Abstract
Porcine epidemic diarrhea virus (PEDV) is a coronavirus that causes emaciation and watery diarrhea in pigs. First identified in Europe in 1977, it eventually spread to Asia and North America, causing deadly outbreaks in neonatal piglets. In the Philippines, PEDV has caused several recorded outbreaks since 2005. However, DNA sequencing studies of local PEDV strains remain few and are limited to gene and gene fragment sequencing. Therefore, to provide updated sequence information about recent PEDV strains in the country, we performed reverse transcription PCR and sequencing of PEDV from swab samples collected from swine farms in the Philippines in 2017. Here, we report the first published whole genome sequence of PEDV from the Philippines as well as CO-26K equivalent (COE) domain sequences of strains from three provinces in Luzon where PEDV was detected in 2017. Sequence analysis suggested that PEDV from both the classical (genotype 1) and pandemic (genotype 2) groups are present in the Philippines, with possible East Asian and North American origins.
Introduction
Porcine epidemic diarrhea (PED) is an enteric disease in pigs caused by porcine epidemic diarrhea virus (PEDV), a member of the family Coronaviridae, order Nidovirales. Characterized by emaciation and watery diarrhea, as well as reduced reproductive performance in breeder pigs, PED can affect pigs of all ages, with disease severity, duration, and mortality decreasing with age. Neonatal piglets are affected the most, with outbreaks causing up to 100% mortality [1, 2].
PEDV is a large, enveloped RNA virus with a single-stranded, positive-sense linear genome approximately 28 kb in length. The genome contains 5’ and 3’ untranslated regions and six genes, namely, ORF1ab, spike (S), ORF3, envelope (E), membrane (M), and nucleoprotein (N). It also features a 5’ cap and a 3’ polyadenylated tail. ORF1 encodes a replicase protein, while a ribosomal frameshift (-1) during translation of ORF 1a and 1b generates products that are post-translationally cleaved into 16 non-structural proteins (nsp 1-16). The S, E, M, and N genes all encode structural proteins, while ORF3 encodes an ion channel protein that regulates virus production. The S gene (encoding the spike protein) is approximately 4.1 kb in length and contains the S1 domain responsible for host receptor binding and antibody-mediated neutralization. Since the spike protein plays a crucial role in infection of cells and the host immune response, its sequence is usually studied to investigate genetic similarities, diversity, and relatedness [1, 2]. Furthermore, a region of the S gene, called the CO-26K equivalent (COE) domain, has been shown to encode a virus-neutralizing epitope that elicits an antibody-mediated immune response in mice [3].
The coronavirus agent CV777 (later considered the classical PEDV strain) was first described by researchers in Belgium in 1978 as the causative agent of the disease – distinct from transmissible gastroenteritis virus (TGEV), which causes similar clinical signs. PED was first reported in Asia in 1982 and has since caused outbreaks in Korea, China, Thailand, and Vietnam. It was first identified in the USA in 2013 and has since spread rapidly to Mexico and Canada [1].
In the Philippines, PED incidences and outbreaks have been recorded in 2005 (Pampanga, Tarlac, Pangasinan, Batangas, Cavite, Laguna, Quezon, and Negros Occidental), 2006 (Bulacan, Rizal, Pampanga, Tarlac, Pangasinan, Batangas, Cavite, Laguna, Bacolod, and General Santos), 2007 and 2010 (Batangas), and 2016 (Pampanga, Tarlac, Batangas, and Agusan). PED outbreaks resulted in approximately 60,000 swine deaths in 2006, 2,179 in 2007, and 17,117 in 2010 [2, 4]. Despite the frequency of these incidences and outbreaks, DNA sequencing studies of local PEDV strains remain few and limited to gene and gene fragment sequencing.
