Abstract
Type I interferon (IFN) is a primary pathogenic factor in systemic lupus erythematosus (SLE). Gain-of-function genetic variants in the type I IFN pathway have been associated with risk of disease. Common polygenic as well as rare monogenic influences on type I IFN have been demonstrated, supporting a complex genetic basis for high IFN in many SLE patients. Both SLE-associated autoantibodies and high type I IFN can be observed in the pre-disease state. Patients with SLE and evidence of high type I IFN have more active disease and a greater propensity to nephritis and other severe manifestations. Despite the well-established association between type I IFN and SLE, the specific triggers of type I IFN production, the mechanisms by which IFNs help perpetuate the cycle of autoreactive cells and autoantibody production are not completely clear. This review provides an updated overview of type I IFN in SLE pathogenesis, clinical manifestations, and current therapeutic strategies targeting this pathway.
Keywords: Type I Interferon, genetics, pathogenesis, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is a chronic, multisystem, autoimmune disease resulting from defects in both the innate and adaptive immune systems [1, 2]. Both genetic and environmental factors are important determinants of the different phenotypes observed in SLE [3]. Type I interferon (IFN) levels are chronically and persistently elevated in blood in approximately 50% of SLE patients [4]. An even greater percentage of patients demonstrate over-expression of type I IFN pathway genes in their peripheral blood cells [5–7], referred to as the “IFN signature”. Type I and Type II IFN can induce many of the same genes, which may explain why more patients demonstrate the signature than have high functional circulating type I IFN in their blood [5]. In addition, other pathways and downstream effectors can induce IFN signature genes [8–10]. Interestingly, a high type I IFN activity is associated with the presence of other cytokines, such as B-cell activating factor (BAFF) and type II IFN [11, 12], specific autoantibodies [13], and certain clinical manifestations, such as lupus nephritis [12].
Despite the well-established association between type I IFN and SLE, the specific triggers of type I IFN production, the mechanisms by which IFNs help perpetuate the cycle of autoreactive cells and autoantibody production, and the clinical relevance of targeting type I IFN in SLE are less clear. In this review, we will provide an updated overview of recent evidence on mechanisms of type I IFN production, genetic associations, the relevance of impaired nucleic acid processing in animal models of lupus and human SLE, as well as the emergence of successful therapeutic agents targeting type I IFN pathways.
Mechanisms of Type I IFN production
Plasmacytoid dendritic cells (pDCs) have been a focus of interest in SLE ([14], reviewed in [15]). Each pDC can produce as many as 109 IFN-α molecules in 12 hours [16]. This striking feature, along with the skewing of blood type I IFNs toward IFN-α over IFN-β in SLE [17], suggest pDCs as the potential cellular IFN source in this disease. Accordingly, pDC deficiency has been shown to ameliorate murine lupus models [18, 19]. However, isolating IFN-α-producing pDCs from SLE patients’ blood and tissue has remained challenging [20, 21]. Although other cell types, including macrophages and fibroblasts, are also a source of type I IFN, these cells primarily synthesize IFN-β [22].
Type I IFN production is mainly triggered by the activation of nucleic acid-binding pattern recognition receptors, including the endosomal toll-like receptors (TLR) 3 4, 7 and 9, the cytosolic sensor cyclic GMP-AMP synthase (cGAS), and the RNA-sensor RIG-I–like receptors (RLRs)-MAVS [23]. Under normal conditions, these nucleic-acid sensing pathways are subject to strict regulation and are required to form an appropriate antiviral response [24], but many SLE patients demonstrate chronic overactivity in these pathways. A recent study suggests this chronic type I IFN pathway activation may actually blunt the response to additional endosomal TLR stimulation, while responses to LPS in patients with chronic type I IFN elevation were augmented [20].
The role of TLR7 in SLE is well established, as its overexpression is associated with severe lupus in mice, and TLR7 inhibition is protective (reviewed in [25]). Conversely, the effect of TLR9 in SLE has remained controversial and a net tolerogenic role has been suggested [26, 27]. In agreement with these observations, recent evidence has shown that the TLR-trafficking chaperone UNC93B1 protects from TLR7-driven autoimmunity in mice, without affecting TLR9 [28]. UNC93B1 limits TLR7 signaling in response to self RNA via binding to the adaptor protein Syntenin-1 and sorting TLR7 into intraluminal vesicles [28]. Similarly, a model of persistent lymphocytic choriomeningitis virus infections in lupus-prone mice has been shown to enhance systemic autoimmunity in a pDC- and endosomal TLR-dependent manner [29]; interestingly, UNC93B13d/3d mutant NZB mice were used to corroborate the requirement for TLR-dependent responses, indirectly suggesting a key role of TLR7 in this model [28, 29].
Mechanisms that impact nucleic acid metabolism are also thought to influence the threshold of activation and triggering of type I IFN production. This is well illustrated in the interferonopathies, a set of monogenic disorders involving genes related to nucleic acid handling (see section on Genetic Influences on Type I IFN in SLE). In addition to the innate nucleic-acid sensors, self-DNA can be recognized by auto-reactive B-cells which may amplify autoreactivity. DNases, particularly the extracellular DNASE1L3, are critical to preserve self-tolerance and development of SLE (reviewed in [30]). Homozygous deletions of DNASE1L3 and coding polymorphisms in humans are associated with SLE [30]. Recently, a Dnase1l3−/− murine model of lupus illustrated the facilitating effect of type I IFN signaling and pDCs to maintaining and amplifying self-reactivity in a TLR-dependent manner [31].
