Abstract
Continuous exploratory use of tree species is threatening the existence of several plants in South America. One of these threatened species is Myracroduron urundeuva, highly exploited due to the high quality and durability of its wood. The chloroplast (cp) has been used for several evolutionary studies as well traceability of timber origin, based on its gene sequences and simple sequence repeats (SSR) variability. Cp genome organization is usually consisting of a large single copy and a small single copy region separated by two inverted repeats regions. We sequenced the complete cp genome from M. urundeuva based on Illumina next-generation sequencing. Our results show that the cp genome is 159,883 bp in size. The 36 SSR identified ranging from mono- to hexanucleotides. Positive selection analysis revealed nine genes related to photosystem, protein synthesis, and DNA replication, and protease are under positive selection. Genome comparison a other Anacardiaceae chloroplast genomes showed great variability in the family. The phylogenetic analysis using complete chloroplast genome sequences of other Anacardiaceae family members showed a close relationship with two other economically important genera, Pistacia and Rhus. These results will help future investigations of timber monitoring and population and evolutionary studies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-00989-1.
Keywords: Aroeira, Brazilian savannah, Conservation, Microsatellite, Tropical tree
Introduction
With the constant anthropogenic disturbances in nature, tropical areas have been extensively degraded by the expansion of agriculture frontiers and cattle breeding. The remaining areas have been isolated in small fragments or resulting in solitary trees. In Brazil, deforestation over last year's reached alarming levels, with perspectives of loss of more than one thousand plant species in the next 30 years in the Brazilian savannah (Crouzeilles et al. 2017).
Myracrodruon urundeuva (Anacardiaceae), commonly known as 'aroeira', is an important tree species with wide distribution in several biomes of Brazil, including Cerrado (Brazilian savannah), Pantanal, Atlantic forest, Caatinga and its transition areas (Carvalho 1994; Lorenzi 2008; Nogueira 2010). In some of the biome hotspots, such as Brazilian savannah, the consequent fragmentation of habitats has greatly reduced the number of individuals of the species. It is possible only to find specimens only in private properties or government protected areas (Moraes et al. 2005). Aroeira is an arboreous tree, dioecious species and pollinated by bees (Santin and Leitão Filho 1991) has great importance due to the wood quality, durability and medicinal properties. It has been used widely in construction and luxury furniture (Almeida et al. 1998; Lorenzi 2008; Viana et al. 2014). During the best growth period some of specimens could reach more than 30 m in height and upto 100 cm of diameter in girth (Nogueira 2010), however the growth is slow and very time-consuming. (Ferretti et al. 1995).
Considering the rapid decline of natural forest populations, conservation studies in 'aroeira' and maintenance of progeny tests have been conducted in order to assess the genetic variability of the species and also to identify the mating systemand other factors that help to understand the population dynamics and also to help the conservation programs (Moraes et al. 2004; Freitas et al. 2006; Viegas et al. 2011; Souza et al. 2018). However, limited genomic and population genetics studies have been undertaken in this species (Viegas et al. 2011; Souza et al. 2018). In this context, the sequencing of chloroplast genomes can play an important role for phylogenetics studies. This can further help in the development of new SSR markers associated to cpDNA for population studies and species identification.
The next generation sequencing (NGS) has significantly increased the availability of sequencing data for non-species model, allowing comparative genomics and phylogenetic studies (Kersten et al. 2016; Yin et al. 2017; Zhang and Chen 2018; Santos and Almeida 2019). The cpDNA are maternally inherent in higher plants (Birky 1995), circular and organized by two inverted repeat regions (IR), separated by two single-copy regions (LSC and SSC regions, large single-copy and small single-copy respectively), with approximately 130 genes related to photosynthesis and carbon fixation (Daniell et al. 2016). Until now more than 4,800 chloroplast genomes for land plants have been submitted in the National Center for Biotechnology Information (NCBI) organelle genome database.
Only eight complete cpDNA genomes are publicaly available in NCBI (data retrieved in August 2020) from Anacardiaceae family consisting of approximately 81 genera and more than 800 species (Pell et al. 2011). We describe here for first time the complete chloroplast genome of M. urundeuva using low coverage Illuminasequencing. We also report the phylogenetic relationships within the family and also characterized SSR associated to cpDNA. Further conducted an evalutionery analysis in order to supliment future studies on population and conservation genetics of the species and related genera.
Materials and methods
Sampling, DNA extraction and construction of libraries
Fresh leaves were collected from M. urundeuva progeny test population maintained as ex-situ conservation site in Fazenda de Ensino, Pesquisa e Extensão da Faculdade de Engenharia de Ilha Solteira, Ilha Solteira, São Paulo, Brazil (20° 20' S, 51° 24' W). Sample collection was authorized by the Institute for Biodiversity Conservation (ICMBio), associated with the Brazilian Ministry of the Environment (MMA) under number SISBIO-52181-1. The leaves were dried in silica and stored in −20 °C until DNA extraction. Total DNA was extracted by CTAB protocol (Doyle and Doyle 1990). Quality of the extracted DNA was verified in 1% agarose gel (with TBE 1X) stained by GelRed (Biotium, Fremont, USA) and quantified by spectophotometer (NanoDrop 1000, Thermofisher Scientific, Wilmington, DE, USA). For sequencing, Illumina libraries were constructed using Nextera DNA library preparation kit using a pool of ten individuals. The librarries were sequenced on MiSeq Sequencing System (Illumina) using a V2 reagent kit of 500 cycles (2 × 250 pb) in a paired-end run.
Chloroplast genome assembly and simple sequence repeats analysis
Sequencing reads were assembled using NOVOPlasty version 3.0 (Dierckxsens et al. 2016) with default parameters. As starting seed, we used Pistacia vera (NC_034998.1) chloroplast genome as input. The annotation was performed in CPGAVAS2 using the option of 2544 plastomes (Shi et al. 2019), followed by manual correction in Geneious 8.1.9 software (https://www.geneious.com). Validation was done by Sanger sequencing using trnH–psbA, trnD–trnT, accD–psaI and trnK–rpd16 regions using available protocols (Hamilton 1999; Scarcelli et al. 2011). The PCR products were visualized on 2% agarose gel and then sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on a 3500 Genetic Analyzer (Applied Biosystems). The circular cp genome map was drawn with OGDRAW (Greiner et al. 2019). The PERF was used for SSR analysis (Avvaru et al. 2018) using criterion for mono- to hexanucleotides with minimum of ten repeats for mononucleotides, four to dinucleotides and three to other motifs. The cp genome sequence has been submitted to GenBank (accession number: MT017571) as well as Sanger validation sequences of the four cp regions (accession numbers: MT955660-MT955669).
