Abstract
Objectives:
Multiple myeloma (MM) is a malignant plasma cell neoplasm, requiring the integration of clinical examination, laboratory and radiological investigations for diagnosis. Detection and isotypic identification of the monoclonal protein(s) and measurement of other relevant biomarkers in serum and urine are pivotal analyses. However, occasionally this approach fails to characterize complex protein signatures. Here we describe the development and application of next generation mass spectrometry (MS) techniques, and a novel adaptation of immunofixation, to interrogate non-canonical monoclonal immunoproteins.
Methods:
Immunoprecipitation immunofixation (IP-IFE) was performed on a Sebia Hydrasys Scan2. Middle-down de novo sequencing and native MS were performed with multiple instruments (21T FT-ICR, Q Exactive HF, Orbitrap Fusion Lumos, and Orbitrap Eclipse). Post-acquisition data analysis was performed using Xcalibur Qual Browser, ProSight Lite, and TDValidator.
Results:
We adapted a novel variation of immunofixation electrophoresis (IFE) with an antibody-specific immunosubtraction step, providing insight into the clonal signature of gamma-zone monoclonal immunoglobulin (M-protein) species. We developed and applied advanced mass spectrometric techniques such as middle-down de novo sequencing to attain in-depth characterization of the primary sequence of an M-protein. Quaternary structures of M-proteins were elucidated by native MS, revealing a previously unprecedented non-covalently associated hetero-tetrameric immunoglobulin.
Conclusions:
Next generation proteomic solutions offer great potential for characterizing complex protein structures and may eventually replace current electrophoretic approaches for the identification and quantification of M-proteins. They can also contribute to greater understanding of MM pathogenesis, enabling classification of patients into new subtypes, improved risk stratification and the potential to inform decisions on future personalized treatment modalities.
Keywords: immunofixation electrophoresis, multiple myeloma, native mass spectrometry, top-down protein sequencing, truncated heavy chains
Introduction
Multiple myeloma (MM) is a hematological malignancy characterized by the uncontrolled expansion of clonal plasma cells that encode for a unique monoclonal immunoglobulin (M-protein) [1]. It is a clinically and molecularly heterogeneous disease [2] with a complex pathogenesis which is almost always preceded by premalignant or more indolent phases, namely monoclonal gammopathy of undetermined significance (MGUS) or smouldering multiple myeloma (SMM). The plasma cells typically secrete one or more M-proteins which are detected in the serum and/or urine using various electrophoretic and immunochemical techniques.
The diagnosis, assessment and risk stratification of a patient suspected of having MM requires an integrated, multifaceted laboratory approach, integral to which is the detection and isotypic identification of the M-protein, the well-defined electrophoretic signature found in the majority of patients presenting with a plasma cell dyscrasia (PCD). The M-protein and the serum free light chain (sFLC) levels are both essential in the differential diagnosis and stratification of MM, and are the most widely used biomarkers for monitoring the course of the disease [3]. The M-protein is isotyped by either immunofixation (IFE), which is considered to be the gold standard, or by immunosubtraction using capillary zone electrophoresis in monoclonal gammopathies involving IgA, IgG, and IgM and where the M-protein is both discrete and conspicuous. However, the complete and unambiguous identification of complex immunoprotein signatures cannot always be achieved using these methods alone.
The increasing use of molecular investigations is expected to become a major contributor for predicting the likelihood of rapid disease progression, responsiveness to specific drug combinations, and monitoring for minimal residual disease (MRD) [4]. For example, recent investigations performed at the Mayo Clinic and the National High Magnetic Field Laboratory have explored alternative strategies including the use of several next-generation mass spectrometry (MS) techniques. In monoclonal gammopathies such as MM and light-chain amyloidosis, top-down and middle-down MS analyses have been used to isotype heavy (HC) and light chains (LC) [5, 6], and to detect low levels of monoclonal proteins from serum that may have otherwise gone undetected [7, 8]. MS techniques have the potential to augment or eventually replace electrophoretic techniques, given their higher sensitivity and specificity.
