Abstract
Recombinant human interferon-β (rhIFN-β) is therapeutically important and new commercially viable approaches are needed for its increased production. In this study, a codon-optimized gene encoding for rhIFN-β(C17S) protein was designed and expressed in E. coli SE1. As a first step of medium optimization, growth of E. coli as a function of different media components was studied. Subsequently, to optimize the media composition, a response surface methodology (RSM) was used. Our results show that optimized medium (15.0 g/L tryptone, 12.3 g/L meat extract, 1.0 g/L MgSO4 and 0.5 g/L thiamine along with minimal medium) obtained in this study provide better growth of recombinant cells and the expression level of recombinant protein was ~ 1.7-fold more than Luria–Bertani medium. The optimized medium may be utilized for the large-scale production of rhIFN-β.
Keywords: Plackett–Burman design, Recombinant human interferon-beta, Response surface methodology, Luria–Bertani medium, Central composite design
Introduction
Human interferon-β (hIFN-β), a 166 amino acid containing hormone protein, is produced in the body by many cell types in response to infection (Reder and Feng 2014; Spolaore et al. 2018). It has diverse biological properties such as antiproliferative, antibacterial, antiviral, and antitumor (Qi et al. 2014). US-FDA has approved two variants of this protein for clinical use: recombinant human interferon-β1a (rhIFN-β1a) and recombinant human interferon-β1b (rhIFN-β1b) for the treatment of multiple sclerosis (Rudick and Goelz 2011; Reder et al. 2014). Recently prokaryotic and eukaryotic expression systems have been used for the commercial production of rhIFN-β proteins (Reder and Feng 2014; Rudick and Goelz 2011). The eukaryotic expression systems produced glycosylated protein rhIFN-β1a which has similar amino acid arrangement to natural protein (Shayesteh et al. 2020). However, prokaryotic expression system produced (IFN-β1b) a non-glycosylated form of hIFN-β (Arregui et al. 2018; Reder and Feng 2014). Although non-glycosylated protein (rhIFN-β1b) exhibit equivalent biologically activity with respect to glycosylated form, generation of this protein (IFN-β1b) using microbial expression system (Escherichia coli; E. coli) offers several advantages and is preferred over other systems (Morowvat et al. 2015; Kusuma et al. 2019). To fulfill the ever-increasing demand of rhIFN-β, there is still a need for the development of new commercially viable approaches for the increased production of rhIFN-β proteins. In this regard, optimization of media composition used for the cultivation of recombinant host cells expressing target recombinant protein has emerged as an attractive approach for increasing the growth of target recombinant protein (Fan et al. 2020; Li et al. 2017; Maldonado et al. 2007; Morowvat et al. 2015; Samarin et al. 2018). For optimizing a number of process parameters, including media composition, response surface methodology (RSM) has been extensively used (Patel et al. 2016, Patil et al. 2016, Morowvat et al. 2015, Kusuma et al. 2019, Papaneophytou and Kontopidis 2014, Katla et al. 2019). In this study, we have used Plackett–Burman (PB) and Central Composite Design (CCD) to optimize the composition of cultivation media for the growth and production of recombinant protein. Further optimized conditions were compared with commercial media (Luria–Bertani).
Materials and methods
Materials
Escherichia coli SE1 has been used for the production of rhIFN-β proteins as prepared in our previous work (Pal et al. 2018). Bradford reagent and markers were acquired from Bio-Rad, Gurgaon, India. Components used in the preparation of medium were purchased from HiMedia (India). All media components were prepared freshly in autoclaved double-distilled water. All other chemicals were used of analytical grade with high quality.
Cultivation
Escherichia coli producing recombinant protein was cultivated at shake-flask level in a rotary shaker (Innova 4230 Incubator, Thermo Fisher Scientific, USA) at 200 rpm and 37 °C temperature, and processed by following the procedure described previously (Pal et al. 2018). Initially inoculum was prepared in a 100 mL flask having 20 mL LB medium, (pH 7.0) with carbenicillin (50 μg/mL) and incubated at 37 °C and 200 rpm to reach OD600 ~ 2. The 1% inoculum was transferred in 250 mL flask containing 50 mL production medium (pH 7.0) and incubated at 200 rpm and 37 °C. Later rhIFN-β protein expression was induced with 1 mM isopropyl-1-thio β-D galactopyranoside (IPTG) once broth OD600 reached 0.6. Fermentation was carried out for 24 h and cells were separated from broth (Morowvat et al. 2015; Pal et al. 2018; Tripathi 2016).
SDS-PAGE and Western blot analysis for expression of rhIFN-β protein
The expressions of rhIFN-β protein in the cell mass were determined through sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as per the protocol developed in our previous study (Pal et al. 2018; Tripathi 2016).
Determination of a relationship between growth of E. coli and wet cell mass
Perkin Elmer Lambda 25 UV–Vis spectrometer was used to monitor growth of E. coli and expressed as either optical density at 600 nm (OD600) or wet cell mass (WCM (g/L)). To determine a relationship between growth of cells (OD600) and WCM of E. coli, the cells were grown as described above. 1 mM IPTG was used to induce expression of recombinant protein and the cultures were divided into 6 parts of 50 mL each (in aseptic condition) and further grown at 200 rpm and 37 °C for 16 h. At a regular time interval, two flasks at a time were withdrawn and processed. The OD600 and the corresponding WCM were noted down for each sample. For obtaining WCM, the cultures were centrifuged in pre-weighed centrifuged bottles (4 °C, 6000xg, 30 min), the cell pellets were collected and WCM was determined using electronic balance. To calculate the relationship between the OD600 and WCM, a standard curve of WCM was plotted against OD600 (Agbogbo et al. 2020, Patil et al. 2017, Poccia et al. 2014).
