Abstract
We created daily concentration estimates for fine particulate matter (PM2.5) at the centroids of each county, ZIP code, and census tract across the western US, from 2008–2018. These estimates are predictions from ensemble machine learning models trained on 24-hour PM2.5 measurements from monitoring station data across 11 states in the western US. Predictor variables were derived from satellite, land cover, chemical transport model (just for the 2008–2016 model), and meteorological data. Ten-fold spatial and random CV R2 were 0.66 and 0.73, respectively, for the 2008–2016 model and 0.58 and 0.72, respectively for the 2008–2018 model. Comparing areal predictions to nearby monitored observations demonstrated overall R2 of 0.70 for the 2008–2016 model and 0.58 for the 2008–2018 model, but we observed higher R2 (>0.80) in many urban areas. These data can be used to understand spatiotemporal patterns of, exposures to, and health impacts of PM2.5 in the western US, where PM2.5 levels have been heavily impacted by wildfire smoke over this time period.
Subject terms: Atmospheric chemistry, Natural hazards
| Measurement(s) | fine respirable suspended particulate matter |
| Technology Type(s) | machine learning |
| Factor Type(s) | elevation • land cover • vehicle emissions • seasonality • spatiotemporal variation |
| Sample Characteristic - Environment | air pollution |
| Sample Characteristic - Location | State of Arizona • State of California • State of Colorado • State of Idaho • State of Montana • State of Nevada • State of New Mexico • State of Oregon • State of Utah • State of Washington • State of Wyoming |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.14161856
Background & Summary
Fine particulate matter (often referred to as PM2.5, meaning particulate matter (PM) which is 2.5 microns in aerodynamic diameter or smaller) air pollution is increasingly associated with numerous adverse health outcomes including, but not limited to, mortality1, respiratory and cardiovascular morbidity2,3, negative birth outcomes4, and lung cancer5. Although PM2.5 concentrations have been declining in many parts of the United States due to policies to limit emissions of air pollutants6, PM2.5 levels have been increasing in parts of the western US7. This increase has been shown to be associated with wildfire smoke7,8, which can cause PM2.5 concentrations that are several times higher than the Environmental Protection Agency’s (EPA’s) daily PM2.5 National Ambient Air Quality Standard (NAAQS) in areas downwind of the wildfires for several days at a time9.
Estimates of PM2.5 concentrations for health studies have traditionally been derived from data from stationary air quality monitors placed in and around populated areas for regulatory purposes. In the US, the EPA’s Federal Reference Method (FRM) PM2.5 monitors often only measure every third or sixth day and most counties do not contain a regulatory air pollution monitor10. There is therefore not enough temporal and spatial coverage from FRM monitors to obtain a good estimate of the air pollution exposures where every person lives.
To improve population exposure assessment of PM2.5, researchers have increasingly been using methods to estimate PM2.5 exposures in the temporal and spatial gaps between regulatory monitoring data using data from satellites (such as aerosol optical depth (AOD) or polygons of smoke plumes or air pollution models11,12) over the past two decades. Each of these data sources has its own benefits and limitations, and researchers are increasingly statistically “blending” information from a combination of data sources to better estimate PM2.5 in space and time. Various methods of blending have been used including spatiotemporal regression kriging13, geographically-weighted regression14, and machine learning methods15–19.
Machine learning methods for estimating PM2.5 concentrations often train large auxiliary datasets, often including satellite AOD, meteorological data, chemical transport model output, and land cover and land use data to provide optimal estimates of PM2.5 where people breathe. These models have been implemented in various locations around the world at city, regional, and national scales20. Some epidemiological questions can only be addressed in longitudinal studies with large sample sizes. Exposure models with large spatial and temporal domains will help enable such studies. Within the US, Di et al. (2016; 2019)16,17, Hu et al. (2017)18 and Park et al. (2020)19 have separately used machine learning algorithms to create fine-resolution daily PM2.5 estimates for the continental US. These models, however, have performed poorly in the western US16,18 and particularly the mountain west17 compared to the rest of the country. Given the increasing trends in PM2.5 concentrations in parts of the western US7,8 and the importance of wildfires as a source of PM2.5 there, it is important to have a model that is tailored to this region to capture the variability in PM2.5 concentrations in space and time in this region.
