Abstract
Error-free progression through mitosis is critical for proper cell division and accurate distribution of the genetic material. The anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase regulates the progression from metaphase to anaphase and its activation is controlled by the cofactors Cdc20 and Cdh1. Additionally, genome stability is maintained by the spindle assembly checkpoint (SAC), which monitors proper attachment of chromosomes to spindle microtubules prior to cell division. We had shown a role for Tank Binding Kinase 1 (TBK1) in microtubule dynamics and mitosis and here we describe a novel role of TBK1 in regulating SAC in breast and lung cancer cells. TBK1 interacts with and phosphorylates Cdc20 and Cdh1 and depletion of TBK1 elevates SAC components. TBK1 inhibition increases the association of Cdc20 with APC/C and BubR1 indicating inactivation of APC/C; similarly, interaction of Cdh1 with APC/C is also enhanced. TBK1 and TTK inhibition reduces cell viability and enhances centrosome amplification and micronucleation. These results indicate that alterations in TBK1 will impede mitotic progression and combining TBK1 inhibitors with other regulators of mitosis might be effective in eliminating cancer cells.
Keywords: Mitosis, TBK1, TTK, SAC, Cancer
INTRODUCTION
Cell cycle checkpoints involve complex regulatory mechanisms that ensure proper genomic maintenance and fidelity of cell division. Especially, error-free progression through mitosis is critical for proper cell division and accurate distribution of the genetic material. Aberrations in cell cycle checkpoints lead to the genesis of multiple disorders and are common in the majority of human cancers 1–5. Studies in the past few decades have delineated the molecular events that govern the accurate replication of the genome during the S-phase. Further, the stringent regulatory mechanisms of the centrosome cycle (commencing at late G1 phase and culminating in mature centrosomes at G2/M phase) and mitosis facilitates the bi-orientation of the chromatids, with the proper attachment of the spindle fibers to the kinetochores. A complex interplay of regulatory molecules including kinases and phosphatases facilitate the proper array of chromosomes on the mitotic plate during metaphase. Once all the chromosomes are attached to the mitotic spindle fibers and properly arranged on the metaphase plate, the ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) degrades securin, activating separase. The active separase then degrades cohesin, resulting in the separation of sister chromatids and facilitating the entry of the cells into anaphase. APC/C thus plays a central role in exit from metaphase, segregation of sister chromatids and entry into anaphase 6.
To prevent improper segregation of chromosomes, cell monitors proper attachment of all chromosomes with spindle fibers through the spindle assembly checkpoint (SAC) to ensure that mitosis is delayed until all the kinetochores are attached. SAC functions through mitotic checkpoint complex (MCC), composed of SAC genes Cdc20, Mad2, BubR1/Mad3, and Bub3 7. Unattached or improperly attached kinetochores to the microtubules are sensed by SAC during mitosis, enhancing the production of active MCC. MCC then associates with APC/CCDC20 to inactivate it, forming the functionally inactive APC/CMCC complex 8. The SAC thus functions by inhibiting the ubiquitin ligase activity of APC/C so that securin remains intact and the sister chromatids are not separated prematurely, leading to aneuploidy 9. Dysfunctional SAC has been shown to cause chromosome miss-aggregation and aneuploidy has been implicated in cancer, birth defects and other human diseases 10–12.
APC/C is a 14 subunit, 1.2 mega-Dalton (MDa) complex 13 and as it is centrally important to cell cycle progression, its activity is tightly regulated. This regulation is achieved in form of transient associations of coactivators Cdc20 and Cdh1 with APC/C 14. Phosphorylation of APC/C, Cdc20 and Cdh1 help in differentially regulating the APC/C activity (by mitotic kinases like Cdk1, Cdk2 and Plk1. APC/C phosphorylation by Cdk1/2 or Plk1 (Polo like kinase 1) increases its binding affinity for Cdc20 and formation of the active APC/CCDC20 complex 15–18. Cdh1 phosphorylation by Cdk1 or Cdk2 on the other hand plays an opposite role and inhibits its binding with and the subsequent activation of APC/C 16,19. Similarly, Cdc20 phosphorylation by Cdk1/2 has also been found to inhibit its binding and activation of APC/C 20–22.
Several kinases 23 and phosphatases 24 are involved in the regulation of APC/C. Our earlier studies showed that the non-canonical IκB kinase, TANK Binding Kinase 1 (TBK1) plays a major role in regulating mitosis. TBK1 regulates interferon signaling and NFκB function and also participates in RalB-mediated inflammatory responses 25. It has been found to be essential for the survival of non-small cell lung cancers (NSCLC) driven by oncogenic KRAS 26. Recent evidence also suggests a role for TBK1 as a driver of cancer progression and it is reported to be overexpressed in breast, colon, lung and pancreatic cancer 26. Our published results demonstrate a role for TBK1 in microtubule dynamics and regulation of mitosis 27. TBK1 was found to phosphorylate and associate with NuMa and CEP170, both of which are essential to mitosis. As TBK1 plays an essential role in both mitosis and cancer progression, we hypothesize that it performs these functions by regulating SAC, which is frequently deregulated in cancer. In the present study we report that TBK1 interacts with and phosphorylates SAC component cdc20 during mitosis. Also, TBK1 inhibition resulted in elevated levels of SAC components BubR1 and Cdc20. TBK1 inhibition also affected the interaction of Cdc20 and Cdh1 with APC/C and accumulation of cells in mitosis. Altogether, we find that TBK1 plays a role in overcoming SAC and this might be a potential mechanism by which it enables cancer progression. We also find that in addition to TBK1, TTK, a kinetochore associated kinase, that is critical to initially phosphorylate and assemble SAC regulators such as Mad and Bub family members to initiate the SAC 28, contributes to the regulation of SAC. The interplay between these kinases and their regulation of SAC appear to be contributing to the proper fidelity of mitotic progression in cancer.
RESULTS
TBK1 and TTK inhibition result in reduced cell viability and proliferation
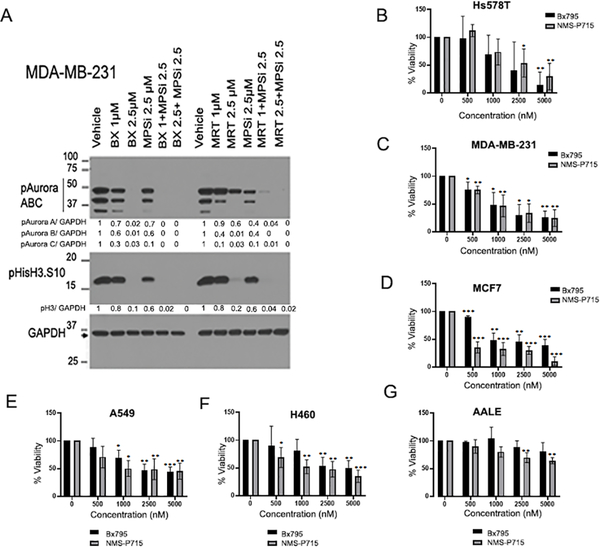
TBK1 and TTK both play prominent roles in regulating mitosis and centrosome biology. Therefore, we examined if inhibition of TBK1 using BX795 or MRT67307, or inhibition of TTK with NMS-P715, alone or in combination, affects the levels of cell cycle regulators and downstream signaling molecules. Interestingly, treatment with the TBK1 inhibitor (BX795 or MRT67307) alone, or in combination with the TTK inhibitor (NMS-P715), suppressed the phosphorylation levels of on all three isoforms of Aurora kinases and Histone H3S10, another mitotic marker (Figure 1A); this is significant because Aurora kinases contribute to EMT and proliferation of multiple cancer cell types 29,30. Further, Aurora Kinase B phosphorylates Hec1 to localize TTK to unattached kinetochores 31. Based on these findings, we examined if TBK1 inhibitor BX795 and TTK inhibitor NMS-P715 influenced the viability of cancer cells by testing increasing concentrations of the inhibitors on three breast cancer cell lines, including the triple-negative Hs578T cells (Figure 1B), triple-negative MDA-MB-231 cells (Figure 1C), and luminal B MCF7 cells (Figure 1D), two lung cancer cell lines A549 (Figure 1E) and H460 (Figure 1F) and an immortalized primary lung epithelial cell line AALE (Figure 1G). Both inhibitors were effective in reducing viability of all the cancer cell lines. At the same time, no significant reduction in viability was observed in AALE cells, especially with BX795; there was a reduction in the viability of AALE cells when high doses of NMS-P715 was used. We then performed an IC50 analysis of Bx795 in combination with increasing dose of NMS-P715 in MDA-MB-231 (Supplementary Figure 1A), A549 (Supplementary Figure 1B) and H460 (Supplementary Figure 1C). IC50 of Bx795 was found to decrease with increasing dose of NMS-P715 in all the three cell lines (Supplementary Figure 1D). These results suggest that TTK and TBK1 may be playing a part in regulating the proliferation of these cells and therefore their inhibition might be effective in specifically killing breast and lung cancer cells while not having a major effect on normal cells.
