Abstract
Isolated brain capillaries are essential for analyzing the changes of protein expressions at the blood–brain barrier (BBB) under pathological conditions. The standard brain capillary isolation methods require the use of at least five mouse brains in order to obtain a sufficient amount and purity of brain capillaries. The purpose of this study was to establish a brain capillary isolation method from a single mouse brain for protein expression analysis. We successfully isolated brain capillaries from a single frozen mouse brain by using a bead homogenizer in the brain homogenization step and combination of cell strainers and glass beads in the purification step. Western blot and proteomic analysis showed that proteins expressed at the BBB in mouse brain capillaries isolated by the developed method were more enriched than those isolated from a pool of five mouse brains by the standard method. By using the developed method, we further verified the changes in expression of BBB proteins in Glut1-deficient mouse. The developed method is useful for the analysis of various mice models with low numbers and enables us to understand, in more detail, the physiology and pathology of BBB.
Keywords: Blood–brain barrier, brain capillary isolation, GLUT1 deficiency syndrome, quantitative protein expression profile, absolute protein expression
Introduction
The blood–brain barrier (BBB) plays a key role in maintaining brain homeostasis for proper neuronal function by forming a physical barrier with tight junction proteins. BBB also serves as a dynamic interface between the peripheral circulation and the central nervous system (CNS) through transport systems such as the ATP-binding cassette (ABC) and solute carrier (SLC) transporters. BBB breakdown has been observed via human epidemiological studies in the hippocampus of the brain before dementia,1 and the expression of P-glycoprotein (P-gp, ABCB1) at the BBB is negatively associated with amyloid deposits in Alzheimer’s disease (AD).2 Genetic studies have revealed that a genetic disruption of glucose transporter 1 (GLUT1, SLC2A1) and creatine transporter (CRT, SLC6A8) leads to the onset of childhood neurodegenerative diseases, GLUT1 deficiency syndromes3 and cerebral creatine deficiency syndromes,4 respectively, due to the reduction of glucose and creatine transport at the BBB. Therefore, BBB dysfunction is thought to be responsible for the initiation and progression of CNS disease.
Genetically engineered mouse models of disease established by homologous recombination in murine embryonic stem cells using CRISPR/Cas9 technology are effective for investigating the physiological and pathological role of BBB. We previously reported that proteomic analysis is a useful technique for revealing protein expression levels in normal physiological conditions as well as under pathological conditions at the BBB.5 To analyze the BBB, brain capillary isolation is an essential method because the volume of brain capillaries is only 0.1% of whole brain.6
In the previous reports, several methods were established for isolating brain capillaries from the brain. The most popular method isolates the brain capillaries from the brain homogenate by density centrifugation using Ficoll or dextran. By using Ficoll or dextran, brain fat contents and brain vessels are easily separated.7,8 However, the brain capillary fraction isolated by the density centrifugation is contaminated by large blood vessels and neuronal cells. Mesh filtration is a standard method for further purifying the brain capillary fraction. Using this method, many BBB transporters, receptors, and tight junction proteins have been identified by quantitative proteomic analysis.9 Other reports use the glass beads column method to purify the brain capillary. Glass beads column method results in reduced contamination of neuronal cells compared to the brain capillary fraction isolated by dextran centrifugation.10 Furthermore, brain capillary endothelial cell can be specifically isolated from mouse brains by single-cell isolation using the fluorescence-activated cell sorting method.11 However, current brain capillary isolation methods have several limitations including requiring at least five mouse brains to allow for enough amount and purity of brain capillary fraction for protein expression analysis (Table S1).7,9,12–18
In many cases of genetically modified mice and pathological mice model, getting the appropriate mouse numbers for brain capillary isolation is quite difficult. Individual differences of protein expression cannot be assessed from the standard method either, as the brain capillary fraction is a mixture of five or more mouse brains. Therefore, an establishment of brain capillary isolation method from a single mouse brain is important for evaluating changes of protein expression profiles at the BBB, and can help to understand the roles of the BBB in CNS diseases.
The purpose of the present study was to establish a brain capillary isolation method from a single mouse brain for protein expression analysis and verify the developed method by examining the changes of protein expression of brain capillary fraction in the Glut1-deficient mouse (Glut1+/− mouse).
Materials and methods
Animal
Eight-week-old C57BL/6N male mice (n = 24) were purchased from Clea (Japan Clea, Tokyo, Japan). Eight-week-old Glut1-deficient male mouse (Glut1+/− mouse) was obtained by in vitro fertilization using frozen sperm and ovum of a wild-type C57BL/6J mouse (Charles River, Yokohama, Japan). Genotyping of all offspring was analyzed using PCR of genomic DNA from the tail, as previously reported19 (Figure S1). The primers used were as follows a, 5ʹ-GGGCCACAGTGATAGAGATAGAATG- 3ʹ;b, 5ʹ -GTGTGACTTTAGGAAACTTACTTCCC- 3ʹ;c, 5ʹ -AAATGGCGTTACTTAAGCTAGCTTGC- 3ʹ.
Primers a and b amplified a DNA fragment from wild-type allele, while primers b and c amplified a fragment from mutated allele. All animals were bred in the Center for Animal Resources and Development (CARD) at the Kumamoto University. Animals were maintained at a 12-h light/dark cycle. All animal experiments were approved by the Institutional Animal Care and Use Committee at Kumamoto University, and they followed the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology and the Animal Research: Reporting in Vivo Experiments guidelines.
Brain capillary isolation (standard method)
Brain capillaries were isolated from the pooled fresh mouse brains (five mice/group) as described previously9 with minor modifications. Five fresh mouse brains were minced 800 times and homogenized with 15 mL of homogenize buffer (101 mM NaCl, 4.6 mM KCl, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 15 mM HEPES, pH 7.4) using a Potter homogenizer (20 strokes). Homogenate was added to 10 mL of homogenize buffer and centrifuged (1000 × g, 10 min, and 4°C). After the supernatant was removed, 15 mL of homogenize buffer was added and the homogenate re-suspended. An equal volume of 32 w/v% dextran in homogenize buffer was added into the suspended sample and mixed by inverting. The samples were then centrifuged (4500 × g, 15 min, 4°C), supernatant collected into another tube, and the supernatant centrifuged (4500 × g, 15 min, 4°C). Kimwipes were used to remove the brain fat content adhered to the wall of the tubes from which the supernatant was removed. The tube pellet was suspended by the suspension buffer (homogenize buffer containing 25 mM NaHCO3, 10 mM glucose, 1 mM pyruvate, and 5 g/L bovine serum albumin) (5 mL × 4) and suspended samples of two tubes were mixed into one tube. The sample was filtered through nylon mesh, loaded to the cut end of the 50 mL tube in the order of 210 µm, 85 µm, and 20 µm. Brain capillaries trapped in the 20 µm mesh were collected by washing the mesh in the 50 mL tube with suspension buffer. The washed buffer was centrifuged (1000 × g, 10 min, 4 °C) and supernatant removed. Pellet was suspended using 500 µL of homogenize buffer. Part of the isolated brain capillary fraction was used for microscopy. Isolated brain capillary fraction was lysed in hypotonic buffer (10 mM Tris-HCl, 10 mM NaCl, 1.5 mM MgCl2, pH 7.4) by sonication and the protein amount was measured by Pierce BCA protein assay kit (Thermo Fisher Scientific, MA, USA).
