD, Gök M, van Kemenade FJ, Wilting SM, et al. Methylation marker analysis of self-sampled cervico-vaginal lavage specimens to triage high-risk HPV-positive women for colposcopy. Int J Cancer. 2014;135(4):880-6. [DOI] [PubMed]
- 93.93. De Strooper LM, Verhoef VM, Berkhof J, Hesselink AT, de Bruin HM, van Kemenade FJ, et al. Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre) cancer in HPV-positive women. Gynecol Oncol. 2016;141(2):341-7. [DOI] [PMC free article] [PubMed]
- 94.94. Van Ostade X, Dom M, Tjalma W, Van Raemdonck G. Candidate biomarkers in the cervical vaginal fluid for the (self-)diagnosis of cervical precancer. Arch Gynecol Obstet. 2018;297(2):295-311. Review. [DOI] [PMC free article] [PubMed]
- 95.95. Verhoef VM, Bosgraaf RP, van Kemenade FJ, Rozendaal L, Heideman DA, Hesselink AT, et al. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): a randomized controlled non-inferiority trial. Lancet Oncol. 2014;15(3):315-22. [DOI] [PubMed]
- 96.96. Aro K, Wei F, Wong DT, Tu M. Saliva Liquid Biopsy for Point-of-Care Applications. Front Public Health. 2017;5:77. [DOI] [PMC free article] [PubMed]
- 97.97. Kaczor-Urbanowicz KE, Martin Carreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT. Saliva diagnostics - Current views and directions. Exp Biol Med (Maywood). 2017;242(5):459-72. Review. [DOI] [PMC free article] [PubMed]
- 98.98. Wingfield M, O’ Herlihy C, Finn MM, Tallon DF, Fottrell PF. Follicular and luteal phase salivary progesterone profiles in women with endometriosis and infertility. Gynecol Endocrinol. 1994;8(1):21-5. [DOI] [PubMed]
- 99.99. Petrelluzzi KF, Garcia MC, Petta CA, Grassi-Kassisse DM, Spadari-Bratfisch RC. Salivary cortisol concentrations, stress and quality of life in women with endometriosis and chronic pelvic pain. Stress. 2008;11(5):390-7. [DOI] [PubMed]