Abstract
Background
Self-reported consanguinity is associated with risk for schizophrenia (SZ) in several inbred populations, but estimates using DNA-based coefficients of inbreeding are unavailable. Further, it is not known whether recessively inherited risk mutations can be identified through homozygosity by descent (HBD) mapping.
Methods
We studied self-reported and DNA-based estimates of inbreeding among Egyptian patients with SZ (n=421, DSM IV criteria) and adult controls without psychosis (n=301), who were evaluated using semi-structured diagnostic interview schedules and genotyped using the Illumina Infinium PsychArray. Following quality control checks, coefficients of inbreeding (F) and regions of homozygosity (ROH) were estimated using PLINK software for HBD analysis. Exome sequencing was conducted in selected cases.
Results
Inbreeding was associated with schizophrenia based on self-reported consanguinity ( χ2=4.506, 1 df, p=0.034) and DNA-based estimates for inbreeding (F); the latter with a significant F x age interaction (β=32.34, p=0.0047). The association was most notable among patients older than age 40 years. Eleven ROH were over-represented in cases on chromosomes 1, 3, 6, 11, and 14; all but one region is novel for schizophrenia risk. Exome sequencing identified six recessively-acting genes in ROH with loss-of-function variants; one of which causes primary hereditary microcephaly.
Conclusions
We propose consanguinity as an age-dependent risk factor for SZ in Egypt. HBD mapping is feasible for SZ in adequately powered samples.
Keywords: genotyping, psychosis, statistical genetics, consanguinity
INTRODUCTION
Schizophrenia (SZ) is a life-long and potentially disabling illness with a 1% lifetime morbid risk, detectable across the globe. Treatment is symptomatic and palliative, failing in over half of patients (Lieberman et al., 2005; Stroup et al., 2006). In view of the unsatisfactory treatment, it is important to understand etiology and pathogenesis of SZ. The relatively high heritability for SZ (twin studies~0.70; single nucleotide polymorphisms, SNP~0.45), motivated a variety of gene mapping efforts (Bulik-Sullivan et al., 2015; Gottesman, 1991; McGue et al., 1983; Zheng et al., 2017). Genome-wide association studies (GWAS) conducted by the Psychiatric Genomics Consortium identified 108 SZ risk loci (Schizophrenia Working Group of the Psychiatric Genomics, 2014); further enlargement of study samples has enabled the identification of even more risk variants (Pardiñas et al., 2018). Rare variants, including copy number variants (CNVs) and de novo mutations have also been implicated (Consortium, 2008; Sebat et al., 2009). Additional risk undoubtedly remains to be identified (So et al., 2011), thus motivating additional genetic studies for this multifactorial, polygenic disorder.
While most GWAS have sought to identify additive risk variants, recessively inherited risk factors have also been proposed for SZ (Gottesman and Shields, 1973). It is relatively easy to map recessively inherited monogenic disorders in populations with historically high rates of inbreeding, because relatively large genomic segments flanking disease mutations are likely to be inherited identical by descent among cases. Homozygous by descent (HBD) analysis was used to map mutations successfully for several hundred rare Mendelian diseases (Alkuraya, 2010; Botstein and Risch, 2003). Although HBD analysis has been used for mapping typical monogenic diseases, it could also be useful for diseases with polygenic or multifactorial inheritance. A linkage study of intellectual disability in consanguineous pedigrees, followed by focused sequencing in regions with suggestive linkage, identified 50 novel mutations and confirmed 23 known disease genes (Najmabadi et al., 2011). This study included a heterogeneous set of families, suggesting success can be achieved despite genetic heterogeneity (Lezirovitz et al., 2008). Thanks to densely spaced genetic markers and innovative analyses, it is no longer necessary to access family members for mapping recessive diseases, because HBD genomic segments can be identified among ostensibly unrelated cases, particularly within inbred populations (Browning and Browning, 2010; Gusev et al., 2009; Purcell et al., 2007). Thus, large inherited homozygous deletions at PCDH10 and DIA1 were reported in one study of autism (Morrow et al., 2008). HBD analysis was used for genetically heterogeneous diseases including non-syndromic intellectual disability associated with variable postnatal microcephaly (Mochida et al., 2009), non-syndromic deafness, and patent ductus arteriosus (Mani et al., 2002; Schraders et al., 2010).
These successes motivated us to test HBD mapping for SZ. Reports from the Middle East, Dagestan, Croatia, Cuba, and India show increased risk for SZ and mood disorders among inbred populations, suggesting the presence of DNA variants acting recessively or have dosage effects (Abaskuliev and Skoblo, 1975; Britvic et al.; Bulaeva et al., 2003; Chaleby and Tuma, 1987; Dobrusin et al., 2008; Ewald et al., 2003; Gindilis et al., 1989; Lerer et al., 2003; Mansour et al., 2010; Rudan et al., 2003). In a consanguineous sample from Egypt, SZ was the most commonly reported chronic psychosis (Okasha, 2004). Approximately one-fifth of marriages in rural regions of Egypt in the Nile Delta are between first cousins(Settin and Algelani, 1997). High population inbreeding rate, along with association between consanguinity and SZ risk we identified in a small sample, suggested genomic regions of homozygosity are more prevalent among consanguineous Egyptian SZ patients versus comparable groups (Mansour et al., 2010). Here, we extend prior analyses by investigating a three-fold larger Egyptian case-control sample, characterizing the full sample using a genome-wide SNP array and a subset of the sample by exome sequencing.
2.0. METHODS
2.1. Research Participants
Egyptian patients and controls were recruited and evaluated as described previously (Mansour et al., 2010). Briefly, apparently unrelated cases were ascertained at Mansoura University Hospital (MUH) Psychiatry outpatient clinics based on clinical diagnosis of SZ (DSM-IV), blind to history of consanguinity. Participants were interviewed by psychiatrists using Arabic version of the Schedule for Clinical Assessment in Neuropsychiatry (Wing et al., 2001). Family histories of consanguinity were obtained using the Arabic version of the Family Interview for Genetics Studies (Mansour et al., 2009). Control individuals (persons without reported psychosis) of similar age to the cases and from the same geographical region were recruited without knowledge of their family structure during the same period as the cases. Individuals (or those with first-degree relatives) with a history of psychosis or bipolar 1 disorder were excluded as controls. Consensus diagnoses were established by ≥2 psychiatrists following a thorough review of available data. Some participants in the present report participated in our earlier study (74 cases, 124 controls) (Mansour et al., 2010).
The study was approved by the Mansoura University Ethics Committee and the University of Pittsburgh Institutional Review Board (IRB). Participants provided written informed consent. Relevant samples and data have been deposited with the NIMH data repository (https://www.nimhgenetics.org), following approval by the IRBs at Mansoura University and the University of Pittsburgh, as well as approval by relevant authorities from both universities.
2.2. Genotyping and Identifying Runs of Homozygosity (ROH)
DNA was extracted from venous blood at the University of Pittsburgh or at the Rutgers University Cell and DNA Repository (RUCDR Infinite Biologics, Piscataway, NJ, USA), using the phenol/chloroform method. Participants were genotyped using the Infinium PsychArray-24 v1.1 BeadChip Kit (“PsychArray”; Illumina Inc., San Diego, CA, USA). The PsychArray includes 571,054 genome-wide markers and ~50,000 markers associated with psychiatric disorders. All statistics were performed using R, unless otherwise stated.
2.2.1. Genotyping Quality Control
Genotypes from SZ (n=421) and adult controls (n=301) were called using GenomeStudio 2.0.1 (Illumina Inc.). Quality control was performed using PLINK v1.9. SNPs were removed from analysis if mapped to non-autosomal regions, were monomorphic, had a non-call rate of >0.05, had a minor allele frequency (MAF)<0.05, violated Hardy-Weinberg expectations in controls (p<0.005), or were not represented on the GRCh37/hg19 human genome reference sequence. Individuals were excluded from analyses when genotyping call rate was<95%, chromosome-X discordant homozygosity rate, or were duplicated. After, there remained 280,746 SNPs and n=674 subjects (n=398 SZ cases and n=276 controls).
2.2.2. Inbreeding Coefficient (F)
The inbreeding coefficient (F) quantifies the expected portion of the genome homozygous by descent. For example, for the progeny of full siblings, F=1/4; for the progeny of first cousins, F=1/8. F was estimated on the entire cohort using PLINK and observed regions of homozygosity (ROH) were identified across the genome.
The association between the genetically determined inbreeding values and SZ was determined using logistic regression of SZ on predictors F, age, sex, and the interaction of age by inbreeding. Furthermore, subjects who had an F≥0.05 were identified, conservatively approximating second cousins or closer relationships, the most common form of consanguineous relationships in this cohort (henceforth referred to as “consanguineous”).