Brain capillary isolation (developed method)
In the developed method, a single mouse brain in 1.5 ml tube was freezed in liquid nitrogen, and kept on −80°C freezer. After melting, a single mouse brain was transferred to a 2 mL screw-cap tube (WATSON, Tokyo, Japan) and 1 mL of homogenize buffer was added along with stainless beads (3.2 mm, 1.8 g, TOMY SEIKO, Tokyo, Japan). The brain was then homogenized by bead homogenizer (Bead Mill 4, Thermo Fisher Scientific) for 60 s at a speed of 3 m/s. Homogenate was transferred to a new 2 mL tube and then centrifuged (1000 × g, 10 min, 4°C). A part of brain homogenate (50 µL) was collected in another tube as a whole brain fraction. The supernatant was removed carefully and up to 1 mL of the homogenize buffer was added to pellet. After suspension, an equal volume of 32 w/v% dextran/homogenize buffer was added into the 2 mL tube and mixed by inverting. The samples were immediately centrifuged (4500 × g, 15 min, 4°C) and the supernatant was collected into new 2 mL tube, while the pellets were kept in on-ice. The supernatant was centrifuged (4500 × g, 15 min, 4 °C) again and the supernatant was discarded. After removing the fat content adhered at the wall of the tube by using Kimwipe, pellets were suspended in the suspension buffer (200 µL × 2), and samples of two tubes were combined into one tube. The samples were filtered through a cell strainer (70 µm) (pluriStrainer-Mini, pluriSelect, Leipzig, Germany) and the strainer mesh was washed by 400 µL suspension buffer for four times. Samples passed through the 70 µm mesh were added to cell strainer filled with 800 mg of glass beads (0.35–0.5 mm, AS ONE, Osaka, Japan) and then washed by 500 µL suspension buffer for 10 times. After washing, glass beads were transferred to a new tube using a spatula and 1 mL of suspension buffer was added and mixed by inverting. The supernatant was then quickly transferred into a new tube. Glass beads were re-added in the 500 µL suspension buffer and mixed by inverting. The supernatant was quickly transferred into the previous tube. The tube was centrifuged (3300 × g, 5 min, 4°C) and supernatant was removed. Pellet was suspended using 100 µL of homogenize buffer. Part of the isolated brain capillary fraction was used for microscopy. Isolated brain capillary fraction was lysed in a hypotonic buffer by sonication and the protein amount was measured using a Pierce BCA protein assay kit.
Western blot analysis
Western blot analysis of isolated brain capillary fraction was performed as described previously.5 Briefly, each sample was separated on an SDS-polyacrylamide gel and blotted onto a PVDF membrane. We used following primary antibodies (Claudin-5, 35-2500, Thermo; Glut1, ab652, abcam; Mdr1, 13978, CST; β-actin, 8H10D10, CST) and HRP conjugated secondary antibodies (Goat anti rabbit IgG HRP, ab6721, abcam; Goat anti mouse IgG HRP, ab6789, abcam). Quantification of band intensity was performed using the ImageJ software (National Institutes of Health, MD, USA).
Multiplexed multiple reaction monitoring (MRM) analysis
Protein amount or quantity of the target molecules in the brain capillary fraction isolated by the standard and developed method were determined simultaneously using the multiplexed MRM analysis as described previously.9,20 Briefly, stable isotope-labeled peptides were spiked in the digested peptide with internal standards. The peptide samples were then desalted and dissolved in 0.1% trifluoroacetic acid for liquid chromatography combined with the tandem mass spectrometry (LC-MS/MS) analysis. Standard samples were prepared as a single dilution series of unlabeled standard peptides at relevant injection amounts (0.5–300 fmol) with 30 fmol of internal standard peptides. The samples were then analyzed using a micro-LC system with a ChromXP C18CL column (Ekisigent, Redwood City, CA, USA) coupled to an electrospray ionization-triple quadrupole mass spectrometer QTRAP6500 (SCIEX, Framingham, MA, USA). The transitions are listed in Table S2. The data were analyzed by Skyline version 19.1 (MacCoss Laboratory, University of Washington, WA, USA). The quantification value was determined as the average value of the peak ratios from three to four sets of transitions.
Quantitative proteomic analysis
Digestion of isolated brain capillary fraction and whole brain fraction using phase transfer surfactant (PTS) method and quantitative proteomic analysis was conducted as previously described.21,22 Proteins were quantified according to the specific peptide data detected by sequential window acquisition of all theoretical fragment-ion spectra (SWATH). Each sample was analyzed by SWATH-MS on a TripleTOF 5600 instrument (SCIEX) interfaced with the DIONEX Ultimate 3000 RSLC nanosystem (Thermo Fisher Scientific). Proteins were identified using Protein Pilot 4.5 (SCIEX) with MS/MS data obtained from the information-dependent acquisition (IDA). Proteins were identified by using the UniProt mouse proteome database. Each peptide peak of SWATH data was extracted by the PeakView Software version 2.1 (SCIEX). The peak area of each protein was calculated by adding all peptide peak area of the protein. The average protein peak area of samples in each group was compared between the two groups.
Data analysis of quantitative proteomic analysis using RNA-Seq database
Among the identified proteins by quantitative proteomic analysis, a cell type marker protein (Endothelial cell, Astrocyte, Neuron, Oligodendrocyte, Microglia) was selected. Maker protein was selected according to the criteria that mRNA expression ratio (expression levels in most highly expressed cell types/expression levels in secondary highly expressed cell types) is over 10-fold. The RNA-sequencing database of mouse brain used has been previously published.23
Statistics analysis
Numerical data are expressed as the mean ± standard deviation. Data normality and equality of variance were determined by the Shapiro–Wilk test and F-test using Origin 2018 b (OriginLab, Northampton, MA, USA) and Microsoft Excel (v15.0, Microsoft, WA, USA), respectively. The statistical significance of differences was determined by Student’s t-test and Welch’s t-test using Microsoft Excel and the Mann–Whitney U-test using Origin 2018 b, as appropriate. During the proteomic analysis, false discovery rate (FDR) of each protein was calculated by Benjamini–Hochberg method using Microsoft Excel.
Results
Establishment of brain capillary isolation method from a single frozen mouse brain
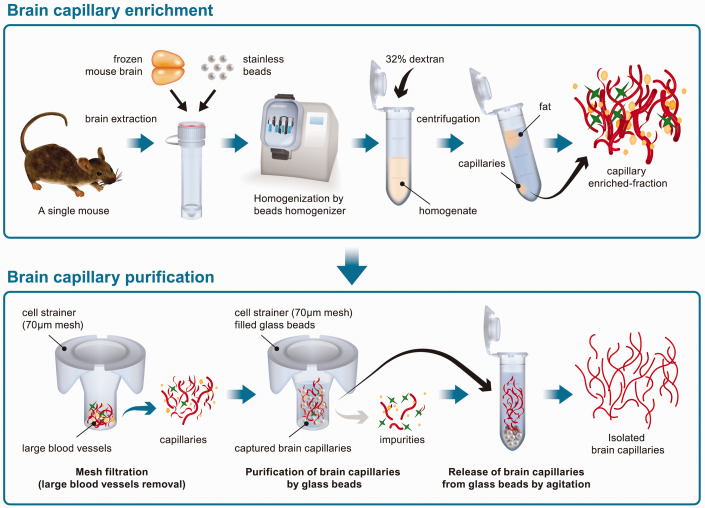
The scheme of the developed isolation of brain capillary from a single frozen mouse brain is shown in Figure 1. Briefly, a single frozen mouse brain was homogenized by a bead homogenizer. Using the bead homogenizer, four mouse brains were rapidly homogenized in parallel. Brain capillaries were concentrated in the pellet by dextran centrifugation. The brain capillaries were then purified by filtering through a cell strainer and capturing them using glass beads.