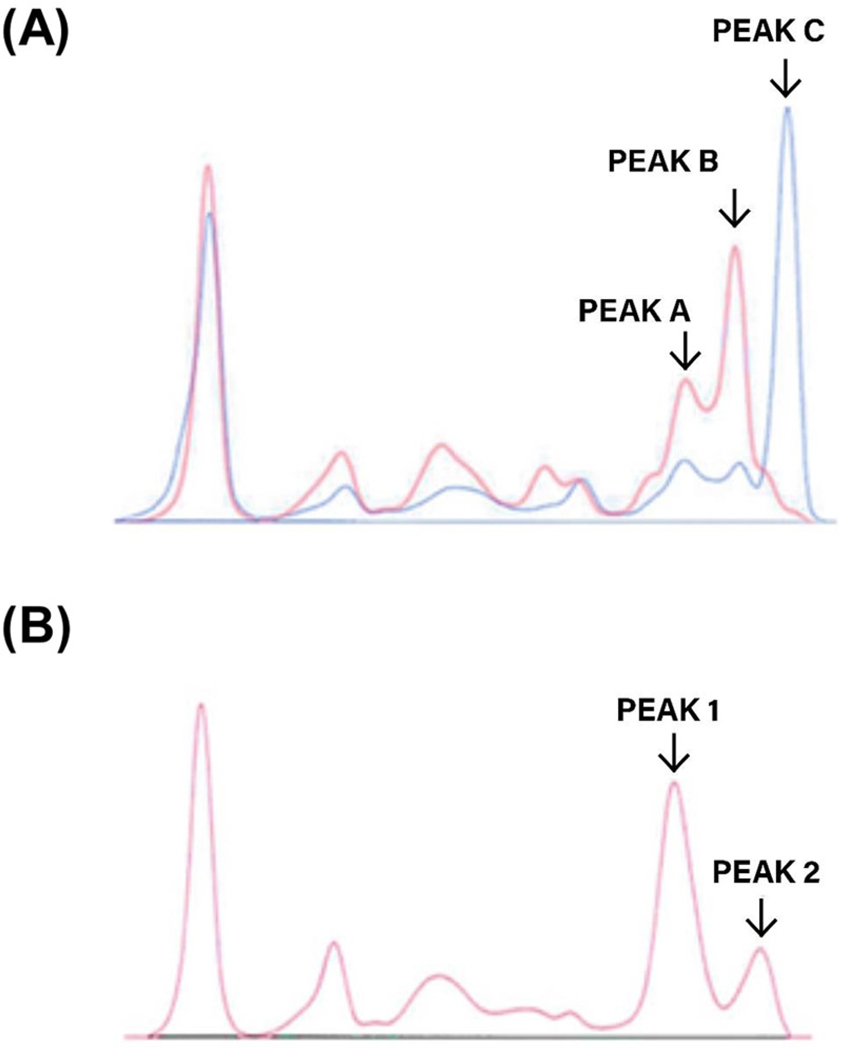
Herein we describe our work in developing and applying new analytical techniques which can be used to more accurately and sensitively evaluate immunoproteins, particularly non-canonical myeloma proteins. These methods were developed in the context of an unusual M-protein, for which the currently available repertoire of protein based investigations was unable to resolve the complex protein signature that was observed. In brief, analysis by capillary electrophoresis revealed a triple gammopathy in the gamma zone, which included two truncated species (Figure 1). The relevant clinical details and disease progression of the patient were previously published as a case report [9], and a succinct summary is provided in the Supplementary Material section of this manuscript. We selected this challenging sample as an example of an aberrant class of malignant immunoproteins that potentially could enhance and expand the current repertoire of immunoprotein analyses.
Figure 1:
Capillary electrophoresis.
(A) Electrophoretogram of serum indicating the gamma zone triple gammopathy at diagnosis (blue) and at terminal relapse (red). At terminal relapse the IgG kappa (peak C) is absent, with ectopic expression of non-canonical M-proteins (truncated gamma HC dimer, peak A; 128 kDa hetero-tetramer, peak B).
(B) Electrophoretogram of terminal relapse sample (post BME reduction) demonstrating the resolution of the truncated gamma HC dimer and the truncated gamma HC’s from the 128 kDa heterotetramer into a single peak(1) and the monoclonal free kappa LC’s and non-covalently bound kappa LC’s into another peak(2).
Firstly, we employed a novel adaptation of IFE which required an antibody-specific immunosubtraction/immunoprecipitation step prior to IFE analysis (IP-IFE). This strategy was adopted to interrogate the clonal signature of the M-protein species present in the gamma zone. Secondly, we leveraged native top-down MS in a novel application to eject subunits from an intact immunoglobulin, providing insight into quaternary structure which complemented the IP-IFE approach. Thirdly, we developed a middle-down de novo sequencing strategy to reveal the primary structure of the unusual immunoprotein complex. On the basis of these studies and in conjunction with routine immunochemical data, we demonstrated that these techniques are effective for proposing conformational structures for myeloma proteins. We also highlight limitations of current methodologies, in contexts where both canonical and non-canonical (truncated) M-proteins are secreted by mutated plasma cells.
Materials and methods
Ethical approval
The research related to human patient samples has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved by the authors’ Clinical Haematology Department. Informed consent was obtained from the individual included in this study.
Immunoprecipitation immunofixation (IP-IFE)
The pre-IFE incubation step precipitates intact immunoglobulin and free HC or LCs corresponding to the antibody added, or to which it is bound. The subsequent IFE with each antibody which had been precipitated demonstrates complete removal of that antibody.
Three 10 μL aliquots of a one in four dilution of the patients’ serum were mixed with 100 μL of Dynamic mask IFE antisera (Sebia) which are optimally concentrated to facilitate the initial precipitation step. A further 10 μL aliquot was mixed with standard Sebia IFE kappa free antiserum, which had been concentrated by a factor of 1.5. A control sample was also selected which contained an IgG kappa M-protein, in the absence of monoclonal free LC’s. The antibody treated samples were incubated for 48 h at 4 °C, centrifuged at 1,500 x g for 10 min and the supernatants decanted from the precipitated antigen-antibody complex.
