SUMMARY
The central pathogen-immune interface in tuberculosis is the granuloma, a complex host immune structure that dictates infection trajectory and physiology. Granuloma macrophages undergo a dramatic transition in which entire epithelial modules are induced and define granuloma architecture. In tuberculosis, relatively little is known about the host signals that trigger this transition. Using the zebrafish-Mycobacterium marinum model, we identify the basis of granuloma macrophage transformation. Single-cell RNA-seq analysis of zebrafish granulomas as well as analysis of Mycobacterium tuberculosis-infected macaques reveal that, even in the presence of robust type 1 immune responses, countervailing type 2 signals associate with macrophage epithelialization. We find that type 2 immune signaling, mediated via stat6, is absolutely required for epithelialization and granuloma formation. In mixed chimeras, stat6 acts cell-autonomously within macrophages, where it is required for epithelioid transformation and incorporation into necrotic granulomas. These findings establish the signaling pathway that produces the hallmark structure of mycobacterial infection.
Graphical Abstract
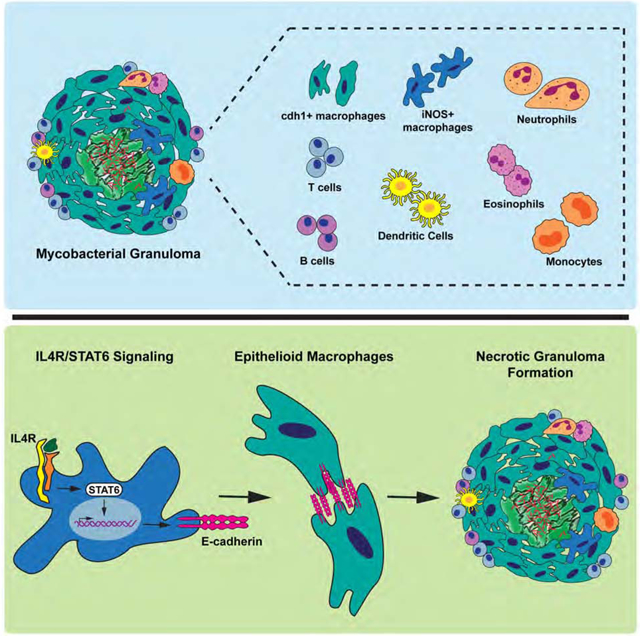
During mycobacterial infections, a non-canonical type 2 immune response underlies the formation and epithelial architecture of the granuloma, the hallmark structure of tuberculosis.
INTRODUCTION
The pathological hallmark of tuberculosis is the formation of the tuberculous granuloma, a structure derived from the aggregation of host immune cells around infecting Mycobacterium tuberculosis (Mtb) (Pagan and Ramakrishnan, 2018). The granuloma serves as a central gateway for the host immune response to infecting mycobacteria, and its structure and properties shape immune interactions, disease progression, and resolution (Ernst et al., 2018; Marakalala et al., 2016; Pagan and Ramakrishnan, 2018). In addition, structural properties of the granuloma affect the efficacy of Mtb therapies, serving as a substantial barrier to the penetration of anti-mycobacterial chemotherapies and shaping the metabolism of Mtb, which in turn alters susceptibility to chemotherapeutics (Blanc et al., 2018; Sarathy et al., 2019).
The central cell type of the granuloma is the macrophage, which aggregates during the formation of the nascent granuloma. These macrophages undergo a morphological transition in which they interdigitate with their neighbors and assume an altered morphology that pathologists have termed epithelioid (Adams, 1976; Pagan and Ramakrishnan, 2018). Epithelioid transformation is a defining feature of tuberculous granulomas (Adams, 1976; Pagan and Ramakrishnan, 2018). We previously found that this transition involves a wholesale epithelialization event that occurs within the granuloma macrophages, and that this macrophage-epithelial transition underlies organized granuloma formation (Cronan et al., 2016). Broad epithelial transcriptional modules are induced, including expression of E-cadherin on granuloma macrophages. Although the granuloma may act to confine infection to circumscribed areas within lung tissue, it also provides a safe harbor in which mycobacteria can persist and replicate (Cronan et al., 2016; Pagan and Ramakrishnan, 2018). Partial disruption of epithelialization and organization can result in improved outcome for infected animals (Cronan et al., 2016; Gautam et al., 2018).
The tuberculous granuloma is a highly organized structure, commonly consisting of an inner core of necrotic cell debris termed caseum in which Mtb resides, surrounded by layers of epithelioid macrophages, while at the edge of the granuloma are clusters of lymphocytes (Adams, 1976; Pagan and Ramakrishnan, 2018). The structural barriers of the granuloma restrict the immune response to mycobacteria (Cronan et al., 2016; Ernst et al., 2018; Kauffman et al., 2018). CD4+ T cells are essential for control of infecting mycobacteria, but CD4+ T cells are restricted to the outer layers of the granuloma, segregated from the bacteria within the macrophage-rich layers (Ernst et al., 2018; Kauffman et al., 2018). Other immune cell types are also restricted from close interactions with the bacterial core, presumably either through decreased recognition of bacteria within the necrotic core or by limiting the ability of the immune cells to migrate to these bacteria (Cronan et al., 2016).
The three-dimensional structure of the granuloma contributes to the therapeutic challenges of granuloma treatment; many chemotherapeutic drugs have difficulty penetrating through the tight layers of cells surrounding the necrotic core and the lipid-rich caseum to the bacteria within (Blanc et al., 2018; Gengenbacher et al., 2017; Prideaux et al., 2015; Sarathy et al., 2019). The granuloma also drives a metabolic shift within mycobacteria as the bacterium must adapt its metabolism to the lipid rich necrotic core as well as the hypoxic environment present within the core (Ehrt et al., 2018; Oehlers et al., 2015; Rittershaus et al., 2013; Rustad et al., 2009). These metabolic shifts subsequently result in alterations to mycobacterial drug susceptibility (Ehrt et al., 2018; Lenaerts et al., 2015; Sarathy et al., 2018; VanderVen et al., 2015).
The signaling that drives the granulomatous response in tuberculosis has long been conjectured. During Mtb infection, it is thought that a type 1 IFN-γ-dependent signaling pathway predominates and contributes to granuloma formation (Ndlovu and Marakalala, 2016). This derived from alterations in granuloma structure in mouse models and human patient samples observed when type 1 signaling is disrupted and the long-standing association of type 1 immune responses with mycobacterial control (Bean et al., 1999; Bustamante, 2020; Cooper et al., 1993; Flynn et al., 1995; Jouanguy et al., 1996). However, in schistosomiasis, granuloma formation is driven by the opposing IL-4 and IL-13 signaling pathways (Jankovic et al., 1999; Pagan and Ramakrishnan, 2018). These dichotomous responses have led to the description of granulomas that are formed by Mtb as well as diseases like sarcoidosis as type 1 granulomas, while Schistosoma granulomas have been termed type 2 granulomas following classical nomenclature of immune polarization. Despite the long-standing association of type 1 immunity with Mtb infection, the signals that drive tuberculous granuloma formation remain unclear.
The difficulty in mapping the determinants of tuberculous granuloma formation and signaling has been driven by the complications of studying the organized granuloma. The complex organization and composition of the fully organized granuloma can only be formed in vivo. However, organized necrotic granulomas fail to form in standard inbred mouse strains infected with Mtb where genetic tools are abundant, but rather form in animal models such as rabbits, guinea pigs, and macaques, in which genetic interrogation is much more difficult (Bucsan et al., 2019b; Flynn, 2006).
The zebrafish-M. marinum model has emerged as a validated model to study the host and pathogen genetics of mycobacterial infection and granuloma formation (Davis et al., 2002). M. marinum is a close relative of the Mtb complex, with a number of conserved virulence loci (Tobin and Ramakrishnan, 2008). Zebrafish infected with this natural pathogen form organized necrotic granulomas that closely mimic granulomas observed in human tuberculosis patients (Parikka et al., 2012; Swaim et al., 2006). Here, we use the zebrafish-M. marinum model to interrogate the genetic basis of macrophage epithelialization and granuloma formation.
Using single cell RNA-seq analysis, we find that, rather than an exclusively type 1 inflammatory response, there is induction of both type 1 and type 2 responses within the granuloma. Consistent with our findings in zebrafish, we find substantial type 2 signaling in granulomas from Mtb-infected macaques and find that type 2 markers localize to epithelialized regions of the granuloma. Contrary to the classical model of tuberculosis wherein tuberculous pathology depends exclusively on a Type 1 immune response, macrophage epithelialization and necrotic granuloma formation is lost in stat6-deficient and IL-4R-deficient animals, in which type 2 responses have been compromised. We find that stat6 function is required specifically in hematopoietic populations and acts cell autonomously during epithelioid transformation. Not only is functional stat6 function required in the formation of new granulomas, but persistent stat6 signaling is required to maintain E-cadherin expression and granuloma structure in established granulomas. These results illustrate a contribution of type 2 immunity in the macrophage epithelioid transition and the formation of defining granuloma pathology in mycobacterial infections.
RESULTS
Single cell RNA-seq reveals a diverse cellular composition in the mycobacterial granuloma
The granulomas that form in adult zebrafish infected with M. marinum recapitulate the structure of granulomas formed in Mtb patients, including central necrosis and macrophage epithelioid differentiation (Parikka et al., 2012; Swaim et al., 2006). We reasoned that the unique advantages of the zebrafish model would allow us to interrogate the determinants of organized granuloma formation in vivo. Classically, the Mtb granuloma has been assumed to be driven by type 1 signaling (Ndlovu and Marakalala, 2016; Sandor et al., 2003). However, data in animal models such as macaques and rabbits as well as in human patient samples has suggested that there may be substantial alternative activation within the granuloma (Huang et al., 2015; Mattila et al., 2013; Mehra et al., 2010; Subbian et al., 2011).
We had previously developed methods to dissect and dissociate granulomas from infected animals. We used these approaches together with single-cell RNA-seq (scRNA-seq) to interrogate the cellular composition and the inflammatory environment within the tuberculous granuloma. Granulomas at a single timepoint within one animal show heterogeneity in composition and trajectory. Some resolve, while others expand, and a broad range of cell-types is apparent upon dissociation and staining. For single cell analysis, we pooled >220 granulomas from 11 animals at a 14-day timepoint in which structured and organized granulomas had formed or were in the process of forming. Using the 10x Genomics Chromium platform, we identified 20 discrete clusters from 9273 cells that reflect the diverse cell types and states present in mature granulomas (Figure 1A, Table S1).
Figure 1. Interrogation of Granuloma Inflammatory State by scRNA-seq Reveals Type 2 Activation and an Inverse Correlation Between Type 1 Responses and Macrophage Epithelialization.
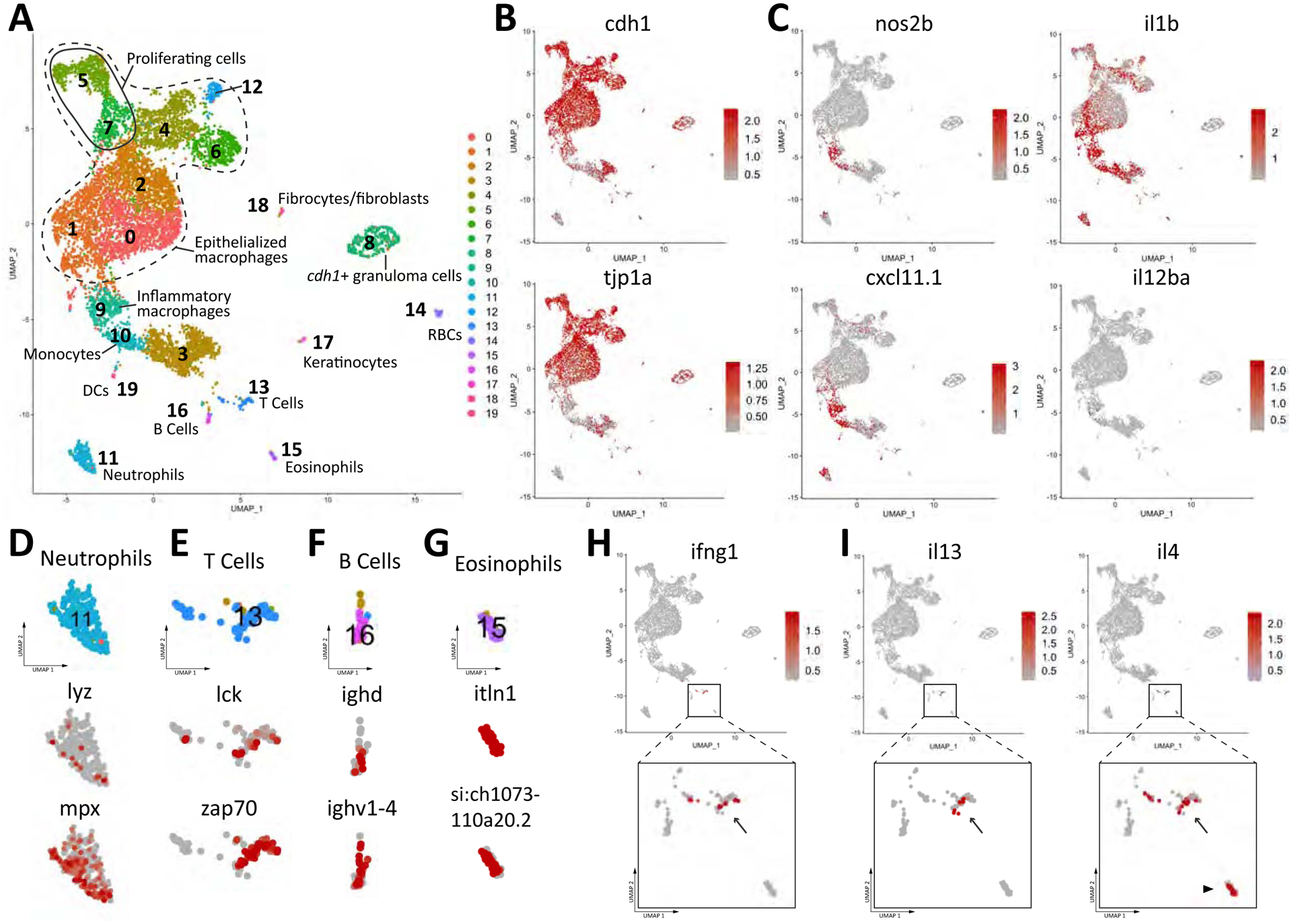
(A) UMAP plot of scRNA-seq data of 9273 cells from dissociated granulomas. (B) Expression map of the epithelial markers E-cadherin and ZO-1 (tjp1a) in cell populations isolated from the granuloma. (C) Expression maps of inflammatory markers within the cell populations of the granuloma. (D-G) Marker gene expression in individual clusters identifying these clusters as (D) neutrophils, (E) T cells, (F) B cells, and (G) Eosinophils. (H and I) Expression of type 1 and type 2 markers, respectively, within T cell (arrow) and eosinophil (arrowhead) populations. (H) IFN-γ. (I) IL-4 and IL-13.
