Abstract
Heat is a major abiotic stress that drastically reduces chickpea yield. This study aimed to identify heat-responsive traits to sustain crop productivity by screening a recombinant inbred line (RILs) population at two locations in India (Ludhiana and Faridkot). The RIL population was derived from an inter-specific cross between heat-tolerant genotype GPF 2 (C. arietinum L.) and heat sensitive accession ILWC 292 (C. reticulatum). The pooled analysis of variance showed highly significant differences for all the traits in RILs and most of the traits were significantly affected by heat stress at both locations. High values of genotypic coefficient of variation (19.52–38.53%), phenotypic coefficient of variation (20.29–39.85%), heritability (92.50–93.90%), and genetic advance as a percentage of mean (38.68–76.74%) have been observed for plant height, number of pods per plant, biomass, yield, and hundred seed weight across the heat stress environments. Association studies and principal component analysis showed a significant positive correlation of plant height, number of pods per plant, biomass, hundred seed weight, harvest index, relative leaf water content, and pollen viability with yield under both timely-sown and late-sown conditions. Path analysis revealed that biomass followed by harvest index was the major contributor to yield among the environments. Both step-wise and multiple regression analyses concluded that number of pods per plant, biomass and harvest index consistently showed high level of contribution to the total variation in yield under both timely-sown and late-sown conditions. Thus, the holistic approach of these analyses illustrated that the promising traits provide a framework for developing heat-tolerant cultivars in chickpea.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-00977-5.
Keywords: Heat stress tolerance, Association analysis, Path coefficient analysis, Regression analysis, Principal component analysis
Introduction
Chickpea (Cicer arietinum L.), originated from South-eastern Turkey (Ladizinsky 1975), is a self-pollinated diploid (2n = 2x = 16) crop having genome size of 738 Mb (Varshney et al. 2013). Among chickpea growing countries, India alone adds to about 70% of the world’s total production (FAOSTAT 2017) and ranks first in terms of production and productivity. Chickpea seeds have high amount of carbohydrates (around 60%) and protein (20–22%). It also contains essential amino acids (Jukanti et al. 2012), vitamins, minerals, and dietary fiber, which helps to improve insulin secretion and control blood sugar levels (Karim and Fattah 2007).
Due to relatively low levels of polymorphism between cultivated chickpea genotypes, interspecific crosses between C. arietinum and C. reticulatum have been the prime focus for genetic studies of agronomic traits (Singh et al. 2008). In spite of its economic importance and its role in human health, productivity has not been increased that much to meet the current demands. The sluggish pace of productivity is due to several abiotic and biotic constraints challenging the crop (Thudi et al. 2014). Among abiotic stresses, heat is considered as one of the major constraints that affects the chickpea production.
The confounding effect of drought stress slows down the progress made on heat tolerance. Studies on the impact of climate change on chickpea production underlined the effect of high temperature on crop development and yield. It is anticipated that climate change will result in temperature rise by 3–4 °C over current levels by 2050 (Basu et al. 2009). Chickpea is usually sown in winter season in the month of October in Northern India. At reproductive stage, it experiences terminal heat stress (> 35 °C) in March/April (Gaur et al. 2007). However, < 10 °C temperature at flowering stage can minimise the grain yield up to 20%. (Chaturvedi et al. 2009). The comparatively narrow genetic base, due to domestication bottleneck of chickpea varieties, is an additional reason for adverse effect on growth and reproductive physiology under high temperature stress (Abbo et al. 2003).
Flowers are the most sensitive organs of plant to heat stress (Toker and Canci 2006). Pollen viability, pod formation and seed set are adversely affected if temperature rises above the threshold level during the flowering or reproductive period, leading to reduction in grain yield and harvest index (Wang et al. 2006; Basu et al. 2009). Heat tolerance is a complex trait and is likely to be multi-genic (Upadhyaya et al. 2011; Kushwah et al. 2020a). Thus, an effective and simple screening method having well-defined traits for selecting heat-tolerant genotypes under field conditions is indispensable for breeding heat-tolerant cultivars (Devasirvatham et al. 2012).
The effects of heat stress during the vegetative and reproductive growth stages have been studied in wheat (Sharma et al. 2005), rice (Weerakoon et al. 2008) and cotton (Cottee et al. 2010), while limited research has been performed in chickpea (Wang et al. 2006). Visual inspection, selection for physiological attributes related to plant response to high temperature, empirical selection for yield and marker assisted selection are four important selection methods used to improve heat tolerance in chickpea through breeding (Howarth 2005; Kushwah et al. 2020b). However, the first step in the breeding process is identification of genetic diversity for economically important traits. Genetic diversity can be assessed by quantifying the variation in morphological traits that are targeted during selection for adaptation to heat stress condition (Krishnamurthy et al. 2011; Upadhaya et al. 2011). Selection of superior heat-tolerant lines are the next step in the breeding process by using a suitable screening environment.
Many breeders use late planting strategy to induce the high levels of heat stress starting from anthesis to the grain filling period (Krishnamurthy et al. 2011). Upadhyaya et al. (2011) evaluated 35 early maturing chickpea germplasm accessions and found large genetic variation for tolerance to heat stress. Devasirvatham et al. (2012) evaluated 167 chickpea genotypes at International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) for heat tolerance over 2 years and observed that most of the heat-tolerant genotypes were unaffected by the temperature variables in terms of yield. Canci and Toker (2009) studied 377 chickpea accessions and concluded that desi chickpeas were usually more heat-tolerant than the kabuli chickpeas. Paul et al. (2018) assessed the genetic variability and identified heat tolerance related traits in a RIL population.
However, understanding of yield contributing traits under heat stress conditions becomes more difficult due to the presence of various mechanisms that plants can adopt to maintain the reproductive growth under stress conditions (Tuberosa and Salvi 2006). Numerous traits such as number of pods per plant, biological yield, hundred seed weight, harvest index, membrane permeability index, relative leaf water content and pollen viability had been studied in some previous studies (Krishnamurthy et al. 2011; Devasirvatham et al. 2012; Paul et al. 2018) to understand the mechanism of heat tolerance. However, no or very less importance or priority was given to traits under stress conditions which resulted in lesser exploitation of the critical traits in heat tolerance breeding (Purushothaman et al. 2016). Consequently, qualitative trait-based breeding approach is being comparatively more emphasized over yield based breeding approach to recognize better stability since grain yield is profoundly influenced by high genotype × environment interactions, thereby exhibiting low heritability (Ludlow and Muchow 1990). Also, qualitative trait-based breeding approach increases the likelihood of crosses which results in additive gene action and reveals high heritability (Wasson et al. 2012). Therefore, this approach has been considered more advantageous in heat tolerance breeding to conquer the environmental effects on yield during heat stress.
Breeding plants for heat tolerance requires the understanding of the type and intensity of heat stress as well as the numerous traits and mechanisms adopted by the plants to maintain the productivity under terminal heat stress such as profuse root system and high water use efficiency without reducing the biomass and grain yield (Purushothaman et al. 2017). Therefore, it becomes crucial to know the component traits responsible for heat tolerance and their importance for understanding the complex mechanism of heat stress. A threshold temperature of 35 °C was found to be a perfect condition to expose the chickpea to heat stress during reproductive phase under field conditions (Krishnamurthy et al. 2011; Devasirvatham et al. 2012; Gaur et al. 2014). The late sowing condition exposes the chickpea to terminal heat stress condition at the time of pod setting (Hamwieh and Imtiaz 2015). Keeping in view the above factors, present study was performed to assess the genetic variability present in the RIL population for a total of 13 traits related to heat stress resilience under heat stress condition.
Material and methods
Population development and variables measured
A total of 204 RILs segregating for heat tolerance related traits from an inter-specific cross between heat-tolerant genotype GPF 2 (C. arietinum L.) and heat sensitive accession ILWC 292 (C. reticulatum) developed using single seed descent methods were evaluated. The RIL population along with parents was planted in alpha lattice design (17 × 12) under timely-sown and late-sown conditions with three replications at two locations, i.e., Ludhiana and Faridkot. The Ludhiana (30.9010° N, 75.8573° E) and Faridkot (30.6769° N, 74.7583° E) locations are categorized as a semi-arid sub-tropical region and semi-arid dry region, respectively. Different heat tolerance related traits were studied in late-sown irrigated condition (January-May 2017), using the timely-sown irrigated condition (November- April 2017) as a control. Late-sown irrigated condition exposed the RIL population to heat stress (> 35 °C) during reproductive stage. During the screening for heat tolerance, irrigation was provided to avoid the confounding effect of drought stress. The daily maximum temperatures for late-sown as well as timely-sown conditions during the reproductive phase at both locations (Ludhiana and Faridkot) were recorded (Fig. 1) for comparison.
Fig. 1.
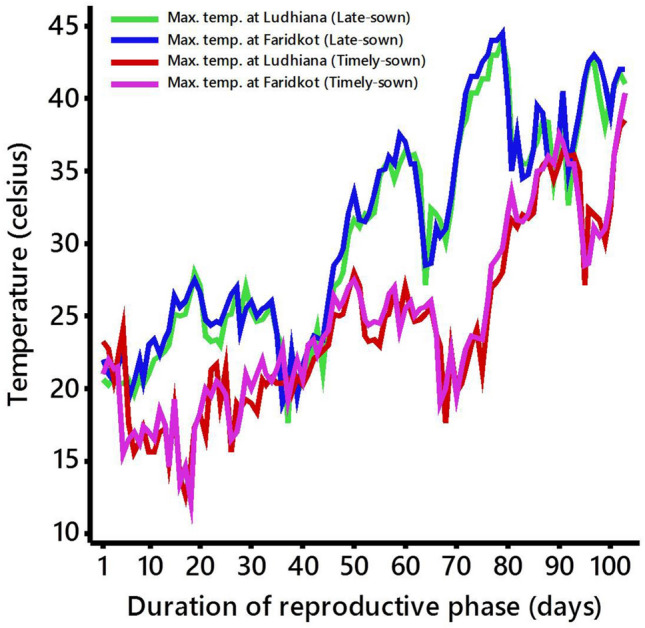
The daily maximum temperatures for late-sown as well as timely-sown conditions during the reproductive phase at both of the locations (Ludhiana and Faridkot)
Phenotypic data were collected for a total of 13 heat tolerance related traits viz., DG, DFI, DFF, DHF, PH, NPP, BIO, YLD, HSW, HI, MPI, RLWC and PV. Randomly five plants were selected to record the observations on PH, NPP, BIO and YLD in each plot, while data on DG, DFI, DFF, DHF and HSW were recorded on a plot basis. HSW was estimated from the plants used for estimating the NPP and YLD by counting a random sample of hundred seeds. HI was calculated as:
Yield and yield contributing traits such as number of pods per plant, biomass, hundred seed weight and harvest index were collected.