The IFE was performed on the Sebia Hydrasys Scan 2. The migration lanes of the separated proteins were overlaid with gamma, kappa (free and bound), lambda (free and bound), and kappa free antisera which resulted in precipitation of any corresponding antigen present in the supernatant. After removal of the unprecipitated soluble proteins the antigen-antibody complex trapped within the gel matrix was stained with Acid Violet.
Sample preparation for mass spectrometry
Immunoglobulins were enriched from serum using a Melon Gel IgG spin purification kit (Thermo Scientific), and enrichment was verified by SDS-PAGE (Supplementary Material, Figure S1). Some samples were reduced with TCEP/GdnCl. Digestion was performed with IdeS protease (Promega), followed by reduction to produce LC, Fd, and Fc subunits [10]. All samples were buffer exchanged in centrifugal filter units prior to MS analysis. Samples for denaturing experiments were exchanged into 5:95 acetonitrile:water containing 0.1% formic acid; samples for native experiments were exchanged into 200 mM ammonium acetate.
Mass spectrometry analysis
Intact mass measurements were acquired under denaturing conditions using an Ultimate 3000 nanoLC (Thermo Scientific) fitted with a polymeric monolithic RP-5H column (100 mm L, 0.5 mm i.d.) online with FTMS (Q Exactive HF, Thermo Scientific). Intact mass analysis under native conditions were performed by direct infusion with a Nanospray Flex ion source (Thermo Scientific) attached to an Orbitrap Eclipse Tribrid (Thermo Scientific), and subunit ejection was performed by modifying the compensation voltage. For de novo sequencing, the subunits resulting from IdeS digestion were separated by reverse phase chromatography, followed by targeted MS2 fragmentation. Additional acquisition parameters are described in the Supplementary Material. Data analysis was performed using Xcalibur Qual Browser (Thermo Scientific), ProSight Lite & TDValidator (Proteinaceous).
Results
Immunoprecipitation immunofixation (IP-IFE)
The application of IP-IFE was utilized in order to probe for the presence of free monoclonal gamma HC’s and to determine if any antibody subtraction/precipitation anomalies existed, which could help explain the uninterpretable immunosubtraction profile identified by capillary electrophoresis. The inspection of the post IP-IFE separations confirmed the presence of several atypical M-proteins (Figure 2). The absence of staining in the lambda lanes is due to the associated immune paresis affecting the polyclonal production of the three main immunoglobulins, IgG, IgA, and IgM.
Figure 2:
Immunoprecipitation immunofixation electrophoresis.
(A) Post IP-IFE (unreduced and post BME reduction)with gamma, kappa, and lambda antisera confirming the presence of free gamma heavy chains which are not precipitated with kappa or lambda and the 128 kDa species. (B) IP-IFE with kappa free antiserum demonstrating the presence of truncated gamma HC’s and the complete removal of free kappa, and both heavy and light chains found in the non-covalent 128 kDa species. The control sample demonstrates that the intact IgG kappa (150 kDa) cannot be subtracted by free kappa.
The post gamma precipitation lanes revealed two free monoclonal kappa LC bands. The non-detection of the free monoclonal kappa LC’s with the kappa free antiserum is due to their minute concentration and the lower avidity of the kappa free antiserum. The post kappa (free and bound) precipitation lanes revealed one small band in the IgG lane. Since this band was also detected in the IgG lane of the post lambda precipitation it most likely represents truncated free gamma HC dimers. The post lambda precipitation lanes revealed two bands with identical mobility to those identified in the post gamma precipitation. However, there was an additional large M-protein detected in the mid-gamma region which immunofixed with gamma, kappa (free and bound), and kappa free antisera.
On the basis of routine electrophoretic investigations it may have been incorrectly concluded that this band was composed of two M-proteins (IgG kappa and free kappa LC’s) having identical mobility. Although this band was absent in the gamma and kappa (free and bound) precipitation lanes, it reacted strongly with kappa free antiserum after precipitation with lambda. However, free kappa was not identified with this mobility after precipitation with gamma.
The post kappa free precipitation lanes are identical to those obtained after kappa (free and bound) precipitation. This IFE signature is not possible in the presence of an intact covalently bound IgG kappa as demonstrated in the control sample (Figure 2B). Only kappa (free and bound) antiserum can precipitate the intact M-protein in the control.
This very rare IP-IFE pattern posits the paradoxical question of how a free LC antiserum can react with LC’s which are not free. The series of antibody precipitations outlined above are indicative of the presence of a non-covalently bound immunoprotein consisting of both truncated gamma HC’s and kappa LC’s. In this proposed species the required target epitopes in the kappa LC’s are exposed which will permit recognition by kappa free antiserum.
When the IP-IFE procedure was repeated after sample reduction with betamercaptoethanol the post gamma precipitation demonstrated that the two free kappa bands were reduced into one monomeric band (Figure 2A). The post kappa precipitation was identical to the unreduced sample whereas the post lambda precipitation revealed the presence of two bands, indicating that the most abundant M-protein in the gamma region had been split into its HC and LC components.