At the cellular level, the predominant cell-types of the granuloma were subsets of macrophages that had undergone epithelial transitions. These cells were discernible by the expression of markers for epithelial cell-cell junctions such as E-cadherin (cdh1) and ZO-1 (tjp1) (Figure 1B, Figure S1 and S2A). In addition to epithelialized macrophages, we detected inflammatory macrophages marked by canonical genes such as iNOS (nos2b), IL1β, CXCL11 (cxcl11.1, (Rougeot et al., 2019)) and IL12 (IL12ba) (Figure 1C and S1). Using canonical markers for other cell types, we were able to identify T cells (lck, zap70), B cells (cd79a, ighv), and neutrophils (lyz, mpx) as well as eosinophils based on genetic signatures identified by scRNA-seq (Baron et al., 2019) (Figures 1D–1G, S1 and S2C–F). The relatively limited numbers of T cells observed is consistent with previous reports that fewer T cells are recruited to zebrafish granulomas than are seen in other models (Swaim et al., 2006). However, as in mammalian models, the T cells present play a role in control of infection (Parikka et al., 2012; Swaim et al., 2006). Finally, we also observed discrete clusters of epithelioid macrophages that appeared to be undergoing proliferation by the expression of proliferating cell nuclear antigen, pcna, (clusters 5 and 7) (Figures 1A and S1). Active proliferation of cell populations within the granuloma was noted in classical works although how this proliferation contributes to the granuloma itself is still an open question (Spector and Lykke, 1966; Spector and Wynne, 1976). A comprehensive list of cell clusters and transcripts is presented in Table S2.
Discrete Type 1 and Type 2 Signatures within Granulomas
We found that there was expression of both canonical type 1-associated transcripts (IFN-γ, IL-12, IL1β) and type 2 signals (IL-4 and IL-13) within the granulomas (Figures 1H and 1I). IFN-γ transcripts in the granuloma were found exclusively in T cells, while IL-12 and IL1β transcripts were produced by populations of inflammatory macrophages within the granuloma. Similarly, IL-4 and IL-13 transcripts were both made by T cells within the granuloma (and largely distinct from the populations of T cells that make IFN-γ (Figure S2B)) while IL-4 transcripts were also produced by eosinophils (Figure 1I). These results demonstrate that, despite the classical view of the granuloma as an exclusively type 1 structure, the mycobacterial granuloma accesses both type 1 and type 2 signaling pathways.
We have previously established that epithelialization is required for organized granuloma formation (Cronan et al., 2016). While mycobacterial granuloma formation has been thought to draw on type 1 signals, we found that populations of macrophages that highly expressed type 1 markers such as iNos, CXCL11, and IL1β did not express markers for epithelialization including E-cadherin and ZO-1 (Figure 2A and 2B). These results suggested that type 1 polarization may be antithetical to macrophage epithelialization within these macrophages themselves. In humans, highly pro-inflammatory signatures are restricted to the innermost layer of macrophages (Marakalala et al., 2016). Previous work in cell culture and helminth infections in vivo have demonstrated that type 2 cytokines can drive E-cadherin and other epithelial marker induction in macrophages, while type 1 cytokines could suppress this response (Van den Bossche et al., 2009; Van den Bossche et al., 2015). Beyond the absence of type 1 markers in the epithelialized macrophages, we observed some characteristics of type 2 polarization. While arginase-1 expression in zebrafish M2 macrophages has not been reported, arginase-2 is associated with M2-skewed macrophages in fish (Edholm et al., 2017), and we observed arginase-2 expression as well as expression of other recently described fish M2-associated markers (Rougeot et al., 2019; Wentzel et al., 2020) in the clusters of epithelialized macrophages (Figures 2C and S2G). As we observed type 2 cytokine signaling within the cells of the granuloma, we wondered if a role for type 2 immunity in the mycobacterial granuloma had been previously overlooked.
Figure 2. Type 1 Macrophage Polarization Negatively Correlates with Macrophage Epithelialization.
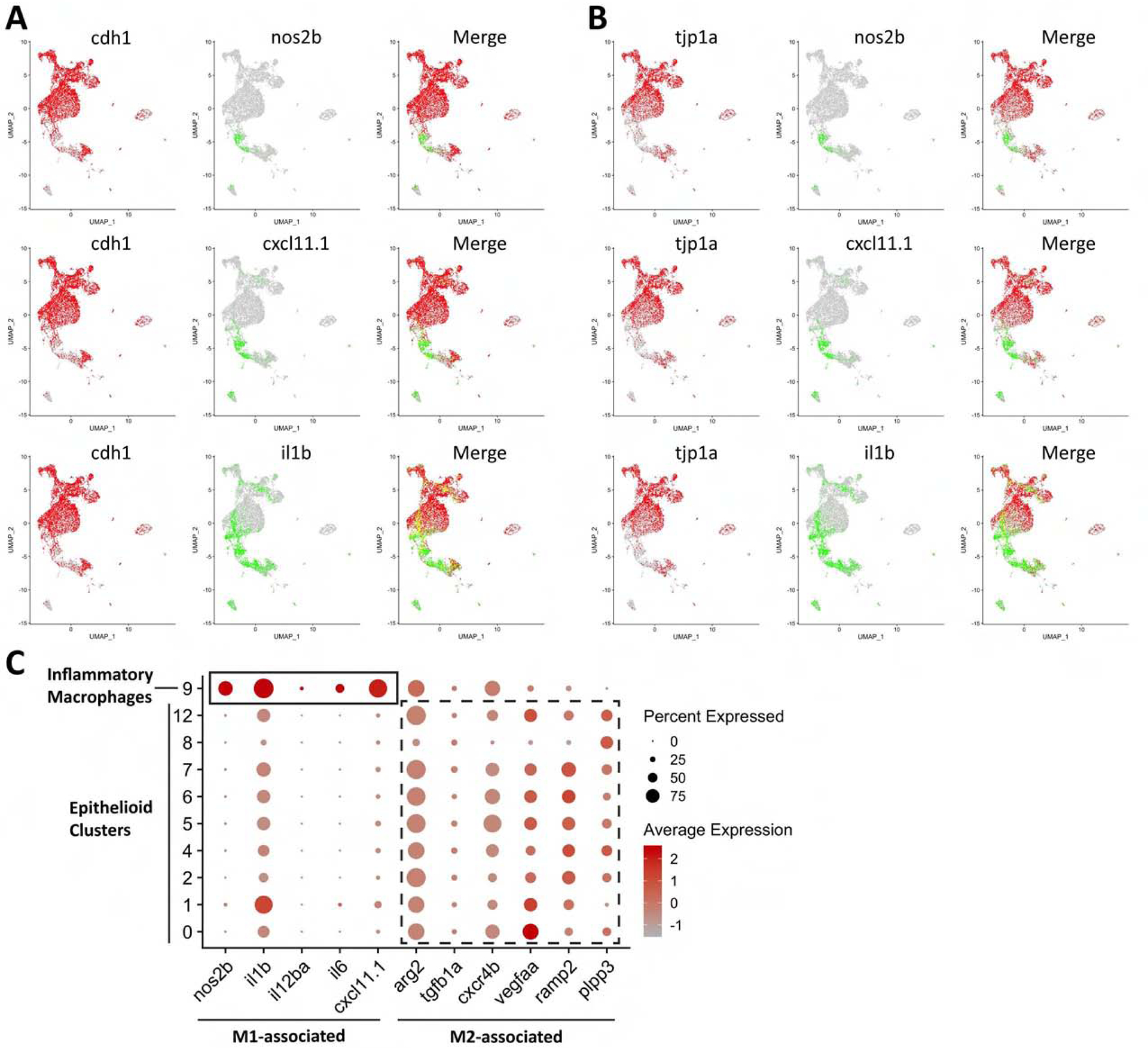
UMAP plots of co-expressed scRNAseq data showing left panels - epithelial markers (A) E-cadherin (cdh1) and (B) ZO-1 (tjp1) in red, center panels - the type 1 markers iNOS (nos2b) CXCL11 (cxcl11.1) and IL1β (il1b) in green and right panels – the overlay of these two populations with co-expression in yellow. The epithelialization markers E-cadherin and ZO-1 and the type 1 markers iNOS, CXCL11, and IL1β are expressed in largely exclusive populations. (C) Dotplot of canonical M1-associated markers (solid box) in the inflammatory macrophage cluster and M2-associated markers (dashed box) in epithelioid clusters. The M1-associated markers are largely expressed in the inflammatory macrophage cluster while M2-associated markers are largely expressed in epithelioid cell clusters.
Alternative Activation is a Feature of Necrotic Granulomas
We investigated if a type 2 immune response was also associated with granuloma formation in other experimental models of organized mycobacterial granuloma formation. Previous work has identified substantive type 2 signals present in well-organized, necrotic granulomas in macaques, humans, and rabbits (Huang et al., 2015; Mattila et al., 2013; Subbian et al., 2011). We assessed whether macrophages with markers of these type 2 signals associated with epithelialization in the granuloma. Macaques infected with Mtb have emerged as a particularly faithful model of human Mtb infection (Bucsan et al., 2019b; Lenaerts et al., 2015). Our previous work established that epithelialization is crucial to organized necrotic granuloma formation (Cronan et al., 2016), and we focused on E-cadherin as a marker of engagement of these granuloma forming pathways. We initially investigated whether E-cadherin was induced within the macrophages in macaque granulomas. Consistent with our previous findings in humans, we found that E-cadherin was induced within CD68-positive macrophages in the granuloma (Figure 3A).
Figure 3. Type 2 Activation Associates with Macrophage Epithelialization in Macaque Granulomas.
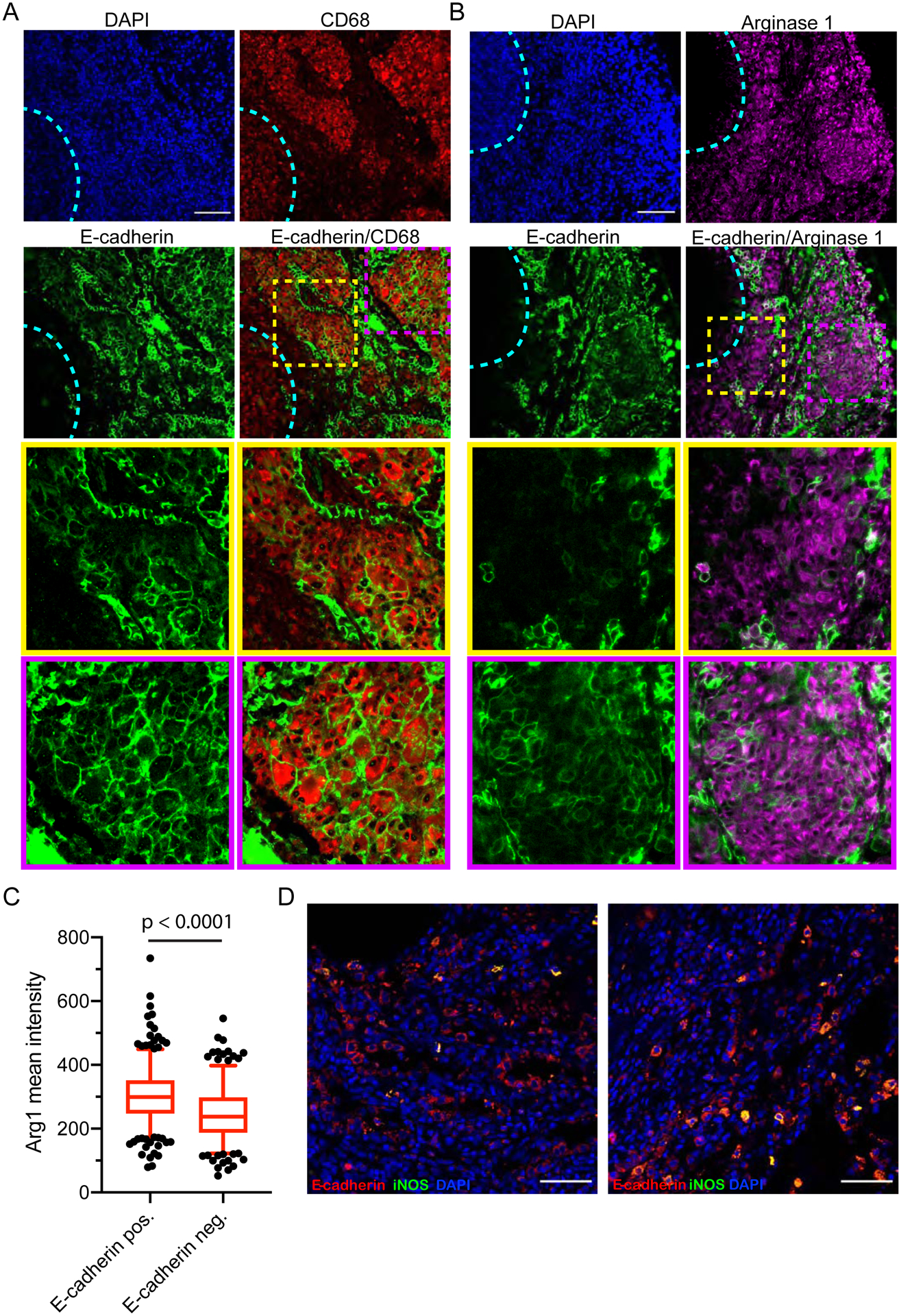
(A) Representative image of a macaque granuloma stained for the epithelial marker E-cadherin (green) and the macrophage marker CD68 (red) demonstrating E-cadherin within the granuloma macrophages in macaque lung granulomas. Nuclei were counterstained with DAPI to identify regions of the granuloma that were necrotic (outlined in the dotted cyan line). Areas in the yellow and purple dotted boxes are shown in close up in the images below, indicated by the yellow and purple borders around each image. Scale bar – 100 μm, images representative of granulomas from 2 animals. (B) Staining of a macaque lung granuloma for the type 2 marker arginase-1 (magenta) and epithelial marker E-cadherin (green) and nuclei (DAPI, blue), revealing the association of E-cadherin with regions of the granuloma with high arginase 1 staining. The necrotic region of the granuloma is indicated with the cyan dotted line. The dotted yellow and purple boxes indicate the regions magnified in the yellow and purple boxes to the right. Scale bar – 100 μm, representative of granulomas from 2 animals. (C) Mean intensity of arginase-1 staining was measured in cells that were E-cadherin positive and cells from regions of the granuloma that were E-cadherin negative. Whiskers indicate 5th and 95th percentile and outliers are displayed as points. Data from 379 E-cadherin-positive cells and 279 cells from E-cadherin negative regions. Results are pooled from 4 granulomas. Populations compared by unpaired t-test. (D) Macaque granulomas co-stained for E-cadherin (red) and iNOS (green). Nuclei are counterstained with DAPI. Arrows indicate E-cadherin-positive cells that are iNOS negative. Two separate granulomas displayed.
To investigate the inflammatory status of the E-cadherin-positive macrophages, we stained macaque granulomas for the type 2 marker arginase-1 along with E-cadherin (Figure 3B). We found that E-cadherin positive cells localized to regions of the granuloma that were arginase-1 positive, consistent with a type 2 response being associated with epithelialization. Arginase staining was low immediately adjacent to the necrotic core but extensive arginase-1 expression was observed in the more distal cell layers from the necrotic core (Figure 3B), consistent with previous findings in macaques (Mattila et al., 2013). We observed little E-cadherin proximal to the necrotic core of the granuloma – presumably where the most inflammatory population of cells resides. We observed consistent co-staining of arginase-1-positive cells with E-cadherin, suggesting it is the macrophages receiving type 2 signals that undergo the characteristic epithelialization. Within multiple epithelialized granulomas, quantitation of arginase-1 levels in E-cadherin-positive cells versus cells in regions of the granuloma that were E-cadherin negative demonstrated that arginase-1 levels were higher in the E-cadherin cells (Figure 3C).