To provide updated sequence information on more-recent PEDV strains, we employed capillary sequencing and next-generation sequencing to sequence the S gene COE domain and the whole genome respectively, of PEDV isolates from Luzon, Philippines. In this paper, we report the existence of genotype 1 and genotype 2 PEDV variant strains in the Philippines, as well as the first published whole genome sequence of PEDV in the country. Study of updated sequence information about local strains, as well as the phylogenetic relationships of these strains to commercially available vaccines and field strains from other countries, will help to improve the monitoring and management of PED in the Philippines.
Materials and methods
Sample collection and processing
A total of 166 porcine rectal swab samples were collected from 46 swine farms from the top backyard-swine-producing provinces in the island group of Luzon, Philippines (Isabela, Batangas, Bulacan, Cagayan, Marinduque, Camarines Sur, Pangasinan, and Palawan). Samples were collected from February to August 2017. Swabs were rinsed with phosphate-buffered saline, and the liquid was extracted and pooled for each farm (for a total of 46 pooled samples). Viral RNA isolation was performed using a QIAGEN Viral RNA Isolation Kit, following the manufacturer’s instructions (QIAGEN, Hilden, Germany).
Reverse transcription PCR
RNA samples were subjected to reverse transcription PCR (RT-PCR) using the PEDV S-F/R primer pair (PEDVF 5’-TTCTGAGTCACGAACAGCCA-3’, PEDVR 5’-CATATGCAGCCTGCTCTGAA-3’) to amplify the 651-bp COE domain of the PEDV S-gene [3]. Reverse transcription PCR was performed using an AccessQuick™ RT-PCR System (Promega Corporation, Wisconsin, USA) as per manufacturer’s instructions, with modifications: one cycle of cDNA synthesis (45°C, 45 min) and initial denaturation (95°C, 2 min), 40 cycles of denaturation (95°C, 1 min), primer annealing (55°C, 1 min), and extension (72°C, 1 min), and a final extension step (72°C, 10 min). RT-PCR products were sized and visualized by electrophoresis in a 2% agarose gel.
Capillary sequencing
RT-PCR products that were positive for the S gene COE domain were submitted to the Philippine Genome Center for capillary sequencing. Amplicons were sequenced in both directions using a BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI PRISM 3130xl Genetic Analyzer (Thermo Fisher Scientific, Massachusetts, USA). Nucleotide sequences were assembled using the DNA Baser sequence assembly software (Heracle Biosoft SRL).
Whole-genome sequencing
RNA samples were submitted to the Philippine Genome Center for whole-genome sequencing. Library preparation for the samples and positive control (a commercially available modified live PEDV P-5V vaccine) was performed using a TruSeq RNA Library Preparation Kit v2 (Illumina, California, USA). Metagenomic shotgun sequencing was done using a MiSeq Reagent Kit v3 (600 cycles) and an Illumina MiSeq instrument (Illumina, California, USA).
Genome assembly and annotation
The Illumina reads were subjected to quality control (QC) and filtering using AfterQC [5]. The filtered reads were mapped to the reference genome (Sus scrofa GCF_000003025.6) using Burrows-Wheeler Aligner (BWA) to identify host sequences [6], and the unmapped reads were extracted. De novo sequence assembly was performed using St. Petersburg Genome Assembler (SPAdes) [7]. The resulting contigs were compared against a database of pre-downloaded viral nucleotide sequences for identification [8] via the Basic Local Alignment Search Tool [9]. Gene prediction and annotation were performed using GeneMarkS [10] and MultAlin [11].
Sequence alignment and phylogenomic/phylogenetic analysis
PEDV sequences were aligned using MAFFT v.7.222 [12] and trimmed using MEGA X [13] and trimAl 1.2 rev59 [14]. The trimmed alignments were tested using jModelTest2 [15] by the Akaike (AIC), corrected Akaike (AICc), Bayesian (BIC), corrected Bayesian information criteria (BICc), and decision theory (DT) methods. The tests were used to determine the best-fit model of nucleotide substitution for phylogenomic/phylogenetic tree generation.