Figure 1: Inhibition of TBK1 and TTK reduces cell proliferation and viability.
(A). MDA-MB-231 cells were treated with indicated doses of TBK1 inhibitor BX795 and TTK inhibitor NMS-P715 alone or in combination for 24 hours. Western blot shows reduced levels of mitotic markers phospho-Aurora kinase and phospho-histone H3-S10, which indicates inhibition of cell proliferation. Normalized quantification for respective proteins is shown below each lane. B-F. MTT assays were performed after treatment with increasing concentrations of TBK1 inhibitor (BX795) or TTK inhibitor (NMS-P715) for 96 hours in the following breast cancer cell lines: HS578T (B); MDA-MB-231 (C); MCF7 (D); and lung cancer cell lines A549 (E); H460 (F); and Immortalized primary lung epithelial cell line AALE (G). Plots are an average of three independent experiments. Error bars represent mean ± S.D (*p<0.05, **p<0.01, ***p<0.001; one-way ANOVA).
TBK1 physically interacts with and phosphorylates Cdc20 and Cdh1
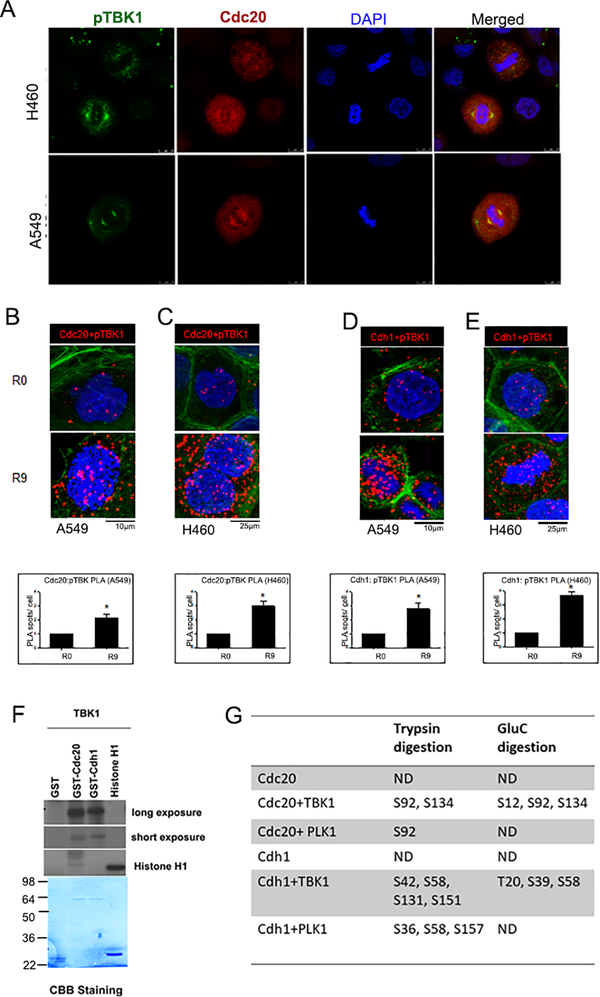
Previous studies from our lab had revealed that TBK1 inhibition resulted in inhibition of mitotic progression resulting in aberrations of the mitotic structures, eventually leading to mitotic catastrophe 27. Since the mitotic spindles were markedly deformed upon inhibition or depletion of TBK1, we examined whether TBK1 associated with components of the SAC machinery. A double immunofluorescence experiment on A549 and H460 lung cancer cells showed that pTBK1 colocalizes with Cdc20 (Figure 2A). This was confirmed using IP-Western blot analysis on A549 cells that were arrested by a double thymidine block (R0) and released from the block for 9hr (R9). There was a marked association of pTBK1 (Supplementary Figure 2A) and total TBK1 (Supplementary Figure 2B) with Cdc20. A proximity ligation assay (PLA)32,33 confirmed a significant association of Cdc20 with pTBK1in both A549 and H460 cells released from thymidine block (Figure 2B and C). In addition, PLA showed increased association of Phospho-TBK1 with Cdh1 9 hr post double thymidine block release as compared to cells in thymidine block (R0) (Figure 2D and E). Since TBK1 is a kinase, in vitro kinase assays were conducted to assess if Cdc20 and Cdh1 act as substrates for TBK1 mediated phosphorylation. Assays were conducted using purified TBK1 protein and GST-Cdc20 or GST-Cdh1 as substrates, using 32P-ATP in the reaction. It was found that TBK1 phosphorylates both Cdc20 as well as Cdh1 (Figure 2F). In vitro kinase assays were also conducted using cold (unlabeled) ATP; reaction products were submitted for mass spectrometry analysis for the identification of the phosphorylation sites. Plk1, one of the key regulators of cell division 34, is known to phosphorylate Cdc20 at Ser 92. Other than the Plk1 mediated phosphorylation sites, TBK1 was found to mediate Cdc20 phosphorylation at Ser 134 in both tryptic and GluC digestion. TBK1 was also found to phosphorylate Cdh1 at Thr 20, Ser 39, Ser 42, Ser 58, Ser131 and Ser 151 (Figure 2G). As a positive control we tested Plk1, one of the key regulators of cell division 34, which is known to phosphorylate Cdc20 at Ser 92, and results confirmed this. Similarly, we also found that Plk1 mediates phosphorylation of Cdh1 at Ser 36, Ser 58 and Ser 157 (Figure 2G). These results suggest that TBK1 physically interacts with Cdc20 and Cdh1 in cells, especially in mitosis, leading to their phosphorylation. It is likely that there are additional residues phosphorylated by TBK1, since mutation of the above sites still yielded phosphorylated bands (data not shown).
Figure 2: TBK1 interacts with and phosphorylates Cdc20 and Cdh1.
(A) Double immunofluorescence for pTBK1 (green) and cdc20 (red) was performed in A549 and H460 cells. Confocal images show a colocalization of pTBK1 with Cdc20 in the cell. (B-C). Proximity ligation assay (PLA) performed on (B) A549 and (C) H460 cells shows interaction of pTBK1 and Cdc20 in cells released from G1/ S block. D-E. PLA performed on (D) A549 and (E) H460 cells shows interaction of pTBK1 and Cdh1 in cells released from G1/ S block. Red dots represent foci of interaction of pTBK1 with Cdc20 or Cdh1, DAPI is shown in blue and phalloidin in green. Plots are an average of two independent experiments. Error bars represent mean ± S.D. (*p<0.05; unpaired two tailed t-test). (F) In vitro kinase assay shows phosphorylation of Cdc20 and Cdh1 by TBK1. (G) Phosphorylation sites were identified using mass spectrometric analysis after trypsin or GluC protease digestions.