Codon usage, nucleotide diversity and positive selection analyses
The codon usage frequency and relative synonymous codon usage (RSCU) was investigated using CodonW software (John Peden, <https://sourceforge.net/projects/codonw/> , version 1.4.2). We included all protein coding genes of M. urundeuva cp genome in the analysis. Relative synonymous codon usage analysis is used to measure codon usage bias and is defined by the ratio of observed frequency of codons to the frequency expected considering equal usage of the synonymous codons for an amino acid (Sharp and Li 1986). RSCU values > 1 are considered as a preferred codon, otherwise the value < 1 are used with less frequency and value equal to 1 means no codon usage bias (Sharp and Li 1987).
The nucleotide diversity (Pi) from Anacardiaceae cp genomes was evaluated for all unique genes extracted with PhyloSuite v. 1.2.2 (Zhang et al. 2020) and aligned by MAFFT v. 7.313 (Katoh and Standley 2013). Following this method the nucleotide diversity was calculated for each unique gene using DnaSP v. 6.12.03 (Rozas et al. 2017).
Positive selection of cp coding genes was evaluated using EasyCodeML software v1.31 (Gao et al. 2019a) assuming codon frequencies estimation (F3 × 4) and site model. A total of 73 coding sequences (CDS) presented in all species analyzed were included in this analysis. The comparison was made between model 1a (nearly neutral) against model 2a (selection), with likelihood ratio test (LRT) selection of critical value of 5.99 at 5% with two degrees of freedom as stated in Gao et al (2019b) and Jeffares et al. (2014). The identification of codons under positive selection was based in Bayes Empirical Bayes (BEB) with a probability threshold of 0.95 for genes with significant LRT p-values (Yang et al. 2005). We also tested the branch-site model considering M. urundeuva as a foreground and the other species as background in attempt to identify variations of ω across the phylogenetic tree. We considered the comparison between model A and model A null (Yang and Nielsen 2002; Zhang et al. 2005) using LRT calculations as before.
Comparative analysis of genome structure
The sequence identity of the cp genomes, from the Myracrodruon clade, were compared with mVISTA with A. occidentale annotated cp genome as a reference against other six cp genome from Anacardiaceae family using shuffle-LAGAN mode (Frazer et al. 2004). We focused to the Myracrodruon clade since Spondias species are a distant group from Myracrodruon genus as revealed by the phylogenetics analysis. The complete analysis including all species is presented in Supplementary Figure S1. Multiple genome alignments were conducted with MAUVE (Darling et al. 2004) to detect rearrangements or inversions. We also examined the borders of LSC, IR and SSC regions with IRscope (Amiryousefi et al. 2018) focused on M. urundeuva clade with other six cp genome resulted from phylogenetics analysis.
Phylogenetics analysis
Ten chloroplast genomes were included in the analysis of Anacardiaceae family including M. urundeuva with Sapindus mukorossi (Sapindaceae) as outgroup (Supplemental table S1). The chloroplast genomes were aligned using MAFFT 7.402 (Katoh and Standley 2013) and the maximum likelihood (ML) analysis was conducted using RAxML 8.2.10 (Stamatakis 2006; Stamatakis et al. 2008) with GTR + G model as well 1000 bootstrap replications in CIPRES Science gateway (Miller et al. 2010).
Results
Genome assembly
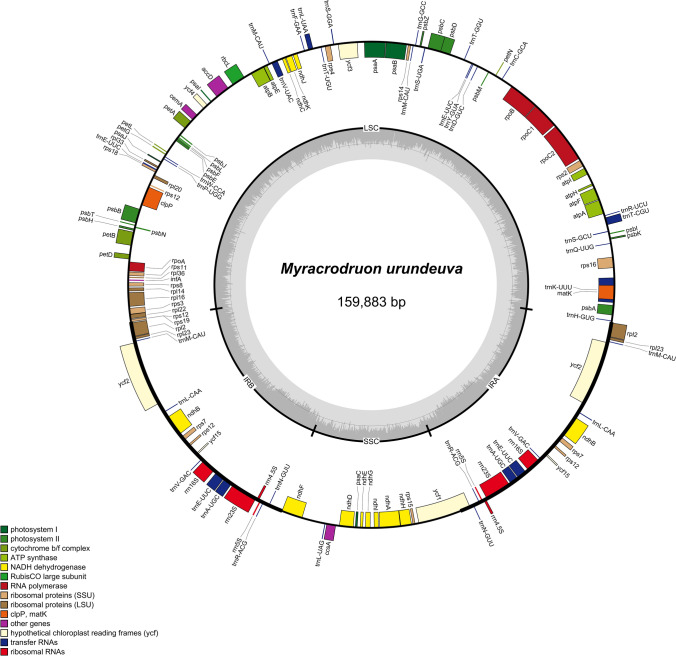
The MiSeq paired-end run generated 17,330,264 of paired-end reads with average 251 bp read length and in total 2.59 Gb data obtained. Considering genome size from other Anacardiaceae genus, such as Pistacia vera of 600 Mb (Motalebipour et al. 2016) and Mangifera indica of 439 Mb (Singh et al. 2016), we obtained a minimum of 4.31 × sequencing coverage for the entire genome size. Raw reads were analyzed using NOVOPlasty software that generated three contigs, ranging from 20,212 bp to 140,786 bp resulting in a final sequence of 159,883 bp for the complete chloroplast genome with average organelle coverage of 2615x. The two IR regions (26,507 bp) were separated by an LSC region (87,772 bp) and a SSC region (19,097 bp) (Fig. 1).
Fig. 1.
Chloroplast genome map of M. urundeuva. Thick lines represent LSC, SSC and IR regions. Inside the circle gene transcription are on clockwise and outside, counterclockwise. Different colors represent different gene groups
The 110 unique genes were identified, including 27 tRNAs, 4 rRNAs and two pseudogenes. It also included four RNA polymerase genes, 20 from ribosome subunits and 46 genes for the photosynthesis, from which seven corresponding to photosystem I, 15 for the photosystem II, six for cythochrome b/f complex, six encoded different subunits of ATP synthase, 11 encoded NADH-dehydrogenase subunits and one encoded the large chain of the ribulose bisphosphate carboxylase (RUBISCO). The other genes are related to acetyl-CoA carboxylase, cythochrome c synthesis, maturase, protease, envelope membrane protein, translational initiation factor genes and component of TIC complex. From these, 15 contained introns (atpF, clpP, ndhA, ndhB, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnE-UUC, trnK-UUU, trnL-UAA, trnT-CGU, trnV-UAC, ycf3). The total GC content for cp genome was 37.8% (Table 1).
Table 1.
