Abstract
Enterovirus D68 (EV-D68) is an atypical non-polio enterovirus that mainly infected the respiratory system of humans, leading to moderate to severe respiratory diseases. In rare cases, EV-D68 can spread to the central nervous system and causes paralysis in infected patients, especially young children and immunocompromised individuals. There is currently no approved vaccine or antiviral available for the prevention and treatment of EV-D68. In this study, we aimed to improve the antiviral potency and selectivity of a previously reported EV-D68 inhibitor, dibucaine, through structure-activity relationship studies. In total, sixty compounds were synthesized and tested against EV-D68 using the viral cytopathic effect (CPE) assay. Three compounds 10a, 12a, and 12c were identified to have significantly improved potency (EC50 < 1 μM) and a high selectivity index (SI > 180) compared to dibucaine against five different strains of EV-D68 viruses. These compounds also showed potent antiviral activity in neuronal cells such as A172 and SH-SY5Y cells, suggesting they might be further developed for the treatment of both respiratory infection as well as neuronal infection.
Keywords: Enterovirus D68, EV-D68, antiviral, quinoline, 2C protein
Graphical Abstract
INTRODUCTION
The human enteroviruses (EVs) are a group of small non-enveloped single-stranded positive-sense RNA viruses that belong to the piconaviridae family.1, 2 Up to date, more than 110 serotypes of enteroviruses have been identified, among which the most notable ones are polioviruses, coxsackieviruses, EV-D68, EV-A71, and rhinoviruses.3 Although poliovirus was nearly eradicated by immunization, no vaccines are available for the non-polio EVs except EV-A71.4 The non-polio enteroviruses such as EV-D68 are of particular concern as they have been shown to be causative pathogens of numerous epidemics of moderate to severe respiratory illness.5–8 Although infections in most cases are mild and self-limiting, EV-D68 infections can lead to serious and life-threatening neurological complications such as acute flaccid myelitis (AFM), most often in infants, young children, and immunocompromised individuals.9 Cases of EV-D68 infection were rarely reported before 2005,10 however, recent years have seen an increasing surge of EV-D68 infections around the world, especially in the United States.6, 11 EV-D68 attracted public attention when an outbreak spread to 49 states in the United States in 2014 that led to more than 1,100 reports of severe respiratory disease and more than 100 cases of acute flaccid myelitis.6 Since then, EV-D68 infection has been continuously reported.6
EV-D68 is an atypical enterovirus and behaves more like a rhinovirus. For example, unlike other enteroviruses that replicate in the gastrointestinal tract at 37°C, EV-D68 is cold adapted and replicates more efficiently at 33°C.12 Individuals with pre-existing respiratory condition such as asthma or chronic obstructive pulmonary disease (COPD) are more likely to develop a severe infection.7 More alarmingly, there has been evidence from both animal studies and human patients that EV-D68 can spread to central nervous systems (CNS) such as spinal cord and cerebrospinal fluid through viremia, causing neurological complications such as AFM.13–18
Due to its global medical and socioeconomic impact, EV-D68 is classified as a priority pathogen by NIAID.19 However, despite decades of research efforts, there is currently no vaccine nor small molecule antiviral exist for EV-D68 infection,2 and treatment is primarily limited to supportive care. To reduce the morbidity and mortality associated with EV-D68 virus infection, there is an urgent need to develop potent and selective small molecule antivirals. Towards this end, we are interested in developing EV-D68 antivirals by targeting the viral 2C protein.
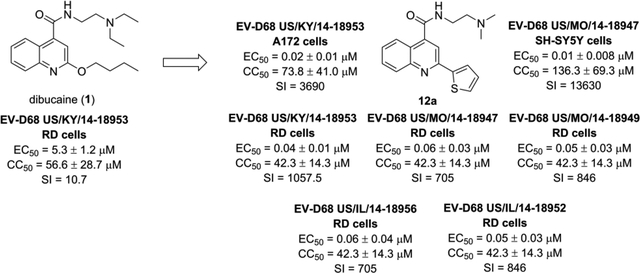
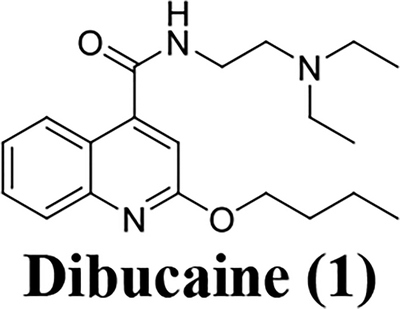
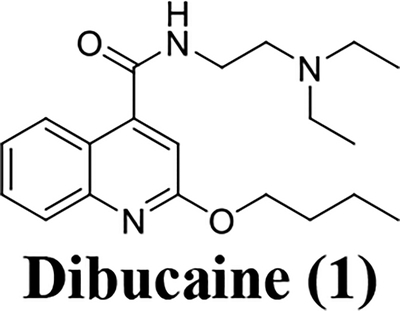
The non-structural protein EV-D68 2C is a multi-functional protein. Genetic studies of 2C have revealed that it has functional roles in viral uncoating, RNA binding and replication, membrane rearrangement, encapsidation of the viral genome, and progeny viral assembly, all of which are essential for viral replication.1, 2 Therefore, EV-D68 2C appears to be an attractive target for anti-enterovirus drug development. Previous phenotypic screenings identified several small molecules such as pirlindole mesylate, fluoxetine, formoterol, dibucaine, and guanidine as EV-D68 antivirals.20–23 Resistance selection through serial viral passage experiments led to the identification of mutations in viral 2C protein that confer resistance to these compounds, suggesting 2C might be the potential drug target. In this study, we first independently verified the antiviral activity and cellular cytotoxicity of these reported 2C protein inhibitors. It was found that all these molecules had only moderate antiviral activity against EV-D68 with a low selectivity index (Table 1), consistent with previously reported results.21, 22 Next, we chose dibucaine as a starting hit compound for structure-activity relationship (SAR) studies due to its modular synthetic accessibility. The aim of this study is therefore to optimize the potency and selectivity index of dibucaine through SAR studies. In total sixty quinoline analogues have been synthesized and tested against EV-D68 (US/KY/14-18953) virus in the primary viral cytopathic effect (CPE) assay. Three compounds (10a, 12a, and 12c) were identified to have significantly improved antiviral activity with EC50 values in the submicromolar range and a selectivity index over 180. We further profiled the broad range antiviral activity of these three compounds against additional four contemporary human EV-D68 viruses. It was found that these three compounds all had potent antiviral activity and a high selectivity index. These three lead compounds also potently inhibited EV-D68 infection in neuronal cells such as A172 and SH-SY5Y cells, suggesting they can be further developed to treat both respiratory and neuronal infections. The mechanism of action of these three compounds was studied by RT-qPCR, western blot, and immunofluorescence imaging. These compounds were found to inhibit viral RNA and protein synthesis, which agrees with the inhibition of viral 2C protein.
Table 1.
Literature reported enterovirus 2C protein inhibitors.
| Structure | EV-D68 (US/KY/14) EC50 (μM)a | RD cells CC50 (μM)b | SI |
|---|---|---|---|
 |
8.4 ± 1.1 | 19.0 ± 1.6 | 2.3 |
 |
1.0 ± 0.1 | 11.9 ± 4.9 | 11.9 |
 |
24.7 ± 1.1 | 146.3 ± 27.8 | 5.9 |
 |
5.3 ± 1.2 | 56.6 ± 28.7 | 10.7 |
 |
219.4 ± 15.0 | >300.0 | > 1.4 |
Antiviral efficacy was determined in the CPE assay with EV-D68 US/KY/14-18953 virus and RD cells.
Cytotoxicity was determined using the neutral red method. The results are the mean ± standard deviation of three repeats. SI = selectivity index (CC50/EC50).
RESULTS AND DISCOSSION
CHEMISTRY
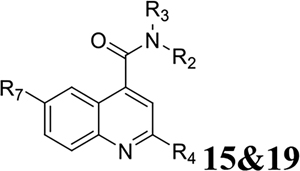
Three positions of dibucaine were varied in the SAR study: the 2-position substituent was substituted with either an alkoxy (O-R1) or an aromatic group (R2); the 4-position was substituted with different amides; and the 6-position was substituted with either F, Cl, or methoxy group. We adapted a stepwise iterative optimization strategy. First, the 4-position substitution was fixed while the 2-position substitution was optimized. Second, the 2-position substitution was fixed while the 4-position substitution was optimized. Third, optimal substitutions from both the 2- and 4-positions were merged. Finally, the 6-position was diversified with the most potent analogs.
The synthesis of quinoline analogs was designed to allow for expeditious late-stage diversification. For example, for varying the 2-position substitution, intermediate 4 was first synthesized by amide coupling. Next, two methods were developed to install alkoxy substitution at the 2-position (Scheme 1, synthesis route 1). Method 1 used sodium metal and excess alcohol, which served as both the reagent and the solvent. This worked for alcohols with a low boiling point such that they can be easily removed during the work up procedure. For alcohols with a high boiling point, method 2 was developed which used DMF as the solvent and two equivalents of sodium hydride and the alcohol. To introduce aromatic substitutions at the 2-position, Suzuki-Miyaura cross coupling was employed using microwave heating (Scheme 1, synthesis route 2). Once the optimal substations at the 2-position were identified, the next step was to optimize the 4-position amide substitution. For this, quinoline 4-carboxylic acids 7, 9, and 11 with optimal 2-position substitutions (isopropoxyl, phenyl, and thienyl) were synthesized. Next, amide coupling was applied to introduce different amines (Scheme 1, synthesis routes 3 & 4). Finally, for analogs with substitutions at the 6-position of quinoline, two synthesis strategies were developed (Scheme 1, synthesis routes 5).24 The first strategy involved Pfitzinger reaction with 5-chloroisatin or 5-methoxylisatin 13 and acetophenone using potassium hydroxide as a base. Coupling of 14 with different amines gave the final product 15. The second strategy involved the reaction of 5-fluoroisatin with malonic acid under refluxing condition in acetic acid to give intermediate 17. Next, a one pot chlorination and amide coupling was achieved by first forming the 2-chloroquinoline-4-carbonyl chloride intermediate through reacting intermediate 17 with thionyl chloride in the presence of catalytic amount of DMF, then different amines were added to convert the acid chloride intermediate to the intermediate 18. Finally, Suzuki-Miyaura cross-coupling was applied to install the 2-position substitution to give the final product 19.
Scheme 1.
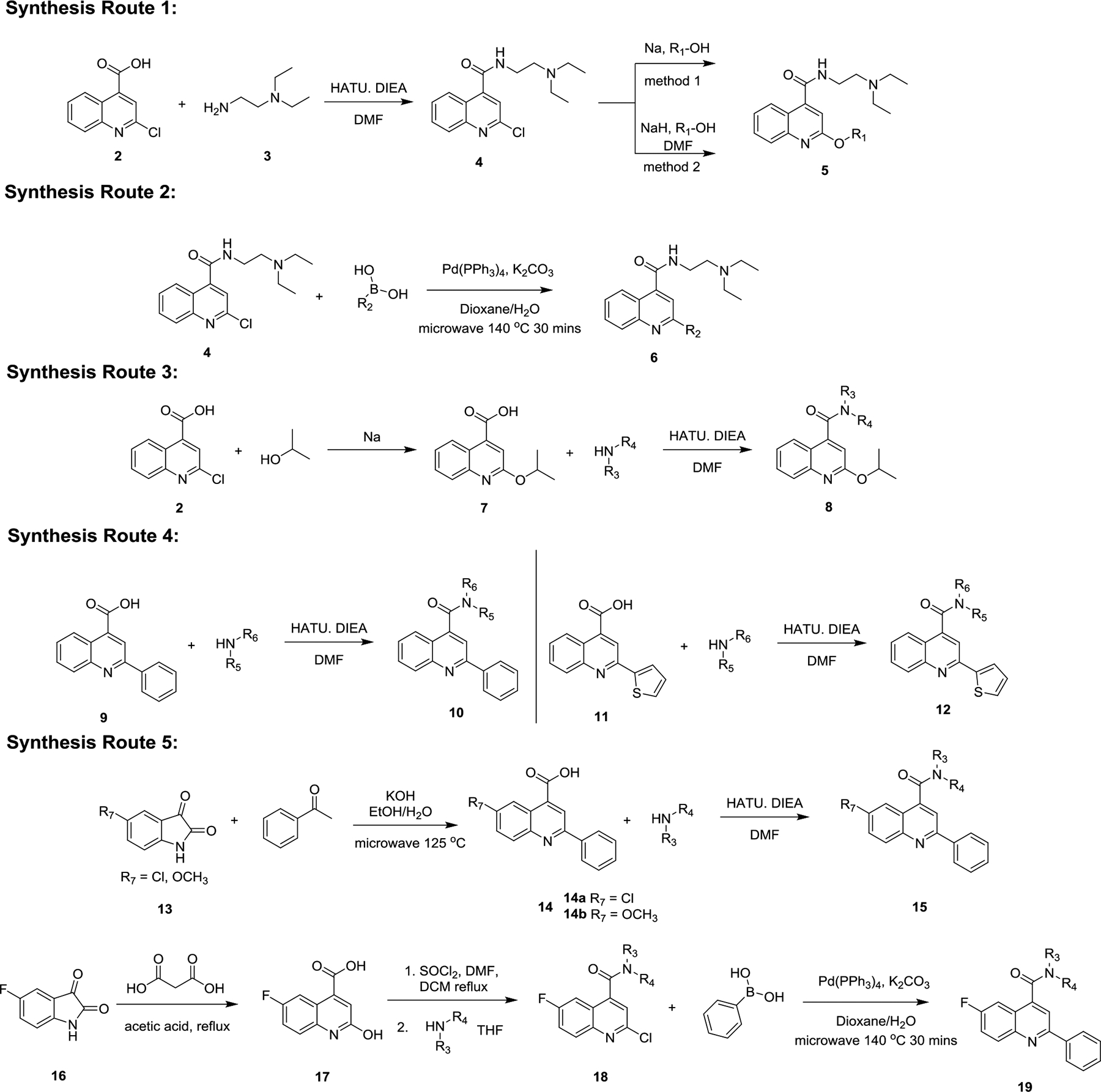
Synthesis routes of dibucaine analogs.
STRUCTURE-ACTIVITY RELATIONSHIP STUDIES
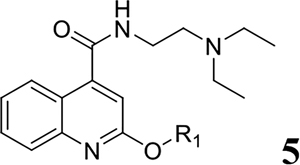
As an initial screening, all compounds were tested for antiviral activity and cellular cytotoxicity in standard viral CPE assay and neutral red cell viability assay, respectively, with the human rhabdomyosarcoma (RD) cells. The CPE assay involved infecting RD cells with the EV-D68 virus (US/KY/14-18953) and monitoring cell viability after 3 days of incubation with and without compound treatment. Dibucaine was included as a positive control. For compound 5 series with different alkoxy substitutions at the 2-position (Table 2), compound 5e with 2-isopropoxy substitution was found to have the highest potency and selectivity (EC50 = 2.5 ± 0.5 μM, CC50 = 111.2 ± 15.4 μM, SI = 44.5). Compound 5k also had improved potency and selectivity compared to dibucaine. Other compounds (5a, 5b, 5c, 5d, 5f, 5g, 5h, 5i, and 5j) either had reduced antiviral activity (5a, 5b, 5g, 5h, 5i, 5j) or reduced selectivity index (5c, 5d, 5f) compared to dibucaine. From these results, it can be concluded that isopropoxy is one of the optimal substitutions at the 2-position of quinoline. Therefore, compound 5e was chosen as a reference compound for following optimization.
Table 2.
SAR study of the 2-position R1 substituent: alkoxy
 | ||||
|---|---|---|---|---|
| Compound ID | R1 | EC50 (μM)a | CC50 (μM)b | SI |
| 5a | 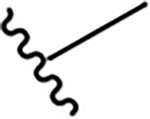 |
52.5 ± 7.1 | > 250.0 | >4.8 |
| 5b | 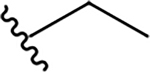 |
5.9 ± 3.2 | 164.9 ± 57.6 | 27.9 |
| 5c | 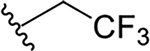 |
3.5 ± 0.4 | 33.1 ± 1.8 | 9.5 |
| 5d | 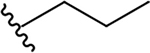 |
4.9 ± 2.0 | 46.7 ± 14.7 | 9.5 |
| 5e | 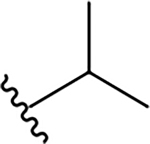 |
2.5 ± 0.5 | 111.2 ± 15.4 | 44.5 |
| Dibucaine 1 | 5.3 ± 1.2 | 56.6 ± 28.7 | 10.7 | |
| 5f | 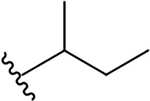 |
2.8 ± 1.3 | 15.7 ± 1.1 | 5.6 |
| 5g | 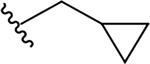 |
7.1 ± 5.0 | 38.5 ± 16.8 | 5.4 |
| 5h | 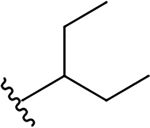 |
> 10 | 9.5 ± 1.1 | N.A.c |
| 5i |  |
6.8 ± 0.5 | 11.7 ± 1.2 | 1.7 |
| 5j | 12.2 ± 1.1 | 143.7 ± 40.5 | 11.8 | |
| 5k |  |
1.1 ± 0.1 | 13.5 ± 2.9 | 12.3 |
Antiviral efficacy was determined in the CPE assay with EV-D68 US/KY/14-18953 virus and RD cells.
Cytotoxicity was determined using the neutral red method.
N.A.= not applicable. The results are the mean ± standard deviation of three repeats. SI = selectivity index (CC50/EC50).
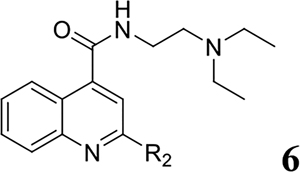
For compound 6 series with aromatic substitutions at the 2-position (Table 3), compounds 6d, 6f, 6g, and 6j had improved antiviral activity and selectivity index compared to compound 5e. Compounds 6e, 6l, 6m, 6r, and 6s had improved antiviral activity compared to 5e, however they were compromised by a lower selectivity index. Other compounds (6a, 6b, 6c, 6h, 6i, 6k, 6n, 6o, 6p, 6q, 6t, and 6u) in this series were less active than 5e and were not further explored. These results suggested unsubstituted thienyl, furanyl, and benzyl are optimal substitutions at the 2-position of quinoline.
Table 3.
