Abstract
The coffee berry borer, the most economically important insect pest of coffee worldwide, is the only insect capable of feeding and reproducing solely on the coffee seed, a food source containing the purine alkaloid caffeine. Twenty-one bacterial species associated with coffee berry borers from Hawai’i, Mexico, or a laboratory colony in Maryland (Acinetobacter sp. S40, S54, S55, Bacillus aryabhattai, Delftia lacustris, Erwinia sp. S38, S43, S63, Klebsiella oxytoca, Ochrobactrum sp. S45, S46, Pantoea sp. S61, Pseudomonas aeruginosa, P. parafulva, and Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75) were found to have at least one of five caffeine N-demethylation genes (ndmA, ndmB, ndmC, ndmD, ndmE), with Pseudomonas spp. S31, S32, S37, S60 and P. parafulva having the full complement of these genes. Some of the bacteria carrying the ndm genes were detected in eggs, suggesting possible vertical transmission, while presence of caffeine-degrading bacteria in frass, e.g., P. parafulva (ndmABCDE) and Bacillus aryabhattai (ndmA) could result in horizontal transmission to all insect life stages. Thirty-five bacterial species associated with the insect (Acinetobacter sp. S40, S54, S55, B. aryabhattai, B. cereus group, Bacillus sp. S29, S70, S71, S72, S73, D. lacustris, Erwinia sp. S38, S43, S59, S63, K. oxytoca, Kosakonia cowanii, Ochrobactrum sp. S45, S46, Paenibacillus sp. S28, Pantoea sp. S61, S62, P. aeruginosa, P. parafulva, Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75, Stenotrophomonas sp. S39, S41, S48, S49) might contribute to caffeine breakdown using the C-8 oxidation pathway, based on presence of genes required for this pathway. It is possible that caffeine-degrading bacteria associated with the coffee berry borer originated as epiphytes and endophytes in the coffee plant microbiota.
Keywords: bark beetle, broca del café, Coffea, Coleoptera (beetles), symbiosis, symbiotes, Hypothenemus hampei, coffee
Introduction
Approximately 80 plant species synthesize purine alkaloids (Ashihara and Crozier, 1999). Among these species, coffee, tea (Camellia spp.), and cacao (Theobroma cacao) are well known for the presence of the purine alkaloid 1,3,7-trimethylxanthine, commonly known as caffeine, which is widely used for its stimulant effects (Cappelletti et al., 2015). Of 125 species in the genus Coffea (Rubiaceae) (Davis et al., 2006, 2011), only three are commercially traded: Coffea arabica, C. canephora (also known as robusta coffee), and C. liberica. All three species contain different levels of caffeine, with mean levels in C. canephora being the highest (2.4%), followed by. C. liberica (1.9%), and C. arabica (1.3%) (Ashihara et al., 2017).
Various roles have been proposed to explain the presence of caffeine in plants: (1) inhibition of competing plants through allelopathic effects (Waller et al., 1986); (2) improved pollinator fidelity, and consequently, plant reproductive success (Wright et al., 2013); and (3) deterrency of insects (Levinson, 1976; Nathanson, 1984; Kim et al., 2006) or plant pathogens (Kim and Sano, 2008; Sano et al., 2013). Indeed, several studies have confirmed a negative effect of caffeine on insects (see Vega et al., 2003 and references therein). An exception is the coffee berry borer, Hypothenemus hampei (Ferrari) (Coleoptera: Curculionidae: Scolytinae), which has evolved an extreme adaptation to caffeine and is the only insect that feeds solely on seeds within the coffee fruit (Vega et al., 2015).
The coffee berry borer infestation cycle begins with an inseminated female entering a coffee fruit, followed by oviposition in galleries built throughout one or both coffee seeds within the fruit. Occasionally, a fruit might be colonized by more than one female. The insect exhibits a skewed sex ratio favoring females as well as sibling mating; therefore, females emerging from the fruit are already inseminated and ready to colonize another fruit (Vega et al., 2015). The insect causes losses in yield and quality which in Brazil alone have been estimated at US$215–358 million per year (Oliveira et al., 2013).
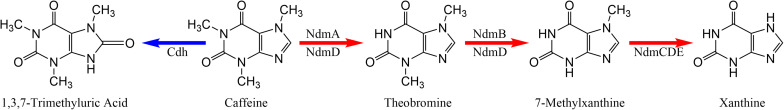
Ceja-Navarro et al. (2015) demonstrated that the adaptation required by the coffee berry borer to survive on a food source containing caffeine (i.e., the coffee seed) involves a caffeine degrading mechanism provided by bacterial symbiotes (we use the term “symbiote” rather than “symbiont” based on the rationale described by Vega and Biedermann, 2020). There are two bacterial mechanisms for caffeine degradation: N-demethylation and C-8 oxidation (Summers et al., 2015). N-demethylation (Figure 1) is the most common pathway for bacterial metabolism of caffeine and requires five genes: ndmA, ndmB, ndmC, ndmD, and ndmE. The enzyme NdmA first removes the N1-methyl group from caffeine to form theobromine (3,7-dimethylxanthine), which is further converted to 7-methyxanthine by NdmB (Summers et al., 2012; Figure 1). 7-Methylxanthine then undergoes N7-demethylation to xanthine by the NdmCDE protein complex (Summers et al., 2013; Figure 1). Both NdmA and NdmB require the presence of NdmD for activity (Summers et al., 2012; Figure 1). NdmE is believed to serve a structural role in the NdmCDE complex and does not have a known catalytic activity (Summers et al., 2013). At least 71 bacterial strains in 27 genera and from many different habitats worldwide have been reported to be involved in caffeine degradation, with Pseudomonas spp. being the most common (Supplementary Table 1). Thus, caffeine degradation is a function useful in diverse ecological contexts and may have evolved several times. The observation that so many bacteria from different parts of the world (Supplementary Table 1) are able to degrade caffeine through a similar mechanism might be explained by an ancient metabolic pathway for methylxanthines or purines in general that is today retained in some bacteria or by horizontal gene transfer.
FIGURE 1.
Metabolic pathways for caffeine degradation by bacteria. N-demethylation is indicated by red arrows. C-8 oxidation is indicated by the blue arrow.
Ceja-Navarro et al. (2015) identified 14 caffeine-degrading bacteria in the alimentary canal of the coffee berry borer and only P. fulva tested positive for ndmA, implying that other caffeine demethylase genes or a different caffeine degradation mechanism (i.e., C-8 oxidation) is present in the other bacterial groups. Elimination of P. fulva from the coffee berry borer caused negative effects on insect fitness (Ceja-Navarro et al., 2015), thereby confirming the critical importance of the bacterial symbiosis.
For the long-term stability of a host-symbiote system, it is beneficial for both players to evolve a reliable mechanism for the transfer of symbiotes from host to host and across generations (Jaenike, 2012). Many host-symbiote systems accomplish this transfer through vertical transmission (i.e., from mother to offspring) (Ebert, 2013). For the coffee berry borer–bacteria system the mode of transmission is unknown. Acquisition of caffeine-degrading bacteria by the coffee berry borer may follow two modes: vertical transmission or horizontal transmission (i.e., among unrelated hosts, including from the environment to the host) (Jaenike, 2012; Ebert, 2013). A combination of these two modes, called mixed-mode transmission, whereby both vertical and horizontal transmission of the same symbiote occurs, may also take place and this has been reported in diverse host-symbiote systems (Ebert, 2013). To assess the mode of transmission, ideally one would study the physical course of transfer of the symbiote from host to host, i.e., the route of transmission. As the observation of the transfer itself is difficult, it is essential to first determine where the symbiotes are found in order to then make an assessment on the route of transmission. For example, if the symbiote is present inside the host’s egg, a transovum route of transmission is indicated, which is a form of vertical transmission. Presence of the symbiotes in the host’s frass allows for potential transfer of symbiotes among unrelated hosts, a form of horizontal transmission. Some routes of transmission are less clearly associated with a mode of transmission and may allow both vertical and horizontal transmission. For example, presence of symbiotes in the female host’s saliva, may allow the mother to transfer symbiotes to her offspring, but this may also happen to unrelated offspring.
Here we focus on (1) the identification of culturable bacteria associated with heads, eggs, larvae, and frass of the coffee berry borer; (2) the detection of caffeine N-demethylation or C-8 oxidation genes in these bacteria; and (3) the in vitro detection of caffeine metabolites for eight bacteria found to have caffeine N-demethylation genes. We also interpret these data with regard to possible modes of transmission of these bacteria. These findings are important in understanding the role of caffeine-degrading bacteria within the coffee agroecosystem, including the plant, the coffee berry borer, and the soil.