Characteristics list of genes identified for M. urundeuva chloroplast genome. Duplicated genes are included into brackets
| Group of genes | Name of genes | |
|---|---|---|
| Protein synthesis and DNA replication | tRNA genes | trnA-UGC (2x), trnC-GCA, trnD-GUC, trnE-UUC (4x), trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU, trnL-CAA (2x), trnL-UAA, trnL-UAG, trnM-CAU (4x), trnN-GUU (2x), trnP-UGG, trnQ-UUG, trnR-ACG (2x), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-CGU, trnT-GGU, trnT-UGU, trnV-GAC (2x), trnV-UAC, trnW-CCA, trnY-GUA |
| rRNA genes | rrn4.5 (2x), rrn5 (2x), rrn16 (2x), rrn23 (2x) | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7 (2x), rps8, rps11, rps12 (2x), rps14, rps15, rps16, rps18, rps19 | |
| Large subunit of ribosome | rpl2 (2x), rpl14, rpl16, rpl20, rpl22, rpl23 (2x), rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3, ycf4 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cythochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH-dehydrogenase | ndhA, ndhB (2x), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Large subunit RUBISCO | rbcL | |
| Other genes | Acetyl-CoA carboxylase | accD |
| Cythochrome c synthesis gene | ccsA | |
| Maturase | matK | |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Translational initiation factor | infA | |
| Component of TIC complex | ycf1 | |
| Pseudogene unknown function | Conserved hypothetical chloroplast ORFs | ycf2 (2x), ycf15 (2x) |
Simple sequence repeats analysis
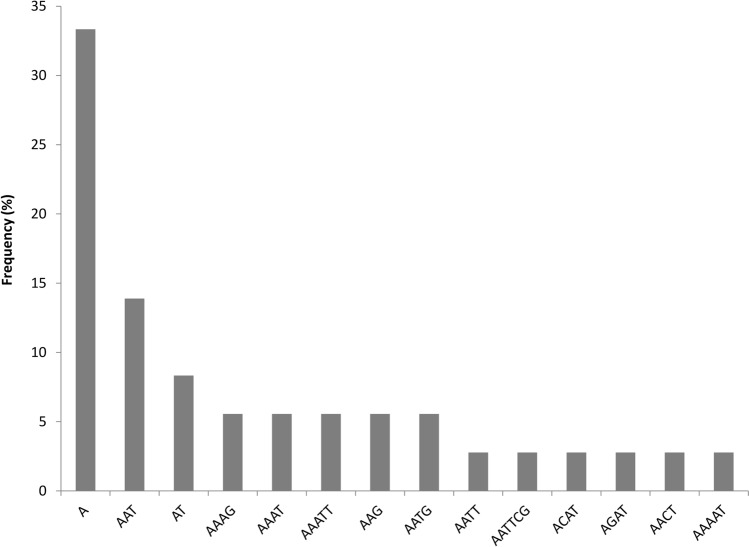
We identified 36 SSRs in the chloroplast genome of M. urundeuva, of which the mononucleotides motifs were the most abundant (33.3%; Fig. 2 and Supplemental Table S2). Second most abundant motif was tetranucleotide repeats (27.7%) and only one hexanucleotide was found. More than 87% of these SSR are present in the intergenic spacer region and introns. The remaining repeats are located within CDS region of ycf1.
Fig. 2.
The frequency of microsatellite motif types in the M. urundeuva chloroplast genome
Codon usage analysis, nucleotide diversity
All protein-coding regions presented 26,006 codons in M. urundeuva chloroplast genome (Table 2). From these, Leucine (Leu) and Cysteine (Cys) with 10.49% and 1.17% were the most and lower representative amino acids, respectively. The RSCU values returned that 31 codons showed codon usage bias (values > 1), which 28 were A/U-ending codons. For codons with RSCU values < 1, the preferential were C/G-ending codons.
Table 2.
Relative synonymous codon usage in M. urundeuva cp genome
| Amino acid | Codon | Number of occurrences | RSCU | Proportion (%) | Amino acid | Codon | Number of occurrences | RSCU | Proportion (%) |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 936 | 1.27 | 5,68 | Tyr | UAU | 748 | 1.59 | 3,61 |
| UUC | 541 | 0.73 | UAC | 192 | 0.41 | ||||
| Leu | UUA | 791 | 1.74 | 10,49 | His | CAU | 478 | 1.48 | 2,48 |
| UUG | 577 | 1.27 | CAC | 168 | 0.52 | ||||
| CUU | 558 | 1.23 | Gln | CAA | 694 | 1.54 | 3,47 | ||
| CUC | 193 | 0.42 | CAG | 208 | 0.46 | ||||
| CUA | 409 | 0.90 | Asn | AAU | 975 | 1.53 | 4,90 | ||
| CUG | 199 | 0.44 | AAC | 300 | 0.47 | ||||
| Ile | AUU | 1075 | 1.46 | 8,47 | Lys | AAA | 1042 | 1.48 | 5,41 |
| AUC | 460 | 0.63 | AAG | 365 | 0.52 | ||||
| AUA | 668 | 0.91 | Asp | GAU | 854 | 1.57 | 4,18 | ||
| Met | AUG | 596 | 1.00 | 2,29 | GAC | 232 | 0.43 | ||
| Val | GUU | 500 | 1.45 | 5,29 | Glu | GAA | 1018 | 1.49 | 5,26 |
| GUC | 176 | 0.51 | GAG | 350 | 0.51 | ||||
| GUA | 513 | 1.49 | Cys | UGU | 226 | 1.49 | 1,17 | ||
| GUG | 188 | 0.55 | UGC | 77 | 0.51 | ||||
| Ser | UCU | 548 | 1.63 | 5,68 | Trp | UGG | 450 | 1.00 | 1,73 |
| UCC | 339 | 1.01 | Arg | CGU | 332 | 1.24 | 3,66 | ||
| UCA | 399 | 1.18 | CGC | 120 | 0.45 | ||||
| UCG | 192 | 0.57 | CGA | 366 | 1.37 | ||||
| Pro | CCU | 408 | 1.55 | 4,05 | CGG | 134 | 0.50 | ||