We also investigated whether E-cadherin was induced in type 1 polarized macrophages. Co-staining for E-cadherin and the type 1 marker iNOS identified many E-cadherin-positive cells within the granuloma that are negative for iNOS (Figure 3D). These findings further support an inverse relationship between type 1 immunity and macrophage epithelialization, consistent with our findings in zebrafish. Taken together with our findings on arginase expression, these results support a potential role for type 2 signaling during macrophage-epithelial transition and granuloma formation during Mtb infection.
Stat6 Signaling is Required for Necrotic Granuloma Formation and Macrophage Epithelialization
To investigate the functional role of type 2 immune responses in the granuloma, we generated zebrafish deficient in the type 2 signaling molecule stat6 by CRISPR/Cas9 mediated knockout (Figure S3A). Stat6 is the canonical transcription factor downstream of IL-4 and IL-13 activation and signaling through the IL4R. stat6 transcripts are expressed broadly in most cell types in the granuloma (Figure S2H). We initially targeted stat6 rather than the upstream IL-4 receptor α (IL4Rα), as il4ra has undergone a tandem duplication event in teleost evolution, complicating any genetic targeting (Zhu et al., 2012). This stat6 mutant allele, termed stat6xt53, has a 10 base pair deletion immediately downstream of the Stat interaction domain and upstream of the Stat coiled coil domain of stat6 resulting in a predicted frameshift and premature truncation of the stat6 protein prior to the DNA binding region (Figures S3B and S3C).
Adult wildtype and stat6-deficient siblings were infected with M. marinum, and granuloma morphology was assessed in these animals. While wildtype (WT) animals formed highly organized necrotic granulomas, consistent with previous descriptions, there was a complete absence of organized, necrotic granuloma formation (Figure 4A) in stat6-deficient animals. In total, we identified organized necrotic granulomas in every WT animal investigated (9 total animals, 54 total organized necrotic granulomas out of 186 total granulomas observed, 8 to 41 granulomas observed per animal) (Figure 4B). However, in stat6 deficient siblings not a single organized necrotic granuloma was identified within a similarly large group of granulomas (8 animals analyzed, 0 organized necrotic granulomas out of 158 infection foci analyzed, 7 to 31 infection foci imaged per animal, mean 19.8) (Figure 4B). The loss of organized granuloma formation in stat6-deficient animals was not due to failure of immune recruitment to the bacteria as staining for the pan-leukocyte marker l-plastin revealed robust recruitment of leukocytes to the bacteria (Figure 4A). Similarly, the lack of organized necrotic core formation was not due to the absence of cell death within the granuloma as nuclear staining revealed numerous pyknotic nuclei at sites of mycobacterial infection in stat6 deficient animals (Figure S4). Taken together, these results demonstrate that contrary to expectation, the mycobacterial granuloma requires Stat6-dependent type 2 immune signals for its organization.
Figure 4. Stat6 is Required for Necrotic Granuloma Formation and Macrophage Epithelialization.
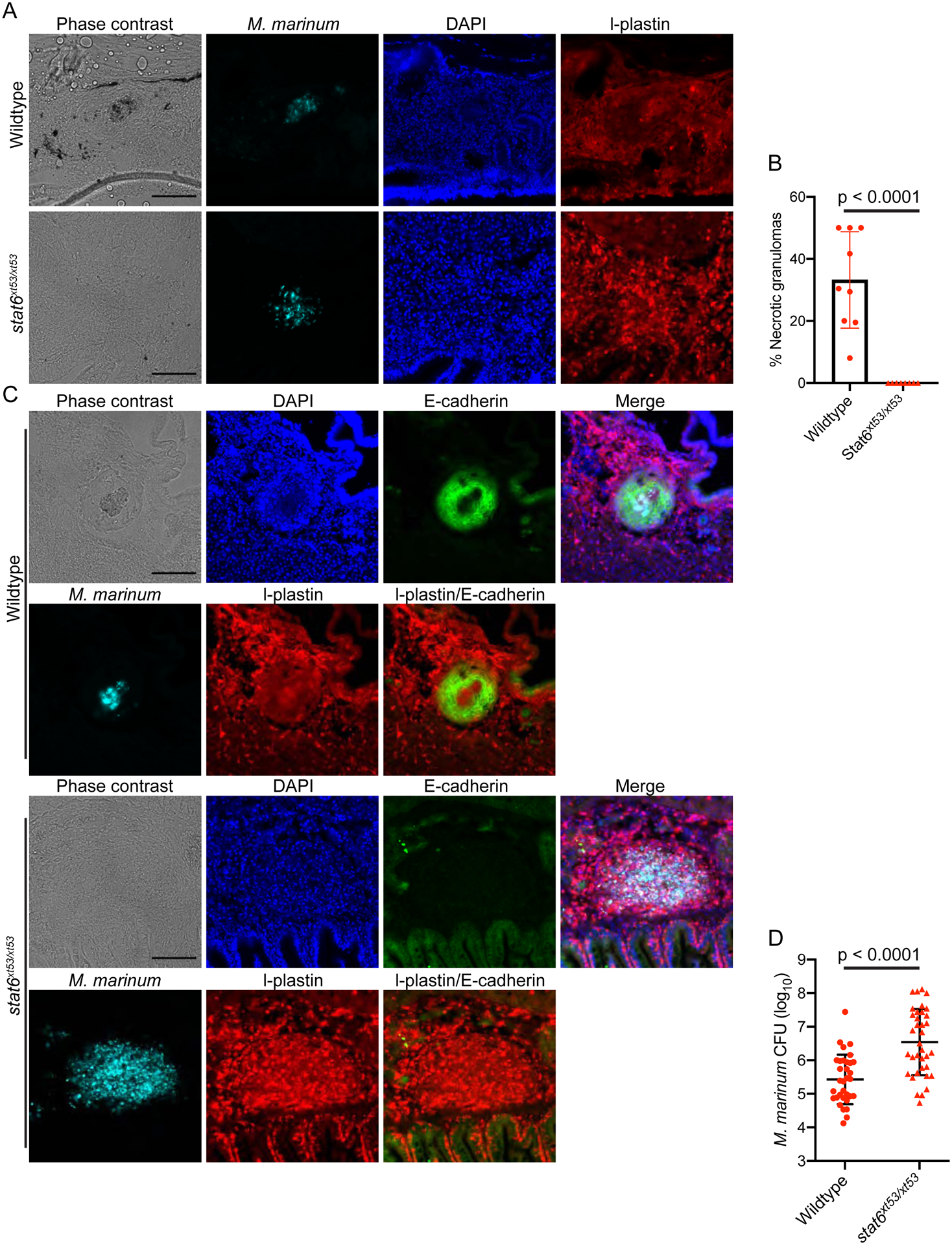
(A) Representative phase contrast and fluorescent images of granulomas in wild type and stat6xt53/xt53 animals infected with cerulean fluorescent protein expressing M. marinum (cyan). Tissue was stained for l-plastin-positive leukocytes (red) and counterstained with DAPI to image nuclei (blue). Scale bar – 100 μm, representative of granulomas from 3 experiments, WT 9 animals total, stat6 8 animals total. (B) Quantitation of the percentage of granulomas or infection foci in each animal that were necrotic for n = 9 animals for WT and n = 8 animals for stat6xt53/xt53 animals. The bar represents mean and each point is the percentage of necrotic granulomas in a single animal. Pooled data from 3 independent experiments. (C) Phase contrast and fluorescent images of granulomas formed in wild type and stat6xt53/xt53 animals infected with cerulean expressing M. marinum (cyan). Granulomas were stained with the pan-leukocyte marker l-plastin (red) and the epithelial marker E-cadherin (green) while nuclei were counterstained with DAPI (blue). Scale bar – 100 μm, representative of results from 3 independent experiments, 9 total animals for WT and 8 total animals for stat6. (D) Bacterial numbers in WT and stat6xt53/xt53 animals enumerated by CFU assay at 2 wpi. Data is pooled from 3 independent experiments from a total of 33 WT animals and 36 stat6xt53/xt53 animals.
We have previously found that a macrophage-epithelial transition is required for formation of organized granulomas (Cronan et al., 2016). This transition involves the induction of classical epithelial markers including E-cadherin and formation of epithelial-like cell junctions within the macrophages of the granuloma. We sought to assess whether the infected foci in stat6 deficient animals underwent this transition. In WT animals, E-cadherin staining revealed a network of E-cadherin positive cell-cell junctions surrounding the necrotic core of the mycobacterial granuloma (Figure 4C). By contrast, stat6 deficient animals exhibited a complete loss of the organized network of E-cadherin-positive adherens junctions that was observed in WT granulomas (Figure 4C). In the stat6-deficient animals, only sporadic E-cadherin positive cells were present at the macrophage aggregate. These few E-cadherin-positive cells were negative for the leukocyte marker l-plastin and morphologically resembled WT epithelial tissues, suggesting that these E-cadherin positive cells were epithelial tissues that had been intercalated into the early macrophage aggregates. Taken together, these results suggest that, contrary to the classical model of the type 1-exclusive Mtb granuloma, the mycobacterial granuloma requires type 2 signals to organize and drive macrophage-epithelial transformation.
To assess the functional consequences of stat6 deficiency within the granuloma, we investigated how lack of stat6 altered bacterial numbers within WT and stat6-deficient animals. Stat6-dependent alternative activation is generally thought to be host-detrimental in mycobacterial infection by reducing IFN-γ and type 1 responses. However, how would changes in the type 1/type 2 balance interact with a complete lack of containment and absence of granulomas? We found that stat6xt53/xt53 animals had markedly enhanced bacterial burden by CFU assay (Figure 4D). This finding indicates that stat6-dependent signaling can play a host beneficial role during infection in animal models where granuloma formation is an important feature.
Granuloma Formation and Epithelialization is Dependent on Stat6 Signaling Within Hematopoietic Populations
Outside of immune populations, Stat6 is known to regulate the response of many other cell types to alternative activation signals (Goenka and Kaplan, 2011). To specifically investigate the requirement for stat6-dependent signals within the hematopoietic compartment, we performed hematopoietic stem cell transplants by transplanting whole kidney marrow (WKM) into HSC-deficient c-myb mutants (Hess et al., 2013). To track donor marrow and their progeny, WT and stat6 deficient animals were crossed to a ubiquitous transgene driven by the ubiquitin B promoter, Tg(ubb:tdTomato)xt23, as well as a fluorescently tagged version of the adherens junction complex member plakoglobin, Gt(jupa-citrine)ct520a (plakoglobin-citrine) to track adherens junctions formation specifically within hematopoietic derived cells (Figure 5A). Juvenile c-myb deficient animals were transplanted with either stat6wt/wt;Tg(ubb:tdtomato)xt23;Gt(jupa-citrine) (referred to as WT WKM) or stat6xt53/xt53;Tg(ubb:tdtomato)xt23;Gt(jupa-citrine) WKM (referred to as stat6 deficient WKM) at 2 weeks post-fertilization and allowed to grow to adulthood.
Figure 5. Hematopoietic Stat6 Signaling is Required for Necrotic Granuloma Formation and Epithelialization.
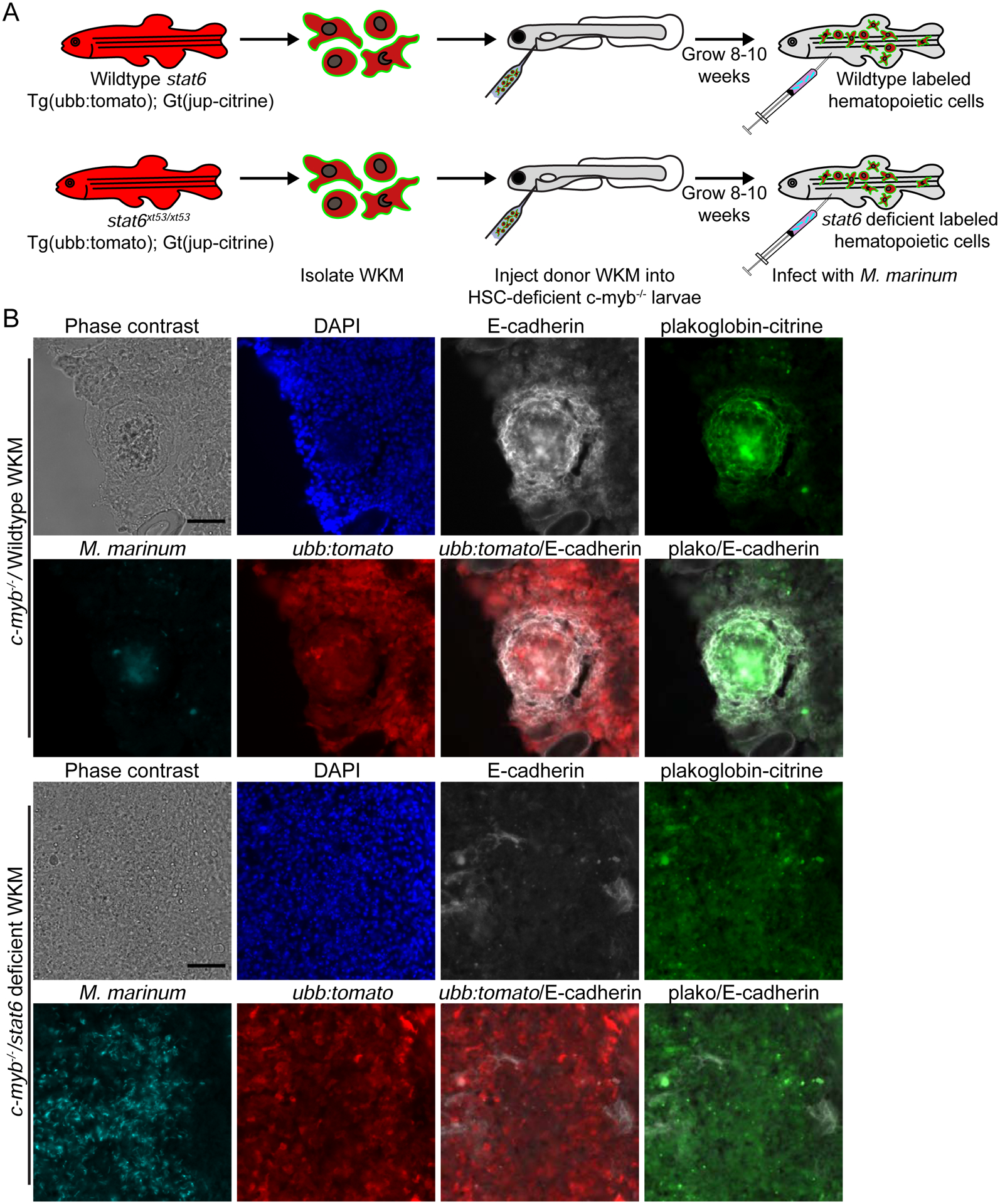
(A) Schematic of the kidney marrow transplant approach used to reconstitute HSC deficient animals with either WT or stat6xt53/xt53 labeled marrow populations. (B) Phase contrast and fluorescent images of granulomas in HSC-deficient myb−/− fish transplanted with either WT; ubb:tomato; plakoglobin-citrine WKM or stat6xt53/xt53; ubb:tomato; plakoglobin-citrine WKM. Fluorescent images show infecting mycobacteria (cyan), nuclei (DAPI, blue), ubiquitious ubb:tomato transgene expressed in hematopoietic cells (red), adherens junctions in hematopoietic cells (plakoglobin-citrine, green) and stained for the epithelial marker E-cadherin (white). Scale bars – 50 μm. Images are representative of results from 5 animals (WT) or 4 animals (stat6) total from 2 independent experiments.