Another example of abnormal handling of nucleic acids was demonstrated by the finding that loss of sumoylation, conjugation of small ubiquitin-like modifiers (SUMOs) to lysines, by SUMO2 and SUMO3 leads to a potent type I IFN response. This is independent of the classic IFN-inducing sensors and transcription factors [32, 33].
The cytosolic nucleic acid sensors are also thought to play critical roles in SLE (reviewed in [24, 33]). Activation of the cGAS-STING pathway is crucial for antiviral immunity and plays an important role in autoimmunity [33]. In this sense, UVB light exposure has been shown to increase type I IFN response both locally and systemically in a c-GAS-dependent manner in mice and humans [34]. Recently, a STING-independent DNA sensing pathway was identified in human cells but not in laboratory mice [35]. The sensor of the STING-independent DNA sensing pathway corresponds to the damage response protein DNA-PK, with the heat shock protein HSPA8 is a downstream target [35]. These data support the idea that cGAS-STING independent pathways could be important in SLE, as does human genetic data which implicates both MDA5 [10] and MAVS [36].
Type I IFNs in the Initiation of SLE
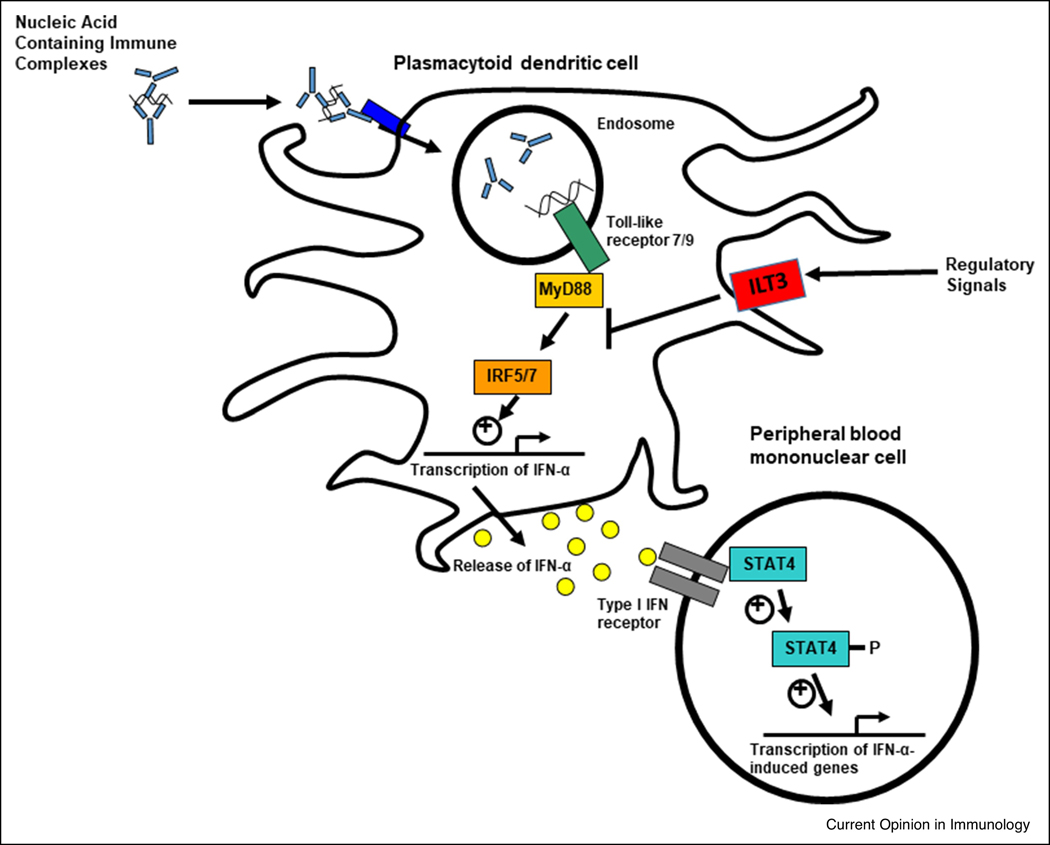
Type I IFNs are implicated in SLE susceptibility by multiple lines of investigation, including genetics, family studies, and induction of SLE by type I IFN treatment [37]. Chronic elevation of type I IFN predisposing to SLE is thought to be due to overproduction, increased sensitivity, and impaired negative regulation (Figure 1). For example, genetic polymorphisms in the interferon regulatory factor (IRF)5 and IRF7 genes are associated with increased type I IFN in circulation [38, 39]. Genes downstream of the type I IFN receptor (IFNAR) have also been implicated in SLE, including the Signal Transducer and Activator Of Transcription 4 (STAT4) [40]. In human studies, SLE patients carrying the STAT4 risk allele have greater type I IFN-induced gene expression for a given amount of IFN compared to patients with the non-risk allele, suggesting that this allele confers increased sensitivity to type I IFN [8]. Supporting this idea, a recent study showed that rs7574865, the most significantly associated SNP in STAT4, is associated with an augmented T-cell response to IFN-α in patients with SLE [41]. Lastly, deficiency in negative regulators of type I IFN has also been suggested by polymorphisms in the suppressive inhibitory molecules immunoglobulin-like transcript 3 (ILT3) receptor expressed on dendritic cells (DCs). Gene variants that reduce expression of the ILT3 receptor are associated with increased circulating type I IFN in patients with SLE [42].