The phylogenomic and phylogenetic trees for the whole-genome and partial gene (COE domain) sequences, respectively, were constructed using RAxMLv8.2.12 [16], with the suggested parameters from jModelTest2 (GTR nucleotide substitution model) and 1,000 bootstrap replicates. The input tree was estimated using BioNJ. The tree was visualized using the Interactive Tree of Life (iTOL) [17].
Results
Observation of clinical signs
Twenty-six out of the 46 farms sampled (56.52%) had pigs that exhibited one or more clinical signs linked to PEDV (diarrhea, emaciation, abortion). Nineteen farms (41.30%) had pigs with diarrhea, 11 (23.91%) had pigs showing emaciation, and two (4.35%) had pigs that experienced abortion. On the other hand, 20 farms (43.48%) had pigs that did not exhibit any PEDV-related clinical signs at the time of sampling (Table 1).
Table 1.
Summary of PEDV-associated clinical signs observed and RT-PCR test results of swine farms from eight selected provinces in Luzon, Philippines
| Farm ID | Province | Number of pigs sampled | RT-PCR | Clinical signs | ||
|---|---|---|---|---|---|---|
| Diarrhea | Emaciation | Abortion | ||||
| SF0117 | Isabela | 6 | - | |||
| SF0217 | Isabela | 6 | + | x | ||
| SF0317 | Isabela | 4 | - | x | ||
| SF0417 | Isabela | 5 | - | x | ||
| SF0517 | Isabela | 2 | - | |||
| SF0617 | Isabela | 2 | - | x | x | x |
| SF0717 | Isabela | 1 | - | x | x | |
| SF0817 | Isabela | 2 | - | |||
| SF0917 | Batangas | 10 | - | x | ||
| SF1017 | Batangas | 5 | - | |||
| SF1117 | Batangas | 5 | - | |||
| SF1217 | Batangas | 6 | - | |||
| SF1317 | Batangas | 6 | + | |||
| SF1417 | Batangas | 6 | - | |||
| SF1517 | Batangas | 6 | - | |||
| SF1617 | Batangas | 13 | - | |||
| SF1717 | Batangas | 4 | - | |||
| SF1817 | Bulacan | 5 | - | |||
| SF1917 | Bulacan | 10 | + | x | ||
| SF2017 | Bulacan | 7 | - | |||
| SF2117 | Bulacan | 8 | + | x | ||
| SF2417 | Cagayan | 2 | - | x | ||
| SF2717 | Cagayan | 2 | - | x | ||
| SF2817 | Cagayan | 2 | - | x | ||
| SF2917 | Cagayan | 1 | - | |||
| SF3017 | Cagayan | 2 | - | |||
| SF3517 | Cagayan | 1 | - | |||
| SF3617 | Cagayan | 1 | - | |||
| SF4017 | Cagayan | 2 | + | x | ||
| SF4917 | Cagayan | 3 | - | x | ||
| SF5017 | Cagayan | 3 | - | x | ||
| SF5117 | Cagayan | 3 | - | |||
| SF5317 | Cagayan | 2 | - | |||
| SF6017 | Marinduque | 1 | - | x | ||
| SF6417 | Marinduque | 2 | - | x | ||
| SF6717 | Marinduque | 2 | - | x | ||
| SF6817 | Marinduque | 1 | + | x | ||
| SF6917 | Marinduque | 1 | - | |||
| SF7417 | Camarines Sur | 4 | - | x | ||
| SF7617 | Camarines Sur | 1 | - | x | ||
| SF7717 | Camarines Sur | 1 | - | x | x | |
| SF9917 | Camarines Sur | 2 | - | x | ||
| SF10117 | Camarines Sur | 2 | - | x | x | |
| SF10817 | Pangasinan | 2 | - | x | ||
| SF11117 | Pangasinan | 2 | - | x | ||
| SF11317 | Palawan | 2 | + | x | x | |
Reverse transcription PCR
PEDV was detected on farms in six out of the eight provinces where samples were obtained. Seven out of 46 pooled swab samples were positive for PEDV by reverse transcription PCR: one from the province of Isabela (SF0217), one from Batangas (SF1317), two from Bulacan (SF1917 and SF2117), one from Cagayan (SF4017), one from Marinduque (SF6817), and one from Palawan (SF11317). Sample SF0217 was from a farm with pigs showing emaciation, samples SF1917, SF4017, and SF6817 were from farms with pigs with diarrhea, sample SF2117 was from a farm with pigs that experienced abortion, and sample SF11317 was from a farm with pigs with both emaciation and diarrhea. On the other hand, sample SF1317 was obtained from a farm with apparently healthy pigs (Table 1).