Adult WKM transplanted animals were subsequently infected with M. marinum, and granuloma formation was assessed. Infected animals transplanted with WT WKM formed necrotic granulomas surrounded by a network of E-cadherin-positive cells. These E-cadherin-positive cells surrounding the necrotic core were hematopoietic in origin, as these cells were marked with the ubiquitous Tomato transgene and E-cadherin-positive junctions were also marked by the fluorescent plakoglobin genetrap expressed within donor WKM tissue (Figure 5B). Stat6 was definitively required in the hematopoietic compartment for granuloma formation. Animals reconstituted with stat6 deficient WKM failed to form necrotic granulomas despite the extensive recruitment of hematopoietic cells labeled by the hematopoietic encoded ubiquitous tdTomato transgene (Figure 5B). Consistent with an absence of necrotic granuloma formation, we found minimal E-cadherin-positive adherens junction formation in granulomas from animals transplanted with stat6-deficient WKM. Of the few E-cadherin-positive adherens junctions that occurred in animals transplanted with stat6-deficient WKM, none of these junctions were labeled by the hematopoietic encoded plakoglobin gene trap or the ubiquitous tdTomato transgene, supporting the conclusion that stat6 is required for epithelialization within the hematopoietic populations composing the granuloma. These results indicate that any E-cadherin-positive cells within the stat6-deficient granulomas are merely epithelium that has been intercalated within this structure, consistent with our l-plastin staining data. Taken together, these results demonstrate that stat6-dependent alternative activation pathways are required for both necrotic granuloma formation and epithelioid transformation of granuloma macrophages.
Stat6 Signaling Functions Cell-Autonomously to Drive Macrophage-Epithelial Transition
The enhanced burden we observed in stat6 deficient animals raised the potential that the absence of necrotic granuloma formation in stat6 deficient animals could be a secondary effect due to exuberant bacterial growth and granuloma breakdown, as has been observed with other cytokines such as TNF (Clay et al., 2008). At multiple earlier timepoints, we were unable to observe such an effect. However, to rule out any possible indirect effects and to formally assess whether stat6 functioned cell autonomously to drive macrophage epithelialization within the granuloma, we generated mixed kidney marrow chimeras in which c-myb-deficient fish that lack HSCs were reconstituted with a mixture of stat6-deficient labeled WKM and wildtype unlabeled WKM (termed WT/stat6) or wildtype labeled WKM with wildtype unlabeled WKM as a control (termed WT/WT). The WKM donor populations were labeled as previously, where cells were ubiquitously labeled with tomato fluorescent protein while any adhesion junctions formed within would be marked by the fluorescently labeled plakoglobin genetrap (Figure 6A).
Figure 6. Stat6 is Required Cell Autonomously for Macrophage Epithelialization and Association with the Epithelialized Regions of the Granuloma.
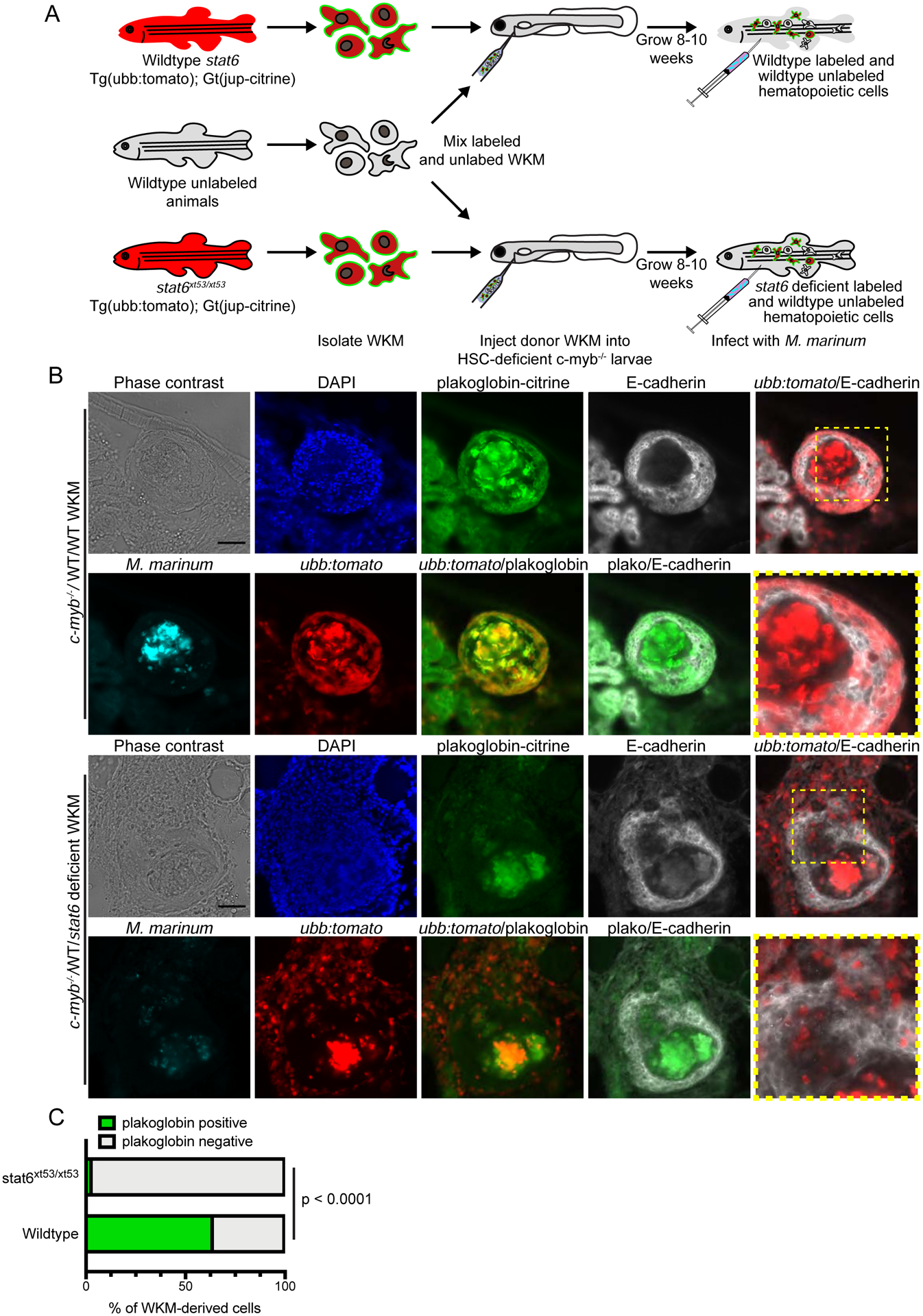
(A) Schematic of the kidney marrow transplant approach used to generate mixed kidney marrow chimeras. (B) Representative phase contrast and fluorescent images of granulomas formed in WT/WT and WT/stat6 animals. Fluorescent images show M. marinum (cyan), nuclear fluorescence (DAPI, blue), hematopoietic cells derived from the labeled transplant cells (ubb:tomato, red), adherens junctions in hematopoietic cells derived from the labeled transplant cells (plakoglobin-citrine, green) and stained for the epithelial marker E-cadherin (white). In a single WT/stat6 animal, very infrequent plakoglobin-positive, stat6-deficient cells were observed, demonstrating that WT cells could rarely rescue epithelialization in trans. Scale bar – 50 μm. Images are representative of results from 4 total animals each for WT and stat6 from 3 independent experiments. (C) Quantitation of adherens junction formation in macrophages from transplanted wildtype or stat6-deficient hematopoietic cells. Adherens junctions within the transplanted cells were quantitated using the genetically encoded plakoglobin genetrap. Pooled data from 578 wildtype cells and 539 stat6-deficient cells from 3 independent experiments. Populations compared by Fisher’s exact test.
We infected WT/WT and WT/stat6 WKM transplant animals with M. marinum and assessed granuloma macrophage epithelialization and granuloma formation. Both WT/WT and WT/stat6 WKM transplant animals were able to form necrotic granulomas, indicating that reconstitution of a population of wild type cells was sufficient to restore necrotic granuloma formation in the presence of stat6 deficient WKM. Although WT/stat6 WKM transplants regained necrotic granuloma formation, these granulomas were predominantly composed of unlabeled WT donor tissue (Figures 6B and 6C). Labeled stat6 deficient populations were largely excluded from the organized area of the necrotic granuloma. In contrast, WT/WT WKM transplants contained unlabeled and labeled populations of wildtype cells that were readily admixed within necrotic granulomas (Figures 6B and 6C). Similarly, visualizing adherens junction formation using the genetically encoded plakoglobin gene trap in stat6 or WT control WKM-derived populations, we found that the stat6 WKM derived cells were almost completely negative for adherens junction formation whereas the labeled WT populations in WT/WT WKM transplants were strongly positive for adherens junction formation (Figures 6B and 6C).
To further investigate granuloma epithelialization in these mixed chimeras, WT/WT and WT/stat6 WKM transplanted animals were stained for E-cadherin to mark the epithelialized network within these necrotic granulomas. In WT/WT mixed chimera animals, we found an organized network of epithelialized macrophages surrounding the necrotic core, consistent with our previous findings. The network was composed of interleaved wildtype labeled and unlabeled tissue (Figure 6B, right panels). The WT/stat6 animals also possessed a network of epithelialized cells surrounding the necrotic core, consistent with these mixed chimeras being able to form necrotic granulomas. However, in agreement with the findings from the plakoglobin gene trap, the network of epithelialized macrophages was composed of the unlabeled WT WKM derived cells while fluorescently labeled stat6 deficient cells localized to defects in the E-cadherin network. These results show that stat6 is required cell autonomously for macrophage epithelioid transformation and involvement in the epithelialized network of cells surrounding the necrotic core.
Acute Stat6 Inhibition in Established Granulomas Results in Macrophage E-cadherin Delocalization
The cell-autonomous effect of stat6 mutations supported a direct role for this pathway in E-cadherin expression or localization. We next sought a non-genetic means to disrupt this pathway acutely. Using an explant technique called Myco-GEM, established granulomas can be excised and maintained in three-dimensional culture while retaining key features of their physiology for up to one week (Cronan et al., 2018). This approach enables high-resolution longitudinal imaging and pharmacological manipulation of these structures. To examine the effect of acute Stat6 inhibition, we applied the Stat6 inhibitor AS1517499 (Nagashima et al., 2007) directly to explanted granulomas and performed continuous imaging over multiple days. In vehicle-treated granulomas, granuloma macrophages maintained high levels of membrane-localized E-cadherin expression throughout the multi-day time-course (Figure 7A–C). In contrast, treatment with the Stat6 inhibitor resulted in delocalization and decreased expression of E-cadherin in the previously epithelialized macrophages (Figures 7A–C, S5, S6A, and Movies S1 and S2). After the loss of E-cadherin expression, macrophages were still viable and survived throughout the time course, retaining active motility (Figure S6A and Movie S2). To assess by a second method that the decreased E-cadherin localization and expression was not a consequence of cell death, we stained treated granulomas with propidium iodide to identify dying cells and found that there was no increase in rates of macrophage cell death within the treated granulomas (Figure S6B).
Figure 7. Acute inhibition of stat6 results in macrophage de-epithelialization.
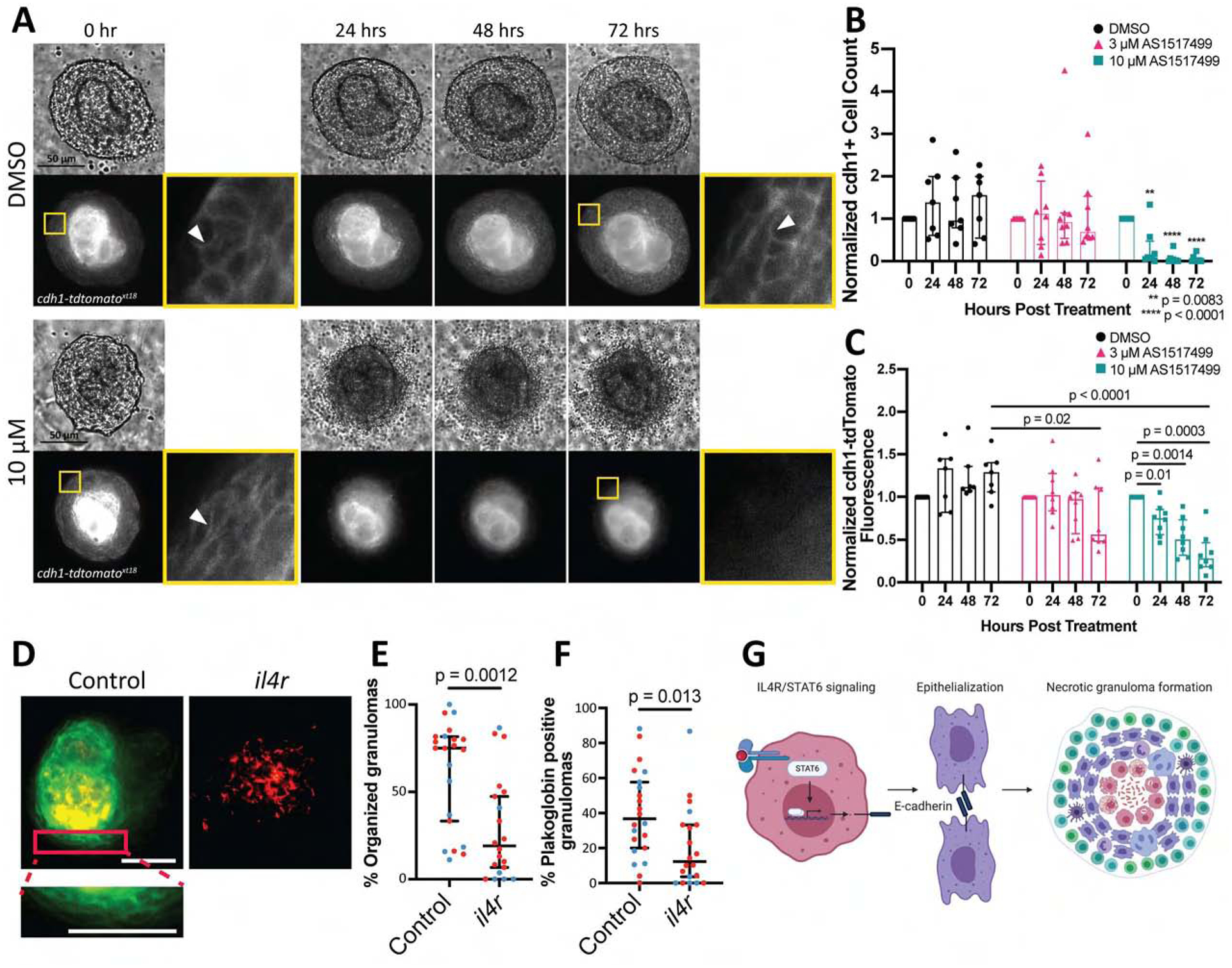
After explant of established, mature granulomas, vehicle or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. (A) Single z-plane, time-lapse images from DMSO (top) and 10 μM AS1517499 Stat6 inhibitor (bottom) treated explant granulomas from E-cadherin knock-in fish line (cdh1-tdtomato)xt18. Granulomas dissected from adult animals at 14 dpi. Yellow boxes show enlarged areas of granuloma cells, white arrowheads indicate E-cadherin fluorescence signals. (B) Number of E-cadherin positive cells, normalized to the cell count at the initial timepoint for each granuloma. n = 7 for DMSO, n = 8 for 3 μM and n = 8 for 10 μM. Bars represent median values with interquartile range and each point represents a single granuloma. P values calculated using Dunnett’s multiple comparison test with the initial timepoint set as the control. (C) Quantitation of Cdh1-tdTomato fluorescence in granulomas treated with vehicle or Stat6 inhibitor, normalized to the fluorescence value at initial timepoint for each granuloma. n = 7 for DMSO, n = 8 for 3 μM and n = 8 for 10 μM. Bars represent median values with interquartile range and each point represents a single granuloma. Dunnett’s multiple comparison test used for p values. Representative of two independent replicates. (D) Representative single z-plane images of granulomas in CLARITY-cleared control and il4r mosaic knockout plakoglobin-citrine (green) gene trap fish (Gt(jupa-citrine))ct520a infected with fluorescent M. marinum (red). Animals infected at 3 wpf and fixed for CLARITY clearing at 12 dpi. Bar, 50 μm. (E) Quantitation of the percentage of organized granulomas in CLARITY cleared control and il4r mosaic knockout plakoglobin-citrine gene trap animals. Median percentage of organized granulomas with 95% CI. (F) Quantitation of the percentage of plakoglobin-positive granulomas in CLARITY-cleared control and il4r mosaic knockout plakoglobin-citrine gene trap animals. Median percentage of plakoglobin positive granulomas with 95% CI. n = 22 for control and n = 20 for il4r mosaic knockout animals. For D-F, a total of 711 aggregates were analyzed for control animals and 446 for il4r knockdown. Data pooled from two independent experiments differentiated by red and blue dots for each animal. (G) Schematic of macrophage epithelialization and granuloma formation.