SAR study of the 2-position R2 substituent: Aromatic Groups
 | ||||
|---|---|---|---|---|
| Compound ID | R2 | EC50 (μM)a | CC50 (μM)b | SI |
| 5e |  |
2.5 ± 0.5 | 111.2 ± 15.4 | 44.5 |
| 6a | 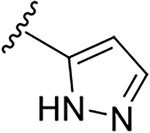 |
19.4 ± 1.2 | 132.3 ± 53.3 | 6.8 |
| 6b |  |
>100.0 | 250.7 ± 28.3 | N.A.c |
| 6c |  |
36.3 ± 1.6 | 132.6 ± 28.7 | 3.7 |
| 6d | 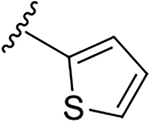 |
0.6 ± 0.5 | 37.4 ± 6.3 | 62.3 |
| 6e | 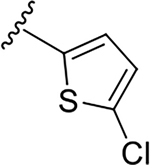 |
0.8 ± 0.6 | 6.1 ± 2.2 | 7.6 |
| 6f | 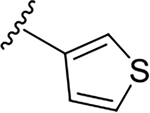 |
1.0 ± 0.4 | 46.9 ± 5.2 | 46.9 |
| 6g | 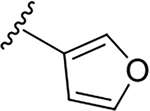 |
1.5 ± 0.9 | 122.5 ± 30.8 | 81.7 |
| 6h | 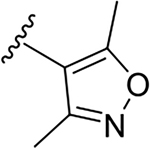 |
>50.0 | 128.6 ± 67.2 | N.A.c |
| 6i | 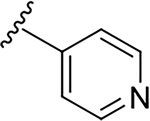 |
6.6 ± 3.3 | 173.5 ± 55.6 | 26.3 |
| 6j | 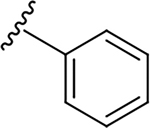 |
0.8 ± 0.3 | 45.1 ± 13.7 | 56.4 |
| 6k | 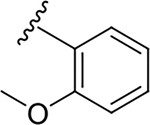 |
13.9 ± 8.1 | 75.9 ± 6.4 | 5.5 |
| 6l | 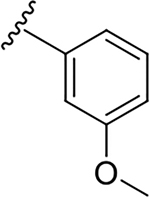 |
1.6 ± 0.8 | 36.5 ± 3.1 | 22.8 |
| 6m | 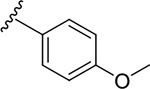 |
2.4 ± 0.2 | 31.6 ± 4.2 | 13.2 |
| 6n | 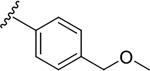 |
17.4 ± 8.9 | 46.6 ± 2.4 | 2.7 |
| 6o | 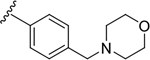 |
>80.0 | 79.2 ± 8.0 | N.A.c |
| 6p | 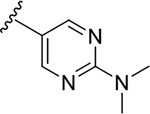 |
24.6 ± 2.2 | 74.2 ± 7.0 | 3.0 |
| 6q | 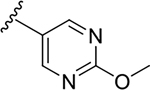 |
87.6 ± 3.1 | 173.5 ± 20.3 | 2.0 |
| 6r | 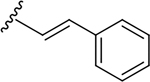 |
0.8 ± 0.2 | 11.5 ± 6.7 | 14.4 |
| 6s | 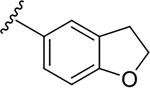 |
1.6 ± 0.1 | 29.5 ± 1.6 | 18.4 |
| 6t | 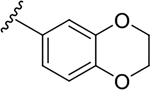 |
9.0 ± 1.3 | 49.9 ± 13.8 | 5.5 |
| 6u |  |
>10.0 | 9.8 ± 0.5 | N.A.c |
Antiviral efficacy was determined in the CPE assay with EV-D68 US/KY/14-18953 virus and RD cells.
Cytotoxicity was determined using the neutral red method.
N.A.= not applicable. The results are the mean ± standard deviation of three repeats. SI = selectivity index (CC50/EC50).
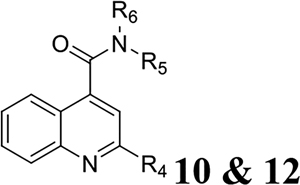
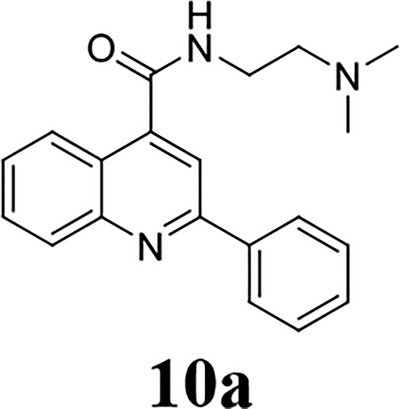
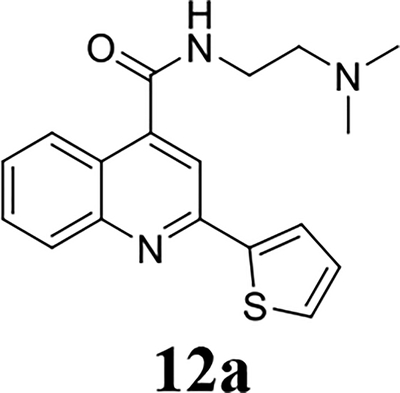
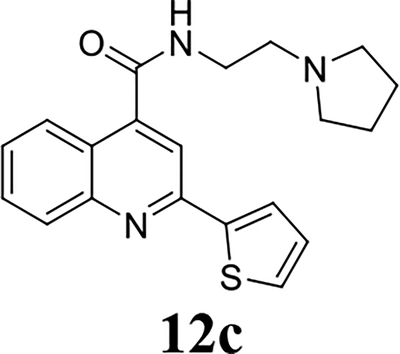
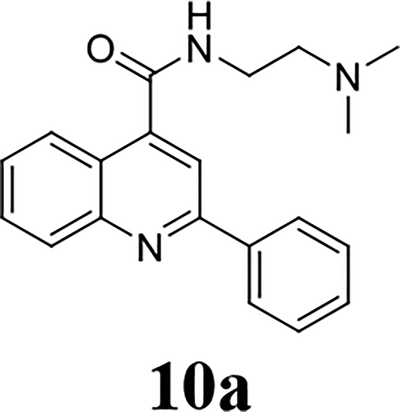
Once the 2-position substitution was optimized, the next step was to optimize the 4-position amide. For compound 8 series with 2-position substitution being isopropoxyl (Table 4), compounds 8a and 8e had improved antiviral activity compared with 5e, however they had a lower selectivity index. All other compounds (8b, 8c, 8d, 8f, 8g, and 8h) had reduced antiviral activity. For compound 10 and 12 series with 2-position being benzyl and thienyl, respectively (Table 5), compound 10a had significantly improved antiviral activity and selectivity index (EC50 = 0.4 ± 0.2 μM, CC50 = 73.7 ± 19.1 μM, SI = 184.3). Similarly, compounds 12a and 12c were also found to be highly potent EV-D68 antivirals (EC50 ≤ 0.1 μM) with a high selectivity index (SI > 600).
Table 4.
SAR study of the 4-position amide substituent
 | ||||
|---|---|---|---|---|
| Compound ID |  |
EC50 (μM)a | CC50 (μM)b | SI |
| 5e |  |
2.5 ± 0.5 | 111.2 ± 15.4 | 44.5 |
| 8a | 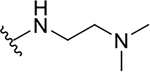 |
1.7 ± 1.2 | 38.5 ± 1.5 | 22.6 |
| 8b | 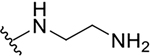 |
3.5 ± 0.3 | 148.8 ± 15.3 | 42.5 |
| 8c | 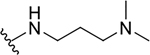 |
11.4 ± 0.7 | 121.7 ± 39.2 | 10.7 |
| 8d | 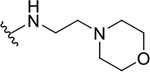 |
17.5 ± 10.2 | > 250.0 | >14.3 |
| 8e | 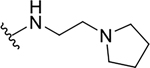 |
1.7 ± 1.2 | 36.9 ± 20.1 | 21.7 |
| 8f | 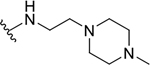 |
25.9 ± 2.1 | 88.8 ± 21.5 | 3.4 |
| 8g | 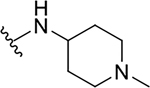 |
17.8 ± 1.1 | 100.7 ± 8.8 | 5.7 |
| 8h | 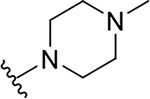 |
> 100.0 | > 300.0 | N.A.c |
Antiviral efficacy was determined in the CPE assay with EV-D68 US/KY/14-18953 virus and RD cells.
Cytotoxicity was determined using the neutral red method.
N.A.= not applicable. The results are the mean ± standard deviation of three repeats. SI = selectivity index (CC50/EC50).
Table 5.
SAR study of the 4-position amide substituent
 | |||||
|---|---|---|---|---|---|
| Compound ID |  |
R4 | EC50 (μM) | CC50 (μM) | SI |
| 6j |  |
 |
0.8 ± 0.3 | 45.1 ± 13.7 | 56.4 |
| 10a | 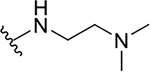 |
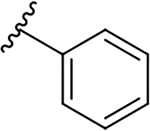 |
0.4 ± 0.2 | 73.7 ± 19.1 | 184.3 |
| 10b | 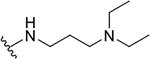 |
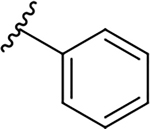 |
9.6 ± 6.7 | 45.4 ± 14.0 | 4.7 |
| 10c | 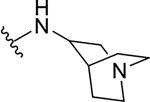 |
 |
3.5 ± 0.3 | 46.5 ± 14.7 | 13.3 |
| 10d | 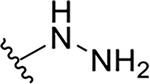 |
 |
100.1 ± 8.5 | > 300.0 | >3 |
| 10e | 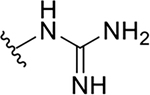 |
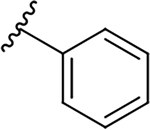 |
34.7 ± 1.4 | >250 | >7.2 |
| 10f | 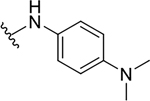 |
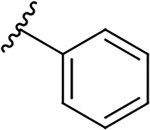 |
>100.0 | >350.0 | >3.5 |
| 10g | 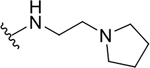 |
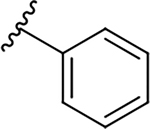 |
0.4 ± 0.1 | 46.7 ± 8.3 | 116.8 |
| 10h | 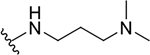 |
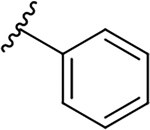 |
3.5 ± 0.6 | 44.9 ± 17.7 | 12.8 |
| 12a | 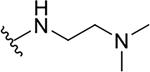 |
 |
0.04 ± 0.01 | 42.3 ± 14.3 | 1057.5 |
| 12b | 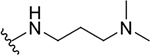 |
 |
0.7 ± 0.3 | 40.5 ± 14.0 | 57.9 |
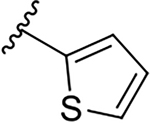
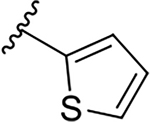
| 12c | 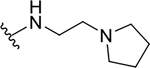 |
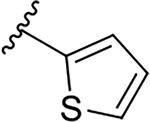 |
0.06 ± 0.03 | 40.0 ± 13.1 | 666.7 |
| 12d | 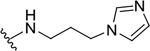 |
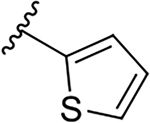 |
8.9 ± 5.8 | 55.7 ± 3.5 | 6.3 |
| 12e |  |
 |
11.9 ± 1.3 | >100.0 | > 8.4 |
Antiviral efficacy was determined in the CPE assay with EV-D68 US/KY/14-18953 virus and RD cells.
Cytotoxicity was determined using the neutral red method. The results are the mean ± standard deviation of three repeats. SI = selectivity index (CC50/EC50).
For compounds 19 and 15 with substitutions at the 6-position (Table 6), none of them had improved potency and selectivity index compared to compound 10a, suggesting the 6-position of quinoline preferred to be unsubstituted.
Table 6.
SAR study of the R7 substituent
 | ||||||
|---|---|---|---|---|---|---|
| Compound ID |  |
R4 | R7 | EC50 (μM) | CC50 (μM) | SI |
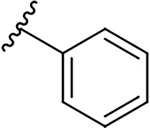
| 10a | 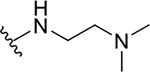 |
 |
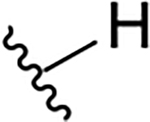 |
0.4 ± 0.2 | 73.7 ± 19.1 | 184.3 |
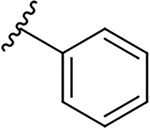
| 19a | 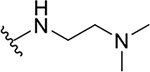 |
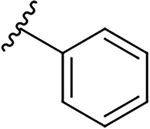 |
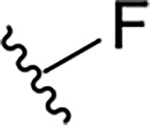 |
0.6 ± 0.3 | 38.8 ± 19.0 | 64.7 |
| 19b | 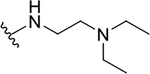 |
 |
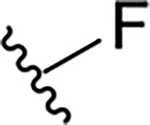 |
1.3 ± 0.3 | 24.9 ± 1.5 | 19.2 |
| 15a | 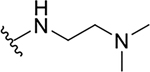 |
 |
 |
1.2 ± 0.2 | 14.3 ± 0.8 | 11.9 |
| 15b | 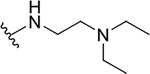 |
 |
 |
1.9 ± 0.1 | 13.5 ± 8.1 | 7.1 |
| 15c | 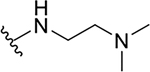 |
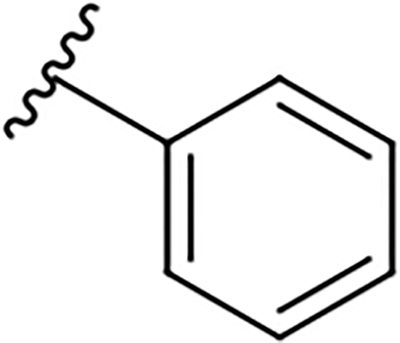 |
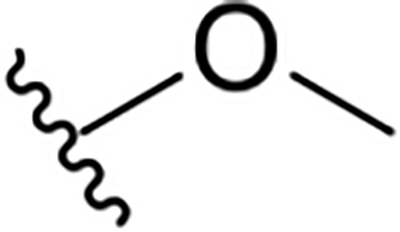 |
8.1 ± 0.6 | 31.8 ± 1.3 | 3.9 |
| 15d | 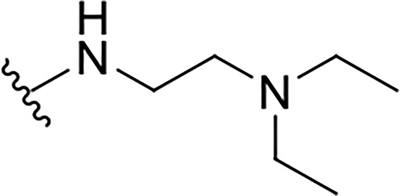 |
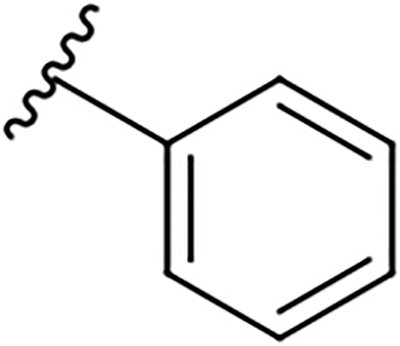 |
 |
12.3 ± 1.1 | 27.5 ± 7.3 | 2.2 |
Overall our SAR study suggest that the optimal substitutions at the 2-position were isopropoxyl, thienyl, furanyl, and benzyl (Fig. 2); the optimal substitutions at the 4-position were N-(2-(diethylamino)ethyl)amide, N-(2-(dimethylamino)ethyl)amide, and N-(2-(pyrrolidin-1-yl)ethyl)amide; the 6-position preferred to be un-substituted.
Figure 2.
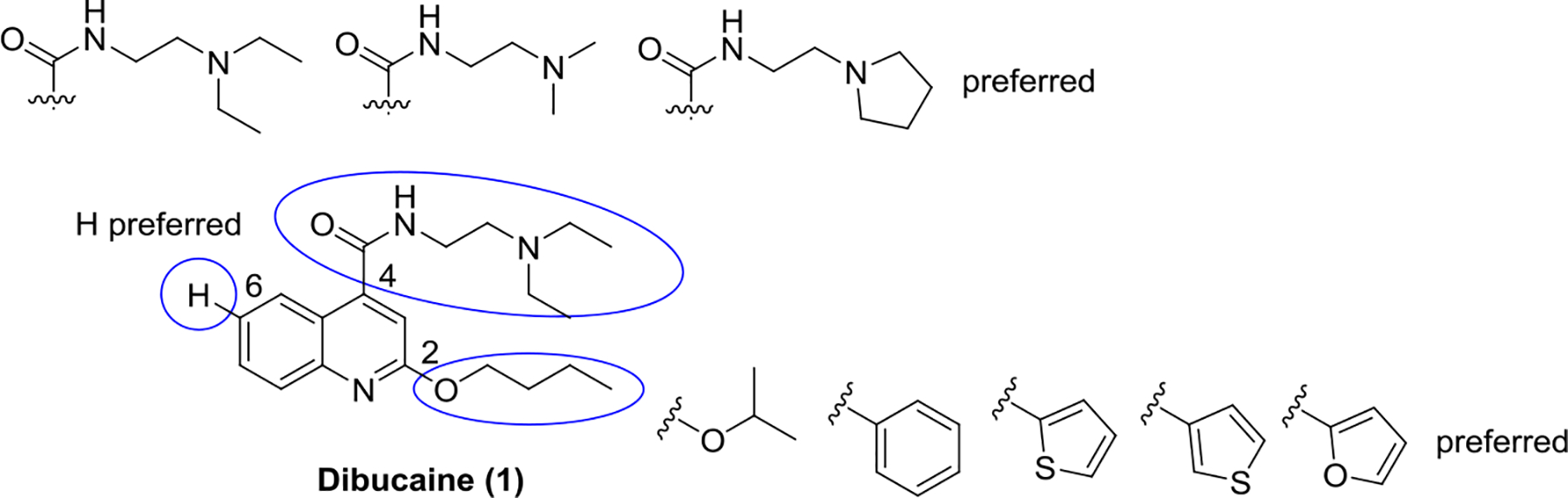
Summary of SAR studies of dibucaine.
BROAD RANGE ANTIVIRAL ACTIVITY
Given the potent antiviral activity and the high selectivity index of lead compounds 10a, 12a, and 12c in inhibiting the EV-D68 US/KY/14-18953 strain, we were interested in exploring their broad range antiviral activity against other contemporary EV-D68 strains. For this, these three compounds were tested against four additional human EV-D68 strains (Table 7). It was found that compounds 10a, 12a, and 12c had similarly potent antiviral activity and high selectivity index against all five EV-D68 strains tested, corroborating that 2C protein is a conserved and high-profile antiviral drug target. The three lead compounds 10a, 12a, and 12c were also well tolerated in two additional cell lines A549 and HeLa (Table 7). Furthermore, as EV-D68 also infects CNS such as spinal cord and brain, we were intrigued to figure out whether these three lead compounds could also inhibit EV-D68 virus replication in the neuronal cells. For this purpose, we infected the human brain glioblastoma A172 cells or the neuroblastoma cell line SH-SY5Y with the EV-D68 virus and quantified viral replication in CPE assay with and without compound treatment (Table 8). Encouragingly, all three compounds had potent antiviral activity against EV-D68 virus replication in neuronal cell lines (EC50 = 0.01 to 0.4 μM). Notably, they were less cytotoxic to A172 and SH-SY5Y cells than the RD cells, resulting in a higher selectivity index. In conclusion, the three lead compounds identified from the SAR studies, 10a, 12a, and 12c inhibits multiple EV-D68 strains with high potency and selectivity index in both muscle cells (RD) and neuronal cells (A172 and SH-SY5Y).
Table 7.