Effect of media components on the growth of Escherichia coli
Recombinant E. coli SE1 producing rhIFN-β was grown in the minimal medium (3.5 g KH2PO4, 5.0 g glucose, 3.5 g (NH4)2HPO4 and 1.0 g MgSO4 for 1 L) containing specified concentrations of particular components. Various carbon sources (10 g/L) screened in this study includes xylose, fructose, glycerol, sucrose, lactose, glucose carboxymethyl cellulose, sorbitol, starch and mannitol. Different nitrogen sources (10 g/L) screened are yeast extract, peptone, meat extract, tryptone, soybean meal, soya-peptone, malt, urea, ammonium acetate, ammonium dihydrogen phosphate (NH4H2PO4), ammonium nitrate (NH4NO3) and sodium nitrate (NaNO3). Sorbitol (10 g/L) was kept as carbon source during the screening of different nitrogen sources in the supplemented medium (Huang et al. 2021; Patel et al. 2018).
Plackett–Burman (PB) design
A computer software Design-Expert® 9 (Stat-Ease Inc., Minneapolis, MN, USA) was employed in this study. To identify the components affecting cell mass production of E. coli, PB design was used. Eleven independent ‘variables’ (peptone, tryptone, meat extract, yeast extract, glycerol, potassium dihydrogen phosphate (KH2PO4), sorbitol, glucose, magnesium sulfate (MgSO4), thiamine and citric acid) were used in 12 experimental trials. Each variable was assessed at 2 different of concentrations (i.e., high (level + 1) and low (level -1)) for their effect on the WCM of E. coli (Table 1). Experiments were done in duplicates and the average value was taken as the response. Variables exerting significant effect on the response were selected using regression analysis, as described (Papaneophytou and Kontopidis 2014; Patel et al. 2020), and used further.
Table 1.
Concentration ranges of the variables used in the Plackett–Burman design
| Factors (g/L) |
Actual levels of coded factors − 1 + 1 |
|
|---|---|---|
| Glucose | 5 | 10 |
| Sorbitol | 5 | 10 |
| Glycerol | 5 | 10 |
| Peptone | 4 | 8 |
| Tryptone | 4 | 8 |
| Meat extract | 5 | 10 |
| Yeast extract | 5 | 10 |
| Potassium dihydrogen phosphate | 4 | 8 |
| Magnesium sulfate | 0.5 | 1 |
| Citric acid | 0.5 | 1 |
| Thiamine | 0.5 | 1 |
Response surface methodology (RSM)
Optimum concentration of selected variables (obtained from the PB design) giving maximum WCM of recombinant E. coli was determined using RSM. Different concentrations of four variables (i.e., tryptone, meat extract, thiamine and MgSO4) were selected and optimized using a CCD (Table 2). A CCD consisting of 16 runs with 6 center points and 8 star points was applied by keeping the other variables constant (5 g/L glucose, 4 g/L KH2PO4 and 1 g/L citric acid) and actual response (from experiment) and predicted response (from software) was compared (two times independently). Second-order polynomial equation (ANOVA) was used to determine interaction between the used variables and the response surface plot was obtained (Huang et al. 2021, Patil et al. 2017).
Table 2.
Concentration ranges of the variables used in the central composite design
| Factors (g/L) | Actual levels of coded factors -1 0 + 1 |
||
|---|---|---|---|
| Tryptone | 5 | 10 | 15 |
| Meat extract | 5 | 10 | 15 |
| Thiamine | 0.5 | 1 | 1.5 |
| Magnesium sulfate | 0.5 | 1 | 1.5 |
Comparison of expression of rhIFN-β
Starter cultures of recombinant E. coli were grown as described above and 1% of the starter culture was used to inoculate either optimized medium or LB medium and the cultures were grown further at 30 °C. 1.0 mM IPTG was used to induce the growing culture when the OD600 = 0.4–0.6 and the cultures were grown further at 20 °C for 32 h (to allow soluble expression of rhIFN-β protein). Cell pellet were collected by centrifuging the cultures (4 °C, 6000×g for 30 min) and suspended in buffer containing 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM β-ME, 0.1% tergitol and lysozyme (10 µg/mL). The cell suspension was sonicated and DNase (1 µg/mL) was added. The suspension was then kept on slow stirring at 4 ºC for 1 h and subjected to centrifugation to separate clear cell lysate (supernatant) and cell debris. SDS-PAGE and Western blot analysis of the samples were done by following the methods described previously (Pal et al 2018; Tripathi 2016).
Results
Naturally occurring hIFN-β contain three Cys residues; one Cys residue at position 17 exists in a free form while Cys 31 and Cys 141 are engaged in a disulphide bond formation (Spolaore et al. 2018). Studies have revealed that this free Cys 17 is primarily responsible for the formation of aggregates during purification and storage of native protein (Mark et al. 1984). It was also reported that substitution of free Cys at position 17 with other amino acid not only results in the variant protein being biologically active but also makes the variant protein more stable (Mark et al. 1984). Thus, in this study, we have used rhIFN-β(C17S) variant.
Expression of rhIFN-β(C17S)
To express rhIFN-β(C17S) protein, a codon-optimization of a gene encoding rhIFN-β(C17S) was done and the recombinant protein was expressed in E. coli SE1 (Pal et al. 2018). E. coli SE l cells were cultivated and the rhIFN-β(C17S) protein was expressed by inducing the culture at 20 °C for 32 h (Pal et al. 2018; Tripathi 2016). In the western blot analysis, a band of ~ 19 kDa was observed in both cell lysate and cell debris fractions, confirming the expression of recombinant protein in E. coli SE l cells (data not shown).