To confirm that stat6 inhibition indeed resulted in a general de-epithelialization of granuloma macrophages, we used an additional marker and infected a gene-trap line in which plakoglobin is directly tagged with the fluorescent protein citrine, Gt(jupa-citrine)ct520a. We observed the same delocalization of plakoglobin upon inhibitor treatment as we saw with E-cadherin (Figure S7) as well as a decrease in overall citrine expression, suggesting a general reprogramming of the epithelioid character of granuloma macrophages upon stat6 inhibition. Thus, there is continuous regulation of macrophage state by stat6 signaling, and acute inhibition can result in dramatic changes to macrophage phenotype and the overall nature of granuloma macrophages.
Inhibition of IL4R results in granuloma defects
Stat6 is the canonical transcription factor on which type 2 signaling associated with IL-4 and IL-13 converges, but there have been reports of Stat6 activation by signaling pathways beyond IL-4/IL-13 (Osorio et al., 2014). To examine the relevant upstream signals required for stat6 activation, we performed knockdown experiments using a pooled sgRNA CRISPR approach in juveniles targeting the receptor subunit, IL4Ra, common to both IL-4 and IL-13 signaling. In order to examine whole animals, we infected juveniles that had been injected with Cas9 and four pooled sgRNAs designed to target both IL4Ra copies in the zebrafish genome or matched sibling controls. This approach has been shown to create biallelic lesions that phenocopy known mutant phenotypes with no or minimal off-target effects (Jao et al., 2013; Wu et al., 2018). We found that CRISPR/Cas9-mediated knockdown of IL4R resulted in decreased granuloma organization, although, as expected, the effect was not as large as the non-mosaic stat6 mutant (Figure 7D–F). These experiments suggest that IL4R signaling upstream of Stat6 is similarly required for classically organized mycobacterial granulomas.
DISCUSSION
Granuloma formation is one of the central hallmarks of Mtb infection and underlies many of the difficulties in clinical treatment. Classically, it had been thought that the formation of the tuberculous granuloma draws exclusively upon type 1 signaling, typified by the IFN-γ signaling (Ndlovu and Marakalala, 2016; Sandor et al., 2003). This central role for type 1 signaling extends from observations that animals models and human patients deficient in type 1 cytokines like IFN-γ and TNF form poorly organized granulomas (Bean et al., 1999; Cooper et al., 1993; Emile et al., 1997; Flynn et al., 1995; Jouanguy et al., 1996). Similarly, CD4+ T-cells have been found to be important to granuloma organization and mycobacterial control (Caruso et al., 1999), effects that are thought to be driven by CD4+ T cell-derived IFN-γ. The association between CD4+ T cells and granuloma formation is also supported by the clinical observation that the extent of granuloma formation in HIV-TB coinfected patients correlates with CD4 T cell counts (Diedrich et al., 2016; Esmail et al., 2018).
However, contrary to the view that type 1 responses alone drive granuloma formation, we found that the mycobacterial granuloma utilizes type 2 signaling to coordinate granuloma formation and macrophage-epithelial transition despite the simultaneous presence of an organized type 1 response. Specifically, formation of the mycobacterial granuloma requires cell-autonomous stat6-dependent signaling to drive epithelialization of granuloma macrophages and orchestrate the formation of the necrotic granuloma (Figure 7G).
The mycobacterial granuloma has generally been viewed as a structure replete with type 1 signals, and we did identify substantial populations of granuloma macrophages with type 1 signatures via scRNA-seq (Figures 1 and 2). However, we also identified specific populations of T cells and eosinophils that expressed IL-4 and IL-13 within the granuloma. Studies in humans and model organisms with necrotic mycobacterial granuloma formation have found disparate results with regard to the type 1 and type 2 signals within the granuloma with some reports describing robust type 2 signaling while others finding that type 1 signaling predominates (Mattila et al., 2013; Mehra et al., 2010; van Crevel et al., 2002). In human TB, while there is robust production of IFN-γ there is also evidence of the presence of Type 2 immune responses, including IL-4 production in response to mycobacterial antigens among PBMCs isolated from patients with active TB (Surcel et al., 1994). Given the increasing recognition of the heterogeneity between granulomas and genetic heterogeneity between individuals, this may in part be due to variability in the host response depending on the state and age of individual granulomas as well as host-dependent effects (Esaulova et al., 2020; Lenaerts et al., 2015; Mehra et al., 2010; Subbian et al., 2011).
In the macaque model, where key aspects of human granuloma heterogeneity are reproduced, we found that epithelialized macrophages predominantly expressed the canonical type 2 marker arginase-1. iNOS-positive macrophages localized near the central inflammatory core of the granuloma, consistent with data from human granuloma specimens (Marakalala et al., 2016). The iNOS-positive macrophages near the core were largely negative for E-cadherin expression.
Classical inbred mouse strains fail to form the extensively organized necrotic granulomas observed in other models. Work within these mouse strains found little or no role for IL-4 and IL-13 signaling on infection outcome (Guler et al., 2015; North, 1998). One set of experiments in BALB/c mice with a high initial starting dose of 106 bacteria showed a host-protective effect of IL-4 depletion, attributed to skewing toward more classical type 1 signals (Buccheri et al., 2007). However, the biology of high-dose infection and the absence of organized granuloma formation in these strains may obscure key aspects of IL4R/Stat6 signaling in these experiments. Indeed, the absence of granuloma and infection phenotype in mice deficient for type 2 signaling may be due to the fact that many inbred mice have a Th1-skewed response in mycobacterial infection that may repress the type 2 response needed to direct macrophage epithelialization and necrotic granuloma formation. Consistent with this idea, we previously established that there is only limited E-cadherin induction within C57BL/6 macrophage aggregates (Cronan et al., 2016). Interestingly, Heitmann et al. demonstrated that overexpression of IL-13 in C57BL/6 mice was sufficient to drive more organized, necrotic granuloma formation (Heitmann et al., 2014). Similarly, necrotic granuloma formation has been observed in disseminated granulomas in mice that lack the type 1 response gene iNos, which is enhanced by blocking IFN-γ or TNF, suggesting that loss of type 1 effectors and cytokines may enable necrotic granuloma formation in mouse models (Reece et al., 2010).
Our findings are also consistent with previous work that demonstrated that the induction of E-cadherin and other epithelial markers in cell culture and in helminth infections requires IL-4, IL-13 and STAT6 (Van den Bossche et al., 2009; Van den Bossche et al., 2015). That the mycobacterial granuloma draws on the same type 2 signaling pathways that drive epithelial markers in cell culture as well as to helminth infection, demonstrates a potentially conserved macrophage epithelialization pathway despite the disparate inflammatory milieu of each of these responses.
Many inflammatory conditions and infections induce type 2 signaling but relatively few of these diseases drive granuloma formation. Thus, despite the requirement for type 2 immune signals for macrophage epithelialization and necrotic granuloma formation, we expect that other cytokines and signaling events are also required for granuloma formation. Work in cell culture demonstrated that other cytokines such as TGFβ and IL-10 can potentiate the induction of E-cadherin by IL-4 or IL-13 (Van den Bossche et al., 2009). Both TGFβ and IL-10 are present in Mtb lesions and in zebrafish granulomas (Bonecini-Almeida et al., 2004; DiFazio et al., 2016; Toossi et al., 1995), suggesting that synergy of IL-4 and IL-13 with these cytokines may be a reason why robust macrophage epithelialization is seen in mycobacterial granulomas but not in other environments where IL-4 and IL-13 predominate alone.
We establish that loss of stat6 results in a complete lack of organized granuloma formation, as well as enhanced bacterial numbers within infected animals. The exuberant growth of mycobacteria within stat6 mutant animals suggests that complete loss of granuloma formation in stat6 mutant animals is a negative outcome for the host. In contrast, partial disruption of granuloma formation by a macrophage-specific dominant negative E-cadherin construct in our previous work found that partial granuloma disruption could be host beneficial (Cronan et al., 2016). This discrepancy may be due to the means used to disrupt granuloma architecture in each approach – that partial disruption of granuloma formation using the dominant negative E-cadherin transgene may be host beneficial by allowing controlled immune access to the bacteria whereas the complete disruption of organized granuloma formation by stat6 deficiency may facilitate bacterial growth by compromising any physical containment or changing the metabolic environment normally present in the granuloma. Although stat6 deficiencies in mouse models generally do not show large differences in burden, presumably because of the absence of organized necrotic granulomas, it remains to be determined whether the stat6 deficiency has additional effects on bacterial burden or inflammatory state that might be obscured by the robust granuloma phenotype. Loss of type 2 immunity in stat6-deficient zebrafish may also push these animals into a hyperinflammatory state characterized by exuberant type 1 activation. Hyperinflammatory states have been associated with worse outcomes in a range of mycobacterial infections (Barber et al., 2019; Kauffman et al., 2021; Mishra et al., 2017; Roca and Ramakrishnan, 2013; Roca et al., 2019; Sakai et al., 2016; Tobin et al., 2012; Tobin et al., 2013). The cell autonomy experiments (Figures 5 and 6) suggest, however, that the epithelialization phenotype is not directly related to changes in inflammatory milieu.
Organized necrotic granuloma formation has been found to alter the metabolism of mycobacteria within the core, altering their chemotherapeutic sensitivity as well as limiting drug diffusion into the core (Gengenbacher et al., 2017; Lenaerts et al., 2015; Prideaux et al., 2015). The loss of macrophage epithelialization and necrotic granuloma formation in stat6 deficient animals raises the possibility that bacteria in these animals may be more accessible to chemotherapy. Treatment of mature granulomas with a small molecule inhibitor of stat6 resulted in dramatic effects on granuloma macrophages within 24 hours, including decreased E-cadherin expression and alterations in granuloma dynamics. Thus, it may be possible to use these stat6 inhibitors as an adjunctive therapy with existing anti-mycobacterial agents to disrupt granuloma barrier function. There are also many anti-mycobacterial agents that are effective in broth culture but are ineffective in vivo likely due to drug pharmacokinetics and tissue penetration or because of metabolic shifts in the bacteria. Disruption of the epithelioid macrophage network may enhance the activity of these normally inactive compounds or may enable increased immune access to the core. However, in any of these adjunctive approaches, it will be important to ensure that granuloma disruption does not enhance bacterial burden or promote dissemination. Deeper understanding of the signaling environment that supports epithelioid granuloma formation, structure, and dynamics will lead to new therapeutic opportunities for tuberculosis.
Limitations of the Study
Although important components of granuloma signaling pathways appear broadly conserved between zebrafish mycobacterial granulomas and non-human primates, the contribution of stat6-mediated signaling to human tuberculosis remains to be more fully examined, particularly in the context of granulomas within the lung environment. Functional and downstream analysis in other animal models where epithelioid, necrotic granulomas form will also provide further details on the conservation of this program. Additionally, the single-cell analysis presented here includes granulomas that were harvested at a single timepoint of infection and does not capture changes in their cell composition and gene expression as they mature or regress. Nonetheless, the stat6 results suggest a fundamental requirement for engagement of this pathway during mycobacterial granuloma formation and epithelialization.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, David Tobin (david.tobin@duke.edu).
Materials Availability:
All materials and lines generated in this study are available from the lead contact.
Data and Code Availability:
The scRNA-seq data generated during this study have been deposited at GEO under accession number GSE161712.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Statement
All zebrafish husbandry and experiments were approved by the Duke University Animal Care and Use Committee (protocol A122-17-05). All procedures in nonhuman primates, including Mtb infection, subsequent sampling and euthanasia were approved by the Tulane National Primate Research Center Institutional Animal Care and Use Committee (IACUC; protocol IDs P3712, P3751, and P3825).
Zebrafish Husbandry
Zebrafish were maintained on a 14 hour light/10 hour dark cycle. Water conditions within the system were maintained at 28 °C between pH 7.0–7.3 and conductivity 600–700 μS. Zebrafish were fed twice per day – once per day with dry food and once per day with Artemia.
Zebrafish Lines
All zebrafish strains used were in the *AB wildtype background. Tg(ubb:tdTomato)xt23, Gt(jupa-citrine)ct520a and (cdh1-tdtomato)xt18 zebrafish lines have been previously described (Cronan et al., 2016; Cronan et al., 2018; Trinh et al., 2011). The stat6xt53 and cmyb mutants were derived in this study as described below.
Bacterial strains
Mycobacterium marinum M strain expressing tdTomato or Cerulean fluorescent proteins have been previously described (Cambier et al., 2014; Oehlers et al., 2015). For infections, single cell aliquots of M. marinum were made and frozen according to the method of (Takaki et al., 2013). Briefly, M. marinum was grown to an OD600 of between 0.7 and 1.0. Single cell suspensions were generated by passing the bacteria through a tuberculin syringe with a 27G needle and subsequent filtration through a 5 μm filter. After bacterial enumeration, aliquots of single cell suspensions were frozen at −80 °C.