2.2.3. The Effect of Inbreeding on Hardy-Weinberg Expectations
For a SNP with minor allele frequency p and major allele frequency q = 1-p in a population, the Hardy-Weinberg principle states the expected genotype frequencies are p2 (minor allele homozygote), q2 (major allele homozygote), and 2pq (heterozygote). These genotype frequencies remain stable across generations in the absence of specific perturbation factors. Non-random mating of an inbred population, however, is one such factor. Deviation from Hardy-Weinberg expectations (HWE) occur due to increased homozygosity of consanguineous individuals, which increases the observed frequencies of the homozygous genotypes in the population relative to HWE expectation (e.g., Table 14.2(Falconer, 1981)).
Population genetic studies routinely remove SNPs that fail HWE because it is likely genotyping errors cause these deviations. To evaluate how this common quality control procedure would affect our results, we investigated the impact of inbreeding for SNPs deviating significantly from HWE utilizing three simulation scenarios that were generated using the attributes of this cohort. For each scenario, HWE tests were performed on different subsets of data, defined by the estimated value of F of the subjects in the Egypt data and a threshold for the maximum F value (maxF, ranging from 0.001 to 0.30). The allele frequencies in the observed Egyptian data were used to simulate genotypes.
Scenario 1 used the estimated F for each individual from the Egyptian sample (for subjects with an estimated F<0, F was set to zero). Scenario 2 was designed to show the effect of sample size on the number of SNPs significantly departing from HWE by keeping the fraction of consanguineous individuals (12.6%) constant as sample size increased. The individuals considered consanguineous were assigned F=0.025, 0.05 or 0.10, while the remaining samples (87.4%) had an assigned F=0. Each scenario was simulated 100 times.
2.2.4. Mapping Regions of Homozygosity (ROH)
SNPs in regions inherited identical-by-descent are homozygous. ROH regions vary in size by locus and individual, although consanguineous individuals are expected to harbor more ROH, on average, than outbred individuals (Mansour et al., 2010). ROH were identified across autosomes using a multi-step process. SNPs passing quality control (280,746) were imputed up to 47,072,716 SNPs on the 1000G reference panel (3.v5) using Michigan Imputation Server (Das et al., 2016). The imputed genotypes were subsequently reduced to 4,914,617 bi-allelic SNPs, following selection for: i)imputation R2>0.81; ii)MAF≥0.05. To avoid redundancy in genotypes, the remaining SNPs were pruned in PLINK with the setting 50 kilobase (kb) window, five SNP sliding window, and maximum linkage disequilibrium (LD) r2<0.9025. This resulted in 1,091,357 SNPs available to calculate ROH.
ROH were determined using the “--homozyg” command (PLINK), which utilizes a sliding window approach, searching for homozygous sections across the genome. Our ROH criteria were: 1000 kb in length, ≥100 SNPs, and at least one SNP was present per 50 kb. The fraction of the genome covered by ROH was 2,659 megabase pairs. To identify overlapping ROH in cases and controls, consensus ROH must contain at least five overlapping ROH segments and at least 95% of the alleles matched (using jointly non-missing/homozygous sites).
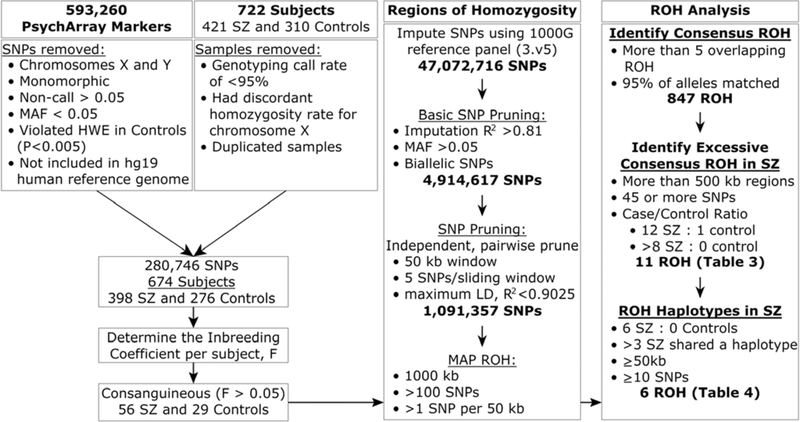
After mapping ROH, common and shared haplotypes were identified among consanguineous cases (F≥0.05). ROH containing haplotypes far more common in SZ than the controls were identified using these criteria: ROH that were at least 500 kb and contained ≥45 SNPs; the ROH were shared by at least eight cases and no controls or at least 12 cases and one control (unadjusted Fisher’s exact test, “FET”, p<0.05); furthermore, these regions had to have at least half (≥3) of the cases presenting with the predominant haplotype (excess haplotype sharing was assessed using FET without correction for multiple testing). The flow chart in Figure 1 depicts data analysis methodology.
Figure 1.
Procedures used to identify significant regions of homozygosity in consanguineous individuals.
2.2.5. Concordance Between Self-Reported Consanguinity and Inbreeding Coefficient
The concordance between self-reported consanguinity and F was compared using Cohen’s Kappa statistic (κ), in SZ and controls.
2.3. Whole Exome Sequencing
Whole exome sequencing (WES) was performed on genomic DNA from a subset of cases. We first sought ROH regions, which were in notable excess in cases versus controls, containing at least 25 SNPs and at least 500 kb using two criteria (unadjusted FET p<0.05): i)ROH observed in ≥8 cases and zero controls or ii)ROH observed in ≥12 cases and one control sample; 11 regions identified. We then selected 12 consanguineous individuals diagnosed with SZ and six non-consanguineous individuals diagnosed with SZ whose genomes matched these criteria for at least one of these regions.
WES was performed in two batches at the Broad Institute (Cambridge, MA, USA) using the Illumina HiSeq 2500 and the Illumina HiSeq 4000, generating an average of 53.8 million paired-end reads of 76bp (14 individuals) and 34.5 million paired-end reads of 151bp (four individuals) per library, respectively. Sequence reads were aligned to human reference genome, GRCh37/hg19, using BWA aln (v0.5.9) with the following parameters: –q5–l 32–k2–o1 (Li and Durbin, 2010). On average, 91% and 93% of the targeted bases in the WES had ≥20X coverage, respectively. Joint variant calling on these libraries was performed by GATK (v3.1–1) (Miller et al., 2010) following best practices (DePristo et al., 2011; Van der Auwera et al., 2013), which yielded 133,300 and 79,897 high-quality variants, respectively, using VCFtools v0.1.11 (Danecek et al., 2011) with the following parameters: --max-alleles2--max missing-count2—min-alleles2--min-mean-depth20--minQ50. Variants were annotated using Variance Effect Predictor (VEP; Ensembl)(McLaren et al., 2016).
2.3.1. Prioritizing Exome Sequencing Variants
In the 12 consanguineous cases with WES data, we sought to characterize deleterious exome variants mapping into ROH of interest. To begin, we extracted WES variants with a mean coverage ≥20X, followed by annotation using VEP (selecting those in canonical transcripts and predicted as “deleterious”). SNPs assigned with multiple consequences were prioritized using the most severe, in the following order: i)stop-gain, ii)frameshift, iii)splice-donor, iv)splice-acceptor, v)start-loss, vi)stop-loss, vii)missense, and viii)all other categories. Variation categories i-vi were referred as protein truncating variants (PTV) and category vii as missense variants (MIS). Next, PTV or MIS variants were identified in genes, which were predicted to be more intolerant to loss-of-function variation (PTV), using the pLI score (Karczewski et al., 2019). pLI reflects how observed genetic variation, which is found within the exons, compares to that expected based on an evolutionary model and the observed variation in the population. Hence, pLI provides a gene-level score of evolutionary constraint of genes. Finally, for each remaining variant, the most severe type was selected based on the following criteria in this order: i)PTV more severe than MIS; ii)pLI; and iii)consequence. The MIS variants were ordered based on a missense variant constraint metric (“MPC”, where a higher value indicates increased gene constraint)(Samocha et al., 2017) or, if MPC values were not available, PolyPhen-2 scores were used as a substitute (“PPH”; PolyPhen is a prediction tool assessing the possible impact of an amino acid substitution on the structure and function of the resulting protein; higher values indicate more severe consequences)(Adzhubei et al., 2010).
Separately, we identified rare WES variants (MAF<0.01 in gnomAD) mapping to recessively-acting genes (n=1,183)(Berg et al., 2013; Blekhman et al., 2008), reasoning consanguineous individuals may harbor deleterious variants within haplosufficient loci.