Structural insight from native mass spectrometry
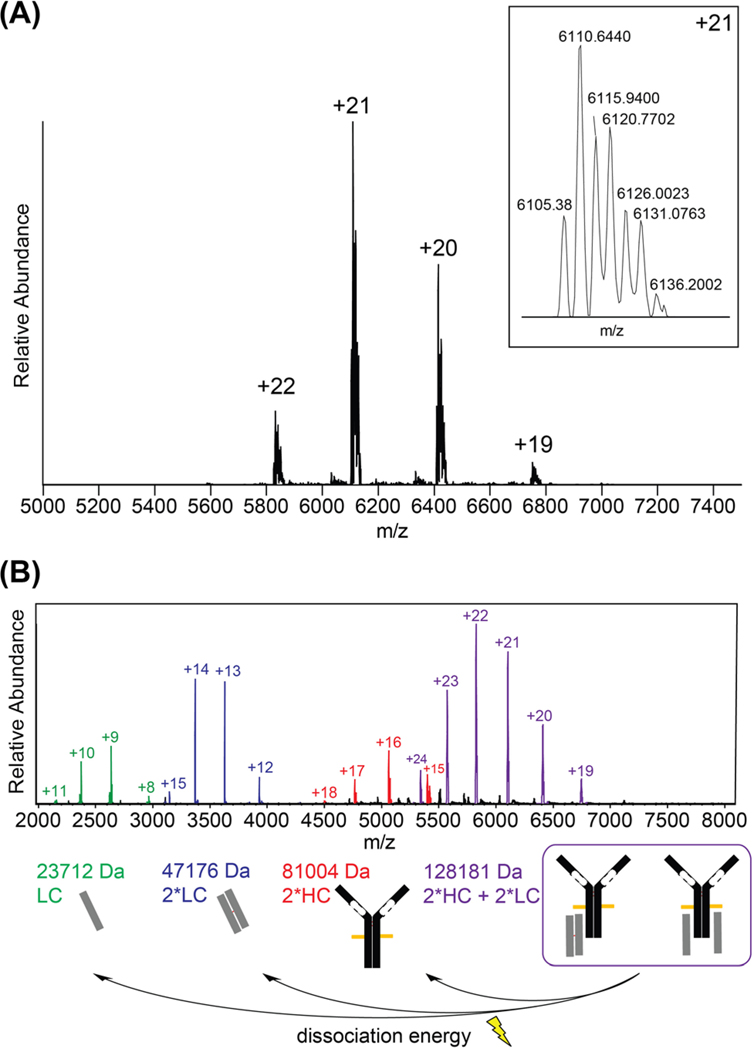
In order to further elucidate the structures indicated by IP-IFE analysis, we applied native MS to investigate the M-protein structures and connectivity within a native buffer environment [11]. The intact native spectrum contained one predominant charge state distribution, with multiple proteoforms evident within each charge state (Figure 3A). These deconvoluted to a cluster of intact masses around 128 kDa (Table S2). Next, we applied a mild dissociation voltage in the ion source region to check the 128 kDa species composition, which was hypothesized to represent a pseudo-intact IgG structure containing truncated HCs. We observed the gas-phase ejection of HC dimers and both LC monomers and dimers (Figure 3B). This result implies that the HC and LC species are non-covalently bound within the hetero-tetramer. By contrast, canonical IgG’s would release no subunits in a low-energy ejection experiment, since the HC and LCs are covalently tethered by disulfide bonds. Our resultant hypothesis is that the pseudo-intact species consists of a mixture of non-covalent complexes including (A) one HC dimer bound to one LC dimer and (B) one HC dimer bound to two LC monomers.
Figure 3:
Native MS analysis of intact immunoproteins.
(A) MS1 spectrum of pseudo-intact IgG complex. (B) MS2 spectrum of subunits ejected from pseudo-intact complex.
Intact mass determination of monoclonal immunoglobulins
High resolution MS (HRMS) coupled to reverse phase liquid chromatography (RPLC) was utilized to precisely determine the intact masses of the monoclonal immunoproteins. Under denaturing conditions, the mixture resolved into four chromatographic peaks (Figure 4) which mirrored the species observed in the ejected native MS spectrum. The deconvoluted masses of these species putatively correlate to free LC’s (peak 1; 23705.1 & 23766.2 Da), an LC homodimer (peak 2; 47175.2 Da), a truncated HC homodimer (peak 3; 81013.0 Da & other glycoforms), and a hetero-tetramer containing two LC’s and two truncated HC’s (peak 4; 128187.6 Da & other glycoforms). Although two unique LC monomers were detected in peak 1, neither matched the presumed monomer mass corresponding to peak 2. After reducing the intact sample, the chromatogram simplified to two major species: an LC monomer (23578.9 Da) and a truncated HC monomer (40515.6 Da). These results support our hypothesis that the species observed in peak 2 and peak 3 of the intact spectrum correlate to LC and HC disulfide-bound homodimers, and are consistent with the approximate MW estimation provided by SDS-PAGE (Supplementary Material, Figure S1).