METHOD DETAILS
Preparation of single cell suspensions, sequencing, and analysis
Wildtype adult *AB animals, both female and male, were infected with approximately 350 fluorescent bacteria (FB) M. marinum by peritoneal injection. Granulomas were dissected from infected animals at 14 dpi as described previously (Cronan et al. 2018). Granulomas were transferred to a 1.5 ml tube and cells were dissociated using 1 ml of 0.05% trypsin/EDTA (ThermoFisher Scientific, 25300054) at 30 °C for 35 minutes with rocking. Cells were collected by centrifugation for 5 min at 300 xg, resuspended in 1x DPBS with 10% FBS on ice, and passed through a 40 μm strainer. Cell viability was measured by trypan blue staining. Single cell samples were submitted to the Molecular Genomics Core at Duke Molecular Physiology Institute for 10X Genomics library preparation and data analysis assistance. Library preparation was done using Chromium Single Cell 3’ GEM, Library and Gel Bead Kit v3. Sequencing was carried out at Duke Center for Genomic and Computational Biology. We followed our standard analytical pipeline for the single cell analysis. Briefly, we demultiplex raw base call (BCL) files generated by an Illumina sequencer into FASTQ before aligning them to the Ensembl zebrafish genome assembly GRCz11.96, performed filtering, barcode and UMI counting using the most current version of 10X’s Cell Ranger software. We then used Seurat (Butler et al., 2018) to perform quality control and linear dimensional reduction. Cells were grouped into an optimal number of clusters for de novo cell type discovery using Seurat’s FindNeighbors() and FindClusters() functions, graph-based clustering approach with visualization of cells being achieved through the use of UMAP plots. Differential expression of relevant cell marker genes is visualized on UMAP plots to reveal specific individual cell types.
Immunostaining of Macaque Tissue
Adult Indian rhesus macaques were exposed to infectious aerosols of Mtb CDC1551 as described earlier (Bucsan et al., 2019a; Gautam et al., 2018), and developed TB disease, characterized by granuloma formation in the lungs. Either at pre-specified endpoints designed to minimize pain and suffering, or at the end of the study, animals were humanely euthanized, and a complete necropsy conducted to retrieve lung samples. Paraffin embedded granuloma tissue was deparaffinized by incubating 2 × 10 min in xylenes followed by rehydration by dipping 20 times in 100% ethanol twice, dipping 20 times in 95% ethanol twice, 20 dips in 70% ethanol and 20 dips in 50% ethanol followed by 3 washes in deionized water. After rehydration, antigen retrieval was performed by pressure cooking for 5 min in 10 mM Tris/1 mM EDTA, pH 9.0. Slides were allowed to cool over an hour and were subsequently blocked in PBS/3% goat serum for 1 hour. After blocking slides were incubated overnight with primary antibody in PBS/3% goat serum. The following antibodies were used for staining – E-cadherin (rabbit anti-E-cadherin, Cell Signaling clone 24E10, cat# 3195S, 1:100) CD68 (mouse anti-CD68, Dako clone KP-1, cat# M0814 1:200) and Arg1 (mouse anti-Arg1, BD Biosciences clone 19, cat# 610708, 1:200). Slides were washed 3 times for 5 min in PBS and incubated for 1–2 hours with AlexaFluor conjugated secondary antibodies in PBS/3% goat serum. Slides were washed 3 times for 5 min in PBS again followed by background reduction and mounting using the Vector TrueView autofluorescence quenching kit with DAPI (Vector labs, cat #SP-8500).
For costaining of iNOS and E-cadherin in macaque lung sections, 5 μm thick sections on slides were baked at 65°C and subsequently de-paraffinized using Xylene followed by re-hydration with decreasing gradations of ethanol and dH2O. Antigen retrieval was carried out by heating the sections in Sodium citrate buffer (10mM, pH 6.0) for 20 min at 95°C. Blocking was done with 3 % BSA in TBST at RT. Rabbit iNOS polyclonal antibody (Thermo Fisher Scientific, USA, 1:20, 2 h at 37°C) and Goat Anti-Human/Mouse E-Cadherin antibody (R&D Systems, USA, 1:20, 2 h at 37°C) were used for identification of iNOS and E-cadherin, respectively. Secondary antibodies- donkey anti-goat IgG (H+L), Alexa Fluor 555 conjugate and chicken anti-rabbit IgG (H+L) and Alexa Fluor 488 conjugate (Thermo Fisher Scientific, USA, 1:400, 1 h at 37°C) were respectively used to label E-Cadherin and iNOS primary antibodies. For nuclear staining, specimens were incubated with DAPI (Thermo Fisher Scientific, USA, 1:5000, 5 min at 37°C) and then mounted with Prolong Diamond Antifade mountant (Thermo Fisher Scientific, USA). The stained sections were then visualized using Zeiss LSM 800 confocal microscope.
Generation of CRISPR-mutant Zebrafish Lines
Cas9 mRNA was made by in vitro transcription of T3TS-nls-zCas9-nls, a gift from Wenbiao Chen (Addgene plasmid # 46757; http://n2t.net/addgene:46757; RRID:Addgene_46757), using the mMessage mMachine T3 kit (ThermoFisher, AM1348). Cas9 protein was purchased from IDT. gRNAs were made according to the method of (Moreno-Mateos et al., 2015). Briefly, gRNA DNA templates were synthesized by annealing and filling in a gRNA target-specific primer with the CRISPRscan universal primer 5’- AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’. Resulting DNA templates for gRNA synthesis were PCR purified and gRNA was synthesized using the MEGAshortscript T7 kit (ThermoFisher) or HiScribe T7 kit (NEB). Targeting template for stat6 exon 5 was made using the target-specific primer 5’- taatacgactcactataGGGATGCCGTTGGTAGGTGGgttttagagctagaa-3’. gRNAs targeting exon 5 of c-Myb were generated using the gene-specific primer 5’- taatacgactcactataGGAAGTACGGCCCCAAACGTgttttagagctagaa-3’.
For injection of stat6-targeting gRNAs, single cell stage eggs were injected with ~1–2 nl of a solution containing 150 ng/μl Cas9 mRNA and 50 ng/μl stat6-specific gRNA. c-Myb editing was achieved by injecting single cell eggs with ~1–2 nl of a solution containing 160 ng/μl Cas9 protein, 50 ng/μl stat6-targeting gRNA in 150 mM KCl. This solution of gRNA and Cas9 protein was heated to 37 °C for 5 min prior to injection. Stat6 injected animals were grown to adult hood and outcrossed to generate F1 progeny. F1 progeny were subsequently screened by Sanger sequencing to identify mutations and a 10 bp deletion resulting in premature frameshift and protein truncation was identified.
Routine genotyping of stat6 mutant animals was performed by high resolution melt assay (HRMA). stat6 HRMA was performed using MeltDoctor HRM Master Mix (Thermo Fisher) along with the stat6-specific primers 5’-TATGCAGTTCCCTCCCTTCG-3’ and 5’- AGCTGATGAAGTGTTTGGCG-3’.
cmyb gRNA injected animals were screened by outcrossing injected animals followed by HRMA based genotyping of their progeny. cmyb mutations were detected using MeltDoctor HRM Master Mix together with the cmyb-specific primers 5’- GGTTTCAGGTGATTGAGTTGGTG-3’ and 5’- CTGCTTTCCGATTCGCCC-3’.
il4r CRISPR Design
Four pooled guide RNAs targeting exactly conserved regions of both predicted il4r orthologues in zebrafish were designed and synthesized following the method of (Wu et al., 2018). Notably, the two annotated orthologues are highly conserved at even the nucleotide level, enabling each guide to target both of the two orthologues. Targeting templates for the il4r used the target-specific primers: 5’ taatacgactcactataGGGCTTGGCAGACGAGTGTGgttttagagctagaaatagc 3’; 5’ taatacgactcactataGGTGATCGGATGTCTTGCACgttttagagctagaaatagc 3’; 5’ taatacgactcactataGGGAAACTTTCATGTTACCTgttttagagctagaaatagc 3’; and 5’ taatacgactcactataGGCCAGGCCGTCTGTGATTCgttttagagctagaaatagc 3’.
WKM transplant in c-Myb Deficient animals
*AB, stat6wt/wt;Tg(ubb:tdtomato);Gt(jupa-citrine) and stat6xt53/xt53; Tg(ubb:tdtomato);Gt(jupa-citrine) animals were euthanized using Tricaine and kidneys were dissected. These donor kidneys were placed in 5 ml of 0.9x PBS/5% fetal bovine serum (FBS) in a GentleMACS C tube and dissociated using a GentleMACS cell dissociator with 2 runs of the program M_Spleen_1. These suspensions were filtered through a 40 μm cell strainer and collected by centrifugation. Cells isolated from each kidney were enumerated using a hemocytometer and the cells were resuspended at a concentration of at least 1 × 105 cells/μl. For mixed WKM chimeras, *AB WKM was mixed with equal cell numbers of either WT WKM or stat6 deficient WKM.
To generate cmyb-deficient animals for transplant recipients, cmyb gRNA injected animals that displayed high degrees of genome editing were incrossed and cmyb deficient animals were identified by the greatly diminished blood cell numbers at 14 days post fertilization (dpf) as has previously been described. These 14 dpf cmyb deficient animals were subsequently injected with ~1/25 of the donor kidney marrow suspension and allowed to grow to adulthood (2–3 months post fertilization). Adult WKM injected animals were subsequently screened for engraftment prior to infection.
Adult Zebrafish Infection
Both males and females were used in roughly equal numbers and sex noted. Adult zebrafish were anesthetized with 120 mg/L Tricaine. Single cell aliquots of M. marinum were thawed and diluted in sterile PBS. Zebrafish were subsequently injected with 5–10 μl of a solution containing between 200–400 fluorescent bacteria. Infected zebrafish were maintained in spawning tanks and were fed daily. Water quality was maintained through daily water changes.
Zebrafish sectioning
Zebrafish were euthanized by immersion in lethal tricaine solution followed by decapitation. A small incision was made in the belly of the zebrafish to enhance permeability of fixative and fish were subsequently fixed in CLARITY hydrogel solution (4% paraformaldehyde, 4% acrylamide, 0.05% bis acrylamide, 0.0025 g/ml VA-044) for 2 days. After fixation, hydrogel was polymerized by incubation at 37 °C for 3 hours. Excess hydrogel was subsequently removed from the tissue and the tissue was equilibrated overnight in 10% sucrose/1x PBS. The tissue was subsequently equilibrated in 20% sucrose/PBS and 30% sucrose/PBS for 1 day each. Incubated for an hour in a 50/50 mix of Neg-50 and 30% sucrose/PBS and frozen in pure Neg-50. Zebrafish were subsequently sectioned at 20 μm on cryostat and stored at −80 °C.
Immunostaining of Zebrafish Tissue
Frozen section slides were dried for 10 min at room temperature prior to washing 3 times in 1x PBS. Slides were blocked for 1 hr in PBS/3% goat serum. After blocking, slides were incubated in primary antibody (mouse anti-E-cadherin, BD Biosciences, clone 36, cat# 610181, 1:100 and rabbit anti-l-plastin 1:2000 (Feng et al., 2012)) in PBS/3% goat serum overnight at 4 °C. Slides were washed 3 times for 5 min per wash and subsequently incubated with AlexaFluor conjugated secondary antibodies (cat# A-21429 and A32728, 1:1000) for 2 hours at room temperature.
Bacterial Burden Determination in Adult Zebrafish
At 2 weeks post infection, wildtype and stat6xt53/xt53 zebrafish were euthanized by Tricaine overdose. Zebrafish were placed with 1 ml of PBS/0.05% Tween-80 in individual 2 ml screwcap tubes with a single steel bead and homogenized by two 15 sec bursts in a bead mill. A dilution series of the resulting homogenates was plated on 7H10 supplemented with 50 μg/ml Hygromycin B, 4 μg/ml Amphoteracin B and 10% OADC supplement. Plates were grown for 2 weeks at 30 °C and counted.
Juvenile Zebrafish infections and CLARITY clearing:
For examination of il4r knockdowns, matched sibling controls or il4r CRISPR animals were infected as juveniles and examined in toto by confocal microscopy after CLARITY processing. Juvenile zebrafish (3 wpf) were anesthetized with 0.016% tricaine and ~ 250 fluorescent M. marinum were injected intraperitoneally per fish using a borosilicate needle. Infected fish were recovered in zebrafish system water and maintained in an incubator set to 28.5°C with a 14h light/dark cycle. At 12 dpi, the animals were euthanized, and CLARITY cleared as described previously with some modifications (Cronan et al., 2015). Briefly, euthanized zebrafish were incubated for 24 h in a solution of 4% PFA, 1% acrylamide, 0.05% bisacrylamide and 0.25% temperature-dependent initiator 2,20-Azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride in 1× phosphate buffered saline (PBS) at 4°C. The hydrogel was then polymerized by incubating for 3 h at 37°C. After polymerization, excess hydrogel was removed and the fish was cleared in 4% SDS in 200 mM boric acid, pH 8.5 at 37°C with shaking for seven days, replacing the SDS solution every other day. After clearing, the tissue was washed twice for 1 day in PBS, 0.1% Triton X-100 at 37°C. The fish was then incubated in refractive-index matching solution overnight before imaging.
Stat6 Inhibition in Zebrafish Granulomas
Adult Gt(jupa-citrine)ct520a and (cdh1-tdtomato)xt18 zebrafish were infected with approximately 400 fluorescent M. marinum by peritoneal injection. Granulomas were dissected from infected animals at 14 dpi and placed in Myco-GEM culture as previously described (Cronan et al., 2018). Granulomas were allowed to settle for 12 hours before application of vehicle (DMSO) or the Stat6 inhibitor AS1517499. Final DMSO concentration was 0.8% and AS1517499 final concentrations were 3 μM and 10 μM. Time-course imaging began at the onset of vehicle or inhibitor application and was done every 30 minutes for 22 hours over 4 consecutive days. Gt(jupa-citrine)ct520a and (cdh1-tdtomato)xt18 fluorescence was measured using ImageJ. Single z-plane images were used for fluorescence analysis of granuloma cells, which does not include the necrotic core and was corrected for background fluorescence. To score the number of E-cadherin-positive macrophages, images were randomized and scored blind.
Propidium Iodide Staining of Zebrafish Granulomas
Propidium iodide (ImmunoChemistry Technologies, LLC) was added to Myco-GEM cultures of vehicle treated (DMSO) or Stat6 inhibited (AS1517499) granulomas after 72 hours of treatment. Granulomas were allowed to stain for 1 hour and final propidium iodide concentration was 0.5% v/v.
QUANTIFICATION AND STATISTICAL ANALYSIS
Arginase Staining Intensity
In macaque sections, Arginase-1 staining intensity was quantitated by thresholding the nucleus by DAPI staining in FIJI/ImageJ. After thresholding, cytoplasmic rings were generated by generating a mask from the thresholded nucleus, dilating the mask, and subtracting the undilated nuclear mask from the dilated mask. The resulting mask was processed by using the analyze particles function in FIJI/ImageJ to ensure that individual objects were the size of single cells. Average Arginase-1 fluorescent intensity was measure in E-cadherin positive cells and compared to cells in E-cadherin-negative regions of the granuloma.