Figure 1.
Schematic diagram of various ways the type I IFN pathway is modulated in SLE. Nucleic acid containing immune complexes formed by SLE-associated autoantibodies are taken up by plasmacytoid dendritic cells. Genetic variants in IRF5 and IRF7 can result in greater type I IFN production, particularly in those patients with autoantibodies. Type I IFN then signals through the IFNAR receptor, and polymorphisms such as the SLE-associated STAT4 allele result in augmented signaling downstream of the receptor. Polymorphisms in regulatory molecules, such as ILT3, can result in defective function and decreased ability to negatively regulate inflammatory pathways. In many patients, this multifactorial process leads to persistent dysregulation of the IFN pathway at multiple locations.
Endogenous stimuli, such as immune complexes involving SLE-associated autoantibodies, act upon a susceptible genetic background to result in type I IFN production. Neutrophils may also provide endogenous type I IFN pathway stimulation via neutrophil extracellular traps (NETs), which can increase IFN-α production by pDCs in a toll-like receptor TLR9-dependent manner [43]. Endogenous retro-elements such as LINE-1 could also activate the type I IFN system in SLE [44] as an additional nucleic acid stimulus.
It seems likely that a feed-forward loop occurs in which type I IFN may participate in the initial breaks in tolerance [17]. Both type I IFN and SLE-associated autoantibodies are elevated in the pre-disease state [45], supporting their role in disease susceptibility and initiation. In the years before diagnosis of SLE, autoantibody specificities increase in number up to the point of diagnosis, suggesting a diversification of the anti-self response in the pre-disease state [46]. These autoantibodies can form nucleic acid immune complexes that stimulate type I IFN production (Figure 1). Type I IFN increases in the pre-disease state, most dramatically beginning approximately 2 years prior to the diagnosis of SLE [45]. The pre-disease studies indicate that many of the cardinal features of the dysregulated immune response in SLE begin years before patients develop clinical manifestations and present for medical care.
Common and Rare Genetic influences on Type I IFN in SLE
SLE has been associated with over 100 genetic risk loci and many of these risk genes encode proteins with functions linked to type I IFN response [47]. Family members of patients with SLE are at higher risk of developing autoimmune diseases [48, 49]. Type I IFN levels are heritable within SLE families [17], suggesting that genetic overactivity in this pathway predisposes to SLE and other IFN-related autoimmune conditions [49]. Polymorphisms in the type I IFN gene locus itself have not been associated with either SLE or higher type I IFN levels, but a diversity of type I IFN pathway and pattern recognition pathway genes pathways have been linked to SLE susceptibility [50]. These SLE-risk polymorphisms fit with an expected gain-of-function model, as they are associated with greater type I IFN pathway activation. In addition to these known SLE susceptibility genes, genome screens have been performed with the goal of identifying new loci associated with elevated type I IFN in SLE patients [51–54]. A genome-wide association study (GWAS) of patients with high versus low levels of type I IFN discovered a distinct panel of genes from those identified in case-control SLE GWAS [52]. One such gene, purine nucleotide phosphatase (PNP), encodes for an enzyme involved in the nucleotide salvage pathway. The polymorphism related to type I IFN in SLE is a coding-change polymorphism that reduces PNP enzymatic function and is also associated with decreased expression of the enzyme. These changes result in an increased propensity to S-phase block, and an increase in type I IFN-induced gene expression [9]. The exact mechanism by which the cell cycle abnormalities result in increased IFN-induced gene expression have not been fully elucidated.
Other rare and low frequency gene variants have also been recently shown to contribute to human SLE via increasing type I IFN production [55]. A recent study demonstrated that BLK regulates type I IFN downstream of TLR7/8 signaling possibly via regulation of BANK1, and variants in both genes can impair their normal function of inhibiting IRF5-mediated type I IFN production. CXorf21 is an IFN-inducible gene associated with SLE in a sex-specific manner, as it escapes X-chromosome inactivation [56]. The CXorf21 function was unknown until recently, when its link to IFN production via endolysosomal TLR signaling was demonstrated [57]. The CXorf21 gene product, TLR adaptor interacting with SLC15A4 on the lysosome (TASL), interacts specifically with SLC15A4 and is necessary to activate IRF5.