Figure 1.
Scheme of the developed brain capillary isolation method from a single mouse brain.
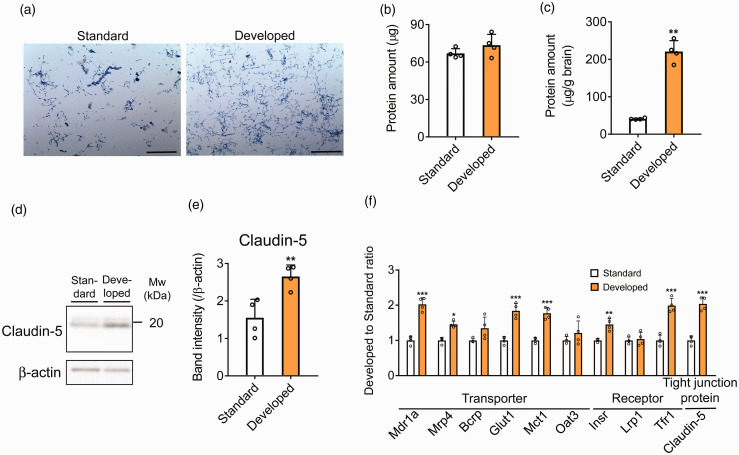
Light microscope images showed that mouse brain capillaries were isolated from a single mouse brain by the developed method as well as the standard method from the pooled five mouse brains (Figure 2(a)). The number of brain capillaries isolated by the developed method was greater than the standard method and seemed to contain less contamination of impurities such as large blood vessels and debris (Figure 2(a)). The amount of brain capillary fraction was measured using the BCA protein assay. Total protein amount of brain capillary fraction isolated by the standard method from five brains and the developed method from a single brain were 66.9 ± 3.3 µg and 73.5 ± 7.5 µg, respectively (Figure 2(b)). The protein recovery amount of brain capillary fraction isolated from 1 g of the brain by the standard and the developed method were 40.9 ± 1.9 µg/g brain and 221 ± 25 µg/g brain, respectively (Figure 2(c)), indicating that the recovery of brain capillary fraction isolated by the developed method was 5.40-fold higher.
Figure 2.
Brain capillary isolation and evaluation of brain capillary enrichment by standard method and developed method. (a) Images of isolated brain capillary by standard method and developed method. Brain capillaries were stained by trypan blue. Scale bar = 250 µm. (b, c) Protein amount of isolated brain capillary fraction by standard method from pooled five mouse brains and those by developed method from a single mouse brain. Total protein amount (b) and protein amount per brain weight (c) were indicated. (d, e) Western blot analysis of protein amount in the brain capillary fraction isolated by standard method and developed method; 1 µg protein of brain capillary fraction was loaded to each lane. Representative Western blot images are shown. Quantification of each band intensity was performed by using ImageJ software. The protein expression was normalized by β-actin as a loading control. (f) Protein amount ratio (developed to standard) of transporters, receptors, and tight junction proteins quantified by MRM; 1 µg protein of brain capillary fraction was injected into the LC-MS/MS. Each data represent the mean ± SD (n = 4). *p < 0.05, **p < 0.01 and ***p < 0.001, significantly different from standard.
The microscope images of brain capillary fraction, concentrated after dextran centrifuge but before mesh filtration, are shown in Figure S2. In samples homogenized using the Potter homogenizer, many brain cells were found still attached to the brain capillaries due to inadequate homogenization (Figure S2, left). By contrast, in samples homogenized by the bead homogenizer, brain capillaries were isolated efficiently from the brain cells (Figure S2, right). Therefore, a better and uniform homogenization of mouse brain by bead homogenizer leads to a higher recovery of brain capillaries from a single mouse brain compared to Potter homogenizer using the standard method.
Enrichment of BBB-related proteins in the brain capillary fractions
Claudin-5 is reported to be predominantly expressed in the mouse brain capillary endothelial cell, according to RNA-seq analysis and immunohistochemistry.23,24 In the Western blot analysis, claudin-5 was detected in both mouse brain capillary fractions. The amount of claudin-5 per 1 µg protein amount of brain capillary fraction isolated by the developed method (D-Bcap) was significantly higher (1.71-fold) than the standard method (S-Bcap) (Figure 2(d) and (e)).
In addition, the absolute protein expressions of the 10 proteins expressed at the BBB including claudin-5, Abc transporters [Abcb1a (Mdr1a), Abcc4 (Mrp4), Abcg2 (Bcrp)], Slc transporters [Slc2a1 (Glut1), Slc16a1 (Mct1), Slc22a8 (Oat3)], and receptors [Insulin receptor (Insr), Low–density lipoprotein receptor–related protein 1 (Lrp1), Transferrin receptor protein 1 (Tfr1)] were measured by targeted proteomics in the same protein amounts of brain capillary fraction using a MRM (Figure 2(f), Table S3). The protein amount of claudin-5 was significantly increased in D-Bcap compared to S-Bcap (2.04-fold) as well as by Western blot. The protein amount ratio of Mdr1a, Mrp4, Glut1, Mct1, Insr, and Tfr1 was significantly increased in D-Bcap compared to S-Bcap (2.03, 1.46, 1.84, 1.77, 1.45 and 1.99, respectively). The protein amount ratio of Bcrp and Oat3 tended to increase in D-Bcap compared to S-Bcap (1.35, p = 0.0571 and 1.21, p = 0.277, respectively). However, the protein amount ratio of Lrp1 remained unchanged in D-Bcap compared to S-Bcap (1.04, p = 0.711). These results suggest that the developed method allows for the isolation of brain capillaries from a single mouse brain with higher recovery and enrichment than by the standard method from pooled five mouse brains.
Uchida et al.9 reported on the absolute quantification of BBB transporters, receptors, and tight junction proteins in isolated mouse brain capillary fractions. We compared the absolute protein amount of proteins in D-Bcap from our study with Uchida’s report (Figure S3). The amount of all proteins quantified by MRM was within 2-folds when compared to the reported protein amount. This indicates that the concentration of brain capillary in our developed method is similar to the previous report.