Effect of media components on the growth of Escherichia coli SE1
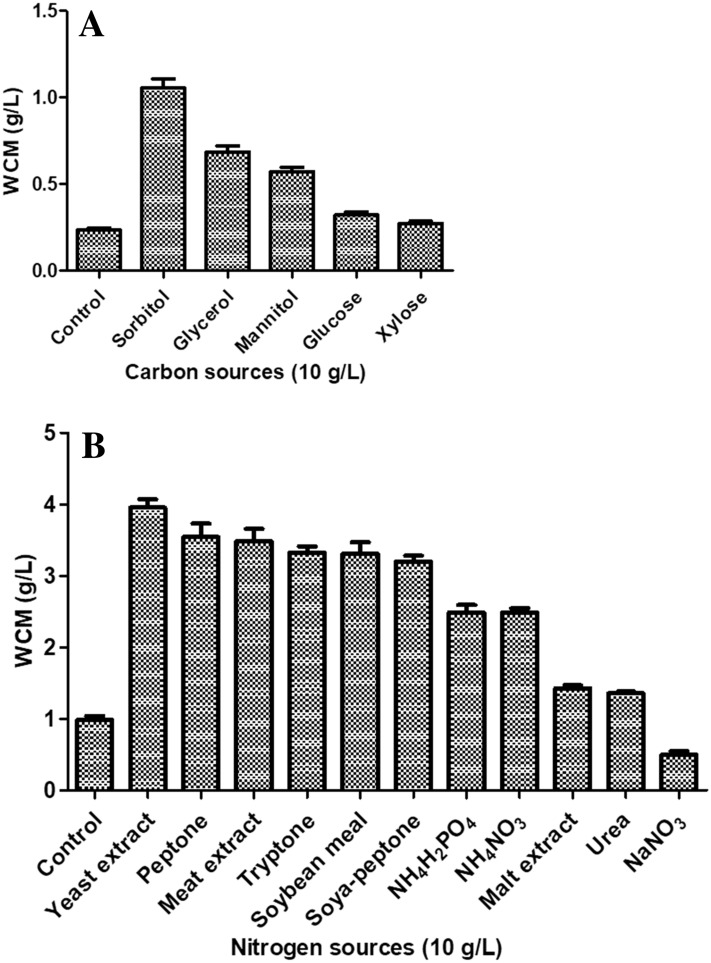
A standard curve was obtained between WCM and OD600 of culture of recombinant E. coli and used to convert OD600 values from the cultures of recombinant E. coli cells to the WCM (g/L) in the subsequent experiments. As a first step of media optimization studies, the effect of media components on the growth of E. coli was studied. The cells were grown in designated media WCM was determined (Fig. 1). Cell mass concentrations were considerably higher when sorbitol, glycerol and mannitol (at a final concentration of 10 g/L) were used in M9 minimal medium (Fig. 1a). A considerable amount of growth was also observed with xylose and glucose. For screening the nitrogen source giving maximum growth, the recombinant E. coli SE1 cells were cultured in minimal medium containing sorbitol (10 g/L) as carbon source and 10 g/L of particular nitrogen source (Fig. 1b). Cell mass concentration was considerably higher when meat extract, yeast extract, peptone, tryptone, soybean meal and soya-peptone (10 g/L) was used as a nitrogen source (Fig. 1b). A substantial amount of cell growth was also observed when NH4H2PO4, NH4NO3, malt extract and urea was used. Based on these results, three carbon sources (sorbitol, glucose and glycerol) and four nitrogen sources (meat extract, tryptone, peptone, and yeast extract), which gave an appreciable amount of cell mass concentration, were selected and used for in the next step of media optimization studies.
Fig. 1.
Effect of media components on the WCM of E. coli SE1. (Panel A—carbon sources and Panel B—nitrogen sources). All quantities are in g/L
Screening of medium components using PB design
PB design was employed to determine the optimum levels of significant variables (i.e., media components) which give maximum cell mass of recombinant E. coli. Three carbon sources and four nitrogen sources were selected from the above experiments (Fig. 1), while remaining four variables (KH2PO4, MgSO4, thiamine and citric acid) were selected from the literature (Shin et al. 2001; Babaeipour et al. 2010, 2007). Experimental design and results for the PB experiments are given in Table 3. Student’s t test for ANOVA was used to screen the variables exhibiting significant effects (Table 4). The F value of 3447.08 indicates significance of our model used and the values of Prob < 0.05 indicate model terms are significant. The statistical significance of our model was also confirmed by the coefficient of determination (R2 = 1) (Table 4).
Table 3.