Antiviral activity of potent leads against different serotypes of EV-D68 in RD cells and cytotoxicity of lead compounds in different cell lines
| Compound ID | Antiviral activity and cytotoxicity in RD cells | Cellular cytotoxicity | |||||
|---|---|---|---|---|---|---|---|
| US/KY/14-18953 (μM) |
US/MO/14–18947 (μM) |
US/MO/14–18949 (μM) |
US/IL/14–18956 (μM) |
UA/IL/14–18952 (μM) |
A549 (μM) |
HeLa (μM) |
|
 |
EC50 = 5.3 ±
1.2 CC50 = 56.6 ± 28.7 SI = 10.7 |
EC50 = 3.5 ±
2.3 CC50 = 56.6 ± 28.7 SI = 16.2 |
EC50 = 2.9 ±
1.8 CC50 = 56.6 ± 28.7 SI = 19.5 |
EC50 = 1.8
CC50 = 56.6 ± 28.7 SI = 31.4 |
EC50 = 2.0 ±
0.7 CC50 = 56.6 ± 28.7 SI = 28.3 |
CC50 = 37.5 ± 4.7 | CC50 = 36.0 ± 4.0 |
 |
EC50 = 0.4 ±
0.2 CC50 = 73.7 ± 19.1 SI = 184.3 |
EC50 = 0.3 ±
0.09 CC50 = 73.7 ± 19.1 SI = 245.7 |
EC50 = 0.6 ± 0.1
CC50 = 73.7 ± 19.1 SI = 122.8 |
EC50 = 0.2 ±
0.07 CC50 = 73.7 ± 19.1 SI = 368.5 |
EC50 = 0.6 ± 0.1
CC50 = 73.7 ± 19.1 SI = 122.8 |
CC50 = 121.1 ± 6.2 | CC50 = 68.7 ± 3.8 |
 |
EC50 = 0.04 ±
0.01 CC50 = 42.3 ± 14.3 SI = 1057.5 |
EC50 = 0.06 ±
0.03 CC50 = 42.3 ± 14.3 SI = 705 |
EC50 = 0.05 ±
0.03 CC50 = 42.3 ± 14.3 SI = 846 |
EC50 = 0.06 ±
0.04 CC50 = 42.3 ± 14.3 SI = 705 |
EC50 = 0.05 ±
0.03 CC50 = 42.3 ± 14.3 SI = 846 |
CC50 = 98.8 ± 3.5 | CC50 = 51.2 ± 2.0 |
 |
EC50 = 0.06 ±
0.03 CC50 = 40.0 ± 13.1 SI = 666.7 |
EC50 = 0.06 ±
0.02 CC50 = 40.0 ± 13.1 SI = 666.7 |
EC50 = 0.05 ±
0.03 CC50 = 40.0 ± 13.1 SI = 800 |
EC50 =
0.05 CC50 = 40.0 ± 13.1 SI = 800 |
EC50 = 0.07 ±
0.03 CC50 = 40.0 ± 13.1 SI = 571.4 |
CC50 = 83.2 ± 4.1 | CC50 = 41.7 ± 4.3 |
Table 8.
Antiviral activity of potent leads against different serotypes of EV-D68 in neuronal cells
| Compound ID | Antiviral activity and cytotoxicity in neuronal cells | |||
|---|---|---|---|---|
| US/KY/14-18953 in A172
cells (μM) |
US/KY/14-18953 in SH-SY5Y
cells (μM) |
US/MO/14–18947 in A172 cells (μM) | US/MO/14–18947 in SH-SY5Y cells (μM) | |
 |
EC50 = 3.7 ±
1.5 CC50 > 30.0 SI > 8.1 |
EC50 = 1.7 ±
0.8 CC50 > 30.0 SI > 17.6 |
EC50 = 1.9 ±
0.1 CC50 > 30.0 SI > 15.8 |
EC50 = 1.9 ±
0.3 CC50 > 30.0 SI > 15.8 |
 |
EC50 = 0.4 ±
0.1 CC50 = 88.4 ± 31.6 SI = 221.0 |
EC50 = 0.4 ±
0.1 CC50 > 300.0 SI > 750.0 |
EC50 = 0.3 ± 0.02
CC50 = 88.4 ± 31.6 SI = 294.7 |
EC50 = 0.2 ± 0.07
CC50 > 300.0 SI > 1500.0 |
 |
EC50 = 0.02 ±
0.01 CC50 = 73.8 ± 41.0 SI = 3690.0 |
EC50 = 0.1 ±
0.05 CC50 = 136.3 ± 69.3 SI = 1363.0 |
EC50 = 0.1 ±
0.01 CC50 = 73.8 ± 41.0 SI = 738.0 |
EC50 = 0.01 ±
0.008 CC50 =136.3 ± 69.3 SI = 13630.0 |
 |
EC50 = 0.05 ±
0.00 CC50 = 65.4 ± 22.2 SI = 1308.0 |
EC50 = 0.1 ±
0.08 CC50 = 93.3 ± 30.7 SI = 933.0 |
EC50 = 0.2 ±
0.02 CC50 = 65.4 ± 22.2 SI = 327.0 |
EC50 =
0.02 CC50 = 93.3 ± 30.7 SI = 4665.0 |
MECHANISM OF ACTION
It was reported that dibucaine inhibits EV-D68 through the inhibition of genome replication by targeting the 2C protein.21 The EV-D68 2C protein is a highly conserved viral protein that plays multiple functions in genome replication. It has been implicated in RNA replication, membrane rearrangements, encapsidation, and uncoating.1 Inhibition of viral 2C protein is expected to inhibit viral RNA and protein synthesis. To test this hypothesis and explore the cellular antiviral mechanism of dibucaine analogs 10a, 12a and 12c, we detected viral capsid protein VP1 expression level by immunofluorescence imaging and western blot, and viral RNA levels by RT-qPCR (Fig. 3). Compared to DMSO control, 3 μM of dibucaine significantly reduced the fluorescent signal of VP1 at 9 hours post infection (hpi) (Fig. 3A). The reduction of VP1 protein level was also independently confirmed in western blot experiment (Fig. 3B), whereas 3 μM of dibucaine nearly completely inhibited VP1 expression (Fig. 3B) while 1 μM dibucaine had no effect. In contrast, when tested at 1 μM, dibucaine analogs, 10a, 12a and 12c, completely blocked the VP1 protein expression (Fig. 3A & 3B) as shown by immunofluorescence imaging and western blot. Similarly, RT-qPCR quantification of viral RNAs showed that 1 μM of dibucaine analogs, 10a, 12a and 12c, completely inhibited the viral RNA synthesis (> 99%), whereas 3 μM of dibucaine reduced viral RNA level by ~80% (Fig. 3C). Taken together, our results confirmed that dibucaine and its analogs share similar mechanism of action, and the analogs had more potent antiviral activity than dibucaine.
Figure 3.
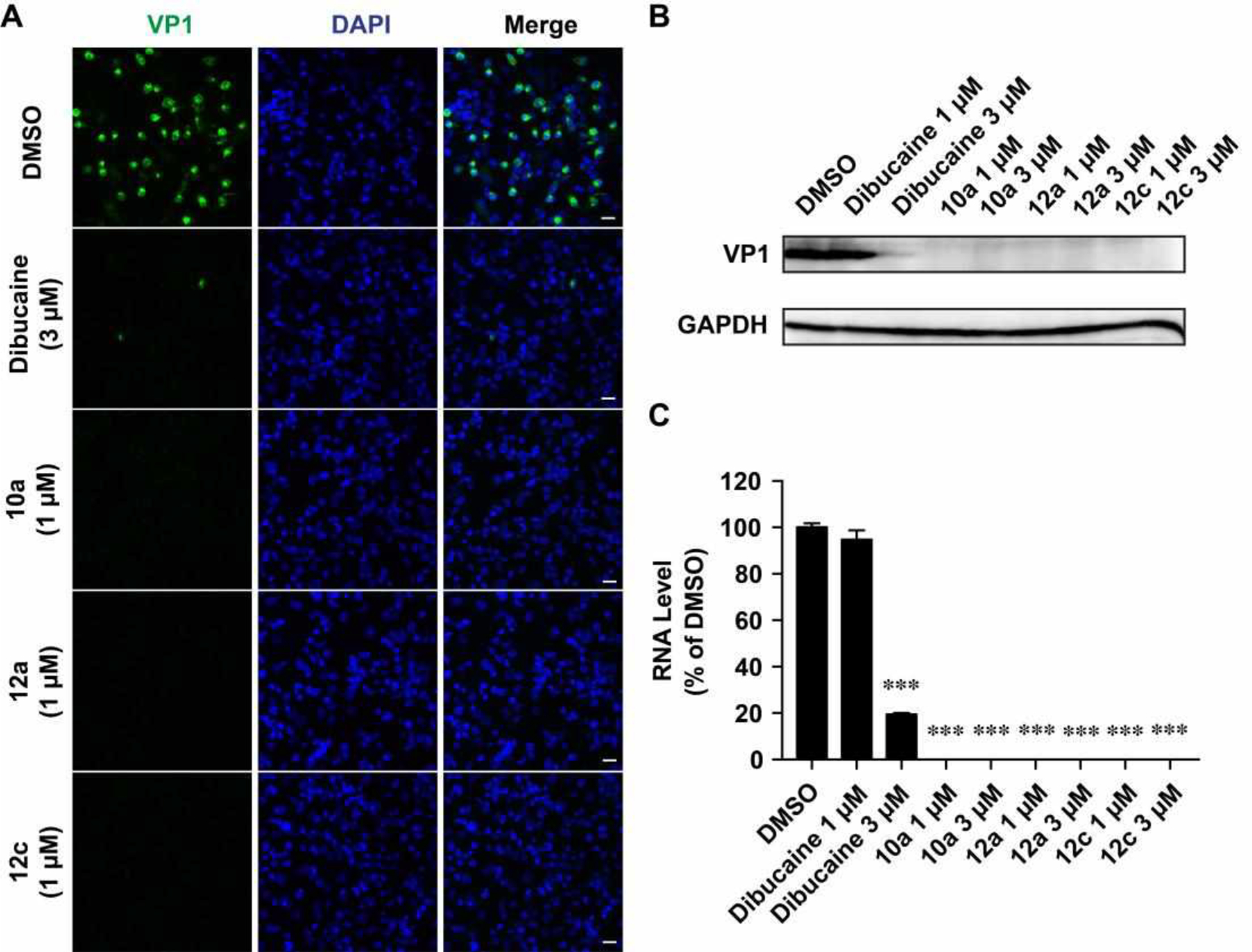
Dibucaine and its analogs 10a, 12a, and 12c reduced the EV-D68 viral protein and RNA levels. (A) Dibucaine and its analogs inhibited EV-D68 replication. RD cells infected with EV-D68 strain US/KY/14-18953 (MOI = 1) were fixed at 9 hpi and stained with anti-VP1 and DAPI for detecting VP1 and nucleus, respectively. (B and C) Dibucaine and its analogs reduced the EV-D68 viral protein and RNA levels. RD cells were infected with EV-D68 strain US/KY/14-18953 (MOI = 1) with treatment of indicated compound. At 9 hpi, cells were harvested for viral protein quantification by western or viral RNA quantification by RT-qPCR. Asterisks indicate statistically significant difference in comparison with the DMSO control (one-way ANOVA analysis by Prism 5, ***P < 0.001). The value is the mean of two independent experiments ± standard deviation.
POTENTIAL SIDE EFFECTS
Dibucaine is a FDA-approved topical anesthetic drug. Its anesthetic activity is due to binding and inhibiting sodium channels within neuronal cell membranes. For the purpose of antiviral development, inhibition of the sodium channel might be an unwanted side effect. As the three lead compounds 10a, 12a, and 12c were derived from dibucaine, we were concerned that they might have anesthetic side effect by inhibiting the sodium channel. To ease this doubt, we chose one representative compound 10a and tested its sodium channel inhibition using the standard sodium flux assay. Dibucaine was included as a positive control. In this assay, rat dorsal root ganglia (DRG) neurons were incubated with various concentrations of testing compounds. Sodium channels were then activated by the sodium channel agonist veratridine. The resulting sodium flux was measured indirectly by quantifying the sodium channel triggered calcium flux, which was measured by a calcium-specific fluorescence dye (Fig. 4). It was found that dibucaine potently inhibited sodium current conductance at both 20 μM and 50 μM, while compounds 10a had minimal to no effect from 2 μM to 50 μM, nearly 100-fold higher than its antiviral EC50 value. These results confirmed that the quinoline lead compounds identified from this study such as 10a are promising candidates with no potential anesthetic side effects, suggesting they are promising for further development. To address the concern that the optimized lead compounds might be promiscuous antivirals, we tested the three lead compounds 10a, 12a, and 12c against two influenza viruses, the influenza A virus A/California/07/2009 (H1N1), and the influenza B virus B/Brisbane/60/2008 (Victoria), in plaque assay.25 It was found that dibucaine and the three lead compounds had no inhibition against these two strains of influenza viruses when tested at 10 μM (Fig. 5), indicating the antiviral activity of the lead compounds 10a, 12a, and 12c, against EV-D68 was not due to non-specific effect of these compounds.
Figure 4.
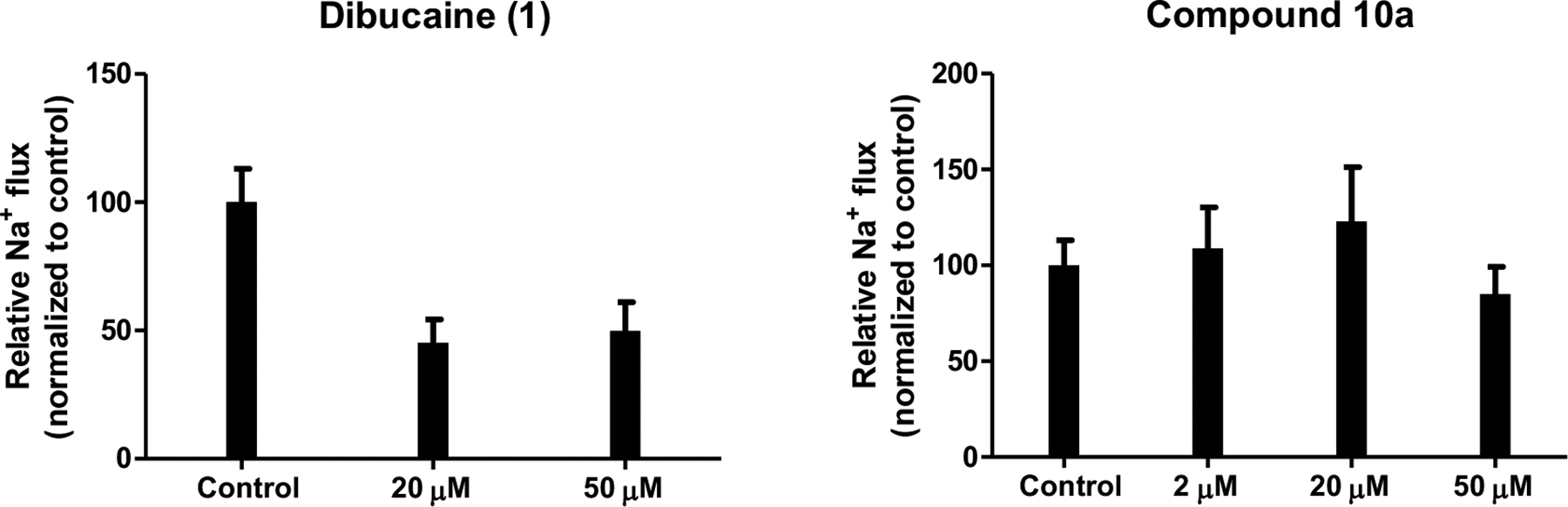
Inhibition of sodium channels by dibucaine and lead compound 10a.
Figure 5.
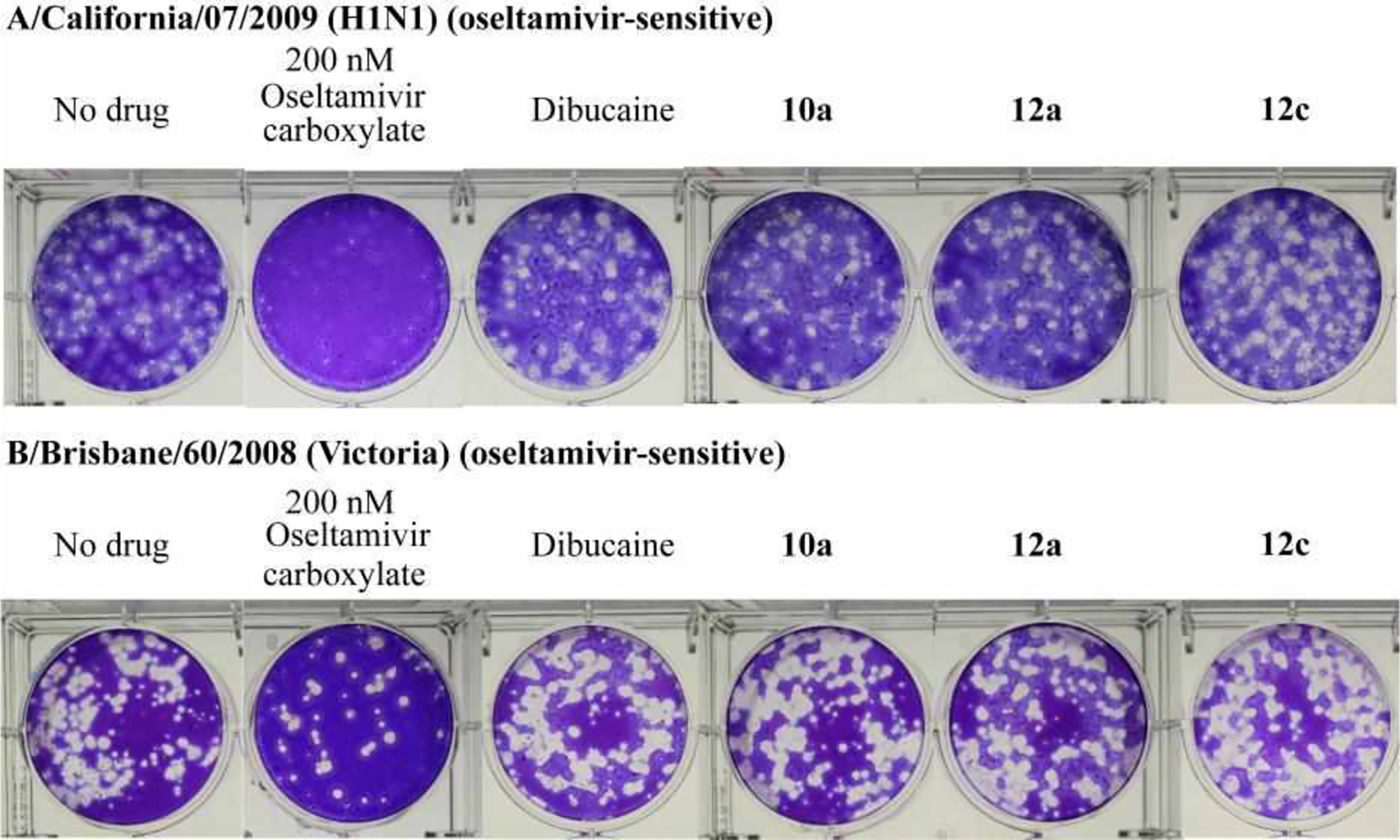
Dibucaine and the optimized lead compounds did not inhibit influenza A and B viruses. Plaque assay was performed using the condition described previously.25 Dibucaine, 10a, 12a, and 12c were tested at 10 μM. Oseltamivir carboxylate was included as a positive control and was tested at 200 nM.