Materials and Methods
Insects
Coffee berry borers were dissected from infested coffee fruits collected in the field in Hawai’i and Mexico and from a colony continuously reared for ca. 15 years on an artificial diet at the United States Department of Agriculture – Agricultural Research Service laboratory in Beltsville, Maryland. Hawaiian specimens were dissected from infested C. arabica berries collected at: (1) Holualoa, Hawai’i (19°38′26″ N; 155°57′19″ W; 44 meters above sea level – masl); (2) two locations in Wood Valley, K’au, Hawai’i (19°16′15″ N; 155°29′19″ W; 795 masl; and 19°15′52″ N; 155°28′00″ W; 638 masl); and (3) Dole Plantation, Waialua, Oahu (21°33′51″ N; 158°04′26″ W; 146 masl). Coffee berry borers from Mexico were dissected from infested C. canephora fruits collected at Finca San Antonio, Cacahoatán, Chiapas (15°00′20″ N; 92°09′12″ W; 562 masl).
Tissue Preparation and Culture
Adult female coffee berry borers (ca. 2 mm long) were sterilized by sonicating in 90% ethanol for 30 s in sterile 1.5 ml Eppendorf tubes placed in a floating foam tube rack placed inside a Branson 2210 ultrasonic cleaner (Branson Ultrasonics Corp., Danbury, CT, United States), followed by soaking in 3.5% sodium hypochlorite for 5 min (no sonication). Insects were then washed twice in nuclease-free water and blotted on a sterile paper towel in the laminar flow hood. Twenty heads from each sampling site (Table 1) were dissected from surface-sterilized adult females and macerated with sterile disposable plastic mini-pestles in 2 ml nuclease-free culture tubes containing 1 ml of nutrient broth (NB; Sigma-Aldrich, St. Louis, MO, United States). The reason for using heads is based on having observed an egg grooming behavior in which adult females smear recently oviposited eggs using her mouthparts (Vega et al., 2017), suggesting they may secrete a substance over the egg. Twenty eggs from each sampling site were cultured with or without surface-sterilization (Table 1). Eggs were sterilized for 1 min by placing in a mixture of 0.5% sodium hypochlorite and 70% ethanol. The sterilizing solution was drawn off with a pipette and replaced with nuclease-free water. Sterilized eggs were left whole or were macerated with sterile disposable plastic mini-pestles in 2 ml nuclease-free culture tubes containing 1 ml of nutrient broth (NB; Sigma-Aldrich, St. Louis, MO, United States). Whole non-sterilized eggs and female larvae (n = 10) from Mexico (Table 1) were directly transferred to NB (see below). Frass (Table 1) was collected with a sterilized metal spatula directly from the surface of artificial media from the Beltsville colony. The effectiveness of the sterilization protocol was assessed by placing a subsample of the nuclease-free water (used to wash the samples after removing the sterilizing solution) on the surface of nutrient agar (NA; Sigma-Aldrich, St. Louis, MO, United States) and observing whether any growth ensued, in which case the samples were discarded.
TABLE 1.
Bacterial identification based on Illumina sequence, with GenBank E-values for genes involved in the caffeine N-demethylation pathway (ndmA, ndmB, ndmC, ndmD, ndmE), coffee berry borer source used for bacterial isolation (eggs, frass, heads, larvae), non-macerated (whole) or macerated tissue, sterile (St) or non-sterile (NSt), sampling site, GenBank accession numbers, and average nucleotide identity (ANI; Chun et al., 2018) analysis.
| Isolate | Illumina Sequence ID | ndmA | ndmB | ndmC | ndmD | ndmE | Source | Whole/Macerated | St/NSt | Sampling Site | GenBank # | ANI |
| S40 | Acinetobacter sp. S40 | ndmC 2E-61 | ndmD 1E-62 | Eggs | Whole | St | Mexico | JACVTU000000000 | 84.19 | |||
| S54 | Acinetobacter sp. S54 | ndmC 2E-61 | ndmD 1E-62 | Eggs | Whole | NSt | Oahu, HI | JACVUI000000000 | 84.20 | |||
| S55 | Acinetobacter sp. S55 | ndmC 2E-61 | ndmD 1E-62 | Eggs | Whole | NSt | Oahu, HI | JACVUJ000000000 | 84.16 | |||
| S34 | Bacillus aryabhattai | ndmA 1E-50 | Eggs | Macerated | St | Mexico | JACVTQ000000000 | 99.53 | ||||
| S35 | Bacillus aryabhattai | ndmA 5E-51 | Frass | N/A | N/A | Beltsville, MD | JACVTR000000000 | 95.93 | ||||
| S65 | Delftia lacustris | ndmA 7E-48 | ndmD 2E-45 | ndmE 1E-40 | Heads | Macerated | St | K’au, HI | JACVUR000000000 | 95.08 | ||
| S66 | Delftia lacustris | ndmA 7E-48 | ndmD 2E-45 | ndmE 1E-40 | Heads | Macerated | St | K’au, HI | JACVUS000000000 | 95.10 | ||
| S67 | Delftia lacustris | ndmA 7E-48 | ndmD 2E-45 | ndmE 1E-40 | Heads | Macerated | St | K’au, HI | JACVUT000000000 | 94.97 | ||
| S38 | Erwinia sp. S38 | ndmA 1E-42 | ndmD 6E-57 | Eggs | Whole | St | Mexico | JACVTS000000000 | 79.12 | |||
| S43 | Erwinia sp. S43 | ndmA 1E-42 | ndmD 2E-59 | Larvae | Whole | NSt | Mexico | JACVTX000000000 | 79.07 | |||
| S63 | Erwinia sp. S63 | ndmD 1E-57 | Heads | Macerated | St | K’au, HI | JACVUP000000000 | 75.50 | ||||
| S69 | Klebsiella oxytoca | ndmA 1E-46 | ndmD 1E-53 | Heads | Macerated | St | Mexico | JACVUU000000000 | 99.10 | |||
| S45 | Ochrobactrum sp. S45 | ndmE 5E-43 | Eggs | Macerated | St | Mexico | JACVTZ000000000 | 83.82 | ||||
| S46 | Ochrobactrum sp. S46 | ndmE 5E-43 | Eggs | Macerated | St | Mexico | JACVUA000000000 | 83.99 | ||||
| S61 | Pantoea sp. S61 | ndmE 5E-40 | Heads | Macerated | St | Holualoa, HI | JACVUO000000000 | 86.23 | ||||
| S68 | Pseudomonas aeruginosa | ndmD 4E-69 | ndmE 6E-52 | Heads | Macerated | St | Mexico | JACVZF000000000 | 99.42 | |||
| S33 | Pseudomonas aeruginosa | ndmD 4E-69 | ndmE 6E-52 | Eggs | Macerated | St | Mexico | JACVTP000000000 | 99.44 | |||
| S36 | Pseudomonas parafulva | ndmA E0 | ndmB E0 | ndmC E0 | ndmD E0 | ndmE 7E-155 | Frass | N/A | N/A | Beltsville, MD | JACVVG000000000 | 98.05 |
| S30 | Pseudomonas sp. S30 | ndmE 8E-45 | Heads | Macerated | St | Beltsville, MD | JACVTO000000000 | 82.20 | ||||
| S31 | Pseudomonas sp. S31 | ndmA E0 | ndmB E0 | ndmC E0 | ndmD E0 | ndmE 4E-154 | Eggs | Whole | NSt | Mexico | JACVVE000000000 | 86.15 |
| S32 | Pseudomonas sp. S32 | ndmA E0 | ndmB E0 | ndmC E0 | ndmD E0 | ndmE 7E-155 | Eggs | Whole | St | Mexico | JACVVF000000000 | 84.66 |
| S37 | Pseudomonas sp. S37 | ndmA E0 | ndmB E0 | ndmC E0 | ndmD E0 | ndmE 7E-155 | Eggs | Whole | St | Mexico | JACVZC000000000 | 89.53 |
| S44 | Pseudomonas sp. S44 | ndmD 2E-65 | ndmE 2E-42 | Eggs | Whole | NSt | Mexico | JACVTY000000000 | 83.88 | |||
| S60 | Pseudomonas sp. S60 | ndmA E0 | ndmB E0 | ndmC E0 | ndmD E0 | ndmE 7E-155 | Heads | Macerated | St | Holualoa, HI | JACVVH000000000 | 84.78 |
| S75 | Pseudomonas sp. S75 | ndmE 8E-45 | Heads | Macerated | St | Beltsville, MD | JACVVA000000000 | 82.14 |
All insects were field collected, except “Beltsville, MD”, which were reared in the laboratory (see column labeled “Sampling Site”).
Samples (macerated sterile heads, whole sterile or non-sterile eggs, macerated sterile eggs, whole non-sterile larvae, and frass) were transferred to 4.5 ml sterile, nuclease-free culture tubes containing 2 ml NB and incubated at 28°C and 225 rpm on a rotary shaker for 48 h. The metagenomic (mixed taxa) NB cultures were archived at −80°C by combining 500 μl of each culture with 125 μl of 80% glycerol in nuclease-free deep-well plates. Ten microliters of each original culture were diluted in 50 μl of molecular-grade water (nuclease-free water) for polymerase chain reaction (PCR) screening analysis. The remainder of the 48 h NB cultures were used for dilution plating on NA (100 μl of each dilution on NA; 24 h at 28°C), to obtain single colony isolates. A total of 938 isolates were obtained: 378 from Mexico; 284 from the Beltsville colony; and 276 from Hawai’i. Single colony isolates were assigned a general morphotype based on macroscopic colony features, picked with a sterile toothpick, transferred to nuclease-free 96-well, deep-well plates containing 500 μl NB and incubated at 28°C and 225 rpm on a rotary shaker for 24 h. NB cultures of the single colony isolates were archived at −80°C (as described previously) in the Sustainable Agricultural Systems Laboratory Collection (USDA, ARS, Beltsville, MD, United States). Ten microliters of each original culture were diluted in 50 μl of nuclease-free water for PCR screening analysis.