SAC components are overexpressed in TBK1 depleted cells
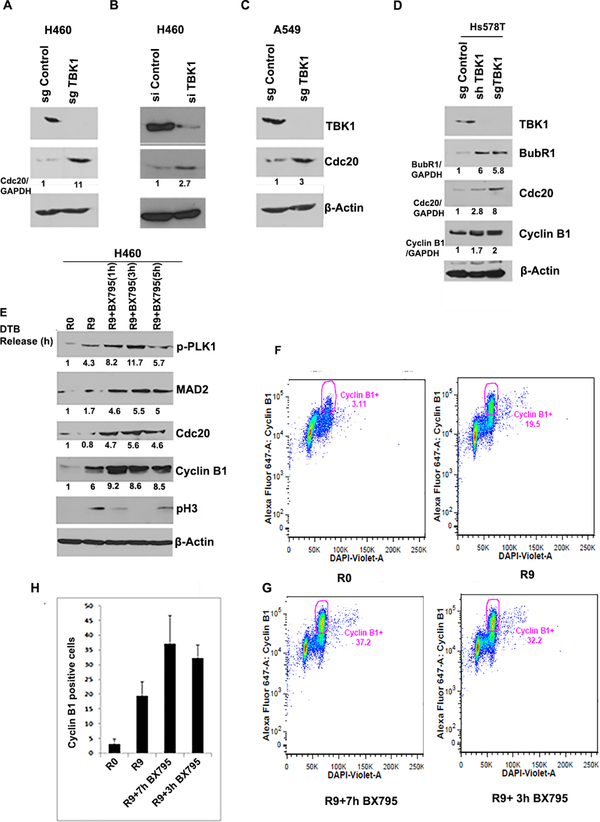
To further expand these studies, TBK1 was knocked out in both lung and breast cancer cell lines using CRISPR/Cas9 genome editing tool. Interestingly, TBK1 depleted cells exhibited an increased level of the SAC component Cdc20 in both lung (Figure 3A- 11 folds; Figure 3B- 3 folds and Figure 3C- 3 folds) and breast cancer cells (Figure 3D- 3 and 8 folds for shTBK1 and sgTBK1 respectively). In addition, protein level of SAC component BubR1 was also found to be enhanced in breast cancer cell line on TBK1 knockdown (Figure 3D- 6 folds). Cyclin B1 protein level was found to be increased in Hs578T (1.7 and 2 fold, respectively, for shTBK1 and SgTBK1). For further confirmation, we synchronized H460 cells through double thymidine block following TBK1 inhibition using BX795. We found an increase in Cdc20 level on TBK1 inhibition (Figure 3E- 5 folds). We also observed an increase in cyclin B1 levels in R9 in comparison to control (Figure 3E- 6 folds). R9 cells treated with BX795 for different time periods were analyzed for SAC components including MAD2, Cdc20 and mitotic markers like pH3 and cyclin B1. Following treatment with BX795, we noted further increase in the levels of Cyclin B1 (9 folds), pH3, Mad2 (5 folds) and Cdc20 (Figure 3E). These observations suggested a possible inactivation of APC/C. We confirmed this by conducting bivariate flow cytometry analysis of cyclin B1 positive cells in the presence of the TBK1 inhibitor BX795. For this, we arrested A549 cells at G1/S boundary by double thymidine block and released it for 9hr with BX795 addition after 2hr (R9+7hr BX795 treatment) and 6hr (R9+3hr BX795) of release. As expected, there was more than two-fold increase in cyclin B1 positive population after 9hr of release (R9) compared to cells at G1/S boundary (R0) (Figure 3F). In addition, we observed a significant increase in the number of Cyclin B1 positive cells following release from double thymidine block in the presence of BX795 (two-fold) compared to non-BX795 treated cells (Figure 3 G and H). This, again, suggested a possible inactivation of APC/C complex.
Figure 3: TBK1 depletion and inhibition upregulates expression of SAC components.
(A-B) TBK1 depletion using (A) CRISPR/ Cas9 tool or (B) siRNA results in increased protein level of Cdc20 in H460 cells. (C) TBK1 depletion using CRISPR/ Cas9 tool results in increased Cdc20 protein level in A549 cells. (D) TBK1 was depleted using CRISPR/ Cas9 or shRNA tool in Hs578T breast cancer cell line. Western blot analysis showed an increase in Cdc20, BubR1 and cyclin B1 on TBK1 depletion. (E) H460 cells synchronized at G1/S and released for indicated time period were treated with BX795. Western blot analysis of cell lysates shows that SAC components Cdc20 and MAD2 are increased on TBK1 inhibition. Levels of Cdc20 and cyclin B1 are also increased. Normalized quantification for respective proteins is shown below each lane. (F-G) A549 cells released from a G1/S block were treated with BX795 for 3 or 7 hours. Bivariate flow cytometry analysis of cyclin B1 positive cells shows increased positive cells in R9 as compared to R0 (F). (G) In the presence of BX795 there is a further increase in cyclin B1 positive cells. (H) Quantification of two experiments shows an increase in cyclin B1 positive cells upon TBK1 inhibition. Error bars represent mean ± S.D.
Inhibition of TBK1 increases association of Cdc20 with APC1 and BubR1
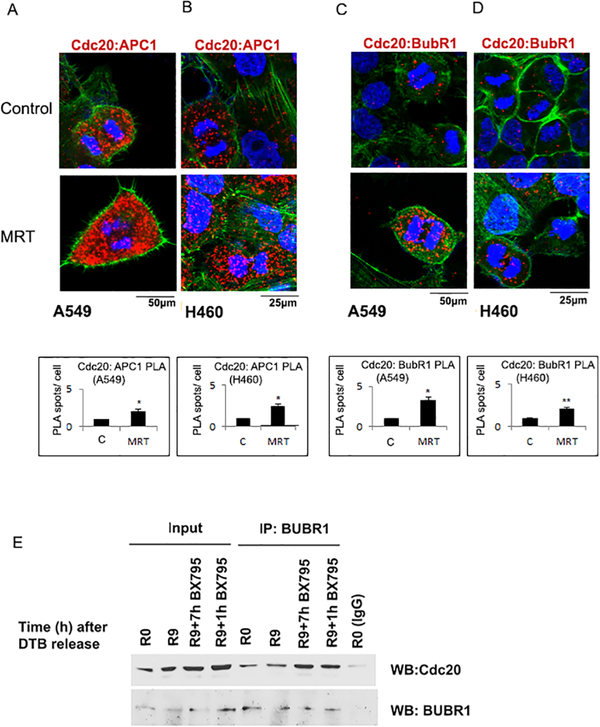
Cdc20 interacts with and activates APC/C 15–18 while Cdc20 phosphorylation by Cdk1/2 has been shown to inhibit its binding with APC/C 20,21. Since we had found that TBK1 phosphorylates Cdc20, we examined how TBK1 inhibition affects the association of Cdc20 with APC/C. A549 and H460 cells were subjected to double thymidine block and treated with the TBK1 inhibitor, MRT67307 35, which has been found to be effective in the past 27. A PLA showed that Cdc20 and APC/C interaction was increased upon TBK1 inhibition in both A549 (Figure 4A) and H460 cells (Figure 4B). We did not observe any change in the APC-Cdc20 interaction in the control AALE cells PLA (Supplementary Figure 3). It is well established that unattached kinetochores are recognized by SAC components, which results in enhanced expression of MCC genes Cdc20, Mad2, BubR1/Mad3 and Bub3 7. The MCC then binds to the active APC/CCdc20 to form the inactive APC/CMCC. Since we had found that TBK1 knockdown and inhibition results in an activation of SAC as indicated by increased levels of MCC proteins- BubR1 (Figure 3D) and MAD2 and (Figure 3E), we examined if TBK1 inhibition led to an increased association of SAC components and Cdc20 to inhibit the active APC/CCdc20. It has been established that BubR1 binding to Cdc20 inhibits activation of APC/C either alone 36 or in combination with Mad2 37. In order to assess if TBK1 inhibition enhances association between BubR1 and Cdc20, we performed a PLA. As expected, A549 and H460 cells treated with TBK1 inhibitor MRT67307 showed an increase in the association of BubR1 and Cdc20, as evident from the increased number of foci in TBK1 inhibitor treated cells (Figure 4 C and D); the single antibody control for this PLA is shown in Supplementary Figure 4. Furthermore, we performed immunoprecipitation on A549 cell lysate using a BubR1 antibody followed by immunoblotting with a Cdc20 antibody. It was found that TBK1 inhibition resulted in an increased interaction between Cdc20 and BubR1 (Figure 4E). Taken together, these results suggest that while interaction of Cdc20 with APC1 increases on TBK1 inhibition, APC/CCdc20 is present as the inactive form of APC/CMCC.
