Abstract
Ivermectin (IVM) is an FDA-approved drug that has shown antiviral activity against a wide variety of viruses in recent years. IVM inhibits the formation of the importin-α/β1 heterodimeric complex responsible for the translocation and replication of various viral species proteins. Also, IVM hampers SARS-CoV-2 replication in vitro; however, the molecular mechanism through which IVM inhibits SARS-CoV-2 is not well understood. Previous studies have explored the molecular mechanism through which IVM inhibits importin-α and several potential targets associated with COVID-19 by using docking approaches and MD simulations to corroborate the docked complexes. This study explores the energetic and structural properties through which IVM inhibits importin-α and five targets associated with COVID-19 by using docking and MD simulations combined with the molecular mechanics generalized Born surface area (MMGBSA) approach. Energetic and structural analysis showed that the main protease 3CLpro reached the most favorable affinity, followed by importin-α and Nsp9, which shared a similar relationship. Therefore, in vitro activity of IVM can be explained by acting as an inhibitor of importin-α, dimeric 3CLpro, and Nsp9, but mainly over dimeric 3CLpro.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, 3CLpro, binding free energy, molecular docking, molecular dynamics simulation
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first discovered in Wuhan, China, in late 2019, causing a pneumonia-like epidemic that rapidly spread worldwide (Wu et al., 2020). In March 2020, the world health organization (WHO) declared SARS-CoV-2 as a pandemic due to its extreme outbreak (Brinks & Ibert, 2020). SARS-CoV2 infects humans, and it is the causative agent of the current viral pandemic COVID-19 (Brinks & Ibert, 2020; Guan et al., 2020; Huang et al., 2020). SARS-CoV-2 belongs to the beta coronavirus groups that are dissimilar to severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East Respiratory coronavirus (MERS-CoV). These two latter viruses produced preceding epidemics (Zhu et al., 2020). To date, about 2.6 million lives have been taken worldwide due to COVID-19, with about 118.0 million people still infected (https://coronavirus.jhu.edu/map.html0p). In the beginning, the virus showed to cause fever, sneezing, coughing, breathing problems, pneumonia, and multiple organ failure leading to death in severe cases (Brinks & Ibert, 2020). More recently, studies showed the likelihood of the viral infection also causing kidney dysfunction and myocardial damage (Kwong et al., 2018; Li, 2020; Nguyen et al., 2016). Therefore, identifying available and inexpensive drugs to treat and decrease the toll of the epidemic caused by COVID-19 is an urgent matter. In this sense, drug repurposing has been one of the best strategies to identify possible therapeutic agents (Borkotoky & Banerjee, 2020; de Oliveira et al., 2020; Khan et al., 2020). The genome of SARS-Cov-2 is deposited into host cells as a positive single-stranded RNA that is translated into polyproteins with the support of the host protein machinery. These polyproteins are then split into structural and non-structural proteins (Nsps) by the main protease (3CLpro) and papain-like protease. Nsps are virus-encoded proteins that are not part of the viral particle but are expressed in infected cells for viral replication. Therefore, structural and Nsps can be employed as potential targets to repurpose drug discovery. Several potential drug targets of SARS-CoV-2 comprise both Nsps and structural proteins such as 3CLpro, papain-like protease, RNA dependent RNA polymerase (RdRp), Nsp 13 helicase, Nsp14 (N-terminal exoribonuclease and C-terminal guanine-N7 methyl transferase), spike monomer, receptor-binding domain (RBD) of spike protein, spike trimer, post-fusion spike protein S2, nucleocapsid (N) protein, and Nsp9 replicase (Gordon et al., 2020; Kong et al., 2020).
IVM is a Food and Drug Administration (FDA)-approved broad-spectrum antiparasitic drug employed in humans to treat different parasitic diseases (Luvira et al., 2014). IVM was submitted to phase III clinical trial in Thailand from 2014 to 2017 to treat dengue viral infection. As a result of this study, it was reported as safe when given as a single daily dose, which resulted in a significant decrease of serum levels of viral carriage (Yamasmith, 2018). It is also reported that IVM decreases HIV-1 and dengue replication in cell cultures by inhibiting the formation of the importin-α/β1 complex, responsible for inhibition of nuclear accumulation of some viral proteins, such as HIV-1 integrase and NS5 polymerase protein (Fraser et al., 2014; Wagstaff et al., 2012). It is suggested that the broad antiviral ability of IVM can be attributed to its capacity to inhibit the host importin-α/β1 complex, intended for nuclear entry of viral proteins, by binding at the importin-α armadillo (ARM) repeat domain (Yang et al., 2020). Recently, Caly and coworkers showed that a single dose of IVM was able to control replication in clinical isolates of SARS-CoV-2 within 24 to 48 h, highlighting the repurposing properties of this drug against COVID-19 (Caly et al., 2020). However, the molecular mechanism at the atomic level of how IVM inhibits the host importin-α/β1 complex and other SARS-CoV-2 targets is yet to be elucidated. Recently, computational studies employed docking and molecular dynamics (MD) simulations to explore the ability of IVM to inhibit mouse importin-α (PDB entry 5FC8) and several SARS-CoV-2 targets. They found that IVM binds to importin-α with moderate affinity and Nsp9 with the highest affinity among several COVID-19 targets (Azam et al., 2020). Another recent study employed docking and MD simulations with the molecular mechanics Poisson-Boltzmann surface area (MMGBSA) approach to explore the affinity of IVM with importin-α and several SARS-CoV-2 targets. They identified IVM bound RdRp-RNA and Nsp13 helicase with a higher affinity than importin-α (Sen Gupta et al., 2020). Both studies provided critical information that can be used to provide more robust results by employing MD simulations combined with the MMGBSA approach. In this contribution, we explore the affinity of IVM for five SARS-Cov2 targets: dimeric 3CLpro, Nsp9 replicase, Nsp13 helicase, RdRp without RNA, and RBD of spike protein, as well as human importin-α, using molecular docking analysis, molecular dynamic MD simulations coupled to the MMGBSA approach, and per-residue decomposition analysis to predict their potential for antiviral treatment of SARS-CoV-2. From the SARS-CoV-2 targets, Nsp9 replicase, Nsp13 helicase and, RdRp were selected because they had been previously identified as potential targets (Azam et al., 2020; Sen Gupta et al., 2020). RBD-spike was chosen because it is crucial in recognizing the human angiotensin-converting enzyme-2 (ACE-2) receptor, which makes it indispensable for viral propagation (Chen & Guo, 2020; Woo et al., 2010). Dimeric 3CLpro was chosen based on previous information showing the importance of this SARS-CoV2 target and differences in its binding affinity in the monomeric versus the dimeric state (Bello 2020; Bello et al., 2020).