3.0. RESULTS
3.0.1. Demographic Features
There were significantly more males in the SZ than in the control group (69% vs 55%, respectively; χ2=12.1, df=1, p=0.0005); the mean age for the cases were younger than the controls (31.4 years, SD=9.79 vs 33.0 years, SD=9.44, respectively; t=2.2, df=667, p=0.027). Among the cases, the age at onset was not significantly different between the consanguineous and the non-consanguineous subjects (21.7 years, SD=6.1 vs 22.2 years, SD=6.9, respectively; t=(−0.49), df=373, p=0.62; Table 1). The prevalence of self-reported consanguinity was greater among cases (χ2=4.9, df=1, p=0.027). There was no significant difference between SZ and control subjects regarding rural/urban residence (p>0.05; Table 1).
TABLE 1.
Demographic features of cases and controls
|
Schizophrenia |
Controls |
|||||||
| n | mean (SD) | n | mean (SD) | t1 | df | p | ||
| Age (years) | 393 | 31.4 (9.8) | 276 | 33.0 (9.4) | 2.2 | 667 | 0.027 | |
| Age at onset (years)2 | consanguineous (F>0.05) | 53 | 21.7 (6.1) | – | – | −0.49 | 373 | 0.62 |
| not consanguineous (F<0.05) | 323 | 22.2 (6.9) | ||||||
|
Schizophrenia |
Controls |
|||||||
| n | percent | n | percent | Chi-square3 | df | p | ||
| Sex Ratio (Male) | 268 | 68.70% | 153 | 55.40% | 12.1 | 1 | 0.0005 | |
| Geographic Region (Urban) | 115 | 30.40% | 68 | 24.70% | 2.7 | 1 | 0.1 | |
| Self-reported Consanguinity (Yes) | 101 | 36.70% | 50 | 18.20% | 4.9 | 1 | 0.027 | |
Two-sided Student’s t test.
Count for age at onset does not equal total cases, due to missing data in 23 individuals (Three consanguineous cases; 20 non-consanguineous cases).
Chi-square test corrected for sex, residential setting, and self-reported familial consanguinity.
F=inbreeding coefficient; n=subject count; SD=standard deviation; df=degrees of freedom; t=test statistic; p=probability.
3.0.2. Identification of Regions of Homozygosity (ROH)
To determine the utility of PLINK’s method of moments approximation for each individual’s inbreeding coefficient, we compared it to another estimator based on the genomic fraction that is ROH. The correlation of these measures was high (r=0.97; Supplemental Figure 1). Thus, the moment approximation for F from PLINK analysis was used for the remainder of this report.
3.0.3. Inbreeding Coefficient (F)
There were 128 cases (32%) and 73 controls (26%) considered to be consanguineous; i.e., second cousins or closer, clinical genetic definition (F>0.0156)(Bittles, 2001). Regression analysis with case status as the outcome indicated significant associations with the SNP-based estimate for F, age, sex, and a significant interaction effect between age and F (Table 2). In view of the significant age by F interaction, the cases and controls were partitioned by age quartiles and the mean F was determined per quartile; there were proportionally more younger cases (χ2=2.86, df=3, p=0.0049; Supplemental Table 1a). Furthermore, the mean F per age quartile was similar for controls but fluctuated in the cases. Specifically, the youngest cases (under 25 years) had lower mean F, and the eldest cases (over 39 years of age) had a mean F comparable to the controls. In addition, the mean F was computed per age quartile in males and females; however, there was no difference in the distribution of individuals per age quartile (χ2=1.2, df=3, p=0.75; Supplemental Table 1b).
TABLE 2.
Age-dependent association of schizophrenia with inbreeding.
| β (SE) | OR | 95% CI (OR) | z | p | |
|---|---|---|---|---|---|
| Inbreeding Coefficient (F) | 32.39 (11.44) | 1.17×1014 | 2.13×104–6.44×1023 | 2.83 | 0.0046 |
| Age (Years) | −0.0067 (0.009) | 0.99 | 0.97–1.01 | −0.76 | 0.58 |
| Sex | −0.62 (0.16) | 0.54 | 0.39–0.74 | −3.78 | 0.00016 |
| Inbreeding Coefficient × Age | −0.87 (0.34) | 0.42 | 0.22–0.80 | −2.61 | 0.0091 |
β=beta regression coefficient, SE=standard error, OR=odds ratio, 95% CI (OR)=the 95% confidence interval of the odds ratio, z=test statistic, p=probability.
3.0.4. Concordance Between Self-Reported Consanguinity and the Inbreeding Coefficient
The concordance between the self-reported measure of consanguinity (yes/no) and the genetically determined inbreeding (F≥0.05) was compared in SZ and controls, separately. Among cases, there was 85% agreement between the self-reported consanguinity and estimated F (z=7.46, κ=0.54, 95% CI=0.43–0.65, p=4.46×10−14). Among controls there was an 88% agreement between the self-report and the estimated F (z=4.94, κ=0.52, 95% CI=0.36–0.67, p=3.82×10−07), indicating no significant case-control differences for self-reported consanguinity.
3.0.5. The Effect of Inbreeding on Hardy-Weinberg Expectations
The effect of inbreeding on genotype frequencies deviating from Hardy-Weinberg expectations was explored in this cohort. While it is expected that some SNPs will depart from HWE due to chance or to genotyping error, loci will also depart from HWE due to inbreeding. Larger sample sizes would be better powered to detect such an effect; smaller sample sizes like ours can detect only larger deviations. This is a challenging problem because it is not possible to know exactly which SNPs are departing HWE due to inbreeding, genotyping error, random chance, or some combination. To explore this challenging problem, we performed three simulation scenarios using parameters of the Egyptian cohort. Genotype frequencies were simulated for each scenario and SNPs were tested for departure from HWE. SNPs with HWE p<0.0005 were considered significant.
Scenario 1 was designed to evaluate random error of observed genotype frequencies by simulating data using the estimated values of F per Egyptian subject and the estimated MAF for the population. If deviations from HWE observed in the Egyptian sample were only due to inbreeding and random sampling, the number of SNPs departing HWE should roughly match what is observed in the simulation. Instead, while the general pattern mirrors the results obtained in the actual data, the counts from Scenario 1 are uniformly lower than expected, presumably due to genotyping error (notably, if we were to simulate data under a no-inbreeding scenario, the number of SNPs departing HWE follows that expected by chance [data not shown]; Supplemental Figure 2a). Scenario 2 highlights the statistical phenomenon of power for detecting SNPs departing HWE due to inbreeding. This scenario tested increasing number of subjects where the proportion of consanguineous individuals was kept constant (12.6%, which was derived from the number of consanguineous individuals we detected in this particular cohort who had F>0.05). Clearly, when the sample size increases, the number of SNPs detected as departing HWE also increases, while the proportion of consanguineous individuals remained constant (Supplemental Figure 2b).
By contrasting results from these scenarios to the actual data (Supplemental Figure 2), we conclude that SNPs depart from HWE due to all of the mechanisms described previously; however, it is not possible to know exactly which SNP(s) are departing HWE due to inbreeding or some other reason. Of note, we did not find SNPs departing HWE that mapped into an ROH listed in Tables 3 and 4.
TABLE 3.
Regions of homozygosity over-represented in schizophrenia cases.
| Region | Chr | Location (base position) | Chromosome Band | Number of samples | Length (kb) | Total SNPs | p | Significant GWAS gene, phenotype1 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | SZ cases | Controls | Total | |||||||
| 1 | 1 | 108,860,573 | 110,323,150 | 1p13.3 | 12 | 1 | 13 | 1462.58 | 111 | < 0.05 | SYPL2, cognitive ability GPR61, chronotype |
| 2 | 14 | 32,438,841 | 32,964,338 | 14q12 | 10 | 0 | 10 | 525.50 | 59 | < 0.05 | AKAP6, SZ, BP, cognition |
| 3 | 1 | 89,045,338 | 89,735,314 | 1p22.2 | 9 | 0 | 9 | 689.98 | 57 | < 0.05 | none |
| 4 | 1 | 161,957,719 | 163,494,784 | 1q23.3 | 9 | 0 | 9 | 1537.07 | 205 | < 0.05 | none |
| 5 | 1 | 165,625,023 | 166,337,053 | 1q24.1 | 9 | 0 | 9 | 712.03 | 83 | < 0.05 | FAM78B, cognition |
| 6 | 14 | 29,895,001 | 31,237,170 | 14q12 | 9 | 0 | 9 | 1342.17 | 129 | < 0.05 | none |
| 7 | 1 | 154,791,128 | 156,378,425 | 1q21.3-q22 | 8 | 0 | 8 | 1587.30 | 103 | < 0.05 | KCNN3-PMVK, SZ, BP ZBTB7B, SZ RIT1, SZ |
| 8 | 1 | 160,513,183 | 161,361,979 | 1q23.3 | 8 | 0 | 8 | 848.80 | 79 | < 0.05 | none |
| 9 | 3 | 190,779,327 | 191,862,817 | 3q28 | 8 | 0 | 8 | 1083.49 | 155 | < 0.05 | AC073365.1, SZ |
| 10 | 6 | 108,323,759 | 109,351,125 | 6q21 | 8 | 0 | 8 | 1027.37 | 87 | < 0.05 | AFG1L, cognition FOXO3, SZ, cognition, neuroticism |
| 11 | 11 | 22,141,171 | 22,647,550 | 11p14.3 | 8 | 0 | 8 | 506.38 | 48 | < 0.05 | none |
Regions of homozygosity were identified that were over-represented in consanguineous schizophrenia cases. SZ=schizophrenia; BP=bipolar disorder; Chr=chromosome; kb=kilobases; p=equivalent to an unadjusted p-value using Fisher’s exact test. Genomic locations based on GRCh37/hg19.