Capillary (Sanger) sequencing
Four partial S gene (COE domain) sequences were obtained – three from the samples (SF1317-Batangas, SF4017-Cagayan, and SF6817-Marinduque) and one from the P-5V vaccine reference control. The sequences from the samples were submitted to GenBank and assigned the accession numbers MK578556-MK578558.
De novo genome sequence assembly
Two whole genome sequences were obtained, namely from SF4017 (Cagayan) and vaccine strain P-5V. The SF4017 sequence assembly had a length of 28,029 bp, with an estimated read coverage of 262.5x, and the sequence assembly of vaccine strain P-5V had a length of 27,986 bp, with an estimated read coverage of 1,665.4x. Both strains were found to have a genome organization identical to that of the classical CV777 strain (5′-ORF1a-ORF1b-S-ORF3-E-M-N-3′). The whole genome sequence of SF4017 was submitted to GenBank and assigned the accession number MK558089. SF4017 is the first published whole genome sequence of PEDV from the Philippines.
Using BLASTn, the SF4017 genome sequence was found to be most similar to that of PEDV isolate MN (KF468752.1), which was isolated in Minnesota, USA, in 2013, with 99.59% identity, whereas the SF4017 genome sequence was found to be 96.53% identical to that of the classical CV777 strain (AF353511.1).
Phylogenetic analysis: COE domain
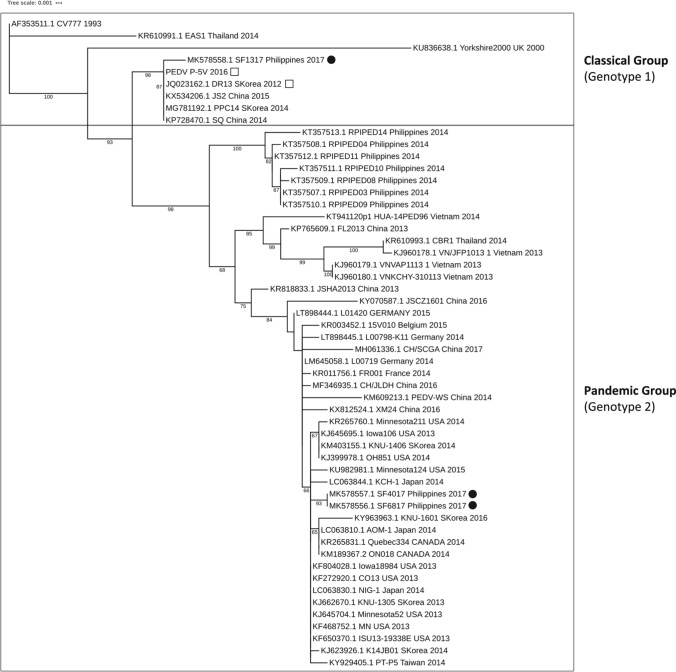
An unrooted phylogenetic tree was constructed using the CO-26K equivalent (COE) domain S gene sequences from three samples from this study (SF-1317, SF-4017, and SF-6817), two vaccine strains (strain DR-13 and the P-5V strain sequenced in this study), seven Philippine strains (RPI/PED03-14) sequenced in 2014, and 42 reference PEDV strains (including the classical CV777) (Fig. 1). The strains can be divided into two groups, the classical group (genotype 1) and the pandemic group (genotype 2) [18]. Strain SF1317 clustered with the classical group, which comprised nine strains in total, the others being classical strain CV777, vaccine strains P-5V, DR13, and EAS1 from Thailand, Yorkshire2000 from the UK, JS2 and SQ from China, and PPC14 from South Korea. On the other hand, SF4017, SF6817, and the seven 2014 Philippine RPI/PED strains clustered in the pandemic (genotype 2) group, along with 36 other strains from North America, Europe, and Asia.