Proteomic evaluation of enrichment of brain cells in the brain capillary fractions
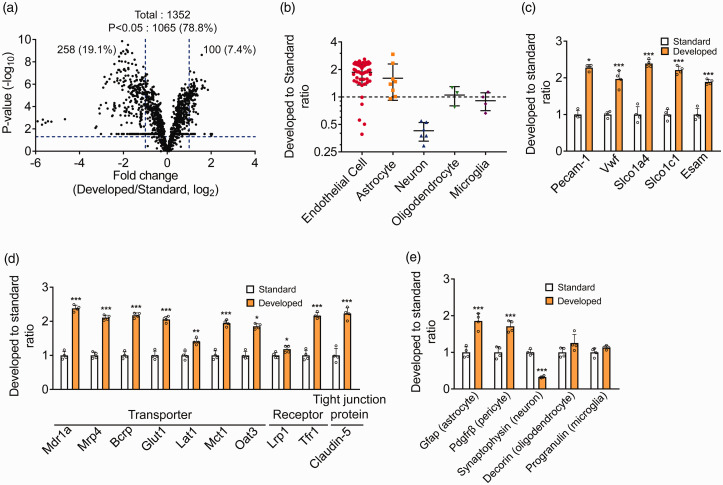
Brain capillary consists of brain capillary endothelial cell surrounded by astrocytes and pericytes. Contamination of neurons and microglia cannot be excluded when using the brain capillary isolation method. To further examine the enrichment of brain capillaries by the developed method, proteomic analysis was performed on the same protein amount of the isolated brain capillary fraction (Figure 3, Table S7). Volcano plot showed that 1352 proteins were quantified in the mouse brain capillary fraction and the levels of 1065 proteins (78.8%) were significantly (p < 0.05) different between the D-Bcap and the S-Bcap (Figure 3(a)). Significant differences were evaluated with false discovery rate (FDR) using Benjamini–Hochberg method; the result showed that 1051 proteins showed significant difference (FDR < 0.05) (Table S4(a)).
Figure 3.
Differences in the protein profile of isolated brain capillary fraction between standard method and developed method. (a) Volcano plot of identified proteins by proteomic analysis of brain capillary fraction isolated by standard method and developed method. The p-values in the t-test were plotted against the fold-change (Developed/Standard) of protein expression for all identified proteins. The horizontal line in the plot represents the criterion of significance (p = 0.05). The vertical lines indicate fold changes (0.5, 2.0). (b) Dot plot of protein amount ratio (Developed to Standard) of marker protein quantified by proteomic analysis. The term “Oligodendrocyte” comprises the oligodendrocyte precursor cell, newly formed oligodendrocyte, and myelinating oligodendrocyte. (c) Protein amount ratio (developed to standard) of brain capillary endothelial cell marker protein (Pecam-1) and four brain capillary endothelial cell-specific proteins. (d, e) Protein amount ratio (developed to standard) of transporters, receptors, and tight junction proteins related to BBB transport function (d) and other cell types marker proteins (e) by proteomic analysis. Each data represent the mean ± SD (n = 4). *p < 0.05, **p < 0.01 and ***p < 0.001, significantly different from standard.
To determine the enrichment of brain cells, proteins selectively expressed in each cell type were extracted and were chosen following the criteria that mRNA expression ratio (FPKM value in most highly expressed cell types/FPKM value in secondary highly expressed cell types) is over 10-fold. Seventy-five proteins were selected as selectively expressed proteins (54 for endothelial cell; 8 for astrocytes; 6 for neurons; 3 for oligodendrocytes; 4 for microglia). The protein amounts ratio of D-Bcap to S-Bcap for 75 proteins is shown in Figure 3(b). Among the 54 proteins selectively expressed in the endothelial cell, 49 proteins indicated a ratio of greater than 1 and the average ratio was 1.86. In addition, among the eight proteins selectively expressed in the astrocytes, seven proteins indicated a ratio greater than 1 and the average ratio was 1.60 (Figure 3(b)). By contrast, the ratio of the six proteins selectively expressed in the neurons was lower than 1 while the average ratio was 0.426. No significant change was observed from 1 for proteins selectively expressed in the oligodendrocyte or microglia (1.05 or 0.908, respectively).
Then, we indicated the amount of brain capillary endothelial cell marker protein (Pecam-1) and four brain capillary endothelial cell-specific proteins (Vwf, Slco1a4, Slco1c1, and Esam), the localization of which has previously been validated using immunostaining25–28 (Figure 3(c)). These proteins were significantly increased in the D-Bcap compared to the S-Bcap. Also, the ratio of 10 transporters, receptors, and tight junction protein expressed at the BBB were 1.18–2.38 (Figure 3(d)). These ratios were almost consistent with those measured by targeted proteomics using an MRM analysis (Figure 2(f)). The ratio of the Gfap (astrocyte marker protein) and Pdgfrβ (pericyte marker protein) was significantly increased (1.86 and 1.71), while of synaptophysin (neuron marker proteins) was significantly decreased in the D-Bcap (0.327) (Figure 3(e)). Oligodendrocyte marker (Decorin) and microglia marker (Progranulin) were not significantly different (1.26 and 1.13). These results suggest that brain capillary endothelial cell, astrocytes and pericytes were concentrated, and contamination of neuronal cells was reduced in the D-Bcap compared to the S-Bcap.
Isolation of brain capillary fraction from Glut1-deficient mouse
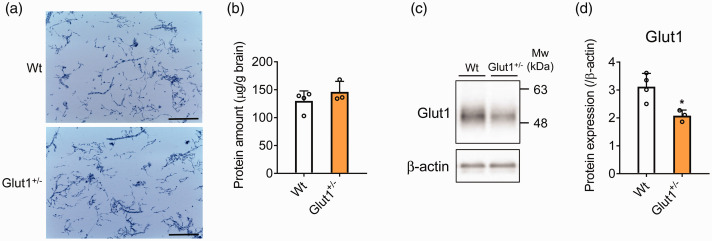
Brain capillaries were isolated from the Glut1-deficient mouse (Glut1+/− mouse) by the developed method in order to examine its versatility. GLUT1 deficiency syndrome is a genetic disease caused by a hetero deficiency of SLC2A1 (GLUT1) gene that reduces glucose uptake into the brain thereby altering BBB barrier function and homeostasis. We isolated brain capillary fraction from a single mouse brain of Glut1+/− mouse and a wild-type littermate (Wt) mouse (Wt; n = 4, Glut1+/−; n = 3). Light microscopic images showed that densities and size of isolated brain capillary were similar between the Wt and the Glut1+/− mouse (Figure 4(a)). The protein amounts of brain capillary fraction from the Glut1+/- mouse were not significantly different from those of Wt (Figure 4(b)). Western blot analysis showed that the Glut1 protein expression was decreased by 33.3% in Glut1+/− mouse (Figure 4(c) and (d)).
Figure 4.
Isolation of brain capillaries and Glut1 protein expression in Glut1+/− mice. (a) Images of isolated brain capillaries from Wt and Glut1+/− mouse by the developed method. Brain capillaries were stained by trypan blue. Scale bar = 250 µm. (b) Protein amount of brain capillary fraction isolated from Wt and Glut1+/− mouse. The protein amount per brain weight was indicated. (c, d) Glut1 protein expression in isolated brain capillary fraction at Wt and Glut1+/− mouse by Western blot analysis. The protein expression was normalized by β-actin as a loading control. The data represents the mean ± SD (Wt: n = 4, Glut1+/−: n = 3). *p < 0.05, significantly different from Wt mice.