Figure 4: APC1 and BubR1 interaction with Cdc20 increases on TBK1 inhibition.
Cells were synchronized at G1/S and released for 9 hours (R9) and treated with TBK1 inhibitor MRT67307 or not for 7 hours, starting 2 hours after release. (A-B) Confocal images of PLA shows an increased association of Cdc20 and APC1 in A. A549 and B. H460 cells after TBK1 inhibition. (C-D) PLA shows an increased association of Cdc20 and BubR1 in C. A549 and D. H460 cells after TBK1 inhibition. Red dots represent foci of interaction of Cdc20 with APC1 or BubR1. DAPI is shown in blue and phalloidin in green. Plots represent quantification of interaction from two independent experiments. A minimum of 100 cells were considered for each quantification. A two-tailed T-test was done for significance. p≤ 0.05 was considered significant. (*=p<0.05 and **=p<0.01) Error bars represent mean ± S.D. (E) Immunoprecipitation- western blot analysis performed on A549 cells, G1/ S synchronized, which were released for 9 hours from the block and treated for the indicated time periods with TBK1 inhibitor show that interaction of Cdc20 with BubR1 increases on TBK1 inhibition. Cell lysates were immunoprecipitated with BubR1 antibody, western blotting was done for Cdc20 to confirm the interaction.
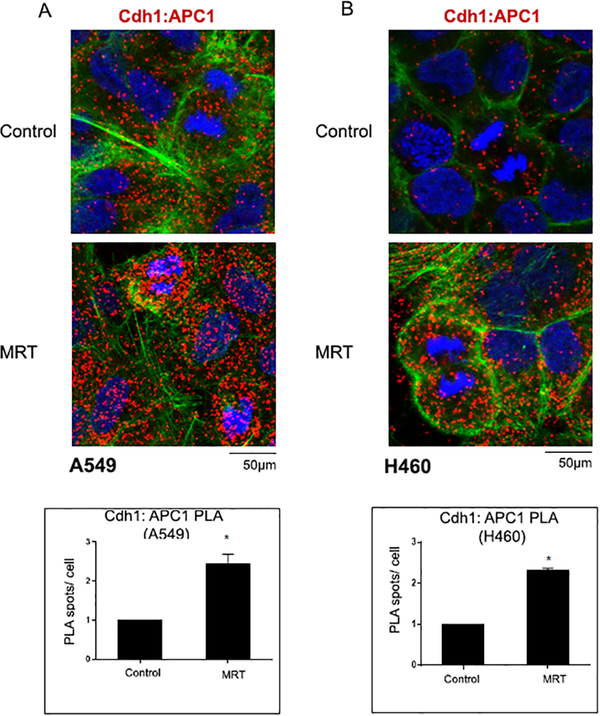
Inhibition of TBK1 increases association of Cdh1 with APC1
Inactivation of APC/CCdh1 is required for the G1 to S transition of the cell 38. There are several mechanisms in place which regulate APC/CCdh1 activity in the cell 14,39. One of these mechanisms is the phosphorylation of Cdh1, which leads to dissociation of Cdh1 from APC 16,40. We had found that TBK1 phosphorylates Cdh1 (Figure 2F, G). We therefore examined whether inhibition of TBK1 would result in an increased association of APC1 with Cdh1. For this, a PLA was performed on A549 and H460 cells after DTB synchronization and treatment with TBK1 inhibitor MRT67307 (Figure 5). A549 cells were also analyzed by PLA after treatment with TBK1 inhibitors amlexanox (Supplementary Figure 5A) and Bx795 (Supplementary Figure 5B). Cells were subjected to a double thymidine block, released for 9 hours and treated with TBK1 inhibitors for the last 7 hr. As expected, foci representing Cdh1 and APC/C interaction were increased upon TBK1 inhibition in both A549 (Figure 5A) and H460 cells (Figure 5B); single antibody control for the PLA is shown in Supplementary Figure 4. This shows that TBK1 activity is necessary for proper progression through mitosis and its inhibition affects the regulatory functions of the APC/C complex, resulting in mitotic aberrations.
Figure 5: Cdh1 and APC1 interaction increases on TBK1 inhibition.
Cells were synchronized at G1/S and released for 9 hours (R9) and treated with TBK1 inhibitor MRT67307 for 7 hours, starting 2 hours after release. (A-B) Confocal images of PLA show an increased association of Cdh1 and APC1 in A549 (A) and H460 (B) cells after TBK1 inhibition. Red dots represent foci of interaction of Cdh1 with APC1. DAPI is shown in blue and phalloidin in green. Plots represent quantification of interaction from two independent experiments. A minimum of 100 cells were considered for each quantification. A two-tailed t-test was done for significance. p≤ 0.05 was considered significant. Error bars represent mean ± S.D.
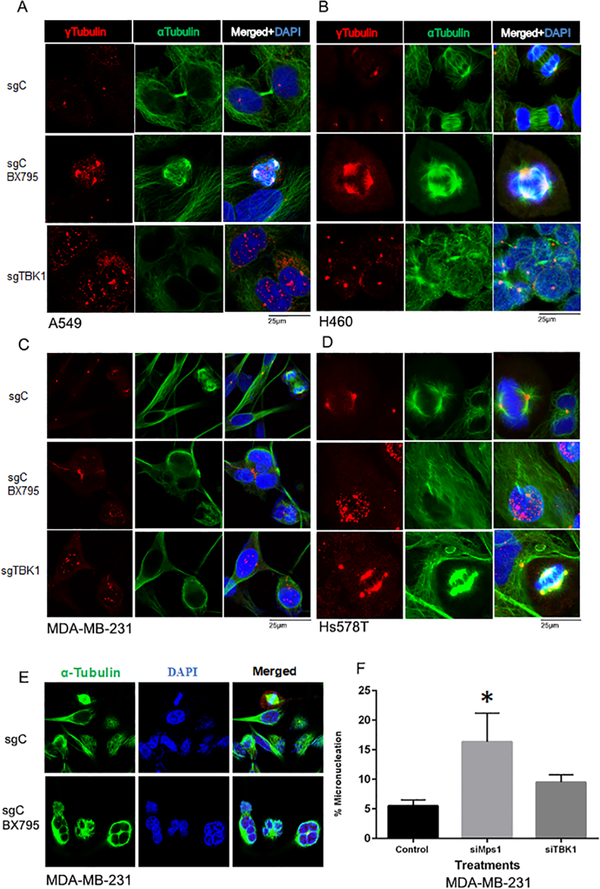
TBK1 depletion and inhibition result in increased mitotic aberrations
It has been shown that TBK1 regulates centrosome homeostasis and mitotic progression by phosphorylating Plk1, NuMA and CEP170 27,41. Therefore, we wanted to study whether TBK1 depletion and inhibition result in centrosome amplification leading to mitotic defects. For this, we did a double immunofluorescence for α and γ tubulin after TBK1 knockdown or inhibition in A549 (Figure 6A), H460 (Figure 6B), MDA-MB-231 (Figure 6C) and HS578T (Figure 6D) cells. Centrosome amplification was evident by γ tubulin staining, which was found to increase upon TBK1 knockdown as well as inhibition in A549, H460, Hs578T and MDA-MB-231cell lines (Figure 6 A–D; quantification is shown in Supplementary Figure 6). Further we observed that TBK1 inhibition in MDA-MB-231 cells resulted in appearance of multinucleated cells (Figure 6E). Studies have shown presence of micronuclei to be a hallmark of chromosome instability. Micronuclei are formed when kinetochores of one or more chromosomes are not properly attached to microtubules 42. DNA in micronuclei is prone to damage as the nuclear envelope of micronuclei is very fragile and prone to breakdown 43. TTK is a SAC kinase and one of its known key regulators 44. Since TBK1 phosphorylates SAC component Cdc20 and plays a key role in maintaining proper microtubule dynamics 27, we examined if TTK and TBK1 contribute to micronuclei formation. For this, we performed micronuclei assay in MDA-MB-231 treated with RNA interference sequences for TTK (Mps1) and TBK1 (Figure 6F). Our results showed that knockdown of TTK resulted in a significant increase in percentage of cells showing micronuclei, by 16.3 %, compared to cells transfected with a control siRNA, where we observed 5.5% of cells with micronuclei. TBK1 knockdown also led to an increase in cells showing micronuclei by 9.5% as compared to cells transfected with control siRNA, however this increase was not found to be significant. These results show that TBK1 and TTK are essential for error free cell division and their reduced activity might result in the formation of micronuclei.