Experimental setup and response of various run in the Plackett–Burman design
| Run | Glucose (g/L) | Sorbitol (g/L) | Glycerol (g/L) | Peptone (g/L) | Tryptone (g/L) | Meat ex. (g/L) | Yeast ex. (g/L) | Potassium dihydrogen phosphate (g/L) | Magnesium sulfate (g/L) | Thiamine (g/L) | Citric acid (g/L) | Response |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WCM (g/L) | ||||||||||||
| 1 | 10 | 10 | 5 | 8 | 8 | 10 | 5 | 4 | 0.5 | 1 | 0.5 | 4.4085 |
| 2 | 5 | 5 | 10 | 4 | 8 | 10 | 5 | 8 | 1 | 1 | 0.5 | 4.68996 |
| 3 | 10 | 5 | 10 | 8 | 4 | 10 | 10 | 8 | 0.5 | 0.5 | 0.5 | 3.54752 |
| 4 | 5 | 10 | 10 | 4 | 8 | 10 | 10 | 4 | 0.5 | 0.5 | 1 | 4.35695 |
| 5 | 10 | 10 | 10 | 4 | 4 | 5 | 10 | 4 | 1 | 1 | 0.5 | 3.7464 |
| 6 | 10 | 10 | 5 | 4 | 4 | 10 | 5 | 8 | 1 | 0.5 | 1 | 4.47178 |
| 7 | 10 | 5 | 5 | 4 | 8 | 5 | 10 | 8 | 0.5 | 1 | 1 | 4.07892 |
| 8 | 10 | 5 | 10 | 8 | 8 | 5 | 5 | 4 | 1 | 0.5 | 1 | 4.2602 |
| 9 | 5 | 5 | 5 | 8 | 4 | 10 | 10 | 4 | 1 | 1 | 1 | 5.05937 |
| 10 | 5 | 5 | 5 | 4 | 4 | 5 | 5 | 4 | 0.5 | 0.5 | 0.5 | 4.25849 |
| 11 | 5 | 10 | 5 | 8 | 8 | 5 | 10 | 8 | 1 | 0.5 | 0.5 | 4.31859 |
| 12 | 5 | 10 | 10 | 8 | 4 | 5 | 5 | 8 | 0.5 | 1 | 1 | 3.98339 |
Table 4.
ANOVA analysis of Plackett–Burman experimental design
| Source | SS | DF | MS | F value |
p value Prob > F |
Significant |
|---|---|---|---|---|---|---|
| Model | 1.78 | 10 | 0.18 | 3447.08 | 0.0133 | |
| A—Glucose | 0.39 | 1 | 0.39 | 7466.99 | 0.0074 | |
| B—Sorbitol | 0.031 | 1 | 0.031 | 596.89 | 0.0260 | |
| C—Glycerol | 0.34 | 1 | 0.34 | 6513.44 | 0.0079 | |
| E—Tryptone | 0.091 | 1 | 0.091 | 1762.35 | 0.0152 | |
| F—Meat ex | 0.30 | 1 | 0.30 | 5740.29 | 0.0084 | |
| G—Yeast ex | 0.078 | 1 | 0.078 | 1498.15 | 0.0164 | |
| H—KH2PO4 | 0.083 | 1 | 0.083 | 1609.43 | 0.0159 | |
| J—MgSO4 | 0.30 | 1 | 0.30 | 5889.81 | 0.0083 | |
| K—Thiamine | 0.047 | 1 | 0.047 | 912.99 | 0.0211 | |
| L—Citric acid | 0.13 | 1 | 0.13 | 2480.43 | 0.0128 | |
| Residual | 5.175E−005 | 1 | 5.175E−005 | |||
| Cor total | 1.78 | 11 |
SS Sum of squares, DF Degrees of freedom, MS Mean sum of squares
The multiple regression equation of the response values for the variables studied are given in the following equation:
| 1 |
Above results suggest that tryptone, meat extract, MgSO4, thiamine and citric acid were most significant variables that influenced WCM production of recombinant E. coli, whereas glucose, sorbitol, glycerol, yeast extract, and KH2PO4 did not had any significant effect (Eq. 1).
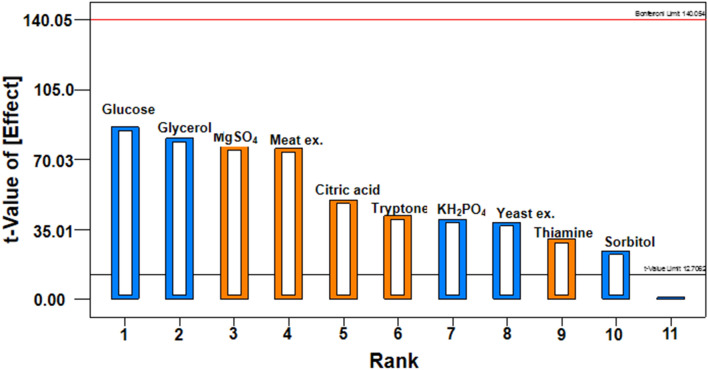
Pareto chart showing the effect of variables on the WCM production of recombinant E. coli SE1 is given in Fig. 2. In the chart, the maximal effect was presented in the left portion and then progress right to the minimal effect. The chart also shows that important variables influencing the WCM production are tryptone, meat extract, MgSO4, thiamine and citric acid. For CCD analysis, thus, we selected tryptone, meat extract, MgSO4 and thiamine.
Fig. 2.
Pareto chart showing the effect of variables on growth of recombinant E. coli SE1 expressing rhIFN-β(C17S)
Optimization by RSM
It is known that PB design is used to screen variables that have significant effect on the response however; in this design, the interactions between the variables cannot be measured (Papaneophytou and Kontopidis 2014). The next step in our media optimization was to find optimum levels of variables selected in the PB design for maximum cell mass production of recombinant E. coli. For this, RSM using a CCD was used and the concentrations of the variables used in this study are given in Table 5.
Table 5.