Proteomic analysis of brain capillary fraction isolated from Glut1+/− mouse
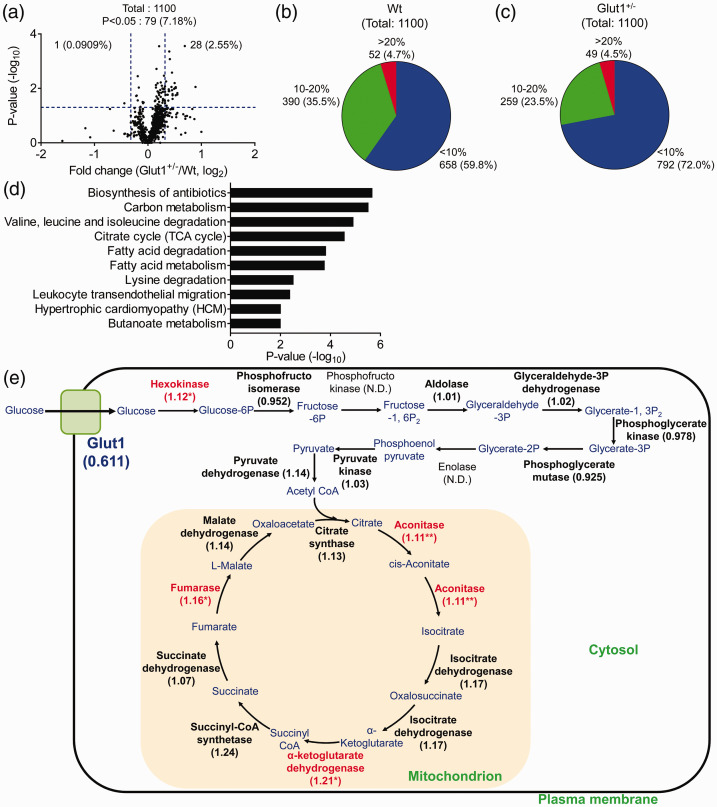
The expressional differences of proteins in brain capillary fraction between Wt and Glut1+/− mice were analyzed using quantitative proteomic analysis (Table S8); 1100 proteins were quantified and 79 proteins were found to be significantly different between the Wt and Glut1+/− mice (Figure 5(a)). Among these proteins, 29 proteins were significantly different with over 1.25-fold change between the Glut1+/− and Wt mice (Table S5). No protein with significant difference, less than 5% of FDR, evaluated by Benjamini–Hochberg method was observed (Table S4(b)). We also evaluated individual differences of each protein expression in the Wt and Glut1+/− mice by analyzing the CV(%) of the protein peak area (Figure 5(b) and (c)). CV(%) of over 95% of the proteins in the identified 1100 proteins were under 20% in both Wt and Glut1+/− mice. These results suggest that the D-Bcap allows for evaluation of individual differences in each protein expression.
Figure 5.
Changes of protein expression in isolated brain capillary fraction from Glut1+/− mouse. (a) Volcano plot of identified proteins. The p-values were plotted against the fold change (Glut1+/−/Wt) of protein expression for all identified proteins. The horizontal line in the plot represents the criterion of significance (p = 0.05). The vertical lines indicate fold changes (0.8, 1.25). (b, c) Distribution of CV(%) of each protein peak area in Wt (b) and Glut1+/− (c) mice quantified by proteomic analysis. (d) Functional annotation clustering and enrichment analysis of proteins for which expression was significantly increased in Glut1+/− mouse by DAVID. The terms giving the 10 lowest p-values in the KEGG pathway. (e) Changes of glycolysis and TCA cycle enzyme in Glut1+/− mouse. Each value indicates the fold change of protein expression and red bold letters indicate the significantly increased enzyme in Glut1+/− mouse. *p < 0.05 and **p < 0.01, significantly different from Wt mice. N.D.; not detected.
Gene ontology analysis using the significantly increased 69 proteins from the Glut1+/− mouse suggests that energy metabolism-related pathways (carbon metabolism, tricarboxylic acid (TCA) cycle, fatty acid degradation, fatty acid metabolism, and butanoate metabolism) were increased in the brain capillary fraction of the Glut1+/− mouse (Figure 5(d)). Since glucose produces ATP by glycolysis, the changes in quantified proteins related to glycolysis and the TCA cycle in the brain capillary fraction of the Glut1+/− mouse are summarized in Figure 5(e). The proteins related to the TCA cycle were significantly or almost significantly (pyruvate dehydrogenase; p = 0.0555, citrate synthase; p = 0.0846, isocitrate dehydrogenase; p = 0.0912, succinyl-CoA synthetase; p = 0.114, succinate dehydrogenase; p = 0.214, malate dehydrogenase; p = 0.0571) increased in Glut1+/− mouse. Some proteins related to glycolysis such as hexokinase, an enzyme that converts glucose to glucose-6-phosphate, were significantly increased in Glut1+/− mouse (1.12-fold), while other glycolysis related enzymes were not significantly changed.
To investigate the expressional changes of BBB transport-related proteins in the Glut1+/− mouse, we extracted transporters that were highly expressed in brain capillary endothelial cell than other cell types based on the RNA-seq database (brain capillary endothelial cell selective protein). The selected 18 proteins were then extracted and deemed as brain capillary endothelial cell selective transporters (Table 1). The expression of Glut1 protein was decreased (38.9%, p = 0.0571) in Glut1+/− mouse compared to the Wt mouse, consistent with the decrease of Glut1 protein expression in Western blot analysis (33.3%, Figure 4(c) and (d)). Meanwhile, other BBB transporters were found not significantly different in Glut1+/− mouse. BBB transport-related receptors and tight junction proteins were also not significantly different in Glut1+/− mouse (Table 1). These results suggest that the expression of BBB transport-related proteins was not affected by the Glut1 reduction.
Table 1.
Changes in protein expression levels of transporters in brain capillary fraction isolated from Wt and Glut1+/− mouse.
| Protein name | Uniprot ID | Fold change (Glut1+/−/Wt) | p-value |
|---|---|---|---|
| ABC transporter | |||
| Abcb1a (Mdr1a/P-gp) | P21447 | 1.04 | 3.64.E-01 |
| Abcc4 (Mrp4) | E9Q236 | 1.04 | 7.61.E-01 |
| Abcc9 | P70170 | 0.875 | 1.85.E-01 |
| Abcg2 (Bcrp) | Q7TMS5 | 1.04 | 4.06.E-01 |
| SLC transporter | |||
| Slc2a1 (Glut1) | P17809 | 0.611 | 5.71.E-02 |
| Slc3a2 | P10852 | 1.04 | 6.32.E-01 |
| Slc6a20a | Q8VDB9 | 1.05 | 6.27.E-01 |
| Slc7a1 | Q09143 | 1.15 | 7.31.E-01 |
| Slc7a5 (Lat1) | Q9Z127 | 1.10 | 5.24.E-01 |
| Slc9a3r2 | Q9JHL1 | 1.24 | 1.12.E-01 |
| Slc16a1 (Mct1) | P53986 | 1.05 | 7.91.E-01 |
| Slc25a5 | P51881 | 1.10 | 2.29.E-01 |
| Slc25a20 | Q9Z2Z6 | 1.00 | 9.99.E-01 |
| Slc25a24 | Q8BMD8 | 1.08 | 2.56.E-01 |
| Slc30a1 | Q60738 | 1.10 | 3.59.E-01 |
| SLCO transporter | |||
| Slco1a4 | Q9EP96 | 0.918 | 4.51.E-01 |
| Slco1c1 | Q9ERB5 | 1.17 | 4.00.E-01 |
| Other transporter | |||
| Mfsd2a | Q9DA75 | 1.14 | 3.14.E-01 |
| Receptor | |||
| Lrp1 | Q91ZX7 | 0.952 | 3.96.E-01 |
| Tfr1 | Q62351 | 1.12 | 4.12.E-01 |
| Tight junction protein | |||
| Claudin-5 | O54942 | 0.943 | 6.17.E-01 |
Note: Each transporter was extracted from the brain capillary endothelial cell (BCEC) selective protein [(expression levels at most highly expressed cell types/expression levels at secondary highly expressed cell types) extracted from the mouse brain RNA-sequencing database is over 1.0-fold].