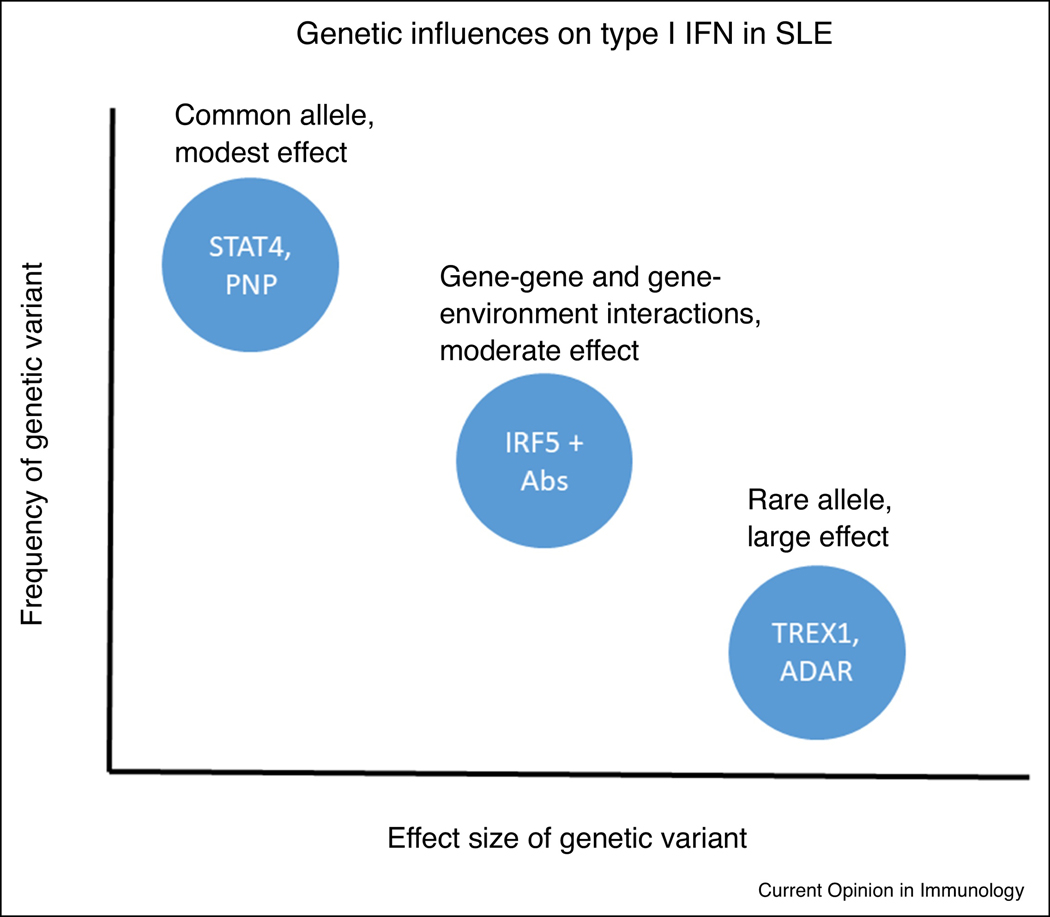
In addition to the above evidence for genetic influence on type I IFN in SLE, monogenic disorders, the so-called interferonopathies, also demonstrate high type I IFN and share some molecular and clinical features of SLE [58, 59]. The gene mutations that give rise to these syndromes include TREX1, SAMHD1, ADAR1, IFIH1, among others, and have a function in nucleic acid handling [58]. The clinical syndromes vary somewhat based upon the specific gene involved, but typical manifestations include central nervous inflammation and calcification, vasculitis, and lupus-like features [58]. Considering the rare single gene interferonopathies with the common alleles that impact type I IFN in SLE, model as outlined in Figure 2 emerges. This model is similar to that observed in complex disease susceptibility. Some polymorphisms are present at a high rate in the general population that individually do not result in a large impact on type I IFN levels. It is possible that these alleles are helpful in our normal immunity and defense against viruses. Some of these alleles will form combinations in gene-gene or gene-environment interactions. Despite these combinations being less common, they may exert a greater influence on IFN levels and consequently associate with increased risk of autoimmunity. An example of such gene-environment interaction in SLE is the IRF5 and IRF7 polymorphisms, which result in much greater type I IFN in patients with SLE-associated autoantibodies [39, 60]. These gene-microenvironment interactions support the idea that the immune complex is needed as the upstream stimulus to observe the full effect of the genotype upon type I IFN.
Figure 2.
Diagram of genetic influences on type I IFN in SLE patients. The X and Y axes represent relative values for effect size and frequency in the population respectively. Common polymorphisms individually exert a modest influence on type I IFN, but when paired with the appropriate gene-gene or gene-environment interaction, would exert a moderate effect on type I IFN. The need for additional factors that assort independently de facto decreases the frequency in the general population. Lastly, some extremely rare monogenic variations have been reported that have a large effect upon the type I IFN pathway and typically lead to the interferonopathy category of diseases, which share some features with SLE.
A recent multi-ancestral SLE case-control study using Immunochip genotype data suggested that in European ancestry genetic risk for SLE manifests as a nonlinear function of the cumulative risk allele load, with a greater effect of some of the alleles when the overall genetic load is high [61]. Younger onset SLE patients had a higher genetic load when considering non-HLA alleles [61], as might be expected. This supports the idea that genetic interactions play a role in SLE, and it is likely that these play out in pathways like the IFN pathway. A recent large family-based inheritance study found similar results, that the inheritance of SLE in childhood onset patients fit better to a more epistatic or interactive model than did adult onset SLE [48].
Epigenetic changes have been reported in SLE, and typically these have been hypomethylation at type I IFN-induced genes [62]. A recent study suggests that this finding is common across a number of other autoimmune diseases as well, including rheumatoid arthritis and Grave’s disease [63]. Whether these epigenetic changes are a cause or a result of type I IFN signaling is not clear, as many of the transcripts are those typical of a type I IFN signature. It is possible that an intrinsic epigenetic sensitivity to type I IFN exists in SLE as well, and this could relate to previous stimulation by IFNs, other cytokines, or other factors.