Genome-wide association studies (GWAS) with significantly associated SNPs identified in prior studies per ROH region. Detailed information for these GWAS is provided in Supplemental Table 2.
Table 4.
Regions of homozygosity having shared haplotypes that are over-represented in schizophrenia cases.
| Major Haplotype | Gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Position | Band | Size (kilobases) | SNP count | Case : Control | N | Case : Control | Symbol | Name |
| 1 | 89,116,300–89,286,673 | 1p22.2 | 170.4 | 11 | 9:0 | 3 | 7:0 | PRKN2a | Protein kinase N2 |
| 1 | 89,530,860–89,721,862 | 1p22.2 | 191 | 14 | 9:0 | 5 | 5:0 | GBP1b; GBP2; GBP4; GBP7 | Guanylate binding proteins |
| 14 | 30,070,659–30,290,431 | 14q12 | 219.8 | 19 | 9:0 | 3 | 7:0 | PRKD1c | Protein kinase D1 |
| 14 | 30,991,559–31,172,582 | 14q12 | 181 | 14 | 9:0 | 4 | 6:0 | G2E3 SCFD1d | G2/M Phase-specific E3 Ubiquitin Ligase SEC1 Family Domain Containing 1 |
| 1 | 146,660,395–146,736,137 | 1q21.1 | 75.7 | 11 | 7:0 | 4 | 4:0 | FMO5e CHD1Lf | Flavin Containing Monooxygenase 5 Chromodomain Helicase DNA-binding Protein 1 Like |
| 12 | 8,698,471–8,820,124 | 12p13.31 | 121.7 | 13 | 6:0 | 4 | 3:0 | AICDA MFAP5 | Activation-induced Cytidine Deaminase Microfibrillar-associated Protein 5 |
ROH in PRKN2 (NM_006256) contained 16 of 22 exons.
ROH in GPB1 (NM_002053) contained one of 11 exons.
ROH in PRKD1 (NM_002742) contained 13 of 18 exons.
ROH in SCFD1 (NM_016106) contained 16 of 25 exons.
ROH in FMO5 (NM_001461) contained 8 of 9 exons.
ROH in CHD1L (NM_004284) contained 7 of 23 exons.
3.0.6. ROH Analysis in Consanguineous Subjects
There were 29 controls and 56 SZ cases with SNP-based F>0.05 and thus were considered “consanguineous” using our definition. Among these individuals, 847 consensus ROH loci were identified, of which 53% (n=448) spanned least 500 kb and were covered by at least 25 SNPs. Furthermore, 66% (n=561) of the consensus ROH spanned at least 100 kb and were covered by ≥10 SNPs. Eleven ROH mapping to chromosomes 1, 3, 6, 11, and 14, were identified as having an excess of ROH in cases versus controls, by our criteria (Table 3). We next investigated whether the 143 autosomal SZ risk loci reported in a GWAS by Pardiñas and colleagues localized to the ROH(Pardiñas et al., 2018). One SNP (rs61937595) mapped into an ROH on chromosome 12. To determine significance, permutation analysis was conducted where regions were identified, randomly and with replacement, in the genome with the same size characteristics as the 11 ROH regions in Table 3. The number of times a region contained one of the 143 SZ risk loci was tabulated. The procedure was repeated 1000 times and an empirical p-value was computed for the simulation data. These ROH loci did not show an excess enrichment for the 143 SZ GWAS loci (p=0.584). The full list of GWAS-significant findings performed in prior mental health studies that map into these 11 ROH are provided in Supplemental Table 2(Davies et al., 2015; Davies et al., 2018; Goes et al., 2015; Hill et al., 2019; Ikeda et al., 2018; Jones et al., 2019; Lam et al., 2017; Lee et al., 2018; Li et al., 2017; Stahl et al., 2019). GWAS findings were obtained from the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) catalog of published GWAS (date accessed 2019–09-17; www.ebi.ac.uk/gwas/).
3.0.7. ROH Haplotype Analysis
We further examined ROH regions (Table 3) with at least half of the cases (≥3) shared a haplotype. Shared haplotypes were identified if they were ≥50 kb and included ≥10 SNPs. Two loci, within 500 kb of one another on 1p22.2, were 170 kb and 191 kb, contained Protein Kinase-N2 (PKN2) and a cluster of Guanylate Binding Protein genes (GBP1, GBP2, GBP4, and GBP7). A 76 kb region on 1q21.1 contained Flavin-Containing Monooxygenase 5 (FMO5) and Chromodomain Helicase DNA-Binding Protein 1-Like (CHD1L). Two loci on 14q12, within 1000 kb of each other, were 220 kb and 181 kb, containing Protein Kinase D1 (PRKD1), G2/M-Phase Specific E3-Ubiquitin Protein Ligase (G2E3) and Sec1 Family-Domain Containing (SCFD1). On chromosome 12p13.31, there was a 122 kb region containing Activation Induced Cytadine Deaminase (AICDA) and Microfibril-Associated Protein-5 (MFAP5; Table 4).
3.1. Whole Exome Sequencing (WES)
3.1.1. Variant Calls, Annotation, and Prioritization
We sequenced 12 consanguineous individuals diagnosed with SZ, prioritizing ROH regions (see Methods), identifying 119,295 unique variants (present in ≥1 individual). Summary metrics of variants generated by PLINK/SEQ (https://atgu.mgh.harvard.edu/plinkseq/) are provided in Supplemental Tables 3a and 3b. The versions of databases used by VEP for annotations are provided in Supplemental Table 3c. There were 118,026 variants present in VEP, mapping to 28,033 canonical gene transcripts. There were 25,246 PTV/MIS variants (9,787 genes) annotated with pLI scores and subsequently reduced to one annotation per variant (Supplemental Table 4a). The PTV/MIS variants were next delineated, using scores for gene constraint (pLI), missense deleteriousness (MPC), or impact of altered amino acid on the resultant protein (PPH). There were 18 PTV mapped into highly constrained genes (pLI>0.995), 118 predicted deleterious MIS variants (MPC>2), and 73 MIS variants predicted as protein-deleterious (PPH>0.9; Supplemental Table 4b).
3.1.2. Exome Variants in ROH loci
Exome variants mapped into ROH established in the consanguineous cohort were identified. Homozygous alternative genotypes localizing to loci within ROH were identified in 12 consanguineous cases. There were 33 unique variants identified with homozygous alternative allele genotypes. Thirty variants were MPC1 and three variants were MPC2 missense variants. The three MPC2 variants map to OSTM1 and SESN1 (described above), along with PVRL4 (the protein product contains two immunoglobulin-like C2-type domains and one immunoglobulin-like V-type domain). The PVRL4 variant has a global allele frequency of 0.08 (gnomeAD) and is tolerant to mutations (pLI<0.5). Autosomal recessive ectodermal dysplasia-syndactyly syndrome type-1 can result from gene mutations in PVRL4; however, this variant was not reported as pathogenic in the ClinVar database (Supplemental Table 5).
Next, we identified exome variants localized to genes implicated in autosomal recessive conditions. From 1,183 curated recessive gene list (Berg et al., 2013; Blekhman et al., 2008), we observed 2,265 variants (760 genes) in 12 subjects. Of these, we focused on rare (MAF<0.01), deleterious (PTV or MPC >2) variants. One subject identified with a homozygous genotype for a variant in ASPM (abnormal spindle homolog, microcephaly-associated). Primary hereditary microcephaly is associated with homozygous mutations in ASPM; however, microcephaly was not reported for this individual.
Furthermore, we sought any variant in the consanguineous (n=12) or the non-consanguineous (n=6) cases with WES data, likely to be deleterious (PTV/MPC>2). We detected a mean of 6.0 (SD=3.7) and 5.9 (SD=3.1) variants homozygous for the alternate allele in the non-consanguineous and consanguineous cases, respectively (W=37.5, p=0.46; not significant, Wilcoxon rank sum test); however, the number of heterozygous deleterious variants were significantly lower in consanguineous cases (mean=22.8, SD=3.9) than non-consanguineous cases (mean=37.7, SD=14.6; W=60, p=0.014).