| CCC | 199 | 0.76 | Ser | AGU | 420 | 1.25 | 2,09 | ||
| CCA | 291 | 1.10 | AGC | 124 | 0.37 | ||||
| CCG | 156 | 0.59 | Arg | AGA | 488 | 1.82 | 2,52 | ||
| Thr | ACU | 495 | 1.55 | 4,92 | AGG | 167 | 0.62 | ||
| ACC | 251 | 0.78 | Gly | GGU | 587 | 1.30 | 6,93 | ||
| ACA | 386 | 1.21 | GGC | 165 | 0.37 | ||||
| ACG | 147 | 0.46 | GGA | 722 | 1.60 | ||||
| Ala | GCU | 622 | 1.77 | 5,40 | GGG | 327 | 0.73 | ||
| GCC | 230 | 0.66 | Stop | UGA | 16 | 0.59 | 0,32 | ||
| GCA | 385 | 1.10 | UAA | 45 | 1.65 | ||||
| GCG | 167 | 0.48 | UAG | 21 | 0.77 |
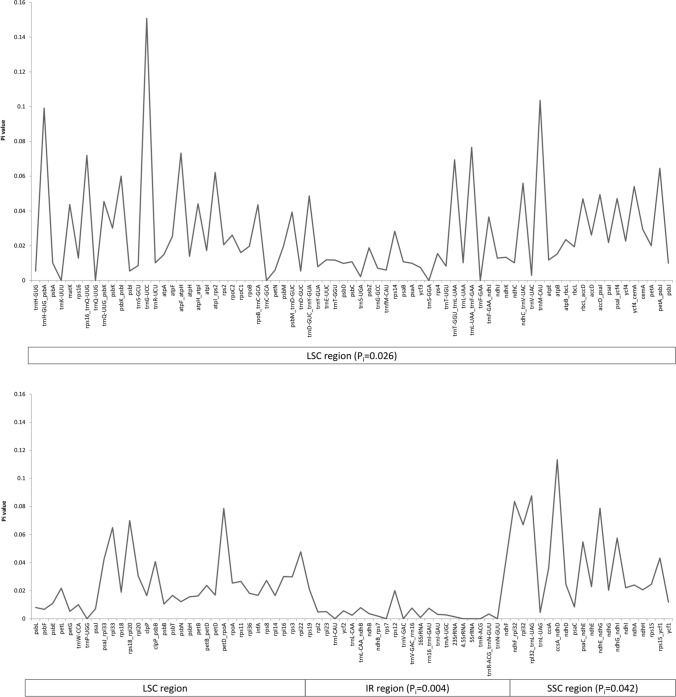
The phylogenetic analysis revealed, that the clade of M. urundeuva is more distant than Spondias genus and subsequently to other genera. The study explored only the nucleotide diversity as well as genome structure of this clade. Considering only genes present in all the species, the nucleotide diversity (Pi) was calculated to determine sequence level of divergence between cp genomes. These values ranged from 0 to 0.15, with the high average level of genetic variation detected for LSC (Pi = 0.026) and SSC regions (Pi = 0.042), followed by IR region (Pi = 0.004). Six gene regions showed high levels of nucleotide diversity (Pi > 0.08), trnH-GUG-psbA (Pi = 0.099), trnG-UCC (Pi = 0.15), trnM-CAU (Pi = 0.103), ndhF-rpl32 (Pi = 0.083), rpl32-trnL-UAG ((Pi = 0.087) and cssA-ndhD (Pi = 0.113).
The selective pressure estimation based in site model among the 73 CDS common in all species revealed, nine genes under positive selection in Anacardiaceae family (ndhB, rpl23, ndhD, rbcL, petD, clpP, rpl2, rpl33 and rpoA) from LRT (p-value < 0.05). The BEB posterior probability found positively selected sites for all genes excluding rpoA (Supplemental Table S3). From these, five are present in the LSC (rbcL, petD, clpP, rpl33, rpoA), three in IR (ndhB, rpl23, rpl2) and one in SSC region (ndhD). Considering their functions, four are related to photosynthesis (ndhB, ndhD, rbcL, petD), four to protein synthesis and DNA replication (rpl23, rpl2, rpl33, rpoA) and one related to other functions (Protease, clpP; Supplemental Table S4). We did not detect any evidence of branch-site selection in the coding genes tested (Fig. 3).
Fig. 3.
Nucleotide diversity across seven chloroplast genomes from M. urundeuva clade including genes and intergenic regions
Comparative analysis of genome structure
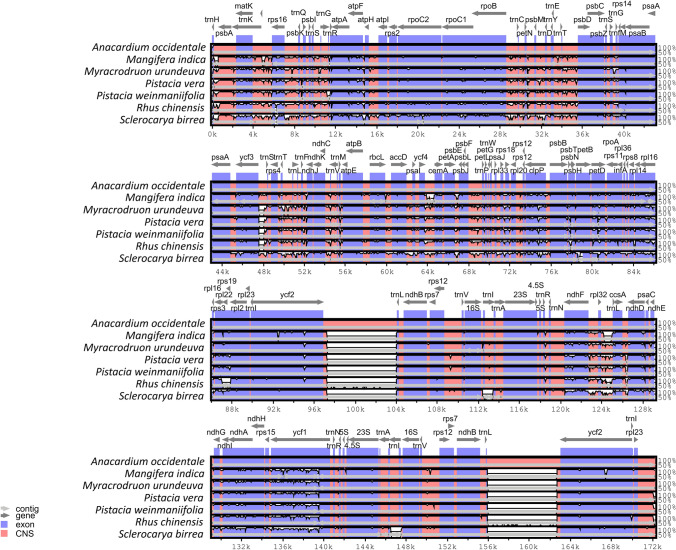
To investigate the structural characteristics within Myracrodruon clade cp genomes, the similarity percentage was plotted using mVISTA with A. occidentale cp genome as a reference while included all Anacardiacae cp genomes with S. mukorossi as a reference (Supplemental Figure S1). The high similarity was detected among Anacardiaceae species, with the coding regions more conserved than non-coding regions (CNS in Fig. 4). Also, there are two insertions (approximately 6,500 pb) in A. occidentale when compared with other Anacardiaceae genomes. These results were also found in MAUVE analysis (Supplemental Figure S2), in addition to an inversion in the M. indica (~ 16,000 bp) and a deletion in the R. chinensis (~ 10,000 bp) when compared among other members of the M. urundeuva clade cp genomes.
Fig. 4.
Genome alignment comparison of seven chloroplast genomes from M. urundeuva clade using mVISTA using A. occidentale as reference. Grey arrows indicate gene orientation, blue indicate exons and light red the conserved non-coding sequences (CNS)
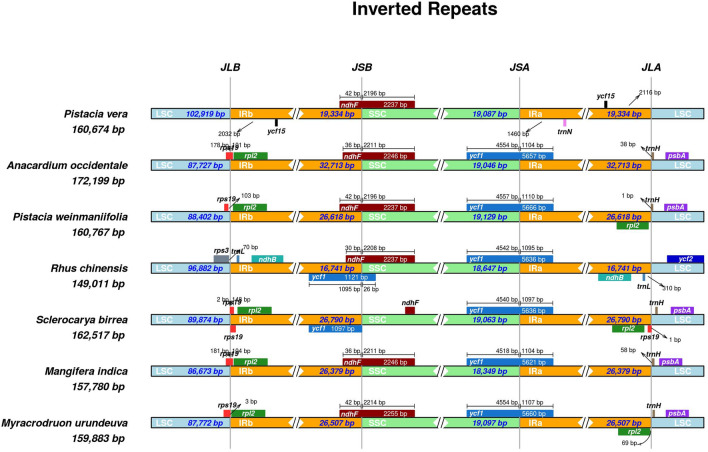
The length of the inverted repeat region and single-copy boundaries were analysed with a variation of IR regions from 16,741 bp for R. chinensis to 32,713 in A. occidentale. The junction of IRb and SSC showed the presence of ndhF gene for all species as well as ycf1 for R. chinensis and S. birrea (duplicated also found in IRa and SSC junction). Other regions exhibited a great variation of genes (Fig. 5).