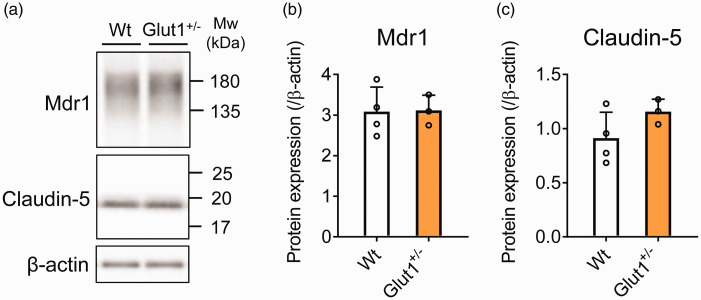
Protein expression of Mdr1 and claudin-5 in brain capillary fraction isolated from Glut1+/− mouse
To validate the changes of BBB proteins expression by proteomic analysis in the Glut1+/− mouse, we performed a Western blot analysis of representative BBB proteins (Figure 6). Expression levels of Mdr1 and claudin-5 proteins in brain capillary fraction were found to be significantly not different between the Glut1+/− and the Wt mice.
Figure 6.
Western blot analysis of brain capillary fraction isolated from Glut1+/− mouse. (a) Representative Western blot images are shown. Quantification of the (b) Mdr1 and (c) claudin-5 band intensity was performed using ImageJ software. The protein expression was normalized by β-actin as a loading control. The data represents the mean ± SD (Wt: n = 4, Glut1+/−: n = 3).
Discussion
In the present study, we established a brain capillary isolation method from a single frozen mouse brain by using bead homogenizer for the brain homogenization and subsequently used a combination of cell strainers and glass beads for its purification (Figure 1). Western blot and proteomic analysis showed that the mouse brain capillary fraction isolated by the developed method was more enriched than that isolated from a pool of five mouse brains using the standard method. This is the first report to isolate brain capillary fractions from a single mouse brain with higher recovery and enrichment for protein expression analysis.
For the brain homogenization step, we used a bead homogenizer in the developed method. Bead homogenizer is used for the homogenization of various samples such as soft tissue, hard tissue, plant, and bacteria.29–32 However, bead homogenizer has never been used for brain homogenization process of the brain capillary isolation. Bead homogenizer could uniformly homogenize the samples. Due to this reason, the number of brain cells in the homogenate is lower when using a bead homogenizer than the Potter homogenizer (Figure S2). The recovery of brain capillaries was increased at the D-Bcap due to a decreased rate of removal of brain capillaries by filtration (Figure 2(c)). Bead homogenizer has an additional advantage where it can homogenize samples in parallel and with short time compared to Potter homogenizer. This also reduces the time required for brain capillary isolation compared to the standard method.
The absolute protein amount of BBB proteins in brain capillary fraction by proteomic analysis was previously reported.9,17 The absolute amount of BBB proteins is useful for evaluating the enrichment of brain capillaries in isolated brain capillary fractions. In our study, the amount of 10 BBB proteins quantified by MRM analysis in the D-Bcap was within the 2-fold range compared to the protein amount of the brain capillary isolation method, reported by Uchida et al. (Figure S3). Therefore, the enrichment of brain capillaries isolated by the developed method is within the degree with previously reported method. These results suggest that the developed method is suitable for the protein expression analysis of BBB.
The procedures of the standard brain capillary isolation method are similar to the ones reported by Uchida et al.9 The present method used 5 mouse brains, while Uchida et al. used 10. The difference in the numbers of brain, therefore, may influence the concentration rate of BBB transport-related proteins in each method compared to the developed Bcap isolation method (Figures 2(f) and S3).
In a previous report, the proteomic analysis of mouse brain capillary fraction isolated by the standard method demonstrated that the brain capillary fraction contains not only the brain capillary endothelial cell but also other brain cell types such as astrocytes, pericytes, and neuronal cells.10 Astrocyte and pericyte, for which the protein amount of each marker protein was significantly increased at D-Bcap compared to S-Bcap (Figure 3(e)), adhere directly to the endothelial cell and play important roles for maintenance of BBB functions.33,34 In the proteomic analysis between isolated brain capillary fraction and whole brain fraction (Table S9), the amounts of astrocyte and pericyte marker proteins were also significantly increased in brain capillary fraction compared with whole brain (Table S6). Astrocyte and pericyte, therefore, keep adhering to the brain capillary endothelial cell and increase with enrichment of brain capillaries in the developed method. In the neuronal cell, the diameter of a nerve cell body is reported to be 4–100 µm35 and part of neuronal cell may be retained on the 20 µm nylon mesh as well as the brain capillaries in a mesh filtration step in the standard method. For the developed method, large blood vessels and neuronal cells over 70 µm diameter are expected to be removed during the first mesh filtration step. The brain capillaries are then purified by using glass beads on the cell strainer for further purification of brain capillaries. During this step, the diameter of the cell strainer is 70 µm. Therefore, neuronal cells under 70 µm are expected to not be retained and be washed out to the follow-through fraction during the washing step. As a result, neuronal cell contaminations were decreased in D-Bcap compared to S-Bcap (Figure 3(e)).
The purity of brain capillaries was estimated by two ways. First, we compared microscopic images pre- and post- brain capillary isolation (Figures 2(a) and S2). Most of the brain cells were removed following isolation, suggesting high purity of capillaries in the fraction. Second, we compared the amounts of brain cell marker proteins between isolated brain capillary fraction and whole brain fraction (Table S6, S9), including synaptophysin (syp, neuron), myelin-oligodendrocyte glycoprotein (mog, oligodendrocyte), and coronin-1a (coro1a, microglia). Their amounts were decreased by 81.7%, 91.8%, and 63.6%, respectively, in the brain capillary fraction compared with whole brain. In contrast, Mdr1a, brain capillary endothelial cells marker, was increased 46.6-fold in the isolated fraction. This suggests that the brain capillaries were highly concentrated in the brain capillary fraction.