PCR Screening of Single Colony Isolates for 16S rRNA and ndmA Genes
Single colony NB dilutions were used as template for PCR amplification of the16S rRNA gene and of the methylxanthine N1-demethylase gene, ndmA. Primers used for 16S rRNA amplification were 27F (5′- AGA GTT TGA TCM TGG CTC AG-3′) and 1392R (5′- ACG GGC GGT GTG TAC A-3′), and for ndmA were CBBcdmF (5′- TGG CAT CCC GTW TGT ACY GT-3′) and CBBcdmR (5′- CTT GKA TAA CRA TTC GCA ACC-3′) (Ceja-Navarro et al., 2015). Twenty-five microliter PCR reactions contained 3 μl of diluted NB isolate culture, 0.4 μM of each primer and 1X AmpliTaq Gold PCR Master Mix (6.25 U AmpliTaq Gold DNA polymerase, 2.5 mM MgCl2, and 200 μM of each dNTP; Applied Biosystems/Life Technologies, Carlsbad, CA, United States). PCR amplification was conducted in an Eppendorf Mastercycler Gradient thermal cycler (Eppendorf, Westbury, NY, United States). The amplification program consisted of an initial 5 min denaturation step at 95° C, followed by 35 cycles at 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min; and a final extension step at 72°C for 10 min. Following amplification, PCR products were electrophoresed on 1% agarose gels containing 1X TAE and 1X GelStar Nucleic Acid Gel Stain (Lonza Rockland Inc., Rockland, ME, United States), and documented using UV transillumination to detect 16S rRNA and ndmA bands.
Dye-Terminator Sequencing of 16S rRNA and ndmA Products
Polymerase chain reaction products from the 16S rRNA and ndmA amplifications were diluted 1:3 with nuclease-free water and direct-sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit according to the manufacturer’s protocol (Applied Biosystems/Life Technologies, Carlsbad, CA, United States). Separate sequencing reactions were run for the respective forward and reverse primers in each set (27F and 1392R, CBBcdmF, and CBBcdmR). Sequencing reactions were purified using the Performa DTR Ultra 96-Well Plate Kit (EdgeBio, Gaithersburg, MD, United States), resuspended in HiDi Formamide (Applied Biosystems/Life Technologies, Carlsbad, CA, United States) and analyzed on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems Inc., Foster City, CA, United States). Sequences were quality checked and aligned using the DNAStar suite of software (DNAStar, Madison, WI, United States), and preliminary identification made using the Basic Local Alignment Search Tool (BLAST) and GenBank nucleotide data bank from the National Center for Biotechnology Information, Bethesda, MD, United States1.
High Throughput Sequencing
Fifty representative single colony isolates were selected for Illumina genomic library preparation using a ranking based on at least one of the following: (1) preliminary 16S rRNA BLAST identification; (2) surface-sterilization or no surface-sterilization; (3) type of insect tissue cultured; (4) sampling site; (5) colony morphotype; (6) whole or macerated tissue; (7) presence of ndmA amplicon; or (8) BLAST identification of ndmA sequence. It is important to note that ndmA was not present in all the isolates when this selection was made, as our interest was focused at casting a wide net representative of as many characteristics as possible. Single colony isolates from glycerol stocks were grown overnight in NB at 28°C and 225 rpm. Cells were pelleted at 10,000 relative centrifugal force for 1 min, and genomic DNA extracted using the Qiagen DNeasy DNA extraction protocol for bacterial cultures (Qiagen, Germantown, MD, United States). Genomic DNA quality was assessed by nanodrop 260/280 ratio, and quantity was measured using a Qubit fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, United States). Libraries were prepared from 1 ng of genomic DNA from 50 coffee berry borer-associated single colony isolates using the Nextera XT DNA Library Prep Kit (Illumina, Inc., San Diego, CA, United States). The libraries were sequenced on a NextSeq by bi-directional multiplex-indexed Illumina sequencing (Nextera XT Index Kit, Illumina, Inc., San Diego, CA, United States) using a NextSeq 500 High Output Kit (150 cycles; Illumina, Inc., San Diego, CA, United States).
Bioinformatics
The quality of the reads was checked by the FASTQC program2. The 50 isolates (4 paired-end libraries for each isolate) were assembled using the SPAdes assembler v3.14.0 (-t 60, -m 500, –careful, -k 17,21,31,41,51) (Bankevich et al., 2012). The read coverage (read mapping back to the contigs) was calculated using Bowtie2 (Langmead and Salzberg, 2012) and the BBtools suite3. Gene prediction on the contigs was performed using the software program Prodigal v2.6.3 (Hyatt et al., 2010). The gene-encoded proteins were annotated by blasting against the refseq of bacterial proteins at the NCBI using the BLASTP program. Gene-encoded proteins were also blasted against known caffeine break-down genes: KAF4561170_ndmA, KAF4561167 ndmB, KAF4561164_ndmC, KAF4561163_ndmD, KAF4561162_ndmE, KAF4561168_cafR, KAF4561173_cafT, KAF4561166_cafP, KAF4561172 frmA, KAF4561161_frmB, KAF4561171_VOC where only high E-values close to E 0.0 were considered to be significant matches. 16S rRNAs were predicted using RNAmmer 1.2 and tRNAs were predicted by tRNAscan-1.4. Bacterial identification was based on the top BLAST hits with the 16S rRNA followed by an analysis of the average nucleotide identity (ANI; Chun et al., 2018; Table 1) between the draft genome generated in this study with the genome of the associated top BLAST. Values > 95% were used as the species boundary following proposed minimal standards by Chun et al. (2018). The Orthologous Average Nucleotide Identity Tool (OrthoANI; Lee et al., 2016; Chun et al., 2018) software was downloaded from4.
Analysis of Cell Metabolism
The 50 bacterial strains sequenced in this study were inoculated from frozen glycerol stocks into a modified M9 minimal medium containing, per liter, 12.8 g Na2HPO4⋅7H2O, 3 g K2HPO4, 2.76 g NH4Cl, 0.5 g NaCl, 493 mg MgSO4, 55.5 mg CaCl2, and 2.5 g caffeine (J. T. Baker, Avantor Performance Materials, Inc., Center Valley, PA, United States) or in nutrient broth (6 g/L peptone and 3 g/L yeast extract; VWR International LLC, Radnor, PA, United States) containing 2.5 g/L caffeine. Aliquots were removed periodically from the cultures and mixed 1:1 (vol:vol) with methanol to stop cell growth and metabolism. Concentrations of caffeine and metabolites were then determined by HPLC as described previously (Summers et al., 2012) using a Shimadzu Prominence series HPLC equipped with a photodiode array. Metabolites were identified based on their retention times and UV spectra when compared with authentic standards.
Results
Bacterial Associates
Assembly statistics for 50 bacterial strains in 14 genera (Acinetobacter, Bacillus, Delftia, Enterococcus, Erwinia, Klebsiella, Kosakonia, Lactococcus, Leuconostoc, Ochrobactrum, Paenibacillus, Pantoea, Pseudomonas, and Stenotrophomonas) isolated from coffee berry borer eggs, frass, head, and larvae are presented in Supplementary Table 2. Based on bioinformatics analysis, 21 bacterial species (Acinetobacter sp. S40, S54, S55, Bacillus aryabhattai, Delftia lacustris, Erwinia sp. S38, S43, S63, Klebsiella oxytoca, Ochrobactrum sp. S45, S46, Pantoea sp. S61, Pseudomonas aeruginosa, P. parafulva, and Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75) contain genes homologous to those involved in caffeine N-demethylation (Table 1); 35 bacterial species (Acinetobacter sp. S40, S54, S55, B. aryabhattai, B. cereus group; Bacillus sp. S29, S70, S71, S72, S73, D. lacustris, Erwinia sp. S38, S43, S59, S63, K. oxytoca, Kosakonia cowanii, Ochrobactrum sp. S45, S46, Paenibacillus sp. S28, Pantoea sp. S61, S62, P. aeruginosa, P. parafulva, Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75, Stenotrophomonas sp. S39, S41, S48, S49) contain genes that may be involved in C-8 oxidation (Supplementary Table 3); and nine bacterial species (Bacillus cereus group, Enterococcus sp. S52, S53, S76, S77, Lactococcus sp. S47, S64, and Leuconostoc sp. S50, S51) did not have any genes known for caffeine breakdown function (Supplementary Table 3).