Figure 6: TBK1 inhibition and depletion results in mitotic aberrations.
TBK1 was either chemically inhibited or depleted using CRISPR/ Cas9 techniques in A549 (A), H460 (B), MDA-MB-231 (C) and Hs578T (D) cells; they were then stained for α tubulin (green) and γ tubulin (red) to study mitotic aberrations, specifically centrosomal amplification. (E) MDA-MB-231 cells were treated with 2.5μM BX795 for TBK1 inhibition and stained with α Tubulin and DAPI show multinucleation. (F) Micronucleation was studied in MDA-MB-231 cells after knockdown of TBK1 or TTK using siRNA transfection. A total of 200 cells were considered and micronuclei counted for each condition for quantification. Plot represents mean of three independent experiments. A one-way ANOVA-test was done for significance. p≤ 0.05 was considered significant. Error bars are mean ± S.D.
DISCUSSION
TBK1 is essential for proper immune response and plays a vital and indispensable role in innate immunity. It functions downstream of the STING pathway and modulates the expression of specific genes downstream of the NFκB pathway, in response to pathogens 45. Apart from its role in the immune system, it has been shown that TBK1 plays a major role in facilitating cellular transformation 46. An unbiased shRNA screen showed that TBK1 was necessary for K-Ras mediated transformation, especially in non-small cell lung cancer 47. Additional studies showed that TBK1 exerts oncogenic functions by promoting survival pathways downstream of Akt. More recently, TBK1 has been found to be a major regulator of mitophagy, by modulating Parkin, PINK1 and optineurin 48,49. It has been reported that PINK1 and Parkin can modulate cell cycle progression by sequestering TBK1 in the mitochondria. In this context, our earlier studies have shown a more direct role for TBK1 in mitosis, where it associated with centrosomal proteins and components of the microtubule machinery. Here, we also showed that it regulates mitosis by phosphorylating CEP170 and NuMa thereby modulating their function 27.
The studies presented here follow up on the earlier observations on the role of TBK1 in mitosis. We had found that the phosphorylation status of TBK1 changes during mitosis, suggesting that it is activated during this phase. Our current results suggest that this activation might be necessary for neutralizing the SAC, once all the chromosomes are properly aligned on the mitotic spindle. Here we show that phosphorylated TBK1 associates with SAC component Cdc20, and this association increases as cells progress through mitosis after a double thymidine block. Phospho-TBK1 also interacts with another APC/C cofactor Cdh1 and the interaction is similarly more in mitotic M phase cells (R9) as compared to cells in S phase (R0). We saw increased protein levels of SAC components on TBK1 chemical inhibition. This coincided with an increased level of cyclin B1, which is a substrate of APC/CCdc20. One of the key mechanisms regulating APC/C activity is phosphorylation of its adaptor proteins, Cdc20 and Cdh1 23. Both of these adaptor proteins have WD40 domains which specifically associate with conserved motifs in APC/C substrates such as the D-box 50, KEN-box 51, A-box 52 or O-box 53. Association with Cdc20 and Cdh1 results in activation of APC/C at distinct time points during the cell cycle. APC/CCdc20 is active during mitosis and facilitates the transition of cells from metaphase to anaphase 54. In the present study our results indicate that TBK1 inhibition leads to an increased association between APC1 and Cdc20. This is in accordance with previous reports which show that Cdc20 phosphorylation interferes with its association with APC/C 23. Further, we show that TBK1 inhibition results in an increased association of Cdc20 with MCC component BubR1. This suggests that MCC associates with active APC/CCdc20, which results in the inactive form of APC/C, ie. APC/CMCC. Similar to Cdc20, phosphorylation of Cdh1 is known to hinder its association with APC/C 23 and we find an increased association of APC1 and Cdh1 on TBK1 inhibition. While these results strongly indicate the TBK1-mediated phosphorylation of Cdc20 and Cdh1 contributes to satisfying SAC, it is possible that indirect regulatory mechanisms might also contribute to the process. For example, TBK1-mediated regulation of Cep170 is necessary for the proper centrosome separation, as we reported earlier 27. Aberrations in the regulation of Cep170 might be contributing to the observed centrosome amplification, which in turn activates SAC. While this is a possibility, our data suggests that TBK1-mediated regulation of Cdc20 and Cdh1 indicate a novel role for TBK1 in facilitating transit through the SAC.
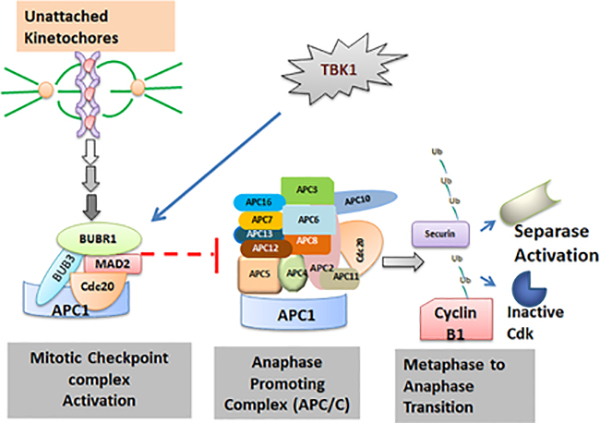
Similar to TBK1, TTK is another important kinase that protects the fidelity of chromosome segregation 28,55. It is a kinetochore associated kinase and its activity has been shown to be essential for the functioning of SAC 56. It plays a role in increasing the rate of formation of MCC 7,57 and it is required for recruitment of PLK1, CENP-E, MAD1 and MAD2 58–61. TTK is rarely found to be mutated in cancer 62, however it is overexpressed in tumors like breast, lung, thyroid etc 63. TTK overexpression correlates with poor prognosis in breast cancer 64,65, however, recent studies have found TBK1 to be overexpressed in a wide range of cancers including that of the breast, colon, lung and pancreas 26. We show here that inhibition of TBK1 and TTK either individually or in combination markedly reduced expression of cell cycle regulators, including Aurora kinases and Phospho-Histone H3. This is significant because Aurora kinases have been reported to contribute to EMT and endocrine resistance 29,30, 29,30, and are currently being tested in clinical trials 66. In addition, mitotic slippage and failure to apoptose are known mechanisms of failure of agents that activate the SAC, including microtubule inhibitors 67. Thus, the fact that a TBK1 inhibitor could downregulate the activity of all the three Aurora kinases is also a translatable observation. We also find that treatment with both these inhibitors reduce the cell viability in breast cancer cell lines across subtypes. Our experiments also suggest that TBK1 and TTK can complement each other to a certain extent. Depletion or inhibition of TBK1 or TTK led to several chromosomal abnormalities and appearance of micronuclei which might eventually lead to mitotic catastrophe31 and cell death. It seems to be a likely possibility that combining TTK and TBK1 inhibitors, or either inhibitor with a mitotic poison, might be a novel and effective strategy to combat certain cancers that overexpress these kinases. Our results from the lung and breast cancer cells support this notion and show that inclusion of TTK inhibitor significantly reduces the IC50 concentration for TBK1 inhibitor. Certain TBK1 inhibitors like Amlexanox have been used as anti-inflammatory agents, and additional studies will be needed to establish if targeting these kinases will be able to eliminate cancer cells selectively without affecting normal cells, and act as viable anti-cancer agents. Our group had previously reported that TBK1 inhibition resulted in inhibition of transition from metaphase to anaphase 27. Phosphorylation is a key mechanism of regulation for major cell cycle effector proteins 68–70. We show here that Cdc20 and Cdh1 are both substrates of TBK1 mediated phosphorylation. SAC function has been found to be compromised in malignant tumors and has been reported to result in aneuploidy and carcinogenesis 71. SAC comes into play to ensure that all the chromosomes are bi-polarly attached to the spindle microtubules, so that they are segregated and divided equally between daughter cells. Mitotic checkpoint complex (MCC) which is composed of Cdc20, Mad2, BubR1 and Bub3 is the effector of SAC. MCC associates with APC/CCdc20 to render it inactive. TBK1 inhibition induced SAC, thus resulting in inactivation of APC/C (Figure 7, Schematic).