Pollen viability test was studied by collecting the pollen samples at the time of 50% flowering. The MPI was determined according to the method of Premchand et al. (1990) as modified by Sairam (1994) using following formula:
where C1 = Initial electrical conductivity at (40 °C); C2 = Final electrical conductivity at (100 °C).
The RLWC was calculated by the formula given by Slavik (1974) using following formula:
where FW = Fresh weight, DW = Dry weight, TW = Turgid weight.
Statistical analyses
All observations were statistically analysed and the mean values of three replications has been used for data analysis. The ANOVA was calculated for individual environment using mixed model analysis to estimate the contribution made by each factor to the total variation using SAS-software version 9.3 (SAS Institute, Cary NC). Broad sense heritability (hb2) was calculated using the following formula given by Allard (1960).
where σ2g = genotypic variance, σ2p = phenotypic variance
GCV and PCV were classified into three categories, i.e., high (> 15%), moderate (10–15%) and low (< 10%). Similarly, heritability was also classified into three categories, i.e., high (> 80%), moderate (50–80%) and low (< 50%).
Expected genetic advance (GA) was figured by the following formula given by Allard (1960).
where k = selection differential (2.06) at 5% selection intensity, hb2 = broad sense heritability, σ2p = phenotypic variance.
The genetic advance as a percentage of the mean was estimated as:
A matrix of Pearson correlation coefficients analysis between the seed yield and its components were computed in order to determine the relationship between the examined traits and seed yield. Multiple linear regression analysis and step wise linear regression analysis procedure was performed to determine the contribution of independent variables on seed yield. Associations among the traits were worked out by using principal component analysis. The path coefficients were analysed according to the method suggested by Dewey and Lu (1959) by using matrix method to know the direct and indirect effects of independent traits on seed yield. The Pearson correlation coefficients analysis, linear regression analysis and principal component analysis were computed with the SAS-software version 9.3 (SAS Institute, Cary NC), while path coefficients analysis was calculated by using INDOSTAT programme.
Results
Phenotypic assessment of RIL population and parents
The maximum temperature data indicated that it crossed the threshold limit and RIL population has been exposed to heat stress during reproductive stage at both the locations (Fig. 1). Significant variation was observed in the RILs as well as the parents for heat stress related traits under timely-sown and late-sown conditions (Table 1). The mean difference between late-sown and timely-sown conditions for DG was 3.79, indicated delayed germination under late-sown condition (Table 1) while, mean difference for DFI, DFF and DHF was − 10.57, − 11.31 and − 11.11, indicated precocity under late-sown condition. Likewise, mean difference for PH, NPP, BIO, YLD and HSW was negative which indicated reduction in the mean values for these traits under late-sown condition. Reduction in HI, low RLWC and low PV was also observed under late-sown condition as mean difference for these traits was negative.
Table 1.
Mean performance of chickpea RIL population for heat stress related traits under timely-sown (TS) and late-sown (LS) conditions (Ludhiana and Faridkot pooled)
| Trait | DG (days) | DFI (days) | DFF (days) | DHF (days) | PH (cm) | NPP | BIO (gm) | YLD (gm) | HSW (gm) | HI (%) | MPI | RLWC (%) | PV (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ILWC 292 (Susceptible parent) |
TS LS |
12.34 15.53 |
90.33 82.82 |
94.26 84.74 |
98.08 88.27 |
42.42 21.82 |
43.53 20.30 |
76.78 38.00 |
27.91 12.35 |
11.27 9.32 |
36.63 32.61 |
42.23 52.71 |
65.28 47.72 |
81.29 69.43 |
|
GPF 2 (Tolerant parent) |
TS LS |
8.12 10.67 |
82.80 75.86 |
86.46 78.76 |
89.78 82.66 |
58.87 43.90 |
68.54 43.96 |
113.32 76.26 |
49.74 33.39 |
16.18 15.85 |
43.55 43.97 |
28.81 38.03 |
88.31 78.61 |
93.09 85.34 |
| Contrast analysis between parents |
TS LS |
44.33** 175.00** |
78.87** 11.53* |
171.50** 19.17** |
171.50** 10.77* |
65.43** 125.85** |
133.10** 117.41** |
55.17** 131.55** |
232.07** 125.87** |
1629.73** 174.08** |
15.81** 22.90** |
150.71** 12.71** |
178.91** 290.43** |
27.36** 36.61** |
|
Mean (RILs) |
TS LS |
9.33 13.12 |
85.99 75.42 |
89.46 78.15 |
93.10 81.99 |
45.68 29.33 |
47.39 29.26 |
81.33 57.03 |
32.14 19.52 |
14.22 13.06 |
38.98 32.85 |
39.42 46.28 |
74.85 62.41 |
84.74 73.55 |
| CV |
TS LS |
5.05 9.23 |
2.01 4.15 |
2.08 3.97 |
1.96 3.86 |
10.47 32.08 |
21.75 31.29 |
15.47 30.29 |
27.07 43.33 |
16.11 22.24 |
16.48 16.10 |
12.12 15.82 |
9.37 17.05 |
7.40 13.59 |
| Range |
TS LS |
8.12–12.34 10.63–16.16 |
78.91–90.33 69.17–83.49 |
82.35–94.26 72.26–86.11 |
86.67–98.08 75.75–90.05 |
33.82–58.87 15.14–48.88 |
25.13–75.07 14.65–48.28 |
51.55–113.70 30.16–90.32 |
14.13–54.69 8.35–38.07 |
9.79–18.42 7.92–18.20 |
22.49–52.86 21.75–44.87 |
28.70–50.76 32.23–59.71 |
59.06–89.94 45.96–79.89 |
70.99–94.77 58.98–93.13 |
| Genotypic Variance |
TS LS |
1.50** 5.09** |
1.96** 6.01** |
2.24** 9.92** |
2.28** 8.77** |
4.05** 45.96** |
18.64** 35.46** |
10.35** 64.94** |
18.16** 53.39** |
20.41** 46.74** |
12.52** 19.04** |
15.49** 27.38** |
6.07** 29.92** |
10.83** 22.49** |
| G X L Variance |
TS LS |
0.51 0.27 |
0.15 0.42 |
0.14 0.36 |
0.20 0.59 |
3.85** 5.11** |
5.82** 6.04** |
4.12** 7.93** |
6.12** 5.15** |
18.35** 5.61** |
5.31** 2.45** |
8.51** 3.70** |
10.17** 5.53** |
5.10** 5.37** |
|
H2 (Broad Sense) |
TS LS |
56.40 59.10 |
35.20 86.00 |
25.10 83.20 |
29.70 84.10 |
79.40 93.90 |
85.30 92.90 |
84.20 93.60 |
88.00 93.50 |
90.40 92.50 |
86.40 88.60 |
88.70 92.30 |
86.60 92.40 |
88.20 92.10 |
| GCV (%) |
TS LS |
4.30 5.69 |
1.08 3.89 |
0.83 3.20 |
0.86 3.21 |
9.74 28.56 |
17.33 27.98 |
13.10 26.53 |
22.96 38.53 |
13.92 19.52 |
14.10 13.90 |
10.15 14.15 |
9.13 15.13 |
6.59 12.37 |
| PCV (%) |
TS LS |
5.73 7.40 |
1.82 4.19 |
1.65 3.51 |
1.58 3.50 |
10.93 29.48 |
18.77 29.03 |
14.27 27.41 |
24.47 39.85 |
14.65 20.29 |
15.17 14.76 |
10.78 14.73 |
9.82 15.74 |
7.02 12.89 |
| GAM (%) |
TS LS |
6.66 9.00 |
1.32 7.43 |
0.86 6.02 |
0.97 6.07 |
17.88 57.00 |
32.97 55.57 |
24.76 52.88 |
44.37 76.74 |
27.27 38.68 |
27.00 26.94 |
19.69 28.00 |
17.51 29.97 |
12.76 24.46 |
* = Significant at 5% probability level, ** = Highly significant at 1% probability level, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height (cm), NPP = Number of pod per plant, BIO = biomass per plant (gm), YLD = yield per plant (gm), HSW = 100-seed weight (gm), HI = harvest index (%), MPI = membrane permeability index, RLWC = relative leaf water content (%), PV = pollen viability (%), CV = coefficient of variation, G X L = genotype by location interaction, H2 = Broad sense heritability, GCV = Genotypic coefficient of variation, PCV = Genotypic coefficient of variation, GAM = Genetic Advancement as percentage of Means at 5% level of significance
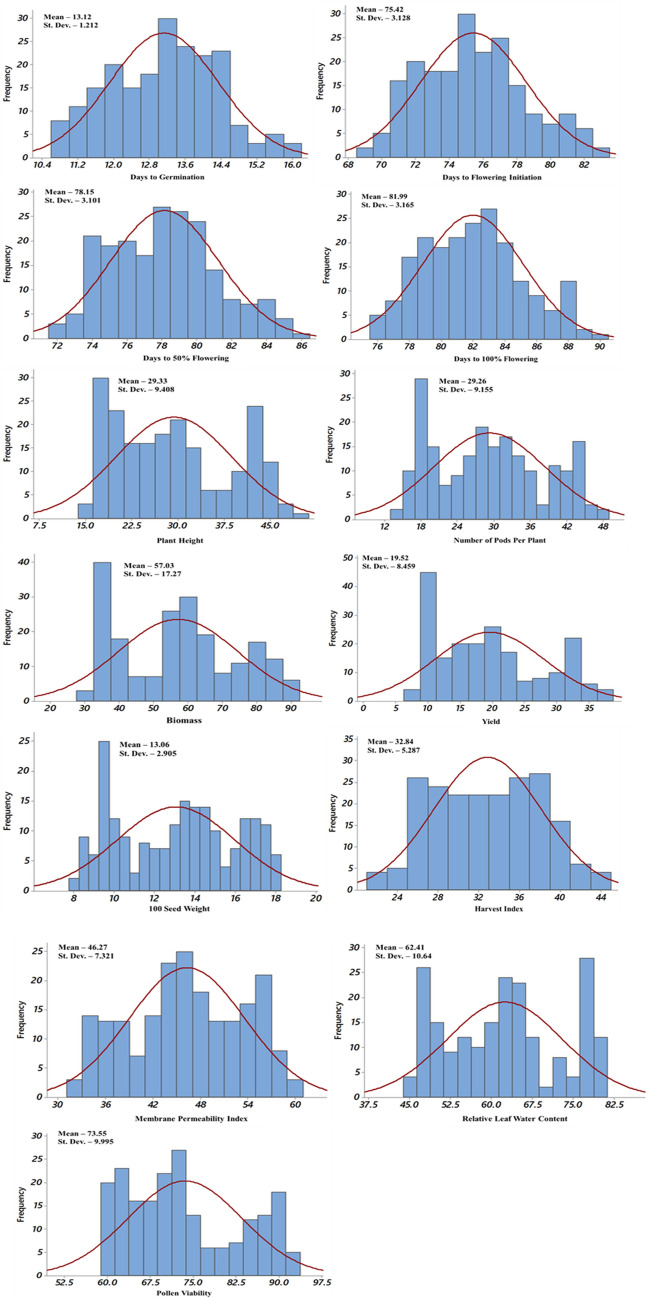
The contrast analysis of parents depicted highly significant differences between parents for all the traits studied under timely-sown and late-sown conditions. The pooled ANOVA for all the traits showed highly significant differences in RILs for genotypic variance for both locations in timely-sown as well as late-sown conditions. Significant differences were also observed for genotype x location (G × L) interaction variance for all the traits, except DG, DFI, DFF and DHF (Table 1). The frequency distributions for most of the traits were normal in both timely-sown as well as late-sown treatments (Figs. 2 and 3).