Granuloma Organization
To quantitate granuloma organization, frozen sections from wildtype and stat6xt53/xt53 animals were stained with DAPI and granulomas were identified by bacterial fluorescence. Every granuloma in a selected section was imaged and DAPI and phase contrast imaging were used to identify the number of organized necrotic granulomas within the animals.
Quantitation of Mixed Kidney Marrow Chimeras
Fluorescent images of granulomas in WT/WT and WT/stat6 animals were acquired. For each image, the region of the granuloma that was E-cadherin positive was identified from E-cadherin staining. Tomato-positive macrophages within the E-cadherin-positive region of granulomas in WT/WT and WT/stat6 animals were scored for the presence or absence of adherens junctions labeled by the genetically encoded plakoglobin-citrine gene.
Statistical Analysis
Single-cell RNAseq data were analyzed using Seurat v3. Statistical analysis was performed using Prism 8 (GraphPad Software). Statistical tests used and the resulting p values are indicated in the figures and figure legends.
Supplementary Material
Figure S1. Transcript Levels Across Granuloma Cell Clusters, Related to Figure 1
Violin plots showing the natural log of transcript numbers measured by sc-RNAseq in granuloma cell clusters.
Figure S2. Expression maps of Selected Transcripts and Canonical Markers in Specific Immune Cell Clusters, Related to Figures 1 and 2
Co-expression UMAP plots for select transcripts and whole UMAP plots for canonical markers. (A) E-cadherin (cdh1) and ZO-1 (tjp1) marks epithelialized macrophages. (B) T-cells expressing type 1 markers (ifng1) are largely distinct from T-cells expressing type 2 markers (il13, il4). Canonical markers of (C) neutrophils, (D) T-cells, (E) B-cells and (F) eosinophils shown in Figure 1D–G. (G) Expression map of arginase2 (arg2) and (H) stat6.
Figure S3. Generation of a stat6-Deficient Zebrafish Line, Related to Figure 4. (A) Schematic of the stat6 genomic locus and the targeting site of the gRNA used to generate the stat6xt53 mutant. (B) Sequence of the WT and Δ10 bp xt53 mutant stat6 sequences. Protein sequences are located above (WT) or below (xt53) the DNAs sequence. (C) Schematic of the frame shift and stop locations within the Stat6 protein.
Figure S4 Infection foci in Stat6-Deficient Zebrafish Are not Deficient in Cell Death, Related to Figure 4. Images of granulomas in stat6xt53/xt53 mutants demonstrating abundant pyknotic nuclei formation in the stat6 mutant. Area within the dotted yellow box is magnified and shown in greyscale within the yellow box on the right hand side. Cyan arrows indicate individual pyknotic nuclei within the stat6 deficient granulomas. Scale bar – 100 μm. Representative of 8 (stat6) total animals from 3 independent experiments.
Figure S5. Still images from Movie S1, Related to Figure 7. Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. Time-lapse images from Movie S1 of a granuloma treated with DMSO (top) and 10 μM AS1517499 (bottom) for the initial 22 hours of treatment, showing a reduction of E-cadherin (cdh1) during Stat6 inhibition. Scale bar is 50μm.
Figure S6. Granuloma macrophages exposed to Stat6 inhibition at 72 hours remain viable and motile with no increase in cell death, Related to Figure 7. Granulomas were dissected from adult (cdh1-tdtomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. (A) Still images from Movie S2, enlarged versions of the Movie S1, showing granuloma treated with 10 μM AS1517499. At timepoint 0, note membrane-localized E-cadherin expression. Pink arrows indicate an E-cadherin negative cell with retained motility, even after leaving the granuloma. White arrowheads indicate cells showing Cdh1-tdTomato delocalization. Scale bar is 50μm. (B) Representative images from DMSO, 3 μM, and 10 μM Stat6 inhibitor-treated explant granulomas stained with propidium iodide for 1 hour at 72 hours post treatment. Granuloma macrophages remain largely unstained and viable. The necrotic core of cell debris is stained brightly, and pink arrowheads indicate examples of propidium iodide staining. Scale bar is 100μm.
Figure S7. Acute inhibition of stat6 results in reduction of plakoglobin localization and fluorescence, Related to Figure 7. After explant of established, mature granulomas, vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. (A) Representative single z-plane, time-lapse images from DMSO (top) and 10 μM AS1517499 Stat6 inhibitor (bottom) treated explant granulomas from plakoglobin-citrine (green) gene trap fish (Gt(jupa-citrine))ct520a. Granulomas were dissected from adult animals at 14dpi. Scale bar is 50 μm. Blue boxes show enlarged areas of granuloma cells, arrowheads indicate plakoglobin fluorescence signals. (B) Quantitation of plakoglobin-citrine fluorescence in granulomas treated with vehicle or 10 μM AS1517499 Stat6 inhibitor, normalized to the fluorescence value at initial timepoint for each granuloma. n = 8 for DMSO and n = 8 for 10 μM. Bars represent median values with interquartile range. Dunnett’s multiple comparison test was used to calculate p-values.
Movie S1. Time-Lapse Imaging of Granulomas During Acute Stat6 Inhibition, Related to Figure S5.
Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. A representative granuloma from DMSO group (top) and 10 μM AS1517499 group (bottom) for the initial 22 hours of treatment, showing a reduction of E-cadherin (Cdh1) during Stat6 inhibition. Brightfield is on the left and cdh1-tdTomato fluorescence is on the right. Scale bar is 50μm.
Movie S2. Close-up of Time-Lapse Imaging of Granuloma Cells During Acute Stat6 Inhibition, Related to Figure S6
Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. A close-up of the representative granuloma from Movie S1 in the 10 μM AS1517499 group during the initial 22 hours of treatment, showing an E-cadherin negative cell with retained motility, even after leaving the granuloma. Time-lapse imaging also shows cells exhibiting Cdh1-tdTomato delocalization. Brightfield is on the left, cdh1-tdTomato fluorescence in the center and merged channels on the right. The yellow box indicates the area that is enlarged in the bottom panels. Only a portion of the 22-hour movie is shown. Scale bar is 50μm.
Table S1. Proportion of cell-types by cluster in scRNA-seq data, related to Figure 1.
Table S2. Global list of transcripts with mean expression value for each cluster, related to Figure 1.
Highlights.
Diverse cell populations and inflammatory cues characterize tuberculous granulomas
Granuloma macrophages that express E-cadherin display type 2 immune signatures
stat6 functions cell autonomously in granuloma macrophage epithelioid transformation
A type 2/Stat6 axis coordinates necrotic granuloma formation and epithelialization
ACKNOWLEDGEMENTS
We would like to acknowledge the contributions of K. Abramson, E. Hocke, and J. Gibson from the Duke Molecular Physiology Institute Molecular Genomics Core for the generation of the single cell libraries and assistance with analysis, E. Hortle and P.M. Cholan for additional technical assistance, C. Pyle for the Myco-GEM propidium idodide staining approach, M. Sweeney for figure artwork created with BioRender.com (Figure 7G), I. Padmanaban and A. Yu for zebrafish care, and J. Coers, A. Pagán, C. Smith, and members of the Tobin laboratory for helpful discussions and feedback. This work was funded by National Institutes of Health grants AI130236, AI125517, AI127115 (D.M.T.), AI111943, AI135726, AI134249, OD011133 (D.K.), OD011104 (D.K. and S.M.), a Vallee Scholar Award (D.M.T.), University of Sydney Fellowship G197581 and New South Wales Health Early-Mid Career Fellowship H18/31086 (S.H.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams DO (1976). The granulomatous inflammatory response. A review. The American journal of pathology 84, 164–192. [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Sakai S, Kudchadkar RR, Fling SP, Day TA, Vergara JA, Ashkin D, Cheng JH, Lundgren LM, Raabe VN, et al. (2019). Tuberculosis following PD-1 blockade for cancer immunotherapy. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CS, Barve A, Muraro MJ, van der Linden R, Dharmadhikari G, Lyubimova A, de Koning EJP, and van Oudenaarden A (2019). Cell Type Purification by Single-Cell Transcriptome-Trained Sorting. Cell 179, 527–542 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, and Britton WJ (1999). Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol 162, 3504–3511. [PubMed] [Google Scholar]
- Blanc L, Sarathy JP, Alvarez Cabrera N, O’Brien P, Dias-Freedman I, Mina M, Sacchettini J, Savic RM, Gengenbacher M, Podell BK, et al. (2018). Impact of immunopathology on the antituberculous activity of pyrazinamide. The Journal of experimental medicine 215, 1975–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonecini-Almeida MG, Ho JL, Boechat N, Huard RC, Chitale S, Doo H, Geng J, Rego L, Lazzarini LC, Kritski AL, et al. (2004). Down-modulation of lung immune responses by interleukin-10 and transforming growth factor beta (TGF-beta) and analysis of TGF-beta receptors I and II in active tuberculosis. Infection and immunity 72, 2628–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccheri S, Reljic R, Caccamo N, Ivanyi J, Singh M, Salerno A, and Dieli F (2007). IL-4 depletion enhances host resistance and passive IgA protection against tuberculosis infection in BALB/c mice. Eur J Immunol 37, 729–737. [DOI] [PubMed] [Google Scholar]
- Bucsan AN, Chatterjee A, Singh DK, Foreman TW, Lee TH, Threeton B, Kirkpatrick MG, Ahmed M, Golden N, Alvarez X, et al. (2019a). Mechanisms of reactivation of latent tuberculosis infection due to SIV coinfection. J Clin Invest 129, 5254–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucsan AN, Mehra S, Khader SA, and Kaushal D (2019b). The current state of animal models and genomic approaches towards identifying and validating molecular determinants of Mycobacterium tuberculosis infection and tuberculosis disease. Pathog Dis 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante J (2020). Mendelian susceptibility to mycobacterial disease: recent discoveries. Hum Genet 139, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, and Ramakrishnan L (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso AM, Serbina N, Klein E, Triebold K, Bloom BR, and Flynn JL (1999). Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. J Immunol 162, 5407–5416. [PubMed] [Google Scholar]
- Clay H, Volkman HE, and Ramakrishnan L (2008). Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity 29, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, and Orme IM (1993). Disseminated tuberculosis in interferon gamma gene-disrupted mice. The Journal of experimental medicine 178, 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan MR, Beerman RW, Rosenberg AF, Saelens JW, Johnson MG, Oehlers SH, Sisk DM, Jurcic Smith KL, Medvitz NA, Miller SE, et al. (2016). Macrophage Epithelial Reprogramming Underlies Mycobacterial Granuloma Formation and Promotes Infection. Immunity 45, 861–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan MR, Matty MA, Rosenberg AF, Blanc L, Pyle CJ, Espenschied ST, Rawls JF, Dartois V, and Tobin DM (2018). An explant technique for high-resolution imaging and manipulation of mycobacterial granulomas. Nat Methods 15, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan MR, Rosenberg AF, Oehlers SH, Saelens JW, Sisk DM, Jurcic Smith KL, Lee S, and Tobin DM (2015). CLARITY and PACT-based imaging of adult zebrafish and mouse for whole-animal analysis of infections. Dis Model Mech 8, 1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, and Ramakrishnan L (2002). Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17, 693–702. [DOI] [PubMed] [Google Scholar]
- Diedrich CR, O’Hern J, Gutierrez MG, Allie N, Papier P, Meintjes G, Coussens AK, Wainwright H, and Wilkinson RJ (2016). Relationship Between HIV Coinfection, Interleukin 10 Production, and Mycobacterium tuberculosis in Human Lymph Node Granulomas. J Infect Dis 214, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFazio RM, Mattila JT, Klein EC, Cirrincione LR, Howard M, Wong EA, and Flynn JL (2016). Active transforming growth factor-beta is associated with phenotypic changes in granulomas after drug treatment in pulmonary tuberculosis. Fibrogenesis Tissue Repair 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm ES, Rhoo KH, and Robert J (2017). Evolutionary Aspects of Macrophages Polarization. Results Probl Cell Differ 62, 3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Schnappinger D, and Rhee KY (2018). Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol 16, 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emile JF, Patey N, Altare F, Lamhamedi S, Jouanguy E, Boman F, Quillard J, Lecomte-Houcke M, Verola O, Mousnier JF, et al. (1997). Correlation of granuloma structure with clinical outcome defines two types of idiopathic disseminated BCG infection. J Pathol 181, 25–30. [DOI] [PubMed] [Google Scholar]
- Ernst JD, Cornelius A, Desvignes L, Tavs J, and Norris BA (2018). Limited Antimycobacterial Efficacy of Epitope Peptide Administration Despite Enhanced Antigen-Specific CD4 T-Cell Activation. J Infect Dis 218, 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaulova E, Das S, Singh DK, Choreno-Parra JA, Swain A, Arthur L, Rangel-Moreno J, Ahmed M, Singh B, Gupta A, et al. (2020). The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell host & microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmail H, Riou C, Bruyn ED, Lai RP, Harley YXR, Meintjes G, Wilkinson KA, and Wilkinson RJ (2018). The Immune Response to Mycobacterium tuberculosis in HIV-1-Coinfected Persons. Annu Rev Immunol 36, 603–638. [DOI] [PubMed] [Google Scholar]
- Feng Y, Renshaw S, and Martin P (2012). Live imaging of tumor initiation in zebrafish larvae reveals a trophic role for leukocyte-derived PGE(2). Current biology : CB 22, 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL (2006). Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect 8, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, and Bloom BR (1995). Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2, 561–572. [DOI] [PubMed] [Google Scholar]
- Gautam US, Foreman TW, Bucsan AN, Veatch AV, Alvarez X, Adekambi T, Golden NA, Gentry KM, Doyle-Meyers LA, Russell-Lodrigue KE, et al. (2018). In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 115, E62–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M, Duque-Correa MA, Kaiser P, Schuerer S, Lazar D, Zedler U, Reece ST, Nayyar A, Cole ST, Makarov V, et al. (2017). NOS2-deficient mice with hypoxic necrotizing lung lesions predict outcomes of tuberculosis chemotherapy in humans. Sci Rep 7, 8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka S, and Kaplan MH (2011). Transcriptional regulation by STAT6. Immunol Res 50, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler R, Parihar SP, Savvi S, Logan E, Schwegmann A, Roy S, Nieuwenhuizen NE, Ozturk M, Schmeier S, Suzuki H, et al. (2015). IL-4Ralpha-dependent alternative activation of macrophages is not decisive for Mycobacterium tuberculosis pathology and bacterial burden in mice. PLoS One 10, e0121070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann L, Abad Dar M, Schreiber T, Erdmann H, Behrends J, McKenzie AN, Brombacher F, Ehlers S, and Holscher C (2014). The IL-13/IL-4Ralpha axis is involved in tuberculosis-associated pathology. J Pathol 234, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess I, Iwanami N, Schorpp M, and Boehm T (2013). Zebrafish model for allogeneic hematopoietic cell transplantation not requiring preconditioning. Proc Natl Acad Sci U S A 110, 4327–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Luo Q, Guo Y, Chen J, Xiong G, Peng Y, Ye J, and Li J (2015). Mycobacterium tuberculosis-Induced Polarization of Human Macrophage Orchestrates the Formation and Development of Tuberculous Granulomas In Vitro. PLoS One 10, e0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Ward JM, Cheever AW, Paul WE, and Sher A (1999). Schistosome-infected IL-4 receptor knockout (KO) mice, in contrast to IL-4 KO mice, fail to develop granulomatous pathology while maintaining the same lymphokine expression profile. J Immunol 163, 337–342. [PubMed] [Google Scholar]