Methods
Preparation of systems
IVM structure was obtained from ChemSpider (http://www.chemspider.com/). The chemical design was optimized at the AM1 level using Gaussian 09 W software (Frisch et al., 2009). The tridimensional structures were retrieved from the PDB database of human importin-α (PDB entry 5H43) and Nsp13 (PDB entry 5RL9), as well as Nsp9 (PDB entry 6WXD), RdRp (PDB entry 7BV2), RBD-spike (PDB entry 6ACG), and 3CLpro (PDB entry 6LU7) of SARS-CoV-2.
Molecular docking
IVM was docked on the six molecular targets using AutoDock Tools 1.5.6 and AutoDock 4.2 programs (Morris et al., 2009). Hydrogen bonds were placed on ligand and protein atoms, and Kollman and Gasteiger partial charges were selected for the receptor and ligand. Ligand location was carried out using a Lamarckian-genetic algorithm. A grid box with a spacing of 0.375 and a size of 70 × 70 × 70 Å was constructed around the center of the binding site for all the five SARS-CoV-2 targets. For importin-α, the grid box was of a similar dimension to SARS-CoV-2 targets but defined at the nuclear localization signal binding domain, the major binding site formed by armadillo repeats 2-4 (Kobe, 1999; Tay et al., 2016). At the end of docking calculations, the complex with the lowest binding free energy was chosen as the starting conformer to perform MD simulations. Validation of the docking method was performed by reproducing the binding mode of histone acetyltransferase KAT8 on importin-α (PDB entry 5H43), 1-(3-fluoro-4-methylphenyl)methane sulfonamide on Nsp13 (PDB entry 5RL9), triphosphate form of remdesivir on RdRp (PDB entry 7BV2) and the inhibitor N3 on 3CLpro (PDB entry 6LU7) with RMSD values lower than 1.0 Å to the experimental binding mode.
Molecular dynamics (MD) simulations
The best complexes predicted from docking were further examined for measuring their thermodynamic stability and binding affinity using MD simulations employing AMBER16 software (Case et al., 2005) with the ff14SB force field (Duan et al., 2003). Ligand parametrization was carried out using AM1-BCC atomic charges and the general Amber force field (Wang et al., 2004). The complexes were placed into a dodecadic box of 12.0 Å filled with a TIP3P water model (Jorgensen et al., 1983). Then, the complexes were neutralized with Na + and Cl- counter ions at 0.15 M. After minimization and equilibration, MD simulations were run for a period of 100 to 150 ns with triplicate repetitions using an NPT ensemble at 310 K. The electrostatic forces were fixed by the PME method (Darden et al., 1993), and a 10 Å cutoff was chosen for the van der Waals forces. The SHAKE algorithm (Van Gunsteren & Berendsen, 1977) was used to set bond lengths at their equilibrium values. The systems' temperature and pressure were maintained by a weak-coupling algorithm (Berendsen et al., 1984). MD simulation results were analyzed using AmberTools16, and the images were constructed using Maestro Schrödinger version 10.5 (Schrodinger, 2016).
Binding free-energy and per-residue decomposition calculations
The MMGBSA (Gohlke & Case, 2004; Miller et al., 2012) method was employed to evaluate the binding free energy (ΔGbind) for the different complexes and calculate the per-residue decomposition free energy. To this, 200 snapshots at time intervals of 100 ps were selected over the last 20 ns of equilibrated simulation time. Before free-energy calculation, all counterions and water molecules were deleted, and a salt concentration of 0.15 M was considered together with the implicit solvation model (Onufriev et al., 2004). ΔGbind and per-residue decomposition free energy evaluation were determined as previously described (Bello & García‐Hernández, 2014) and correspond to triplicate experiments.