Fig. 5.
Comparison of junction sites from LSC (Long Single Copy), SSC (Short Single copy) and IRs (Inverted Repeats) regions. JLB (LSC/IRb), JSB (IRb/SSC), JSA (SSC/IRa) and JLA (IRa/LSC) are the junction sites of each region
Phylogenetic analysis
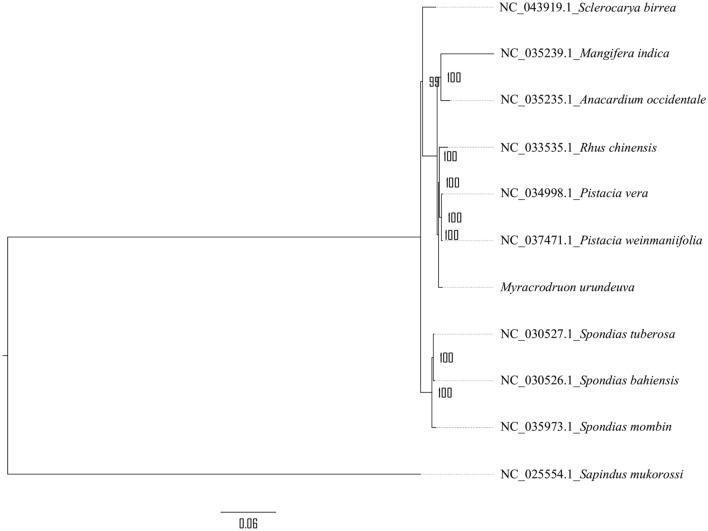
In this study, we included nine publicly available chloroplast genomes from Anacardiaceae family and S. mukorossi as outgroup. Phylogenetic analyses were performed using ML analysis with the complete sequence of chloroplast genomes (Fig. 6). In the ML tree, all nodes showed bootstrap values higher than 99%. The results showed a close relationship of M. urundeuva with Pistacia and Rhus as well as Mangifera and Anacardium genera. The other genera, Spondias and Sclerocarya are more distant related.
Fig. 6.
Maximum likelihood tree of Anacardiaceae species based in complete chloroplast genomes. Number on nodes corresponds to bootstrap support values
Discussion
The complete sequencing of cp genomes from tropical trees can aid studies of evolution and traceability timber. The study of plastid genomes plays an important role of phylogenetics in angiosperms (Moore et al. 2007). Despite that the rapid development of sequencing technologies the availability of chloroplast genomes y remains scarce in Anacardiaceae family. The assembly of cp genome based on short reads here showed as a alternative for several plant without prior genomic information (Santos and Almeida 2019; Souza et al. 2019; Khan et al. 2019).
The characterization of genome structure, as well as repeat markers, is important, since chloroplast markers have been used for population studies and traceability of genetic materials (Finkeldey et al. 2010; Blanc-Jolivet and Lisebach 2015; Phumichai et al. 2015; Nowakowska et al. 2015; Schroeder et al. 2016; Yue et al. 2018). In Hevea brasiliensis, cp SSR exhibited greatest variability in Brazilian genetic stocks comparing with other world populations (Phumichai et al. 2015). To avoid illegal logging of valuable trees, cp markers contribute in determining haplotypes for origin of timber (Blanc-Jolivet and Lisebach 2015; Schroeder et al. 2016). Moreover, a combination of cp SSR and haplotypes also contributed to the understanding of the origin and formation of a basis for future studies of genetic improvement in pears (Yue et al. 2018).
In M. urundeuva, the identification of microsatellite loci in the intergenic spacer region and introns can show a potential polymorphism since coding regions showed to be conserved across other genomes. There is a predominance of mononucleotides, followed by tetranucleotides. As described in other studies, the number of SSR identified for other Anacardiaceae chloroplast genomes range from 53 in Spondias bahiensis to 57 in Mangifera indica, where the most representative motifs are related to A/T repeats, with a predominance of mononucleotides repeats (Jo et al. 2017; Santos and Almeida 2019). These high rates of A/T are related to great content of these bases in these chloroplast genomes, as found in this study. When considering other families, such as Oleaceae, the number of SSR can reach more than 250, as reported for Syringa pinnatifolia (Zhang et al. 2019). Therefore, the characterization of SSR markers, as well as the sequence of M. urundeuva cp genome, will contribute to future population studies and timber origin control in 'aroeira' and related species.
Codon bias is a phenomenon that occurs when synonymous codons are used at different frequencies related to a more efficient mechanism of translation influenced by mutation pressure and natural selection (Hershberg and Petrov 2008; Machado et al. 2017; Zhang et al. 2018a, b). In M. urundeuva cp genome there is a great number of codon usage bias (values > 1) for A/U-ending codons. When analyzing 12 chloroplast genomes of Solanum, a codon usage bias analysis showed that the most preferred are A/U-ending codons and there are significant RSCU differences between wild and cultivated species (Zhang et al. 2018a). Similar results obtained in this study have been also reported in other plants (Li et al. 2019; Zhang et al. 2019). Considering the nucleotide diversity, we identified four gene regions with high levels of nucleotide diversity (Pi > 0.08). These loci can be used in phylogenetic and evolution studies, as evidenced by studies of molecular identification (DNA barcode; trnH-psbA) and phylogenetic studies from angiosperms (Scarcelli et al. 2011; Bolson et al. 2015).