Figure 4:
LCMS analysis of intact immunoproteins under denaturing conditions.
(A) The total ion chromatogram shows four distinct peaks (P1–4). (B) FTMS spectra showing (P1) free LC’s, (P2) disulfide-bound LC dimers, (P3) disulfide-bound HC dimers, and (P4) a pseudo-intact heterotetramer.
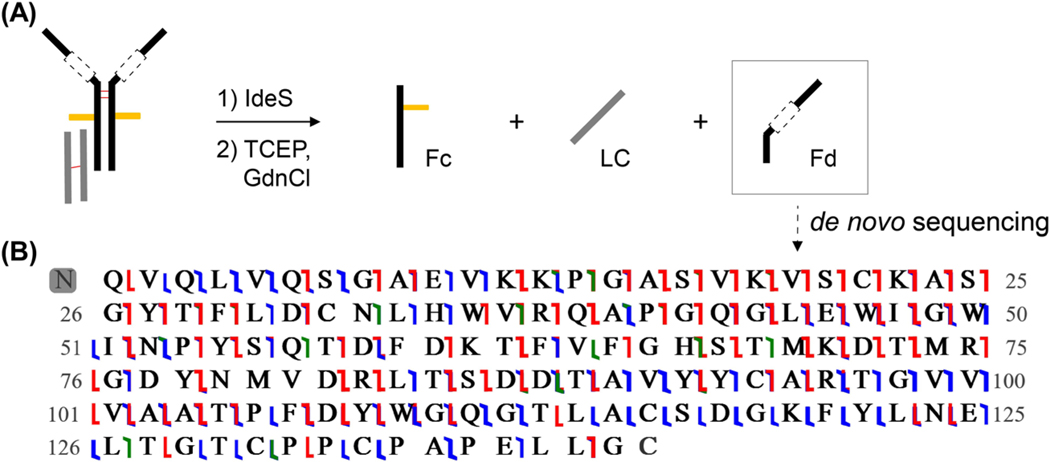
In order to further localize the HC truncation, the sample of ‘intact’ IgG was treated with IdeS protease, which selectively cleaves the IgG HC at a site N-terminal to the hinge region, thereby liberating the Fd and Fc subunits (Figure 5). Treatment with IdeS protease was followed by TCEP reduction. LCMS analysis demonstrated three chromatographic peaks, corresponding to the Fc, LC, and Fd subunits. The deconvoluted mass of the LC was 23578.9 Da, the masses associated with the Fc were 25174.7 & 25336.7 Da – representing different glycoforms – and the mass of the Fd was lower than hypothesized at 15334.5 Da. These results localized the truncation to the Fd subunit.
Figure 5:
(A) IdeS reaction products & (B) graphical fragment map depicting the resulting sequence from de novo sequencing of the Fd subunit, with a combined sequence coverage of 91% from multiple fragmentation modes. Blue, red, and green flags represent b/y, c/z, and a/x ions, respectively. Leucine and isoleucine residues could not be differentiated in this analysis.
De novo sequence analysis using middle-down mass spectrometry
Having localized the truncation to the Fd subunit, we sought to precisely characterize the sequence abnormalities and to assess whether either the VH or CH1 region remains intact in this species. Since the VH region amino acid sequence is derived from VDJ recombination, standard database search approaches are incapable of completely identifying this atypical species. Accordingly, we applied a middle-down MS approach to deduce the Fd sequence by de novo sequencing. Using RPLC in-line with HRMS, the reduced Fd subunit was isolated and subjected to multiple fragmentation techniques, including collision-induced dissociation, higher-energy collisional dissociation, electron-transfer dissociation, electron-transfer/higher-energy collisional dissociation, and ultraviolet photo dissociation. In combination, the resulting fragmentation spectra provided high sequence coverage of the Fd subunit (Figure 5). The theoretical monoisotopic mass for the proposed sequence is 15334.3 Da (within 10 ppm error from observed mass), and 91% residue cleavages were observed (15 ppm tolerance). This high resolution data can differentiate between amino acids with very similar mass, such as glutamine and lysine, however it cannot distinguish the isomeric residues leucine and isoleucine. The only observed posttranslational modification is an N-terminal pyroglutamate, which is expected when a sequence is initiated with a glutamine residue [12].
The analysis of the obtained sequence reveals several important features. It appears to contain an intact VH region, which comprises framework regions consistent with known VH patterns in the UniProt database. The sequence includes the expected cysteine residues at positions 22 and 94 which form the VH structural loop. In contrast, the CH1 region is almost completely absent; there are only 15 connecting residues present between the VH and the hinge region. The hinge region sequence responsible for HC dimerization (CPPCPAPELLG) was confirmed by fragmentation data, but crucially we did not observe a cysteine residue at position 103 of the CH1(hinge) which is requisite for disulfide bond formation between the Fd and the cysteine residue at position 106 of the kappa LC. These results provide a clear rationale for the lack of covalent bonds between LC and HCs, contributing to the unusual fragmentation patterns observed by native MS.