Fig. 1.
Phylogenetic tree of porcine epidemic diarrhea virus (PEDV) strains generated using the COE domain sequences of 10 Philippine strains (including three from this study), two vaccine strains, and 42 isolates from across Asia, Europe, and North America. The phylogenetic tree was generated using the maximum-likelihood method and bootstrapping with 1,000 replicates. Vaccine strains are indicated by empty squares, while the 2017 Philippine strains are indicated by black circles. The scale bar and horizontal branch lengths indicate the mean number of nucleotide substitutions per site
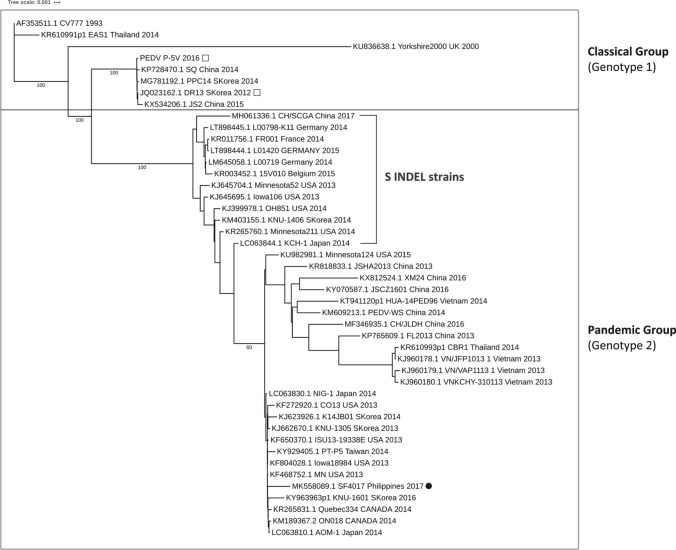
Phylogenomic analysis
An unrooted phylogenomic tree was constructed using the full-length genome sequence of the local strain SF4017 and the same set of sequences used for the phylogenetic analysis of the COE domain, except for the seven 2014 Philippine RPI/PED strains (which have S gene sequences only) (Fig. 2). The tree features a topology similar to that observed with the COE domain sequences, with strains divided into the classical (genotype 1) and pandemic (genotype 2) groups.
Fig. 2.
Phylogenomic tree of porcine epidemic diarrhea virus (PEDV) strains generated using the whole genome sequences of SF4017, two vaccine strains, and 42 isolates from across Asia, Europe, and North America. The phylogenomic tree was generated using the maximum-likelihood method and bootstrapping with 1,000 replicates. Vaccine strains are indicated by empty squares, while the Philippine strain SF4017 is indicated by a black circle. The scale bar and horizontal branch lengths indicate the mean number of nucleotide substitutions per site
Discussion
Several PED outbreaks have been recorded in the Philippines over the past years (in 2005, 2006, 2007, 2010, and 2016); however, sequence analysis of locally circulating PEDV strains is lacking, with only a handful of studies and sequences published. Prior to this study, the only publicly available set of PEDV S gene sequences (in GenBank) dates back to 2014, and comes from a single study [19]. While a report on sequencing a 753-bp portion of the S1 gene from samples collected from provincial PEDV hotspots in the Philippines (Batangas, Pampanga, Tarlac, and Agusan) was published in 2015 [20], no such sequences are publicly available at present. In addition, there were no publicly available Philippine PEDV genome sequences prior to this study.