4.0. DISCUSSION
We find inbreeding is associated with risk for SZ in Mansoura, Egypt, using self-reported consanguinity, and SNP-based estimates of inbreeding. Furthermore, we detect a significant interaction of the coefficient of inbreeding (F) with age. These results are consistent with our prior analyses that used a subset of these subjects and short tandem repeat polymorphisms (STRPs) to determine F (Mansour et al., 2010). Other groups report associations between autozygosity (alleles/regions HBD by consanguineous matings), and an increased risk for SZ, autism, and depression/suicidal behavior (Gamsiz et al., 2013; Keller et al., 2012; Maguire et al., 2018; Melhem et al., 2017; Wang et al., 2010). Previous work on either inbred or outbred cohorts report conflicting results. A whole-genome heterozygosity association study in 178 unrelated Caucasian SZ and 144 controls indicated ROH on chromosomes 1q23.3, 2q31.1, 17q21.31, and 18q12.3 were more common in cases (Lencz et al., 2007). Another study, of two independent samples of Ashkenazi Jewish and Japanese ancestry, indicated consistent association between SZ risk and homozygous fragments in the HLA region on chromosome six (Mukherjee et al., 2014). An increased prevalence of SZ subtypes were reported in genetically-isolated communities in Daghestan, Russia, where investigators demonstrated significant linkage on chromosomes 22q11 (LOD=4.4) and 17p11-p12 (LOD=3.7)(Bulayeva et al., 2007). Conversely, a recent study reported on 39,830 Caucasian individuals, selected without regard to self-reported consanguinity, did not identify excess ROH in these regions (Johnson et al., 2016). The discordant results from prior studies suggest well-powered analyses need conducted in inbred populations.
In our sample, the relationship between inbreeding and SZ risk is age-dependent; to our knowledge, this relationship has not been reported. In our previous study using a subset of the current study’s subjects, the mean age was balanced (Mansour et al., 2010). Nevertheless, we examined the mean age of individuals from the earlier study versus independent individuals from the current study, and found the subjects from the current study were significantly older (earlier study mean age=27 years, SD=7 and current study mean age=32 years, SD=9; t=(−8.1), df=442, p<0.0001). In all samples, age quartiles were examined in cases and controls separately. The controls have a consistent level of inbreeding across age quartiles; however, younger controls (≤25 years) exhibit the lowest mean F (Supplemental Table 1), which may reflect falling rates of consanguinity in Egypt in recent years, especially in urban areas (Shawky et al., 2011). Cases over age 40 have a mean inbreeding coefficient similar to controls, which was unexpected. The apparently low levels of inbreeding could reflect inadequate sample size, higher mortality in the children of consanguineous families, or it could be explained by increased mortality of consanguineous individuals diagnosed with SZ. Public health programs for psychiatric patients were instituted in Egypt only in the 1990s (Okasha, 2004), and these programs likely benefit younger cases more than older cases. Additionally, there was a hepatitis C viral epidemic in Egypt during the 1950s-1980s (Elgharably et al., 2017); conceivably, older, consanguineous individuals diagnosed with both SZ and hepatitis C could have even higher mortality.
Consanguinity is present in a major proportion of the world’s population. It is important to investigate whether the risk of polygenic diseases like SZ, conferred by inbreeding, could be attributed to inherited DNA variation. We therefore tested the feasibility of HBD analyses in a case-control sample from Egypt, reasoning family-based linkage analyses might not enable sufficient power to definitively identify risk-conferring loci. In our sample, inbreeding was determined using over one-million informative genetic markers; the estimate of inbreeding levels of controls is consistent with prior estimates of self-reported average consanguinity in the Nile Delta region (Mansour et al., 2010; Settin and Algelani, 1997). Among the associated risk loci, we find chromosome 1q23.3 overrepresented in the cases (Table 3). We also identified one consanguineous patient harboring a homozygous variant in ASPM (abnormal spindle homolog, microcephaly-associated). Variations in ASPM are a common cause of autosomal recessive primary microcephaly (Nicholas et al., 2009). ASPM exhibits significant allelic heterogeneity; there are 209 pathogenic variations in HGMD professional (2019.1). Among deleterious, exome variants, we found less heterozygous variants in the consanguineous subjects, as is expected for individuals who have a higher degree of consanguinity.
Some limitations of the study should be noted. Some ROH could be due to structural variations masquerading as homozygous genotypes. However, there was a 97% correlation between the SNP-based estimate of F and the ROH fraction (Supplemental Figure 1); thus, we do not expect this to be a substantial factor. Furthermore, in view of the relatively young age of the controls, not all individuals passed the age of risk for SZ (Ochoa et al., 2012). Still, not more than three controls are likely to be misclassified thus; moreover, misclassification would underestimate the observed associations with consanguinity. The current study utilized a subset of earlier samples (74 cases and 124 controls) along with newly recruited subjects, the present results do not represent a replication of our prior report; similarly, independent replication of the F x age interaction need to be sought.
In conclusion, we report consanguinity is an age-dependent risk factor for SZ in the Northern Delta region of Egypt. HBD analysis using larger samples may enable fine mapping for chromosomal regions contributing to the observed risk.
Supplementary Material
ACKNOWLEDGEMENTS
We thank participants and their families. This project used University of Pittsburgh HSCRF Genomics Research Core. The Genotype-Tissue Expression (GTEx) Project was supported by Common Fund of the Office of the Director of the National Institutes of Health, and NCI/NHGRI/NHLBI/NIDA/NIMH/NINDS. Data used for analyses described in this manuscript were obtained from: the GTEx Portal on 04-18-2019 (phs000424.v7.p2).
FUNDING
This work was supported by National Institutes of Health (grant number MH93246 and D43-TW009114 to VLN and grant number R37-MH057881 to BD) and Stanley Medical Research Institute (grant number 07R-1712 to VN).
Footnotes
CONFLICT OF INTEREST
The Authors have declared no conflicts of interest in relation to the subject of this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abaskuliev A, Skoblo G, 1975. Inbreeding, endogamy and exogamy among relatives of schizophrenia patients. Genetika 11(3), 145–148. [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR, 2010. A method and server for predicting damaging missense mutations. Nat Methods 7(4), 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya FS, 2010. Homozygosity mapping: One more tool in the clinical geneticist’s toolbox. Genet Med. [DOI] [PubMed] [Google Scholar]
- Berg JS, Adams M, Nassar N, Bizon C, Lee K, Schmitt CP, Wilhelmsen KC, Evans JP, 2013. An informatics approach to analyzing the incidentalome. Genet Med 15(1), 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles A, 2001. Consanguinity and its relevance to clinical genetics. Clin Genet 60(2), 89–98. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M, 2008. Natural selection on genes that underlie human disease susceptibility. Curr Biol 18(12), 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N, 2003. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 33 Suppl, 228–237. [DOI] [PubMed] [Google Scholar]