Discussion
Typical immunoglobulins have a hetero-tetrameric structure which is assembled by the covalent linkage of two HC’s followed by successive attachment of two LC’s to the constant region of the HC’s [13]. In the normal individual the antigenic stimulus of B lineage cells can generate upwards of 1016 possible unique sequences which constitute the polyclonal immunoglobulin repertoire [13,14]. However, in MM, the plasma cell (end stage B cell) clone characteristically secretes a single complete immunoglobulin molecule and/or free monoclonal LC’s, and is oftentimes accompanied by concomitant immuneparesis.
The entry of immunoglobulins into the secretory pathway is carefully regulated by quality control mechanisms that ensure incompletely folded, misfolded, or truncated proteins are retained, degraded and not expressed in the peripheral blood circulation [15]. Complete HC’s are therefore never secreted without an associated LC, but when deletions occur in the HC, particularly involving the CH1 domain, the transportation of truncated HC’s can be facilitated into the circulation [16]. Deletions, insertions and point mutations are most likely acquired during somatic hypermutation in the germinal center B cells [17]. The CH1 region of the IgG antibody is an intrinsically disordered domain which requires its cognate partner, the CL domain, to fold the CH1 into the structure observed in IgG antibodies [14,18]. When the CH1 domain is missing, LC’s can attach non-covalently to the CH2 or CH3 domain of the HC [13].
The current standard immunochemical and electrophoretic techniques are satisfactory for the characterization of structurally normal M-proteins but are inadequate when non-canonical immunoglobulins are present. Similarly, protein sequencing by a standard bottom-up proteomics approach can be successful in concert with homology database searching, but this approach falls short for novel antibody sequences which are not represented in databases. For the M-proteins described herein, methodological limitations were observed with bottom-up sequencing and with SDS-PAGE electrophoresis and Western blot analysis of the unreduced sample, due to the sequence novelty and structural instability of the 128 kDa hetero-tetrameric species (data not shown). The presence of Tris/glycine and SDS in the polyacrylamide gels resulted in dissociation of non-covalent bonds, altered migration and inaccurate MW determination of the pseudo-intact IgG. In addition, the total IgG concentration was overestimated by immunoturbidimetry because the standard curve is derived from an intact immunoglobulin calibrant. Also, in the case of the 128 kDa species, the standard IFE approach demonstrated co-incident staining for gamma, kappa, and kappa free antisera which could have been reported incorrectly as an intact IgG kappa M-protein comigrating alongside monoclonal free kappa LC’s. The IP-IFE approach provided some degree of clarification, but in doing so demonstrated anomalous antisera reactivity, which suggested the presence of a truncated non-covalent M-protein in the mid-gamma zone.
A modification of this IP-IFE technique, which involves initial precipitation with combined kappa and lambda antisera [19], has recently been described in the context of HC disease. This approach is appropriate in patients with HC disease but would have failed to detect the 128 kDa species in this sample. Fundamentally, the abovementioned methodologies cannot distinguish between covalent and non-covalent structures.
Despite these limitations, further corroborative evidence for the MS results can be gleaned from post-hoc analysis of the Freelite® and Hevylite® assays (results recorded in Table S1). The Hevylite® assay targets unique conformational junctional epitopes which bridge the HC (CH1 domain) and the LC (CL domain) of the intact immunoglobulin molecule. The monoclonal gamma HC, (Figure 1A, peak A), and the 128 kDa, CH1 deficient species (Figure 1A, peak B) were therefore unreactive with the IgG kappa/IgG lambda antisera. This assay also confirmed the absence of an intact IgG kappa (150 kDa) M-protein at terminal relapse.
The Freelite® assay targets epitopes that are found in many areas of the constant domain as well as in part of the variable domain, all of which are exposed in the non-covalently bound hetero-tetrameric 128 kDa species. The large concentration of non-covalently bound LC’s are therefore measured as ‘free’, resulting in a total ‘free’ LC concentration which consists of a small amount of circulating unbound free LC’s. This explains why only minute amounts of Bence Jones protein (BJP) were detected in the urine during the course of the patients’ disease. Regrettably, immunoglobulin gene sequencing was not possible due to the unavailability of cellular DNA at the time of this analysis. This limitation made the interpretation of the mass spectrometric data more challenging and necessitated the use of de novo sequencing.
The new MS techniques outlined above and others under development [5, 20, 21] will provide clinicians with distinct advantages over the current gel and capillary based electrophoretic methods for patients presenting with paraproteinaemia. These include increased resolution, improved turnaround time [22] and the ability to accurately stratify therapeutic responses in the setting of monoclonal antibody treatment when there is co-migration with the patients’ endogenous IgG kappa M-protein [1, 23]. They also will provide a greater understanding of the pathogenesis of MM which may help to stratify patients into new categories and potentially guide future personalized treatment modalities [24]. In addition, native MS and middle-down de novo sequencing provide distinctive insight into novel protein structures and sequences that is complementary to more traditional MS approaches.