In this study, six out of the seven farms that were positive for PEDV via RT-PCR (SF0217, SF1917, SF2117, SF4017, SF6817, and SF11317) had pigs that exhibited clinical signs associated with PEDV. On one of these farms (SF2117), the only clinical sign that was observed was abortion, which has recently been linked to PEDV outbreaks [21, 22]. On the other hand, one farm (SF1317) was positive for PEDV even though the pigs were apparently healthy and had no history of diarrhea or other clinical signs associated with PEDV. This may have been due to sampling at an early stage of infection before the onset of clinical signs, as PED has an incubation period of 1 to 8 days, depending on environmental conditions [1]. It is also possible that the infection was subclinical; a low viral load may not have been enough to cause disease, but PEDV may still be detectable owing to the sensitivity of the RT-PCR assay. Pigs may be seropositive for PEDV (i.e., have been infected) despite exhibiting no clinical signs [23].
The sequencing results show that the PEDV strains sequenced in this study belong to two groups. SF1317 (from Batangas) clustered in the classical group (genotype 1), while SF4017 (from Cagayan) and SF6817 (from Marinduque) clustered in the pandemic/virulent group (genotype 2). Based on the S gene COE domain sequences, the three strains are genetically distinct from the RPI/PED 2014 Philippine isolates, suggesting a separate introduction event. Phylogenetic analysis of the strains sequenced in this study suggested a possible East Asian origin of SF1317 and a possible North American/East Asian origin of SF-6817 (Fig. 1).
Strain SF4017 was found to be most closely related to PEDV strain MN (KF468752.1), suggesting a possible USA origin. Strain MN was one of the three representative strains of PEDV responsible for the 2013 PEDV outbreak in the USA in 2013 [24]. Sequence alignment of the 2014 MN strain and the 2017 SF4017 strain showed 114 nucleotide differences (substitutions). Thirty-four of the 114 nucleotide substitutions resulted in amino acid replacements: 14 in ORF 1a, four in ORF 1b, 11 in the spike (S) gene, one in ORF3, two in the membrane (M) protein gene, and two in the nucleocapsid (N) protein gene (Table 2). The envelope (E) protein amino acid sequences are identical.
Table 2.
Genomic variants found in the local SF4017 strain that result in amino acid substitutions. The reported variant positions are relative to the closest database match of S4017, the PEDV isolate MN (GenBank accession no. KF468752.1) from Minnesota, USA
| Gene | Nucleotide substitution a |
Amino acid substitution a |
|---|---|---|
| ORF1a |
T392C G832T C2151A G2155A T3038G A3103G T3138G G3508A T3542C T4196C C4827G G4889A T5518C A7963G |
F131S G278C D717E V719I V1013G N1035D D1046E V1170I I1181T L1399S F1609L S1630N F1840L T2655A |
| ORF1b |
G310T G3378T G3940A G775A |
A174S L1196F D1384N S2662N |
| S |
T437G T469C C677T T1006G T1129G A1183G T1456C, C1457A C2639T A3295G T3769G |
L146R Y157H S226F L336V L377V I395V S486H A880V T1099A S1257A |
| ORF3 | T358C | F120L |
| M |
T125C A595G |
V42A I199V |
| N |
T1127C C1237A |
I376T P413T |
a Numerical positions are relative to the respective genes
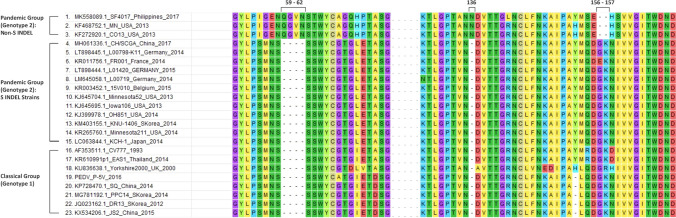
Like some highly virulent PEDV strains such as strain MN and Colorado-2013 [25], SF4017 is distinct from the less virulent PEDV S INDEL strains (such as OH851, Minnesota52, and KNU-1406) and classical strains (such as CV777 and P-5V) in that it has an amino acid deletion mutation at position 156-157 (GK) and insertion mutations at positions 59–62 (QGVN) and 136 (N) (relative to CV777) (Fig. 3). This distinction can be observed further in the phylogenomic tree, where SF4017 clusters with strain MN and Colorado 2013 in a subgroup that is separate from the S INDEL strains (Fig. 2).