CONCLUSIONS
The magnitude and severity of EV-D68 outbreak in recent years underscores a need for effective antivirals, not only to be used for prophylaxis, but also for the treatment of EV-D68 infection. In this study, we chose a previously reported hit compound, dibucaine, as a starting point for the structure-activity relationship studies. Dibucaine was identified as an EV-D68 antiviral from a drug repurposing phenotypic screening. However, the moderate antiviral activity and a low selectivity index of dibucaine impeded its clinical use as an antiviral. As such, we aim to optimize the potency and selectivity index of dibucaine through structure-activity relationship studies. This effort led to the identification of three lead compounds 10a, 12a, and 12c, with significantly improved antiviral efficacy and selectivity index (EC50 < 1 μM, SI > 180). The antiviral mechanism of action of these three compounds were investigated by immunofluorescence staining, western blot, and RT-qPCR assays. Collectively, all three compounds inhibited viral protein and RNA synthesis, consistent with the proposed mechanism of 2C protein inhibition. Nevertheless, to confirm the mechanism of action, serial viral passage experiments need to be designed to select resistant mutant in viral 2C protein. Alternatively, direct binding needs to be measured between lead compounds and viral 2C protein. The lead compounds might either affect the function of viral 2C protein directly or disrupt essential interactions between viral 2C protein and host factors. Such experiments are ongoing and will be reported when available. To our knowledge, these three compounds represent the most potent and select EV-D68 2C inhibitors reported so far. Of particular highlights are the easy synthesis accessibility of these compounds which can be made in two steps from commercial available materials, broad range antiviral activity against all five contemporary EV-D68 strains, and consistent antiviral activity across different cell lines including muscle cells (RD) and neuronal cells (A172 and SH-SY5Y). It has been shown that recent circulating EV-D68 strains such as the ones tested in this study acquired the fitness of replication in neuronal cells. Therefore it is important for an antiviral drug candidate to show consistent antiviral activity in different cell lines. It should be pointed out that although EV-D68 infection has a direct link with AFM,26 it is not clear whether AFM is a result of viral replication in the CNS (spinal cord and brain) or due to inflammation caused by viral infection. Therefore it is desired to develop both blood-brain-barrier (BBB) permeable and impermeable EV-D68 antivirals and test them in mouse model studies to dissect whether BBB permeability is essential for AFM treatment. The compounds reported herein represent not only lead candidates for further translational development into EV-D68 antivirals, but also valuable chemical probes to help understand the structure, function, and mechanism of EV-D68 2C protein. Accordingly, future efforts will focus on optimizing the in vitro and in vivo pharmacokinetic properties of this series of compounds and evaluating their in vivo antiviral efficacy in mouse model studies. On another hand, the structure of the EV-D68 2C protein is currently unknown. The inhibitors discovered herein might be able to stabilize the 2C protein and help facilitate its structure determination by either NMR or X-ray crystallography. A high-resolution crystal structure of EV-D68 2C is highly desired as it will be invaluable in guiding the rational design of next-generation of 2C inhibitors.
EXPERIMENTAL SECTION
Chemistry.
Chemicals were ordered from commercial sources and were used without further purification. Synthesis procedures for reactions described in Scheme 1 were shown below. All final compounds were purified by flash column chromatography. 1H and 13C NMR spectra were recorded on a Bruker-400 NMR spectrometer. Chemical shifts are reported in parts per million referenced with respect to residual solvent (CD3OD) 3.31 ppm, (DMSO-d6) 2.50 ppm, and (CDCl3) 7.24 ppm or from internal standard tetramethylsilane (TMS) 0.00 ppm. The following abbreviations were used in reporting spectra: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets; ddd, doublet of doublet of doublets. All reactions were carried out under N2 atmosphere unless otherwise stated. HPLC-grade solvents were used for all reactions. Flash column chromatography was performed using silica gel (230–400 mesh, Merck). High-resolution mass spectra were obtained using the positive ESI method for all the compounds, obtained in an Ion Cyclotron Resonance (ICR) spectrometer. Low-resolution mass spectra were obtained using an ESI technique on a 3200 Q Trap LC/MS/MS system (Applied Biosystems). The purity was assessed by using a Shimadzu LC-MS with a Waters XTerra MS C-18 column (part no. 186000538), 50 mm × 2.1 mm, at a flow rate of 0.3 mL/min; λ = 250 and 220 nm; mobile phase A, 0.1% formic acid in H2O, and mobile phase B′, 0.1% formic in 60% 2-propanol, 30% CH3CN, and 9.9% H2O. All compounds submitted for antiviral CPE assay, cytotoxicity assay, and mechanistic studies were confirmed to be >95.0% purity by LC-MS traces.
Synthesis procedures.
General procedure of amide coupling.
To a DMF solution of carboxylic acid (1 mmol) was added HATU (1 mmol) and DIEA (1.2 mmol). After stirring for 2 minutes, amine (1 mmol) was added. The resulting solution was stirred overnight at room temperature. The reaction was diluted with dichloromethane and extracted with aqueous NaHCO3 solution, followed by brine. The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (1–10% CH3OH/CH2Cl2) to give the final product.
General procedure of Suzuki-Miyaura cross coupling.
To a solution of 2-chloroquinoline (1 mmol) and boronic acid (1mmol) in 1,4-dioxane in a microwave reaction vial was added an aqueous solution of K2CO3 (2 mmol). The resulting solution was purged with N2 for 5 minutes. The catalyst, Pd(PPh3)4 (0.1 mmol), was added in one portion. The vial was capped and heated to 140 °C for 30 minutes with microwave irradiation. After cooling down to room temperature, the reaction solution was diluted with CH2Cl2 and extracted with aqueous NaHCO3 solution, followed by brine. The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (1–10% CH3OH/CH2Cl2) to give the final product.
Compounds 4, 8, 10, 12, and 15 were synthesized using the general procedure of amide coupling.
Compounds 6 and 19 were synthesized using the general procedure of Suzuki-Miyaura cross coupling.
Procedure for the synthesis of compound 5 through method 1.
Sodium metal (10 mmol) was cut into small pieces and was added to the alcohol solution. The solution was stirred at room temperature under N2 until all the sodium metal was dissolved (heating to 60 °C when necessary). 2-chloro-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (4) (1 mmol) was added in one portion and the resulting solution was heated to reflux overnight. Reaction was quenched with H2O and excess of alcohol was removed under reduced pressure. The crude product was diluted with CH2Cl2 and extracted with aqueous NaHCO3 solution, followed by brine. The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (1–10% CH3OH/CH2Cl2) to give the final product.
Procedure for the synthesis of compound 5 through method 2.
To a solution of alcohol (2 mmol) in DMF was added NaH (3 mmol), and the mixture was stirred at room temperature for 1 hour. A solution of 2-chloro-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (4) (1 mmol) in DMF was added dropwise, and the resulting solution was stirred at 80 °C for 4 hours. The reaction mixture was quenched with aqueous NaHCO3 solution, and extracted with CH2Cl2. The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (1–10% CH3OH/CH2Cl2) to give the final product.
Procedure for the synthesis of compound 14.
To a solution of isatin (1 mmol) and acetophenone (1 mmol) in ethanol was added the aqueous solution of KOH (10 mmol). The reaction mixture was heated to 125 °C under microwave irradiation for 20 minutes. After cooling down to room temperature, the reaction was quenched with HCl. The precipitate was collected by filtration, washed sequentially with water, ethyl acetate, and dichloromethane. The crude product was purified by flash column chromatography (10–20% CH3OH/CH2Cl2) to give the final product.
Procedure for the synthesis of compound 17.
A solution of 5-fluoroisatin (1 mmol) and malonic acid (3 mmol) in acetic acid was refluxed for 16 hours. Acetic acid was removed under reduced pressure, and the residue was diluted with water. Insoluble solid was collected by filtration and was suspended in saturated NaHCO3 aqueous solution. The mixture was filtered and the filtrate was acidified to pH 1 using concentrated HCl (12 N). Precipitate formed was filtered and washed with water. The resulting solid was further purified by flash column chromatography (10–20% CH3OH/CH2Cl2) to give the final product.
General procedure for the synthesis of compound 18.
To a solution of 6-fluoro-2-hydroxyquinoline-4-carboxylic acid (17) (1 mmol) in CH2Cl2 was added a few drops of DMF and thionyl chloride (4 mmol). The mixture was refluxed for 3 hours. Solvent was removed under reduced pressure and the resulting acid chloride was dissolved in THF. Amine (3 mmol) was added and the reaction mixture was stirred at room temperature overnight. Solvent was removed under reduced pressure and the resulting residue was dissolved in CH2Cl2. The solution was extracted with aqueous NaHCO3 solution, followed by brine. The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude product was purified by flash column chromatography (1–10% CH3OH/CH2Cl2) to give the final product.
2-chloro-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (4).
Yield: 82%. 1H NMR (400 MHz, CD3OD-d4) δ 8.18 (d, J = 8.9 Hz, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.79 (t, J = 7.7 Hz, 1H), 7.63 (t, J = 8.3 Hz, 1H), 7.58 (s, 1H), 3.62 (t, J = 6.9 Hz, 2H), 2.89 (t, J = 6.9 Hz, 2H), 2.78 (q, J = 7.2 Hz, 4H), 1.14 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, CD3OD) δ 168.19, 151.20, 149.28, 146.61, 132.35, 129.29, 128.99, 126.69, 124.63, 121.29, 52.30, 48.20, 37.90, 11.18. C16H20ClN3O, EI-MS: m/z (M+H+): 306.8 (calculated), 306.8 (found).
N-[2-(diethylamino)ethyl]-2-methoxyquinoline-4-carboxamide (5a).
Yield: 72%. 1H NMR (400 MHz, CD3OD-d4) δ 8.06 (dd, J = 8.4, 1.4 Hz, 1H), 7.85 (dd, J = 8.5, 1.5 Hz, 1H), 7.66 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.43 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.02 (s, 1H), 4.07 (s, 3H), 3.66 – 3.56 (m, 2H), 2.90 (t, J = 7.0 Hz, 2H), 2.81 (q, J = 7.2 Hz, 4H), 1.16 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, CD3OD) δ 169.71, 163.13, 148.38, 146.29, 131.18, 128.50, 126.32, 125.79, 122.73, 112.16, 54.12, 52.35, 48.32, 37.75, 11.13. C17H23N3O2, HRMS (ESI): m/z (M+H+): 302.1869 (calculated), 302.1863 (found).
N-[2-(diethylamino)ethyl]-2-ethoxyquinoline-4-carboxamide (5b).
Yield: 84%. 1H NMR (400 MHz, DMSO-d6) δ 8.72 – 8.61 (m, 1H), 8.08 (dd, J = 8.3, 1.4 Hz, 1H), 7.87 – 7.74 (m, 1H), 7.68 (ddd, J = 8.3, 6.8, 1.5 Hz, 1H), 7.44 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 6.99 (d, J = 11.4 Hz, 1H), 4.49 (q, J = 7.0 Hz, 2H), 3.46 – 3.32 (m, 2H), 2.75 – 2.52 (m, 6H), 1.39 (t, J = 7.0 Hz, 3H), 1.01 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.03, 160.91, 146.40, 145.71, 130.05, 130.00, 127.08, 125.61, 125.58, 124.51, 124.41, 121.43, 110.87, 61.50, 53.37, 51.29, 46.57, 14.39, 11.62. C18H25N3O2, HRMS (ESI): m/z (M+H+): 316.2025 (calculated), 316.2020 (found).
N-[2-(diethylamino)ethyl]-2-(2,2,2-trifluoroethoxy)quinoline-4-carboxamide (5c).
Yield: 71%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 – 8.67 (m, 1H), 8.14 (ddd, J = 8.3, 1.5, 0.6 Hz, 1H), 7.86 (ddd, J = 8.5, 1.4, 0.7 Hz, 1H), 7.75 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.52 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.17 (s, 1H), 5.20 (q, J = 9.1 Hz, 2H), 3.53 – 3.33 (m, 2H), 2.76 – 2.52 (m, 6H), 1.01 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO): δ 165.68, 158.94, 146.77, 145.60, 130.55, 127.19, 125.76, 125.41, 125.37, 122.65, 122.10, 110.08, 61.91, 61.56, 61.22, 60.87, 51.30, 46.57, 37.17, 11.62. C18H22F3N3O2, HRMS (ESI): m/z (M+H+): 370.1742 (calculated), 370.1737 (found).
N-[2-(diethylamino)ethyl]-2-propoxyquinoline-4-carboxamide (5d).
Yield: 87%. 1H NMR (400 MHz, DMSO-d6) δ 8.80 – 8.64 (m, 1H), 8.08 (dd, J = 8.3, 1.4 Hz, 1H), 7.79 (dd, J = 8.4, 1.2 Hz, 1H), 7.68 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.44 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.02 (s, 1H), 4.39 (t, J = 6.6 Hz, 2H), 3.51 – 3.35 (m, 2H), 2.90 – 2.52 (m, 6H), 1.79 (p, J = 7.1 Hz, 2H), 1.14 – 0.91 (m, 9H). 13C NMR (101 MHz, DMSO) δ 166.08, 161.08, 146.40, 145.54, 130.00, 127.08, 125.57, 124.42, 121.42, 110.94, 67.22, 51.08, 48.57, 46.56, 21.75, 11.30, 10.43. C19H27N3O2, HRMS (ESI): m/z (M+H+): 330.2182 (calculated), 330.2176 (found).
N-[2-(diethylamino)ethyl]-2-(propan-2-yloxy)quinoline-4-carboxamide (5e).
Yield: 80%. 1H NMR (400 MHz, CD3OD-d4) δ 8.04 (dd, J = 8.4, 1.4 Hz, 1H), 7.81 (dd, J = 8.6, 1.2 Hz, 1H), 7.65 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.41 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 6.96 (s, 1H), 5.55 (p, J = 6.2 Hz, 1H), 3.59 (dd, J = 7.6, 6.4 Hz, 2H), 2.86 (dd, J = 7.7, 6.3 Hz, 2H), 2.77 (q, J = 7.2 Hz, 4H), 1.41 (d, J = 6.2 Hz, 6H), 1.15 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, MeOD) δ 169.79, 162.31, 148.50, 146.28, 131.06, 128.49, 126.24, 125.57, 122.54, 112.77, 69.59, 52.39, 48.26, 37.88, 22.30, 11.28. C19H27N3O2, HRMS (ESI): m/z (M+H+): 330.2182 (calculated), 330.2176 (found).
2-(butan-2-yloxy)-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (5f).
Yield: 72%. 1H NMR (400 MHz, DMSO-d6) δ 8.85 – 8.67 (m, 1H), 8.06 (dd, J = 8.3, 1.4 Hz, 1H), 7.77 (dd, J = 8.5, 1.2 Hz, 1H), 7.67 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.43 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 6.99 (s, 1H), 5.35 (q, J = 6.2 Hz, 1H), 3.57 – 3.36 (m, 2H), 2.98 – 2.52 (m, 6H), 1.81 – 1.61 (m, 2H), 1.33 (d, J = 6.2 Hz, 3H), 1.06 (d, J = 7.5 Hz, 6H), 0.95 (t, J = 7.4 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 166.16, 160.74, 146.42, 129.98, 127.06, 125.50, 124.33, 121.26, 111.41, 72.37, 46.54, 28.38, 19.17, 9.62. C20H29N3O2, HRMS (ESI): m/z (M+H+): 344.2338 (calculated), 344.2333 (found).
2-(cyclopropylmethoxy)-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (5g).
Yield: 71%. 1H NMR (400 MHz, DMSO-d6) δ 8.79 – 8.64 (m, 1H), 8.08 (dd, J = 8.3, 1.4 Hz, 1H), 7.78 (dd, J = 8.4, 1.2 Hz, 1H), 7.67 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.44 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 7.03 (s, 1H), 4.28 (d, J = 7.2 Hz, 2H), 3.53 – 3.37 (m, 2H), 2.88 – 2.57 (m, 6H), 1.38 – 1.25 (m, 1H), 1.04 (t, J = 7.1 Hz, 6H), 0.66 – 0.51 (m, 2H), 0.45 – 0.32 (m, 2H). 13C NMR (101 MHz, DMSO) δ 166.10, 161.00, 146.36, 130.02, 127.05, 125.56, 124.43, 121.40, 110.99, 70.30, 54.90, 51.03, 46.57, 11.17, 9.96, 3.19. C20H27N3O2, HRMS (ESI): m/z (M+H+): 342.2182 (calculated), 342.2176 (found).
N-[2-(diethylamino)ethyl]-2-(pentan-3-yloxy)quinoline-4-carboxamide (5h).
Yield: 77%. 1H NMR (400 MHz, DMSO-d6) δ 8.74 – 8.62 (m, 1H), 8.06 (dd, J = 8.3, 1.4 Hz, 1H), 7.75 (dd, J = 8.5, 1.2 Hz, 1H), 7.66 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.42 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 6.97 (s, 1H), 5.30 (p, J = 6.0 Hz, 1H), 3.47 – 3.32 (m, 2H), 2.72 – 2.51 (m, 6H), 1.82 – 1.61 (m, 4H), 1.01 (t, J = 7.1 Hz, 6H), 0.92 (t, J = 7.4 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.08, 162.24, 161.18, 146.36, 145.74, 129.93, 127.03, 125.51, 124.25, 121.33, 111.19, 76.67, 51.23, 46.57, 37.00, 35.74, 30.73, 25.67, 11.52, 9.46. C21H31N3O2, HRMS (ESI): m/z (M+H+): 358.2495 (calculated), 358.2489 (found).
2-(cyclopentyloxy)-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (5i).
Yield: 82%. 1H NMR (400 MHz, DMSO-d6) δ 8.75 – 8.60 (m, 1H), 8.08 (d, J = 8.2 Hz, 1H), 7.78 (d, J = 8.4 Hz, 1H), 7.67 (t, J = 7.7 Hz, 1H), 7.42 (t, J = 7.9 Hz, 1H), 6.94 (s, 1H), 5.65 – 5.54 (m, 1H), 3.49 – 3.31 (m, 2H), 2.80 – 2.53 (m, 6H), 2.09 – 1.95 (m, 2H), 1.83 – 1.68 (m, 4H), 1.68 – 1.54 (m, 2H), 1.02 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.07, 160.68, 146.48, 145.48, 129.93, 127.16, 125.54, 124.32, 121.29, 111.30, 77.53, 54.89, 51.17, 46.57, 36.90, 32.36, 23.48, 11.43. C21H29N3O2, HRMS (ESI): m/z (M+H+): 356.2338 (calculated), 356.2333 (found).
N-[2-(diethylamino)ethyl]-2-(2-ethoxyethoxy)quinoline-4-carboxamide (5j).