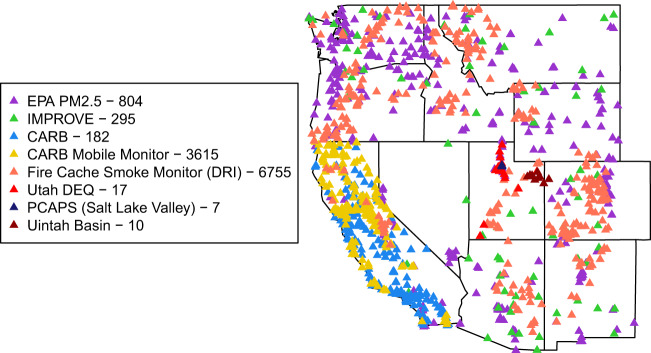
The dataset we describe here improves upon previous daily estimates of PM2.5 concentrations from machine learning models in the following ways: (1) use of a more extensive monitoring station network that captures more spatial locations (Fig. 1), some of which are closer to wildfires, a key driver of PM2.5 in the western US, (2) retaining high PM2.5 monitoring observations and estimating for years with many wildfires to train on, thus likely allowing our models to better predict the high PM2.5 values that occur during wildfire episodes, (3) employing robust spatial cross-validation techniques and 10% of monitoring locations that were completely left out of training on which to evaluate our models, (4) creating a long time series of daily PM2.5 concentration estimates at three geographic levels (county, ZIP code, and census tract) that can be applied in environmental health studies and (5) making the data available in a public repository. The data are available as daily PM2.5 concentration estimates at census tract, ZIP code, and county scales with the aim that they be used by researchers to understand the societal impacts of air pollution exposure in the western US, where wildfires are a significant contributor to PM2.5 concentrations.
Fig. 1.
PM2.5 Monitoring Locations by Source of Monitoring Data. This map shows the locations of all PM2.5 monitors used to train the machine learning models that created the daily PM2.5 surfaces.
Methods
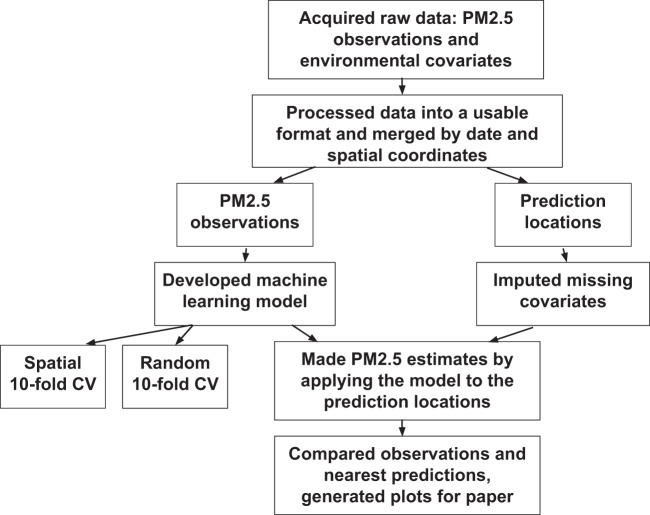
An overview of our methods is shown in Fig. 2.
Fig. 2.
Flowchart diagram of methods to create daily PM2.5 concentration estimates for ZIP codes, census tracts, and counties throughout the western US from 2008–2018.
Study area
Our study area includes 11 western US states: Arizona, California, Colorado, Idaho, Montana, Nevada, New Mexico, Oregon, Utah, Washington, and Wyoming (Fig. 1). Our temporal domain includes all days between January 1, 2008 and December 31, 2018.
PM2.5 Measurements
Regulatory monitoring for PM2.5 is done by the US EPA and their data is freely available on their website. In the western US, however, these monitoring sites do not represent all regions very well. We did an extensive search for as many PM2.5 monitoring data within our spatial and temporal study area as we could find, including from state governments and university researchers. We downloaded PM2.5 data (including parameter codes 88101, 88500, 88502, 81104) from all of the following sources: EPA Air Now data21, EPA IMPROVE Network22, California Air Resources Board (stationary and mobile monitoring networks)23, Federal Land Manager Environmental Database24, Fire Cache Smoke Monitor Archive25, Utah State University26, Utah Department of Environmental Quality27, and the University of Utah28. These data provided a more comprehensive set of locations and time points of PM2.5 measurement throughout the western US than are included in most PM2.5 modelling studies. The EPA IMPROVE monitors capture air quality information in more rural areas22. PM2.5 data in the Fire Cache Smoke Monitor Archive25 includes U.S. Forest Service monitors that were deployed to capture air quality impacts during wildfire events – the high number of geographic locations for these monitors in Fig. 1 (Fire Cache Smoke Monitor (DRI)) is because these are mobile monitors that are placed for each wildfire event and our data is showing the number of geographic locations for a given dataset rather than unique monitors. Any data that was repeated from multiple sources were removed. Also, all observations above 850 µg/m3 (N = 52 or 0.002% of our training observations), many of which were above 1000 µg/m3, were removed due to concern about the validity of these observations. This yielded a total of 1,589,462 daily PM2.5 observations, which represent 7,753 locations and 4,006 days. Descriptive statistics for the PM2.5 observations are in Supplementary Table 1.