Next generation technologies are already making an impact in various ways at the cellular, proteomic, and molecular level. This has resulted in the superior detection of small numbers of malignant plasma cells, leading to enhanced detection of MRD following treatment, or early relapse. Dysregulation of protein production, the pathways involved, and changes in clonal architecture can now be identified, all combining to give critical insight into the biology of MM development, disease progression and treatment responses with associated development of drug resistance [25,26]. In summary, our work has demonstrated the unique ability of next-generation technologies such as native/top-down MS, and IP-IFE, to shed light on otherwise elusive non-canonical monoclonal immunoproteins. As with many next generation technologies, those reported in this manuscript are complex, and at present are beyond the capabilities of health care laboratories other than those at the leading edge of translational research. However, it can be anticipated that further standardization and refinement of methodologies and instrumentation will make such an approach more accessible, helping to inform its clinical utility in diagnosis, risk stratification, treatment strategies, and detection of MRD and/or early relapse. We would therefore recommend their utilization for the identification and characterization of truncated immunoglobulin HCs, and when the evaluation of immunochemical and electrophoretic data is inexplicable or ambiguous in the clinical setting of a PCD.
Supplementary Material
Acknowledgments:
We thank our collaborators at Thermo Scientific who assisted with data acquisition on the Orbitrap Eclipse MS, including Kristina Srzentić, Romain Huguet, Christopher Mullen, and Philip Remes. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P41 GM108569 and by the National Cancer Institute CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by the National Science Foundation Cooperative Agreement No. DMR-1644779 and the State of Florida. LFS is a Gilliam Fellow of the Howard Hughes Medical Institute.
Research funding: National Science Foundation, Directorate for Mathematical and Physical Sciences, Division of Materials Research, DMR-1644779. U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute, CCSG P30 CA060553. U.S. Department of Health and Human Services, National Institutes of Health, National Institute of General Medical Sciences, P41GM108569.
Footnotes
Competing interests: NLK is involved with Proteinaceous, Inc., whose software was used for some of the data analysis.
Informed consent: Informed consent was obtained from all individuals included in this study.
Ethical approval: The research related to human patient samples has been complied with all the relevant national regulations, institutional policies and in accordance with the tenets of the Helsinki Declaration, and has been approved by the authors’ Clinical Haematology Department.
Supplementary Material: The online version of this article offers supplementary material (https://doi.org/10.1515/cclm-2020-1072).
Contributor Information
W. Ian Deighan, Department of Clinical Chemistry, Altnagelvin Hospital, Londonderry, BT47 6SB, UK.
Valerie J. Winton, Proteomics Center of Excellence, Northwestern University, Evanston, IL, USA..
Rafael D. Melani, Proteomics Center of Excellence, Northwestern University, Evanston, IL, USA..
Lissa C. Anderson, Ion Cyclotron Resonance Program, National High Magnetic Field Laboratory, Tallahassee, FL, USA..
John P. McGee, Chemical and Biological Engineering, Northwestern University, Evanston, IL, USA..
Luis F. Schachner, Department of Chemistry, Northwestern University, Evanston, IL, USA..
David Barnidge, Laboratory Medicine and Pathology, Mayo Clinic College of Medicine, Rochester, MN, USA.
David Murray, Laboratory Medicine and Pathology, Mayo Clinic College of Medicine, Rochester, MN, USA.
H. Denis Alexander, Northern Ireland Centre for Stratified Medicine, Biomedical Sciences Research Institute, Ulster University, Londonderry, UK.
David S. Gibson, Northern Ireland Centre for Stratified Medicine, Biomedical Sciences Research Institute, Ulster University, Londonderry, UK
Michael J. Deery, Cambridge Centre for Proteomics, University of Cambridge, Cambridge, UK
Feargal P. McNicholl, Department of Haematology, Altnagelvin Area Hospital, Londonderry, UK
Joseph McLaughlin, Northern Ireland Centre for Stratified Medicine, Biomedical Sciences Research Institute, Ulster University, Londonderry, UK.
Neil L. Kelleher, Proteomics Center of Excellence & Departments of Chemistry and Molecular Biology, Northwestern University, Evanston, IL, USA
Paul M. Thomas, Proteomics Center of Excellence, Northwestern University, 2170 Campus Drive, Evanston, IL 60208, USA.