Yield: 62%. 1H NMR (400 MHz, DMSO-d6) δ 8.78 – 8.64 (m, 1H), 8.09 (ddd, J = 7.4, 5.8, 1.4 Hz, 1H), 7.79 (dd, J = 8.4, 1.4 Hz, 1H), 7.68 (dddd, J = 8.4, 6.9, 3.2, 1.5 Hz, 1H), 7.44 (dddd, J = 8.3, 6.9, 4.0, 1.3 Hz, 1H), 7.02 (d, J = 18.9 Hz, 1H), 4.63 – 4.41 (m, 2H), 3.83 – 3.70 (m, 2H), 3.51 (q, J = 7.0 Hz, 2H), 3.47 – 3.31 (m, 2H), 2.83 – 2.54 (m, 6H), 1.13 (t, J = 7.0 Hz, 3H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.02, 160.86, 146.27, 145.66, 130.06, 127.09, 125.60, 124.53, 121.50, 110.92, 68.03, 65.61, 65.13, 61.51, 51.13, 46.57, 15.09, 14.39. C20H29N3O3, HRMS (ESI): m/z (M+H+): 360.2287 (calculated), 360.2282 (found).
2-(benzyloxy)-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (5k).
Yield: 84%. 1H NMR (400 MHz, CD3OD-d4) δ 8.07 (dd, J = 8.4, 1.4 Hz, 1H), 7.86 (dd, J = 8.5, 1.3 Hz, 1H), 7.66 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.56 – 7.47 (m, 2H), 7.47 – 7.41 (m, 1H), 7.41 – 7.33 (m, 2H), 7.33 – 7.24 (m, 1H), 7.08 (s, 1H), 5.54 (s, 2H), 3.60 (t, J = 7.0 Hz, 2H), 2.90 (t, J = 7.0 Hz, 2H), 2.80 (q, J = 7.2 Hz, 4H), 1.15 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, MeOD) δ 169.71, 162.46, 148.30, 146.41, 138.44, 131.23, 129.43, 129.25, 128.95, 128.58, 126.31, 125.89, 122.84, 112.31, 68.92, 52.32, 48.36, 37.71, 11.07. C23H27N3O2, HRMS (ESI): m/z (M+H+): 378.2182 (calculated), 378.2176 (found).
N-[2-(diethylamino)ethyl]-2-(1H-pyrazol-5-yl)quinoline-4-carboxamide (6a).
Yield: 71%. 1H NMR (400 MHz, CD3OD-d4) δ 8.25 – 8.02 (m, 3H), 7.84 – 7.72 (m, 2H), 7.59 (ddd, J = 8.3, 6.9, 1.2 Hz, 1H), 7.12 (d, J = 2.3 Hz, 1H), 3.60 (dd, J = 8.0, 6.3 Hz, 2H), 2.79 (dd, J = 7.9, 6.4 Hz, 2H), 2.68 (q, J = 7.2 Hz, 4H), 1.12 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, MeOD) δ 169.88, 149.49, 144.51, 131.44, 130.09, 128.27, 126.43, 125.09, 117.66, 105.41, 52.51, 48.14, 38.33, 11.68. C19H23N5O, HRMS (ESI): m/z (M+H+): 338.1981 (calculated), 338.1975 (found).
N-[2-(diethylamino)ethyl]-2-(1-methyl-1H-pyrazol-4-yl)quinoline-4-carboxamide (6b).
Yield: 78%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 – 8.71 (m, 1H), 8.52 (s, 1H), 8.20 (s, 1H), 8.14 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 7.97 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.86 (s, 1H), 7.74 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.53 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 3.94 (s, 3H), 3.52 – 3.42 (m, 2H), 2.82 – 2.70 (m, 2H), 2.70 – 2.53 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.66, 151.51, 147.97, 143.17, 137.82, 130.62, 129.93, 128.64, 125.95, 125.50, 122.83, 122.68, 116.48, 51.29, 46.60, 38.83, 36.97, 11.45. C20H25N5O, HRMS (ESI): m/z (M+H+): 352.2137 (calculated), 352.2132 (found).
N-[2-(diethylamino)ethyl]-2-(1-methyl-1H-pyrazol-5-yl)quinoline-4-carboxamide (6c).
Yield: 65%. 1H NMR (400 MHz, DMSO-d6) δ 8.87 – 8.79 (m, 1H), 8.24 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.11 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.97 (s, 1H), 7.83 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.66 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.58 (d, J = 2.0 Hz, 1H), 7.10 (d, J = 2.0 Hz, 1H), 4.35 (s, 3H), 3.53 – 3.43 (m, 2H), 2.80 – 2.70 (m, 2H), 2.70 – 2.54 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.24, 148.85, 147.21, 143.44, 139.95, 137.91, 130.39, 129.31, 127.42, 125.53, 122.95, 118.36, 108.21, 51.23, 46.60, 39.91, 37.00, 11.37. C20H25N5O, HRMS (ESI): m/z (M+H+): 352.2137 (calculated), 352.2132 (found).
N-[2-(diethylamino)ethyl]-2-(thiophen-2-yl)quinoline-4-carboxamide (6d).
Yield: 82%. 1H NMR (400 MHz, DMSO-d6) δ 9.00 – 8.88 (br s, 1H), 8.26 – 8.15 (m, 2H), 8.09 (d, J = 3.7 Hz, 1H), 8.06 – 7.94 (m, 1H), 7.87 – 7.72 (m, 2H), 7.62 – 7.55 (m, 1H), 7.27 – 7.21 (m, 1H), 3.70 – 3.46 (m, 2H), 3.04 – 2.57 (m, 6H), 1.11 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.51, 151.61, 147.64, 144.25, 143.00, 130.33, 130.02, 128.77, 128.59, 127.65, 126.72, 125.56, 123.31, 115.64, 50.81, 46.55, 36.25, 10.60. C20H23N3OS, HRMS (ESI): m/z (M+H+): 354.1640 (calculated), 354.1629 (found).
2-(5-chlorothiophen-2-yl)-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (6e).
Yield: 85%. 1H NMR (400 MHz, CD3OD-d4) δ 8.15 (ddd, J = 8.4, 1.4, 0.7 Hz, 1H), 8.09 – 7.95 (m, 2H), 7.75 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.70 (d, J = 4.0 Hz, 1H), 7.56 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.05 (d, J = 4.0 Hz, 1H), 3.70 (dd, J = 7.4, 6.4 Hz, 2H), 3.04 (t, J = 6.9 Hz, 2H), 2.93 (q, J = 7.2 Hz, 4H), 1.22 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, CD3OD) δ 169.95, 152.46, 149.54, 144.76, 143.89, 134.84, 131.61, 130.19, 128.90, 128.25, 127.47, 126.36, 124.77, 115.92, 52.32, 48.43, 37.57, 10.76. C20H22ClN3OS, HRMS (ESI): m/z (M+H+): 388.1250 (calculated), 389.1245 (found).
N-[2-(diethylamino)ethyl]-2-(thiophen-3-yl)quinoline-4-carboxamide (6f).
Yield: 86%. 1H NMR (400 MHz, CD3OD-d4) δ 8.21 (dd, J = 3.0, 1.3 Hz, 1H), 8.14 (dd, J = 8.4, 1.4 Hz, 1H), 8.09 – 8.01 (m, 1H), 7.98 (s, 1H), 7.88 (dd, J = 5.1, 1.3 Hz, 1H), 7.71 (ddd, J = 8.5, 6.9, 1.5 Hz, 1H), 7.58 – 7.45 (m, 2H), 3.60 (t, J = 7.0 Hz, 2H), 2.87 (t, J = 7.0 Hz, 2H), 2.74 (q, J = 7.2 Hz, 4H), 1.12 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, CD3OD) δ 169.90, 154.33, 149.58, 144.07, 142.93, 131.35, 130.15, 128.03, 127.74, 127.72, 126.84, 126.30, 124.57, 118.37, 52.36, 48.22, 37.91, 11.19. C20H23N3OS, HRMS (ESI): m/z (M+H+): 354.1640 (calculated), 354.1629 (found).
N-[2-(diethylamino)ethyl]-2-(furan-3-yl)quinoline-4-carboxamide (6g).
Yield: 84%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 – 8.73 (m, 1H), 8.62 (dd, J = 1.6, 0.8 Hz, 1H), 8.17 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.03 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.94 (s, 1H), 7.88 – 7.82 (m, 1H), 7.77 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.58 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 7.23 (dd, J = 1.9, 0.8 Hz, 1H), 3.58 – 3.37 (m, 2H), 2.79 – 2.69 (m, 2H), 2.69 – 2.55 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 167.03, 151.43, 148.29, 145.25, 143.98, 143.83, 130.52, 129.42, 127.49, 126.98, 125.99, 123.66, 117.30, 109.35, 51.80, 47.07, 37.56, 11.95. C20H23N3O2, HRMS (ESI): m/z (M+H+): 338.1869 (calculated), 338.1864 (found).
N-[2-(diethylamino)ethyl]-2-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-4-carboxamide (6h).
Yield: 68%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 – 8.75 (m, 1H), 8.23 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.08 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.84 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.76 – 7.54 (m, 2H), 3.59 – 3.39 (m, 2H), 2.76 – 2.68 (m, 2H), 2.71 (s, 3H), 2.68 – 2.56 (m, 4H), 2.52 (s, 3H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 168.81, 166.82, 159.08, 150.56, 148.27, 143.91, 130.74, 129.59, 127.62, 125.96, 123.20, 118.84, 115.81, 51.80, 47.03, 37.63, 12.89, 11.90. C21H26N4O2, HRMS (ESI): m/z (M+H+): 367.2134 (calculated), 367.2122 (found).
N-[2-(diethylamino)ethyl]-2-(pyridin-4-yl)quinoline-4-carboxamide (6i).
Yield: 85%. 1H NMR (400 MHz, DMSO-d6) δ 8.92 – 8.82 (m, 1H), 8.83 – 8.71 (m, 2H), 8.38 – 8.22 (m, 4H), 8.18 (ddd, J = 8.5, 1.3, 0.7 Hz, 1H), 7.86 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.70 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 3.55 – 3.43 (m, 2H), 2.81 – 2.70 (m, 2H), 2.68 – 2.54 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.36, 153.40, 150.49, 147.86, 145.07, 143.76, 130.50, 129.72, 127.89, 125.61, 124.08, 121.28, 116.65, 51.30, 46.60, 37.18, 11.51. C21H24N4O, HRMS (ESI): m/z (M+H+): 349.2028 (calculated), 349.2022 (found).
N-[2-(diethylamino)ethyl]-2-phenylquinoline-4-carboxamide (6j).
Yield: 92%. 1H NMR (400 MHz, DMSO-d6) δ 8.88 – 8.77 (m, 1H), 8.34 – 8.22 (m, 3H), 8.20 – 8.05 (m, 2H), 7.82 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.70 – 7.45 (m, 4H), 3.48 (q, J = 6.5 Hz, 2H), 2.82 – 2.69 (m, 2H), 2.69 – 2.54 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.61, 155.74, 147.93, 143.29, 138.24, 130.13, 129.84, 129.47, 128.88, 127.23, 126.98, 125.52, 123.40, 116.58, 51.32, 46.63, 38.21, 11.55. C22H25N3O, HRMS (ESI): m/z (M+H+): 348.2076 (calculated), 348.2069 (found).
N-[2-(diethylamino)ethyl]-2-(2-methoxyphenyl)quinoline-4-carboxamide (6k).
Yield: 88%. 1H NMR (400 MHz, DMSO-d6) δ 8.69 (t, J = 5.7 Hz, 1H), 8.23 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.10 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.92 (s, 1H), 7.86 – 7.74 (m, 2H), 7.63 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.49 (ddd, J = 8.3, 7.3, 1.8 Hz, 1H), 7.21 (dd, J = 8.4, 1.0 Hz, 1H), 7.13 (td, J = 7.4, 1.0 Hz, 1H), 3.86 (s, 3H), 3.45 (q, J = 6.4 Hz, 2H), 2.75 – 2.67 (m, 2H), 2.67 – 2.54 (m, 4H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.76, 157.01, 155.98, 147.98, 141.83, 131.03, 130.89, 129.76, 129.38, 128.17, 126.91, 125.39, 123.05, 120.78, 120.65, 112.00, 55.69, 54.90, 51.20, 46.59, 37.06, 11.53. C23H27N3O2, HRMS (ESI): m/z (M+H+): 378.2182 (calculated), 378.2174 (found).
N-[2-(diethylamino)ethyl]-2-(3-methoxyphenyl)quinoline-4-carboxamide (6l).
Yield: 77%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 (t, J = 5.7 Hz, 1H), 8.26 (ddd, J = 8.4, 1.4, 0.6 Hz, 1H), 8.19 – 8.02 (m, 2H), 7.92 – 7.76 (m, 3H), 7.63 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.49 (ddd, J = 8.1, 7.5, 0.5 Hz, 1H), 7.10 (ddd, J = 8.2, 2.6, 1.0 Hz, 1H), 3.89 (s, 3H), 3.47 (q, J = 6.5 Hz, 2H), 2.78 – 2.66 (m, 2H), 2.66 – 2.53 (m, 4H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.58, 159.80, 155.52, 147.84, 143.30, 139.74, 130.12, 129.98, 129.50, 127.02, 125.52, 123.49, 119.64, 116.71, 115.59, 112.43, 55.29, 51.36, 46.64, 37.23, 11.62. C23H27N3O2, HRMS (ESI): m/z (M+H+): 378.2182 (calculated), 378.2174 (found).
N-[2-(diethylamino)ethyl]-2-(4-methoxyphenyl)quinoline-4-carboxamide (6m).
Yield: 81%. 1H NMR (400 MHz, DMSO-d6) δ 8.80 – 8.78 (m, 1H), 8.37 – 8.25 (m, 2H), 8.23 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.11 – 7.99 (m, 2H), 7.78 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.58 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.18 – 7.04 (m, 2H), 3.85 (s, 3H), 3.48 (q, J = 6.5 Hz, 2H), 2.83 – 2.2.78 (m, 2H), 2.68 – 2.63 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 167.19, 161.30, 155.87, 148.40, 143.61, 131.14, 130.48, 129.73, 129.17, 126.98, 125.96, 123.55, 116.54, 114.74, 55.79, 51.79, 49.06, 47.12, 12.00. C23H27N3O2, HRMS (ESI): m/z (M+H+): 378.2182 (calculated), 378.2174 (found).
N-[2-(diethylamino)ethyl]-2-[4-(methoxymethyl)phenyl]quinoline-4-carboxamide (6n).
Yield: 71%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 (t, J = 5.7 Hz, 1H), 8.39 – 8.21 (m, 3H), 8.17 – 8.05 (m, 2H), 7.81 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.62 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 7.57 – 7.43 (m, 2H), 4.51 (s, 2H), 3.54 – 3.41 (m, 2H), 3.35 (s, 3H), 2.76 – 2.67 (m, 2H), 2.66 – 2.56 (m, 4H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.61, 155.49, 147.93, 143.31, 140.18, 137.33, 130.12, 129.44, 127.83, 127.15, 126.94, 125.53, 123.40, 116.48, 73.21, 57.66, 51.36, 46.64, 37.22, 11.60. C24H29N3O2, HRMS (ESI): m/z (M+H+): 392.2338 (calculated), 392.2329 (found).
N-[2-(diethylamino)ethyl]-2-{4-[(morpholin-4-yl)methyl]phenyl}quinoline-4-carboxamide (6o).
Yield: 77%. 1H NMR (400 MHz, DMSO-d6) δ 8.85 – 8.82 (m, 1H), 8.36 – 8.20 (m, 3H), 8.15 – 8.05 (m, 2H), 7.81 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.62 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.56 – 7.42 (m, 2H), 3.60 (t, J = 4.6 Hz, 4H), 3.55 (s, 2H), 3.53 – 3.38 (m, 2H), 2.80 – 2.69 (m, 2H), 2.69 – 2.54 (m, 4H), 2.45 – 2.31 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 167.11, 156.12, 148.42, 143.68, 140.28, 137.54, 130.58, 129.90, 129.85, 127.62, 127.37, 126.00, 123.84, 117.02, 66.68, 62.57, 53.69, 51.77, 47.11, 11.99. C27H34N4O2, HRMS (ESI): m/z (M+H+): 447.2760 (calculated), 447.2751 (found).
N-[2-(diethylamino)ethyl]-2-[2-(dimethylamino)pyrimidin-5-yl]quinoline-4-carboxamide (6p).
Yield: 77%. 1H NMR (400 MHz, CDCl3) δ 9.07 (s, 2H), 8.16 (ddd, J = 8.3, 1.4, 0.6 Hz, 1H), 8.09 (ddd, J = 8.5, 1.3, 0.7 Hz, 1H), 7.73 (s, 1H), 7.70 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.49 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 6.98 – 6.91 (m, 1H), 3.61 (dt, J = 6.2, 5.2 Hz, 2H), 3.27 (s, 6H), 2.80 – 2.68 (m, 2H), 2.58 (q, J = 7.1 Hz, 4H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 167.49, 162.29, 156.86, 153.20, 148.93, 143.28, 130.27, 129.89, 126.86, 125.17, 123.25, 120.01, 114.83, 51.37, 46.81, 37.58, 37.42, 11.93. C22H28N6O, HRMS (ESI): m/z (M+H+): 393.2403 (calculated), 393.2377 (found).
N-[2-(diethylamino)ethyl]-2-(2-methoxypyrimidin-5-yl)quinoline-4-carboxamide (6q).
Yield: 79%. 1H NMR (400 MHz, DMSO-d6) δ 9.45 (s, 2H), 8.82–8.80 (m, 1H), 8.33 – 8.17 (m, 2H), 8.11 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.82 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 7.64 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 4.02 (s, 3H), 3.64 – 3.36 (m, 2H), 2.75 – 2.73 (m, 2H), 2.68 – 2.63 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.84, 166.11, 158.84, 151.95, 148.28, 144.16, 130.85, 129.84, 127.74, 126.28, 126.05, 123.95, 116.48, 55.51, 51.77, 47.07, 37.58, 11.89. C21H25N5O2, HRMS (ESI): m/z (M+H+): 380.2087 (calculated), 380.2077 (found).
N-[2-(diethylamino)ethyl]-2-[(E)-2-phenylethenyl]quinoline-4-carboxamide (6r).
Yield: 71%. 1H NMR (400 MHz, DMSO-d6) δ 8.82 – 8.74 (m, 1H), 8.18 (ddd, J = 8.3, 1.5, 0.6 Hz, 1H), 8.04 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.90 (d, J = 17.0 Hz, 2H), 7.82 – 7.68 (m, 3H), 7.59 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 7.52 (d, J = 16.4 Hz, 1H), 7.47 – 7.40 (m, 2H), 7.40 – 7.32 (m, 1H), 3.48 (q, J = 6.5 Hz, 2H), 2.82 – 2.71 (m, 2H), 2.70 – 2.56 (m, 4H), 1.05 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.67, 155.32, 147.96, 142.95, 136.09, 134.72, 130.05, 129.03, 128.90, 128.86, 128.30, 127.31, 126.69, 125.50, 123.38, 117.38, 51.28, 46.60, 37.03, 11.44. C24H27N3O, HRMS (ESI): m/z (M+H+): 374.2232 (calculated), 374.2228 (found).