Experimental setup of CCD and responses in the various runs
| Run | A | B | C | D | WCM (g/L) |
|---|---|---|---|---|---|
| 1 | 5 | 15 | 1.5 | 1.5 | 0.40044 |
| 2 | 10 | 10 | 1 | 1 | 1.88932 |
| 3 | 10 | 10 | 1 | 1 | 1.95678 |
| 4 | 5 | 5 | 0.5 | 0.5 | 0.644759 |
| 5 | 10 | 15 | 1 | 1 | 1.129 |
| 6 | 10 | 5 | 1 | 1 | 0.952602 |
| 7 | 10 | 10 | 1 | 1 | 1.92456 |
| 8 | 15 | 5 | 0.5 | 0.5 | 2.32763 |
| 9 | 10 | 10 | 1 | 1 | 1.99765 |
| 10 | 10 | 10 | 1 | 1 | 1.89657 |
| 11 | 10 | 10 | 1.5 | 1 | 3.67115 |
| 12 | 10 | 10 | 0.5 | 1 | 3.46201 |
| 13 | 10 | 10 | 1 | 0.5 | 0.938676 |
| 14 | 5 | 15 | 0.5 | 0.5 | 1.73931 |
| 15 | 5 | 15 | 0.5 | 1.5 | 2.34921 |
| 16 | 5 | 5 | 1.5 | 0.5 | 1.3743 |
| 17 | 15 | 15 | 1.5 | 0.5 | 3.64574 |
| 18 | 15 | 5 | 0.5 | 1.5 | 2.35084 |
| 19 | 15 | 5 | 1.5 | 0.5 | 3.93419 |
| 20 | 15 | 15 | 0.5 | 1.5 | 4.82165 |
| 21 | 5 | 5 | 0.5 | 1.5 | 1.5226 |
| 22 | 5 | 10 | 1 | 1 | 1.35329 |
| 23 | 15 | 15 | 1.5 | 1.5 | 3.18324 |
| 24 | 15 | 15 | 0.5 | 0.5 | 4.8192 |
| 25 | 5 | 5 | 1.5 | 1.5 | 1.22868 |
| 26 | 15 | 5 | 1.5 | 1.5 | 3.82091 |
| 27 | 15 | 10 | 1 | 1 | 3.18348 |
| 28 | 10 | 10 | 1 | 1 | 1.95643 |
| 29 | 5 | 15 | 1.5 | 0.5 | 0.346689 |
| 30 | 10 | 10 | 1 | 1.5 | 0.879306 |
A: Tryptone, B: Meat extract, C: Thiamine and D: Magnesium sulfate. All values are in g/L
ANOVA was conducted (Table 6) with the response function and the relationship and interaction between the variables was determined using second-order polynomial equation (Eq. 2):
| 2 |
Table 6.
ANOVA analysis of central composite design
| Source | SS | DF | MS | F value | p value Prob > F |
|
|---|---|---|---|---|---|---|
| Model | 44.99 | 14 | 3.21 | 61.16 | < 0.0001 | Significant |
| A—Tryptone | 24.80 | 1 | 24.80 | 471.97 | < 0.0001 | |
| B—Meat ex | 1.02 | 1 | 1.02 | 19.35 | 0.0005 | |
| C—Thiamine | 0.33 | 1 | 0.33 | 6.25 | 0.0245 | |
| D—MgSO4 | 0.034 | 1 | 0.034 | 0.65 | 0.4314 | |
| AB | 0.99 | 1 | 0.99 | 18.76 | 0.0006 | |
| AC | 0.63 | 1 | 0.63 | 11.96 | 0.0035 | |
| AD | 0.24 | 1 | 0.24 | 4.50 | 0.0509 | |
| BC | 5.84 | 1 | 5.84 | 111.13 | < 0.0001 | |
| BD | 0.012 | 1 | 0.012 | 0.23 | 0.6393 | |
| CD | 0.30 | 1 | 0.30 | 5.66 | 0.0311 | |
| A2 | 0.53 | 1 | 0.53 | 10.12 | 0.0062 | |
| B2 | 1.55 | 1 | 1.55 | 29.59 | < 0.0001 | |
| C2 | 7.95 | 1 | 7.95 | 151.21 | < 0.0001 | |
| D2 | 2.13 | 1 | 2.13 | 40.51 | < 0.0001 | |
| Residual | 0.79 | 15 | 0.053 | |||
| Lack of fit | 0.78 | 10 | 0.078 | 45.81 | 0.0003 | |
| Pure error | 8.510E−003 | 5 | 1.702E−003 | |||
| Cor total | 45.77 | 29 |
SS Sum of squares, DF Degrees of freedom, MS Mean sum of squares
where A, B, C and D are tryptone, meat extract, thiamine and MgSO4, respectively.
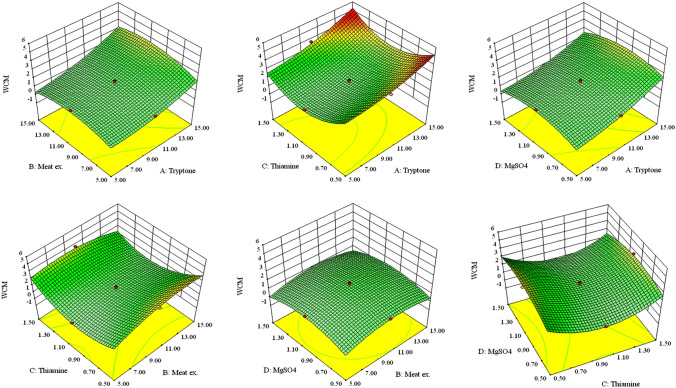
Results suggest that variables A, B and C are significant (p < 0.05) for the cell mass production (Table 6). The interactions between tryptone (A) and meat extract (B), tryptone (A) and thiamine (C), meat extract (B) and thiamine (C) and thiamine (C) and MgSO4 (D) were significant (as was evident by the p values) (Table 6). These results indicate that the concentrations of tryptone, meat extract and thiamine in the cultivation media have substantial influence on WCM of E. coli SE1. p values for the model (p < 0.0001) and for lack of fit (0.0003) suggest a good fit of obtained data with the model, which is also confirmed by the coefficient of determination value (R2 = 0.9828). To study the interaction among different variables and to determine the optimum levels of the variables for E. coli WCM production, Eq. (2) was also expressed as 3D response surface plot for each pair of variables (A—tryptone, B—meat extract, C—thiamine, and D—MgSO4), while other variables were held constant (Fig. 3).