Clinical associations with high serum IFN levels in SLE patients
There is substantial evidence that type I IFNs are important in propagating ongoing disease activity in SLE, and not just an initial susceptibility factor. Previous studies have demonstrated that elevated type I IFN in blood is associated with increased disease activity in cross-sectional studies [5, 6, 64]. Despite this robust association, longitudinal studies generally do not support the idea that IFN levels fluctuate predictably with changes in SLE disease activity [65, 66]. These findings suggest that type I IFN levels demarcate groups of patients that are more or less likely on average to have higher disease activity and flares at any given time, but that type I IFN may not be highly informative as a longitudinal biomarker. This is supported by a recent study in which elevations of type I IFN in patients who are in remission was associated with a higher risk of relapse [67].
A recent study has shown that pregnancy is associated with downregulation of the type I IFN signature in both healthy and uncomplicated SLE pregnancies; these changes typically persist through late pregnancy and the post-partum period. In complicated SLE pregnancies, the IFN signature frequently remains elevated [68], suggesting that persistent activation of the type I IFN pathway in pregnancy may contribute to adverse pregnancy outcomes [68].
It is clear that ongoing chronic type I IFN stimulation shapes the clinical phenotype and overall disease activity in SLE patients. Increased type I IFN in circulation is associated with the presence of lupus nephritis, mucocutaneous manifestations, arthritis, and autoantibodies including anti-Ro, anti-Sm, anti-ribunucleoprotein (RNP), and anti-double-stranded (anti-dsDNA) antibodies [12, 69, 70]. High IFN signature has been associated with mucocutaneous disease activity, possibly mediated by UVB-induced keratinocyte apoptosis. A recent study showed that acute exposure to UVB light stimulates a robust local and systemic type I IFN response in humans and mice, in a cGAS-dependent manner [34].
As previously mentioned, neutrophils may mediate type I IFN production via release of NETs with interferogenic properties. In particular, the low-density granulocyte (LDGs) subset of neutrophils are elevated in SLE patients and have the greatest type I IFN signature among neutrophils, suggesting that LDGs are highly responsive to IFN [71]. Moreover, subsets with the LDG population may correlate with specific clinical features of lupus and atherosclerotic disease [71].
Non-α/β type I IFNs in SLE
Although much of the research on IFNs in SLE has focused on IFN-α and IFN-β, recent studies have shown a role for IFN-κ, another member of the type I IFN family. IFN-κ is mainly expressed in keratinocytes after exposure to UV light, hence, it has been suggested as a key mediator involved in photosensitivity and other cutaneous manifestations of SLE in humans and murine models [72, 73]. Additionally, the presence of genetic associations between IFNK gene variants and cutaneous phenotypes in SLE have been described [74]. Overall, these findings support a role for IFN-κ in the pathogenesis of cutaneous lupus mediating UV light toxicity. IFN-κ could be a potential therapeutic target to modulate type I IFN response in SLE, particularly in patients with predominantly cutaneous manifestations. A direct role of other type I IFNs in SLE is less clear.
Therapeutic strategies targeting type I IFN
Current standard of care treatment of SLE involves the use of corticosteroids and immunosuppressive agents that are associated with a wide range of potential adverse effects [75]. Type I IFN has been considered as a potential target to reduce chronic inflammation and end-organ damage in SLE [75]. Various therapeutics agents which target specific aspects of the type I interferon pathway are currently in different phases of clinical development [76]. For instance, IFN-α kinoid, a vaccine designed to induce neutralizing antibodies against IFN-α, was shown to reduce the IFN gene signature with a satisfactory safety profile in a recent phase IIb study [77]. Phase III studies are being planned. A phase I study evaluating a promising monoclonal antibody against a pDC-specific receptor that inhibits type I IFN production (blood DC antigen 2, BDCA2) was also recently published and further trials are ongoing [78]. Inhibitors of the Janus Kinases (JAK) have recently shown efficacy in cutaneous and articular manifestations of SLE. While these kinase inhibitors block multiple cytokine signals, it is likely that the efficacy of JAK inhibition in SLE relates at least in part to an impact on type I IFN signaling [79, 80].
Anifrolumab is a monoclonal antibody that blocks both IFN-α and IFN-β signal by binding to the type I IFN receptor. Interestingly, previous anti-IFN-α antibodies had not shown sufficient efficacy in phase II trials to be further developed [81, 82], while anifrolumab was taken forward to phase III trials. This could be partially due to the difference in mechanism of action, as the anti-IFN-α antibodies would not interrupt IFN-β signaling, in contrast to the complete type I IFN blockade achieved by anifrolumab. IFN-α predominates in the blood in SLE [17], but IFN-β could still play important roles in the tissue [83]. This year, results from the second of two phase III Anifrolumab clinical trials showed a statistically significant reduction in disease activity and corticosteroid use at week 52 compared with placebo in SLE patients with moderate-to-severe disease activity [84]. The first phase III trial of this agent met several secondary endpoints but failed to meet its primary endpoint [85]. Taken together, the data from all of the phase II and phase III studies support efficacy of this agent in SLE [86]. The fact that only one of the two anifrolumab phase III studies met the primary endpoint, along with many other failed SLE trials in the recent past, underscores the difficulty in SLE trial design and conduct in general [75].
Conclusion
As our understanding of SLE pathogenesis grows, our ability to directly target the underlying pathogenic mechanisms in clinical practice continues to improve. Ideally, our enhanced understanding of human disease biology and pathogenesis, including genetic susceptibility, clinical symptoms, and immunological dysfunction will allow for more specific and effective therapy, and eventually a personalized medicine approach in SLE [87].