Fig. 2.
Graphical representations of RILs for various traits in chickpea under timely-sown condition
Fig. 3.
Graphical representations of RILs for various traits in chickpea under late-sown condition
Genetic variability, heritability and genetic advance
Usually, the estimates of PCV were higher than that of GCV (Paul et al. 2018) which are in accordance with our results. The values of GCV and PCV were high for PH, NPP, BIO, YLD and HSW (19.52–39.85%), moderate for HI, MPI, RLWC and PV (12.37–15.74%) and low for DG, DFI, DFF and DHF (3.20–7.40%) in case of late-sown environment (Table 1). In case of timely-sown environment, the values of GCV and PCV were low to moderate for all the traits. Broad sense heritability was found to be high for traits like PH, NPP, BIO, YLD, HSW, HI, MPI, RLWC and PV (79.40–90.40%) and low to moderate (25.10–56.40%) for traits like DG, DFI, DFF and DHF in case of timely-sown environment. However, in case of late-sown environment, high heritability (83.20–93.90%) was observed for all the traits except DG (59.10%) which showed moderate heritability. The GAM for all the traits varied from 0.86 to 44.37% in timely-sown condition and from 6.02 to 76.74% in late-sown condition (Table 1). The value of GAM was observed moderately high for all the traits, except DG, DFI, DFF and DHF in case of timely-sown environment. In contrast, high value of GAM was recorded for traits like PH, NPP, BIO, YLD and HSW, moderate for HI, MPI, RLWC and PV and low for DG, DFI, DFF and DHF in late-sown condition.
Trait association analysis and path analysis
The worth of independent secondary traits in the selection process can be understood by their significant association with a dependent trait like yield. Results of trait association in timely-sown as well as in late-sown condition indicated that the yield exhibited significantly high positive correlation with PH, NPP, BIO, HSW, HI, RLWC and PV consistently at Ludhiana and Faridkot locations (Tables 2, 3). Significant but inconsistent negative correlation of yield was found with DG, DFI, DFF and DHF, while highly negative association was observed with MPI under timely-sown as well as in late-sown condition at both the locations. A significantly high positive association was also observed among the traits like PH, NPP, BIO, YLD, HSW, RLWC and PV consistently in timely-sown as well as late-sown condition at both the locations.
Table 2.
Phenotypic correlation coefficient among different characters in chickpea in timely-sown condition
| DFI | DFF | DHF | PH | NPP | BIO | YLD | HSW | HI | MPI | RLWC | PV | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
0.22** 0.06 0.20** |
0.22** 0.05 0.13** |
0.21** 0.05 0.13** |
− 0.14** 0.04 − 0.08** |
− 0.07 − 0.03 − 0.07* |
− 0.10* 0.01 − 0.08** |
− 0.07 − 0.05 − 0.07* |
− 0.11** − 0.01 0.00 |
− 0.03 − 0.08* − 0.03 |
0.06 0.05 0.04 |
− 0.08* − 0.05 − 0.12** |
− 0.08 − 0.03 − 0.14** |
|
Ludhiana Faridkot Pooled |
DFI |
0.97** 0.97** 0.96** |
0.92** 0.95** 0.93** |
− 0.14** − 0.06 − 0.12** |
− 0.09* − 0.11** − 0.11** |
− 0.12** − 0.08* − 0.12** |
− 0.12** − 0.09* − 0.11** |
− 0.15** − 0.06 − 0.07** |
− 0.10* − 0.06 − 0.06* |
0.09* 0.11** 0.09** |
− 0.14** − 0.11** − 0.16** |
− 0.11** − 0.08* − 0.14** |
|
|
Ludhiana Faridkot Pooled |
DFF |
0.96** 0.97** 0.97** |
− 0.14** − 0.05 − 0.09** |
− 0.09* − 0.09* − 0.09** |
− 0.12** − 0.07 − 0.10** |
− 0.11** − 0.08* − 0.10** |
− 0.15** − 0.05 − 0.09** |
− 0.10* − 0.06 − 0.07* |
0.09* 0.10* 0.09** |
− 0.15** − 0.10* − 0.13** |
− 0.11** − 0.08* − 0.11** |
||
|
Ludhiana Faridkot Pooled |
DHF |
− 0.11** − 0.07 − 0.09** |
− 0.07 − 0.10* − 0.09** |
− 0.10* − 0.08* − 0.10** |
− 0.10* − 0.08* − 0.09** |
− 0.13** − 0.05 − 0.08** |
− 0.08 − 0.05 − 0.06* |
0.08* 0.10* 0.09** |
− 0.16** − 0.11** − 0.14** |
− 0.09* − 0.08* − 0.10** |
|||
|
Ludhiana Faridkot Pooled |
PH |
0.46** 0.60** 0.53** |
0.71** 0.81** 0.77** |
0.56** 0.54** 0.54** |
0.41** 0.43** 0.39** |
0.29** 0.00 0.10** |
− 0.41** − 0.48** − 0.43** |
0.33** 0.44** 0.41** |
0.49** 0.44** 0.47** |
||||
|
Ludhiana Faridkot Pooled |
NPP |
0.93** 0.90** 0.91** |
0.89** 0.85** 0.87** |
0.77** 0.74** 0.74** |
0.64** 0.40** 0.51** |
− 0.80** − 0.79** − 0.79** |
0.49** 0.76** 0.63** |
0.78** 0.73** 0.74** |
|||||
|
Ludhiana Faridkot Pooled |
BIO |
0.90** 0.80** 0.85** |
0.75** 0.73** 0.71** |
0.58** 0.21** 0.37** |
− 0.76** − 0.72** − 0.74** |
0.50** 0.69** 0.61** |
0.79** 0.66** 0.72** |
||||||
|
Ludhiana Faridkot Pooled |
YLD |
0.85** 0.86** 0.84** |
0.87** 0.74** 0.80** |
− 0.85** − 0.92** − 0.88** |
0.57** 0.90** 0.73** |
0.89** 0.86** 0.85** |
|||||||
|
Ludhiana Faridkot Pooled |
HSW |
0.76** 0.58** 0.67** |
− 0.81** − 0.82** − 0.81** |
0.46** 0.80** 0.60** |
0.77** 0.75** 0.70** |
||||||||
|
Ludhiana Faridkot Pooled |
HI |
− 0.74** − 0.70** − 0.71** |
0.50** 0.69** 0.59** |
0.79** 0.68** 0.69** |
|||||||||
|
Ludhiana Faridkot Pooled |
MPI |
− 0.43** − 0.88** − 0.65** |
− 0.78** − 0.84** − 0.78** |
||||||||||
|
Ludhiana Faridkot Pooled |
RLWC |
0.51** 0.82** 0.70** |
* = Significant at 5% probability level, ** = Highly significant at 1% probability level, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
Table 3.