Predictor variables
The predictor variables for the machine learning ensemble are listed in Online-only Table 1 and described in more detail below.
Online-only Table 1.
Input variables and their descriptions/sources and names in the training/validation and prediction datasets.
| Variable | Source/Description |
|---|---|
| Spatial variables | |
| Coordinates in degrees (Longitude, Latitude) | |
| State | |
| Region within the western US | Northwest = WA, OR; Southwest = CA, NV; Four corners = AZ, CO, NM, UT; and Northern mountain states = WY, MT, ID |
| Population Density | US Census 201044; spatial resolution = census tract level |
| Percent urban landcover in circular buffers of radius 1 km, 5 km, 10 km (%) | Derived from the USGS National Map Program NLCD, derived from 2011 Landsat imagery43 |
| Lengths of arterial and collector roads in circular buffers of radius 100 m, 250 m, 500 m, 1000 m | Derived from the US Federal Highway Administration National Highway Planning Network46 |
| Elevation (m) | USGS 3D Elevation Program, National Elevation Database (NED)42 |
| Temporal variables | |
| Date | |
| Season | Summer = June-August, Fall = September-November, Winter = December-February, Spring = March-May |
| Cosine of month | Cosine (2*pi*month/12) |
| Cosine of day of year | Cosine (2*pi*day of year/365) |
| Cosine of day of week | Cosine (2*pi*day of week/7) |
| Day of week | |
| Year | |
| Study period – binary indicator of middle study period | Years 2013–2016 |
| Study period – binary indicator of late study period | Years 2017–2018 |
| Spatio-temporal variables | |
| PM2.5 observations (µg/m3) | Federal and state air quality databases |
| Aerosol optical depth | NASA MAIAC29; spatial resolution = 1 km, temporal resolution = generally two observations per day are found but additional ones are possible near the edges of the scan |
| Chemical transport model simulation | US EPA CMAQ38; spatial resolution = 12 km, temporal resolution = daily, only available for 2008–2016 |
| Vegetation index | NASA MODIS NDVI45; spatial resolution = 1 km, temporal resolution = monthly |
| Active fire points within the last week, weighted inversely by distance (up to 500 km) | Derived from NASA MODIS thermal anomalies56; spatial resolution = 1 km, temporal resolution = daily |
| Indicator for whether there was at least one fire in the last week within 500 km | Derived from NASA MODIS thermal anomalies56; spatial resolution = 1 km, temporal resolution = daily |
| Temperature 2 m above the ground (K) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Relative humidity 2 m above the ground (%) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Dew point temperature 2 m above the ground (K) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Height of the planetary boundary layer (m) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Surface pressure (Pa) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Pressure reduced to mean sea level (Pa) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| East-West and North-South components of windspeed 10 m above the ground (m/s) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Vertical component of windspeed, 700 and 850 mb (m/s) | North American Mesoscale Analysis (NAM)35; spatial resolution = 12 km, temporal resolution = 6 hours |
| Interaction indicators between region and time span | 4 regions (see above), 3 timespans(see above) |
Satellite AOD is a measure of particle loading in the atmosphere from the ground to the satellite. We obtained daily estimates of AOD from the MODIS Terra and Aqua combined Multi-angle Implementation of Atmospheric Correction (MAIAC) dataset29. This is the finest resolution (1 km) AOD dataset currently available and was available for our whole time period and spatial domain. After downloading each Hierarchical Data Format (HDF) file from the online repository, we calculated the average daily AOD values at each location and took the value from the nearest neighbour at each PM2.5 monitoring location. MAIAC AOD has been shown to better predict PM2.5 than coarser resolution AOD30 and has been used in many studies in various geographic regions in blended models to predict daily PM2.531–33.
We obtained meteorological data from the North American Mesoscale (NAM) Analysis meteorological model34 because it includes all of the standard meteorological variables which play a role in PM2.5 levels, including planetary boundary layer height which can be important to help scale AOD values to ground-level estimates of PM2.535. We calculated 24-hour averages from 6-hour average data for planetary boundary layer height, temperature at 2-meter elevation, relative humidity at 2 meters, dew point temperature at 2 meters, U- (east-west) and V- (north-south) components of wind speed at 10 meters, surface pressure, pressure reduced to mean sea level, vertical wind velocity at an altitude of 850 mb, and vertical wind velocity at an altitude of 700 mb. NAM has 12 km spatial resolution. The NAM data was retrieved using the rNOMADS package36.
PM2.5 concentration estimates from chemical transport models have been shown to be an important input to machine learning models for PM2.515,17. We obtained daily estimates of PM2.5 at 12 km spatial resolution from runs of the CMAQ (Community Multi-scale Air Quality) model from the U.S. EPA for the years 2008–2016, the years for which the CMAQ estimates are available37.