- Britvic D, Aleksic-Shihabi A, Titlic M, Dolic K, 2010. Schizophrenia spectrum psychosis in a Croatian Genetic Isolate: Genealogical Reconstructions. Psychiatr Danub 22(1), 51–56. [PubMed] [Google Scholar]
- Browning SR, Browning BL, 2010. High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet 86(4), 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulaeva KB, Pavlova TA, Kurbanov RM, Leal S, Bulaev OA, 2003. Genetic and epidemiological studies in Dagestan highland isolate]. Genetika. 39(3), 413–422. [PubMed] [Google Scholar]
- Bulayeva KB, Glatt SJ, Bulayev OA, Pavlova TA, Tsuang MT, 2007. Genome-wide linkage scan of schizophrenia: a cross-isolate study. Genomics 89(2), 167–177. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM, Consortium, R., Consortium, P.G., 3, G.C.f.A.N.o.t.W.T.C.C.C., 2015. An atlas of genetic correlations across human diseases and traits. Nat Genet 47(11), 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleby K, Tuma TA, 1987. Cousin marriages and schizophrenia in Saudi Arabia. British Journal of Psychiatry 150, 547–549. [DOI] [PubMed] [Google Scholar]
- Consortium, T.I.S., 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455(7210), 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Group GPA, 2011. The variant call format and VCFtools. Bioinformatics 27(15), 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C, 2016. Next-generation genotype imputation service and methods. Nat Genet 48(10), 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, Hofer E, Ibrahim-Verbaas CA, Kirin M, Lahti J, van der Lee SJ, Le Hellard S, Liu T, Marioni RE, Oldmeadow C, Postmus I, Smith AV, Smith JA, Thalamuthu A, Thomson R, Vitart V, Wang J, Yu L, Zgaga L, Zhao W, Boxall R, Harris SE, Hill WD, Liewald DC, Luciano M, Adams H, Ames D, Amin N, Amouyel P, Assareh AA, Au R, Becker JT, Beiser A, Berr C, Bertram L, Boerwinkle E, Buckley BM, Campbell H, Corley J, De Jager PL, Dufouil C, Eriksson JG, Espeseth T, Faul JD, Ford I, Gottesman RF, Griswold ME, Gudnason V, Harris TB, Heiss G, Hofman A, Holliday EG, Huffman J, Kardia SL, Kochan N, Knopman DS, Kwok JB, Lambert JC, Lee T, Li G, Li SC, Loitfelder M, Lopez OL, Lundervold AJ, Lundqvist A, Mather KA, Mirza SS, Nyberg L, Oostra BA, Palotie A, Papenberg G, Pattie A, Petrovic K, Polasek O, Psaty BM, Redmond P, Reppermund S, Rotter JI, Schmidt H, Schuur M, Schofield PW, Scott RJ, Steen VM, Stott DJ, van Swieten JC, Taylor KD, Trollor J, Trompet S, Uitterlinden AG, Weinstein G, Widen E, Windham BG, Jukema JW, Wright AF, Wright MJ, Yang Q, Amieva H, Attia JR, Bennett DA, Brodaty H, de Craen AJ, Hayward C, Ikram MA, Lindenberger U, Nilsson LG, Porteous DJ, Raikkonen K, Reinvang I, Rudan I, Sachdev PS, Schmidt R, Schofield PR, Srikanth V, Starr JM, Turner ST, Weir DR, Wilson JF, van Duijn C, Launer L, Fitzpatrick AL, Seshadri S, Mosley TH Jr., Deary IJ, 2015. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol Psychiatry 20(2), 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, Hagenaars SP, Ritchie SJ, Marioni RE, Fawns-Ritchie C, Liewald DCM, Okely JA, Ahola-Olli AV, Barnes CLK, Bertram L, Bis JC, Burdick KE, Christoforou A, DeRosse P, Djurovic S, Espeseth T, Giakoumaki S, Giddaluru S, Gustavson DE, Hayward C, Hofer E, Ikram MA, Karlsson R, Knowles E, Lahti J, Leber M, Li S, Mather KA, Melle I, Morris D, Oldmeadow C, Palviainen T, Payton A, Pazoki R, Petrovic K, Reynolds CA, Sargurupremraj M, Scholz M, Smith JA, Smith AV, Terzikhan N, Thalamuthu A, Trompet S, van der Lee SJ, Ware EB, Windham BG, Wright MJ, Yang J, Yu J, Ames D, Amin N, Amouyel P, Andreassen OA, Armstrong NJ, Assareh AA, Attia JR, Attix D, Avramopoulos D, Bennett DA, Bohmer AC, Boyle PA, Brodaty H, Campbell H, Cannon TD, Cirulli ET, Congdon E, Conley ED, Corley J, Cox SR, Dale AM, Dehghan A, Dick D, Dickinson D, Eriksson JG, Evangelou E, Faul JD, Ford I, Freimer NA, Gao H, Giegling I, Gillespie NA, Gordon SD, Gottesman RF, Griswold ME, Gudnason V, Harris TB, Hartmann AM, Hatzimanolis A, Heiss G, Holliday EG, Joshi PK, Kahonen M, Kardia SLR, Karlsson I, Kleineidam L, Knopman DS, Kochan NA, Konte B, Kwok JB, Le Hellard S, Lee T, Lehtimaki T, Li SC, Lill CM, Liu T, Koini M, London E, Longstreth WT Jr., Lopez OL, Loukola A, Luck T, Lundervold AJ, Lundquist A, Lyytikainen LP, Martin NG, Montgomery GW, Murray AD, Need AC, Noordam R, Nyberg L, Ollier W, Papenberg G, Pattie A, Polasek O, Poldrack RA, Psaty BM, Reppermund S, Riedel-Heller SG, Rose RJ, Rotter JI, Roussos P, Rovio SP, Saba Y, Sabb FW, Sachdev PS, Satizabal CL, Schmid M, Scott RJ, Scult MA, Simino J, Slagboom PE, Smyrnis N, Soumare A, Stefanis NC, Stott DJ, Straub RE, Sundet K, Taylor AM, Taylor KD, Tzoulaki I, Tzourio C, Uitterlinden A, Vitart V, Voineskos AN, Kaprio J, Wagner M, Wagner H, Weinhold L, Wen KH, Widen E, Yang Q, Zhao W, Adams HHH, Arking DE, Bilder RM, Bitsios P, Boerwinkle E, Chiba-Falek O, Corvin A, De Jager PL, Debette S, Donohoe G, Elliott P, Fitzpatrick AL, Gill M, Glahn DC, Hagg S, Hansell NK, Hariri AR, Ikram MK, Jukema JW, Vuoksimaa E, Keller MC, Kremen WS, Launer L, Lindenberger U, Palotie A, Pedersen NL, Pendleton N, Porteous DJ, Raikkonen K, Raitakari OT, Ramirez A, Reinvang I, Rudan I, Dan R, Schmidt R, Schmidt H, Schofield PW, Schofield PR, Starr JM, Steen VM, Trollor JN, Turner ST, Van Duijn CM, Villringer A, Weinberger DR, Weir DR, Wilson JF, Malhotra A, McIntosh AM, Gale CR, Seshadri S, Mosley TH Jr., Bressler J, Lencz T, Deary IJ, 2018. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun 9(1), 2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43(5), 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrusin M, Weitzman D, Levine J, Kremer I, Rietschel M, Maier W, Belmaker RH, 2008. The rate of consanguineous marriages among parents of schizophrenic patients in the Arab Bedouin population in Southern Israel. World J Biol Psychiatry, 1–3. [DOI] [PubMed] [Google Scholar]
- Elgharably A, Gomaa AI, Crossey MM, Norsworthy PJ, Waked I, Taylor-Robinson SD, 2017. Hepatitis C in Egypt - past, present, and future. Int J Gen Med 10, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald H, Kruse TA, Mors O, 2003. Genome wide scan using homozygosity mapping and linkage analyses of a single pedigree with affective disorder suggests oligogenic inheritance. Am J Med Genet 120B(1), 63–71. [DOI] [PubMed] [Google Scholar]
- Falconer DS, 1981. Introduction to quantitative genetics, 2nd ed. Longmans., London, New York. [Google Scholar]
- Gamsiz ED, Viscidi EW, Frederick AM, Nagpal S, Sanders SJ, Murtha MT, Schmidt M, Triche EW, Geschwind DH, State MW, Istrail S, Cook EH, Devlin B, Morrow EM, Consortium, S.S.C.G., 2013. Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am J Hum Genet 93(1), 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindilis VM, Gainullin RG, Shmaonova LM, 1989. [Genetico-demographic patterns of the prevalence of various forms of endogenous psychoses]. Genetika 25(4), 734–743. [PubMed] [Google Scholar]
- Goes FS, McGrath J, Avramopoulos D, Wolyniec P, Pirooznia M, Ruczinski I, Nestadt G, Kenny EE, Vacic V, Peters I, Lencz T, Darvasi A, Mulle JG, Warren ST, Pulver AE, 2015. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet 168(8), 649–659. [DOI] [PubMed] [Google Scholar]
- Gottesman I, 1991. Schizophrenia Genesis: The Origins of Madness. WH Freeman, New York. [Google Scholar]
- Gottesman II, Shields J, 1973. Genetic theorizing and schizophrenia. Br J Psychiatry 122(566), 15–30. [DOI] [PubMed] [Google Scholar]