Fig. 3.
3D response surface plots showing interaction between variables and their optimum values for production (WCM) of E. coli SE1 expressing rhIFN-β(C17S). All quantities are in g/L
Validation experiment
The model used in the optimization studies predicted that maximum cell mass concentration can be obtained when the composition of the medium minimal contain 15.0 g/L tryptone, 12.3 g/L meat extract, 1.0 g/L MgSO4 and 0.5 g/L thiamine. To confirm this, E. coli SE1 producing rhIFN-β(C17S) was grown in the optimized medium and WCM production was determined experimentally and compared with the values obtained from our model. All the experiments were done in duplicate and the variation was within ± 5%. A cell mass concentration (WCM) of 5.53 g/L was obtained when the optimized medium was used in the experiment, which is in agreement with the value predicted in our model (5.6 g/L). These data validate the model used for the media optimization studies.
Comparison of expression of rhIFN-β protein
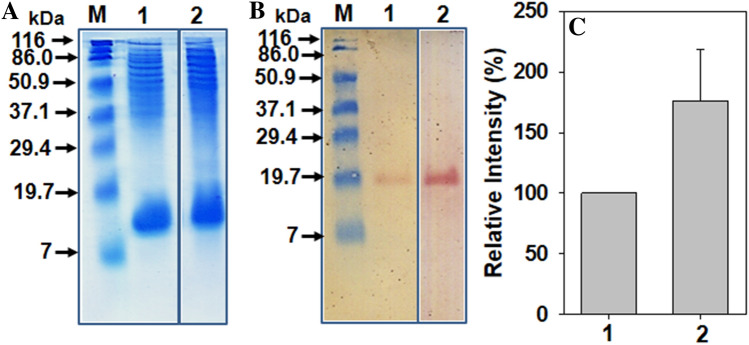
Recombinant strains of E. coli are routinely cultured in LB medium. To see the effect of optimized medium on expression of recombinant protein, the recombinant E. coli SE1 cells were separately grown both in optimized and LB medium, and the expression of active (soluble) recombinant protein was analyzed by SDS-PAGE and Western blot analysis (Fig. 4a and 4b). A band of ~ 19 kDa was observed in Western blot analysis of the cell supernatant (cell lysate) fractions of cells grown in both the media. Densitometry analysis revealed more expression of the recombinant protein (~ 1.7-fold) in the E. coli SE1 when grown in optimized medium as compared to LB medium (Fig. 4c).
Fig. 4.
SDS-PAGE (Panel a), Western blot (Panel b) and densitometric analysis (Panel c) of cell lysate samples of E. coli-producing recombinant protein. Lane 1 and bar 1—cells grown in LB medium, Lane 2 and bar 2—cells grown in optimized medium
Discussion
The rhIFN-β protein is not only a drug of choice for the treatment of MS but also is a potential therapeutic agent for the treatment of several other disease conditions in humans (Reder and Feng 2014). Because of its cost effectiveness, production of rhIFN-β using E. coli is favored over the other expression platforms (Rosano and Ceccarelli 2014). In this study, rhIFN-β(C17S) protein was cloned and expressed in E. coli SE1. As the pStaby–E. coli SE1 is an antibiotic-free expression system, it is a cost effective approach that offers high yield of recombinant proteins (Pal et al. 2018; Stevens et al. 2008; Bey et al. 2013; Collins et al. 2013). The pStaby–E. coli SE1 is used by many researchers to produce a variety of recombinant proteins (Pal et al. 2018; Stevens et al. 2008; Bey et al. 2013; Collins et al. 2013).
Different statistical approaches are developed and utilized to optimize the medium components (Kusuma et al. 2019; Papaneophytou and Kontopidis 2014; Katla et al. 2019). Using RSM, in this study, we have optimized the cultivation medium composition for maximal growth of E. coli expressing rhIFN-β(C17S). The yield of soluble rhIFN-β protein using the optimized medium was found to be ~ 1.7-fold more than LB medium. LB medium contains yeast extract (5 g/L), casein hydrolysate (10 g/L) and NaCl (10 g/L). The relative higher expression of rhIFN-β protein, when the cells were grown in the optimized medium, as compared to LB medium, could be due to the higher concentration of thiamine used in the optimized medium. It is important to note that E. coli (SE1) strain which is derived from E. coli K12 strain, is deficient in synthesis of vitamin B complex, and hence, external supplementation of thiamine could result in increased propagation of E. coli cells, thereby resulting in higher cell mass accumulation and higher expression of rhIFN-β protein. The other factors, such as higher content of tryptone and addition of meat extract in the optimized media, could have also positively affected the expression of rhIFN-β in the optimized medium.