Results and discussion
Docking results
Interactions between IVM and importin-α
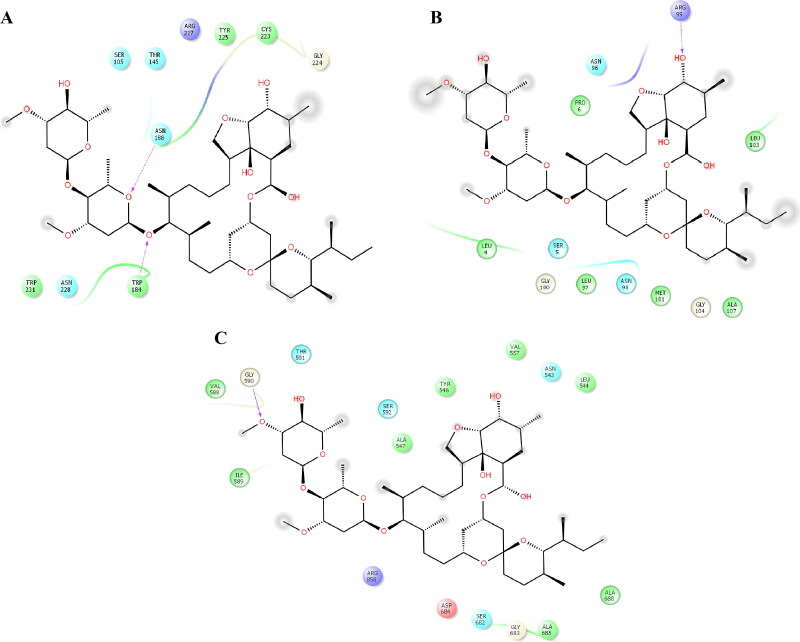
Importin-α is built of two domains; an importin-β binding (IBB) domain and an NLS-binding domain, which consists of 10 armadillo (ARM) repeats (Gorlich et al., 1996; Kobe, 1999). The LS-binding domain represents the major binding site, covering ARM repeats 2-4, and the minor site covers ARM repeats 6-8 (Pumroy & Cingolani, 2015). Importin-α is involved in the molecular recognition of different substrates through binding of the NLS-binding domain. The IVM docked complex with importin-α was shown to have one H-bond with Ser149. Polar contacts were established with Ser105, Asn146, Gln109, and Asn228. Hydrophobic interactions took place with Leu104, Pro110, Trp142, Trp184, and Trp231 (Figure 1A, supplementary material). Of these residues, Leu104, Arg106, Glu107, Trp142, Asn146, Ser149, Trp184, and Trp231 were also observed stabilizing the complex between importin and histone acetyltransferase KAT8 (PDB entry 5H43), which was bound at the major binding site that covered ARM repeats 2-4.
Figure 1.
Complexes between ivermectin with importin-α, Nsp9 replicase, Nsp13 helicase, and RdRp of SARS-CoV-2 through MD simulations. Interaction of ivermectin with importin-α (A), Nsp9 replicase (B), and RdRp (C).
Interactions between IVM and Nsp9, Nsp13, RdRP, spike protein, and 3CLpro
The Nsp9 replicase is an Nsp encoded by ORF1a, which has no vital function, but it is linked to viral RNA synthesis (Littler et al., 2020); therefore, it can be an indispensable target for drug discovery by inhibiting viral progression. Nsp9 replicase is built by a single folded beta-barrel exclusive to this protein, different from the single-domain proteins. Its crystallographic structure highlights the importance of its dimeric state for developing its biologic functions. Nsp9 replicase binds to the RNA, then to active Nsp8 protein by binding on it, a process that is crucial for its operation (Sutton et al., 2004). IVM was docked with Nsp9 replicase through one H-bond with Asn95 and four polar contacts with Thr35, Asn33, Thr67, Asp95, and Asn98. In comparison, hydrophobic contacts took place with Leu4, Phe40, Val41, Trp53, Ile65, Tyr66, Ile91, Leu94, Leu97, and Met 101 (Figure 1B, supplementary material). Of these residues, Leu4, Leu97, and Met101 were observed to form protein-protein interactions in the dimeric state of Nsp9 replicase present in the crystallographic complex (PDB entry 6WXD).
Nsp13 helicase is a multi-functional protein that is a conserved and essential viral replication component, representing another important target for antiviral drug discovery (Jia et al., 2019). In the case of Nsp13 helicase, IVM was docked by 2 H-bonds with Ser289 and His464. Polar contacts took place via 14 neutral and charged residues: Glu261, Gly285, Thr286, Gly287, Lys288, His290, Lsy320, Arg442, Arg465, Gly538, Ser539, Glu540, Arg443, and Lys569. Hydrophobic contacts were coordinated by only three residues: Pro284, Ala316, and Tyr324 (Figure 1C, supplementary material). Among these residues, Glu261, Gly285, Thr286, Gly287, His290, and Lsy320 were observed in the co-crystallized complex between 1-(3-fluoro-4-methylphenyl) methanesulfonamide and Nsp13 at the ADP site (PDB entry 5RL9).
RdRP plays an essential role in viral replication in host cells. The conserved structure of the RdRP core and the connected motifs are pivotal in its viral catalytic function and represent potential targets for drug discovery (Shu & Gong, 2016). In the complex with RdRp, IVM was docked by 1 H-bond with Asp845. Polar interactions were observed with polar and charged residues: Asn497, Lys545, Arg836, Asp845, Ser861, Glu857, Arg858, and Asp865, whereas hydrophobic interactions were coordinated by Tyr546, Ala547, Ile548, Ala840, Ile847, Val848, and Leu862 (Figure 1D, supplementary material). From these residues, Lys545 was observed in the co-crystallized complex between the triphosphate form of remdesivir on RdRp (PDB entry 7BV2).