- Gusev A, Lowe JK, Stoffel M, Daly MJ, Altshuler D, Breslow JL, Friedman JM, Pe’er I, 2009. Whole population, genome-wide mapping of hidden relatedness. Genome Res 19(2), 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WD, Weiss A, Liewald DC, Davies G, Porteous DJ, Hayward C, McIntosh AM, Gale CR, Deary IJ, 2019. Genetic contributions to two special factors of neuroticism are associated with affluence, higher intelligence, better health, and longer life. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Takahashi A, Kamatani Y, Momozawa Y, Saito T, Kondo K, Shimasaki A, Kawase K, Sakusabe T, Iwayama Y, Toyota T, Wakuda T, Kikuchi M, Kanahara N, Yamamori H, Yasuda Y, Watanabe Y, Hoya S, Aleksic B, Kushima I, Arai H, Takaki M, Hattori K, Kunugi H, Okahisa Y, Ohnuma T, Ozaki N, Someya T, Hashimoto R, Yoshikawa T, Kubo M, Iwata N, 2018. Genome-Wide Association Study Detected Novel Susceptibility Genes for Schizophrenia and Shared Trans-Populations/Diseases Genetic Effect. Schizophr Bull. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Bjelland DW, Howrigan DP, Abdellaoui A, Breen G, Borglum A, Cichon S, Degenhardt F, Forstner AJ, Frank J, Genovese G, Heilmann-Heimbach S, Herms S, Hoffman P, Maier W, Mattheisen M, Morris D, Mowry B, Müller-Mhysok B, Neale B, Nenadic I, Nöthen MM, O’Dushlaine C, Rietschel M, Ruderfer DM, Rujescu D, Schulze TG, Simonson MA, Stahl E, Strohmaier J, Witt SH, Sullivan PF, Keller MC, Consortium, S.W.G.o.t.P.G., 2016. No Reliable Association between Runs of Homozygosity and Schizophrenia in a Well-Powered Replication Study. PLoS Genet 12(10), e1006343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS, Tuke MA, Yaghootkar H, Sharp SA, Jie Y, Thompson WD, Harrison JW, Dawes A, Byrne EM, Tiemeier H, Allebrandt KV, Bowden J, Ray DW, Freathy RM, Murray A, Mazzotti DR, Gehrman PR, Lawlor DA, Frayling TM, Rutter MK, Hinds DA, Saxena R, Weedon MN, 2019. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun 10(1), 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K, Francioli L, Tiao G, Cummings B, Alfoldi J, Wang Q, Collins R, et al. , 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of- function intolerance across human protein-coding genes, bioRxiv [Google Scholar]
- Keller MC, Simonson MA, Ripke S, Neale BM, Gejman PV, Howrigan DP, Lee SH, Lencz T, Levinson DF, Sullivan PF, Consortium, S.P.G.-W.A.S., 2012. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet 8(4), e1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Trampush JW, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, Christoforou A, Reinvang I, DeRosse P, Lundervold AJ, Steen VM, Espeseth T, Raikkonen K, Widen E, Palotie A, Eriksson JG, Giegling I, Konte B, Roussos P, Giakoumaki S, Burdick KE, Payton A, Ollier W, Chiba-Falek O, Attix DK, Need AC, Cirulli ET, Voineskos AN, Stefanis NC, Avramopoulos D, Hatzimanolis A, Arking DE, Smyrnis N, Bilder RM, Freimer NA, Cannon TD, London E, Poldrack RA, Sabb FW, Congdon E, Conley ED, Scult MA, Dickinson D, Straub RE, Donohoe G, Morris D, Corvin A, Gill M, Hariri AR, Weinberger DR, Pendleton N, Bitsios P, Rujescu D, Lahti J, Le Hellard S, Keller MC, Andreassen OA, Deary IJ, Glahn DC, Malhotra AK, Lencz T, 2017. Large-Scale Cognitive GWAS Meta-Analysis Reveals Tissue-Specific Neural Expression and Potential Nootropic Drug Targets. Cell Rep 21(9), 2597–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linner R, Fontana MA, Kundu T, Lee C, Li H, Li R, Royer R, Timshel PN, Walters RK, Willoughby EA, Yengo L, Alver M, Bao Y, Clark DW, Day FR, Furlotte NA, Joshi PK, Kemper KE, Kleinman A, Langenberg C, Magi R, Trampush JW, Verma SS, Wu Y, Lam M, Zhao JH, Zheng Z, Boardman JD, Campbell H, Freese J, Harris KM, Hayward C, Herd P, Kumari M, Lencz T, Luan J, Malhotra AK, Metspalu A, Milani L, Ong KK, Perry JRB, Porteous DJ, Ritchie MD, Smart MC, Smith BH, Tung JY, Wareham NJ, Wilson JF, Beauchamp JP, Conley DC, Esko T, Lehrer SF, Magnusson PKE, Oskarsson S, Pers TH, Robinson MR, Thom K, Watson C, Chabris CF, Meyer MN, Laibson DI, Yang J, Johannesson M, Koellinger PD, Turley P, Visscher PM, Benjamin DJ, Cesarini D, 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50(8), 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK, 2007. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A 104(50), 19942–19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerer B, Segman RH, Hamdan A, Kanyas K, Karni O, Kohn Y, Korner M, Lanktree M, Kaadan M, Turetsky N, Yakir A, Kerem B, Macciardi F, 2003. Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol Psychiatry 8(5), 488–498. [DOI] [PubMed] [Google Scholar]
- Lezirovitz K, Pardono E, de Mello Auricchio MT, de Carvalho ESFL, Lopes JJ, Abreu-Silva RS, Romanos J, Batissoco AC, Mingroni-Netto RC, 2008. Unexpected genetic heterogeneity in a large consanguineous Brazilian pedigree presenting deafness. Eur J Hum Genet 16(1), 89–96. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R, 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5), 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, Yi Q, Li C, Li X, Shen J, Song Z, Ji W, Wang M, Zhou J, Chen B, Liu Y, Wang J, Wang P, Yang P, Wang Q, Feng G, Liu B, Sun W, Li B, He G, Li W, Wan C, Xu Q, Wen Z, Liu K, Huang F, Ji J, Ripke S, Yue W, Sullivan PF, O’Donovan MC, Shi Y, 2017. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet 49(11), 1576–1583. [DOI] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK, 2005. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med 353(12), 1209–1223. [DOI] [PubMed] [Google Scholar]
- Maguire A, Tseliou F, O’Reilly D, 2018. Consanguineous Marriage and the Psychopathology of Progeny: A Population-wide Data Linkage Study. JAMA Psychiatry 75(5), 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Meraji SM, Houshyar R, Radhakrishnan J, Mani A, Ahangar M, Rezaie TM, Taghavinejad MA, Broumand B, Zhao H, Nelson-Williams C, Lifton RP, 2002. Finding genetic contributions to sporadic disease: a recessive locus at 12q24 commonly contributes to patent ductus arteriosus. Proc Natl Acad Sci U S A 99(23), 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour H, Fathi W, Klei L, Wood J, Chowdari K, Watson A, Eissa A, Elassy M, Ali I, Salah H, Yassin A, Tobar S, El-Boraie H, Gaafar H, Ibrahim NE, Kandil K, El-Bahaei W, El-Boraie O, Alatrouny M, El-Chennawi F, Devlin B, Nimgaonkar VL, 2010. Consanguinity and increased risk for schizophrenia in Egypt. Schizophr Res 120(1–3), 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour H, Klei L, Wood J, Talkowski M, Chowdari K, Fathi W, Eissa A, Yassin A, Salah H, Tobar S, El-Boraie H, Gaafar H, Elassy M, Ibrahim NE, El-Bahaei W, Elsayed M, Shahda M, El Sheshtawy E, El-Boraie O, El-Chennawi F, Devlin B, Nimgaonkar VL, 2009. Consanguinity associated with increased risk for bipolar I disorder in Egypt. Am J Med Genet B Neuropsychiatr Genet 150B(6), 879–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Gottesman II, Rao DC, 1983. The transmission of schizophrenia under a multifactorial threshold model. American Journal of Human Genetics 35(6), 1161–1178. [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F, 2016. The Ensembl Variant Effect Predictor. Genome Biol 17(1), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem NM, Hamdan S, Klei L, Wood S, Zelazny J, Frisch A, Weizman A, Carmel M, Michaelovsky E, Farbstein I, Wasserman D, El-Heib M, Ferrell R, Apter A, Devlin B, Brent D, 2017. Runs of homozygosity, copy number variation, and risk for depression and suicidal behavior in an Arab Bedouin kindred. Psychiatr Genet 27(5), 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH, 2010. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86(5), 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA, 2009. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet 85(6), 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, Al-Saad S, Ware J, Joseph RM, Greenblatt R, Gleason D, Ertelt JA, Apse KA, Bodell A, Partlow JN, Barry B, Yao H, Markianos K, Ferland RJ, Greenberg ME, Walsh CA, 2008. Identifying autism loci and genes by tracing recent shared ancestry. Science 321(5886), 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Guha S, Ikeda M, Iwata N, Malhotra AK, Pe’er I, Darvasi A, Lencz T, 2014. Excess of homozygosity in the major histocompatibility complex in schizophrenia. Hum Mol Genet 23(22), 6088–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Püttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, Darvish H, Wrogemann K, Hadavi V, Lipkowitz B, Esmaeeli-Nieh S, Wieczorek D, Kariminejad R, Firouzabadi SG, Cohen M, Fattahi Z, Rost I, Mojahedi F, Hertzberg C, Dehghan A, Rajab A, Banavandi MJ, Hoffer J, Falah M, Musante L, Kalscheuer V, Ullmann R, Kuss AW, Tzschach A, Kahrizi K, Ropers HH, 2011. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478(7367), 57–63. [DOI] [PubMed] [Google Scholar]
- Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K, Hampshire D, Ahmed M, Bond J, Di Benedetto D, Fichera M, Romano C, Dobyns WB, Woods CG, 2009. The molecular landscape of ASPM mutations in primary microcephaly. J Med Genet 46(4), 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa S, Usall J, Cobo J, Labad X, Kulkarni J, 2012. Gender differences in schizophrenia and first-episode psychosis: a comprehensive literature review. Schizophr Res Treatment 2012, 916198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okasha A, 2004. Focus on psychiatry in Egypt. Br J Psychiatry. 185, 266–272. [DOI] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Byrne EM, Dannlowski U, Eley TC, Hayward C, Martin NG, McIntosh AM, Plomin R, Porteous DJ, Wray NR, Caballero A, Geschwind DH, Huckins LM, Ruderfer DM, Santiago E, Sklar P, Stahl EA, Won H, Agerbo E, Als TD, Andreassen OA, Bækvad-Hansen M, Mortensen PB, Pedersen CB, Børglum AD, Bybjerg-Grauholm J, Djurovic S, Durmishi N, Pedersen MG, Golimbet V, Grove J, Hougaard DM, Mattheisen M, Molden E, Mors O, Nordentoft M, Pejovic-Milovancevic M, Sigurdsson E, Silagadze T, Hansen CS, Stefansson K, Stefansson H, Steinberg S, Tosato S, Werge T, Collier DA, Rujescu D, Kirov G, Owen MJ, O’Donovan MC, Walters JTR, Consortium:, G., Consortium:, C., Consortium, G., Consortium, C., 2018. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50(3), 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudan I, Rudan D, Campbell H, Carothers A, Wright A, Smolej-Narancic N, Janicijevic B, Jin L, Chakraborty R, Deka R, Rudan P, 2003. Inbreeding and risk of late onset complex disease. J Med Genet 40(12), 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Kosmicki JA, Karczewski KJ, O’Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, Neale BM, Daly MJ, 2017. Regional missense constraint improves variant deleteriousness prediction. bioRxiv, 148353. [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C., 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510), 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, Lee K, Oostrik J, Huygen PL, Ali G, Hoefsloot LH, Veltman JA, Cremers FP, Basit S, Ansar M, Cremers CW, Kunst HP, Ahmad W, Admiraal RJ, Leal SM, Kremer H, 2010. Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet 86(2), 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE, 2009. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet 25(12), 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settin A, Algelani A, 1997. Carrier and gene frequency of β-thalassemia depicted from consanguinity data of family cases and controls. Egypt Journal of Pediatrics 14, 129–136. [Google Scholar]
- Shawky R, El-Awady M, Elsayed S, Hamadan G, 2011. Consanguineous matings among Egyptian population. Egyptian Journal of Medical Human Genetics 12(2), 157–163. [Google Scholar]
- So HC, Gui AH, Cherny SS, Sham PC, 2011. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 35(5), 310–317. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, de Leeuw CA, Steinberg S, Pavlides JMW, Trzaskowski M, Byrne EM, Pers TH, Holmans PA, Richards AL, Abbott L, Agerbo E, Akil H, Albani D, Alliey-Rodriguez N, Als TD, Anjorin A, Antilla V, Awasthi S, Badner JA, Baekvad-Hansen M, Barchas JD, Bass N, Bauer M, Belliveau R, Bergen SE, Pedersen CB, Boen E, Boks MP, Boocock J, Budde M, Bunney W, Burmeister M, Bybjerg-Grauholm J, Byerley W, Casas M, Cerrato F, Cervantes P, Chambert K, Charney AW, Chen D, Churchhouse C, Clarke TK, Coryell W, Craig DW, Cruceanu C, Curtis D, Czerski PM, Dale AM, de Jong S, Degenhardt F, Del-Favero J, DePaulo JR, Djurovic S, Dobbyn AL, Dumont A, Elvsashagen T, Escott-Price V, Fan CC, Fischer SB, Flickinger M, Foroud TM, Forty L, Frank J, Fraser C, Freimer NB, Frisen L, Gade K, Gage D, Garnham J, Giambartolomei C, Pedersen MG, Goldstein J, Gordon SD, Gordon-Smith K, Green EK, Green MJ, Greenwood TA, Grove J, Guan W, Guzman-Parra J, Hamshere ML, Hautzinger M, Heilbronner U, Herms S, Hipolito M, Hoffmann P, Holland D, Huckins L, Jamain S, Johnson JS, Jureus A, Kandaswamy R, Karlsson R, Kennedy JL, Kittel-Schneider S, Knowles JA, Kogevinas M, Koller AC, Kupka R, Lavebratt C, Lawrence J, Lawson WB, Leber M, Lee PH, Levy SE, Li JZ, Liu C, Lucae S, Maaser A, MacIntyre DJ, Mahon PB, Maier W, Martinsson L, McCarroll S, McGuffin P, McInnis MG, McKay JD, Medeiros H, Medland SE, Meng F, Milani L, Montgomery GW, Morris DW, Muhleisen TW, Mullins N, Nguyen H, Nievergelt CM, Adolfsson AN, Nwulia EA, O’Donovan C, Loohuis LMO, Ori APS, Oruc L, Osby U, Perlis RH, Perry A, Pfennig A, Potash JB, Purcell SM, Regeer EJ, Reif A, Reinbold CS, Rice JP, Rivas F, Rivera M, Roussos P, Ruderfer DM, Ryu E, Sanchez-Mora C, Schatzberg AF, Scheftner WA, Schork NJ, Shannon Weickert C, Shehktman T, Shilling PD, Sigurdsson E, Slaney C, Smeland OB, Sobell JL, Soholm Hansen C, Spijker AT, St Clair D, Steffens M, Strauss JS, Streit F, Strohmaier J, Szelinger S, Thompson RC, Thorgeirsson TE, Treutlein J, Vedder H, Wang W, Watson SJ, Weickert TW, Witt SH, Xi S, Xu W, Young AH, Zandi P, Zhang P, Zollner S, Adolfsson R, Agartz I, Alda M, Backlund L, Baune BT, Bellivier F, Berrettini WH, Biernacka JM, Blackwood DHR, Boehnke M, Borglum AD, Corvin A, Craddock N, Daly MJ, Dannlowski U, Esko T, Etain B, Frye M, Fullerton JM, Gershon ES, Gill M, Goes F, Grigoroiu-Serbanescu M, Hauser J, Hougaard DM, Hultman CM, Jones I, Jones LA, Kahn RS, Kirov G, Landen M, Leboyer M, Lewis CM, Li QS, Lissowska J, Martin NG, Mayoral F, McElroy SL, McIntosh AM, McMahon FJ, Melle I, Metspalu A, Mitchell PB, Morken G, Mors O, Mortensen PB, Muller-Myhsok B, Myers RM, Neale BM, Nimgaonkar V, Nordentoft M, Nothen MM, O’Donovan MC, Oedegaard KJ, Owen MJ, Paciga SA, Pato C, Pato MT, Posthuma D, Ramos-Quiroga JA, Ribases M, Rietschel M, Rouleau GA, Schalling M, Schofield PR, Schulze TG, Serretti A, Smoller JW, Stefansson H, Stefansson K, Stordal E, Sullivan PF, Turecki G, Vaaler AE, Vieta E, Vincent JB, Werge T, Nurnberger JI, Wray NR, Di Florio A, Edenberg HJ, Cichon S, Ophoff RA, Scott LJ, Andreassen OA, Kelsoe J, Sklar P, 2019. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51(5), 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroup TS, Lieberman JA, McEvoy JP, Swartz MS, Davis SM, Rosenheck RA, Perkins DO, Keefe RS, Davis CE, Severe J, Hsiao JK, 2006. Effectiveness of olanzapine, quetiapine, risperidone, and ziprasidone in patients with chronic schizophrenia following discontinuation of a previous atypical antipsychotic. Am J Psychiatry 163(4), 611–622. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA, 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11.10.11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LS, Hranilovic D, Wang K, Lindquist IE, Yurcaba L, Petkovic ZB, Gidaya N, Jernej B, Hakonarson H, Bucan M, 2010. Population-based study of genetic variation in individuals with autism spectrum disorders from Croatia. BMC Med Genet 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing J, Sarotorius N, et al. , 2001. A reference manual for SCAN. Cambridge University Press. [Google Scholar]
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Pourcain BS, Warrington NM, Finucane HK, Price AL, Bulik-Sullivan BK, Anttila V, Paternoster L, Gaunt TR, Evans DM, Neale BM, Consortium, E.G.a.L.E.E.E., 2017. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33(2), 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.