Caffeine Demethylation Genes
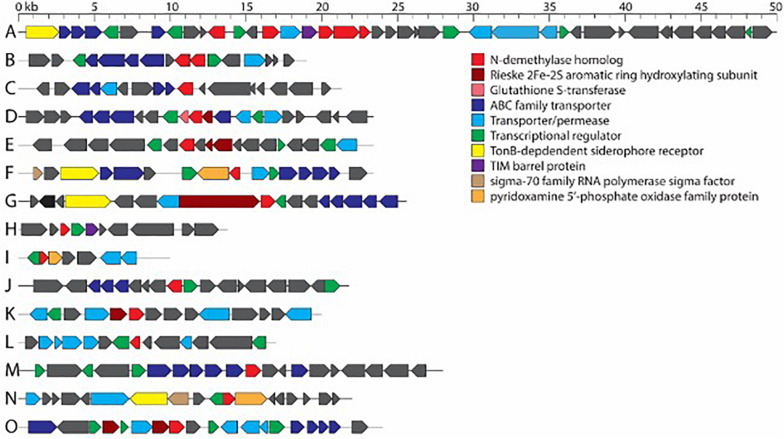
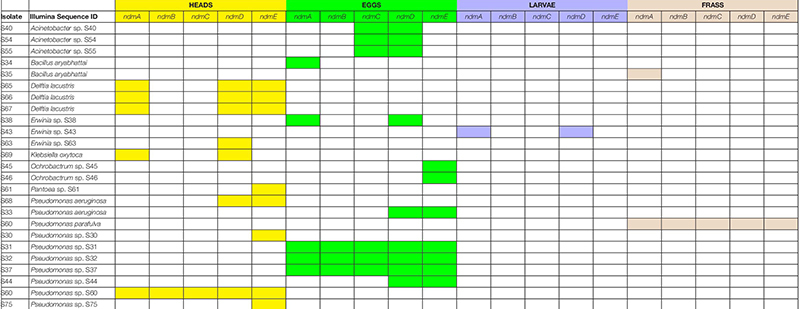
The full complement of caffeine N-demethylation genes (ndmABCDE) was detected in (1) P. parafulva isolated from coffee berry borer frass from Beltsville; (2) Pseudomonas sp. S60 isolated from macerated sterilized heads from Hawai’i; (3) Pseudomonas sp. S31 isolated from whole non-sterilized coffee berry borer eggs from Mexico; and (4) Pseudomonas sp. S32, S37 isolated from whole sterilized eggs from Mexico (Table 1). In each case, the ndmABCDE genes were found in a gene cluster (Figure 2), with gene arrangement identical to the alkylxanthine (Alx) gene cluster encoding N-demethylase enzymes and which has been reported in Pseudomonas sp. CES (Summers et al., 2020) and Pseudomonas sp. NCIM 5235 (Retnadhas and Gummadi, 2018). A summary for the specific insect tissue from which the 21 bacterial species involved in caffeine N-demethylation were isolated is presented in Table 2. Nineteen bacterial species were isolated from only one source, i.e., heads, eggs, larvae, or frass (Table 2). Two bacterial species were isolated from two different sources: B. aryabhattai (eggs and frass), and P. aeruginosa (heads and eggs) (Table 2).
FIGURE 2.
Consensus gene clusters containing N-demethylase genes from bacterial isolates in this work. (A) ndmABCDE cluster from Pseudomonas sp. S31, S32, S37, S60, and Pseudomonas parafulva. (B) ndmAD cluster from Erwinia sp. S38, S43. (C) ndmA cluster from Bacillus aryabhattai. (D) ndmC gene cluster from Acinetobacter sp. S40, S54, S55. (E) ndmD gene cluster from Acinetobacter sp. S40, S54, S55. (F) ndmE gene cluster from Ochrobactrum sp. S45, S46. (G) ndmD gene cluster from Pseudomonas aeruginosa. (H) ndmE gene cluster from Pseudomonas aeruginosa. (I) ndmE gene cluster from Pseudomonas sp. S30, S75. (J) ndmD gene cluster from Erwinia sp. S63. (K) ndmD gene cluster from Pseudomonas sp. S44. (L) ndmE gene cluster from Pseudomonas sp. S44. (M) ndmA gene cluster from Delftia lacustris. (N) ndmD gene cluster from D. lacustris. (O) ndmE gene cluster from D. lacustris. Genes in each cluster are listed in Supplementary Table 4.
TABLE 2.
Coffee berry borer source (heads, eggs, larvae, frass) in which bacteria with caffeine N-demethylation genes (ndmA, ndmB, ndmC, ndmD, ndmE) were detected.
|
Colored boxes across bacterial species indicate which genes were detected. Refer to Table 1 for more details on sampling sites, etc.
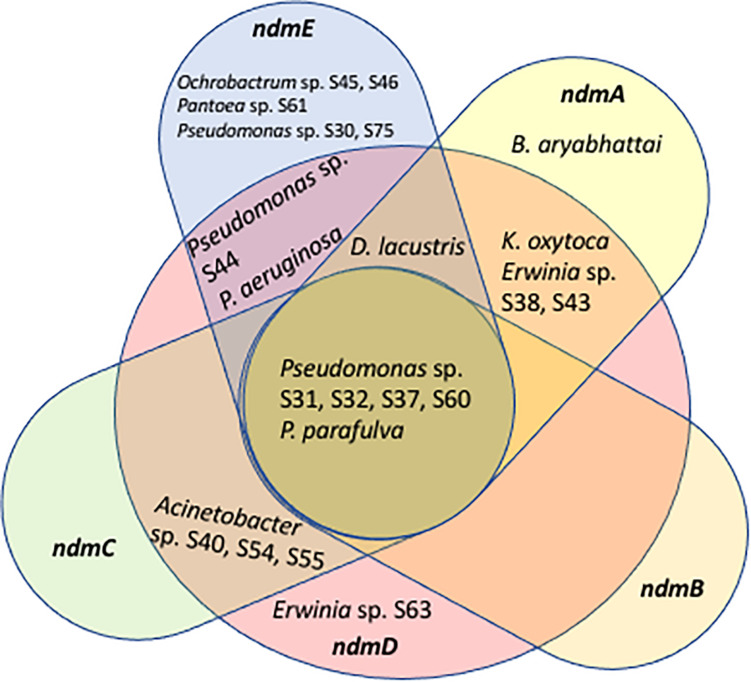
Some of the ndm genes were also detected in Acinetobacter sp. S40, S54, S55 (ndmCD), B. aryabhattai (ndmA), D. lacustris (ndmADE), Erwinia sp. S38, S43 (ndmAD), Erwinia sp. S63 (ndmD), K. oxytoca (ndmAD), Ochrobactrum sp. S45, S46 (ndmE), Pantoea sp. S61 (ndmE), P. aeruginosa (ndmDE), Pseudomonas sp. S30, S75 (ndmE), and Pseudomonas sp. S44 (ndmDE) (Table 1). A Venn diagram showing how the genes involved in the caffeine N-demethylation pathway are shared among the various bacterial species is shown in Figure 3.
FIGURE 3.
Venn diagram showing genes involved in the caffeine N-demethylation pathway shared among Acinetobacter sp. S40, S54, S55, Bacillus aryabhattai, Delftia lacustris, Erwinia sp. S38, S43, S63, Klebsiella oxytoca, Ochrobactrum sp. S45, S46, Pantoea sp. S61, Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75, P. aeruginosa, and P. parafulva.
Aside from ndmAD in Erwinia sp. S38, S43, which were found adjacent to each other (Figure 2), no other ndm gene homologs were in the same gene cluster. Rather, these genes were scattered throughout the genome (Figure 2). However, most of the homologs were surrounded by similar genes such as ABC family and other transporters, permeases, and GntR, LysR, and MarR transcriptional regulators (Figure 2; Supplementary Table 4).
While Pseudomonads are common caffeine degraders, two Acinetobacter sp. strains, a Klebsiella/Rhodococcus mixture, three Pantoea species, an Ochrobactrum sp., and S. maltophilia have been previously reported to degrade caffeine (Supplementary Table 1). Thus, the diversity observed here is not surprising but adds additional genera, such as Bacillus, Delftia, and Erwinia to the list of potential caffeine-degrading bacteria.
Analysis of Cell Metabolism
Of the 50 strains tested grown in M9 minimal media containing 0.25% caffeine as the sole carbon source to detect metabolites, only eight grew and produced sufficient metabolites to facilitate their detection by HPLC (Table 3). In each case, we detected theobromine and 7-methylxanthine in the growth medium, indicating that caffeine is metabolized by N-demethylation. The remaining 42 strains did not grow when caffeine was provided as the sole carbon and nitrogen source.
TABLE 3.