References
- 1.Mills JR, Kohlhagen MC, Willrich MA, Kourelis T, Dispenzieri A, Murray DL. A universal solution for eliminating false positives in myeloma due to therapeutic monoclonal antibody interference. Blood 2018;132:670–2. [DOI] [PubMed] [Google Scholar]
- 2.Lê GN, Bones J, Coyne M, Bazou D, Dowling P, O’Gorman P, et al. Current and future biomarkers for risk-stratification and treatment personalisation in multiple myeloma. Mol Omics 2019;15:7–20. [DOI] [PubMed] [Google Scholar]
- 3.San Miguel JF, Gutiérrez NC, Mateo G, Orfao A. Conventional diagnostics in multiple myeloma. Eur J Canc 2006;42:1510–9. [DOI] [PubMed] [Google Scholar]
- 4.Zajec M, Langerhorst P, VanDuijn M, Gloerich J, Russcher H, van Gool A, et al. Mass spectrometry for identification, monitoring, and minimal residual disease detection of M-proteins. Clin Chem 2020;66:421–33. [DOI] [PubMed] [Google Scholar]
- 5.He L, Anderson LC, Barnidge DR, Murray DL, Dasari S, Dispenzieri A, et al. Classification of plasma cell disorders by 21 Tesla Fourier transform ion cyclotron resonance top-down and middle-down MS/MS analysis of monoclonal immunoglobulin light chains in human serum. Anal Chem 2019;91:3263–9. [DOI] [PubMed] [Google Scholar]
- 6.Barnidge DR, Dasari S, Botz CM, Murray DH, Snyder MR, Katzmann JA, et al. Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J Proteome Res 2014;13:1419–27. [DOI] [PubMed] [Google Scholar]
- 7.Saadalla AM, Singh A, Barnidge D, Kohlhagen M, Merlini G, Falk RH, et al. High sensitivity M-protein detection in a case of light-chain cardiac amyloidosis without evidence of plasma cell dyscrasia. Am J Hematol 2019;94:619–21. [DOI] [PubMed] [Google Scholar]
- 8.Thoren KL. Mass spectrometry methods for detecting monoclonal immunoglobulins in multiple myeloma minimal residual disease. Semin Hematol 2018;55:41–3. [DOI] [PubMed] [Google Scholar]
- 9.Deighan WI, O’Kane MJ, McNicholl FP, Keren DF. Multiple myeloma and multiple plasmacytomas associated with free gamma heavy chain, free kappa light chain and IgGk paraproteins: an unusual triple gammopathy. Ann Clin Biochem 2016;53:706–11. [DOI] [PubMed] [Google Scholar]
- 10.von Pawel-Rammingen U, Johansson BP, Björck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBOJ 2002;21:1607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGee JP, Melani RD, Goodwin M, McAlister G, Huguet R, Senko MW, et al. Voltage rollercoaster filtering of low-mass contaminants during native protein analysis. J Am Soc Mass Spectrom 2020;31:763–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelius D, Jing K, Lueras A, Rehder DS, Dillon TM, Vizel A, et al. Formation of pyroglutamic acid from N-terminal glutamic acid in immunoglobulin gamma antibodies. Anal Chem 2006;78:2370–6. [DOI] [PubMed] [Google Scholar]
- 13.Hendershot L, Bole D, Köhler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol 1987;104:761–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brewer JW, Hendershot LM. Building an antibody factory: a job for the unfolded protein response. Nat Immunol 2005;6:23. [DOI] [PubMed] [Google Scholar]
- 15.Reddy PS, Corley RB. The contribution of ER quality control to the biologic functions of secretory IgM. Immunol Today 1999;20:582–8. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y-K, Brewer JW, Hellman R, Hendershot LM. BiPand immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell 1999;10:2209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens T, Klein U, Küppers R. Frequent occurrence of deletions and duplications duringsomatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci Unit States Am 1998;95:2463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, et al. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell 2009;34:569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulli F, Napodano C, Pocino K, Cuccaro A, Hohaus S, Basile U. Heavy chain disease: our experience. Clin Chem Lab Med 2017;56:e10–e2. [DOI] [PubMed] [Google Scholar]
- 20.Mills JR, Barnidge DR, Murray DL. Detecting monoclonal immunoglobulins in human serum using mass spectrometry. Methods 2015;81:56–65. [DOI] [PubMed] [Google Scholar]
- 21.Ge F, Tao S, Bi L, Zhang Z, Zhang XE. Proteomics: addressing the challenges of multiple myeloma. Acta Biochim Biophys Sin 2011;43:89–95. [DOI] [PubMed] [Google Scholar]
- 22.Mills JR, Kohlhagen MC, Dasari S, Vanderboom PM, Kyle RA, Katzmann JA, et al. Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin Chem 2016;62:1334–44. [DOI] [PubMed] [Google Scholar]
- 23.McCudden CR, Voorhees PM, Hainsworth SA, Whinna HC, Chapman JF, Hammett-Stabler CA, et al. Interference of monoclonal antibody therapies with serum protein electrophoresis tests. Clin Chem 2010;56:1897–9. [DOI] [PubMed] [Google Scholar]
- 24.Murray D, Dispenzieri A, Kourelis T, Kohlhagen M, Barnidge D, Dasari S, et al. Going off the “gold-standard”: replacing electrophoretic methods with mass spectrometry for plasma cell disorders. Clin Lymphoma, Myeloma & Leukemia 2017;17:e18–e9. [Google Scholar]
- 25.Dutta AK, Hewett DR, Fink JL, Grady JP, Zannettino AC. Cutting edge genomics reveal new insights into tumour development, disease progression and therapeutic impacts in multiple myeloma. Br J Haematol 2017;178:196–208. [DOI] [PubMed] [Google Scholar]
- 26.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer 2012;12:335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.