Abstract
Summary
Despite the improvement in variant detection algorithms, visual inspection of the read-level data remains an essential step for accurate identification of variants in genome analysis. We developed BamSnap, an efficient BAM file viewer utilizing a graphics library and BAM indexing. In contrast to existing viewers, BamSnap can generate high-quality snapshots rapidly, with customized tracks and layout. As an example, we produced read-level images at 1000 genomic loci for >2500 whole-genomes.
Availability and implementation
BamSnap is freely available at https://github.com/parklab/bamsnap.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
With the advances in sequencing technology, an increasingly large number of genomes are being sequenced. Although algorithms for alignment of sequenced reads and identification of genomic variation have improved substantially over the years, the resulting set of variants still contain false positives due to inaccurate alignment (especially near repetitive sequences), incomplete reference sequence, insufficient sequence coverage, PCR duplicates, DNA contamination and other reasons (Dou et al., 2018; Li, 2014). To ascertain the accuracy of called variants (single nucleotide variants, indels, rearrangement breakpoints, etc.), visual inspection of the read configuration is critical, as it reveals features whose contributions may not have been optimally weighted for a specific variant in the algorithm. For instance, the impact of a nearby low-complexity region—which can induce mistaken indel calls—is variable depending on the context and is difficult to incorporate into an algorithm, but it becomes apparent in manual inspection.
For visualizing aligned reads, several methods have been proposed. BamView (Carver et al., 2010) from the Sanger Institute is an interactive Java application; Integrative Genomics View (IGV) (Robinson et al., 2011) from the Broad Institute is coded in Java and JavaScript and supports a graphic user interface for multiple data types including BAM files; PyBamViewer (Gymrek, 2014) is a web-based viewer which provides base-wise alignment plot similar to the samtools view (Li et al., 2009); and pileup.js (Vanderkam et al., 2016) is a web-based JavaScript viewer with similar features as IGV. But because these tools were created for interactive exploration, they are designed to deal with a small number of BAM files at a time. Accessing a large number of BAM files to generate read alignment maps in real time often results in memory errors or is time-consuming. It is possible to write custom scripts to capture images from interactive viewers, but it is slow and cumbersome.
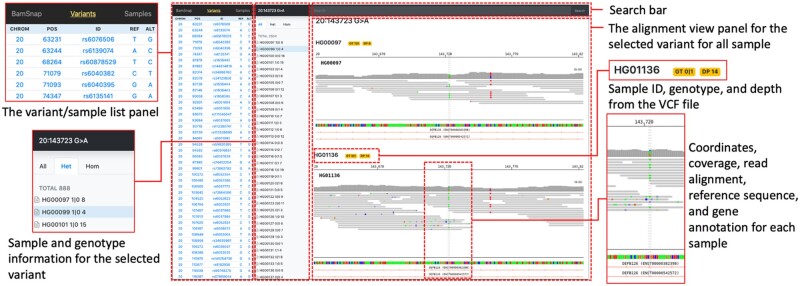
Here, we describe BamSnap, a highly efficient viewer to generate read-level view across thousands BAM or CRAM files at a time. BamSnap supports visualization of genomic tracks for reference sequences, mapped reads (paired or unpaired), read depth (coverage), variants and gene annotations (Fig. 1). The web viewer generated by BamSnap can be easily customized and incorporated into a data portal without the need to access the original alignment files.
Fig. 1.
BamSnap viewer. A read alignment view of the chr20:143,723 G > A variant was generated across 2504 samples from the 1000 Genomes Project
2 Basic usage and features
Users can run BamSnap on sorted and indexed BAM files and VCF (or gzip/bgzip-compressed VCF) files with multi-threading, using simple commands as illustrated here:
bamsnap -bam NA12877.bam NA12878.bam NA12879.bam -vcf trio.vcf -out test -thread 20
Various options for layout and input/output format are available. BamSnap has the following distinguishing features compared to existing tools (a detailed comparison is in Supplementary Table S1):
A snap-image based viewer: it does not require access to the BAM files to show the alignment and enables fast loading and navigation; the images can be easily shared and/or published.
Batch processing: images can be generated for multiple variants and multiple BAM (or CRAM) files with a single command.
High-quality images: PNG, SVG and PDF are supported.
Flexible layout: the image layout (color set, margin, track height, etc.) can be customized by modifying the configuration file.
Additional tracks: UCSC browser and ENSEMBL annotation tracks (e.g. conservation score, mappability) can be appended.
Command line interface (CLI): CLI offers additional flexibility and facilitates repetitive tasks and automated pipelines.
Jupiter notebook implementation: Users can implement the alignment image in Jupiter notebook using a Python API.
Multi-threading: an almost linear speedup with the increase in the number of cores used.
3 Implementations
BamSnap is a Python-based CLI application; for a web viewer, it utilizes Python application programming interface (API). The workflow consists of two parts: (i) generation of a snap image from BAM files using Pillow (v2.0.0), a Python image library based on the GD graphic library, and (ii) a web viewer implementation. To quickly fetch data from specific genomic regions of interest, it uses Pytabix (v0.1) and Pysam library (v0.11.2.2), which use indexed BAM (or CRAM) and VCF files. The BamSnap view is implemented using HTML5 and Web 2DGL-based Pixi.js for fast and effective viewing in a web browser. It supports the latest versions of the major browsers available at this time: Chrome 84+, Firefox 79+, Safari 13+ and Microsoft Edge 84+.
4 An example use case
We generated images for 1000 common variants (±100 bp neighborhood) across 2504 samples with whole genome sequencing data (27 high- and 2477 low-coverage) (1000 Genomes Project Consortium et al., 2015). We ran BamSnap on a 3.8 GHz Intel Xeon CPU with 20 GB RAM and 30 threads, and obtained images of 2.5 million genomic sites in ∼3.8 h. The average times of image generation for a single site were 1.63 s and 0.15 s, respectively, for high- and low-coverage. The results are available at https://github.com/parklab/bamsnap.
5 Conclusion
We built a lightweight visualization tool for generation of publication-quality graphics based on BAM (or CRAM) and VCF files. Researchers and clinicians can use BamSnap to inspect a large number of candidate variants quickly and share their images in a HTML wrapper without having to share the raw data. Developers can easily implement BamSnap in their own systems using CLI and API.
Funding
This work was supported by National Institutes of Health [U01MH106883].
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Minseok Kwon, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
Soohyun Lee, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
Michele Berselli, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
Chong Chu, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
Peter J Park, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
References
- 1000 Genomes Project Consortium. et al. (2015) A global reference for human genetic variation. Nature, 526, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. et al. (2010) BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinf. Oxf. Engl., 26, 676–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y. et al. (2018) Detecting somatic mutations in normal cells. Trends Genet., 34, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M. (2014) PyBamView: a browser-based application for viewing short read alignments. Bioinformatics, 30, 3405–3407. [DOI] [PubMed] [Google Scholar]
- Li H. et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics, 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T. et al. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderkam D. et al. (2016) pileup.js: a JavaScript library for interactive and in-browser visualization of genomic data. Bioinformatics, 32, 2378–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.