Strains with caffeine N-demethylation genes and/or metabolites isolated from coffee berry borers, coffee plantation soils, and coffee plants.
| Coffee berry borer (present paper) | Coffee berry borer (Ceja-Navarro et al., 2015) | Coffee plantation soils | Coffee plants |
| Pseudomonas aeruginosa (S68) | Brachybacterium rhamnosum | Acinetobacter sp. (Yamaoka-Yano and Mazzafera, 1998) | Pseudomonas sp. (Baker et al., 2012) |
| Pseudomonas parafulva | Enterobacter sp. | Coryneform bacterium (Yamaoka-Yano and Mazzafera, 1998) | P. monteilii (Arimurti et al., 2018) |
| Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75 | Jonesiae | Flavobacterium sp. (Yamaoka-Yano and Mazzafera, 1998) | P. putida (Nunes and de Melo, 2006) |
| Acinetobacter sp. S40*, S54*, S55* | Kosakonia cowanii | Pseudomonas sp.** (Dash and Gummadi, 2006) | |
| Bacillus aryabhattai* | Microbacterium binotii | Pseudomonas sp. (Gokulakrishnan et al., 2007) | |
| Delftia lacustris* | Novosphigobium sp. | Pseudomonas alcaligenes (Sarath Babu et al., 2005) | |
| Erwinia sp. S38*, S43*, S63* | Ochrobactrum sp. | Pseudomonas putida (Yamaoka-Yano and Mazzafera, 1998) | |
| Klebsiella oxytoca* | Pantoea eucalypti | Pseudomonas putida** (Yamaoka-Yano and Mazzafera, 1999) | |
| Ochrobactrum sp. S45*, S46* | Pantoea septica | Serratia marcescens** (Mazzafera et al., 1996) | |
| Pantoea sp. S61*, S62 | Pantoea vagans | ||
| Bacillus sp. S72, | Pseudomonas sp. | ||
| Stenotrophomonas sp. S39 | Pseudomonas fluorescens | ||
| Pseudomonas fulva | |||
| Stenotrophomonas maltophilia |
In column one, Pseudomonas sp. S32, S37, S60 and P. parafulva, had the full complement of N-demethylation genes (ndmABCDE) and caffeine breakdown metabolites were detected by HPLC; Pseudomonas sp. S31 had the full complement of N-demethylation genes but metabolites were not detected; an asterisk denotes that one, two, or three N-demethylation genes were identified, but no N-demethylation metabolites were detected, i.e., N-demethylation is not being expressed; no N-demethylation genes were detected in Bacillus sp. S72, Pantoea sp. S62, and Stenotrophomonas sp. S39, but metabolites were detected; ndmD and ndmE, as well as N-demethylation metabolites, were detected in Pseudomonas aeruginosa. Of the 50 strains isolated from the coffee berry borer, eight listed in bold in the first column were the only ones capable of growing in M9 minimal media containing 0.25% caffeine as the sole carbon source. Among all the bacteria listed in the second column, N-demethylation was only positively determined for P. fulva (Ceja-Navarro et al., 2015). Two asterisks in the third column indicates N-demethylation; otherwise, the caffeine breakdown mechanism was not determined. The caffeine breakdown mechanism was not determined for bacteria isolated from coffee plants (fourth column).
Discussion
Distribution of Caffeine-Degrading Genes
The presence of the full complement of ndm genes in several Pseudomonads was expected, given that N-demethylation is the most commonly observed caffeine degradation mechanism in bacteria (Supplementary Table 1) and that Pseudomonas is the most common caffeine-degrading genus worldwide (Supplementary Table 1). In contrast to these strains, only a subset of the genes was found in several bacterial strains. It is possible that the bacterial cells use other undiscovered genes and enzymes for caffeine metabolism or that the similarity of the homologous genes is lower than our cutoff threshold. To minimize false positives, E-values close to E 0.0 with high sequence similarity to known ndm genes were considered, since ndm genes have sequence similarity to proteins that might not be involved in caffeine breakdown. Similarly, the portion of the genome containing the relevant ndm and caffeine breakdown genes in these strains may not have been sequenced due to what Baptista and Kissinger (2019) refer to as “troublesome characteristics”, including extreme nucleotide bias, long homopolymeric runs, repetitive sequences, mobile elements, etc.
Another possibility for the low number of strains with a full complement of ndm genes (Figure 3) is that some strains may be able to carry out one step of the caffeine degradation pathway and rely on other strains to carry out additional steps. For example, the D. lacustris, Erwinia sp. S38, S43, and K. oxytoca strains contain ndmAD homologs, which may enable them to convert caffeine to the less-toxic theobromine. Acinetobacter sp. S40, S54, S55 contains ndmCD genes, which could potentially convert 7-methylxanthine to xanthine, which is formed during the biosynthesis of caffeine in coffee plants (Ashihara et al., 1996). Therefore, drivers for bacteria to carry out individual steps in the process may be to reduce toxicity of caffeine or to use metabolites already present, such as theobromine or 7-methylxanthine. Curiously, none of the strains with only a subset of genes contain ndmB homologs. However, given that as few as two mutations to the ndmA active site are enough to change the activity from N1- to N3-demethylation (Kim et al., 2019) some of the genes identified as ndmA may have a broader positional specificity than those which have already been characterized. Alternatively, there may be unculturable bacteria with ndmB homologs that we were not able to detect using the approach described here. NdmE is a glutathione-S-transferase homolog and is believed to play a structural role in the NdmCDE complex; there is no known catalytic role for NdmE (Summers et al., 2013). Thus, strains that only contain ndmE, such as Ochrobactrum sp. S45, S46 and Pantoea sp. S61, may not have any N-demethylation activity but may have a ndmE homolog that carries out a different function in the cell.
We note that the presence of ndm gene homologs does not indicate that the strains can completely degrade caffeine, or even that those genes are involved in caffeine metabolism. However, there are no other reports of N-demethylase enzymes capable of degrading caffeine and the NdmABCDE enzymes have only exhibited activity toward caffeine and related methylxanthines. These enzymes are part of the Rieske non-heme iron oxygenase family, which contains enzymes responsible for degradation of aromatic compounds toluene, naphthalene, biphenyl, and the herbicide dicamba (Gibson and Parales, 2000). Thus, there may be other aromatic compounds toward which these gene products are active. Regardless, the presence of ndm gene homologs does give insight into possible mechanisms and provides the basis for further studies to elucidate caffeine degradation by bacteria.
In each of the eight strains grown in M9 minimal media with 0.25% caffeine as the sole carbon and nitrogen source (Table 3) and which produced sufficient metabolites for HPLC detection, we detected the production of theobromine and 7-methylxanthine, indicating an active N-demethylation pathway. The P. parafulva and Pseudomonas sp. S32, S37, S60 containing the full set of ndm genes were found among this group. The other four strains that grew were Bacillus sp. S72, Pantoea sp. S62, and Stenotrophomonas sp. S39 among which no ndm genes were detected, and P. aeruginosa, in which only ndmD and ndmE were present. The ability of these strains to carry out N-demethylation of caffeine indicates that they may possess different genes encoding enzymes of similar function, the ndm genes were simply incompletely sequenced or not sequenced (Baptista and Kissinger, 2019), or that the ndm gene E-values did not meet our cutoff threshold. To date, no other genes/enzymes have been shown to carry out N-demethylation of caffeine or other methylxanthines in bacteria.
The 42 strains that did not grow when caffeine was provided as the sole carbon and nitrogen source may not have all of the genes necessary to completely degrade caffeine. Another explanation is that the genes were not induced under the growth conditions. It may be possible for strains with ndmAD gene combinations to convert caffeine to theobromine, but the single methyl group removed would likely not enable cells to grow. Additionally, very little is known about the regulation of the caffeine-degrading genes. Studies have indicated that these genes are regulated very tightly and only expressed under certain conditions. For example, P. putida CBB5 can express the ndm genes when grown with 0.25% caffeine and 0.4% soytone, but expression is greatly reduced when soytone is replaced with yeast nitrogen base (YNB) (Yu et al., 2009). In contrast, the caffeine dehydrogenase expression in Pseudomonas sp. CBB1 is not inhibited by YNB but is almost completely repressed by soytone (Yu et al., 2008). Thus, the absence of metabolites does not indicate a lack of genes, but observation of caffeine metabolites does provide evidence for specific enzymes. Indeed, we did not observe any caffeine degradation from any of the 50 strains when grown in nutrient broth supplemented with 0.25% caffeine, indicating that there are specific circumstances in which the ndm genes are expressed.
We also analyzed caffeine dehydrogenase genes (cdhA, cdhB, cdhC) involved in C-8 oxidation, the second pathway of caffeine break down (Figure 1; Supplementary Table 3). The C-8 oxidation pathway involves the oxidation of caffeine to 1,3,7-trimethyluric acid (TMU; Figure 1; Supplementary Table 3), which is further degraded in a pathway homologous to uric acid metabolism. The caffeine dehydrogenase enzyme, encoded by genes cdhABC, carries out the first step in this pathway (Yu et al., 2008). Further steps are catalyzed by trimethyluric acid-degrading genes (tmuM, tmuH, tmuD) (Mohanty et al., 2012). The peptide sequence of the cdhA gene, which forms one subunit of the heterotrimeric caffeine dehydrogenase is similar to xanthine dehydrogenases, aldehyde dehydrogenases, and carbon monoxide dehydrogenases (Yu et al., 2008). Similarly, the tmuM, tmuH, and tmuD genes have homologs involved in uric acid metabolism. It is possible that the genes listed in Supplementary Table 3 are instead involved in xanthine and uric acid metabolism. However, xanthine dehydrogenase is not known to metabolize caffeine, nor have the tmu genes shown activity toward trimethyluric acid. To date, we do not have experimental evidence of TMU formation from caffeine by any of these strains, indicating that either the homologs we detected do not metabolize caffeine or that the conditions for their expression have not yet been identified. Cloning and expression of the genes and characterization of the enzymes they encode would be necessary to determine if these genes are specific for caffeine.