Phenotypic correlation coefficient among different characters in chickpea in late-sown condition
| DFI | DFF | DHF | PH | NPP | BIO | YLD | HSW | HI | MPI | RLWC | PV | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
0.63** 0.26** 0.46** |
0.61** 0.30** 0.48** |
0.59** 0.30** 0.48** |
− 0.10* − 0.09* − 0.07* |
− 0.10* − 0.10* − 0.07** |
− 0.11** − 0.10* − 0.09** |
− 0.10* − 0.09* − 0.08** |
− 0.08* − 0.09* − 0.07* |
− 0.07 − 0.05 − 0.05 |
0.10* 0.09* 0.09** |
− 0.08 − 0.10* − 0.09** |
− 0.07 − 0.10* − 0.06* |
|
Ludhiana Faridkot Pooled |
DFI |
0.98** 0.90** 0.92** |
0.97** 0.86** 0.92** |
− 0.21** − 0.24** − 0.14** |
− 0.19** − 0.24** − 0.13** |
− 0.20** − 0.21** − 0.15** |
− 0.20** − 0.23** − 0.16** |
− 0.18** − 0.24** − 0.15** |
− 0.15** − 0.22** − 0.14** |
0.20** 0.23** 0.18** |
− 0.18** − 0.26** − 0.22** |
− 0.18** − 0.21** − 0.11** |
|
|
Ludhiana Faridkot Pooled |
DFF |
0.99** 0.96** 0.97** |
− 0.19** − 0.26** − 0.19** |
− 0.18** − 0.27** − 0.18** |
− 0.19** − 0.24** − 0.19** |
− 0.19** − 0.25** − 0.20** |
− 0.17** − 0.26** − 0.19** |
− 0.15** − 0.23** − 0.17** |
0.20** 0.25** 0.21** |
− 0.18** − 0.28** − 0.23** |
− 0.17** − 0.24** − 0.16** |
||
|
Ludhiana Faridkot Pooled |
DHF |
− 0.20** − 0.26** − 0.18** |
− 0.19** − 0.27** − 0.17** |
− 0.20** − 0.24** − 0.18** |
− 0.20** − 0.26** − 0.19** |
− 0.17** − 0.27** − 0.18** |
− 0.16** − 0.23** − 0.16** |
0.20** 0.25** 0.20** |
− 0.19** − 0.28** − 0.23** |
− 0.18** − 0.24** − 0.15** |
|||
|
Ludhiana Faridkot Pooled |
PH |
0.88** 0.91** 0.90** |
0.96** 0.96** 0.96** |
0.96** 0.95** 0.95** |
0.91** 0.90** 0.90** |
0.80** 0.72** 0.76** |
− 0.85** − 0.92** − 0.88** |
0.87** 0.91** 0.87** |
0.92** 0.87** 0.90** |
||||
|
Ludhiana Faridkot Pooled |
NPP |
0.93** 0.94** 0.93** |
0.96** 0.96** 0.96** |
0.92** 0.93** 0.92** |
0.86** 0.79** 0.82** |
− 0.85** − 0.93** − 0.89** |
0.84** 0.91** 0.86** |
0.88** 0.88** 0.88** |
|||||
|
Ludhiana Faridkot Pooled |
BIO |
0.97** 0.96** 0.96** |
0.92** 0.92** 0.92** |
0.76** 0.68** 0.72** |
− 0.86** − 0.93** − 0.90** |
0.88** 0.92** 0.89** |
0.90** 0.88** 0.89** |
||||||
|
Ludhiana Faridkot Pooled |
YLD |
0.93** 0.95** 0.94** |
0.88** 0.85** 0.87** |
− 0.87** − 0.95** − 0.91** |
0.87** 0.93** 0.89** |
0.94** 0.89** 0.91** |
|||||||
|
Ludhiana Faridkot Pooled |
HSW |
0.84** 0.84** 0.84** |
− 0.86** − 0.92** − 0.89** |
0.87** 0.90** 0.88** |
0.86** 0.89** 0.87** |
||||||||
|
Ludhiana Faridkot Pooled |
HI |
− 0.79** − 0.78** − 0.78** |
0.75** 0.76** 0.75** |
0.81** 0.75** 0.78** |
|||||||||
|
Ludhiana Faridkot Pooled |
MPI |
− 0.80** − 0.93** − 0.86** |
− 0.83** − 0.91** − 0.85** |
||||||||||
|
Ludhiana Faridkot Pooled |
RLWC |
0.82** 0.89** 0.83** |
* = Significant at 5% probability level, ** = Highly significant at 1% probability level, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
Path analysis explained that BIO and HI were the major contributors towards YLD under timely-sown as well as late-sown environment at both the locations (Ludhiana and Faridkot). BIO was observed to have the highest positive indirect effect on YLD through NPP, followed by PH and HSW under timely-sown environment and through PH, followed by NPP, HSW, PV and RLWC under late-sown environment at both the locations (Tables 4, 5).
Table 4.
Direct (in bold and highlighted) and indirect contribution of various traits on grain yield in RIL population of chickpea under timely-sown environment
| DG | DFI | DFF | DHF | PH | NPP | BIO | HSW | HI | MPI | RLWC | PV | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
0.009 − 0.005 0.006 |
0.002 − 0.001 0.003 |
0.002 − 0.001 0.002 |
0.002 − 0.001 0.002 |
− 0.001 0.000 − 0.001 |
− 0.001 0.000 − 0.001 |
− 0.001 0.000 − 0.001 |
− 0.001 0.000 0.001 |
− 0.001 0.001 0.000 |
0.000 0.000 0.000 |
− 0.001 0.000 − 0.001 |
− 0.001 0.000 − 0.002 |
|
Ludhiana Faridkot Pooled |
DFI |
0.005 0.003 − 0.024 |
0.021 0.017 − 0.051 |
0.021 0.016 − 0.048 |
0.021 0.015 − 0.046 |
− 0.003 − 0.001 0.013 |
− 0.002 − 0.003 0.010 |
− 0.003 − 0.002 0.011 |
− 0.004 − 0.002 0.003 |
− 0.003 − 0.003 0.005 |
0.002 0.003 − 0.007 |
− 0.004 − 0.003 0.013 |
− 0.002 − 0.004 0.014 |
|
Ludhiana Faridkot Pooled |
DFF |
0.004 − 0.004 0.022 |
0.015 − 0.020 0.074 |
0.015 − 0.021 0.079 |
0.014 − 0.020 0.077 |
− 0.002 0.001 − 0.016 |
− 0.001 0.004 − 0.014 |
− 0.002 0.002 − 0.014 |
− 0.003 0.002 − 0.008 |
− 0.002 0.004 − 0.009 |
0.002 − 0.003 0.010 |
− 0.002 0.004 − 0.017 |
− 0.002 0.005 − 0.016 |
|
Ludhiana Faridkot Pooled |
DHF |
− 0.007 0.003 − 0.007 |
− 0.029 0.011 − 0.023 |
− 0.030 0.012 − 0.025 |
− 0.031 0.013 − 0.025 |
0.003 − 0.001 0.006 |
0.002 − 0.003 0.004 |
0.003 − 0.002 0.005 |
0.005 − 0.001 0.002 |
0.003 − 0.002 0.002 |
− 0.003 0.002 − 0.003 |
0.005 − 0.002 0.006 |
0.003 − 0.003 0.005 |
|
Ludhiana Faridkot Pooled |
PH |
0.001 − 0.001 0.006 |
0.001 0.001 0.008 |
0.001 0.001 0.006 |
0.001 0.001 0.007 |
− 0.008 − 0.007 − 0.032 |
− 0.004 − 0.004 − 0.018 |
− 0.006 − 0.006 − 0.025 |
− 0.004 − 0.003 − 0.016 |
− 0.003 0.000 − 0.006 |
0.004 0.004 0.018 |
− 0.002 − 0.004 − 0.015 |
− 0.004 − 0.004 − 0.018 |
|
Ludhiana Faridkot Pooled |
NPP |
− 0.002 0.000 0.005 |
− 0.002 − 0.001 0.012 |
− 0.002 − 0.001 0.011 |
− 0.002 − 0.001 0.011 |
0.011 0.002 − 0.034 |
0.024 0.004 − 0.060 |
0.023 0.004 − 0.056 |
0.019 0.003 − 0.048 |
0.016 0.002 − 0.036 |
− 0.020 − 0.004 0.052 |
0.011 0.003 − 0.040 |
0.019 0.003 − 0.049 |
|
Ludhiana Faridkot Pooled |
BIO |
− 0.056 0.008 − 0.085 |
− 0.063 − 0.051 − 0.141 |
− 0.058 − 0.045 − 0.114 |
− 0.047 − 0.052 − 0.119 |
0.358 0.317 0.495 |
0.489 0.362 0.581 |
0.516 0.399 0.627 |
0.428 0.307 0.492 |
0.344 0.111 0.293 |
− 0.442 − 0.323 − 0.525 |
0.224 0.306 0.408 |
0.426 0.301 0.504 |
|
Ludhiana Faridkot Pooled |
HSW |
− 0.003 0.001 0.001 |
− 0.004 − 0.004 − 0.001 |
− 0.004 − 0.003 − 0.001 |
− 0.003 − 0.003 − 0.001 |
0.011 0.017 0.007 |
0.017 0.028 0.011 |
0.017 0.028 0.011 |
0.020 0.037 0.014 |
0.017 0.022 0.010 |
− 0.018 − 0.032 − 0.012 |
0.010 0.031 0.009 |
0.017 0.030 0.010 |
|
Ludhiana Faridkot Pooled |
HI |
− 0.033 − 0.041 − 0.001 |
− 0.063 − 0.070 − 0.050 |
− 0.055 − 0.066 − 0.061 |
− 0.044 − 0.057 − 0.048 |
0.170 0.013 0.092 |
0.344 0.169 0.308 |
0.335 0.101 0.240 |
0.414 0.222 0.374 |
0.502 0.364 0.514 |
− 0.410 − 0.277 − 0.413 |
0.243 0.277 0.327 |
0.412 0.283 0.393 |
|
Ludhiana Faridkot Pooled |
MPI |
− 0.001 − 0.011 − 0.001 |
− 0.002 − 0.036 − 0.008 |
− 0.002 − 0.032 − 0.008 |
− 0.002 − 0.033 − 0.007 |
0.011 0.114 0.032 |
0.017 0.177 0.051 |
0.018 0.168 0.049 |
0.018 0.179 0.052 |
0.017 0.158 0.047 |
− 0.021 − 0.208 − 0.059 |
0.009 0.198 0.041 |
0.018 0.196 0.051 |
|
Ludhiana Faridkot Pooled |
RLWC |
− 0.001 − 0.008 − 0.003 |
− 0.002 − 0.026 − 0.005 |
− 0.002 − 0.023 − 0.004 |
− 0.002 − 0.023 − 0.004 |
0.002 0.069 0.008 |
0.005 0.111 0.011 |
0.004 0.104 0.011 |
0.005 0.115 0.011 |
0.005 0.103 0.011 |
− 0.005 − 0.130 − 0.012 |
0.010 0.136 0.017 |
0.005 0.124 0.013 |
|
Ludhiana Faridkot Pooled |
PV |
− 0.001 − 0.002 − 0.003 |
− 0.001 − 0.005 − 0.003 |
− 0.001 − 0.005 − 0.002 |
− 0.001 − 0.005 − 0.002 |
0.006 0.011 0.006 |
0.010 0.018 0.008 |
0.010 0.016 0.008 |
0.010 0.018 0.007 |
0.010 0.017 0.007 |
− 0.010 − 0.021 − 0.008 |
0.006 0.020 0.007 |
0.012 0.022 0.010 |
Residual effect of Ludhiana = 0.075; Faridkot = 0.078; Pooled = 0.075, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
Table 5.