Figure 7: Schematic representing mechanism of cell cycle regulation by TBK1.
Unattached kinetochores result in the activation of mitotic checkpoint complex, which leads to inhibition of activation of APC/C and the subsequent onset of anaphase. TBK1 mediated phosphorylation of Cdc20 or Cdh1 inhibits the formation of MCC and drives the cell through mitosis.
Cdh1 is another adaptor which renders specificity to the ubiquitin ligase function of APC/C. APC/CCDH1 is active during late mitosis and regulates the G1 transition 14. Similar to Cdc20, phosphorylation of Cdh1 hinders its association with APC/C 23. Therefore, expectedly, we observed an increase in association of APC1 and Cdh1 on TBK1 inhibition. It has been previously established that inactivation of APC/C Cdh1 is necessary for G1 to S transition of the cell. This suggests that TBK1 can potentially bypass the G1/S checkpoint.
Our studies and other groups have previously shown that TBK1 is a key regulator of centrosome homeostasis and is required for mitosis progression 27,41. We show in this study that TBK1 inhibition or depletion results in centrosome amplification and mitotic defects. In addition, micronuclei formation, which is a hallmark of chromosome instability, is increased on TTK and TBK1 knockdown. In conclusion, while TTK is a well-established regulator of SAC, we show a novel role for TBK1 in SAC regulation. Altogether, our study shows that while TBK1 is essential for mitotic progression, it has the potential to bypass both the G1/S and the spindle assembly checkpoints by which the cell ensures genetic fidelity. Thus, we propose that this might be the mechanism through which TBK1 acts as a cancer driver as suggested by its over-expression in various cancers.
MATERIALS & METHODS
Cell Culture and mitotic arrest
Human non-small cell lung adenocarcinoma cell lines A549 and H460, immortalized primary lung epithelial cell line AALE and breast cancer cell lines MCF7, Hs578T and MDA-MB-231 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). A549 cells were maintained in Ham’s F12K medium (Cellgro, Corning) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO), H460 cells were maintained in RPMI 1640 (Gibco, Life Technologies) containing 10% fetal bovine serum, MCF7, Hs579T and MDA-MB-231 were maintained in DMEM (Corning) containing 10% fetal bovine serum, AALE was maintained in BEGM medium (Lonza). To synchronize cells in G1/S boundary, cells were subjected to double-thymidine block (DTB) using standard protocols 72. In brief, cells were treated with 2mM thymidine (Sigma-Aldrich) for 18 hours after which they were washed and incubated with fresh medium for 9 hours. For the second thymidine block, cells were incubated with 2mM thymidine for 18 hours and then released with complete medium for indicated time points.
Plasmids and Reagents
Cells were treated with BX795 (#s1274, selleckchem), MRT67307 (#SML0702, Millipore Sigma) or amlexanox (#4857, Tocris) to inhibit TBK1 and NMS-P715 (#475949, Millipore Sigma) to inhibit TTK Plasmids expressing GST-Cdh1 and GST-Cdc20 were kind gift from Dr. Lixin Wan. Phospho (S172) TBK1 (#5483), phospho PLK1 (#5472S), phospho histone H3 (#3377), TTK (#3255T), APC1 (#13329), pAurora A (Thr288), B (Thr232), C (Thr 198) (#2914S) and total TBK1 (#3013S) antibodies were purchased from Cell Signaling, and antibody against Cdc20 (SC-13162) was purchased from Santa Cruz Biotechnology. β-actin (#A1978, clone AC-15) and α-tubulin (#T6074) antibodies were purchased from Sigma Chemical Co. Cyclin B1 antibody (#5472S) was purchased from BD Biosciences. pTTK antibody (#44–1325G) was purchased from Novex. BubR1 (ab70544), Mad2 (ab97777) and Cdh1 (ab89535) antibodies were purchased from Abcam. γ-tubulin antibody (# PA5–34815) was purchased from Invitrogen. Control siRNA (sc-37007) was purchased from Santa Cruz Biotechnology and TBK1 siRNA (# 4457298, ID: s761) was purchased from Ambion.
Lysate Preparation and Western Blot Analysis
Cells were washed twice with cold 1x PBS, scraped off the plates, collected by centrifugation for 5 minutes at 4,000 rpm, and lysed in M2 lysis buffer (20mM Tris-HCl pH6.0, 0.5% NP-40, 250mM NaCl, 3mM EGTA, and 3mM EDTA) containing protease inhibitors as described in our previous work 73. After lysis, protein concentration was measured using Bradford assay (BioRad) and equal amounts of proteins were resolved on 8 or10% SDS-page polyacrylamide gels and transferred onto nitrocellulose membranes using BioRad semi-dry transfer unit. Immunoblotting was then performed with the indicated antibodies. Interaction between proteins in vivo was analyzed by immunoprecipitation–western blot analyses with 250μg of lysate and 2μg of the indicated antibody as described previously 74. Western blotting images are representative of at least two independent experiments.
Immunofluorescence
A549, H460, MDA-MB-231 and Hs578T cells were seeded on eight-well glass chamber slides at a density of 5,000 cells per well. After indicated treatments cells were fixed with 10% buffered formalin for 20 mins or with chilled methanol at −20°C for 5 minutes, followed by cell permeabilization with PBS containing 0.25% Triton X-100 in case of formalin fixation. Cells were then blocked using 5% goat serum for an hour and incubated with primary antibody overnight. Primary antibodies used were phospho-TBK1 (1:200), Alpha tubulin (1:2,000), Gamma tubulin (1:250) and Cdc20 (1:200). Anti-rabbit Alexa Fluor-488 or anti-mouse Alexa Fluor-594 (Molecular Probes) was used as secondary antibody. DAPI was used to stain the nuclei. Cells were visualized with a DM16000 inverted Leica TCS SP8 tandem scanning confocal microscope with 63x/1.40NA oil immersion objective. Images and Z-stacks were produced with three cooled photomultiplier detectors and analyzed with the LAS AF software version 1.6.0 build 1016 (Leica Microsystems, Germany).
Lentiviral sgRNA production and infection
TBK1 guide RNA (gRNA) was cloned into pLentiCRISPRV2 backbone vector using the BsmBI (NEB) restriction enzyme. For lentivirus production, 1×106 HEK293FT cells were seeded in a 100mm dish. Lentiviral vectors expressing sgRNAs specific for control sequences or for TBK1 (5μg) were transfected into 293FT cells along with the packaging plasmids pSPAX2 (10μg) and pMD2.G (10μg) using Fugene HD (Roche). Culture supernatants containing lentivirus were collected 48 and 72 h after transfection. Virus was pooled and stored at −80 °C. Cells (seeded at the density of 120,000 cells per well of 6 well plate) were spin-infected using the supernatant containing virus in polybrene (8ug/ml) containing medium, by centrifuging at 2000rpm for 1h at RT, to enhance the efficacy of infection. After two rounds of infection, cells were allowed to grow for another 48h before selection using puromycin (1–2μg/ml).