For strains such as Ochrobactrum sp. S45, S46, Pantoea sp. S61, and Pseudomonas sp. S30, S75 that have ndmE and cdh homologs, it is more likely that they metabolize caffeine through the C-8 oxidative route rather than the N-demethylation route because NdmE has no known catalytic activity. Two B. aryabhattai strains contain the full set of C-8 oxidative genes, but only the ndmA gene from the N-demethylation pathway. Because this strain does not contain an ndmD homolog necessary for N-demethylation, it would most likely use the oxidative route, as well.
Possible involvement of the cdh and tmu genes (Supplementary Table 3) in the breakdown of caffeine and subsequent products needs to be experimentally determined to ascertain whether this role could be occurring in the coffee berry borer. It is interesting that ndm genes were not identified in K. cowanii, although it has CdhABC and TmuMD; Ceja-Navarro et al. (2015) identified K. cowanii as a caffeine-degrading bacterium in the coffee berry borer.
It is also important to note that there might be other unculturable bacteria that are capable of degrading caffeine, although that was not the focus of this paper.
Pseudomonas and Delftia in the Coffee Berry Borer
The genus Pseudomonas comprises ca. 150 species (Pereira et al., 2018) and in the present study, Pseudomonas aeruginosa, P. parafulva, and Pseudomonas sp. S30, S31, S32, S37, S44, S60, S75 were found to have caffeine N-demethylation genes, with five of them having the full complement of caffeine demethylating genes (ndmABCDE; Tables 1, 2 and Figure 3): P. parafulva (frass), Pseudomonas sp. S60 (heads), and Pseudomonas sp. S31, S32, S37 (eggs). Pseudomonas aeruginosa had ndmDE (heads, eggs) and Pseudomonas sp. S30, S75 only had ndmE (heads) (Tables 1, 2 and Figure 3).
Mariño et al. (2018) reported 392 bacterial genera in the microbiome of eggs and whole adult female coffee berry borers, with a preponderance of Pseudomonas and Pantoea. Pseudomonas spp. represented 48.5% of the microbiota, with P. chloroaphis being common in laboratory-reared insects and P. putida being more common in field-collected insects. Two other Pseudomonas species identified in the microbiome were P. viridiflava and P. fulva.
This is the first report of potential caffeine N-demethylation genes in D. lacustris (ndmADE). Intriguingly, as for Pseudomonas sp. S30, S60, S75, the detection was made from sterilized, macerated heads. Delftia has been isolated from the coffee berry borer by Mariño et al. (2018).
Possible Modes of Bacterial Transmission
Transmission of a symbiote from the mother to its offspring is known as vertical transmission (Jaenike, 2012; Ebert, 2013; Kucuk, 2020) and could be mediated internally or externally by the ovum and referred to as transovum transmission (Solter, 2006). One type of transovum transmission is transovarial transmission involving the internal invasion of the egg by the symbiote, as has been reported for many insects (Zchori-Fein et al., 2006; Skaljac et al., 2010). We detected eight bacteria with N-demethylation genes in sterile eggs: Acinetobacter sp. S40 (ndmCD), B. aryabhattai (ndmA), Erwinia sp. S38 (ndmAD), Ochrobactrum sp. S45, S46 (ndmE), P. aeruginosa (ndmDE), and Pseudomonas sp. S32, S37 (ndmABCDE) (Table 1). It is unlikely that Acinetobacter sp. S40, B. aryabhattai, Erwinia sp. S38, and Ochrobactrum sp. S45, S46 are transovarially transmitted and detection might have been a result of contamination or imperfect sterilization. Transovarial transmission would be rather surprising for these multiple free-living bacteria which readily grow on standard cultivation media and are otherwise not consistently associated with the host. Using fluorescent in situ hybridization, Sayaka Aoki (University of Hawai’i; personal communication) has detected Pseudomonas spp. with at least one caffeine degrading gene inside coffee berry borer eggs.
The other type of ovum mediated transmission involves inoculation of the egg shell via frass or secretions containing the symbiote, which is ingested upon egg hatching (Hosokawa et al., 2012; Salem et al., 2015b). As stated above, we have observed an egg grooming behavior in which female coffee berry borers appear to smear her eggs shortly after oviposition (Vega et al., 2017). A similar situation has been reported in Adomerus stinkbugs, which smear a secretion containing a symbiote over the eggs (Hosokawa et al., 2013). The detection of ndmABCDE in Pseudomonas sp. S60 isolated from macerated sterilized heads (Table 1) could be a mechanism for vertical transmission if the hatching embryos acquire the bacterial symbiote by eating the egg shell or becoming otherwise contaminated with the symbiote. This possible type of vertical transmission needs further study.
Coffee berry borer larvae could acquire Acinetobacter sp. S54, S55 (ndmCD) and Pseudomonas sp. S31 (ndmABCDE), which were isolated from non-sterile eggs (Table 1), from the egg shell as the larvae hatches. These bacteria could also contaminate the entire environment within the coffee berry. The coffee berry borer rectum ends with the anus, which is an opening independent of the genital orifice (gonopore) (Alba-Alejandre et al., 2019). Both orifices are very close to each other, with the anal opening being above the genital orifice. Due to the large size of coffee berry borer eggs, it is very likely that as they are being oviposited they will place pressure on the rectum, which could expel fecal matter that would then contaminate the egg shell.
In addition, the presence of ndmABCDE in P. parafulva and ndmA in B. aryabhattai isolated from coffee berry borer frass from the Beltsville colony (Table 1) suggests that potential caffeine demethylating bacteria could contaminate the egg shell or be acquired from the environment (i.e., horizontal transmission) via bacterial dissemination from frass through insect contact, probing, and subsequent spread in galleries. The acquisition of bacteria through coprophagy (Salem et al., 2015a; Onchuru et al., 2018) cannot be ruled out for the coffee berry borer. Erwinia sp. S43 (ndmAD) was detected in non-sterile larvae from Mexico (Table 1), suggesting they could be acquiring the bacterium inside the gallery. Horizontal transmission of caffeine-degrading bacteria also needs further study.
Caffeine-Degrading Bacteria in Soil and Plants
Caffeine-degrading functions might also be expressed by bacteria isolated from soil in coffee plantations, as has been shown for Pseudomonas sp. (Dash and Gummadi, 2006; Gokulakrishnan et al., 2007), P. alcaligenes (Sarath Babu et al., 2005), P. putida (Yamaoka-Yano and Mazzafera, 1998, 1999), and Serratia marcescens (Mazzafera et al., 1996; Supplementary Table 1). In addition, endophytic bacteria in coffee plants might also be capable of degrading caffeine. Vega et al. (2005) identified 19 bacterial genera as endophytes in coffee plants from Colombia, Hawai’i, and Mexico, with Bacillus, Burkholderia, Clavibacter, Curtobacterium, Escherichia, Micrococcus, Pantoea, Pseudomonas, Serratia, and Stenotrophomonas isolated from the seed. Pseudomonas sp., P. chloroaphis, and P. putida were identified as endophytes in various coffee plant tissues sampled (Vega et al., 2005) and P. monteilii has been isolated from coffee pulp waste (Arimurti et al., 2018). A coffee plant endophytic Pseudomonas sp. isolated in India was capable of breaking down 98.6% of the caffeine added to a nutrient broth, and six additional unidentified endophytic bacteria were also capable of catabolizing caffeine with a lower efficiency (Baker et al., 2012). Two P. putida strains endophytic in C. arabica and Coffea canephora leaves, stems, and roots in Brazil were capable of degrading caffeine (Nunes and de Melo, 2006).
For the coffee berry borer, the presence of bacteria on the surface of the fruit, or endophytically in the pulp or seed (Vega et al., 2005), might result in the acquisition of caffeine-degrading bacteria as the colonizing female infests fruit and seeds. A similar situation has been reported for 13 bark beetle species in the genus Dendroctonus, which acquire bacterial members of their microbiome from the environment, including their host trees (Hernández-García et al., 2017).
Conclusion
The detection of genes involved in the two mechanisms for bacterial metabolism of caffeine, i.e., N-demethylation and C-8 oxidation, has revealed the complexity of culturable bacterial symbiotes of the coffee berry borer. The results could serve as a basis for in-depth studies aimed at determining if these bacteria become established in the alimentary canal of the insect and whether their elimination reduces insect fitness, as has been shown for P. fulva (Ceja-Navarro et al., 2015). In addition, the study serves as a first attempt to determine possible mechanisms for vertical and horizontal transmission of some of the genes involved in N-demethylation. The isolation and identification of culturable bacteria for an assessment of caffeine-breakdown capabilities would be useful in understanding the coffee berry borer microbial diversity in various coffee-growing regions as well as various coffee- growing regimes such as Arabica vs robusta and shaded coffee vs coffee grown at full sun.