Direct (highlighted) and indirect contribution of various traits on grain yield in RIL population of chickpea under late-sown environment
| DG | DFI | DFF | DHF | PH | NPP | BIO | HSW | HI | MPI | RLWC | PV | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
0.006 0.006 0.014 |
0.003 0.003 0.008 |
0.003 0.003 0.008 |
0.003 0.003 0.008 |
− 0.001 − 0.001 − 0.001 |
− 0.001 − 0.001 − 0.001 |
− 0.001 − 0.001 − 0.001 |
− 0.001 − 0.001 − 0.001 |
− 0.001 0.000 − 0.001 |
0.001 0.001 0.002 |
0.000 − 0.001 − 0.001 |
− 0.001 − 0.001 − 0.001 |
|
Ludhiana Faridkot Pooled |
DFI |
0.023 − 0.002 − 0.053 |
0.039 − 0.003 − 0.092 |
0.039 − 0.003 − 0.087 |
0.039 − 0.003 − 0.089 |
− 0.011 0.001 0.020 |
− 0.009 0.001 0.019 |
− 0.010 0.001 0.021 |
− 0.009 0.001 0.022 |
− 0.009 0.001 0.022 |
0.011 − 0.001 − 0.025 |
− 0.009 0.001 0.030 |
− 0.010 0.001 0.016 |
|
Ludhiana Faridkot Pooled |
DFF |
− 0.036 0.025 0.070 |
− 0.063 0.056 0.115 |
− 0.063 0.056 0.121 |
− 0.063 0.055 0.119 |
0.016 − 0.018 − 0.036 |
0.015 − 0.018 − 0.035 |
0.016 − 0.016 − 0.035 |
0.014 − 0.017 − 0.035 |
0.015 − 0.016 − 0.037 |
− 0.017 0.016 0.039 |
0.014 − 0.018 − 0.042 |
0.015 − 0.016 − 0.032 |
|
Ludhiana Faridkot Pooled |
DHF |
0.007 − 0.023 − 0.033 |
0.013 − 0.051 − 0.054 |
0.013 − 0.052 − 0.055 |
0.013 − 0.053 − 0.056 |
− 0.003 0.016 0.015 |
− 0.003 0.017 0.015 |
− 0.003 0.015 0.015 |
− 0.003 0.016 0.016 |
− 0.003 0.014 0.016 |
0.004 − 0.015 − 0.017 |
− 0.003 0.017 0.019 |
− 0.003 0.015 0.014 |
|
Ludhiana Faridkot Pooled |
PH |
− 0.024 − 0.011 − 0.014 |
− 0.049 − 0.029 − 0.038 |
− 0.048 − 0.029 − 0.053 |
− 0.048 − 0.028 − 0.049 |
0.184 0.092 0.176 |
0.164 0.085 0.165 |
0.178 0.089 0.171 |
0.169 0.085 0.166 |
0.154 0.072 0.153 |
− 0.169 − 0.087 − 0.166 |
0.166 0.087 0.162 |
0.174 0.086 0.168 |
|
Ludhiana Faridkot Pooled |
NPP |
− 0.020 − 0.007 − 0.011 |
− 0.039 − 0.020 − 0.029 |
− 0.038 − 0.020 − 0.040 |
− 0.039 − 0.020 − 0.037 |
0.146 0.058 0.130 |
0.164 0.063 0.138 |
0.154 0.059 0.134 |
0.153 0.059 0.133 |
0.145 0.054 0.125 |
− 0.150 − 0.061 − 0.132 |
0.143 0.060 0.127 |
0.150 0.060 0.131 |
|
Ludhiana Faridkot Pooled |
BIO |
− 0.057 − 0.078 − 0.045 |
− 0.104 − 0.175 − 0.110 |
− 0.101 − 0.171 − 0.142 |
− 0.102 − 0.170 − 0.135 |
0.395 0.597 0.476 |
0.385 0.583 0.471 |
0.410 0.614 0.488 |
0.381 0.573 0.464 |
0.323 0.467 0.411 |
− 0.381 − 0.592 − 0.466 |
0.370 0.591 0.456 |
0.379 0.584 0.463 |
|
Ludhiana Faridkot Pooled |
HSW |
0.001 0.002 0.006 |
0.001 0.006 0.020 |
0.001 0.005 0.025 |
0.001 0.005 0.024 |
− 0.005 − 0.017 − 0.080 |
− 0.005 − 0.017 − 0.081 |
− 0.005 − 0.017 − 0.081 |
− 0.005 − 0.018 − 0.085 |
− 0.005 − 0.016 − 0.078 |
0.005 0.017 0.081 |
− 0.005 − 0.017 − 0.080 |
− 0.005 − 0.017 − 0.079 |
|
Ludhiana Faridkot Pooled |
HI |
− 0.018 − 0.012 − 0.015 |
− 0.051 − 0.092 − 0.063 |
− 0.051 − 0.089 − 0.078 |
− 0.050 − 0.089 − 0.074 |
0.184 0.256 0.225 |
0.195 0.279 0.233 |
0.174 0.246 0.218 |
0.195 0.285 0.239 |
0.220 0.324 0.259 |
− 0.189 − 0.283 − 0.233 |
0.175 0.280 0.226 |
0.189 0.282 0.231 |
|
Ludhiana Faridkot Pooled |
MPI |
0.006 − 0.014 − 0.002 |
0.012 − 0.039 − 0.004 |
0.012 − 0.038 − 0.005 |
0.012 − 0.038 − 0.005 |
− 0.042 0.124 0.015 |
− 0.042 0.126 0.015 |
− 0.043 0.125 0.015 |
− 0.043 0.124 0.015 |
− 0.040 0.114 0.014 |
0.046 − 0.130 − 0.015 |
− 0.040 0.127 0.015 |
− 0.041 0.126 0.014 |
|
Ludhiana Faridkot Pooled |
RLWC |
0.005 − 0.002 0.003 |
0.014 − 0.006 0.009 |
0.014 − 0.006 0.010 |
0.014 − 0.006 0.010 |
− 0.055 0.017 − 0.026 |
− 0.053 0.017 − 0.026 |
− 0.055 0.017 − 0.026 |
− 0.055 0.017 − 0.026 |
− 0.048 0.015 − 0.024 |
0.053 − 0.017 0.026 |
− 0.061 0.018 − 0.028 |
− 0.052 0.017 − 0.025 |
|
Ludhiana Faridkot Pooled |
PV |
− 0.018 0.017 − 0.002 |
− 0.042 0.049 − 0.012 |
− 0.041 0.048 − 0.017 |
− 0.042 0.048 − 0.016 |
0.161 − 0.160 0.064 |
0.157 − 0.162 0.063 |
0.159 − 0.162 0.063 |
0.153 − 0.163 0.062 |
0.147 − 0.148 0.059 |
− 0.153 0.164 − 0.061 |
0.147 − 0.164 0.060 |
0.171 − 0.170 0.067 |
Residual effect of Ludhiana = 0.055; Faridkot = 0.081; Pooled = 0.073, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
Regression analysis
Multiple linear regression is used to predict the outcome of a response variable (grain yield) from several explanatory variables. The analysis revealed that PH, NPP, BIO and HI had contributed positively, while DHF, MPI, HSW and RLWC contributed negatively to the variation in YLD in late-sown environment (Table 7, Figure S1). However in timely-sown environment, DFF, BIO and HI contributed majorly and positively, while DG, DHF, PH, MPI and PV contributed negatively to the variation in YLD (Table 6. Figure S2). Overall, multiple linear regression depicted that NPP, BIO and HI had greater contribution to the total variation of YLD in heat stress environment. Based on the estimated results the prediction equation for seed yield (YLD as dependent variable) was formulated using the chickpea plant variables (as independent variables) as follow:
Table 7.