The spike proteins form a peak shape on the surface of SARS-CoV-2 and represent a fundamental research interest, as little is known about its attachment and entry into the host cell (Walls et al., 2020). Spike proteins are represented by two subunits (S1 and S2). The S1 has divergent sequences, whereas the S2 is highly conserved. These spike proteins have heptad replications of hydrophobic domains that aid fusion into the host cell. The cell entry process is regulated by the spike proteins, mainly through binding to the angiotensin-converting enzyme-2 (ACE-2) receptor on the host exterior and facilitating viral infection. This role demonstrates the vast possibilities of targets in the search for effective compounds to inhibit viral infection. The docked complex between IVM and RBD-spike protein showed one H-bond with Asn487. Five polar contacts had polar, charged, and neutral residues: Arg389, Lys403, Gln479, Gln484, and Gly482, whereas hydrophobic interactions took place with Leu441, Tyr439, Tyr481, Phe483, and Tyr491 (Figure 2A, supplementary material). Among these residues, Gly482, Asn487, and Tyr491 formed interactions with ACE-2 in the co-crystallized complex between trimeric spike protein and ACE-2 (PDB entry 6ACG).
Figure 2.
MD simulation complexes between ivermectin and RBD of spike protein and 3CLpro of SARS-CoV-2. Interaction of ivermectin with RBD-spike protein (A). Interaction of ivermectin with subunit 1 of 3CLpro (B) and subunit 2 of 3CLpro (C).
The 3CLpro is also known as the main protease due to its dominant role in the post-translational machinery of the replicase (Kanchan et al., 2003). The 3CLpro of SARS-CoV-2 proteolytically slices the pp1a and pp1ab polyproteins of ORF a/b into useful proteins, a critical stage during viral replication, suggesting that it could be a significant target for decreasing the impact of COVID-19 (Zhang et al., 2020). The complex between IVM and subunit 1 of 3CLpro displayed 2 H-bonds by a polar residue of subunit 2 (Thr304) (Figure 2B, supplementary material). This complex also formed nine polar interactions with polar, charged, and neutral residues at subunit 1 (His41, Ser46, Asn142, His164, Glu166, Thr169, Gly170, Asp187, Arg188, and Gln189) and two polar residues of subunit 2 (Ser1 and Gln306). Hydrophobic interactions took place via seven hydrophobic residues of subunit 1 (Leu27, Met49, Leu50, Cys145, Met165, Leu167, and Pro168) and two hydrophobic interactions with residues of subunit 2 (Phe305 and Val303). Subunit2 of the 3CLpro/IVM complex formed one H-bond with Glu166 (Figure 2C, supplementary material). This complex also formed ten polar interactions with His41, Asn142, Ser144, His163, His164, His172, Arg188, Asp187, Gln189, and Gln192; 1 polar residue of subunit 1 (Ser1); and seven hydrophobic interactions with residues of subunit 2 (Cys44, Met49, Tyr54, Phe140, Leu141, Cys145, Met165, and Val186). Of these residues, His41, Met49, Asn142, Ser144, Cys145, His163, His164, Glu166, Pro168, His172, Asp187, Gln189, and Gln192 were also observed in the co-crystallized complex between inhibitor N3 and 3CLpro (PDB entry 6LU7).
MD simulations
The convergence of MD simulations
MD simulations allowed us to observe that IVM remained bound to importin-α and four of the five SARS-Cov-2 targets, except for Nsp13 helicase. IVM dissociated from the docked complex with Nsp13 helicase after 50 ns of MD simulation; therefore, it was excluded from further analysis. Evaluation of convergence for the complexes between IVM and importin-α, Nsp9 replicase, and dimeric 3CLpro showed that the root-mean-square deviation (RMSD) and radius of gyration (Rg) reached stable values for importin-α-IVM, Nsp9 replicase-IVM, and dimeric 3CLpro-IVM between 10 and 40 ns. In contrast, RdRp-IVM and RBD-spike-protein-IVM came with stable RMSD and Rg values between 80 and 150 ns (Figure 3 and 4, supplementary material). Based on this analysis, the first 80 ns were discarded from the MD simulation for additional research in all five systems.
MD simulations of the importin-α-IVM complex
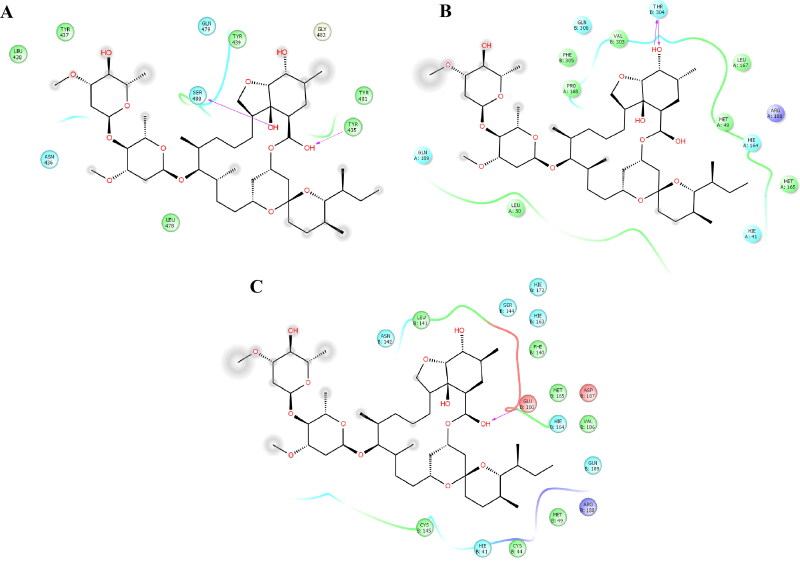
The most populated conformation of the importin-α-IVM complex showed that two H-bonds stabilized the ligand with Trp184 and Asn188. Polar contacts were maintained with Ser105, Thr145, Arg227, and Asn228. Hydrophobic interactions took place with Trp184, Cys223, Tyr225, and Trp231 (Figure 1A). Among these residues, Ser105, Trp184, Asn188, and Trp231 were present to stabilize the complex between importin-α and histone acetyltransferase KAT8 at the major binding site covering ARM repeats 2-4. This interaction map appeared more optimized based on the distribution of interactions than predicted by docking procedures (Figure 1A, supplementary material). Some of the residues stabilizing IVM and mouse importin-α (Trp184, Asn228, and Trp231) were also observed in a previous theoretical study combining docking and MD simulation to monitor the prevalence of the docked complex (Sen Gupta et al., 2020).