It is well established that maximum growth of recombinant host cells (expressing the gene of interest) during cultivation is a first and crucial step to obtain higher yield of target proteins (Maldonado et al. 2007; Morowvat et al. 2015; Huang et al. 2012). By modulating cultivation media compositions, one can increase the growth of recombinant cells and can obtain a better yield of target proteins (Maldonado et al. 2007; Morowvat et al. 2015). It is thus recommended that the concentration of medium components should be carefully selected such that the final medium not only contains all the necessary components, but also the concentration of these components should be optimal to avoid adverse effects on the growth of recombinant host cells during cultivation (Maldonado et al. 2007; Morowvat et al. 2015; Huang et al. 2012). Thus, optimization of the cultivation media component has emerged as an attractive approach for increasing the growth of recombinant host cells (Maldonado et al. 2007; Morowvat et al. 2015; Huang et al. 2012; Zhang et al. 2017).
Conclusion
In this study, we report medium composition optimization strategy that included one factor at a time, PB design and CCD for the production of cell mass and recombinant protein overexpression in E. coli. In nutshell, the expression system and the optimized medium reported in this study may be employed in the commercial production of rhIFN-β.
Abbreviations
- CCD
Central composite design
- rhIFN-β(C17S)
Recombinant human interferon-beta variant containing C17S substitution
- PB
Plackett–Burman
- LB medium
Luria–Bertani medium
- RSM
Response surface methodology
- IPTG
Isopropyl-1-thio β-D galactopyranoside
- Cys
Cysteine
Author contributions
AHP and UCB were in-charge of the experiments and paper writing. DP, GP and PD performed experimental studies and participated in paper writing. SHN and GP performed review and editing. All the authors read and approved the final manuscript.
Funding
This work was supported in partial by the research grants to AHP from the Department of Biotechnology (New Delhi) (BT/PR23283/MED/30/1953/2018) and NIPER, SAS Nagar (NPLC-AHP). DP and GP are thankful to the University Grants Commission and Department of Biotechnology, New Delhi, Govt. of India for support.
Availability of data and material
Not applicable.
Declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Dharam Pal and Gopal Patel have contributed equally to this work.
Contributor Information
Dharam Pal, Email: dharampaldadiya@gmail.com.
Gopal Patel, Email: gopal87patel@gmail.com.
Prakashkumar Dobariya, Email: dobariya.prakash@yahoo.in.
Shivraj Hariram Nile, Email: nileshivraj@yahoo.com.
Abhay H. Pande, Email: apande@niper.ac.in
Uttam Chand Banerjee, Email: ucbanerjee@niper.ac.in.
References
- Agbogbo FK, Ramsey P, et al. Upstream, development of Escherichia coli fermentation process with PhoA promoter using design of experiments (DoE) J Ind Microbiol. 2020;7:789–799. doi: 10.1007/s10295-020-02302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arregui MB, Callum GJM, et al. Production of biologically active feline interferon beta in insect larvae using a recombinant baculovirus. 3 Biotech. 2018;8:341. doi: 10.1007/s13205-018-1369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaeipour V, Shojaosadati SA, et al. Enhancement of human γ-Interferon production in recombinant E. coli using batch cultivation. Appl Biochem Biotechnol. 2010;160:2366–2376. doi: 10.1007/s12010-009-8718-5. [DOI] [PubMed] [Google Scholar]
- Babaeipour V, Shojaosadati SA, et al. Over-production of human interferon-γ by HCDC of recombinant Escherichia coli. Process Biochem. 2007;42:112–117. doi: 10.1016/j.procbio.2006.07.009. [DOI] [Google Scholar]
- Bey RF, Simonson RR et al (2013) Rotavirus subunit vaccines and methods of making and use there of. WO2013123219A1
- Chen X, Li Y, et al. Application of response surface methodology in medium optimization for spore production of Coniothyrium minitans in solid-state fermentation. World J Microbiol Biotechnol. 2005;21:593–599. doi: 10.1007/s11274-004-3492-6. [DOI] [Google Scholar]
- Collins T, Barroca MJ, et al. Optimising the production of a silk-elastin-like protein in E. coli: overcoming acetate accumulation and plasmid instability. Microbiotec. 2013;2013:2013. [Google Scholar]
- Fan G, Zhu Y, et al. Optimization of fermentation conditions for the production of recombinant feruloyl esterase from Burkholderia pyrrocinia B1213. 3 Biotech. 2020;10:216. doi: 10.1007/s13205-020-02198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli A, Shojaosadati SA, et al. Effect of parallel feeding of oxidizing agent and protein on fed-batch refolding process of recombinant interferon beta-1b. Process Biochem. 2011;46(3):796–800. doi: 10.1016/j.procbio.2010.11.007. [DOI] [Google Scholar]
- Huang C-J, Lin H, et al. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- Huang C, Feng Y, et al. Production, immobilization and characterization of beta-glucosidase for application in cellulose degradation from a novel Aspergillus versicolor. Int J Biol Macromol. 2021;177:437–446. doi: 10.1016/j.ijbiomac.2021.02.154. [DOI] [PubMed] [Google Scholar]