The strategies of plants against the adversities of environment may lead sequence evolution of cp genomes, with some genes under positive selection in numerous plant lineages (Rockenbach et al. 2016; Wu et al. 2020). The selective pressure can be measured as a ratio (ω) of the nonsynonymous substitution rate (dN) to the synonymous substitutions rate (dS) and using the site-model, we can assess the variability at different sites of a gene by comparing different models of evolution, such nearly neutral against selection by a LRT (Nielsen and Yang 1998; Jeffares et al. 2014; Wu et al. 2020). We did not detect evidence of branch-site selection, which reflects that M. urundeuva is not evolving faster than the other species in the family. On the other hand, significant evidence from site-model selection indicates that there are nine genes which are under positive selection, with the most genes under purifying selection in Anacardiaceae family. Several plastid genes were reported to be under selective pressure from different lineages of angiosperms, such clpP and accD (Erixon and Oxelman 2008; Rockenbach et al. 2016). In our study, we found that clpP gene has at least three codons under positive selection, but none related to accD gene. Furthermore, approximately 45% of genes from photosystem are under positive selection (ndhB, ndhD, rbcL and petD). In Chrysosplenium and Oryza genera, several genes related to photosynthesis (such rbcL and psbB) are associated to the environment due to high levels of UV radiation, which may lead to DNA damages and mutations, resulting in high mutation levels (Gao et al. 2019b; Wu et al. 2020). In fact, rbcL was proposed as a DNA barcoding marker in association with matK from Consortium for the Barcode of Life (CBOL) Plant Working Group (CBOL 2009). Dong et al. (2015) also reported high levels of discrimination success in plants based in rbcL and matK markers, and also proposed ycf1 as a new universal DNA barcoding marker, but in this work, the LRT revealed that ycf1 was not significant in the family. Thus, in addition to the genes already proposed as discriminating species in the literature, there is a need for intraspecific investigation of these positively selected genes identified in this work at the population level to assess their potential as markers for use in species identification.The mVISTA analysis showed that the coding regions are more conserved than non-coding regions. The presence of insertions in A. occidentale as well as the inverted region in M. indica and the deletion in R. chinensis shows that the family exhibits a great variability in the cp genome evolution. On the other hand, Pistacia and Sclerocarya genus found to be more conserved with M. urundeuva. If we consider the IR boundaries, there is still a variation in the size of the IR regions with R. chinensis (~ 16,000 bp) to A. occidentale (32,000 bp). In Leguminosae family species, Souza et al (2019) do not identified such variation, with the IR regions close to 26,000 bp. Other species, such Plantago ovata has has more than 37 kb of IR region (Asaf et al. 2020). These contraction and expansion of IR borders are considered as a key of plastomes sizes variations, where these variations could arise markers of distinctive evolutionary lineages (Raubeson et al. 2007; Niu et al. 2018). In case when considering the Myracrodruon clade (containing Sclerocarya, Mangifera, Anacardium, Rhus and Pistacia), the closest lengths to the species studied were found to P. weinmannifolia (26,618 bp), S. birrea (26,790 bp) and M. indica (26,379 bp). Also, there is variation even within Pistacia species and so, they reflect on the different cp genome sizes found in the family.
The phylogenetic relationships of Anacardiaecae members points that M. urundeuva is in a well-supported clade together Pistacia and Rhus with Mangifera and Anacardium as a sister group. On the other hand, the position of Spondias was confirmed as being more distant from the other genera in Anacardiaceae (Santos and Almeida 2019). Based on morphological and molecular traits, the phylogenetic studies show P. vera and P. weinmannifolia in different sections and as a sister group of Rhus (Yi et al. 2007, 2008; Al-Saghir 2010). Through molecular markers, Weeks et al. (2014) confirmed that Pistacia is sister group of Rhus and that Spondias (S. tuberosa and S. mombin) is a distant group (basal position) from other members of Anacardiaceae, being closely related to Asian Spondias species. Moreover, based on nuclear ITS1 and chloroplast trnL-F sequences, Astronium urundeuva (synonym of M. urundeuva) has grouped in Anacardioideae subfamily along with Schinus, Pistacia and Rhus, and Spondias in the Spondioideae subfamily (Wheeler and Madeira, 2007). In an extensive study of Schinus genus based on the external and internal transcribed spacers (ETS and ITS), cp intergenic spacer (trnL-trnF) and cp intron (rps16), confirmed that Myracrodruon (M. urundeuva and M. balansae) is a sister group of Astronium (A. fraxinifolium and A. lecontei) genus, but not directly related to Schinus as reported earlier (Silva-Luz et al. 2019). These results and other previous studies with cp genomes of the family Anacardiaceae (Lee et al. 2016; Jo et al. 2017; Xu et al. 2019; Zhang et al. 2018b; Zheng et al. 2018; Santos and Almeida 2019), support that M. urundeuva is closely related to Pistacia group. Thus, the use of Pistacia as a reference genome (Zeng et al. 2019) for M. urundeuva and R. chinensis can represent an alternative for genomic studies in this family.
Conclusions
In this study we reported for the first time the complete cp genome for M. urundeuva based on Illumina sequencing. The characteristics of genes and regions are compatible with other cp family published genomes, as well as GC content. Also, the SSR analysis shows a great potential for future populations and timber traceability studies as well as the evidence of positive selected genes can be useful for further phylogenetic studies. Genome comparisons showed a wide variability across Anacardiaceae cp genomes, with insertions, deletions, and inverted regions on other members of the family. The phylogenetic analysis based on complete cp genome indicated that Myracrodruon is closely related to Rhus and Pistacia and confirmed to be more distant than Brazilian endemic genus Spondias. These results may help in the development of new markers, as well as the use of the Pistacia as a reference genome for other genomic studies in Myracrodruon and related genera.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful for Dr. José Cambuim for support during sample collections.
Author contributions
BCR designed the research, carried analysis, and wrote the manuscript. MLTM contributed new reagents and collection of samples. CLM and MLTM revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by National Council for Scientific and Technological Development (CNPq; grant number 303103/2017–5 by MLTM).
Data availability
The datasets generated during and/or analysed during the current study are available in the GenBank repository under accessions numbers: MT017571 and MT955660-MT955669.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Sample collection was authorized by the Institute for Biodiversity Conservation (ICMBio), associated with the Brazilian Ministry of the Environment (MMA) under number SISBIO-52181-1.