The main reason that PM2.5 concentrations have been increasing in the western US, while they have been decreasing in other regions, is the increasing number and magnitude of wildfires in that region7,8. To include information on proximity to active fires in our machine learning models, we collected daily data about fire detection locations and size from the MODIS Thermal Anomalies/Fire Daily L3 Global 1 km product (MOD14A1 and MYD14A1)38. As fires in closer proximity are likely to influence PM2.5 more than fires farther away, we calculated the number of active fires in radial buffers of 25, 50, 100, and 500 km radii around each monitoring location, on the current day as well as on each of the previous seven days. We then calculated an inverse-distance-weighted average for each temporal lag. Finally, we created an indicator variable for whether there were one or more fires within 500 km of a monitor in the last week.
Elevation can influence PM2.5 concentrations. For example, PM2.5 can accumulate in mountain valleys during persistent cold air pools (commonly referred to as inversions) during winter39. We obtained elevation data from the 3D Elevation Program of the U.S. Geological Society, with has a resolution of 1 arc-second, which is approximately 30 m north/south and varies east/west with latitude40.
Surrounding land cover can be a proxy for air pollution emissions not from wildfires, such as emissions from traffic and industry in urban areas, and lack of particulate air pollution emissions in more vegetated areas. We used the land cover class information from the Landsat-derived National Land Cover Dataset (NLCD) 201141, which has a spatial resolution of 30 m and uses circa 2011 Landsat satellite data to classify each pixel into one of a variety of land cover codes. We used the NLCD 2011 to calculate the percentage of urban development (codes 22, 23, and 24) within buffer radii of 1 km, 5 km, and 10 km around each monitor. We also included population density as areas with higher population have more sources of air pollution emissions. Population density was obtained from the 2010 U.S. Census42. For a time-varying measure of vegetation abundance, we used monthly measures of the Normalized Difference Vegetation Index (NDVI) from the MODIS satellite product MOD13A343 which has a 1 km spatial resolution.
As another, potentially more specific proxy indicator of emissions from vehicles, we calculated the sum of all road lengths of type “Arterial” and “Collector” within 100, 250, 500, and 1000 m buffers of each monitoring location. Arterial roads are high-capacity urban roads, including highways. Collector roads are low-to-moderate capacity roads. The road data came from the National Highways Planning Network44, which contains spatial information on over 450,000 miles of highways in the United States.
To account for seasonality in PM2.5 data, we created the following predictor variables: cosine of day-of-week, cosine of day-of-year, and cosine of month. The use of cosine of these terms ensures that day/month values at the end and beginning of the week and year align.
We also created indicator variables to represent spatial and temporal variation in the data that could not be explained by any of the other spatial, temporal, or spatiotemporal variables. Use of nested levels of spatiotemporal variables can help to capture nonlinear spatiotemporal effects. Temporal variable nesting consisted of variables to indicate the periods 2008–2012, 2013–2016, and 2017–2018; year; season; cosine of month; and cosine of day of year. Spatial variable nesting consisted of dummy variables for region (within the 11 western states: northwest (i.e., WA, OR), southwest (i.e., CA, NV), four corners (i.e., AZ, CO, NM, UT), and northern mountain states (i.e., WY, MT, ID)); state; latitude; and longitude. We also included interaction terms for time period (grouping of years) and region. This type of nesting has been referred to as a “multiresolution basis”45.
Data merging
We created seven datasets that all required merging the above datasets together: one dataset to train the model (with the 2008–2016 dataset able to be subset from the full 2008–2018 dataset) and six prediction datasets, with three spatial levels of prediction (county, ZIP code, and census tract) for each of the two models (2008–2016 which included CMAQ as a predictor variable, and 2008–2018 which did not include CMAQ as a predictor variable). The training dataset merged all predictor variables to each 24-hour average PM2.5 monitoring observation by linking the data temporally (using date) and spatially (by selecting the nearest spatial observation for each predictor variable using latitude and longitude). Similarly, the prediction datasets were created by spatially (by nearest latitude and longitude) and temporally (by date) linking all predictor variables to the centroid of each county, ZIP code, and census tract for each day in the study domain.
Machine learning modelling
We employed ensemble machine learning to model PM2.5 exposures across the western US. Specifically, we used a generalized linear model (GLM) to combine the results from two machine learning algorithms: a random forest model and a gradient boosting model. These models performed best on preliminary analyses of random subsets of our dataset, which aligns with a previous study that found that tree-based models (using random forest, gradient boosting, and cubist algorithms) performed best in air pollution modelling46. Then, we used the same random subsets of the data to tune hyperparameters for each algorithm via a grid-search (Supplementary Information 1).