N-[2-(diethylamino)ethyl]-2-(2,3-dihydro-1-benzofuran-5-yl)quinoline-4-carboxamide (6s).
Yield: 86%. 1H NMR (400 MHz, DMSO-d6) δ 8.79 (t, J = 5.8 Hz, 1H), 8.32 – 8.16 (m, 2H), 8.16 – 7.98 (m, 3H), 7.77 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.57 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 6.93 (d, J = 8.3 Hz, 1H), 4.63 (t, J = 8.7 Hz, 2H), 3.47 (q, J = 6.5 Hz, 2H), 3.38 – 3.22 (m, 2H), 2.78 – 2.69 (m, 2H), 2.69 – 2.54 (m, 4H), 1.04 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.73, 161.49, 155.68, 147.93, 142.99, 130.81, 129.96, 129.17, 128.31, 127.69, 126.37, 125.47, 124.14, 123.02, 116.15, 109.11, 71.58, 51.32, 46.65, 37.13, 28.83, 11.53. C24H27N3O2, HRMS (ESI): m/z (M+H+): 390.2182 (calculated), 390.2172 (found).
N-[2-(diethylamino)ethyl]-2-(2,3-dihydro-1,4-benzodioxin-6-yl)quinoline-4-carboxamide (6t).
Yield: 91%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 (t, J = 5.6 Hz, 1H), 8.23 (ddd, J = 8.4, 1.5, 0.7 Hz, 1H), 8.12 – 7.98 (m, 2H), 7.88 – 7.70 (m, 3H), 7.58 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 7.03 (d, J = 8.4 Hz, 1H), 4.33 (s, 4H), 3.49 (q, J = 6.5 Hz, 2H), 2.80 – 2.72 (m, 2H), 2.72 – 2.56 (m, 4H), 1.05 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.66, 155.06, 147.87, 145.21, 143.72, 142.92, 131.53, 130.01, 129.30, 126.62, 125.48, 123.16, 120.45, 117.42, 116.19, 115.86, 64.37, 64.10, 54.90, 51.20, 46.64, 36.97, 11.36. C24H27N3O3, HRMS (ESI): m/z (M+H+): 406.2131 (calculated), 406.2118 (found).
2-{[1,1’-biphenyl]-4-yl}-N-[2-(diethylamino)ethyl]quinoline-4-carboxamide (6u).
Yield: 77%. 1H NMR (400 MHz, DMSO-d6) δ 8.85 (t, J = 5.8 Hz, 1H), 8.48 – 8.36 (m, 2H), 8.28 (dd, J = 8.5, 1.4 Hz, 1H), 8.20 (s, 1H), 8.15 (dd, J = 8.6, 1.2 Hz, 1H), 7.93 – 7.86 (m, 2H), 7.86 – 7.73 (m, 3H), 7.64 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.51 (dd, J = 8.4, 6.9 Hz, 2H), 7.46 – 7.34 (m, 1H), 3.50 (q, J = 6.5 Hz, 2H), 2.81 – 2.71 (m, 2H), 2.71 – 2.54 (m, 4H), 1.05 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.64, 155.25, 147.99, 143.28, 141.40, 139.32, 137.17, 130.16, 129.48, 129.01, 127.87, 127.80, 127.10, 126.99, 126.72, 125.55, 123.45, 116.50, 51.31, 46.65, 38.21, 11.51. C28H29N3O, HRMS (ESI): m/z (M+H+): 424.2389 (calculated), 424.2379 (found).
2-(propan-2-yloxy)quinoline-4-carboxylic acid (7).
Compound 7 was synthesized using synthesis route 1 method 1. Yield: 84 %. 1H NMR (400 MHz, DMSO-d6) δ 8.49 (ddd, J = 8.2, 1.5, 0.6 Hz, 1H), 7.74 (ddd, J = 8.3, 1.4, 0.6 Hz, 1H), 7.62 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.40 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.11 (s, 1H), 5.47 (p, J = 6.2 Hz, 1H), 1.35 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 168.92, 161.27, 147.24, 145.76, 129.89, 127.44, 126.85, 124.50, 122.38, 113.17, 68.10, 22.27. C13H13NO3, EI-MS: m/z (M-H+): 230.3 (calculated), 230.3 (found).
N-[2-(dimethylamino)ethyl]-2-(propan-2-yloxy)quinoline-4-carboxamide (8a).
Yield: 80%. 1H NMR (400 MHz, DMSO-d6) δ 8.78 – 8.58 (m, 1H), 8.06 (ddd, J = 8.3, 1.5, 0.6 Hz, 1H), 7.79 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.68 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.45 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 5.59 – 5.43 (m, 1H), 3.43 (td, J = 6.7, 5.7 Hz, 2H), 2.57 – 2.43 (m, 2H), 2.25 (s, 6H), 1.38 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.09, 160.46, 146.46, 145.65, 129.97, 127.08, 125.49, 124.40, 121.32, 111.33, 67.88, 57.86, 45.09, 37.13, 21.79. C17H23N3O2, HRMS (ESI): m/z (M+H+): 302.1869 (calculated), 302.1863 (found).
N-(2-aminoethyl)-2-(propan-2-yloxy)quinoline-4-carboxamide (8b).
Compound 8b was synthesized by first coupling intermediate 7 with N-Boc-ethylenediamine, followed by TFA deprotection. Yield: 72%. (TFA salt). 1H NMR (400 MHz, DMSO-d6) δ 8.92 (t, J = 5.6 Hz, 1H), 8.18 – 7.96 (m, 4H),7.78 (d, J = 8.2 Hz, 1H), 7.67 (t, J = 7.7 Hz, 1H), 7.47 – 7.37 (m, 1H), 7.22 (s, 1H), 5.58 – 5.40 (m, 1H), 3.65 – 3.49 (m, 2H), 3.13 – 2.97 (m, 2H),1.37 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.74, 160.55, 146.55, 144.59, 130.01, 127.15, 125.56, 124.45, 121.21, 112.28, 67.92, 38.42, 37.08, 21.79. C15H19N3O2, HRMS (ESI): m/z (M+H+): 274.1556 (calculated), 274.1550 (found).
N-[3-(dimethylamino)propyl]-2-(propan-2-yloxy)quinoline-4-carboxamide (8c).
Yield: 93%. 1H NMR (400 MHz, DMSO-d6) δ 8.74 (t, J = 5.6 Hz, 1H), 7.99 (ddd, J = 8.2, 1.5, 0.6 Hz, 1H), 7.77 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.67 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.44 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 6.94 (s, 1H), 5.51 (p, J = 6.2 Hz, 1H), 3.33 (td, J = 6.9, 5.6 Hz, 2H), 2.46 – 2.36 (m, 2H), 2.23 (s, 6H), 1.72 (dq, J = 8.5, 7.0 Hz, 2H), 1.37 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.03, 160.46, 146.43, 145.61, 129.96, 127.09, 125.34, 124.40, 121.29, 111.32, 67.87, 56.35, 44.73, 37.21, 26.51, 21.78. C18H25N3O2, HRMS (ESI): m/z (M-H+): 316.2025 (calculated), 316.2017 (found).
N-[2-(morpholin-4-yl)ethyl]-2-(propan-2-yloxy)quinoline-4-carboxamide (8d).
Yield: 70%. 1H NMR (400 MHz, DMSO-d6) δ 8.71 (t, J = 5.8 Hz, 1H), 8.23 – 8.10 (m, 1H), 7.84 (dd, J = 8.4, 1.2 Hz, 1H), 7.73 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.49 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 6.97 (s, 1H), 5.57 (p, J = 6.2 Hz, 1H), 3.66 (t, J = 4.6 Hz, 4H), 3.50 (q, J = 6.3 Hz, 2H), 2.77 – 2.70 (m, 3H), 2.60 – 2.53 (m, 3H), 1.43 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.06, 160.47, 146.43, 145.92, 129.97, 127.05, 125.58, 124.30, 121.33, 111.15, 67.86, 66.27, 57.15, 53.20, 38.21, 36.16, 21.77. C19H25N3O3, HRMS (ESI): m/z (M-H+): 344.1974 (calculated), 344.1967 (found).
2-(propan-2-yloxy)-N-[2-(pyrrolidin-1-yl)ethyl]quinoline-4-carboxamide (8e).
Yield: 72%. 1H NMR (400 MHz, DMSO-d6) δ 8.84 (t, J = 5.7 Hz, 1H), 8.08 (ddd, J = 8.3, 1.5, 0.6 Hz, 1H), 7.79 (ddd, J = 8.3, 1.3, 0.6 Hz, 1H), 7.69 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.44 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 7.03 (s, 1H), 5.52 (p, J = 6.2 Hz, 1H), 3.54 (q, J = 6.3 Hz, 2H), 2.92 (t, J = 6.5 Hz, 2H), 2.86–2.84 (m, 4H), 1.90 – 1.71 (m, 4H), 1.38 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.71, 160.92, 146.93, 145.75, 130.45, 127.54, 125.97, 124.84, 121.73, 112.01, 68.36, 54.37, 53.88, 49.05, 37.78, 23.44, 22.24. C19H25N3O2, HRMS (ESI): m/z (M+H+): 328.2025 (calculated), 328.2018 (found).
N-[2-(4-methylpiperazin-1-yl)ethyl]-2-(propan-2-yloxy)quinoline-4-carboxamide (8f).
Yield: 84%. 1H NMR (400 MHz, DMSO-d6) δ 8.68 (t, J = 5.7 Hz, 1H), 8.07 (dd, J = 8.3, 1.4 Hz, 1H), 7.78 (dd, J = 8.4, 1.3 Hz, 1H), 7.68 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.44 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 6.92 (s, 1H), 5.50 (p, J = 6.2 Hz, 1H), 3.44 (t, J = 6.2 Hz, 2H), 2.79 (s, 4H), 2.70 – 2.52 (m, 5H), 2.52 – 2.41 (m, 4H), 1.37 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.10, 160.47, 146.43, 145.76, 130.00, 127.07, 125.56, 124.38, 121.29, 111.23, 67.89, 56.15, 53.58, 50.83, 43.85, 36.35, 21.79. C20H28N4O2, HRMS (ESI): m/z (M+H+): 357.2291 (calculated), 357.2284 (found).
N-(1-methylpiperidin-4-yl)-2-(propan-2-yloxy)quinoline-4-carboxamide (8g).
Yield: 90%. 1H NMR (400 MHz, DMSO-d6) δ 8.70 (d, J = 7.6 Hz, 1H), 7.98 (ddd, J = 8.2, 1.5, 0.6 Hz, 1H), 7.78 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.67 (ddd, J = 8.4, 6.9, 1.5 Hz, 1H), 7.44 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 6.93 (s, 1H), 5.51 (p, J = 6.2 Hz, 1H), 3.93 – 3.81 (m, 1H), 3.00 – 2.82 (m, 2H), 2.30 (s, 3H), 2.30 – 2.20 (m, 2H), 1.95 – 1.85 (m, 2H), 1.71 – 1.55 (m, 2H), 1.37 (d, J = 6.2 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 165.50, 160.42, 146.41, 145.47, 129.94, 127.10, 125.26, 124.41, 121.29, 111.43, 67.84, 53.52, 45.58, 45.02, 30.45, 21.78. C19H25N3O2, HRMS (ESI): m/z (M+H+): 328.2025 (calculated), 328.2018 (found).
4-(4-methylpiperazine-1-carbonyl)-2-(propan-2-yloxy)quinoline (8h).
Yield: 78%. 1H NMR (400 MHz, DMSO-d6) δ 7.80 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.70 (ddd, J = 8.3, 6.9, 1.5 Hz, 1H), 7.64 (ddd, J = 8.1, 1.5, 0.6 Hz, 1H), 7.47 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 6.88 (s, 1H), 5.50 (p, J = 6.2 Hz, 1H), 3.89 – 3.85 (m, 1H), 3.71 – 3.55 (m, 1H), 3.20 – 3.07 (m, 2H), 2.50 – 2.46 (m, 1H), 2.42 – 2.38 (m, 1H), 2.27 – 2.23 (m, 1H), 2.19 (s, 3H), 2.13 – 2.00 (m, 1H), 1.38 (t, J = 5.7 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 165.56, 161.06, 146.72, 146.10, 130.75, 127.76, 125.20, 125.08, 121.23, 110.58, 68.47, 55.19, 54.63, 46.88, 45.99, 41.46, 38.70, 22.24. C18H23N3O2, HRMS (ESI): m/z (M+H+): 314.1869 (calculated), 314.1866 (found).
N-[2-(dimethylamino)ethyl]-2-phenylquinoline-4-carboxamide (10a).
Yield: 93%. 1H NMR (400 MHz, DMSO-d6) δ 8.82 (t, J = 5.7 Hz, 1H), 8.39 – 8.29 (m, 2H), 8.25 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.20 – 8.07 (m, 2H), 7.83 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.65 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.63 – 7.45 (m, 3H), 3.50 (td, J = 6.7, 5.7 Hz, 2H), 2.55 – 2.53 (m, 2H), 2.26 (s, 6H). 13C NMR (101 MHz, DMSO) δ 167.08, 156.21, 148.38, 143.84, 138.72, 130.60, 130.32, 129.96, 129.36, 127.73, 127.52, 125.94, 123.90, 117.01, 58.50, 45.70, 37.86. C20H21N3O, HRMS (ESI): m/z (M+H+): 320.1763 (calculated), 320.1758 (found).
N-[3-(diethylamino)propyl]-2-phenylquinoline-4-carboxamide (10b).
1H NMR (400 MHz, DMSO-d6) δ 8.91 (t, J = 5.6 Hz, 1H), 8.40 – 8.27 (m, 2H), 8.19 (dd, J = 8.4, 1.4 Hz, 1H), 8.17 – 8.07 (m, 2H), 7.82 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.64 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.60 – 7.42 (m, 3H), 3.42 (q, J = 6.5 Hz, 2H), 2.66 – 2.52 (m, 6H), 1.77 (p, J = 7.0 Hz, 2H), 0.99 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.55, 155.76, 147.92, 143.28, 138.23, 130.12, 129.85, 129.52, 128.87, 127.25, 127.03, 125.31, 123.40, 116.56, 49.76, 46.27, 37.62, 26.07, 11.22. C23H27N3O, HRMS (ESI): m/z (M+H+): 362.2232 (calculated), 362.2221 (found).
N-{1-azabicyclo[2.2.2]octan-3-yl}-2-phenylquinoline-4-carboxamide (10c).
Yield: 81%. 1H NMR (400 MHz, DMSO-d6) δ 8.89 (d, J = 7.0 Hz, 1H), 8.39 – 8.27 (m, 2H), 8.18 – 8.04 (m, 3H), 7.82 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.64 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.61 – 7.43 (m, 3H), 4.15 – 4.01 (m, 1H), 3.26 – 3.13 (m, 1H), 2.90 – 2.77 (m, 1H), 2.77 – 2.58 (m, 4H), 2.09 – 1.98 (m, 1H), 1.90 – 1.75 (m, 1H), 1.70 – 1.56 (m, 2H), 1.43 – 1.29 (m, 1H). 13C NMR (101 MHz, DMSO) δ 166.71, 155.73, 147.82, 143.52, 138.24, 130.11, 129.85, 129.51, 128.89, 127.30, 127.09, 125.17, 123.47, 116.60, 53.87, 47.16, 46.80, 46.33, 25.81, 25.62, 19.96. C23H23N3O, HRMS (ESI): m/z (M+H+): 358.1919 (calculated), 358.1913 (found).
2-phenylquinoline-4-carbohydrazide (10d).
Yield: 82%. Compound 10d was synthesized through the ester intermediate. Briefly, to a solution of ethyl 2-phenylquinoline-4-carboxylate (1 mmol) in ethanol was added hydrazine monohydrate (2 mmol). The mixture was heated under reflux to 130 °C in a sealed tube for 12 hours. Solvent was removed in vacuo and the product was purified by flash column chromatography (5–15% CH3OH/CH2Cl2). 1H NMR (400 MHz, DMSO-d6) δ 10.03 (s, 1H), 8.35 – 8.27 (m, 2H), 8.25 (dd, J = 8.4, 1.4 Hz, 1H), 8.17 – 8.08 (m, 2H), 7.82 (ddd, J = 8.4, 6.8, 1.5 Hz, 1H), 7.64 (ddd, J = 8.3, 6.8, 1.3 Hz, 1H), 7.61 – 7.45 (m, 3H), 4.73 (s, 2H). 13C NMR (101 MHz, DMSO) δ 165.87, 155.74, 147.90, 141.69, 138.23, 130.18, 129.87, 129.52, 128.91, 127.28, 127.07, 125.41, 123.61, 116.93. C16H13N3O, HRMS (ESI): m/z (M+H+): 264.1137 (calculated), 264.1129 (found).
N-carbamimidoyl-2-phenylquinoline-4-carboxamide (10e).
Compound 10e was synthesized through the acid chloride intermediate. Briefly, to a solution of intermediate 9 (1 mmol) in dichloromethane was added four drops of DMF, followed by oxalyl chloride (1.2 mmol). The mixture was stirred at room temperature for 1 hour and the solvent was removed in vacuo. THF was added to dissolve the acid chloride intermediate. In a separate round bottom flask, to a NaOH aqueous solution (15 eq) was added guanidine HCl (15 mmol). After 30 minutes, the acid chloride solution in THF was added dropwise to the aqueous solution of neutralized guanidine. The resulting mixture was stirred at room temperature overnight. THF was removed in vacuo and the resulting solution was extracted with dichloromethane. The DCM layer was separated and acidified with HCl. Final product was purified by flash column chromatography (10–20% CH3OH/CH2Cl2). Yield: 89% (HCl salt). 1H NMR (400 MHz, DMSO-d6) δ 12.92 (s, 1H), 8.92 (br s, 2H), 8.74 (br s, 2H), 8.59 (s, 1H), 8.48 – 8.37 (m, 2H), 8.31 (dd, J = 8.5, 1.2 Hz, 1H), 8.20 (dd, J = 8.5, 1.1 Hz, 1H), 7.89 (ddd, J = 8.3, 6.8, 1.4 Hz, 1H), 7.73 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.68 – 7.47 (m, 3H). 13C NMR (101 MHz, DMSO) δ 167.43, 155.83, 155.66, 147.70, 138.25, 137.61, 130.85, 130.33, 129.41, 128.96, 128.16, 127.78, 124.89, 122.53, 119.08. C17H14N4O, HRMS (ESI): m/z (M+H+): 291.1246 (calculated), 291.1239 (found).
N-[4-(dimethylamino)phenyl]-2-phenylquinoline-4-carboxamide (10f).
Yield: 89%. 1H NMR (400 MHz, CDCl3-d) δ 8.26 (s, 1H), 8.14 – 8.09 (m, 1H), 8.09 – 7.98 (m, 3H), 7.76 (s, 1H), 7.72 – 7.65 (m, 1H), 7.65 – 7.58 (m, 2H), 7.52 – 7.39 (m, 4H), 6.86 – 6.74 (m, 2H), 2.99 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 165.35, 156.68, 148.57, 148.55, 143.23, 138.55, 130.31, 129.89, 129.78, 128.99, 127.52, 127.50, 127.40, 125.20, 123.33, 122.06, 116.39, 113.07, 40.97. C24H21N3O, HRMS (ESI): m/z (M+H+): 368.1763 (calculated), 368.1750 (found).