Consent for publication
All authors approved the manuscript for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Almeida SPD, Proença CEB, Sano SM, Ribeiro JF. Cerrado: espécies vegetais úteis. Planaltina (DF): EMBRAPA-CPAC; 1998. p. 464. [Google Scholar]
- Al-Saghir MG. Phylogenetic analysis of the genus Pistacia L. (Anacardiaceae) based on morphological data. Asian J Plant Sci. 2010;9:28–35. [Google Scholar]
- Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- Asaf S, Khan AL, Lubna, , et al. Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci Rep. 2020;10:3881. doi: 10.1038/s41598-020-60803-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvaru AK, Sowpati DT, Mishra RK. PERF: an exhaustive algorithm for ultra-fast and efficient identification of microsatellites from large DNA sequences. Bioinformatics. 2018 doi: 10.1093/bioinformatics/btx721. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes—mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Jolivet C, Liesebach M. Tracing the origin and species identity of Quercus robur and Quercus petraea in Europe: a review. Silvae Genetica. 2015;64(4):182–193. [Google Scholar]
- Bolson M, Smidt EdC, Brotto ML, Silva-Pereira V. ITS and trnH-psbA as efficient DNA barcodes to identify threatened commercial woody angiosperms from Southern Brazilian Atlantic rainforests. PLoS ONE. 2015;10(12):e0143049. doi: 10.1371/journal.pone.0143049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho PER. Espécies florestais brasileiras: recomendações silviculturais, potencialidade e uso da madeira. Colombo: Empresa Brasileira de Pesquisa Agropecuária - Centro Nacional de Pesquisas Florestais; 1994. p. 640. [Google Scholar]
- CBOL Plant Wording Group A DNA barcode for land plants. PNAS. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzeilles R, Feltran-Barbieri R, Ferreira MS, Strassburg BBN. Hard times for the Brazilian environment. Nat Ecol Evol. 2017;1:1213. doi: 10.1038/s41559-017-0303-7. [DOI] [PubMed] [Google Scholar]
- Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Xu C, Li C, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Erixon P, Oxelman B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS ONE. 2008;3:e1386. doi: 10.1371/journal.pone.0001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti AR, Kageyama PY, Arboez GF, Santos JD, Barros M, Lorza RF, Oliveira C. Classificação das espécies arbóreas em grupos ecofisiológicos para revegetação com nativas no estado de São Paulo. Florestar Estatístico, São Paulo. 1995;3(7):73–77. [Google Scholar]
- Finkeldey R, Leinemann L, Gailing O. Molecular genetic tools to infer the origin of forest plants and wood. Appl Microbiol Biotechnol. 2010;85:1251–1258. doi: 10.1007/s00253-009-2328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas MLM, Aukar APA, Sebbenn AM, Moraes MLT, Lemos EGM (2006) Variação genética em progênies de Myracrodruon urundeuva F.F. and M.F. Allemão em três sistemas de cultivo. Rev Árvore, Viçosa 30(3):319–329
- Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LZ, Liu YL, Zhang D, et al. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun Biol. 2019;2:278. doi: 10.1038/s42003-019-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. Organellar genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MB. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol. 1999;8:521–523. [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Tomiczek B, Sojo V, dos Reis M. A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. In: Peacock C, editor. Parasite genomics protocols. Methods in molecular biology. New York: Humana Press; 2015. pp. 65–90. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim H-W, Kim Y-K, Sohn J-Y, Cheon S-H, Kim KJ. The complete plastome sequences of Mangifera Indica L. (Anacardiaceae) Mitochondrial DNA Part B. 2017;2(2):698–700. doi: 10.1080/23802359.2017.1390407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B, Faivre RP, Mader M, Le Paslier MC, Bounon R, Berard A, Vettori C, Schroeder H, Leplé JC, Fladung M. Genome sequences of Populus Tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS ONE. 2016;11:e0147209. doi: 10.1371/journal.pone.0147209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Asaf S, Khan AL, et al. Complete chloroplast genomes of medicinally important Teucrium species and comparative analyses with related species from Lamiaceae. PeerJ. 2019;7:e7260. doi: 10.7717/peerj.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kim I, Kim J-K, Park JY, Joh HJ, Park H-S, Lee HO, Lee S-C, Hur Y-L, Yang T-L. The complete chloroplast genome sequence of Rhus Chinensis Mill (Anacardiaceae) Mitochondrial DNA Part B Resources. 2016;1(1):696–697. doi: 10.1080/23802359.2016.1222250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Pan Z, Gao S, He Y, Xia Q, Jin Y, Yao H. Analysis of synonymous codon usage of chloroplast genome in Porphyra umbilicalis. Genes Genom. 2019;41:1173. doi: 10.1007/s13258-019-00847-1. [DOI] [PubMed] [Google Scholar]
- Lorenzi H. Árvores Brasileiras. Manual de identificação e cultivo de plantas arbóreas nativas do Brasil. 5.ed. Nova Odessa (SP): Instituto Plantarum de Estudos da Flora Ltda; 2008. p. 384. [Google Scholar]
- Machado HE, Lawrie DS and Petrov DA (2017) Strong selection at the level of codon usage bias: evidence against the Li-Bulmer model. bioRxiv. doi: 10.1101/106476 [DOI] [PMC free article] [PubMed]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA (pp 1–8)
- Moore MJ, Bell CD, Soltis P, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes MLT, Kageyama PY, Sebbenn AM (2005) Diversidade e estrutura genética espacial em duas populações de Myracrodruon urundeuva Fr. All sob diferentes condições antrópicas. Rev. Árvore, Viçosa, 29(2):281–289
- Moraes MLT, Kageyama PY, Sebbenn AM. Correlated matings in dioecious tropical tree, Myracrodruon urundeuva fr. all. For Genet. 2004;11(1):55–61. [Google Scholar]
- Motalebipour EZ, Kafkas S, Khodaeiaminjan M, Çoban N, Gözel H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: development of novel SSR markers and genetic diversity in Pistacia species. BMC Genom. 2016;17(1):998. doi: 10.1186/s12864-016-3359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y-T, et al. Combining complete chloroplast genome sequences with target loci data and morphology to resolve species limits in Triplostegia (Caprifoliaceae) Mol Phylogenetics Evol. 2018;129:15–26. doi: 10.1016/j.ympev.2018.07.013. [DOI] [PubMed] [Google Scholar]
- Nogueira JCB. Reflorestamento misto com essências nativas: a mata ciliar. São Paulo: Instituto Florestal; 2010. p. 148. [Google Scholar]
- Nowakowska JA, Oszako T, Tereba A, Konecka A. Forest tree species traced with a DNA-based proof for illegal logging case in Poland. In: Pontarotti P, editor. Evolutionary biology: biodiversification from genotype to phenotype. Cham: Springer International Publishing; 2015. pp. 373–388. [Google Scholar]
- Pell SK, Mitchell JD, Miller AJ, Lobova TA. Anacardiaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Flowering plants. Eudicots. Sapindales, Curcubitales, Myrtales. Berlin: Springer; 2011. pp. 7–50. [Google Scholar]