The bacteria involved in the N-demethylation metabolic pathway for caffeine degradation in the coffee berry borer (Tables 1, 2 and Figure 3) likely originated as epiphytes and endophytes in the coffee plant, as a caffeine-degrading mechanism is necessary in order for the bacteria to thrive on coffee. Vega et al. (2005) identified 19 genera of endophytic bacteria in coffee plants from Colombia, Hawai’i, and Mexico, including genera potentially capable of breaking down caffeine, e.g., Enterobacter, Klebsiella, Pantoea, Pseudomonas, Serratia, and Stenotrophomonas (Table 3). Many of the bacteria associated with coffee plants might be coffee plant mutualists, deriving nutrition as well as a substrate from the plant and providing caffeine-degrading functions on leaves and fruits falling on the soil, and within the soil itself, thus recycling nutrients (Kennedy, 1999) and possibly lowering caffeine concentrations in the soil. Eight bacterial species isolated from soil in coffee plantations are capable of breaking down caffeine (Table 3). The bacterial composition in coffee fruits, leaves and roots needs to be identified and compared to the bacterial community in the soil, as well as in the coffee berry borer, in order to understand shared bacterial composition and functional redundancy (Kennedy, 1999; Nannipieri et al., 2017) in terms of caffeine-degrading genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA659418.
Author Contributions
FEV, SE, AS, and JEM conceived and designed the coffee berry borer experiments. FEV, SE, and AS performed the coffee berry borer experiments. MBM and RMS conceived, designed, and performed the experiments related to caffeine metabolites. JS performed the bioinformatics analysis. FEV, SE, JS, RMS, and MBM analyzed the data. All authors interpreted the results and provided significant comments and inputs to the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Jose “Pepe” Miranda for providing coffee berry borer-infested coffee berries from K’au, Hawai’i; Mónica Santín-Durán, Jo Ann Van Kessel, and Seon-Woo Kim (USDA, ARS) for sequencing support; and Javier Alba-Tercedor (University of Granada, Spain) and Meredith Blackwell (University of South Carolina) for answering questions relevant to this article.
Funding. This study was supported by the United States Department of Agriculture, Agricultural Research Service Project Number 8042-22000-287-00D and by University of Alabama research funds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.644768/full#supplementary-material
At least 71 bacterial strains in 27 genera have been reported to be capable of caffeine breakdown, with Pseudomonas spp. being the most common.
Assembly statistics for 50 bacterial strains isolated from the coffee berry borer.
Bacteria isolated from the coffee berry borer, indicating which strains might have C-8 oxidation activity (cdhA, cdhB, cdhC, tmuM, tmuH, tmuD), coffee berry borer source used for bacterial isolation (eggs, frass, heads, larvae), non-macerated (whole) or macerated tissue, sterile (St) or non-sterile (NSt), sampling site, GenBank accession numbers, and average nucleotide identity (ANI; Chun et al., 2018) analysis. All insects were field collected, except “Beltsville, MD”, which were reared in the laboratory (see column labeled “Sampling Site”).
Arrangement of gene clusters.
References
- Alba-Alejandre I., Alba-Tercedor J., Vega F. E. (2019). Anatomical study of the coffee berry borer (Hypothenemus hampei) using micro-computed tomography. Sci. Rep. 9:17150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimurti S., Ardyati T., Nurani Y., Siswoyo T. A., Suharjono S. (2018). Degradation of caffeine by Pseudomonas monteilii KRM9. Malays. J. Microbiol. 14 55–60. [Google Scholar]
- Ashihara H., Crozier A. (1999). Biosynthesis and metabolism of caffeine and related purine alkaloids in plants. Adv. Bot. Res. 30 117–205. 10.1016/s0065-2296(08)60228-1 [DOI] [Google Scholar]
- Ashihara H., Mizuno K., Yokota T., Crozier A. (2017). “Xanthine alkaloids: occurrence, biosynthesis, and function in plants,” in Progress in the Chemistry of Organic Natural Products, Vol. 105 eds Kinghorn A. D., Falk H., Gibbons S., Koyabashi J. (Cham: Springer; ), 1–88. 10.1007/978-3-319-49712-9_1 [DOI] [PubMed] [Google Scholar]
- Ashihara H., Monteiro A. M., Gillies F. M., Crozier A. (1996). Biosynthesis of caffeine in leaves of coffee. Plant Physiol. 111 747–753. 10.1104/pp.111.3.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Sahana S., Rakshith D., Kavitha H. U., Kavitha K. S., Satish S. (2012). Biodecaffeination by endophytic Pseudomonas sp. isolated from Coffea arabica L. J. Pharm. Res. 5 3654–3657. [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista R. P., Kissinger J. C. (2019). Is reliance in an inaccurate genome sequence sabotaging your experiments? PLoS Pathog. 15:e1007901. 10.1371/journal.ppat.1007901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti S., Daria P., Sani G., Aromatario M. (2015). Caffeine: cognitive and physical performance enhancer or psychoactive drug? Curr. Neuropharmacol. 13 71–88. 10.2174/1570159x13666141210215655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceja-Navarro J. A., Vega F. E., Karaoz U., Hao Z., Jenkins S., Lim H. C., et al. (2015). Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 6:7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Oren A., Ventosa A., Christensen H., Ruiz Arahal D., da Costa M. S.,, et al. (2018). Proposed minimal standards for the use of genome data for taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Dash S. S., Gummadi S. N. (2006). Biodegradation of caffeine by Pseudomonas sp. NCIM 5235. Res. J. Microbiol. 1 115–123. 10.3923/jm.2006.115.123 [DOI] [Google Scholar]
- Davis A. P., Govaerts R., Bridson D. M., Stoffelen P. (2006). An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot. J. Linn. Soc. 152 465–512. 10.1111/j.1095-8339.2006.00584.x [DOI] [Google Scholar]
- Davis A. P., Tosh J., Ruch N., Fay M. F. (2011). Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data: implications for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linn. Soc. 167 357–377. 10.1111/j.1095-8339.2011.01177.x [DOI] [Google Scholar]
- Ebert D. (2013). The epidemiology and evolution of symbionts with mixed-mode transmission. Annu. Rev. Ecol. Evol. Syst. 44 623–643. 10.1146/annurev-ecolsys-032513-100555 [DOI] [Google Scholar]
- Gibson D. T., Parales R. E. (2000). Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11 236–243. 10.1016/s0958-1669(00)00090-2 [DOI] [PubMed] [Google Scholar]
- Gokulakrishnan S., Chandraraj K., Gummadi S. N. (2007). A preliminary study of caffeine degradation by Pseudomonas sp. GSC 1182. Int. J. Food Microbiol. 113 346–350. 10.1016/j.ijfoodmicro.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Hernández-García J. A., Briones-Roblero C. I., Rivera-Orduña F. N., Zuñiga G. (2017). Revealing the gut bacteriome of Dendroctonus bark beetles (Curculionidae: Scolytinae): diversity, core members and co-evolutionary patterns. Sci. Rep. 7:13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T., Hironaka M., Mukai H., Inadomi K., Suzuki N., Fukatsu T. (2012). Mothers never miss the moment: a fine-tuned mechanism for vertical symbiont transmission in a subsocial insect. Anim. Behav. 83 293–300. 10.1016/j.anbehav.2011.11.006 [DOI] [Google Scholar]
- Hosokawa T., Hironaka M., Mukai H., Inadomi K., Suzuki N., Fukatsu T. (2013). Diverse strategies for vertical symbiont transmission among subsocial stinkbugs. PLoS One 8:e65081. 10.1371/journal.pone.0065081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J. (2012). Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 27 226–232. 10.1016/j.tree.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Kennedy A. C. (1999). Bacterial diversity in agroecosystems. Agric. Ecosyst. Environ. 74 65–76. 10.1016/b978-0-444-50019-9.50007-8 [DOI] [Google Scholar]
- Kim J. H., Kim B. H., Brooks S., Kang S. Y., Summers R. M., Song H. K. (2019). Structural and mechanistic insights into caffeine degradation by the bacterial N-demethylase complex. J. Mol. Biol. 431 3647–3661. [DOI] [PubMed] [Google Scholar]
- Kim Y.-S., Sano H. (2008). Pathogen resistance of transgenic tobacco plants producing caffeine. Phytochemistry 69 882–888. 10.1016/j.phytochem.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Kim Y.-S., Uefuji H., Ogita S., Sano H. (2006). Transgenic tobacco plants producing caffeine: a potential new strategy for insect pest control. Transgenic Res. 15 667–672. 10.1007/s11248-006-9006-6 [DOI] [PubMed] [Google Scholar]
- Kucuk R. A. (2020). Gut bacteria in the Holometabola: a review of obligate and facultative symbionts. J. Insect Sci. 20:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Kim Y. O., Park S.-C., Chun J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66 1100–1103. [DOI] [PubMed] [Google Scholar]