For the machine learning modelling, we performed both random and spatial 10-fold cross-validation so that our models would not overfit the training data47. We performed random 10-fold cross-validation, in which a random 10% of all observations are in each fold and the data is trained on 9 folds and tested on the left-out fold and then repeated 10 times such that every fold is a left-out fold once, for comparison to other studies. We performed spatial 10-fold cross-validation, whereby all observations from a given monitoring site are within the same fold, as a more appropriate tool for evaluating the accuracy of a model when predicting PM2.5 at new (left-out) locations, such as those in our prediction set48. Before either kind of 10-fold cross-validation, we also removed 10% of the monitoring locations for a testing data set, which was not used in model development. The spatial-folds analysis used a spatial held-out 10% (random 10% of the locations), while the random-folds analysis used a random held-out 10% (10% of the observations regardless of location). Most previous studies to create daily PM2.5 estimates using machine learning15–18 present results from only random 10-fold cross-validation, which violates the assumption of independence between folds because of repeated observations (on different days) from the same locations (PM2.5 monitor locations). Additionally, they do not provide performance metrics for predictions on a completely left-out test set. Their performance metrics are therefore overly optimistic for how their models will perform in their output data.
We used the metrics root-mean-squared error (RMSE) and R2 to report accuracy, for both the 10-fold cross-validation and for the left-out testing data set, for both spatial folds and random folds. Note that our spatial-folds metrics are different than spatial R2 that has been reported in other papers such as Di et al., (2019)17.
All analyses were run using R49, and all machine learning models utilized the R packages caret50 and caret ensemble51. Variable importance was calculated using the “permutation” importance algorithm in the caret package.
Our main model and prediction data set provide daily PM2.5 estimates by county, ZIP code, and census tract for 2008–2016. Because of our interest in the years 2017 and 2018, when there were many large wildfires in the western US, we created another model and prediction dataset for the 11-year period of 2008–2018 because we did not have CMAQ PM2.5 concentration output for 2017 and 2018. This separate ensemble machine learning model for 2008–2018 does not include CMAQ PM2.5 as a predictor variable but still created a consistent dataset of daily PM2.5 predictions for those 11 years.
We observed that our models, like most models, underpredict at very high values (e.g., above 200 µg/m3). We hypothesized that some of the higher values were being generated by a fundamentally different process than the lower values, most likely because of wildfires. We therefore did a sensitivity analysis in which we examined whether we would observe better performance by having different models for low values that are not likely influenced by wildfires and another for high values which are likely influenced by wildfires. This did not yield better performance and therefore these models did not create prediction datasets, but for the reader interested in this, a more detailed description of the split analysis (“high” versus “low”) can be found in Supplementary Information 2.
Daily PM2.5 prediction creation
After merging the prediction input datasets by spatially and temporally linking all predictor variables to the centroid of each county, ZIP code, and census tract for each day in the study domain, we observed some missingness in the predictors that required imputation to create daily PM2.5 predictions. We observed missingness for fewer than 1% of the location-days within each state, except for the meteorological variables within Wyoming, for which nearly 10% of the location-days were missing. We used the missRanger52 package in R to impute the missing data for each state, based on all the available data for that state for all years in the given model (2008–2016 for the model including CMAQ PM2.5 and 2008–2018 otherwise).
Post-imputation, we applied the full models (trained with the entire training dataset including the left-out 10% testing data) to the prediction input datasets to make the final daily PM2.5 predictions at each county, ZIP code, and census tract within our 11-state study domain for the 9- and 11-year time periods.
We performed additional validation of our prediction data set that is separate from the accuracy estimates provided above of the machine learning model against the observations on which it was trained and tested. To assess the spatiotemporal alignment of our predicted values with observed values, we compared each observation to its spatially nearest prediction on the same day and calculated RMSE and R2 values for these. This is the most realistic estimate of our model’s performance overall.
Data Records
Table 1 lists the names and descriptions of the datafiles that are available on figshare with a doi of 10.6084/m9.figshare.1256849653. We provide files by state for the predictions using the model with CMAQ PM2.5 (Ensemble_preds_with_CMAQ_[state].RData), and files by state for the predictions using the model without CMAQ PM2.5 (Ensemble_preds_no_CMAQ_[state].RData). All prediction data sets have predictions at three spatial resolutions: county, ZIP code, and census tract. Within the prediction data sets, the variable ranger_preds refers to the predictions made by the Ranger algorithm, the variable xgbt_preds to the predictions made by the extreme gradient boosting tree (XGBT) algorithm, and Ens_preds to the predictions made by the GLM ensemble of Ranger and XGBT. Missing_vars indicates if there was at least one missing input variable (and subsequent imputation) for that row of data.
Table 1.