Fig. 3.
Multiple sequence alignment of representative classical/genotype 1 porcine epidemic diarrhea virus (PEDV) strains and pandemic/genotype 2 (non-S INDEL and S-INDEL) strains, showing characteristic insertion-deletion mutations of S INDEL strains at positions 59-62 (QGVN), 136 (N), and 156-157 (GK)
PED vaccines are commercially available for use in Asia. In the Philippines, as of 2017, a modified live P-5V (genotype 1) vaccine is registered and commercially available [26]. An oral live attenuated DR-13 (genotype 1) vaccine was also once available in the country, in 2011 [27, 28], but has now been withdrawn from use. Despite their availability, PED vaccines are not widely used in the Philippines because their efficacy varies between herds, and vaccination does not prevent severe acute outbreaks. Moreover, vaccinated pig herds have been reported to respond poorly to feedback (a considerably more effective disease management method) when compared to unvaccinated herds [29]. Phylogenetic analysis of the spike protein COE-domain sequences indicates that the vaccine strains P-5V and DR13 are closely related to the 2017 SF1317 strain (from Batangas) but distantly related to the 2014 RPI/PED strains, the 2017 SF4017 strain (from Cagayan), and the SF6817 strain (from Marinduque). Since the spike protein plays a key role in the host immune response to PEDV infection, sequence differences may affect the ability of a vaccine strain to confer effective immunity against infection with a heterologous field strain. This may explain the reported differences in the efficacy of these genotype 1 strain vaccines in the Philippines.
The continued emergence of variant strains of PEDV highlights the need for continuous development of vaccines based on phenotypically and genotypically identical field strains [1, 2] to ensure efficacy. Thus, identification and genetic characterization of circulating PEDV strains is necessary to guide efforts in the development of effective vaccines, as well as proper administration of these vaccines based on circulating strains in different regions of the Philippines.
Sequencing plays a critical role in the development and evaluation of molecular diagnostic kits, especially in the case of RNA viruses such as PEDV, where multiple variant strains continue to emerge from different regions across the globe [2]. Comparing nucleotide sequences of local variant strains with those of the RT-PCR primers being utilized for diagnostics ensures that testing kits and protocols remain accurate relative to currently circulating strains. Sequence information about field strains can also be valuable for the development and evaluation of immunological assays such as virus neutralization test and ELISA – the sequence information can guide in the selection of appropriate antibodies against matching strains of circulating PEDV.
Our findings provide evidence of the existence of both classical (genotype 1) and pandemic/virulent (genotype 2) strains of PEDV in the Philippines and show their phylogenetic relationship to commercially available vaccine strains and strains from other countries. The findings of the study can be used as a reference for future efforts in vaccine and diagnostic kit development/evaluation, as well as in monitoring, prevention, management, and control of PED in the Philippines. It is recommended that more-extensive sampling and sequencing work be done (both in Luzon and other parts of the Philippines) to further monitor and characterize locally circulating PEDV strains, as well as to better understand the epidemiology of PED in the Philippines.