Highlights:
Type I IFN is implicated in SLE pathogenesis by multiple lines of evidence, including genetics, gene expression, and induction of SLE by IFN treatment.
Nucleic-acid sensing pathways influence the threshold of activation and triggering of type I IFN production. Genetic mutations and molecules targeting these pathways have been used in the development of animal models of lupus.
Abnormalities in extracellular processing of DNA can enhance type I IFN production, with a potential key role of DNASE1L3 as demonstrated by the development of lupus-like disease in the Dnase1l3−/− mice.
High type I IFN levels are associated with active nephritis, mucocutaneous involvement, hematological manifestations, low complement levels and the presence of autoantibodies.
Anifrolumab, a monoclonal antibody that blocks the type I IFN receptor, has shown therapeutic benefit in SLE. Multiple studies on other promising therapeutic agents directly or indirectly targeting the type I IFN pathway are underway.
Acknowledgements:
Grants: Appenzeller S: Fundação de Amparo à Pesquisa do Estado São Paulo-Brasil (FAPESP 2008/02917-0 and 2016/23269-3), Conselho Nacional Pesquisa Desenvolvimento-Brasil CNPq (300447/2009-4 and 471343/2011-0 and 302205/2012-8 and 473328/2013-5 and 157534/2015-4). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. Niewold TB: Colton Center for Autoimmunity, NIH (AR060861, AR057781, AR065964, AI071651), the Lupus Research Foundation, and the Lupus Research Alliance
Disclosure of Interest
Grants: Appenzeller S: Fundação de Amparo à Pesquisa do Estado São Paulo-Brasil (FAPESP 2008/02917-0 and 2016/23269-3), Conselho Nacional Pesquisa Desenvolvimento-Brasil CNPq (300447/2009-4 and 471343/2011-0 and 302205/2012-8 and 473328/2013-5 and 157534/2015-4). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. Niewold TB: Colton Center for Autoimmunity, NIH (AR060861, AR057781, AR065964, AI071651), the Lupus Research Foundation, and the Lupus Research Alliance
Disclosures:
TBN has received research grants from EMD Serono and Janssen, Inc., and has consulted for Thermo Fisher and Inova, all unrelated to the current manuscript. Other authors have no conflict of interest to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsokos GC, Systemic lupus erythematosus. N Engl J Med, 2011. 365(22): p. 2110–21. [DOI] [PubMed] [Google Scholar]
- 2.Weckerle CE and Niewold TB, The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol, 2011. 40(1): p. 42–9.* Largest study to date of functional type I IFN levels in lupus patients, demonstrating differences between ancestral backgrounds and strong association with serological profiles.
- 3.Ghodke-Puranik Y and Niewold TB, Immunogenetics of systemic lupus erythematosus: A comprehensive review. J Autoimmun, 2015. 64: p. 125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weckerle CE, et al. , Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum, 2011. 63(4): p. 1044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baechler EC, et al. , Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A, 2003. 100(5): p. 2610–5.* Early study documenting the IFN signature in SLE, also makes the point that type I and type II IFNs are difficult to separate by their downstream signatures.
- 6.Bennett L, et al. , Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med, 2003. 197(6): p. 711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow MK, Kirou KA, and Wohlgemuth J, Microarray analysis of interferon-regulated genes in SLE. Autoimmunity, 2003. 36(8): p. 481–90. [DOI] [PubMed] [Google Scholar]
- 8.Kariuki SN, et al. , Cutting edge: Autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol, 2009. 182(1): p. 34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghodke-Puranik Y, et al. , Lupus-Associated Functional Polymorphism in PNP Causes Cell Cycle Abnormalities and Interferon Pathway Activation in Human Immune Cells. Arthritis Rheumatol, 2017. 69(12): p. 2328–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson T, et al. , Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. Journal of immunology, 2011. 187(3): p. 1298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritterhouse LL, et al. , B lymphocyte stimulator levels in systemic lupus erythematosus: higher circulating levels in African American patients and increased production after influenza vaccination in patients with low baseline levels. Arthritis Rheum, 2011. 63(12): p. 3931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oke V, et al. , High levels of circulating interferons type I, type II and type III associate with distinct clinical features of active systemic lupus erythematosus. Arthritis Res Ther, 2019. 21(1): p. 107.** Recent study that simultaneously compares type I, type II, and type III IFN levels in SLE patients along with clinical associations with these IFNs.