Data Files provided at 10.6084/m9.figshare.1256849653.
| File Name(s) | Name of the Data Frame(s) in the .RData File | Years of Data | Variables |
|---|---|---|---|
| Ensemble_preds_no_CMAQ_[state].RData | DF | 2008–2016 | County_FIPS, Tract_code, ZCTA5_code, Lon, Lat, Date, ranger_preds, xgbt_preds, Missing_vars, Ens_pred |
| Ensemble_preds_with_CMAQ_[state].RData | DF | 2008–2018 | County_FIPS, Tract_code, ZCTA5_code, Lon, Lat, Date, CMAQ_ranger_preds, CMAQ_xgbt_preds, Missing_vars, Ens_pred |
Because much of our input data derive from sources that are not ours, we cannot share out input data for our ML models at this time, but this data can be made available upon request. Most of our data sources are available free from government online databases, and therefore someone wishing to replicate our findings could download the data and follow our scripts, which we provide.
Technical Validation
Performance metrics on training and testing data
Table 2 shows the performance metrics (RMSE and R2) of our ensemble machine learning models with spatial folds and random folds. The results for the training data are based on 10-fold cross-validation, whereas the testing RMSE and R2 are for the completely left-out 10% of the data, a good test of how our model will do predicting at locations on which it did not train.
Table 2.
Performance metrics (RMSE and R2) of our models using various subsets of data and types of CV folds.
| Model | Training 10-fold CV RMSE (µg/m3) and R2 | Testing (left out 10%) RMSE (µg/m3) and R2 |
|---|---|---|
| 2008–2016 with CMAQ, spatial folds | 5.061 (0.659) | 5.420 (0.589) |
| 2008–2018 without CMAQ, spatial folds | 6.576 (0.598) | 6.599 (0.593) |
| 2008–2016 with CMAQ, random folds | 4.482 (0.732) | 4.642 (0.715) |
| 2008–2018 without CMAQ, random folds | 5.482 (0.719) | 5.954 (0.680) |
| 2008 – 2016 without CMAQ, random folds* | 4.747 (0.702) | |
| 2008–2016 with CMAQ, no folds or testing set | 1.726 (0.960) | |
| 2008–2018 without CMAQ, full model, no folds or testing set | 2.027 (0.961) |
*This test was performed to show the impact of including CMAQ in the model.
Overall, our models including CMAQ PM2.5 (2008–2016 models) perform better (have lower RMSE and higher R2 values) than models without CMAQ PM2.5 (2008–2018 models). This may be due to the additional information provided by the CMAQ output or could be because the models without CMAQ PM2.5 include two additional years of data that have more days with high PM2.5, which are much harder to predict accurately than lower values. For comparison, results for a random-folds model without CMAQ PM2.5 on the years 2008–2016 performed slightly worse than the model for years 2008–2016 with CMAQ PM2.5 included as a predictor variable.
The spatial-folds performance metrics are worse than the random-folds performance metrics. This is not surprising because the spatial folds do not allow for observations from the same location to be in multiple folds, therefore the models are predicting at locations that they did not train on, whereas random folds have likely trained on observations at all locations, thus are more likely to predict values better for those locations. Using solely random folds can therefore be misleading as to the performance of the models given that these kinds of models predict at locations without monitoring data and thus not in the training data48. Thus, we posit that most of the models presented in the literature previously that use random folds CV are reporting R2 values that are likely higher than their predictive performance at non-sampled locations. Henceforth all results in this section refer to the spatial-folds analysis.
Performance of our models on our completely left-out testing data set provide worse metrics than their training (10-fold CV) counterparts. Some of the discrepancy between training and testing set results is because the testing data set was not used to inform the development of the model; some of the discrepancy is because of random chance of a given monitoring site being in the testing data set. Given that most previous machine learning air pollution modelling studies do not report metrics for completely left-out testing data15–19, their results on the accuracy of their models’ predictions may be misleading. For comparison, the performance metrics for our full models (without any cross-validation folds) on the 2008–2016 and the 2008–2018 datasets are, respectively, RMSE = 1.726 µg/m3 and R2 = 0.960; RMSE = 2.027 µg/m3 and R2 = 0.961. These are much better performance metrics than any of those in Table 2 because all models overfit the data they are trained on. Full models train on all observations with none left out and therefore are not realistic representations of how accurately the model will predict at locations outside of the training set.