- Jao LE, Wente SR, and Chen W (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 110, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, et al. (1996). Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med 335, 1956–1961. [DOI] [PubMed] [Google Scholar]
- Kauffman KD, Sakai S, Lora NE, Namasivayam S, Baker PJ, Kamenyeva O, Foreman TW, Nelson CE, Oliveira-de-Souza D, Vinhaes CL, et al. (2021). PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci Immunol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman KD, Sallin MA, Sakai S, Kamenyeva O, Kabat J, Weiner D, Sutphin M, Schimel D, Via L, Barry CE 3rd, et al. (2018). Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol 11, 462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts A, Barry CE 3rd, and Dartois V (2015). Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev 264, 288–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala MJ, Raju RM, Sharma K, Zhang YJ, Eugenin EA, Prideaux B, Daudelin IB, Chen PY, Booty MG, Kim JH, et al. (2016). Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nature medicine 22, 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, Via LE, Barry CE 3rd, Klein E, Kirschner DE, et al. (2013). Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol 191, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra S, Pahar B, Dutta NK, Conerly CN, Philippi-Falkenstein K, Alvarez X, and Kaushal D (2010). Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas. PLoS One 5, e12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BB, Lovewell RR, Olive AJ, Zhang G, Wang W, Eugenin E, Smith CM, Phuah JY, Long JE, Dubuke ML, et al. (2017). Nitric oxide prevents a pathogen-permissive granulocytic inflammation during tuberculosis. Nat Microbiol 2, 17072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, and Giraldez AJ (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S, Yokota M, Nakai E, Kuromitsu S, Ohga K, Takeuchi M, Tsukamoto S, and Ohta M (2007). Synthesis and evaluation of 2-{[2-(4-hydroxyphenyl)-ethyl]amino}pyrimidine-5-carboxamide derivatives as novel STAT6 inhibitors. Bioorg Med Chem 15, 1044–1055. [DOI] [PubMed] [Google Scholar]
- Ndlovu H, and Marakalala MJ (2016). Granulomas and Inflammation: Host-Directed Therapies for Tuberculosis. Front Immunol 7, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RJ (1998). Mice incapable of making IL-4 or IL-10 display normal resistance to infection with Mycobacterium tuberculosis. Clin Exp Immunol 113, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlers SH, Cronan MR, Scott NR, Thomas MI, Okuda KS, Walton EM, Beerman RW, Crosier PS, and Tobin DM (2015). Interception of host angiogenic signalling limits mycobacterial growth. Nature 517, 612–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio EY, Travi BL, da Cruz AM, Saldarriaga OA, Medina AA, and Melby PC (2014). Growth factor and Th2 cytokine signaling pathways converge at STAT6 to promote arginase expression in progressive experimental visceral leishmaniasis. PLoS Pathog 10, e1004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan AJ, and Ramakrishnan L (2018). The Formation and Function of Granulomas. Annu Rev Immunol 36, 639–665. [DOI] [PubMed] [Google Scholar]
- Parikka M, Hammaren MM, Harjula SK, Halfpenny NJ, Oksanen KE, Lahtinen MJ, Pajula ET, Iivanainen A, Pesu M, and Ramet M (2012). Mycobacterium marinum Causes a Latent Infection that Can Be Reactivated by Gamma Irradiation in Adult Zebrafish. PLoS Pathog 8, e1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, Chen C, Kaya F, Weiner DM, Chen PY, et al. (2015). The association between sterilizing activity and drug distribution into tuberculosis lesions. Nature medicine 21, 1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece ST, Loddenkemper C, Askew DJ, Zedler U, Schommer-Leitner S, Stein M, Mir FA, Dorhoi A, Mollenkopf HJ, Silverman GA, et al. (2010). Serine protease activity contributes to control of Mycobacterium tuberculosis in hypoxic lung granulomas in mice. J Clin Invest 120, 3365–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus ES, Baek SH, and Sassetti CM (2013). The normalcy of dormancy: common themes in microbial quiescence. Cell host & microbe 13, 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca FJ, and Ramakrishnan L (2013). TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca FJ, Whitworth LJ, Redmond S, Jones AA, and Ramakrishnan L (2019). TNF Induces Pathogenic Programmed Macrophage Necrosis in Tuberculosis through a Mitochondrial-Lysosomal-Endoplasmic Reticulum Circuit. Cell 178, 1344–1361 e1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeot J, Torraca V, Zakrzewska A, Kanwal Z, Jansen HJ, Sommer F, Spaink HP, and Meijer AH (2019). RNAseq Profiling of Leukocyte Populations in Zebrafish Larvae Reveals a cxcl11 Chemokine Gene as a Marker of Macrophage Polarization During Mycobacterial Infection. Front Immunol 10, 832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Sherrid AM, Minch KJ, and Sherman DR (2009). Hypoxia: a window into Mycobacterium tuberculosis latency. Cellular microbiology 11, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, Ganusov VV, and Barber DL (2016). CD4 T Cell-Derived IFN-gamma Plays a Minimal Role in Control of Pulmonary Mycobacterium tuberculosis Infection and Must Be Actively Repressed by PD-1 to Prevent Lethal Disease. PLoS Pathog 12, e1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor M, Weinstock JV, and Wynn TA (2003). Granulomas in schistosome and mycobacterial infections: a model of local immune responses. Trends Immunol 24, 44–52. [DOI] [PubMed] [Google Scholar]
- Sarathy J, Blanc L, Alvarez-Cabrera N, O’Brien P, Dias-Freedman I, Mina M, Zimmerman M, Kaya F, Ho Liang HP, Prideaux B, et al. (2019). Fluoroquinolone Efficacy against Tuberculosis Is Driven by Penetration into Lesions and Activity against Resident Bacterial Populations. Antimicrobial agents and chemotherapy 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy JP, Via LE, Weiner D, Blanc L, Boshoff H, Eugenin EA, Barry CE 3rd, and Dartois VA (2018). Extreme Drug Tolerance of Mycobacterium tuberculosis in Caseum. Antimicrobial agents and chemotherapy 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector WG, and Lykke AW (1966). The cellular evolution of inflammatory granulomata. J Pathol Bacteriol 92, 163–167. [DOI] [PubMed] [Google Scholar]
- Spector WG, and Wynne KM (1976). Proliferation of macrophages in inflammation. Agents Actions 6, 123–126. [DOI] [PubMed] [Google Scholar]
- Subbian S, Tsenova L, Yang G, O’Brien P, Parsons S, Peixoto B, Taylor L, Fallows D, and Kaplan G (2011). Chronic pulmonary cavitary tuberculosis in rabbits: a failed host immune response. Open Biol 1, 110016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surcel HM, Troye-Blomberg M, Paulie S, Andersson G, Moreno C, Pasvol G, and Ivanyi J (1994). Th1/Th2 profiles in tuberculosis, based on the proliferation and cytokine response of blood lymphocytes to mycobacterial antigens. Immunology 81, 171–176. [PMC free article] [PubMed] [Google Scholar]
- Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, and Ramakrishnan L (2006). Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infection and immunity 74, 6108–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki K, Davis JM, Winglee K, and Ramakrishnan L (2013). Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nature protocols 8, 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, and Ramakrishnan L (2008). Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cellular microbiology 10, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, et al. (2012). Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148, 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Roca FJ, Ray JP, Ko DC, and Ramakrishnan L (2013). An enzyme that inactivates the inflammatory mediator leukotriene b4 restricts mycobacterial infection. PLoS One 8, e67828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toossi Z, Gogate P, Shiratsuchi H, Young T, and Ellner JJ (1995). Enhanced production of TGF-beta by blood monocytes from patients with active tuberculosis and presence of TGF-beta in tuberculous granulomatous lung lesions. J Immunol 154, 465–473. [PubMed] [Google Scholar]
- Trinh LA, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, et al. (2011). A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev 25, 2306–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Crevel R, Ottenhoff TH, and van der Meer JW (2002). Innate immunity to Mycobacterium tuberculosis. Clin Microbiol Rev 15, 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bossche J, Bogaert P, van Hengel J, Guerin CJ, Berx G, Movahedi K, Van den Bergh R, Pereira-Fernandes A, Geuns JM, Pircher H, et al. (2009). Alternatively activated macrophages engage in homotypic and heterotypic interactions through IL-4 and polyamine-induced E-cadherin/catenin complexes. Blood 114, 4664–4674. [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, Laoui D, Naessens T, Smits HH, Hokke CH, Stijlemans B, Grooten J, De Baetselier P, and Van Ginderachter JA (2015). E-cadherin expression in macrophages dampens their inflammatory responsiveness in vitro, but does not modulate M2-regulated pathologies in vivo. Sci Rep 5, 12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, et al. (2015). Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog 11, e1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzel AS, Petit J, van Veen WG, Fink IR, Scheer MH, Piazzon MC, Forlenza M, Spaink HP, and Wiegertjes GF (2020). Transcriptome sequencing supports a conservation of macrophage polarization in fish. Sci Rep 10, 13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Lam II, Clay H, Duong DN, Deo RC, and Coughlin SR (2018). A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev Cell 46, 112–125 e114. [DOI] [PubMed] [Google Scholar]
- Zhu LY, Pan PP, Fang W, Shao JZ, and Xiang LX (2012). Essential role of IL-4 and IL-4Ralpha interaction in adaptive immunity of zebrafish: insight into the origin of Th2-like regulatory mechanism in ancient vertebrates. J Immunol 188, 5571–5584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Transcript Levels Across Granuloma Cell Clusters, Related to Figure 1
Violin plots showing the natural log of transcript numbers measured by sc-RNAseq in granuloma cell clusters.
Figure S2. Expression maps of Selected Transcripts and Canonical Markers in Specific Immune Cell Clusters, Related to Figures 1 and 2
Co-expression UMAP plots for select transcripts and whole UMAP plots for canonical markers. (A) E-cadherin (cdh1) and ZO-1 (tjp1) marks epithelialized macrophages. (B) T-cells expressing type 1 markers (ifng1) are largely distinct from T-cells expressing type 2 markers (il13, il4). Canonical markers of (C) neutrophils, (D) T-cells, (E) B-cells and (F) eosinophils shown in Figure 1D–G. (G) Expression map of arginase2 (arg2) and (H) stat6.
Figure S3. Generation of a stat6-Deficient Zebrafish Line, Related to Figure 4. (A) Schematic of the stat6 genomic locus and the targeting site of the gRNA used to generate the stat6xt53 mutant. (B) Sequence of the WT and Δ10 bp xt53 mutant stat6 sequences. Protein sequences are located above (WT) or below (xt53) the DNAs sequence. (C) Schematic of the frame shift and stop locations within the Stat6 protein.
Figure S4 Infection foci in Stat6-Deficient Zebrafish Are not Deficient in Cell Death, Related to Figure 4. Images of granulomas in stat6xt53/xt53 mutants demonstrating abundant pyknotic nuclei formation in the stat6 mutant. Area within the dotted yellow box is magnified and shown in greyscale within the yellow box on the right hand side. Cyan arrows indicate individual pyknotic nuclei within the stat6 deficient granulomas. Scale bar – 100 μm. Representative of 8 (stat6) total animals from 3 independent experiments.
Figure S5. Still images from Movie S1, Related to Figure 7. Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. Time-lapse images from Movie S1 of a granuloma treated with DMSO (top) and 10 μM AS1517499 (bottom) for the initial 22 hours of treatment, showing a reduction of E-cadherin (cdh1) during Stat6 inhibition. Scale bar is 50μm.
Figure S6. Granuloma macrophages exposed to Stat6 inhibition at 72 hours remain viable and motile with no increase in cell death, Related to Figure 7. Granulomas were dissected from adult (cdh1-tdtomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. (A) Still images from Movie S2, enlarged versions of the Movie S1, showing granuloma treated with 10 μM AS1517499. At timepoint 0, note membrane-localized E-cadherin expression. Pink arrows indicate an E-cadherin negative cell with retained motility, even after leaving the granuloma. White arrowheads indicate cells showing Cdh1-tdTomato delocalization. Scale bar is 50μm. (B) Representative images from DMSO, 3 μM, and 10 μM Stat6 inhibitor-treated explant granulomas stained with propidium iodide for 1 hour at 72 hours post treatment. Granuloma macrophages remain largely unstained and viable. The necrotic core of cell debris is stained brightly, and pink arrowheads indicate examples of propidium iodide staining. Scale bar is 100μm.
Figure S7. Acute inhibition of stat6 results in reduction of plakoglobin localization and fluorescence, Related to Figure 7. After explant of established, mature granulomas, vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. (A) Representative single z-plane, time-lapse images from DMSO (top) and 10 μM AS1517499 Stat6 inhibitor (bottom) treated explant granulomas from plakoglobin-citrine (green) gene trap fish (Gt(jupa-citrine))ct520a. Granulomas were dissected from adult animals at 14dpi. Scale bar is 50 μm. Blue boxes show enlarged areas of granuloma cells, arrowheads indicate plakoglobin fluorescence signals. (B) Quantitation of plakoglobin-citrine fluorescence in granulomas treated with vehicle or 10 μM AS1517499 Stat6 inhibitor, normalized to the fluorescence value at initial timepoint for each granuloma. n = 8 for DMSO and n = 8 for 10 μM. Bars represent median values with interquartile range. Dunnett’s multiple comparison test was used to calculate p-values.
Movie S1. Time-Lapse Imaging of Granulomas During Acute Stat6 Inhibition, Related to Figure S5.
Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. A representative granuloma from DMSO group (top) and 10 μM AS1517499 group (bottom) for the initial 22 hours of treatment, showing a reduction of E-cadherin (Cdh1) during Stat6 inhibition. Brightfield is on the left and cdh1-tdTomato fluorescence is on the right. Scale bar is 50μm.
Movie S2. Close-up of Time-Lapse Imaging of Granuloma Cells During Acute Stat6 Inhibition, Related to Figure S6
Granulomas were dissected from adult (cdh1-tdTomato)xt18 animals at 14 dpi. Vehicle (DMSO) or the Stat6 inhibitor AS1517499 was applied during Myco-GEM culture. A close-up of the representative granuloma from Movie S1 in the 10 μM AS1517499 group during the initial 22 hours of treatment, showing an E-cadherin negative cell with retained motility, even after leaving the granuloma. Time-lapse imaging also shows cells exhibiting Cdh1-tdTomato delocalization. Brightfield is on the left, cdh1-tdTomato fluorescence in the center and merged channels on the right. The yellow box indicates the area that is enlarged in the bottom panels. Only a portion of the 22-hour movie is shown. Scale bar is 50μm.
Table S1. Proportion of cell-types by cluster in scRNA-seq data, related to Figure 1.
Table S2. Global list of transcripts with mean expression value for each cluster, related to Figure 1.
Data Availability Statement
The scRNA-seq data generated during this study have been deposited at GEO under accession number GSE161712.