MD simulations between IVM and Nsp9, RdRP, spike protein, and 3CLpro
IVM was coupled in the Nsp9 replicase-IVM complex through one H-bond with Arg99 and five polar contacts with polar, charged, and neutral residues: Ser5, Asn96, Asn98, Gly100, and Gly104. Hydrophobic contacts were established with Leu4, Pro6, Leu97, Met101, Leu103, and Ala107 (Figure 1B and Figure 5A, supplementary material). Of these residues, Leu4, Ser5, Pro6, Leu97, Gly100, Met101, Leu103, Gly104, and Ala107 also interacted at the protein-protein interface of the dimeric state of Nsp9 (PDB entry 6WXD). Interestingly, these interactions were more optimized than those previously observed through docking calculations (Figure 1B, supplementary material). This stable conformation of IVM at the protein-protein interface suggests that the inhibitory activity of IVM could occur by inhibiting the dimerization of Nsp9 replicase since dimer formation has been related to the enhancement of nucleic acid binding and viral replication (Qiu & Xu, 2020). Some of the interactions present in the docked Nsp9-IVM complex (Pro6 and Gly100) were also identified in a previous theoretical study (Azam et al., 2020).
In the RdRp-IVM complex, the ligand was stabilized by one H-bond with Gly590. Polar interactions took place with polar and charged residues: Asp543, Gly590, Thr591, Ser592, Ser682, Gly683, Asp684, and Arg858. Hydrophobic interactions were established by Ala547, Val557, Val588, Ile589, Ala685, and Ala688 (Figure 1C). Of these interactions, Val557, Ser682, and Ala688 were present in the co-crystallized complex with the triphosphate form of remdesivir (PDB entry 7BV2). Comparing the complex predicted by docking (Figure 1D, supplementary material) with the MD simulation result indicated that the simulation optimized interactions, allowing the identification of more interactions present experimentally through x-ray experiments.
The complex between IVM and RBD-spike protein demonstrated that one H-bond stabilized the ligand with Ser480. Asn436, Gln479, and Gly482 stabilized polar contacts. Tyr435, Tyr437, Leu438, Tyr439, and Leu478 stabilized hydrophobic interactions (Figure 1A). These interactions were dissimilar to those predicted by docking studies (Figure 2A, supplementary material), sharing only interactions with Tyr439, Gln479, Tyr481, and Gly482. Of the residues observed stabilizing the RBD-spike-IVM complex during simulations, Tyr436, Gln479, and Gly482 were present in the experimental complex between the spike protein and ACE-2 (PDB entry 6ACG).
2 H-bonds stabilized the complex of IVM at subunit 1 of 3CLpro (3CLpro-sub1-IVM) by Thr304 of chain B. Polar interactions took place with polar, charged, and neutral residues (His41, His164, and Gln189) and one polar residue from chain B (Gln306). Hydrophobic interactions were stabilized by five hydrophobic residues (Met49, Leu50, Met165, Leu167, and Pro168) and two hydrophobic interactions of chain B with Phe305 and Val303 (Figures 2B and 5B, supplementary material). In the case of subunit 2, IVM formed one H-bond with Glu166. Polar interactions occurred with polar and charged residues: His41, Asn142, Ser144, His163, His164, His172, Asp187, Arg188, and Gln189. Hydrophobic interactions were formed with Cys44, Met49, Phe140, Leu141, Cys145, Met165, and Val186 (Figures 2B and 5C, supplementary material). Some key residues in IVM stabilization, His41, Met49, Asn142, Ser144, Cys145, His163, His164, Glu166, Pro168, His172, Asp187, and Gln189, were also present in the experimental complex with the inhibitor N3 (PDB entry 6LU7).
Binding free-energy calculations
Changes in the binding free energy (ΔGbind) were determined for the different systems using the MMGBSA approach. Table 1 shows that all the complexes were thermodynamically favorable and guided mainly through van der Waals energy (ΔEvdw). Although all systems showed good electrostatic contributions (ΔEele), the importin-α-IVM complex was the most favorable electrostatic contribution. However, the higher polar solvation energy (ΔGpol, sol) compared with the electrostatic contribution contributes to eliminating the electrostatic contribution from the ΔGbind.
Table 1.