- Phumichai C, Phumichai T, Wongkaew A. Novel chloroplast microsatellite (cpSSR) markers for genetic diversity assessment of cultivated and wild hevea rubber. Plant Mol Biol Rep. 2015;33:1486. doi: 10.1007/s11105-014-0850-x. [DOI] [Google Scholar]
- Raubeson LA, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenbach K, Havird JC, Monroe JG, Triant DA, Taylor DR, Sloan DB. Positive selection in rapidly evolving plastid-nuclear enzyme complexes. Genetics. 2016;204(4):1507–1522. doi: 10.1534/genetics.116.188268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Leitão SDA, Filho HF. Restabelecimento e revisão taxonômica do gênero Myracrodruon Freire Allemão (Anacardiaceae) Rev Bras Bot. 1991;14:133–145. [Google Scholar]
- Santos V, Almeida C. The complete chloroplast genome sequences of three Spondias species reveal close relationship among the species. Genet Mol Biol. 2019;42(1):132–138. doi: 10.1590/1678-4685-GMB-2017-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d’Anfray A, Vigouroux Y, Pintaud JC. A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLoS ONE. 2011;6:e19954. doi: 10.1371/journal.pone.0019954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H, Cronn R, Yanbaev Y, Jennings T, Mader M, Degen B, Kersten B. Development of molecular markers for determining continental origin of wood from white oaks (Quercus L. sect. Quercus) PLoS One. 2016;11(6):e0158221. doi: 10.1371/journal.pone.0158221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1–2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47(W1):W65–W73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Luz CLD, Pirani JR, Mitchell JD, Daly D, Capelli NDV, Demarco D, Pell SK, Plunkett GM. Phylogeny of Schinus L. (Anacardiaceae) with a new infrageneric classification and insights into evolution of spinescence and floral traits. Mol Phylogenet Evol. 2019;133:302–351. doi: 10.1016/j.ympev.2018.10.013. [DOI] [PubMed] [Google Scholar]
- Singh NK, Mahato AK, Jayaswal PK, Singh A, et al. Origin, diversity and genome sequence of mango (Mangifera indica L.) Indian J Hist Sci. 2016;51(22):355–368. doi: 10.16943/ijhs/2016/v51i2.2/48449. [DOI] [Google Scholar]
- Souza DCL, Rossini BC, Souza FB, Sebbenn AM, Marino CL, Moraes MLT. Development of microsatellite markers for Myracrodruon urundeuva (FF and MF Allemão), a highly endangered species from tropical forest based on next-generation sequencing. Mol Biol Rep. 2018;45(1):71–75. doi: 10.1007/s11033-017-4142-z. [DOI] [PubMed] [Google Scholar]
- Souza UJBd, Nunes R, Targueta CP, et al. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae - Caesalpinioideae): comparative analysis with related Mimosoid species. Sci Rep. 2019;9:14206. doi: 10.1038/s41598-019-50620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A fast bootstrapping algorithm for the RAxML web-servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Viana G, Calou I, Bandeira MA, Galvão W, Brito G. Myracrodruon urundeuva Allemão, a Brazilian medicinal species, presents neuroprotective effects on a Parkinson’s disease model, in rats. Eur Neuropsychopharmacol. 2014;24(2):230–231. [Google Scholar]
- Viegas MP, Silva CLSP, Moreira JP, Cardin LT, Azevedo VCR, Ciampi AY, Freitas MLM, Moraes MLT, Sebbenn AM (2011) Diversidade genética e tamanho efetivo de duas populações de Myracrodruon urundeuva Fr. All., sob conservação ex situ. Rev. Árvore, Viçosa 35(4):769–779
- Weeks A, Zapata F, Pell SK, Daly DC, Mitchell JD, Fine PV. To move or to evolve: contrasting patterns of intercontinental connectivity and climatic niche evolution in "Terebinthaceae" (Anacardiaceae and Burseraceae) Front Genet. 2014;5:409. doi: 10.3389/fgene.2014.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GS, Madeira PT. Phylogeny within the Anacardiaceae predicts host range of potential biological control agents of Brazilian peppertree. Biol Control. 2017;108:22–29. doi: 10.1016/j.biocontrol.2017.01.017. [DOI] [Google Scholar]
- Wu Z, Liao R, Yang T, et al. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae) BMC Genomics. 2020;21:621. doi: 10.1186/s12864-020-07045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-H, Zhang D-X, Sun H, Wang X-R, Xiang Q-H, Wang W, Guan W-B. The complete chloroplast genome sequences of Pistacia chinensis Bunge a potential bioenergy tree. Mitochondrial DNA Part B Resour. 2019;4(1):1774–1775. [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaption at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yi T, Miller AJ, Wen J. Phylogeny of Rhus (Anacardiaceae) based on sequences of nuclear NIA-i3 intron and chloroplast trnCtrnD. Syst Bot. 2007;32:379–391. [Google Scholar]
- Yi T, Wen J, Golan-Goldhirsh A, Parfitt DE. Phylogenetics and reticulate evolution in Pistacia (Anacardiaceae) Am J Bot. 2008;95(2):241–251. doi: 10.3732/ajb.95.2.241. [DOI] [PubMed] [Google Scholar]
- Yin D, Wang Y, Zhang X, Ma X, He X, Zhang J. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci Rep. 2017;7:11649. doi: 10.1038/s41598-017-12026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Zheng X, Zong Y, Jiang S, Hu C, Yu P, Liu G, Cao Y, Hu H, Teng Y. Combined analyses of chloroplast DNA haplotypes and microsatellite markers reveal new insights into the origin and dissemination route of cultivated pears native to East Asia. Front Plant Sci. 2018;9:591. doi: 10.3389/fpls.2018.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Tu XL, Dai H, et al. Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol. 2019;20(1):79. doi: 10.1186/s13059-019-1686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang L, Wang W, Zhang Z. Differences in codon usage bias between photosynthesis-related genes and genetic system-related genes of chloroplast genomes in cultivated and wild solanum species. Int J Mol Sci. 2018;19:3142. doi: 10.3390/ijms19103142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chen Z, Liu C. The complete plastid genome of marula (Sclerocarya birrea) Mitochondrial DNA Part B Resour. 2018;4(1):1111–1113. [Google Scholar]
- Zhang Y, Chen C. The complete chloroplast genome sequence of the medicinal plant Fagopyrum dibotrys (Polygonaceae) Mitochondrial DNA Part B. 2018;3(2):1087–1089. doi: 10.1080/23802359.2018.1483761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jiang Z, Su H, Zhao H, Cai J. The complete chloroplast genome sequence of the endangered species Syringa pinnatifolia (Oleaceae) Nord J Bot. 2019 doi: 10.1111/njb.02201. [DOI] [Google Scholar]
- Zhang D, Gao F, Li WX, et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Res. 2020;20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zheng W, Li K, Wang W, Xu X. The complete chloroplast genome of the threatened Pistacia weinmannifolia, an economically and horticulturally important evergreen plant. Conserv Genet Resour. 2018;10:535. doi: 10.1007/s12686-017-0871-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the GenBank repository under accessions numbers: MT017571 and MT955660-MT955669.