2-phenyl-N-[2-(pyrrolidin-1-yl)ethyl]quinoline-4-carboxamide (10g).
1H NMR (400 MHz, DMSO-d6) δ 8.91 (s, 1H), 8.30 (ddd, J = 21.3, 8.5, 1.6 Hz, 3H), 8.21 – 8.09 (m, 2H), 7.83 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.72 – 7.46 (m, 4H), 3.56 (q, J = 6.4 Hz, 2H), 2.79 (t, J = 6.8 Hz, 2H), 2.67 (s, 5H), 1.76 (p, J = 3.0 Hz, 4H). 13C NMR (101 MHz, DMSO) δ 166.66, 155.73, 147.90, 143.28, 138.22, 130.11, 129.83, 129.47, 128.87, 127.25, 126.98, 125.50, 123.42, 116.58, 54.38, 53.52, 38.22, 38.14, 23.15. C22H23N3O, HRMS (ESI): m/z (M+H+): 346.1919 (calculated), 346.1914 (found).
N-[3-(dimethylamino)propyl]-2-phenylquinoline-4-carboxamide (10h).
Yield: 87%. 1H NMR (400 MHz, DMSO-d6) δ 8.88 (t, J = 5.6 Hz, 1H), 8.37 – 8.25 (m, 2H), 8.19 (dd, J = 8.4, 1.4 Hz, 1H), 8.16 – 8.03 (m, 2H), 7.82 (ddd, J = 8.4, 6.8, 1.4 Hz, 1H), 7.64 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.61 – 7.43 (m, 3H), 3.53 – 3.32 (m, 2H), 2.38 (t, J = 7.1 Hz, 2H), 2.20 (s, 6H), 1.77 (p, J = 7.1 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 166.54, 155.77, 147.91, 143.30, 138.24, 130.12, 129.84, 129.51, 128.88, 127.26, 127.05, 125.33, 123.42, 116.56, 56.63, 45.02, 37.52, 26.81. C21H23N3O, HRMS (ESI): m/z (M+H+): 334.1919 (calculated), 334.1913 (found).
N-[2-(dimethylamino)ethyl]-2-(thiophen-2-yl)quinoline-4-carboxamide (12a).
Yield: 88%. 1H NMR (400 MHz, DMSO-d6) δ 8.79 (t, J = 5.7 Hz, 1H), 8.14 (ddd, J = 8.4, 1.5, 0.6 Hz, 1H), 8.10 (s, 1H), 8.06 (dd, J = 3.8, 1.1 Hz, 1H), 8.01 (ddd, J = 8.5, 1.3, 0.6 Hz, 1H), 7.85 – 7.70 (m, 2H), 7.59 (ddd, J = 8.2, 6.8, 1.3 Hz, 1H), 7.24 (dd, J = 5.0, 3.7 Hz, 1H), 3.48 (td, J = 6.7, 5.7 Hz, 2H), 2.57 – 2.50 (m, 2H), 2.26 (s, 6H). 13C NMR (101 MHz, DMSO) δ 166.43, 151.58, 147.59, 144.27, 143.54, 130.32, 129.99, 128.76, 128.60, 127.58, 126.74, 125.50, 123.36, 115.35, 57.94, 45.15, 37.28. C18H19N3OS, HRMS (ESI): m/z (M+H+): 326.1327 (calculated), 326.1324 (found).
N-[3-(dimethylamino)propyl]-2-(thiophen-2-yl)quinoline-4-carboxamide (12b).
Yield: 92%. 1H NMR (400 MHz, DMSO-d6) δ 8.91 (t, J = 5.6 Hz, 1H), 8.12 (s, 1H), 8.11 – 8.04 (m, 2H), 8.01 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.82 – 7.69 (m, 2H), 7.60 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.24 (dd, J = 5.0, 3.7 Hz, 1H), 3.40 (td, J = 7.0, 5.6 Hz, 2H), 2.46 (t, J = 7.2 Hz, 2H), 2.26 (s, 6H), 1.86 – 1.68 (m, 2H). 13C NMR (101 MHz, DMSO) δ 166.38, 151.63, 147.61, 144.27, 143.40, 130.32, 130.01, 128.80, 128.60, 127.65, 126.77, 125.36, 123.36, 115.43, 56.37, 44.69, 37.38, 26.50. C19H21N3OS, HRMS (ESI): m/z (M+H+): 340.1484 (calculated), 340.1472 (found).
N-[2-(pyrrolidin-1-yl)ethyl]-2-(thiophen-2-yl)quinoline-4-carboxamide (12c).
Yield: 88%. 1H NMR (400 MHz, DMSO-d6) δ 8.84 (t, J = 5.7 Hz, 1H), 8.25 – 8.16 (m, 1H), 8.14 (s, 1H), 8.07 (dd, J = 3.8, 1.1 Hz, 1H), 8.01 (ddd, J = 8.4, 1.3, 0.6 Hz, 1H), 7.84 – 7.70 (m, 2H), 7.58 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.24 (dd, J = 5.0, 3.7 Hz, 1H), 3.52 (q, J = 6.6 Hz, 2H), 2.73 (t, J = 6.7 Hz, 2H), 2.66 – 2.54 (m, 4H), 1.74 (p, J = 3.3 Hz, 4H). 13C NMR (101 MHz, DMSO) δ 166.45, 151.61, 147.60, 144.29, 143.57, 130.32, 129.99, 128.75, 128.60, 127.60, 126.68, 125.57, 123.38, 115.37, 54.42, 53.53, 38.24, 23.19. C20H21N3OS, HRMS (ESI): m/z (M+H+): 352.1484 (calculated), 352.1473 (found).
N-[3-(1H-imidazol-1-yl)propyl]-2-(thiophen-2-yl)quinoline-4-carboxamide (12d).
Yield: 82%. 1H NMR (400 MHz, DMSO-d6) δ 8.91 (t, J = 5.5 Hz, 1H), 8.16 – 8.04 (m, 3H), 8.01 (dd, J = 8.6, 1.2 Hz, 1H), 7.84 – 7.73 (m, 2H), 7.69 (d, J = 1.1 Hz, 1H), 7.60 (ddd, J = 8.2, 6.9, 1.3 Hz, 1H), 7.31 – 7.20 (m, 2H), 6.92 (t, J = 1.1 Hz, 1H), 4.11 (t, J = 6.9 Hz, 2H), 3.41 – 3.27 (m, 2H), 2.05 (p, J = 6.8 Hz, 2H). 13C NMR (101 MHz, DMSO) δ 166.59, 151.64, 147.59, 144.25, 143.29, 137.38, 130.38, 130.08, 128.81, 128.61, 128.44, 127.74, 126.87, 125.34, 123.28, 119.41, 115.49, 43.77, 36.51, 30.56. C20H18N4OS, HRMS (ESI): m/z (M+H+): 363.1280 (calculated), 363.1270 (found).
2-(thiophen-2-yl)quinoline-4-carbohydrazide (12e).
Yield: 77%. Compound 12e was synthesized by following the same procedure as compound 10e. 1H NMR (400 MHz, DMSO-d6) δ 9.98 (s, 1H), 8.13 (dd, J = 8.4, 1.4 Hz, 1H), 8.09 (s, 1H), 8.05 (dd, J = 3.7, 1.2 Hz, 1H), 8.03 – 7.96 (m, 1H), 7.82 – 7.73 (m, 2H), 7.60 (ddd, J = 8.3, 6.9, 1.3 Hz, 1H), 7.24 (dd, J = 5.0, 3.7 Hz, 1H), 4.71 (s, 2H). 13C NMR (101 MHz, DMSO) δ 165.64, 151.55, 147.57, 144.20, 141.88, 130.38, 130.05, 128.79, 128.65, 127.64, 126.77, 125.44, 123.56, 115.79. C14H11N3OS, HRMS (ESI): m/z (M+H+): 270.0701 (calculated), 270.0696 (found).
6-chloro-2-phenylquinoline-4-carboxylic acid (14a).
Yield: 50%. 1H NMR (400 MHz, DMSO-d6) δ 8.74 (d, J = 2.4 Hz, 1H), 8.49 (s, 1H), 8.30 – 8.17 (m, 2H), 8.10 (d, J = 9.0 Hz, 1H), 7.80 (dd, J = 9.0, 2.4 Hz, 1H), 7.62 – 7.45 (m, 3H). 13C NMR (101 MHz, DMSO) δ 167.00, 156.26, 146.91, 137.48, 136.03, 132.38, 131.77, 130.58, 130.16, 128.98, 127.22, 124.32, 124.26, 120.54. C16H10ClNO2, EI-MS: m/z (M-H+): 282.7 (calculated), 282.5 (found).
6-methoxy-2-phenylquinoline-4-carboxylic acid (14b).
Yield: 57%. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (s, 1H), 8.30 – 8.19 (m, 2H), 8.13 (d, J = 2.8 Hz, 1H), 8.06 (d, J = 9.2 Hz, 1H), 7.64 – 7.36 (m, 4H), 3.91 (s, 3H). 13C NMR (101 MHz, DMSO) δ 167.68, 158.30, 153.16, 144.76, 138.05, 135.41, 131.36, 129.49, 128.92, 126.84, 124.98, 122.45, 119.75, 103.67, 55.44. C17H13NO3, EI-MS: m/z (M-H+): 278.3 (calculated), 278.3 (found).
6-chloro-N-[2-(dimethylamino)ethyl]-2-phenylquinoline-4-carboxamide (15a).
Yield: 82 %. 1H NMR (400 MHz, DMSO-d6) δ 8.91 (t, J = 5.7 Hz, 1H), 8.37 (dd, J = 2.4, 0.5 Hz, 1H), 8.34 – 8.26 (m, 2H), 8.21 (s, 1H), 8.13 (dd, J = 9.0, 0.5 Hz, 1H), 7.82 (dd, J = 9.0, 2.4 Hz, 1H), 7.61 – 7.49 (m, 3H), 3.52 (q, J = 6.4 Hz, 2H), 2.62 (t, J = 6.5 Hz, 2H), 2.34 (s, 6H). 13C NMR (101 MHz, DMSO) δ 166.59, 156.74, 146.88, 142.67, 138.27, 132.06, 132.03, 131.12, 130.58, 129.40, 127.78, 124.94, 124.61, 118.12, 58.13, 45.32, 37.48. C20H20ClN3O, HRMS (ESI): m/z (M+H+): 354.1373 (calculated), 354.1362 (found).
6-chloro-N-[2-(diethylamino)ethyl]-2-phenylquinoline-4-carboxamide (15b).
Yield: 82%. 1H NMR (400 MHz, CD3OD-d4) δ 8.27 (d, J = 2.3 Hz, 1H), 8.21 – 8.10 (m, 2H), 8.10 – 7.98 (m, 2H), 7.68 (dd, J = 9.0, 2.4 Hz, 1H), 7.58 – 7.42 (m, 3H), 3.70 (t, J = 6.7 Hz, 2H), 3.05 (t, J = 6.7 Hz, 2H), 2.94 (q, J = 7.2 Hz, 4H), 1.23 (t, J = 7.2 Hz, 6H). 13C NMR (101 MHz, CD3OD) δ 169.14, 158.31, 147.83, 142.19, 139.23, 133.92, 131.95, 131.82, 130.85, 129.71, 128.36, 125.10, 125.03, 119.03, 52.06, 48.17, 37.24, 10.58. C22H24ClN3O, HRMS (ESI): m/z (M+H+): 382.1686 (calculated), 382.1675 (found).
N-[2-(dimethylamino)ethyl]-6-methoxy-2-phenylquinoline-4-carboxamide (15c).
Yield: 90%. 1H NMR (400 MHz, DMSO-d6) δ 8.80 (t, J = 5.7 Hz, 1H), 8.34 – 8.21 (m, 2H), 8.08 (s, 1H), 8.04 (d, J = 9.2 Hz, 1H), 7.62 (d, J = 2.8 Hz, 1H), 7.55 (dd, J = 8.2, 6.4 Hz, 2H), 7.49 (tt, J = 9.2, 2.1 Hz, 2H), 3.89 (s, 3H), 3.50 (q, J = 6.3 Hz, 2H), 2.56 (t, J = 6.6 Hz, 2H), 2.27 (s, 6H). 13C NMR (101 MHz, DMSO) δ 166.74, 157.66, 153.22, 144.10, 141.74, 138.34, 131.07, 129.40, 128.83, 126.91, 124.50, 122.46, 116.93, 103.45, 57.93, 55.41, 45.11, 37.12. C21H23N3O2, HRMS (ESI): m/z (M+H+): 350.1869 (calculated), 350.1859 (found).
N-[2-(diethylamino)ethyl]-6-methoxy-2-phenylquinoline-4-carboxamide (15d).
Yield: 92%. 1H NMR (400 MHz, DMSO-d6) δ 8.93 – 8.80 (m, 1H), 8.35 – 8.19 (m, 2H), 8.13 (s, 1H), 8.04 (d, J = 9.2 Hz, 1H), 7.65 (d, J = 2.9 Hz, 1H), 7.60 – 7.53 (m, 2H), 7.53 – 7.41 (m, 2H), 3.89 (s, 3H), 3.52 (q, J = 6.4 Hz, 2H), 2.94 – 2.78 (m, 2H), 2.78 – 2.56 (m, 4H), 1.07 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.87, 157.68, 153.25, 144.17, 141.22, 138.37, 131.10, 129.43, 128.83, 126.92, 124.54, 122.38, 117.13, 103.57, 55.41, 51.03, 46.68, 36.73, 11.13. C23H27N3O2, HRMS (ESI): m/z (M+H+): 378.2182 (calculated), 378.2170 (found).
6-fluoro-2-hydroxyquinoline-4-carboxylic acid (17).
Yield: 42%. 1H NMR (400 MHz, DMSO-d6) δ 8.01 (dd, J = 10.6, 2.9 Hz, 1H), 7.56 – 7.18 (m, 2H), 7.00 (s, 1H). 13C NMR (101 MHz, DMSO) δ 166.37, 160.77, 158.28, 155.92, 139.53, 139.50, 136.29, 125.63, 119.11, 118.87, 117.63, 117.54, 116.64, 116.54, 111.46, 111.21. C10H6FNO3, EI-MS: m/z (M-H+): 206.2 (calculated), 206.0 (found).
2-chloro-N-[2-(dimethylamino)ethyl]-6-fluoroquinoline-4-carboxamide (18a).
Yield: 52%. 1H NMR (400 MHz, DMSO-d6) δ 8.85 (t, J = 5.8 Hz, 1H), 8.10 (dd, J = 9.2, 5.5 Hz, 1H), 8.03 (dd, J = 10.2, 2.9 Hz, 1H), 7.81 (ddd, J = 9.2, 8.2, 2.9 Hz, 1H), 7.69 (s, 1H), 3.46 (q, J = 6.2 Hz, 2H), 2.55 – 2.43 (m, 2H), 2.25 (s, 6H). 13C NMR (101 MHz, DMSO) δ 164.63, 161.54, 159.09, 148.88, 148.85, 145.25, 145.19, 144.79, 131.24, 131.14, 124.39, 124.28, 121.31, 121.06, 120.91, 109.72, 109.48, 57.81, 45.06, 37.27. C14H15ClFN3O, EI-MS: m/z (M+H+): 296.7 (calculated), 296.5 (found).
2-chloro-N-[2-(diethylamino)ethyl]-6-fluoroquinoline-4-carboxamide (18b).
Yield: 48%. 1H NMR (400 MHz, DMSO-d6) δ 8.81 (t, J = 5.8 Hz, 1H), 8.06 (ddd, J = 22.1, 9.7, 4.2 Hz, 2H), 7.79 (ddd, J = 9.2, 8.3, 2.9 Hz, 1H), 7.67 (s, 1H), 3.41 (q, J = 6.3 Hz, 2H), 2.65 – 2.51 (m, 6H), 0.99 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 164.60, 161.54, 159.08, 148.88, 148.86, 145.37, 145.31, 144.79, 131.23, 131.14, 124.36, 124.26, 121.33, 121.08, 120.86, 109.76, 109.52, 51.34, 46.40, 37.36, 11.64. C16H19ClFN3O, EI-MS: m/z (M+H+): 324.8 (calculated), 324.4 (found).
N-[2-(dimethylamino)ethyl]-6-fluoro-2-phenylquinoline-4-carboxamide (19a).
Yield: 81%. 1H NMR (400 MHz, DMSO-d6) δ 8.87 (t, J = 5.7 Hz, 1H), 8.39 – 8.25 (m, 2H), 8.25 – 8.14 (m, 2H), 8.06 (dd, J = 10.4, 2.9 Hz, 1H), 7.83 – 7.70 (m, 1H), 7.67 – 7.46 (m, 3H), 3.51 (q, J = 6.3 Hz, 2H), 2.60 – 2.49 (m, 2H), 2.28 (s, 6H). 13C NMR (101 MHz, DMSO) δ 166.17, 161.27, 158.83, 155.38, 155.35, 145.23, 142.51, 142.45, 137.98, 132.38, 132.29, 129.89, 128.89, 127.21, 124.24, 124.13, 120.32, 120.07, 117.47, 109.17, 108.94, 57.93, 45.10, 37.29. C20H20FN3O, HRMS (ESI): m/z (M+H+): 338.1669 (calculated), 338.1661 (found).
N-[2-(diethylamino)ethyl]-6-fluoro-2-phenylquinoline-4-carboxamide (19b).
Yield: 88%. 1H NMR (400 MHz, DMSO-d6) δ 8.94 – 8.78 (m, 1H), 8.39 – 8.25 (m, 2H), 8.25 – 8.13 (m, 2H), 8.07 (dd, J = 10.4, 2.9 Hz, 1H), 7.74 (td, J = 8.8, 2.9 Hz, 1H), 7.64 – 7.45 (m, 3H), 3.57 – 3.39 (m, 2H), 2.85 – 2.50 (m, 6H), 1.03 (t, J = 7.1 Hz, 6H). 13C NMR (101 MHz, DMSO) δ 166.17, 161.27, 158.83, 155.39, 155.36, 145.25, 142.46, 137.98, 132.38, 132.29, 129.90, 128.89, 127.19, 124.23, 124.12, 120.34, 120.08, 117.49, 109.21, 108.98, 51.31, 46.53, 37.22, 11.53. C22H24FN3O, HRMS (ESI): m/z (M+H+): 366.1982 (calculated), 366.1969 (found).
Cell lines and Viruses
Rhabdomyosarcoma (RD, ATCC, CCL-136), A172 (ATCC, CRL-1620), A549 (ATCC, CCL-185), HeLa (ATCC, CCL-2), and SH-SY5Y (ATCC, CRL-2266) were maintained in a 37 °C in a 5% CO2 atmosphere. RD and A172 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S). SH-SY5Y were cultured in 10% FBS and 1% P/S with 50% DMEM and 50% F-12 medium. All of the following EV-D68 strains used in this study were purchased from ATCC: US/KY/14-18953 (ATCC, NE-49132), US/MO/14-18947 (ATCC, NR-49129), US/MO/14-18949 (ATCC, NR-49130), US/IL/14-18952 (ATCC, NR-49131), US/IL/14-18956 (ATCC, NR-49133).