Binding free energies of complexes between IVM and importin-α and four SARS-CoV-2 targets (in units of kcal/mol).
| System | ΔEvdw | ΔEele | ΔGpol, sol | ΔGnpol, sol | ΔGmmgbsa |
|---|---|---|---|---|---|
| Importin-α-IVM | −40.54 (5.3) | −22.49 (5.88) | 36.77 (6.34) | −4.81 (0.70) | −31.09 (5.0) |
| Nsp9-IVM | −41.03 (4.2) | −7.49 (6.27) | 20.64 (4.7) | −4.79 (0.50) | −32.68 (4.2) |
| RdRp-IVM | −37.89 (4.9) | −12.11 (7.6) | 37.13 (7.6) | −4.39 (0.70) | −17.26 (5.0) |
| RBD-spike-IVM | −36.59 (3.4) | −11.40 (5.3) | 29.55 (4.7) | −4.45 (0.32) | −22.90 (2.8) |
| 3CLpro-sub1-IVM | −57.74 (7.6) | −11.21 (4.3) | 38.47 (8.2) | −7.54 (1.0) | −38.03 (7.7) |
| 3CLpro-sub2-IVM | −50.75 (3.6) | −13.38 (4.0) | 32.37 (4.0) | −5.80 (0.40) | −37.57 (4.0) |
Table 1 also shows that the coupling of IVM on both subunits of dimeric 3CLpro reached the most favorable ΔGbind, followed by the importin-α-IVM and Nsp9-IVM complexes, which showed a similar affinity for ligands. In contrast, the RdRp-IVM and RBD-spike-IVM complexes showed less favorable ΔGbind values. This result contrasts with those previously reported by docking and MD simulations with the MMPBSA approach exhibiting the highest affinity of RdRp to IVM concerning importin-α and monomeric 3CLpro (Sen Gupta et al., 2020), which may be because they employed the RdRp with RNA and monomeric 3CLpro. In contrast, we used RdRp without RNA and dimeric 3CLpro. Overall, our result indicates that the inhibitory activity of IVM can be by targeting importin-α and two targets of SARS-CoV2 (3CLpro and Nsp9).
Per-residue free-energy decomposition
Analysis of the residues that contributed the most to the ΔGbind (Table 2) for all the systems showed that Trp184, Asn188, Arg227, and Trp231 were the primary sources of ΔGbind of the importin-α-IVM complex. Among these residues, Asn188 and Trp184 participated in forming H-bonds, and Arg227 and Trp231 formed polar and non-polar interactions (Figure 1A). In the Nsp9-IVM complex, Leu4, Pro6, Leu97, Arg99, Gly100, Met101, Leu103, and Gly104 were the main contributors to the affinity. Arg99 formed one H-bond of these residues, and the other residues formed polar and non-polar interactions (Figure 1B). The RdRp-IVM complex observed that only Ile589 and Gly590 contributed appreciably to the global affinity. In fact, for the stabilization of this system, it was observed that although a high number of residues stabilized it, most of them contributed energies lower than −0.5 kcal/mol.
Table 2.
Per-residue free energy decomposition for complexes between IVM and importin-α and four SARS-CoV-2 targets (values kcal/mol).
| Residue | IMP-IVM | Residue | Nsp9-IVM | Residue | RdRp-IVM | Residue | Spike-IVM | Residue | 3CLpro-sub1-IVM | Residue | 3CLpro-sub1-IVM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ser105 | −0.287 | Leu4 | −1.455 | Asn543 | −0.247 | His41 | −1.024 | His41 | −0.769 | ||
| Thr145 | −0.387 | Ser5 | −0.646 | Tyr546 | −0.235 | Tyr435 | −2.686 | Met49 | −1.691 | Cys44 | −0.195 |
| Trp184 | −4.02 | Pro6 | −1.293 | Ala547 | −0.397 | Asn436 | −0.285 | Leu50 | −1.997 | Met49 | −1.776 |
| Asn188 | −1.386 | Asn96 | −0.374 | Val557 | −0.561 | Tyr437 | −0.148 | His164 | 0.267 | Phe140 | −0.569 |
| Cys223 | −0.222 | Leu97 | −2.158 | Val588 | −0.271 | Leu438 | −2.051 | Met165 | −0.885 | Leu141 | −1.327 |
| Gly224 | −0.720 | Asn98 | −0.149 | Ile589 | −1.749 | Tyr439 | −0.179 | Leu167 | −0.218 | Asn142 | −1.252 |
| Tyr225 | −0.157 | Arg99 | −2.093 | Gly590 | −0.847 | Leu478 | −0.901 | Pro168 | −0.907 | Ser144 | −0.662 |
| Arg227 | −1.81 | Gly100 | −1.684 | Thr591 | −0.568 | Gln479 | −1.486 | Arg188 | −0.216 | Cys145 | −0.502 |
| Asn228 | −0.70 | Met101 | −1.026 | Ser592 | −0.305 | Ser480 | −2.576 | Gln189 | −1.525 | His163 | −0.191 |
| Trp231 | −2.7 | Leu103 | −2.532 | Ser682 | −0.136 | Tyr481 | −0.459 | Val303(B) | −0.987 | His164 | −0.365 |
| Gly104 | −1.192 | Gly683 | −0.167 | Gly482 | −0.33 | Thr304(B) | −1.671 | Met165 | −3.634 | ||
| Ala107 | −0.502 | Asp684 | −0.212 | Phe305(B) | −2.712 | Glu166 | −2.262 | ||||
| Ala685 | −0.551 | Gln306(B) | −0.479 | His172 | −0.346 | ||||||
| Ala688 | −0.317 | Val186 | −0.173 | ||||||||
| Arg858 | −0.292 | Asp187 | −0.443 | ||||||||
| Arg188 | −0.228 | ||||||||||
| Gln189 | −0.843 |
In the RBD-spike-IVM complex, Tyr435, Leu438, Gln479, and Ser480 were the major contributors to the affinity, from which Ser480 formed one H-bond, and the rest of the residues formed polar and non-polar interactions (Figure 2A). In the case of 3CLpro-sub1-IVM, His41, Met49, Leu50, Gln189, Thr304, and Phe305 contributed the most to the ΔGbind. Of these residues, Thr304 formed one H-bond, and the remaining amino acids established polar and non-polar interactions. For the 3CLpro-sub2-IVM system, Met49, Leu141, Asn142, Met165, and Glu166 were the primary sources of the ΔGbind values. Only Glu166 formed one H-bond with IVM of these residues, and the remaining residues formed polar and hydrophobic interactions.