In vitro kinase assays
In vitro kinase reaction was carried out as described previously 27. Reaction buffer (50 mM HEPES, pH 7.9, 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, and 10 mM β-glycerophosphate) containing GST-Cdc20 or GST-Cdh1 as substrate (2ug), 100 μM ATP, and 10 μCi γ32P-ATP was incubated with 100 ng enzymatically active TBK1 (Invitrogen/Life Technologies) at 30°C for 45 min. Histone H1 was used as positive control. Samples were then boiled in SDS sample loading buffer and resolved by polyacrylamide gel electrophoresis and dried using a vacuum gel dryer. The phosphorylation of Cdc20, Cdh1 and Histone H1 was visualized by autoradiography.
Identification of Cdc20 and Cdh1 amino acid residues phosphorylated by TBK1 by mass spectrometry
In-vitro kinase assay was performed as described above. Cdc20 and Cdh1 were incubated either alone or with enzymatically active recombinant TBK1 or PLK1 for identification of amino acid residues phosphorylated by both the kinases. Samples were then boiled for 10 minutes in SDS sample loading dye and resolved by polyacrylamide gel electrophoresis. Gels were then stained by coomassie brilliant blue. After destaining, gel slices were treated with TCEP and iodoacetamide to alkylate and reduce the proteins. After in-gel digestion using trypsin or GluC, peptides were extracted and concentrated using vaccum centrifugation. Samples were then analyzed using mass spectrometry according to established protocol 27.
Proximity ligation assay
Proximity ligation assays (PLA) were performed using Duolink system (#DUO92013, Sigma) according to manufacturer’s instructions. Briefly, A549 or H460 cells were cultured in 8 chamber slides (#154534, Thermoscientific, Nunc, Lab-Tek). Cells were given a double thymidine block, released for 9 hours and treated with 2.5μM MRT during the last 7 hours of release. Cells were fixed in 10% buffered formalin for 25 minutes and permeabilized with 0.5% Triton X-100 in PBS for 10 minutes. Blocking was done for 1 hour at room temperature with 5% normal goat serum. Primary antibodies were prepared in 5% normal goat serum at desired concentrations (cdc20–1:100, APC1–1:100, BubR1–1:100, pTBK1–1:100, Cdh1–1:100). Incubation with combination of primary antibodies was done overnight at 4°C. After incubation, the slides were washed twice for 5 minutes with wash buffer A (0.02M Tris, 0.15M NaCl, 0.05% Tween 20, pH7.4). To detect interaction of the two proteins, secondary antibodies conjugated with unique oligonucleotides- PLA probe PLUS for rabbit (#DUO92002, Sigma) and MINUS for mouse (#DUO92004, Sigma) were used. Cells were incubated with secondary antibodies diluted in 5% normal goat serum for 1 hour at 37°C. Following this, slides were washed twice with wash buffer A for 5 minutes each. The ligation solution consists of two oligonucleotides which are bound to the PLUS and MINUS PLA probes conjugated to secondary antibodies by ligase enzyme. This conjugation forms a closed circle if the two proteins being assessed are in close proximity to each other. Ligation is carried out at 37°C for 30 minutes. After ligation, the slides were washed twice in wash buffer A for 2 minutes each. The amplification solution consisting of a polymerase, nucleotides and fluorescently labelled oligonucleotides was added and rolling circle amplification reaction was carried out at 37°C for 120 minutes. Following this, the slides are washed twice with buffer B (0.2M Tris, 4.24gm Tris base, 0.1M NaCl, pH7.5) for 10 minutes each and then with 0.1% wash buffer B for 1 minute. After this, cells were incubated with Alexa Fluor 488- phalloidin (diluted 1:100 in 1%BSA) for 1 hour at room temperature in dark. Finally, cells were washed once with PBS for 5 minutes, and mounted using DAPI mounting medium. Imaging was done using Leica SP8 confocal microscope (Leica Microsystems, Germany); fluorescent spots indicate foci of interaction. Quantification was done using ImageJ software and the plot represents PLA intensity for a minimum of 50 cells for each condition.
Micronuclei assay
A density of 3 × 104 MDA-MB-231 cells were plated in 4-well chamber slides with 0.5 mL of DMEM, 1x medium and incubated overnight. Cells were transfected with silencer negative control siRNA #1 or 50 nM of siRNA constructs for Mps1 or TBK1 and incubated for 66 hrs. Cells were fixed in 4% paraformaldehyde for 10 min., washed 3 times with 1× PBS and permeabilized with 0.01% Triton-X 100/ PBS for 10 min. Then, cells were washed with 1× PBS as described above prior to the staining with DAPI (1 μg/mL). Slides were allowed to seal overnight at room temperature. Pictures were taken at ×40 magnification using an Olympus BX60 fluorescence microscope. A total of two hundred cells were counted on each treatment; micronuclei were counted to obtain the percentage of micronucleation.
Statistical Analysis
Data are reported as the means ± standard deviation. Comparisons were carried out using unpaired Student’s two-tailed t test or one-way ANOVA. All statistical tests were carried out using PRISM version 5.03 (GraphPad Software, La Jolla, CA, USA), statistics software. P < 0.05 was accepted as significant. *p<0.05, **p<0.01 and ***p<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support of the Analytical Microscopy and Flow Cytometry Core Facilities at the Moffitt Cancer Center. This study was supported by the grant U54 CA163068 from the NIH.
ABBREVIATIONS
- TBK1
TANK binding kinase1
- MPS1
Monopolar spindle1
- SAC
spindle assembly checkpoint
- MCC
mitotic checkpoint complex
REFERENCES
- 1.Nojima H Cell cycle checkpoints, chromosome stability and the progression of cancer. Hum Cell 10, 221–230 (1997). [PubMed] [Google Scholar]
- 2.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Bower JJ et al. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene 29, 4787–4799, doi: 10.1038/onc.2010.232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor SS & McKeon F Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89, 727–735, doi: 10.1016/s0092-8674(00)80255-x (1997). [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH & Kastan MB Cell cycle control and cancer. Science 266, 1821–1828, doi: 10.1126/science.7997877 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Thornton BR & Toczyski DP Precise destruction: an emerging picture of the APC. Genes Dev 20, 3069–3078, doi: 10.1101/gad.1478306 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Musacchio A & Salmon ED The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8, 379–393, doi: 10.1038/nrm2163 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Izawa D & Pines J The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 517, 631–634, doi: 10.1038/nature13911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pines J Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 12, 427–438, doi: 10.1038/nrm3132 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Vogel C, Kienitz A, Muller R & Bastians H The mitotic spindle checkpoint is a critical determinant for topoisomerase-based chemotherapy. J Biol Chem 280, 4025–4028, doi: 10.1074/jbc.C400545200 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Musacchio A The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biol 25, R1002–1018, doi: 10.1016/j.cub.2015.08.051 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bower JJ et al. Patterns of cell cycle checkpoint deregulation associated with intrinsic molecular subtypes of human breast cancer cells. NPJ Breast Cancer 3, 9, doi: 10.1038/s41523-017-0009-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang LF, Zhang Z, Yang J, McLaughlin SH & Barford D Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513, 388–393, doi: 10.1038/nature13543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters JM The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7, 644–656, doi: 10.1038/nrm1988 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Lahav-Baratz S, Sudakin V, Ruderman JV & Hershko A Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci U S A 92, 9303–9307, doi: 10.1073/pnas.92.20.9303 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M & Peters JM Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11, 1555–1569, doi: 10.1091/mbc.11.5.1555 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golan A, Yudkovsky Y & Hershko A The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J Biol Chem 277, 15552–15557, doi: 10.1074/jbc.M111476200 M111476200 [pii] (2002). [DOI] [PubMed] [Google Scholar]