The regression coefficient (b), standard error (SE), t-value and probability of the estimated variables in predicting chickpea seed yield by the multiple linear regression analysis late-sown condition
| Location | Entered variable | Regression coefficient (b) | SE | t-value | p-value |
|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
0.080 − 0.001 0.033 |
0.02629 0.02337 0.01879 |
3.03** − 0.04 1.76 |
0.0026 0.9660 0.0778 |
|
Ludhiana Faridkot Pooled |
DFI |
0.072 0.036 − 0.011 |
0.04664 0.02221 0.01840 |
1.54 1.64 − 0.59 |
0.1234 0.1010 0.5532 |
|
Ludhiana Faridkot Pooled |
DFF |
− 0.147 0.029 0.068 |
0.06874 0.04471 0.03672 |
− 2.13* 0.64 1.85 |
0.0333 0.5218 0.0648 |
|
Ludhiana Faridkot Pooled |
DHF |
− 0.039 − 0.106 − 0.150 |
0.05081 0.03881 0.03246 |
− 0.76 − 2.74** − 4.61** |
0.4463 0.0063 < .0001 |
|
Ludhiana Faridkot Pooled |
PH |
0.229 0.058 0.109 |
0.01783 0.01462 0.01159 |
12.83** 3.99** 9.38** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
NPP |
0.191 0.063 0.094 |
0.01559 0.01415 0.01061 |
12.28** 4.43** 8.89** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
BIO |
0.1845 0.306 0.273 |
0.01219 0.01148 0.00875 |
15.18** 26.68** 31.24** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
HSW |
− 0.156 − 0.099 − 0.157 |
0.03670 0.04415 0.03070 |
− 4.27** − 2.26* − 5.13** |
< .0001 0.0244 < .0001 |
|
Ludhiana Faridkot Pooled |
HI |
0.321 0.504 0.451 |
0.01540 0.01431 0.01095 |
20.85** 35.26** 41.22** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
MPI |
− 0.066 − 0.131 − 0.095 |
0.00788 0.00996 0.00667 |
− 8.35** − 13.20** − 14.17** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
RLWC |
− 0.059 − 0.031 − 0.047 |
0.00648 0.01017 0.00572 |
− 9.12** − 2.99** − 8.20** |
< .0001 0.0029 < .0001 |
|
Ludhiana Faridkot Pooled |
PV |
0.047 − 0.069 − 0.001 |
0.00791 0.00911 0.00623 |
5.92** − 7.55** − 0.21 |
< .0001 < .0001 0.8319 |
* = Significant at 5% probability level, ** = Significant at 1% probability level, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
Table 6.
The regression coefficient (b), standard error (SE), t-value and probability of the estimated variables in predicting chickpea seed yield by the multiple linear regression analysis under timely-sown condition
| Location | Entered Variable | Regression coefficient (b) | SE | t-value | p-value |
|---|---|---|---|---|---|
|
Ludhiana Faridkot Pooled |
DG |
− 0.064 − 0.120 − 0.170 |
0.05477 0.04640 0.03587 |
− 1.17 − 2.58* − 4.74** |
0.2434 0.0102 < .0001 |
|
Ludhiana Faridkot Pooled |
DFI |
0.054 0.001 − 0.075 |
0.05420 0.05998 0.03918 |
1.00 0.02 − 1.91 |
0.3167 0.9867 0.0562 |
|
Ludhiana Faridkot Pooled |
DFF |
0.017 0.097 0.184 |
0.07775 0.07765 0.05599 |
0.22 1.25 3.29** |
0.8257 0.2101 0.0010 |
|
Ludhiana Faridkot Pooled |
DHF |
− 0.115 − 0.163 − 0.222 |
0.05054 0.06071 0.04187 |
− 2.27 − 2.68** − 5.30** |
0.0236 0.0076 < .0001 |
|
Ludhiana Faridkot Pooled |
PH |
− 0.028 − 0.003 − 0.030 |
0.01432 0.01246 0.00966 |
− 1.95 − 0.22 − 3.09** |
0.0512 0.8258 0.0020 |
|
Ludhiana Faridkot Pooled |
NPP |
0.015 − 0.003 − 0.009 |
0.01416 0.01268 0.00984 |
1.07 − 0.23 − 0.96 |
0.2852 0.8144 0.3373 |
|
Ludhiana Faridkot Pooled |
BIO |
0.387 0.351 0.393 |
0.01409 0.01399 0.01009 |
27.48** 25.11** 38.97** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
HSW |
0.014 0.127 0.008 |
0.03136 0.04057 0.02572 |
0.43 3.12** 0.27 |
0.6675 0.0019 0.7852 |
|
Ludhiana Faridkot Pooled |
HI |
0.732 0.616 0.672 |
0.01104 0.01254 0.00852 |
66.27** 49.06** 78.84** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
MPI |
− 0.088 − 0.276 − 0.204 |
0.01341 0.01690 0.01081 |
− 6.53** − 16.33** − 18.90** |
< .0001 < .0001 < .0001 |
|
Ludhiana Faridkot Pooled |
RLWC |
0.012 0.019 0.010 |
0.00600 0.01039 0.00530 |
1.93 1.88 1.85 |
0.0537 0.0607 0.0653 |
|
Ludhiana Faridkot Pooled |
PV |
− 0.033 − 0.062 − 0.056 |
0.01274 0.01193 0.00855 |
− 2.55* − 5.24** − 6.59** |
0.0109 < .0001 < .0001 |
* = Significant at 5% probability level, ** = Significant at 1% probability level, DG = days to germination, DFI = days to flowering initiation, DFF = days to 50% flowering, DHF = days to 100% flowering, PH = plant height, NPP = Number of pod per plant, BIO = biomass per plant, YLD = yield per plant, HSW = 100-seed weight, HI = harvest index, MPI = membrane permeability index, RLWC = relative leaf water content, PV = pollen viability
The regression equation of YLD and other yield related traits in timely-sown environment for pooled condition:
YLD = 4.61 − 0.170DG + 0.184DFF − 0.222DHF − 0.030PH + 0.393BIO + 0.672HI − 0.204MPI − 0.056PV; R2 = 0.9988, Adjusted R2 = 0.9988.
The regression equation of YLD and yield related traits in late-sown environment for pooled condition:
YLD = − 9.051 − 0.150DHF + 0.109PH + 0.094NPP + 0.273BIO − 0.157HSW + 0.451HI − 0.131MPI − 0.047RLWC; R2 = 0.9979, Adjusted R2 = 0.9979.
In stepwise linear regression analysis, MPI was entered at first in the model in timely-sown condition and explained 77% of the variation in response trait like YLD, followed by BIO and HI. While in late-sown environment, BIO contributed 93% to the variation in response trait like YLD, followed by HI and PH. Thus, stepwise linear regression depicted that BIO and HI consistently showed a very high level of contribution to the total variation of YLD in late-sown as well as timely-sown conditions (Table S1, S2).
Principal component analysis
The PCA provides information about the traits elucidating the maximum variability present in the population under specific environment. Results from PCA analysis revealed that the first three principal components explained 80.76% and 91.52% of the total phenotypic variability under timely-sown and late-sown environment, respectively. The principal component analysis revealed that PH, NPP, BIO, HSW, HI, MPI, RLWC and PV were the main contributing traits in PC1 (Table S3, S4; Fig. 4). MPI was one of the major contributing traits in PC1 but negatively correlated with YLD under timely-sown as well as late-sown environment.
Fig. 4.
Biplots based on principal component analysis (PCA) showing the relationship of yield (YLD) under timely-sown and late-sown environment
Discussion
Heat stress reduces the seed yield at higher rate during pod development as compared to heat stress during vegetative phase or early flowering (Wang et al. 2006). Thus, late-sown condition was proved to be an ideal condition for screening of heat tolerance as the temperature goes beyond the threshold limit (> 35 °C) at the time of reproductive phase (Fig. 1). At both the locations (Ludhiana and Faridkot), the crop was exposed to heat stress as temperature goes beyond the threshold limit during the reproductive stage, indicated that both the locations were ideal for evaluation of RIL population for heat tolerance. However, similar temperature was also observed for 10 to 15 days out of 100 days which has no significant effect on results, as shown in Fig. 1.
The contrast analysis depicted highly significant differences between the parents for all the traits under late-sown and timely-sown conditions which validated the experimental conditions for screening of RILs for heat tolerance related traits (Table 1). It also indicated that the RIL population developed by using these parents were perfect for studying the heat stress. In earlier studies also, contrast analysis of parents showed significant differences for all the traits studied, except biomass under heat stress environment (Paul et al. 2018; Devasirvatham et al. 2013). The mean difference between late-sown and timely-sown conditions indicated delayed germination while, mean difference for DFI, DFF and DHF indicated precocity under late-sown condition (Table 1).
All the morphological and physiological traits were significantly affected by heat stress environment, except HSW and HI which were moderately affected. Low pollen viability could be one of the major causes of reduced seed yield due to heat stress. Significant differences were observed for yield and yield contributing traits and physiological traits in RIL population and its parents in late-sown as compared to timely-sown condition (Table 1). Studies conducted in the past had also reported the significant differences for most of the morphological and physiological traits between the heat stress and non-stress environments (Krishnamurthy et al. 2011; Devasirvatham et al. 2013; Paul et al. 2018). Low pollen viability and pollen sterility could be one of the major causes of reduced seed yield due to poor pod setting in the RILs during pre-anthesis heat stress environment (Porch and Jahn 2001; Devasirvatham et al. 2010). Sakata and Higashitani (2008) also observed that poor pod setting during pre-anthesis high temperature stress was associated with low pollen viability, indehiscent anthers and other anther abnormalities. Whereas, poor pod setting during post-anthesis high temperature stress was related to poor pollen germination, pollen tube growth and fertilization (Kakani et al. 2002).
Wang et al. (2006) also concluded that post-anthesis high temperature stress reduced the seed yield in chickpea by reducing the number of pods per plant and hundred seed weight. Anther and stigma were the most sensitive organs to heat stress in chickpea (Nayyar et al. 2005). Devasirvatham et al. (2013) concluded that low pollen viability was the major reason of sterility under high temperature stress during anthesis in chickpea which results in poor pod setting percentage. Thus, pollen viability can be one of the useful indicators which help to know the genetic variations present among the genotypes for heat tolerance at reproductive phase.
A number of morphological traits like PH, YLD and HSW and physiological traits such as RLWC, MPI were adversely affected by the late-sown condition as compared to timely-sown condition which was in agreement with the results of Kumar et al. 2017a, b. The present study indicated that reduction in RLWC due to increase in transpiration was associated with reduced water availability which was frequently associated with heat stress under field conditions. Due to this, both source and sink relationship for photosynthate assimilates was disturbed during heat stress which caused reduction in seed yield (Simões-Araújo et al. 2003; Devasirvatham et al. 2012).