Conclusion
Previous experimental studies demonstrated that IVM could inhibit SARS-CoV-2 replication in vitro; however, IVM's molecular mechanism inhibits binding to importin-α, and other SARS-CoV-2 receptors have not yet been elucidated. Recently, docking studies suggested the possible molecular mechanism through which IVM could inhibit mouse importin-α and some key SARS-CoV-2 targets (Nsp9 replicase, RdRp, and Nsp13 helicase), paving the way to the development of more robust studies. In this research, we explored the ability of IVM to inhibit human importin-α and five SARS-CoV-2 targets: dimeric 3CLpro, Nsp9 replicase, Nsp13 helicase, RdRp, and RBD-spike protein, using docking analysis, MD simulations coupled to the MMGBSA approach, and per-residue decomposition analysis. Hydrophobic and hydrophilic interactions guided interactions between IVM and importin-α, dimeric 3CLpro, and Nsp9, hydrophilic interactions being more critical for IVM binding at the central binding groove of importin-α. Per-residue free energy analysis let us identify hot-spot residues for importin-α-IVM (Trp184, Asn188, Arg227, and Trp231), 3CLpro-IVM (His41, Met49, Leu50, Leu141, Asn142, Met165, Glu166, and Gln189), and Nsp9-IVM (Leu4, Pro6, Leu97, Arg99, Gly100, Met101, Leu103, and Gly104), which contribute significantly to the affinity. MMGBSA results revealed that dimeric 3CLpro has the highest relationship among the different COVID-19 targets, followed by importin-α and Nsp9, which showed similar IVM affinity. This result indicates that the inhibitory activity of IVM maybe by targeting IMP and two essential targets of SARS-CoV2 (dimeric 3CLpro and Nsp9).
Supplementary Material
Funding Statement
The work was supported by grants from CONACYT (CB-A1-S-21278) and SIP/IPN (20210516).
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Borkotoky, S., & Banerjee, M. (2020). A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam, F., Taban, I. M., Eid, E. E., Iqbal, M., Alam, O., Khan, S., Mahmood, D., Anwar, M. J., Khalilullah, H., & Khan, M. U. (2020). An in-silico analysis of ivermectin interaction with potential SARS-CoV-2 targets and host nuclear importin α. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello, M. (2020, December). Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach. Journal of Molecular Graphics & Modelling, 101, 107762. 10.1016/j.jmgm.2020.107762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello, M., & García‐Hernández, E. (2014). Ligand entry into the calyx of β-lactoglobulin. Biopolymers, 101(7), 744–757. 10.1002/bip.22454 [DOI] [PubMed] [Google Scholar]
- Bello, M., Martínez-Muñoz, A., & Balbuena-Rebolledo, I. (2020). Identification of saquinavir as a potent inhibitor of dimeric SARS-CoV2 main protease through MM/GBSA. Journal of Molecular Modeling, 26(12), 1–11. 10.1007/s00894-020-04600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. Journal of Chemical Physics, 81(8), 3684–3690. [Google Scholar]
- Brinks, V., & Ibert, O. (2020, July). From corona virus to corona crisis: The value of an analytical and geographical understanding of crisis. Tijdschrift Voor Economische en Sociale Geografie, 111(3), 275–287. 10.1111/tesg.12428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., & Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26(16), 1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. L. Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, parthenogenesis. Journal of Virology, 92, 418423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald-an N.Log(N) method for Ewald sums in large systems. Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- de Oliveira, O. V., Rocha, G. B., Paluch, A. S., & Costa, L. T. (2020). Repurposing approved drugs as inhibitors of SARS-CoV-2S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y., Wu, C., Chowdhury, S., Lee, M. C., Xiong, G., Zhang, W., Yang, R., Cieplak, P., Luo, R., Lee, T., Caldwell, J., Wang, J., & Kollman, P. (2003). A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry, 24(16), 1999–2012. 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- Fraser, J. E., Watanabe, S., Wang, C., Chan, W. K. K., Maher, B., Lopez- Denman, A., Hick, C., Wagstaff, K. M., Mackenzie, J. M., Sexton, P. M., Vasudevan, S. G., & Jans, D. A. (2014). A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. The Journal of Infectious Diseases, 210(11), 1780–1791. 10.1093/infdis/jiu319 [DOI] [PubMed] [Google Scholar]
- Frisch, M. J. T., G. W., Schlegel, H. B., & Scuseria, G. E. (2009). Gaussian 09, Revision D.01. Gaussian Gaussian Inc. [Google Scholar]
- Gohlke, H., & Case, D. A. (2004). Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf . Journal of Computational Chemistry, 25(2), 238–250. 10.1002/jcc.10379 [DOI] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O'Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., … Krogan, N. J. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., Henklein, P., Laskey, R. A., & Hartmann, E. (1996). A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. The EMBO Journal, 15(8), 1810–1817. 10.1002/j.1460-2075.1996.tb00530.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W.-J., Ni, Z.-Y., Hu, Y., Liang, W.-H., Ou, C.-Q., He, J.-X., Liu, L., Shan, H., Lei, C.-L., Hui, D. S. C., Du, B., Li, L.-J., Zeng, G., Yuen, K.-Y., Chen, R.-C., Tang, C.-L., Wang, T., Chen, P.-Y., Xiang, J., … Zhong, N.-S. (2020, April). Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine, 382(18), 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020, February). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England), 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Z., Yan, L., Ren, Z., Wu, L., Wang, J., Guo, J., Zheng, L., Ming, Z., Zhang, L., Lou, Z., & Rao, Z. (2019). Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Research, 47(12), 6538–6550. 10.1093/nar/gkz409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., Klein, M. L., (1983). Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kanchan, A., John, Z., Parvesh, W., Jeroen, R. M., & Rolf, H. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science, 300, 1763–1767. [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y.-J., Chinnasamy, S., & Wei, D.-Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B. (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nature Structural Biology, 6(4), 388–397. 10.1038/7625 [DOI] [PubMed] [Google Scholar]