- Li J, Sun C, et al. Optimization of the secretory expression of recombinant human C-reactive protein in Pichia pastoris. 3 Biotech. 2017;7:291. doi: 10.1007/s13205-017-0917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado LMTP, Hernández VEB, et al. Optimization of culture conditions for a synthetic gene expression in Escherichia coli using response surface methodology: the case of human interferon beta. Biomol Eng. 2007;24:217–222. doi: 10.1016/j.bioeng.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Mark DF, Lu SD, et al. Site-specific mutagenesis of the human fibroblast interferon gene. Proc Natl Acad Sci. 1984;81:5662–5666. doi: 10.1073/pnas.81.18.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowvat MH, Babaeipour V, et al. Optimization of fermentation conditions for recombinant human interferon beta production by Escherichia coli using the response surface methodology. Jundishapur J Microbiol. 2015;8(4):e16236. doi: 10.5812/jjm.8(4)2015.16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Tripathy RK, et al. Antibiotic-free expression system for the production of human interferon-beta protein. 3 Biotech. 2018;8(1):36. doi: 10.1007/s13205-017-1056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaneophytou CP, Kontopidis G. Statistical approaches to maximize recombinant protein expression in Escherichia coli: a general review. Protein Expr Purif. 2014;94:22–32. doi: 10.1016/j.pep.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Patel G, Biswas K, et al. Bioreactor studies for the growth and production of mycophenolic acid by Penicillium brevicompactum. Biochem Eng J. 2018;140:77–84. doi: 10.1016/j.bej.2018.09.007. [DOI] [Google Scholar]
- Patel G, Patil MD, et al. Production of mycophenolic acid by Penicillium brevicompactum using solid state fermentation. Appl Biochem Biotechnol. 2016;182(1):97–109. doi: 10.1007/s12010-016-2313-3. [DOI] [PubMed] [Google Scholar]
- Patel G, Khobragade TP, et al. Optimization of media and culture conditions for the production of tacrolimus by Streptomyces tsukubaensis in shake flask and fermenter level. Biocatal Agric Biotechnol. 2020;29:101803. doi: 10.1016/j.bcab.2020.101803. [DOI] [Google Scholar]
- Patil MD, Dev M. Surfactant-mediated permeabilization of Pseudomonas putida KT2440 and use of the immobilized permeabilized cells in biotransformation. Process Biochem. 2017;63:113–121. doi: 10.1016/j.procbio.2017.08.002. [DOI] [Google Scholar]
- Patil MD, Shinde KD, et al. Use of response surface method for maximizing the production of arginine deiminase by Pseudomonas putida. Biotechnol Rep. 2016;10:29–37. doi: 10.1016/j.btre.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peubez I, Chaudet N, et al. Antibiotic-free selection in E. coli: new considerations for optimal design and improved production. Microb Cell Fact. 2010;9:65. doi: 10.1186/1475-2859-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poccia ME, Beccaria AJ, et al. Modelling the microbial growth of two Escherichia coli strains in a multi-substrate environment. Brazilian J Chem Eng. 2014;31:347–354. doi: 10.1590/0104-6632.20140312s00002587. [DOI] [Google Scholar]
- Qi M, Lu D, et al. Antitumor effect of recombinant human interferon-β adenovirus on esophageal squamous cell cancer in vitro. Dis Esophagus. 2014;27(2):196–201. doi: 10.1111/dote.12081. [DOI] [PubMed] [Google Scholar]
- Reder AT, Feng X. How type I interferons work in multiple sclerosis and other diseases: some unexpected mechanisms. J Interf Cytokine Res. 2014;34:589–599. doi: 10.1089/jir.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudick RA, Goelz SE. Beta-interferon for multiple sclerosis. Exp Cell Res. 2011;317:1301–2131. doi: 10.1016/j.yexcr.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Samarin ZE, Abolghasemi S, et al. Response surface optimization of the expression conditions for synthetic human interferon a -2b gene in Escherichia coli. Indian J Pharm Sci. 2018;80(3):470–479. doi: 10.4172/pharmaceutical-sciences.1000380. [DOI] [Google Scholar]
- Shayesteh M, Ghasemi F, et al. Design, construction, and expression of recombinant human interferon beta gene in CHO-s cell line using EBV-based expression system. Res Pharm Sci. 2020;15(2):144–153. doi: 10.4103/1735-5362.283814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CS, Hong MS, et al. High-level production of recombinant human IFN-α2a with co-expression of tRNA in high-cell-density cultures of Escherichia coli. Biotechnol Bioprocess Eng. 2001;6:301–305. doi: 10.1007/BF02931994. [DOI] [Google Scholar]
- Sohoni SV, Nelapati D, et al. Optimization of high cell density fermentation process for recombinant nitrilase production in E. coli. Bioresour Technol. 2015;188:202–208. doi: 10.1016/j.biortech.2015.02.038. [DOI] [PubMed] [Google Scholar]
- Spolaore B, Forzato G, et al. Site-specific derivatization of human interferon β-1a at lysine residues using microbial transglutaminase. Amino Acids. 2018;7:1–10. doi: 10.1007/s00726-018-2563-1. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Suzuki SM, et al. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc Natl Acad Sci. 2008;105:12780–12784. doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer CY, Milinkovitch MC. Separate-component-stabilization system for protein and DNA production without the use of antibiotics. Biotechniques. 2005;38:775–781. doi: 10.2144/05385RR02. [DOI] [PubMed] [Google Scholar]
- Tabandeh F, Khodabandeh M, et al. Response surface methodology for optimizing the induction conditions of recombinant interferon beta during high cell density culture. Chem Eng Sci. 2008;63:2477–2483. doi: 10.1016/j.ces.2008.02.003. [DOI] [Google Scholar]
- Tripathi NK. Production and purification of recombinant proteins from Escherichia coli. ChemBioEng Rev. 2016;3(3):116–133. doi: 10.1002/cben.201600002. [DOI] [Google Scholar]
- Zhang Y, Wei X, et al. Optimization of culturing conditions of recombined Escherichia coli to produce umami octopeptide-containing protein. Food Chem. 2017;227:78–84. doi: 10.1016/j.foodchem.2017.01.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.