Lrp1 was not concentrated in the proteomic analysis and the MRM analysis in D-Bcap compared to S-Bcap (Figures 2(f) and 3(d)). Although the Lrp1 plays a role in BBB function,36 Lrp1 is highly expressed in oligodendrocytes, microglia, and astrocytes compared to the brain capillary endothelial cell according to the mouse brain RNA-seq database.23 Therefore, Lrp1 may not be enriched in the brain capillary fractions isolated by the developed method.
GLUT1 deficiency syndrome is a genetic disease caused by SLC2A1 (GLUT1) gene mutation that reduces glucose uptake into the brain. In our study, metabolic pathway-related enzymes were increased in the Glut1+/− mouse (Figure 5(d) and (e)). Therefore, metabolic pathway-related proteins may be increased as compensation for ATP production in response to the decreased brain glucose uptake in the brain capillary of Glut1+/− mouse.
Glut1+/− mouse is reported to exhibit a typical epileptic spike.37 In our study, BBB transporters were not significantly different in Glut1+/− mouse (Table 1). Therefore, neuronal abnormalities of Glut1+/− mouse may be induced by the brain glucose uptake deficiency. The increase of brain glucose uptake is, therefore, the most effective strategy for treating GLUT1 deficiency syndrome and improving brain parenchymal function. In fact, in recent GLUT1 gene therapy, neuronal function and behavior were found to be improved due to the increased GLUT1 protein expression at the BBB.38
The ketogenic diet is also reported to improve the movement disorders of GLUT1 deficiency syndrome patients.39 Ketone body is a nutrient used as an energy source for the brain instead of glucose. Ketone body is transported into the brain across the monocarboxylate transporter 1 (Mct1, Slc16a1) expressed in brain capillary endothelial cell. Our study showed that Mct1 was not significantly altered in the Glut1+/− mouse (1.05-fold, Table 1). In a previous report, Mct1 protein expression level at the BBB depended on the blood ketone body concentration.40 Therefore, in GLUT1 deficiency syndrome patients, an increase of blood ketone body concentration by a ketogenic diet induces an increase in Mct1 protein expression at the BBB in a similar fashion to a healthy person and serves as an effective treatment for glucose deficiency.
Generating enough models through genetic modification is difficult due to increased associated cost, time, and burden. In these cases, the developed method is useful as the numbers of the samples is reduced. In a previous report, at least 5 mouse brains were required for one sample and 15 mice were needed to collect n = 3 samples (Table S1). The developed method enables them to get an n = 3 sample from three mouse brains. The developed method also contributes to the reduction of the experimental animal, which is one of the principles of 3Rs (replacement, reduction, and refinement).
Analyzing the protein expression of individual mice in the same group is important for understanding the differences of reaction to various environmental and genetic backgrounds. With the standard method, individual differences of protein expression cannot be analyzed as the S-Bcap is a mixture of five or more mouse brain capillaries. On the other hand, the developed method can isolate brain capillaries from a single mouse. Therefore, the developed method can evaluate the individual differences of protein expression in each of the mouse brain capillary fractions (Figure 5(b) and (c)).
The BBB function changes during pathological conditions such as cerebral ischemia and brain trauma. It has been reported that brain capillaries have been isolated and analyzed from ischemic stroke rat model using homogenizer and dextran centrifugation method.41 In our developed method, isolated brain capillaries were less fragmented and better retained their vessels’ form than that seen in the standard method (Figure 2(a)). Therefore, this developed method could be used to investigate the molecular changes of BBB during cerebral ischemia and brain trauma model mouse.
In the present study, brain capillaries were isolated from a frozen brain. Some previous brain capillary extraction protocols suggested using fresh brain tissues.15,42 The standard method used in the present study can isolate capillaries from either fresh or frozen brain tissue, and the fundamental principle of isolation is the same as that of the developed methods; homogenization, dextran centrifugation, and filtration. Therefore, the developed method may be able to isolate capillaries from fresh brains as well. We reported that stroke of homogenization by Potter homogenizer was different between fresh mouse brain42 and frozen mouse brain.43 Therefore, the optimization of bead homogenizer is necessary for applying the developed method to fresh brain tissues.
In conclusion, we established a brain capillary isolation method from a single frozen mouse brain. Also, we proved the developed method can apply to the analysis of genetically modified mouse. This method is useful for the analysis of various mice models, where it may be difficult to generate enough number of littermates. By using this method, physiological and pathological roles of the BBB can be studied in more detail thus further elucidating the association between BBB function and CNS diseases.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-3-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-4-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-5-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: We are grateful for partial financial support in the form of Grant-in-Aid for Scientific Research(B), Mishima Kaiun Memorial Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and JST CREST Grant Number JP171024167, Japan.
Declaration of conflicting interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Ohtsuki S is a full professor at Kumamoto University and is also a director of Proteomedix Frontiers. This study was not supported by the company, and his position at the company did not influence the design of the study, the collection of the data, the analysis or interpretation of the data, the decision to submit the manuscript for publication, or the writing of the manuscript, and did not present any financial conflicts. The other authors declare no competing interests.
Authors’ contributions: All authors, Ogata S, Ito S, Masuda T and Ohtsuki S, contributed to study design and manuscript revision. Ogata S conducted experiments and performed data analysis. Ogata S, Ito S. and Ohtsuki S wrote the manuscript. All authors provided the final approval of the submitted manuscript.
Supplemental material: Supplemental material for this article is available online.