- 18.Kraft C et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J 22, 6598–6609, doi: 10.1093/emboj/cdg627 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaspersen SL, Charles JF & Morgan DO Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 9, 227–236, doi: 10.1016/s0960-9822(99)80111-0 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M & Hershko A Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun 271, 299–304, doi: 10.1006/bbrc.2000.2622 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Craney A et al. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc Natl Acad Sci U S A 113, 1540–1545, doi: 10.1073/pnas.1522423113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labit H et al. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J 31, 3351–3362, doi: 10.1038/emboj.2012.168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533, 260–264, doi: 10.1038/nature17973 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kataria M & Yamano H Interplay between Phosphatases and the Anaphase-Promoting Complex/Cyclosome in Mitosis. Cells 8, doi: 10.3390/cells8080814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien Y et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157–170, doi: 10.1016/j.cell.2006.08.034 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Shen RR & Hahn WC Emerging roles for the non-canonical IKKs in cancer. Oncogene 30, 631–641, doi: 10.1038/onc.2010.493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai S et al. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun 6, 10072, doi: 10.1038/ncomms10072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pachis ST & Kops G Leader of the SAC: molecular mechanisms of Mps1/TTK regulation in mitosis. Open Biol 8, doi:rsob.180109 [pii] 10.1098/rsob.180109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Assoro AB et al. The mitotic kinase Aurora--a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERalpha(+) breast cancer cells. Oncogene 33, 599–610, doi: 10.1038/onc.2012.628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opyrchal M et al. Aurora-A mitotic kinase induces endocrine resistance through down-regulation of ERalpha expression in initially ERalpha+ breast cancer cells. PLoS One 9, e96995, doi: 10.1371/journal.pone.0096995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu T et al. Phosphorylation of Microtubule-binding Protein Hec1 by Mitotic Kinase Aurora B Specifies Spindle Checkpoint Kinase Mps1 Signaling at the Kinetochore. J Biol Chem 288, 36149–36159, doi: 10.1074/jbc.M113.507970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weibrecht I et al. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics 7, 401–409, doi: 10.1586/epr.10.10 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Soderberg O et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods 3, 995–1000, doi: 10.1038/nmeth947 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Lee SY, Jang C & Lee KA Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev Reprod 18, 65–71, doi: 10.12717/DR.2014.18.1.065 DR-18-65 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark K et al. Novel cross-talk within the IKK family controls innate immunity. Biochem J 434, 93–104, doi: 10.1042/BJ20101701 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Tang Z, Bharadwaj R, Li B & Yu H Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1, 227–237 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Fang G Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell 13, 755–766, doi: 10.1091/mbc.01-09-0437 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M & Zhang P The function of APC/CCdh1 in cell cycle and beyond. Cell Div 4, 2, doi:1747–1028-4–2 [pii] 10.1186/1747-1028-4-2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesin JA & Orr-Weaver TL Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol 24, 475–499, doi: 10.1146/annurev.cellbio.041408.115949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukas C et al. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature 401, 815–818, doi: 10.1038/44611 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Dominguez-Brauer C et al. Targeting Mitosis in Cancer: Emerging Strategies. Mol Cell 60, 524–536, doi: 10.1016/j.molcel.2015.11.006 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Soto M, Garcia-Santisteban I, Krenning L, Medema RH & Raaijmakers JA Chromosomes trapped in micronuclei are liable to segregation errors. J Cell Sci 131, doi:jcs.214742 [pii] 10.1242/jcs.214742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatch EM, Fischer AH, Deerinck TJ & Hetzer MW Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60, doi:S0092–8674(13)00709–5 [pii] 10.1016/j.cell.2013.06.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chmielewska AE, Tang NH & Toda T The hairpin region of Ndc80 is important for the kinetochore recruitment of Mph1/MPS1 in fission yeast. Cell Cycle 15, 740–747, doi: 10.1080/15384101.2016.1148842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678, doi: 10.1038/nature07317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou YH et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell 41, 458–470, doi: 10.1016/j.molcel.2011.01.019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbie DA et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112, doi: 10.1038/nature08460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo JM et al. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci Adv 4, eaav0443, doi: 10.1126/sciadv.aav0443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter B et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 113, 4039–4044, doi: 10.1073/pnas.1523926113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang G, Yu H & Kirschner MW Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell 2, 163–171, doi:S1097–2765(00)80126–4 [pii] 10.1016/s1097-2765(00)80126-4 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Pfleger CM & Kirschner MW The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev 14, 655–665 (2000). [PMC free article] [PubMed] [Google Scholar]
- 52.Littlepage LE & Ruderman JV Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev 16, 2274–2285, doi: 10.1101/gad.1007302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Araki M, Yu H & Asano M A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev 19, 2458–2465, doi:gad.1361905 [pii] 10.1101/gad.1361905 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao R et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci U S A 113, E2570–2578, doi: 10.1073/pnas.1604929113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch A, Maia A, Janssen A & Medema RH Molecular basis underlying resistance to Mps1/TTK inhibitors. Oncogene 35, 2518–2528, doi: 10.1038/onc.2015.319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss E & Winey M The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol 132, 111–123, doi: 10.1083/jcb.132.1.111 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abrieu A et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106, 83–93, doi: 10.1016/s0092-8674(01)00410-x (2001). [DOI] [PubMed] [Google Scholar]
- 58.Liu ST et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell 14, 1638–1651, doi: 10.1091/mbc.02-05-0074 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin-Lluesma S, Stucke VM & Nigg EA Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267–2270, doi: 10.1126/science.1075596 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Vigneron S et al. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15, 4584–4596, doi: 10.1091/mbc.e04-01-0051 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong OK & Fang G Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J Cell Biol 170, 709–719, doi: 10.1083/jcb.200502163 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X & Winey M The MPS1 family of protein kinases. Annu Rev Biochem 81, 561–585, doi: 10.1146/annurev-biochem-061611-090435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matloff ET, Brierley KL & Chimera CM A clinician’s guide to hereditary colon cancer. Cancer J 10, 280–287, doi: 10.1097/00130404-200409000-00002 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Maia AR et al. Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann Oncol 26, 2180–2192, doi: 10.1093/annonc/mdv293 (2015). [DOI] [PubMed] [Google Scholar]
- 65.King JL et al. TTK promotes mesenchymal signaling via multiple mechanisms in triple negative breast cancer. Oncogenesis 7, 69, doi: 10.1038/s41389-018-0077-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falchook GS, Bastida CC & Kurzrock R Aurora Kinase Inhibitors in Oncology Clinical Trials: Current State of the Progress. Semin Oncol 42, 832–848, doi: 10.1053/j.seminoncol.2015.09.022 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Henriques AC et al. Mitosis inhibitors in anticancer therapy: When blocking the exit becomes a solution. Cancer Lett 440–441, 64–81, doi: 10.1016/j.canlet.2018.10.005 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Foley EA & Kapoor TM Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14, 25–37, doi:nrm3494 [pii] 10.1038/nrm3494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lara-Gonzalez P, Westhorpe FG & Taylor SS The spindle assembly checkpoint. Curr Biol 22, R966–980, doi:S0960–9822(12)01189-X [pii] 10.1016/j.cub.2012.10.006 (2012). [DOI] [PubMed] [Google Scholar]
- 70.London N & Biggins S Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 15, 736–747, doi:nrm3888 [pii] 10.1038/nrm3888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teixeira JH et al. An overview of the spindle assembly checkpoint status in oral cancer. Biomed Res Int 2014, 145289, doi: 10.1155/2014/145289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen G & Deng X Cell Synchronization by Double Thymidine Block. Bio Protoc 8, doi: 10.21769/BioProtoc.2994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dasgupta P et al. Disruption of the Rb--Raf-1 interaction inhibits tumor growth and angiogenesis. Molecular and cellular biology 24, 9527–9541, doi: 10.1128/MCB.24.21.9527-9541.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S, Nath N, Adlam M & Chellappan S Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18, 3501–3510, doi: 10.1038/sj.onc.1202684 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.