- Levinson H. Z. (1976). The defensive role of alkaloids in insects and plants. Experientia 32 408–411. 10.1007/bf01920763 [DOI] [Google Scholar]
- Mariño Y. A., Ospina O. E., Verle Rodrigues J. C., Bayman P. (2018). High diversity and variability in the bacterial microbiota of the coffee berry borer (Coleoptera: Curculionidae), with emphasis on Wolbachia. J. Appl. Microbiol. 125 528–543. 10.1111/jam.13768 [DOI] [PubMed] [Google Scholar]
- Mazzafera P., Olsson O., Sandberg G. (1996). Degradation of caffeine and related methylxanthines isolated from soil under coffee cultivation. Microb. Ecol. 31 199–207. [DOI] [PubMed] [Google Scholar]
- Mohanty S. K., Yu C. L., Das S., Louie T. M., Gakhar L., Subramanian M. (2012). Delineation of the caffeine C-8 oxidation pathway in Pseudomonas sp. strain CBB1 via characterization of a new trimethyluric acid monooxygenase and genes involved in trimethyluric acid metabolism. J. Bacteriol. 194 3872–3882. 10.1128/jb.00597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannipieri P., Ascher J., Ceccherini M. T., Landi L., Pietramellara G., Renella G. (2017). Microbial diversity and soil functions. Eur. J. Soil Sci. 68 12–26. [Google Scholar]
- Nathanson J. (1984). Caffeine and related methylxanthines: possible naturally occurring pesticides. Science 226 184–187. 10.1126/science.6207592 [DOI] [PubMed] [Google Scholar]
- Nunes F. V., de Melo I. S. (2006). Isolation and characterization of endophytic bacteria of coffee plants and their potential in caffeine degradation. WIT Trans. Biomed. Health 10 293–297. [Google Scholar]
- Oliveira C. M., Auad A. M., Mendes S. M., Frizzas M. R. (2013). Economic impact of exotic insect pests in Brazilian agriculture. J. Appl. Entomol. 137 1–15. 10.1111/jen.12018 [DOI] [Google Scholar]
- Onchuru T. O., Martinez A. J., Ingham C. S., Kaltenpoth M. (2018). Transmission of mutualistic bacteria in social and gregarious insects. Curr. Opin. Insect Sci. 2 50–58. 10.1016/j.cois.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Pereira R. P. A., Peplies J., Mushi D., Brettar I., Höfle M. G. (2018). Pseudomonas-specific NGS assay provides insight into abundance and dynamics of Pseudomonas species including P. aeruginosa in a cooling tower. Front. Microbiol. 9:1958. 10.3389/fmicb.2018.01958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retnadhas S., Gummadi S. N. (2018). Identification and characterization of oxidoreductase component (NdmD) of methylxanthine oxygenase system in Pseudomonas sp. NCIM 5235. Appl. Microbiol. Biotechnol. 102 7913–7926. 10.1007/s00253-018-9224-x [DOI] [PubMed] [Google Scholar]
- Salem H., Florez L., Gerardo N., Kaltenpoth M. (2015a). An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. R. Soc. B 282:20142957. 10.1098/rspb.2014.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H., Onchuru T. O., Bauer E., Kaltenpoth M. (2015b). Symbiont transmission entails the risk of parasite infection. Biol. Lett. 11:20150840. 10.1098/rsbl.2015.0840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Kim Y.-S., Choi Y.-E. (2013). Like cures like: caffeine immunizes plants against biotic stress. Adv. Bot. Res. 68 273–300. [Google Scholar]
- Sarath Babu V. R., Patra S., Thakur M. S., Karanth N. G., Varadaraj M. C. (2005). Degradation of caffeine by Pseudomonas alcaligenes CFR 1708. Enzyme Microb. Technol. 37 617–624. 10.1016/j.enzmictec.2005.03.022 [DOI] [Google Scholar]
- Skaljac M., Zanic K., Goreta Ban S., Kontseladov S., Ghanim M. (2010). Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol. 10:142. 10.1186/1471-2180-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter L. F. (2006). Transmission as a predictor of ecological host specificity with a focus on vertical transmission of microsporidia. J. Invertebr. Pathol. 92 132–140. 10.1016/j.jip.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Summers R., Shao J., Mock M. B., Yu C. L., Vega F. E. (2020). Draft genome of Pseudomonas sp. strain CES, containing the entire alkylxanthine gene cluster for caffeine breakdown. Microbiol. Resour. Announc. 9:e00484-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers R. M., Louie T. M., Yu C. L., Gakhar L., Louie K. C., Subramanian M. (2012). Novel, highly specific N-demethylases enable bacteria to live on caffeine and related purine alkaloids. J. Bacteriol. 194 2041–2049. 10.1128/jb.06637-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers R. M., Mohanty S. K., Gopishetty S., Subramanian M. (2015). Genetic characterization of caffeine degradation by bacteria and its potential applications. Microb. Biotechnol. 8 369–378. 10.1111/1751-7915.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers R. M., Seffernick J. L., Quandt E. M., Yu C. L., Barrick J. E., Subramanian M. V. (2013). Caffeine junkie: an unprecedented glutathione S-transferase-dependent oxygenase required for caffeine degradation by Pseudomonas putida CBB5. J. Bacteriol. 195 3933–3939. 10.1128/jb.00585-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F. E., Biedermann P. H. W. (2020). On interactions, associations, mycetangia, mutualists and symbiotes in insect-fungus symbioses. Fungal Ecol. 44:100909. 10.1016/j.funeco.2019.100909 [DOI] [Google Scholar]
- Vega F. E., Blackburn M. B., Kurtzman C. P., Dowd P. F. (2003). Identification of a coffee berry borer-associated yeast: does it break down caffeine? Entomol. Exp. Appl. 107 19–24. 10.1046/j.1570-7458.2003.00034.x [DOI] [Google Scholar]
- Vega F. E., Infante F., Johnson A. J. (2015). “The genus Hypothenemus, with emphasis on H. hampei, the coffee berry borer,” in Bark Beetles: Biology and Ecology of Native and Invasive Species, eds Vega F. E., Hofstetter R. W. (San Diego, CA: Academic Press; ), 427–494. 10.1016/b978-0-12-417156-5.00011-3 [DOI] [Google Scholar]
- Vega F. E., Pava-Ripoll M., Posada F., Buyer J. S. (2005). Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45 371–380. [DOI] [PubMed] [Google Scholar]
- Vega F. E., Simpkins A., Rodríguez-Soto M. M., Infante F., Biedermann P. H. W. (2017). Artificial diet sandwich reveals subsocial behavior in the coffee berry borer Hypothenemus hampei (Coleoptera: Curculionidae: Scolytinae). J. Appl. Entomol. 141 470–476. 10.1111/jen.12362 [DOI] [Google Scholar]
- Waller G. R., Kumari D., Friedman J., Friedman N., Chou C.-H. (1986). “Caffeine autotoxicity in Coffea arabica L.,” in The Science of Allelopathy, eds Putnam A. R., Tang C.-S. (New York, NY: John Wiley & Sons; ), 243–269. [Google Scholar]
- Wright G. A., Baker D. D., Palmer M. J., Stabler D., Mustard J. A., Power E. F., et al. (2013). Caffeine in floral nectar enhances a pollinator’s memory of reward. Science 339 1202–1204. 10.1126/science.1228806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka-Yano D. M., Mazzafera P. (1998). Degradation of caffeine by Pseudomonas putida isolated from soil under coffee cultivation. Allelopathy J. 5 23–34. [Google Scholar]
- Yamaoka-Yano D. M., Mazzafera P. (1999). Catabolism of caffeine and purification of a xanthine oxidase responsible for methyluric acids production in Pseudomonas putida L. Rev. Microbiol. 30 62–70. 10.1590/s0001-37141999000100013 [DOI] [Google Scholar]
- Yu C. L., Kale Y., Gopishetty S., Louie T. M., Subramanian M. (2008). A novel caffeine dehydrogenase in Pseudomonas sp. strain CBB1 oxidizes caffeine to trimethyluric acid. J. Bacteriol. 190 772–776. 10.1128/jb.01390-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. L., Louie T. M., Summers R., Kale Y., Gopishetty S., Subramanian M. (2009). Two distinct pathways for metabolism of theophylline and caffeine are coexpressed in Pseudomonas putida CBB5. J. Bacteriol. 191 4624–4632. 10.1128/jb.00409-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zchori-Fein E., Borad C., Harari A. R. (2006). Oogenesis in the date stone beetle, Coccotrypes dactyliperda, depends on symbiotic bacteria. Physiol. Entomol. 31 164–169. 10.1111/j.1365-3032.2006.00504.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
At least 71 bacterial strains in 27 genera have been reported to be capable of caffeine breakdown, with Pseudomonas spp. being the most common.
Assembly statistics for 50 bacterial strains isolated from the coffee berry borer.
Bacteria isolated from the coffee berry borer, indicating which strains might have C-8 oxidation activity (cdhA, cdhB, cdhC, tmuM, tmuH, tmuD), coffee berry borer source used for bacterial isolation (eggs, frass, heads, larvae), non-macerated (whole) or macerated tissue, sterile (St) or non-sterile (NSt), sampling site, GenBank accession numbers, and average nucleotide identity (ANI; Chun et al., 2018) analysis. All insects were field collected, except “Beltsville, MD”, which were reared in the laboratory (see column labeled “Sampling Site”).
Arrangement of gene clusters.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA659418.