Cytopathic effect (CPE) assay
For antiviral CPE assays, cells were grown to approximately 90% confluency (one day after seeding in a 96 well plate). For infection, growth media was removed and cells were washed with PBS supplemented with magnesium and calcium. Cells were infected with EV-D68 strains diluted in DMEM with 2% FBS and 30 mM MgCl2 and transferred to a 33 °C incubator in a 5% CO2 atmosphere.20 After 1 h, P/S was added to a final concentration of 1% followed by the addition of compounds. Plates were gently shaken on an orbital shaker for about 5 min before incubation. For RD, A172, and SH-SY5Y cells, virus was added at a viral titer that resulted in complete CPE after 3 days of incubation (approximately an MOI of 0.01, 0.3, and 1 for RD, A172, and SH-SY5Y, respectively). Media was aspirated and a 66 μg/ml solution of neutral red dye was used to stain viable cells in each well. Absorbance at 540 nm was measured using a Multiskan FC Microplate Photometer (ThermoFisher Scientific). The EC50 values were calculated from best-fit dose response curves using GraphPad Prism, and all EC50 values reported were done in triplicates.
Cytotoxicity assay
The cytotoxicity of each compound was determined using the neutral red cell viability assay. The assay was performed under similar conditions (incubation temperature, time, and media) as the CPE, but excluded viral infection. Data acquisition and analysis (CC50) was performed similarly to the antiviral CPE assay, and all values are from triplicate experiments.
Western blotting
RD cells were infected with EV-D68 strain US/KY/14-18953 (MOI = 1). Total proteins were extracted at 9 hpi using RAPI lysis buffer [50 mM Tris (pH 8.0), 1% NP-40, 0.1% SDS, 150 mM NaCl, 0.5% Sodium deoxycholate, 5 mM EDTA, 10 mM NaF, 10 mM NaPPi, 2 mM phenyl-methylsulfonyl, and 1 mM PMSF]. Equal amount of extracted total proteins were separated by electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane. Viral protein VP1 or host GAPDH was recognized by rabbit anti-VP1 (GeneTex: GTX132313; 1:3,000 dilution) or mouse anti-GAPDH antibody (EMD Millipore: MAB374; 1:3,000 dilution), respectively, followed by detection using Horse radish peroxidase (HRP)-conjugated secondary antibody (ThermoFisher Scientific: 32430 or 656120; 1:3,000 dilutions) and Supersignal West Femto substrate (ThermoFisher Scientific).
RNA extraction and quantitative Real-time PCR (RT-qPCR).
RD cells were infected with EV-D68 strain US/KY/14-18953 (MOI = 1). Total RNA was extracted at 9 hpi using Trizol reagents (ThermoFisher Scientific). After removing genomic DNA by RQ1 RNase-Free DNase (Promega), 1.2 μg of total RNA was used to synthesize first strand of cDNA of viral RNA and host mRNA using SuperScript III reverse transcriptase (ThermoFisher Scientific) and oligo (dT)18. Viral RNA was amplified on a QuantStudio 5 Real-Time PCR System (ThermoFisher Scientific) using FastStart Universal SYBR Green Master (Rox) (Roche) and virus-specific primers: D68-F (5’-CGCTGAACTTGGCGTGGTCC-3’) and D68-R (5’-GGCTGCCCTGCTAAGAAAATTCTCC-3’). GAPDH was amplified to serve as a control using GAPDH-specific primers (GAPDH-F: 5’-ACACCCACTCCTCCACCTTTG-3’ and GAPDH-R: 5’-CACCACCCTGTTGCTGTAGCC-3’). The amplification conditions were: 95 °C for 10 min; 40 cycles of 15 sec at 95 °C and 60 sec at 60 °C. Melting curve analysis was performed to verify the specificity of each amplification.
Immunostaining
RD cells were infected with EV-D68 strain US/KY/14-18953 (MOI = 1). At 9 hpi, infected cells were fixed with 4% formaldehyde for 10 min followed by permeabilization with 0.2% Triton X-100 for another 10 min. After blocking with 10% bovine serum, cells were stained with rabbit anti-VP1 antibody (GeneTex: GTX132313) and followed by staining with anti-rabbit secondary antibody conjugated to Alexa-488 (ThermoFisher Scientific). Nucleus were stained with 300 nM DAPI (ThermoFisher Scientific) after secondary antibodies incubation. Fluorescent images were acquired using a Leica SP5-II spectral Confocal Microscope (Leica).
Sodium flux assay
Calcium imaging in acutely dissociated dorsal root ganglion (DRG) neurons.
Dorsal root ganglion neurons were prepared as previously described27 and were loaded for 30 minutes at 37°C with 3 μM Fura-2AM (Cat# F1221, Thermo Fisher, stock solution prepared at 1mM in DMSO, 0.02% pluronic acid, Cat#P-3000MP, Life technologies) to follow changes in intracellular calcium([Ca2+]c) in a standard bath solution containing 139 mM NaCl, 3 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 10 mM Na HEPES, pH 7.4, 5 mM glucose exactly as previously described28 Fluorescence imaging was performed with an inverted microscope, NikonEclipseTi-U (Nikon Instruments Inc., Melville, NY), using objective Nikon Fluor 4X and a Photometrics cooled CCD camera Cool SNAP ES2 (Roper Scientific, Tucson, AZ) controlled by Nis Elements software (version 4.20, Nikon Instruments). The excitation light was delivered by a Lambda-LS system (Sutter Instruments, Novato, CA). The excitation filters (340 ± 5 and 380 ± 7) were controlled by a Lambda 10 to 2 optical filter change (Sutter Instruments). Fluorescence was recorded through a 505-nm dichroic mirror at 535 ± 25 nm. To minimize photobleaching and phototoxicity, the images were taken every ~10 seconds during the time-course of the experiment using the minimal exposure time that provided acceptable image quality. The changes in [Ca2+]c were monitored by following a ratio of F340/F380, calculated after subtracting the background from both channels.
Concentration-Dependent Sodium-channel activated calcium imaging
DRG neurons were incubated overnight with testing compounds or Dibucaine at concentrations of 2 μM, 20 μM, and 50 μM as indicated. Following overnight incubation, cells were imaged via calcium imaging protocol with the following specifications: after a 1-minute baseline measurement, sodium channel agonist, Veratridine was added (30 μM) and indirect sodium influx was measured by obtained fluorescence ratio of 340 nm/380 nm of resulting sodium-channel triggered calcium influx. Total data collection time for each coverslip was 7 minutes in length.
Figure 1.
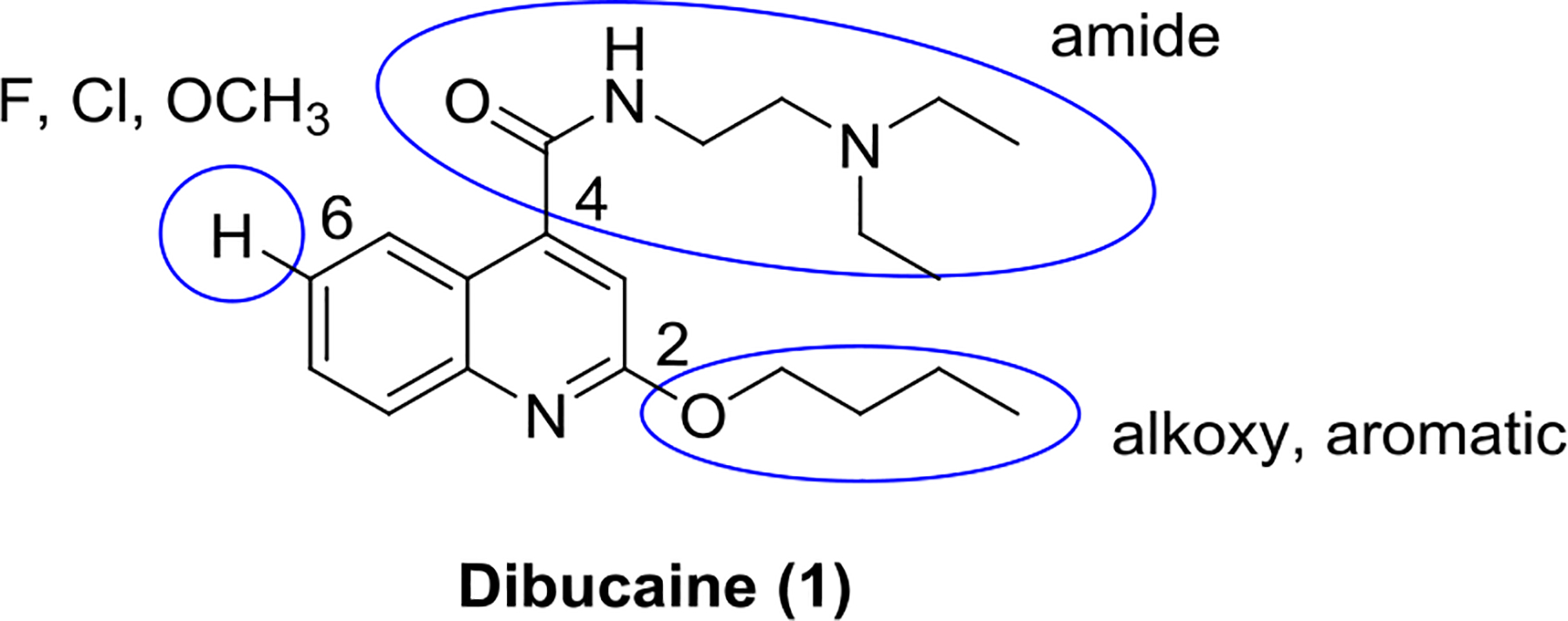
Chemical structure of dibucaine. Three positions, 2, 4, and 6, were examined in the current SAR studies and were circled.
ACKNOWLEDGEMENTS
This research is supported by startup funding from the University of Arizona and NIH grants AI119187 and AI144887 to J.W. R.K. is supported by National Institutes of Health Awards (1R01NS098772, 1R01DA042852, and R01AT009716). S.S.B. is supported by funding through the Spirit of Inquiry Grant from the University of Arizona Honors College.
ABBREVIATIONS USED
- EV-D68
enterovirus D68
- AFM
acute flaccid myelitis
Footnotes
The authors declare no competing financial interest.
Supporting information: Molecular Formula Strings.
REFERENCES
- 1.Van der Linden L; Wolthers KC; van Kuppeveld FJM Replication and inhibitors of enteroviruses and parechoviruses. Viruses 2015, 7, 4529–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggen J; Thibaut HJ; Strating JRPM; van Kuppeveld FJM The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Micrbiol 2018, 16, 368–381. [DOI] [PubMed] [Google Scholar]
- 3.Glyn Stanway TH, Knowles Nick J., Hyypiä Timo. Molecular and biological basis of picornavirus taxonomy. In Molecular Biology of Picornavirus, Wimmer B. L. S. a. E., Ed. ASM Press: Washington DC, 2002; pp 17–24. [Google Scholar]
- 4.Mao Q; Wang Y; Bian L; Xu M; Liang Z EV-A71 vaccine licensure: a first step for multivalent enterovirus vaccine to control HFMD and other severe diseases. Emerg. Microbes Infect 2016, 5, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyler KL Acute viral encephalitis. N. Engl. J. Med 2018, 379, 557–566. [DOI] [PubMed] [Google Scholar]
- 6.Holm-Hansen CC; Midgley SE; Fischer TK Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis 2016, 16, e64–e75. [DOI] [PubMed] [Google Scholar]
- 7.Midgley CM; Watson JT; Nix WA; Curns AT; Rogers SL; Brown BA; Conover C; Dominguez SR; Feikin DR; Gray S; Hassan F; Hoferka S; Jackson MA; Johnson D; Leshem E; Miller L; Nichols JB; Nyquist AC; Obringer E; Patel A; Patel M; Rha B; Schneider E; Schuster JE; Selvarangan R; Seward JF; Turabelidze G; Oberste MS; Pallansch MA; Gerber SI; Group E-DW Severe respiratory illness associated with a nationwide outbreak of enterovirus D68 in the USA (2014): a descriptive epidemiological investigation. Lancet Respir. Med 2015, 3, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oermann CM; Schuster JE; Conners GP; Newland JG; Selvarangan R; Jackson MA Enterovirus d68. A focused review and clinical highlights from the 2014 U.S. Outbreak. Ann. Am. Thorac. Soc 2015, 12, 775–781. [DOI] [PubMed] [Google Scholar]
- 9.Huang HI; Shih SR Neurotropic enterovirus infections in the central nervous system. Viruses 2015, 7, 6051–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khetsuriani N; Lamonte-Fowlkes A; Oberst S; Pallansch MA; Centers for Disease, C.; Prevention. Enterovirus surveillance--United States, 1970–2005. MMWR Surveill Summ 2006, 55, 1–20. [PubMed] [Google Scholar]
- 11.Abedi GR; Watson JT; Nix WA; Oberste MS; Gerber SI Enterovirus and parechovirus surveillance - United States, 2014–2016. MMWR Morb. Mortal. Wkly. Rep 2018, 67, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberste MS; Maher K; Schnurr D; Flemister MR; Lovchik JC; Peters H; Sessions W; Kirk C; Chatterjee N; Fuller S; Hanauer JM; Pallansch MA Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J. Gen. Virol 2004, 85, 2577–2584. [DOI] [PubMed] [Google Scholar]
- 13.Hurst BL; Evans WJ; Smee DF; Van Wettere A; Tarbet EB Evaluation of antiviral therapies in respiratory and neurological disease models of Enterovirus D68 infection in mice. Virology 2019, 526, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hixon AM; Clarke P; Tyler KL Evaluating treatment efficacy in a mouse model of enterovirus D68-associated paralytic myelitis. J. Infect. Dis 2017, 216, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H; Wang J; Li B; Guo L; Li H; Song J; Yang Z; Li H; Fan H; Huang X; Long H; Cheng C; Chu M; He Z; Yu W; Li J; Gao Y; Ning R; Li N; Yang J; Wu Q; Shi H; Sun M; Liu L A novel neutralizing antibody specific to the DE loop of VP1 can inhibit EV-D68 infection in mice. J. Immunol 2018, 201, 2557–2569. [DOI] [PubMed] [Google Scholar]
- 16.Hixon AM; Yu G; Leser JS; Yagi S; Clarke P; Chiu CY; Tyler KL A mouse model of paralytic myelitis caused by enterovirus D68. PLoS Pathog. 2017, 13, e1006199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greninger AL; Naccache SN; Messacar K; Clayton A; Yu G; Somasekar S; Federman S; Stryke D; Anderson C; Yagi S; Messenger S; Wadford D; Xia D; Watt JP; Van Haren K; Dominguez SR; Glaser C; Aldrovandi G; Chiu CY A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012–14): a retrospective cohort study. Lancet Infect. Dis 2015, 15, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafolie J; Labbé A; L’Honneur AS; Madhi F; Pereira B; Decobert M; Adam MN; Gouraud F; Faibis F; Arditty F; Marque-Juillet S; Guitteny MA; Lagathu G; Verdan M; Rozenberg F; Mirand A; Peigue-Lafeuille H; Henquell C; Bailly J-L; Archimbaud C; Chacé A; Corlouer C; Mercier J-C; Cotillon M; Magdoud El Alaoui F; Epaud R; Nathanson S; Coutard A; Rochette E; Brebion A; Chambon M; Regagnon C; De Pontual L; Carbonnelle E; Poilane I; Benoist G; Gault E; Millet-Zerner V; Kuentz M; Gallet S; Macchi V; Ducrocq S; Epelbaum S; Lambert C; Faye A; Soudée-Mayer S; Titomanlio L; Bonacorsi S; Cointe A; Cloix I; Raobison A-H; Boutry M; Tavani F Assessment of blood enterovirus PCR testing in paediatric populations with fever without source, sepsis-like disease, or suspected meningitis: a prospective, multicentre, observational cohort study. Lancet Infect. Dis 2018, 18, 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed March 15th, 2019).
- 20.Smee DF; Evans WJ; Nicolaou KC; Tarbet EB; Day CW Susceptibilities of enterovirus D68, enterovirus 71, and rhinovirus 87 strains to various antiviral compounds. Antiviral Res. 2016, 131, 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulferts R; de Boer SM; van der Linden L; Bauer L; Lyoo HR; Mate MJ; Lichiere J; Canard B; Lelieveld D; Omta W; Egan D; Coutard B; van Kuppeveld FJM Screening of a library of FDA-approved drugs identifies several enterovirus replication inhibitors that target viral protein 2C. Antimicrob. Agents Chemother 2016, 60, 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo J; Kye S; Quinn KK; Cooper P; Damoiseaux R; Krogstad P Discovery of structurally diverse small-molecule compounds with broad antiviral activity against enteroviruses. Antimicrob. Agents Chemother 2015, 60, 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulferts R; van der Linden L; Thibaut HJ; Lanke KH; Leyssen P; Coutard B; De Palma AM; Canard B; Neyts J; van Kuppeveld FJ Selective serotonin reuptake inhibitor fluoxetine inhibits replication of human enteroviruses B and D by targeting viral protein 2C. Antimicrob. Agents Chemother 2013, 57, 1952–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baragana B; Norcross NR; Wilson C; Porzelle A; Hallyburton I; Grimaldi R; Osuna-Cabello M; Norval S; Riley J; Stojanovski L; Simeons FR; Wyatt PG; Delves MJ; Meister S; Duffy S; Avery VM; Winzeler EA; Sinden RE; Wittlin S; Frearson JA; Gray DW; Fairlamb AH; Waterson D; Campbell SF; Willis P; Read KD; Gilbert IH Discovery of a quinoline-4-carboxamide derivative with a novel mechanism of action, multistage antimalarial activity, and potent in vivo efficacy. J. Med. Chem 2016, 59, 9672–9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y; Hu Y; Xu S; Zhang Y; Musharrafieh R; Hau RK; Ma C; Wang J In vitro pharmacokinetic optimizations of AM2-S31N channel blockers led to the discovery of slow-binding inhibitors with potent antiviral activity against drug-resistant influenza A viruses. J. Med. Chem 2018, 61, 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyda A; Stelzer-Braid S; Adam D; Chughtai AA; MacIntyre CR The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV-D68) - what is the evidence for causation? Eurosurveillance 2018, 23, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francois-Moutal L; Wang Y; Moutal A; Cottier KE; Melemedjian OK; Yang X; Wang Y; Ju W; Largent-Milnes TM; Khanna M; Vanderah TW; Khanna R A membrane-delimited N-myristoylated CRMP2 peptide aptamer inhibits CaV2.2 trafficking and reverses inflammatory and postoperative pain behaviors. Pain 2015, 156, 1247–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brittain JM; Duarte DB; Wilson SM; Zhu W; Ballard C; Johnson PL; Liu N; Xiong W; Ripsch MS; Wang Y; Fehrenbacher JC; Fitz SD; Khanna M; Park CK; Schmutzler BS; Cheon BM; Due MR; Brustovetsky T; Ashpole NM; Hudmon A; Meroueh SO; Hingtgen CM; Brustovetsky N; Ji RR; Hurley JH; Jin X; Shekhar A; Xu XM; Oxford GS; Vasko MR; White FA; Khanna R Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca(2)(+) channel complex. Nat. Med 2011, 17, 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]