The pooled ANOVA revealed significant differences for all the traits in RILs for genotypic variance across the environments, suggesting the presence of good amount of variations among the RILs (Table 1). Significant genotypic variation for various physiological and morphological traits were also observed by Tongden et al. (2006) and Kumar et al. (2017a, b) under controlled as well as late-sown conditions. Significant variation was also observed due to genotype × location (G × L) interaction for all the traits except DG, DFI, DFF, DHF, which were consistently non-significant (Table 1). Genotypic variances among the RILs were significant and consistent across the environments as well as highly interactive with the locations and could be the reason for significant variances due to both genotypes and G × L interaction (Upadhyaya et al. 2011). Paul et al. (2018) also observed highly significant genotypic and G × E interaction variances for pooled analysis of two heat stress environments for almost all the traits studied. Upadhyaya et al. (2011) also reported a highly significant genetic and G × E interaction variance across the heat stress environments which were in agreement with our results.
The value of GCV and PCV components facilitates to assess the extent of genetic variation present in a population for various traits (Paul et al. 2018). High values of GCV and PCV for the traits like PH, NPP, BIO, YLD and HSW in late-sown condition, while low to moderate in timely-sown condition explained the large effect of heat stress revealing genetic variation in RILs (Table 1). The results indicated that the RILs performed differently in terms of their tolerance to heat stress under high temperature. However, the values of GCV and PCV for DG, DFI, DFF and DHF were low and showed similar magnitude in both non-stress and heat stress environment indicating that these traits were less influenced by heat stress (Table 1). High values of GCV and PCV for total number of seeds per plot and grain yield, moderate for hundred seed weight, while low for days to 50% flowering in heat stress environment was observed by Paul et al. (2018) which was in agreement with our results.
High heritability was observed for all the traits, except DG which showed moderate heritability in case of late-sown environment. While in case of timely-sown condition, high heritability was found for PH, NPP, BIO, YLD, HSW, HI, MPI, RLWC and PV, while low to moderate heritability was observed for DG, DFI, DFF and DHF (Table 1). It is remarkable that consistently moderate to high heritability was observed for all the traits studied except for a few. Heritability facilitates understanding the magnitude of genetic expression under specific environment (Kumar et al. 2017a, b). In this study, higher heritability was observed under heat stress environment for yield and yield related traits as compared to non-stress environment at both Ludhiana and Faridkot locations, indicating that these traits were less influenced by environmental fluctuations. This implicated that these traits were expressed more distinctly under heat stress condition, and thus, selection will be more promising for these traits as also explained by Krishnamurthy et al. (2011). Wide range of genotypic variability in the RILs led to higher broad sense heritability under heat stress environment. High heritability for the traits such as PH, NPP, BIO, YLD, HSW, HI, MPI, RLWC and PV under abiotic stress conditions in chickpea was also observed in previous studies (Vadez et al. 2012; Varshney et al. 2014; Paul et al. 2018).
It is noteworthy that value of GAM for the traits like PH, NPP, BIO, YLD, HSW, HI, MPI, RLWC and PV were consistently high or moderately high, while DG, DFI, DFF and DHF were consistently low across the environments at both Ludhiana and Faridkot locations (Table 1). High value of GAM for yield and yield contributing traits except biomass and low value of GAM for days to 50% flowering was also recorded by Paul et al. (2018) which was in accordance with our results. Traits having high heritability combined with high genetic advance explained the major effect of additive genes for these traits. Thus, the selection should be based on yield per se and other important traits like PH, NPP, BIO, HSW, HI, MPI, RLWC and PV which were indirectly correlated with yield and have high heritability along with high genetic advances.
A strong positive association has been observed between PH, NPP, BIO, HSW, HI, RLWC and PV with YLD which revealed significance of these traits for determining yield across the environments (Tables 2, 3). It appears that these traits are the major contributors for enhancing YLD in chickpea. A positive and significant association of grain yield was observed with plant height, number of pods per plant, biomass and harvest index in some previous studies (Jivani et al. 2013; Purushothaman et al. 2017; Paul et al. 2018).
A significant negative association of yield was found with DG, DFI, DFF, DHF and MPI, however, it was inconsistent across the environments (Mallu et al. 2015). Genotypes showing early flowering had comparatively extended period of reproductive phase which resulted into more seed yield before the onset of stress, consequently seed yield was negatively correlated with days to flowering (Zaman-Allah et al. 2011; Maqbool et al. 2016; Purushothaman et al. 2017; Paul et al. 2018).
Path analysis showed that BIO followed by HI had the highest direct effect on YLD, while BIO contributed the highest positive indirect effect on YLD through NPP under timely-sown environment and through PH under late-sown environment (Tables 4, 5). These traits also expressed the significant and positive correlation with yield. The combined results of correlation and path analysis concluded that these traits were significant to devise selection indices for screening high yielding heat-tolerant genotypes in chickpea. In some previous studies, path analysis revealed that the number of pods per plant, number of seeds per plot, harvest index, biomass, hundred seed weight and plant height was the major direct contributor to grain yield (Renukadevi and Subbalakshmi 2006; Thakur and Sirohi 2009; Jivani et al. 2013; Paul et al. 2018). The BIO and HI were found to have the high negative indirect effect on YLD through MPI under timely-sown and late-sown environment at both locations.
Multiple linear regression depicted the major positive contribution of NPP, BIO and HI and major negative contribution of MPI to the total variation of YLD consistently across the environments (Tables 6, 7). Likewise, stepwise linear regression represents that BIO and HI consistently showed a high level of contribution to the total variation of yield among the environments (Table S1, S2). Hence, the combination of these traits can be a good strategy for creating selection index for screening of chickpea genotypes under heat stress conditions. In previous studies, regression analysis revealed that number of pods per plant, number of seeds per plot and number of filled pods are the major contributor towards yield (Zaman-Allah et al. 2011; Paul et al. 2018).
The principal component analysis depicts that PH, NPP, BIO, HSW, HI, MPI, RLWC and PV consistently were the main contributing traits for yield under timely-sown and late-sown environment (Table S3, S4). Thus, these traits were clustered together and contributed to maximum variability for YLD in both timely-sown and late-sown environment. Hence, phenotypic selection for these traits will be helpful for the monitoring of chickpea genotypes under heat stress. In previous studies, number of pods per plant, harvest index and biomass were observed to be the main contributing traits under heat stress conditions revealed by principal component analysis which was in agreement with our findings (Canci and Toker 2009; Krishnamurthy et al. 2011; Paul et al. 2018).
Conclusion
This study illustrated the presence of large genetic variation in RILs for yield and yield contributing traits under heat stress environments. Among these traits, PH, NPP, BIO and YLD were the most affected traits due to heat stress. High value of GCV and PCV along with high heritability and GAM were recorded for PH, NPP, BIO, YLD and HSW across the heat stress environments. Path analysis and both multiple and step-wise regression analysis depicted that NPP, BIO and HI were the major contributor to YLD consistently across the environments, while correlation studies and principal component analysis showed positive association of PH, NPP, BIO, HSW, HI, RLWC and PV with YLD across the environments. Thus, a holistic approach across these analyses identified NPP, BIO, and HI as the key traits, which could be used for improving chickpea yield through indirect selection for developing heat-tolerant cultivars. Large genetic variability present in RILs can be used for developing heat-tolerant cultivars in heat tolerance breeding programme in chickpea.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- ANOVA
Analysis of variance
- BIO
Biomass
- DHF
Days to 100% flowering
- DFF
Days to 50% flowering
- DFI
Days to flowering initiation
- DG
Days to germination
- GAM
Genetic advance as percent of mean
- G × E
Genotype × environment interaction
- GCV
Genotypic coefficient of variation
- HI
Harvest index
- HSW
Hundred seed weight
- MAS
Marker assisted selection
- MPI
Membrane permeability index
- NPP
Number of pods per plant
- PCV
Phenotypic coefficient of variation
- PH
Plant height
- PV
Pollen viability
- PCA
Principal component analysis
- RILs
Recombinant inbred lines
- RLWC
Relative leaf water content
- YLD
Yield
Author’s contribution
AK, GS, SS and IS designed and conducted the experiments. DB, AK, SB and SV performed the data analysis. AK, DB, SV and SS prepared and edited the final manuscript.
Funding
The INSPIRE research grant provided to the AK by Department of Science and Technology (DST), New Delhi, India and research grant provided under the project ‘Incentivizing Research in Agriculture’ by Indian Council of Agricultural Research, New Delhi to SS for carrying out the research are highly acknowledged.
Code availability
Not applicable.
Declarations
Conflict of interest
All authors declare that there are no conflicts of interest.
Ethics approval
Not applicable.