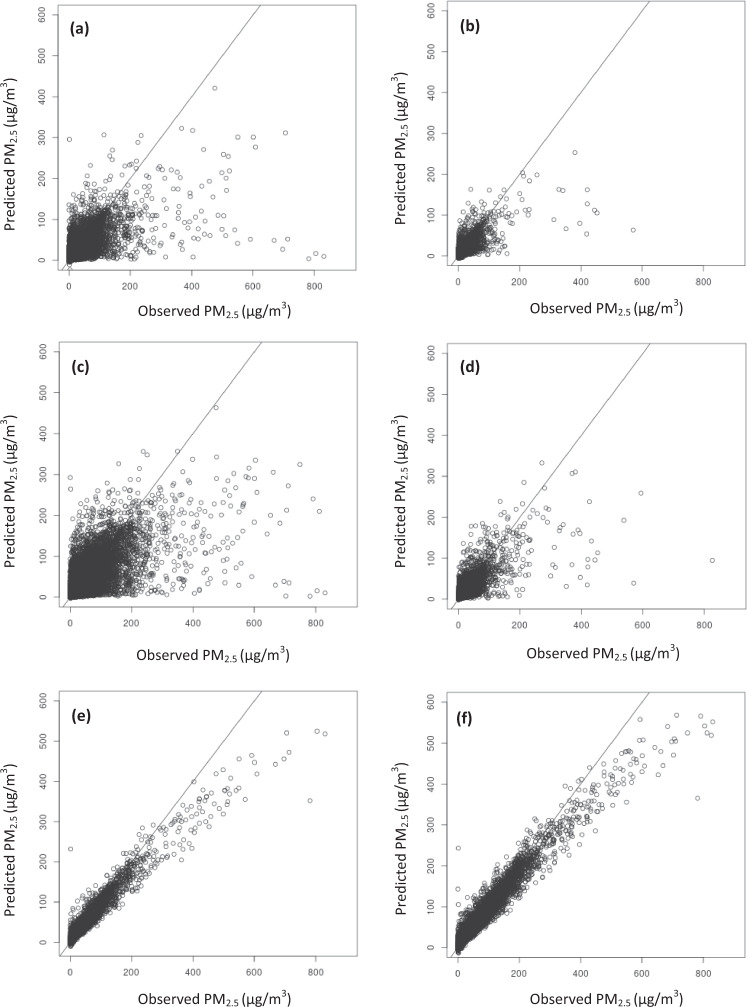
The predicted-versus-observed plots in Fig. 3 demonstrate that the model trained on all of the data (Fig. 3e,f) has the greatest agreement between predictions and observations, which is expected when machine learning models are fit without cross-validation because they overfit to the data used. These figures also show that there were many more high values in the years 2017 and 2018 (on the non-CMAQ model plots, Fig. 3c,d,f). Also, all models tend to underpredict values of PM2.5 higher than 200 µg/m3, which is likely because there are fewer high values than low values in the training set. Many previous studies using similar methodology only present the accuracy of their PM2.5 predictions up to 60 µg/m3 without documenting the range of values trained on16,17 or training on observations that had a much smaller range (up to 136.8 µg/m3)18. All of these models additionally perform worse at high concentrations, similar to ours, due to few observations to train on at those values. The Di et al., (2016)16 and Di et al., (2019)17 papers dismiss the importance of their models’ poor performance at values above 60 µg/m3 because they state that there are few days in locations in the U.S. that experience PM2.5 concentrations above that value. This implies that days affected by wildfires are either not important to predict well and/or that these previous models should not be used to predict PM2.5 on days in locations affected by wildfire smoke. All of these previous models were trained solely on EPA AQS without inclusion of the additional monitoring data from the USFS and other sources that we included so that our models would have those higher values on which to train. Although we do show underprediction at these higher values, training our model on a larger range of input values than previous studies allows us to predict values that are likely closer to the “true” concentration to which populations were exposed. It should be noted the two Di et al. papers16,17 only present performance metrics for all values below 60 µg/m3 and do not indicate what the range of values were that were used to train their model. The Hu et al. (2017)18 and Park et al. (2019)19 models were only trained on values up to 136.8 µg/m3.
Fig. 3.
Predicted-versus-observed plots of daily PM2.5 values from our ensemble machine learning models using spatial 10-fold cross validation. All lines in Fig. 3 depict the y = x lines in which predictions are equal to observed values. The subplots refer to (a) spatial 10-fold cross-validation training dataset for the 2008–2016 model (90% of the data) (includes CMAQ PM2.5), (b) spatial 10-fold cross-validation testing dataset (10% of the data) for the 2008–2016 model (includes CMAQ PM2.5), (c) spatial 10-fold cross-validation training dataset for the 2008–2018 model (90% of the data) (does not include CMAQ PM2.5), (d) spatial 10-fold cross-validation testing dataset (10% of the data) for the 2008–2018 model (does not include CMAQ PM2.5), (e) all observations in the 2008–2016 model (includes CMAQ PM2.5) with no CV (the full model), (f) all observations in the 2008–2018 model (does not include CMAQ PM2.5) with no CV (the full model).
Table 3 shows the RMSE (and R2) values of our models on different levels of PM2.5, years, states, and seasons using spatial folds. RMSE (and R2) values from the random-folds analysis of the same models are in Supplementary Table 2.
Table 3.