- 13.Wampler Muskardin TL and Niewold TB, Type I interferon in rheumatic diseases. Nature Reviews Rheumatology, 2018. 14(4): p. 214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronnblom L and Alm GV, A pivotal role for the natural interferon alpha-producing cells (plasmacytoid dendritic cells) in the pathogenesis of lupus. J Exp Med, 2001. 194(12): p. F59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reizis B, Plasmacytoid Dendritic Cells: Development, Regulation, and Function. (1097–4180 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronnblom L and Alm GV, Systemic lupus erythematosus and the type I interferon system. Arthritis Res Ther, 2003. 5(2): p. 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewold TB, et al. , High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun, 2007. 8: p. 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baccala R, et al. , Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc Natl Acad Sci U S A, 2013. 110(8): p. 2940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sisirak V, et al. , Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J Exp Med, 2014. 211(10): p. 1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thanarajasingam U, et al. , Type I Interferon Predicts an Alternate Immune System Phenotype in Systemic Lupus Erythematosus. ACR Open Rheumatol, 2019. 1(8): p. 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Der E, et al. , Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol, 2019. 20(7): p. 915–927.** Study of single cell gene expression that documents type I IFN signatures in single cells in the lupus kidney, including distinct molecular signatures that differentiated histological classes of lupus nephritis and a type I IFN signature that correlated with responsiveness to treatment
- 22.Kumaran Satyanarayanan S, et al. , IFN-β is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat Commun, 2019. 10(1): p. 3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowl JT, et al. , Intracellular Nucleic Acid Detection in Autoimmunity. Annual review of immunology, 2017. 35: p. 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlee M and Hartmann G, Discriminating self from non-self in nucleic acid sensing. Nature Reviews Immunology, 2016. 16(9): p. 566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kono DH, Baccala R, and Theofilopoulos AN, TLRs and interferons: a central paradigm in autoimmunity. Current opinion in immunology, 2013. 25(6): p. 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilstra JS, et al. , B cell–intrinsic TLR9 expression is protective in murine lupus. The Journal of Clinical Investigation, 2020. 130(6): p. 3172–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen SR, et al. , Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity, 2006. 25(3): p. 417–28. [DOI] [PubMed] [Google Scholar]
- 28.Majer O, et al. , UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity. Nature, 2019. 575(7782): p. 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Quintial R, et al. , Lupus acceleration by a MAVS-activating RNA virus requires endosomal TLR signaling and host genetic predisposition. PLoS One, 2018. 13(9): p. e0203118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soni C and Reizis B, DNA as a self-antigen: nature and regulation. Current opinion in immunology, 2018. 55: p. 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soni C, et al. , Plasmacytoid Dendritic Cells and Type I Interferon Promote Extrafollicular B Cell Responses to Extracellular Self-DNA. (1097–4180 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowl JT and Stetson DB, SUMO2 and SUMO3 redundantly prevent a noncanonical type I interferon response. Proceedings of the National Academy of Sciences of the United States of America, 2018. 115(26): p. 6798–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowl JT, et al. , Intracellular Nucleic Acid Detection in Autoimmunity. Annu Rev Immunol, 2017. 35: p. 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skopelja-Gardner S, et al. , The early local and systemic Type I interferon responses to ultraviolet B light exposure are cGAS dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burleigh K, et al. , Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol, 2020. 5(43). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pothlichet J, et al. , A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med, 2011. 3(3): p. 142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niewold TB, Interferon alpha as a primary pathogenic factor in human lupus. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research, 2011. 31(12): p. 887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niewold TB, et al. , Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum, 2008. 58(8): p. 2481–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salloum R, et al. , Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum, 2010. 62(2): p. 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remmers EF, et al. , STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med, 2007. 357(10): p. 977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagberg N, et al. , The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann Rheum Dis, 2018. 77(7): p. 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen MA, et al. , Functional genetic polymorphisms in ILT3 are associated with decreased surface expression on dendritic cells and increased serum cytokines in lupus patients. Annals of the rheumatic diseases, 2013. 72(4): p. 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Romo GS, et al. , Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med, 2011. 3(73): p. 73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavragani CP, et al. , Defective regulation of L1 endogenous retroelements in primary Sjogren’s syndrome and systemic lupus erythematosus: Role of methylating enzymes. J Autoimmun, 2018. 88: p. 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munroe ME, et al. , Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Annals of the rheumatic diseases, 2016. 75(11): p. 2014–2021.** Study that documents pre-disease increases in type I and type II IFNs in the context of pre-clinical autoantibody formation.
- 46.Arbuckle MR, et al. , Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med, 2003. 349(16): p. 1526–33. [DOI] [PubMed] [Google Scholar]
- 47.Niewold TB, Advances in lupus genetics. Current opinion in rheumatology, 2015. 27(5): p. 440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinicato NA, et al. , Familial aggregation of childhood and adulthood-onset Systemic Lupus Erythematosus. Arthritis Care Res (Hoboken), 2019. [DOI] [PubMed] [Google Scholar]
- 49.Niewold TB, et al. , Familial aggregation of autoimmune disease in juvenile dermatomyositis. Pediatrics, 2011. 127(5): p. e1239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghodke-Puranik Y and Niewold TB, Genetics of the type I interferon pathway in systemic lupus erythematosus. International journal of clinical rheumatology, 2013. 8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kariuki SN, et al. , Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther, 2010. 12(4): p. R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kariuki SN, et al. , Genetic analysis of the pathogenic molecular sub-phenotype interferon-alpha identifies multiple novel loci involved in systemic lupus erythematosus. Genes and immunity, 2015. 16(1): p. 15–23.* Large genome-wide genetic screen of the type I IFN trait in SLE patients which identified novel genes associated with high IFN in SLE.