Acknowledgments
This study is part of the project titled “Profiling of Economically Important Diseases of Swine and Cattle in the Philippines for Enhanced Disease Management, Surveillance, and Control” funded by the United States Department of Agriculture through the United States Public Law 480 (PL-480) Title-I Program, and administered through the Department of Agriculture – Philippine Council on Agriculture and Fisheries (DA-PCAF). We thank the provincial veterinarians of participating provinces for providing valuable assistance in sample collection. We also thank the Philippine Genome Center for providing DNA sequencing and bioinformatics training and support.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging swine virus. Virol J. 2015;12(1):193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4(2):166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang SH, Bae JL, Kang TJ, Kim J, Chung GH, Lim CW, et al. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells. 2002;14(2):295–299. [PubMed] [Google Scholar]
- 4.Morales RG, Umandal AC, Lantican CA. Emerging and re-emerging diseases in Asia and the Pacific, with special emphasis on porcine epidemic diarrhoea. Conference OIE. 2007;2007:185–189. [Google Scholar]
- 5.Chen S, Huang T, Zhou Y, Han Y, Xu M, Gu J. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017;18(S3):80. doi: 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.BLAST Virus Reference Genome Database. Maryland: The United States National Library of Medicine. 2018—[cited 12 May 2020]. ftp://ftp.ncbi.nlm.nih.gov/blast/db/. Accessed 12 May 2020
- 9.Basic Local Alignment Search Tool (2017) Maryland: the United States National Library of Medicine. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 12 May 2020
- 10.Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29(12):2607–2618 [DOI] [PMC free article] [PubMed]
- 11.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh S. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 8;44(W1):W242–W245 [DOI] [PMC free article] [PubMed]
- 18.Sun M, Ma J, Wang Y, Wang M, Song W, Zhang W, et al. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J Clin Microbiol. 2015;53(5):1484–1492. doi: 10.1128/JCM.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YK, Cho YY, An BH, Lim SI, Lim JA, Cho IS, et al. Molecular characterization of the spike and ORF3 genes of porcine epidemic diarrhea virus in the Philippines. Arch Virol. 2016;161(5):1323–1328. doi: 10.1007/s00705-016-2758-2. [DOI] [PubMed] [Google Scholar]
- 20.Paraguison-Alili R, Domingo CY. Phylogenetic tracking of current porcine epidemic diarrhea virus (PEDV) strains in the Philippines. Arch Virol. 2016;161(9):2601–2604. doi: 10.1007/s00705-016-2938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olanratmanee E, Kunavongkrit A, Tummaruk P. Impact of porcine epidemic diarrhea virus infection at different periods of pregnancy on subsequent reproductive performance in gilts and sows. Anim Reprod Sci. 2010;122(1–2):42–51. doi: 10.1016/j.anireprosci.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Lin J-D, Lin C-F, Chung W-B, Chiou M-T, Lin C-N (2016) Impact of mated female nonproductive days in breeding herd after porcine epidemic diarrhea virus outbreak. PLoS One. 10.1371/journal.pone.0147316 [DOI] [PMC free article] [PubMed]
- 23.Koike N, Mai TN, Shirai M, Kubo M, Hata K, Marumoto N, et al. Detection of neutralizing antibody against porcine epidemic diarrhea virus in subclinically infected finishing pigs. J Vet Med Sci. 2018;80(11):1782–1786. doi: 10.1292/jvms.18-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YW, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, et al (2013) Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4(5):e00737–13 [DOI] [PMC free article] [PubMed]
- 25.Lin CM, Saif LJ, Marthaler D, Wanga Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lantin, et al. Philippine veterinary products directory. 19. Pasig: Medicomm Pacific Inc; 2017. [Google Scholar]
- 27.Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lantin, et al. Philippine veterinary products directory. 16. Pasig: Medicomm Pacific Inc; 2017. [Google Scholar]
- 29.Sanchez RE (2014) Management of porcine epidemic diarrhea. University of the Philippines Los Baños. https://www.dsm.com/content/dam/dsm/anh/en_US/documents/Porcine_Epidemic_Diarrhea.pdf. Accessed 12 May 2020