References
- 1.Montagne A, Barnes SR, Sweeney MD, et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015; 85: 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelgesang S, Cascorbi I, Schroeder E, et al. Deposition of Alzheimer's beta-amyloid is inversely correlated with P-glycoprotein expression in the brains of elderly non-demented humans. Pharmacogenetics 2002; 12: 535–541. [DOI] [PubMed] [Google Scholar]
- 3.Gras D, Roze E, Caillet S, et al. GLUT1 deficiency syndrome: an update. Rev Neurol 2014; 170: 91–99. [DOI] [PubMed] [Google Scholar]
- 4.Schulze A.Creatine deficiency syndromes. Mol Cel Biochem 2003; 244: 143–150. [PubMed] [Google Scholar]
- 5.Ogata S, Ito S, Masuda T, et al. Changes of blood-brain barrier and brain parenchymal protein expression levels of mice under different insulin-resistance conditions induced by high-fat diet. Pharm Res 2019; 36: 141. [DOI] [PubMed] [Google Scholar]
- 6.Pardridge WM.Blood-brain barrier genomics. Stroke 2007; 38(2 Suppl): 686–690. [DOI] [PubMed] [Google Scholar]
- 7.Chun HB, Scott M, Niessen S, et al. The proteome of mouse brain microvessel membranes and basal lamina. J Cereb Blood Flow Metab 2011; 31: 2267–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang S, Hsuchou H, Kastin AJ, et al. Diet-induced obesity suppresses expression of many proteins at the blood-brain barrier. J Cereb Blood Flow Metab 2014; 34: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida Y, Tachikawa M, Obuchi W, et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood-brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS 2013; 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yousif S, Marie-Claire C, Roux F, et al. Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res 2007; 1134: 1–11. [DOI] [PubMed] [Google Scholar]
- 11.Czupalla CJ, Yousef H, Wyss-Coray T, et al. Collagenase-based single cell isolation of primary murine brain endothelial cells using flow cytometry. Bio Protoc 2018; 8: e3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Zepeda D, Taghi M, Smirnova M, et al. LC-MS/MS-based quantification of efflux transporter proteins at the BBB. J Pharm Biomed Anal 2019; 164: 496–508. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal S, Uchida Y, Mittapalli RK, et al. Quantitative proteomics of transporter expression in brain capillary endothelial cells isolated from P-glycoprotein (P-gp), breast cancer resistance protein (Bcrp), and P-gp/Bcrp knockout mice. Drug Metab Dispos 2012; 40: 1164–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Feteisi H, Al-Majdoub ZM, Achour B, et al. Identification and quantification of blood-brain barrier transporters in isolated rat brain microvessels. J Neurochem 2018; 146: 670–685. [DOI] [PubMed] [Google Scholar]
- 15.Hoshi Y, Uchida Y, Tachikawa M, et al. Quantitative atlas of blood-brain barrier transporters, receptors, and tight junction proteins in rats and common marmoset. J Pharma Sci 2013; 102: 3343–3355. [DOI] [PubMed] [Google Scholar]
- 16.Kubo Y, Ohtsuki S, Uchida Y, et al. Quantitative determination of luminal and abluminal membrane distributions of transporters in porcine brain capillaries by plasma membrane fractionation and quantitative targeted proteomics. J Pharma Sci 2015; 104: 3060–3068. [DOI] [PubMed] [Google Scholar]
- 17.Uchida Y, Ohtsuki S, Katsukura Y, et al. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J Neurochem 2011; 117: 333–345. [DOI] [PubMed] [Google Scholar]
- 18.Al-Majdoub ZM, Al Feteisi H, Achour B, et al. Proteomic quantification of human blood-brain barrier SLC and ABC transporters in healthy individuals and dementia patients. Mol Pharm 2019; 16: 1220–1233. [DOI] [PubMed] [Google Scholar]
- 19.Ohtsuki S, Kikkawa T, Hori S, et al. Modulation and compensation of the mRNA expression of energy related transporters in the brain of glucose transporter 1-deficient mice. Biol Pharm Bull 2006; 29: 1587–1591. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura K, Hirayama-Kurogi M, Ito S, et al. Large-scale multiplex absolute protein quantification of drug-metabolizing enzymes and transporters in human intestine, liver, and kidney microsomes by SWATH-MS: comparison with MRM/SRM and HR-MRM/PRM. Proteomics 2016; 16: 2106–2117. [DOI] [PubMed] [Google Scholar]
- 21.Kuno T, Hirayama-Kurogi M, Ito S, et al. Effect of intestinal flora on protein expression of drug-metabolizing enzymes and transporters in the liver and kidney of germ-free and antibiotics-treated mice. Mol Pharm 2016; 13: 2691–2701. [DOI] [PubMed] [Google Scholar]
- 22.Masuda T, Tomita M, Ishihama Y.Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J Proteome Res 2008; 7: 731–740. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita K, Sasaki H, Furuse M, et al. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 1999; 147: 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao B, Stieger B, Noe B, et al. Localization of the organic anion transporting polypeptide 2 (Oatp2) in capillary endothelium and choroid plexus epithelium of rat brain. J Histochem Cytochem 1999; 47: 1255–1264. [DOI] [PubMed] [Google Scholar]
- 26.Nasdala I, Wolburg-Buchholz K, Wolburg H, et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J Biol Chem 2002; 277: 16294–16303. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama D, Kusuhara H, Taniguchi H, et al. Functional characterization of rat brain-specific organic anion transporter (Oatp14) at the blood-brain barrier: high affinity transporter for thyroxine. J Biol Chem 2003; 278: 43489–43495. [DOI] [PubMed] [Google Scholar]
- 28.Yu CH, Yhee JY, Kim JH, et al. Increased expression of vascular endothelial growth factor in neo-vascularized canine brain tissue. Canad J Veter Res 2012; 76: 62–68. [PMC free article] [PubMed] [Google Scholar]
- 29.Bzducha-Wrobel A, Blazejak S, Kawarska A, et al. Evaluation of the efficiency of different disruption methods on yeast cell wall preparation for beta-glucan isolation. Molecules 2014; 19: 20941–20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubacq S.Performing efficient sample preparation with hard tumor tissue: Precellys® bead-beating homogenizer solution. Nat Meth 2016; 13: i–iii. [Google Scholar]
- 31.Rodríguez-Gutiérrez G, Wood S, Fernández-Bolaños Guzmán J, et al. Determination of 3,4-dihydroxyphenylglycol, hydroxytyrosol and tyrosol purified from olive oil by-products with HPLC in animal plasma and tissues. Food Chem 2011; 126: 1948–1952. [DOI] [PubMed]
- 32.Kesanakurti P, Belton M, Saeed H, et al. Screening for plant viruses by next generation sequencing using a modified double strand RNA extraction protocol with an internal amplification control. J Virol Methods 2016; 236: 35–40. [DOI] [PubMed]
- 33.Abbott NJ, Ronnback L, Hansson E.Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci 2006; 7: 41–53. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee S, Naik UP.Pericyte-endothelial cell interaction: a survival mechanism for the tumor vasculature. Cell Adhes Migrat 2012; 6: 157–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sher G. Handbook of Neuroevolution Through Erlang. Springer, 2012, p.47.
- 36.Storck SE, Meister S, Nahrath J, et al. Endothelial LRP1 transports amyloid-beta(1-42) across the blood-brain barrier. J Clin Invest 2016; 126: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin-Valencia I, Good LB, Ma Q, et al. Glut1 deficiency (G1D): epilepsy and metabolic dysfunction in a mouse model of the most common human phenotype. Neurobiol Dis 2012; 48: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura S, Osaka H, Muramatsu SI, et al. Gene therapy for a mouse model of glucose transporter-1 deficiency syndrome. Mol Genet Metab Rep 2017; 10: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandu C, Burloiu CM, Barca DG, et al. Ketogenic diet in patients with GLUT1 deficiency syndrome. Maedica 2019; 14: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierre K, Parent A, Jayet PY, et al. Enhanced expression of three monocarboxylate transporter isoforms in the brain of obese mice. J Physiol 2007; 583(Pt 2): 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KA, Kim D, Kim JH, et al. Autophagy-mediated occludin degradation contributes to blood-brain barrier disruption during ischemia in bEnd.3 brain endothelial cells and rat ischemic stroke models. Fluids Barriers CNS 2020; 17: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtsuki S, Yamaguchi H, Kang YS, et al. Reduction of L-type amino acid transporter 1 mRNA expression in brain capillaries in a mouse model of Parkinson's disease. Biol Pharm Bull 2010; 33: 1250–1252. [DOI] [PubMed] [Google Scholar]
- 43.Yamasaki Y, Kobayashi K, Okuya F, et al. Characterization of P-glycoprotein humanized mice generated by chromosome engineering technology: its utility for prediction of drug distribution to the brain in humans. Drug Metab Dispos 2018; 46: 1756–1766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-3-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-4-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-5-jcb-10.1177_0271678X20941449 for Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis by Seiryo Ogata, Shingo Ito, Takeshi Masuda and Sumio Ohtsuki in Journal of Cerebral Blood Flow & Metabolism