- 53.Koldobskaya Y, et al. , Gene-expression-guided selection of candidate loci and molecular phenotype analyses enhance genetic discovery in systemic lupus erythematosus. Clinical & developmental immunology, 2012. 2012: p. 682018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghodke-Puranik Y, et al. , Novel genetic associations with interferon in systemic lupus erythematosus identified by replication and fine-mapping of trait-stratified genome-wide screen. Cytokine, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang SH, et al. , Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat Commun, 2019. 10(1): p. 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odhams CA, et al. , Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in Systemic Lupus Erythematosus. Nat Commun, 2019. 10(1): p. 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz LX, et al. , TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7–9. Nature, 2020. 581(7808): p. 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee-Kirsch MA, The Type I Interferonopathies. Annu Rev Med, 2017. 68: p. 297–315. [DOI] [PubMed] [Google Scholar]
- 59.Uggenti C, Lepelley A, and Crow YJ, Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu Rev Immunol, 2019. 37: p. 247–267. [DOI] [PubMed] [Google Scholar]
- 60.Niewold TB, et al. , IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Annals of the rheumatic diseases, 2012. 71(3): p. 463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langefeld CD, et al. , Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun, 2017. 8: p. 16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coit P, et al. , Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun, 2013. 43: p. 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S, et al. , Genome-Wide DNA Methylation Profiles Reveal Common Epigenetic Patterns of Interferon-Related Genes in Multiple Autoimmune Diseases. Front Genet, 2019. 10: p. 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirou KA, et al. , Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum, 2005. 52(5): p. 1491–503. [DOI] [PubMed] [Google Scholar]
- 65.Connelly KL, et al. , Longitudinal association of type 1 interferon-induced chemokines with disease activity in systemic lupus erythematosus. Sci Rep, 2018. 8(1): p. 3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petri M, et al. , Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus, 2009. 18(11): p. 980–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathian A, et al. , Ultrasensitive serum interferon-alpha quantification during SLE remission identifies patients at risk for relapse. Ann Rheum Dis, 2019. 78(12): p. 1669–1676. [DOI] [PubMed] [Google Scholar]
- 68.Hong S, et al. , Longitudinal Profiling of the Blood Transcriptome in Healthy and Lupus Pregnancy. J Exp Med, 2019.* Recent study of the interferon signature in healthy and lupus pregnancies that suggests type I IFN may contribute to adverse preganancy outcome in SLE.
- 69.Bengtsson AA, et al. , Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus, 2000. 9(9): p. 664–71. [DOI] [PubMed] [Google Scholar]
- 70.Grondal G, et al. , Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin Exp Rheumatol, 2000. 18(5): p. 565–70. [PubMed] [Google Scholar]
- 71.Mistry P, et al. , Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A, 2019. 116(50): p. 25222–25228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarkar MK, et al. , Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann Rheum Dis, 2018. 77(11): p. 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolf SJ, et al. , Ultraviolet light induces increased T cell activation in lupus-prone mice via type I IFN-dependent inhibition of T regulatory cells. J Autoimmun, 2019. 103: p. 102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harley ITW, et al. , The role of genetic variation near interferon-kappa in systemic lupus erythematosus J Biomed Biotechnol, 2010. 2010: p. Article ID 706825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postal M, et al. , Drugs in early clinical development for Systemic Lupus Erythematosus. Expert Opin Investig Drugs, 2016. 25(5): p. 573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niewold TB, Targeting type I interferon in systemic lupus erythematosus. Nature Reviews Rheumatology, 2016. 12(7): p. 377–U13. [DOI] [PubMed] [Google Scholar]
- 77.Houssiau FA, et al. , IFN-α kinoid in systemic lupus erythematosus: results from a phase IIb, randomised, placebo-controlled study. Annals of the Rheumatic Diseases, 2020. 79(3): p. 347. [DOI] [PubMed] [Google Scholar]
- 78.Furie R, et al. , Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J Clin Invest, 2019. 129(3): p. 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fetter T, et al. , Selective Janus Kinase 1 Inhibition Is a Promising Therapeutic Approach for Lupus Erythematosus Skin Lesions. Front Immunol, 2020. 11: p. 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wallace DJ, et al. , Baricitinib for systemic lupus erythematosus: a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet, 2018. 392(10143): p. 222–231. [DOI] [PubMed] [Google Scholar]
- 81.Khamashta M, et al. , Sifalimumab, an anti-interferon-alpha monoclonal antibody, in moderate to severe systemic lupus erythematosus: a randomised, double-blind, placebo-controlled study. Annals of the rheumatic diseases, 2016. 75(11): p. 1909–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kalunian KC, et al. , A Phase II study of the efficacy and safety of rontalizumab (rhuMAb interferon-alpha) in patients with systemic lupus erythematosus (ROSE). Annals of the rheumatic diseases, 2016. 75(1): p. 196–202. [DOI] [PubMed] [Google Scholar]
- 83.Gao L, et al. , Bone marrow mesenchymal stem cells from patients with SLE maintain an interferon signature during in vitro culture. Cytokine, 2020. 132: p. 154725. [DOI] [PubMed] [Google Scholar]
- 84.Morand EF, et al. , Trial of Anifrolumab in Active Systemic Lupus Erythematosus. N Engl J Med, 2020. 382(3): p. 211–221.** Successful phase III trial of type I IFN pathway blockade in lupus using the IFNAR blocking antibody anifrolumab.
- 85.Furie RA, et al. , Type I interferon inhibitor anifrolumab in active systemic lupus erythematosus (TULIP-1): a randomised, controlled, phase 3 trial. The Lancet Rheumatology, 2019. 1(4): p. e208–e219. [DOI] [PubMed] [Google Scholar]
- 86.Salmon JE and Niewold TB, A Successful Trial for Lupus - How Good Is Good Enough? N Engl J Med, 2020. 382(3): p. 287–288. [DOI] [PubMed] [Google Scholar]
- 87.Wampler Muskardin TL, et al. , Lessons from precision medicine in rheumatology. Mult Scler, 2020. 26(5): p. 533–539.*=of importance, **= of considerable importance