- Kong, R., Yang, G., Xue, R., Liu, M., Wang, F., Hu, J., Guo, X., & Chang, S. (2020). COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics (Oxford, England), 36(20), 5109–5111. 10.1093/bioinformatics/btaa645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong, J. C., Schwartz, K. L., Campitelli, M. A., Chung, H., Crowcroft, N. S., Karnauchow, T., Katz, K., Ko, D. T., McGeer, A. J., McNally, D., Richardson, D. C., Rosella, L. C., Simor, A., Smieja, M., Zahariadis, G., & Gubbay, J. B. (2018). Acute myocardial infarction after laboratory-confirmed influenza infection. New England Journal of Medicine, 378(4), 345–353. 10.1056/NEJMoa1702090 [DOI] [PubMed] [Google Scholar]
- Littler, D. R., Gully, B. S., Colson, R. N., & Rossjohn, J. (2020). Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9. iScience, 23(7), 101258. https://doi.org/ 10.1016/j.isci.2020.101258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. (2020). Caution on kidney dysfunctions of COVID-19 patients. SSRN Electronic Journal, 2020.02.08.20021212. Mar. 10.1101/2020.02.08.20021212 [DOI] [Google Scholar]
- Luvira, V., Watthanakulpanich, D., & Pittisuttithum, P. (2014). Management of Strongyloides stercoralis: A puzzling parasite. International Health, 6(4), 273–281. 10.1093/inthealth/ihu058 [DOI] [PubMed] [Google Scholar]
- Miller, B. R., McGee, T. D., Swails, J. M., Homeyer, N., Gohlke, H., & Roitberg, A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, J. L., Yang, W., Ito, K., Matte, T. D., Shaman, J., & Kinney, P. L. (2016). Seasonal influenza infections and cardiovascular disease mortality. JAMA Cardiology, 1(3), 274–281. 10.1001/jamacardio.2016.0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufriev, A., Bashford, D., & Case, D. A. (2004). Exploring protein native states and large‐scale conformational changes with a modified generalized born model. Proteins: Structure, Function, and Bioinformatics, 55(2), 383–394. 10.1002/prot.20033 [DOI] [PubMed] [Google Scholar]
- Pumroy, R. A., & Cingolani, G. (2015). Diversification of importin-a isoforms in cellular trafficking and disease states. Biochemical Journal, 466(1), 13–28. 10.1042/BJ20141186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y., & Xu, K. (2020). Functional studies of the coronavirus nonstructural proteins. STEMedicine, 1(2), e39–e39. 10.37175/stemedicine.v1i2.39 [DOI] [Google Scholar]
- Schrödinger, LLC . (2016). Maestro, version 10.5. 2016–1.
- Sen Gupta, P. S., Biswal, S., Panda, S. K., Ray, A. K., & Rana, M. K. (2020). Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. Journal of Biomolecular Structure and Dynamics, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, B., & Gong, P. (2016). Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proceedings of the National Academy of Sciences, 113(28), E4005–E4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, G., Fry, E., Carter, L., Sainsbury, S., Walter, T., Nettleship, J., Berrow, N., Owens, R., Gilbert, R., Davidson, A., Siddell, S., Poon, L. L. M., Diprose, J., Alderton, D., Walsh, M., Grimes, J. M., & Stuart, D. I. (2004). The nsp9 Replicase Protein of SARS-coronavirus, structure and functional insights. Structure (London, England : 1993), 12(2), 341–353. 10.1016/j.str.2004.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M. Y. F., Smith, K., Ng, I. H. W., Chan, K. W. K., Zhao, Y., Ooi, E. E., Lescar, J., Luo, D., Jans, D. A., Forwood, J. K., & Vasudevan, S. G. (2016). The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathogens, 12(9), e1005886. 10.1371/journal.ppat.1005886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gunsteren, W. F., & Berendsen, H. J. C. (1977). Algorithms for macromolecular dynamics and constraint dynamics. Molecular Physics, 34(5), 1311–1327. e 10.1080/00268977700102571 [DOI] [Google Scholar]
- Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., & Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin a/b-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochemical Journal, 443(3), 851–856. 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., Veesler, D., & Alexandra, C. W. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Woo, P. C., Huang, Y., Lau, S. K., & Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses, 2(8), 1804–1820. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasmith, E. (2018). Efficacy and safety of ivermectin against dengue infection: A phase III, randomized, double-blind, placebo-controlled trial [Paper presentation]. 34th Annual Meeting the Royal College of Physicians of Thailand. Internal Medicine and One Health. [Google Scholar]
- Yang, S. N. Y., Atkinson, S. C., Fraser, J. E., Wang, C., Maher, B., Roman, N., Forwood, J. K., Wagstaff, K. M., Borg, N. A., & Jans, D. A. (2019). Novel flavivirus antiviral that targets the host nuclear transport importin a/b1 heterodimer. Cells, 8(3), 281. 10.3390/cells8030281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.