Availability of data and material
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbo S, Berger JD, Turner NC. Evolution of cultivated chickpea: four genetic bottlenecks limit diversity and constrain crop adaptation. Funct Plant Biol. 2003;30:1081–1087. doi: 10.1071/FP03084. [DOI] [PubMed] [Google Scholar]
- Allard RW. Principles of plant breeding. New York: John Wiley and Sons Inc; 1960. [Google Scholar]
- Basu PS, Ali M, Chaturvedi SK (2009) Terminal heat stress adversely affects chickpea productivity in northern India- Strategies to improve thermo tolerance in the crop under climate change. Workshop Proceedings, impact of climate change on agriculture, pp. 23–25
- Canci H, Toker C. Evaluation of yield criteria for drought and heat resistance inchickpea (Cicer arietinum L.) J Agron Crop Sci. 2009;195(1):47–54. [Google Scholar]
- Chaturvedi SK, Mishra DK, Vyas P, Mishra N. Breeding for cold tolerance in chickpea. Trends Biosci. 2009;2:1–6. [Google Scholar]
- Cottee NS, Tan DKY, Bange MP, Cothren JT, Campbell LC. Multi-level determination of heat tolerance in cotton (Gossypium hirsutum L.) under field conditions. Crop Sci. 2010;50:2553–2564. [Google Scholar]
- Devasirvatham V, Tan DKY, Trethowan RM, Gaur PM, Mallikarjuna N (2010) Impact of high temperature on the reproductive stage of chickpea. Proceeding of the 15th Australian Society of Agronomy Conference, Lincoln, New Zealand
- Devasirvatham V, Gaur PM, Mallikarjuna M, Tokachichu RN, Trethowan RM, Tan DKY. Effect of high temperature on the reproductive development of chickpea genotypes under controlled environments. Funct Plant Biol. 2012;139:1009–1018. doi: 10.1071/FP12033. [DOI] [PubMed] [Google Scholar]
- Devasirvatham V, Gaur PM, Mallikarjuna N, Tokachichu R, Trethowan R, Tan DKY. Reproductive biology of chickpea response to heat stress in the field is associated with the performance in controlled environments. Field Crops Res. 2013;142:9–19. [Google Scholar]
- Dewey DR, Lu KH. A correlation and path-coefficient analysis of components of crested wheatgrass seed production. Agron J. 1959;51:515–518. [Google Scholar]
- FAOSTAT (2017) Statistical Database of the United Nation Food and Agriculture Organization (FAO) Statistical Division Rome Available at: http://www.fao.org/faostat/en/#home. Accessed Sep 2017
- Gaur PM, Srinivasan S, Gowda CLL, Rao BV. Rapid generation advancement in chickpea. J SAT Agric Res. 2007;3(1):1–3. [Google Scholar]
- Gaur PM, Jukanti AK, Srinivasan S, et al. Climate change and heat stress tolerance in chickpea. In: Tuteja N, Gill SS, et al., editors. Climate change and plant abiotic stress tolerance. Weinheim: Wiley; 2014. pp. 839–856. [Google Scholar]
- Hamwieh A, Imtiaz M. Identifying water-responsive and drought-tolerant chickpea genotypes. Crop Pasture Sci. 2015;66:1003–1011. [Google Scholar]
- Howarth CJ. Genetic improvements of tolerance to high temperature. In: Ashraf M, Harris PJC, editors. Abiotic stresses-plant resistance through breeding and molecular approaches. USA: The Haworth Press; 2005. pp. 277–300. [Google Scholar]
- Jivani JV, Mehta DR, Pithia MS, Madariya RB, Mandavia CK. Variability analysis and multivariate analysis in chickpea (Cicer arietinum L.) Electron J Plant Breed. 2013;4:1284–1291. [Google Scholar]
- Jukanti AK, Gaur PM, Gowda CLL, Chibbar RN. Chickpea: nutritional properties and its benefits. Br J Nutr. 2012;108:11–26. doi: 10.1017/S0007114512000797. [DOI] [PubMed] [Google Scholar]
- Kakani VG, Prasad PVV, Craufurd PQ, Wheeler TR. Response of in vitro pollen germination and pollen tube growth of groundnut (Arachis hypogaea L.) genotypes to temperature. Plant Cell Environ. 2002;25:1651–1661. [Google Scholar]
- Karim MF, Fattah QA. Growth analysis of chickpea cv bari chhola-6 as affected by foliar spray of growth regulators. Bangladesh J Bot. 2007;36(2):105–110. [Google Scholar]
- Krishnamurthy L, Gaur PM, Basu PS, et al. Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L) germplasm. Plant Genet Resour. 2011;9:59–61. [Google Scholar]
- Kumar J, Gupta DS, Gupta S, Dubey S, Gupta P, Kumar S. Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 2017;36:1187–1213. doi: 10.1007/s00299-017-2127-y. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kumar A, Kumar RR, Roy RK, Agrawal T. Genetic variability of chickpea genotypes under heat stress condition: correlation and path analysis: based analysis. Ind J Ecol. 2017;44(4):59–64. [Google Scholar]
- Kushwah A, Gupta S, Bindra S, et al. Gene pyramiding and multiple character breeding. In: Singh M, et al., editors. Chickpea crop wild relatives for enhancing genetic gains. Elsevier Academic Press; 2020. pp. 131–165. [Google Scholar]
- Kushwah A, Bindra S, Singh I, et al. Advances in chickpea breeding and genomics for varietal development and trait improvement in India. In: Gosal SS, Wani SH, et al., editors. Accelerated plant breeding. Springer; 2020. pp. 31–66. [Google Scholar]
- Ladizinsky G. A new Cicer from Turkey. Notes Roy Bot Gard. 1975;34:201–202. [Google Scholar]
- Ludlow MM, Muchow RC. A critical evaluation of traits for improving crop yields in water limited environments. Adv Agron. 1990;43:107–153. [Google Scholar]
- Mallu TS, Nyende AB, Rao NG, Odeny DA, Mwangi SG. Assessment of interrelationship among agronomic and yield characters of chickpea. Int J Agric Crop Sci. 2015;8(2):128–135. [Google Scholar]
- Maqbool MA, Aslam M, Ali H, Shah TM. Evaluation of advanced chickpea (Cicer arietinum L) accessions based on drought tolerance indices and SSR markers against different water treatments. Pak J Bot. 2016;48:1421–1429. [Google Scholar]
- Nayyar H, Bains T, Kumar S. Low temperature induced floral abortion in chickpea: relationship with abscisic acid and cryoprotectants in reproductive organs. Environ Exp Bot. 2005;53:39–47. [Google Scholar]
- Paul PJ, Samineni S, Sajja SB, et al. Capturing genetic variability and selection of traits for heat tolerance in a chickpea recombinant inbred line (RIL) population under field conditions. Euphytica. 2018;214:27. [Google Scholar]
- Porch TG, Jahn M. Effects of high-temperature stress on microsporogenesis in heat-sensitive and heat-tolerant genotypes of Phaseolus vulgaris. Plant Cell Environ. 2001;24:723–731. [Google Scholar]
- Premchand GS, Sangroka T, Ogatta S. Cell membrane stability as indicators of drought tolerance as affected by applied nitrogen in soybean. J Agric Sci. 1990;11:563–566. [Google Scholar]
- Purushothaman R, Krishnamurthy L, Upadhyaya HD, Vadez V, Varshney RK. Shoot traits and their relevance in terminal drought tolerance of chickpea (Cicer arietinum L) Field Crops Res. 2016;197:10–27. doi: 10.1016/j.fcr.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman R, Krishnamurthy L, Upadhyaya HD, Vadez V, Varshney R. Genotypic variation in soil water use and root distribution and their implications for drought tolerance in chickpea. Funct Plant Biol. 2017;44:235–252. doi: 10.1071/FP16154. [DOI] [PubMed] [Google Scholar]
- Renukadevi P, Subbalakshmi B. Correlations and path coefficient analysis in chickpea. Legume Res. 2006;29:201–204. [Google Scholar]
- Sairam RK. Effect of moisture stress on physiological activities of two contrasting wheat genotypes. Indian J Exp Biol. 1994;32:584–593. [Google Scholar]
- Sakata T, Higashitani A. Male sterility accompanied with abnormal anther development in plants–genes and environmental stresses with special reference to high temperature injury. Int J Plant Dev Biol. 2008;2:42–51. [Google Scholar]
- Sharma KD, Pannu RK, Behl RK (2005) Effect of early and terminal heat stress on biomass portioning chlorophyll stability and yield of different wheat genotypes. Proceeding of the international conference on sustainable crop production in stress environments: management and genetic options, pp. 187–94
- Simões-Araújo JL, Rumjanek NG, Margis-Pinheiro M. Small heat shock proteins genes are differentially expressed in distinct varieties of common bean. Braz J Plant Physiol. 2003;15(1):33–41. [Google Scholar]
- Singh R, Sharma P, Varshney RK, Sharma SK, Singh NK. Chickpea improvement: role of wild species and genetic markers. Biotechnol Genet Eng Rev. 2008;25:267–314. doi: 10.5661/bger-25-267. [DOI] [PubMed] [Google Scholar]
- Slavik B. Methods of studying plant water relations. Berlin and New York: Springer-Verlag; 1974. [Google Scholar]
- Thakur SK, Sirohi A. Correlation and path coefficient analysis in chickpea (Cicer arietinum L) Legume Res. 2009;32(1):1–6. [Google Scholar]
- Thudi M, Upadhyaya HD, Rathore A, et al. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE. 2014;9(5):e96758. doi: 10.1371/journal.pone.0096758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker C, Canci H (2006) Selection for drought and heat resistance in chickpea under terminal drought conditions. 4th international food legumes research conference, food legumes for nutritional security and sustainable agriculture, pp. 18–22
- Tongden C, Basant M, Chakraborty U. Screening of thermotolerant cultivars of chickpea using cell membrane stability test and biochemical markers. J Hill Res. 2006;19:52–58. [Google Scholar]
- Tuberosa R, Salvi S. Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci. 2006;11:405–412. doi: 10.1016/j.tplants.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Upadhaya HD, Dronavalli N, Gowda CLL, Singh S. Identification and evaluation of chickpea germplasm for tolerance to heat stress. Crop Sci. 2011;51:2079–2094. [Google Scholar]
- Vadez V, Krishnamurthy L, Thudi M, et al. Assessment of ICCV 2 9 JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol Breed. 2012;30(1):9–21. [Google Scholar]
- Varshney RK, Mohan SM, Gaur PM, et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotech Adv. 2013;10:1016–1022. doi: 10.1016/j.biotechadv.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Thudi M, Nayak SN, et al. Genetic dissection of drought tolerance in chickpea (Cicerarietinum L) Theor Appl Genet. 2014;127(2):445–462. doi: 10.1007/s00122-013-2230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gan YT, Clarke F, McDonald CL. Response of chickpea yield to high temperature stress during reproductive development. Crop Sci. 2006;46:2171–2178. [Google Scholar]
- Wasson AP, Richards RA, Chatrath R, et al. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J Exp Bot. 2012;63:3485–3498. doi: 10.1093/jxb/ers111. [DOI] [PubMed] [Google Scholar]
- Weerakoon WMW, Maruyama A, Ohba K. Impact of humidity on temperature induced grain sterility in rice (Oryza sativa L) J Agron Crop Sci. 2008;194:134–140. [Google Scholar]
- Zaman-Allah M, Jenkinson DM, Vadez V. Chickpea genotypes contrasting for seed yield under terminal drought stress in the field differ for traits related to the control of water use. Funct Plant Biol. 2011;38:270–281. doi: 10.1071/FP10244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.