RMSE and R2 values for the 10-fold spatial cross-validation training set (90% of the data) and the 10% left-out testing set for both the 2008–2016 and 2008–2018 ensemble models.
| 2008–2016 Ensemble Model Training RMSE (µg/m3) (and R2) | 2008–2016 Ensemble Model Testing RMSE (µg/m3) (and R2) | 2008–2018 Ensemble Model Training RMSE (µg/m3) (and R2) | 2008–2018 Ensemble Model Testing RMSE (µg/m3) (and R2) | |
|---|---|---|---|---|
| By PM2.5 Level (µg/m3) | ||||
| Below 35 | 3.346 (0.682) | 3.905 (0.575) | 4.201 (0.523) | 4.151 (0.522) |
| Below 60 | 3.664 (0.714) | 4.179 (0.612) | 4.651 (0.568) | 4.499 (0.569) |
| Below 150 | 4.010 (0.720) | 4.526 (0.621) | 5.154 (0.615) | 5.009 (0.612) |
| Below 300 | 4.319 (0.706) | 4.646 (0.631) | 5.635 (0.626) | 5.376 (0.641) |
| Below 500 | 4.618 (0.690) | 5.247 (0.603) | 6.044 (0.618) | 6.118 (0.616) |
| Below 1000 | 5.061 (0.659) | 5.420 (0.589) | 6.576 (0.598) | 6.599 (0.593) |
| By Year | ||||
| 2008 | 4.037 (0.787) | 4.818 (0.689) | 4.920 (0.680) | 4.903 (0.661) |
| 2009 | 3.755 (0.744) | 4.416 (0.637) | 4.603 (0.614) | 4.611 (0.602) |
| 2010 | 3.537 (0.707) | 3.834 (0.634) | 4.372 (0.559) | 4.013 (0.579) |
| 2011 | 4.016 (0.723) | 4.264 (0.642) | 4.840 (0.598) | 4.396 (0.615) |
| 2012 | 5.459 (0.684) | 8.006 (0.504) | 6.180 (0.581) | 8.456 (0.449) |
| 2013 | 4.990 (0.683) | 5.927 (0.588) | 5.836 (0.562) | 6.022 (0.576) |
| 2014 | 4.816 (0.668) | 5.175 (0.572) | 5.728 (0.530) | 5.359 (0.540) |
| 2015 | 5.881 (0.667) | 5.107 (0.687) | 6.723 (0.571) | 5.423 (0.645) |
| 2016 | 7.239 (0.449) | 5.296 (0.520) | 7.794 (0.370) | 5.627 (0.454) |
| 2017 | N/A | N/A | 9.348 (0.684) | 8.868 (0.669) |
| 2018 | N/A | N/A | 8.553 (0.687) | 10.435 (0.619) |
| By State | ||||
| Arizona | 3.139 (0.614) | 4.094 (0.374) | 3.795 (0.469) | 3.894 (0.418) |
| California | 4.753 (0.719) | 4.026 (0.756) | 6.577 (0.632) | 5.197 (0.724) |
| Colorado | 5.940 (0.499) | 3.539 (0.472) | 9.143 (0.351) | 3.758 (0.432) |
| Idaho | 7.016 (0.663) | 11.539 (0.546) | 7.789 (0.605) | 10.034 (0.607) |
| Montana | 5.428 (0.648) | 5.192 (0.593) | 7.642 (0.636) | 7.183 (0.595) |
| Nevada | 3.639 (0.679) | 3.630 (0.634) | 4.273 (0.497) | 3.809 (0.568) |
| New Mexico | 3.628 (0.536) | 9.732 (0.028) | 3.732 (0.388) | 10.823 (0.006) |
| Oregon | 5.239 (0.597) | 10.063 (0.362) | 8.441 (0.612) | 12.081 (0.436) |
| Utah | 4.667 (0.638) | 4.979 (0.647) | 5.223 (0.495) | 4.584 (0.665) |
| Washington | 5.642 (0.583) | 4.409 (0.586) | 6.814 (0.529) | 8.412 (0.515) |
| Wyoming | 6.983 (0.516) | 3.859 (0.531) | 6.405 (0.418) | 3.839 (0.465) |
| By Season | ||||
| Fall | 5.960 (0.598) | 6.465 (0.520) | 7.896 (0.599) | 8.505 (0.561) |
| Spring | 3.168 (0.611) | 3.227 (0.561) | 3.871 (0.444) | 3.293 (0.543) |
| Summer | 5.745 (0.635) | 5.760 (0.571) | 7.771 (0.605) | 7.629 (0.591) |
| Winter | 4.682 (0.767) | 5.632 (0.658) | 5.787 (0.639) | 5.620 (0.650) |
The models with CMAQ PM2.5 (2008–2016 models) always perform better than the models without CMAQ PM2.5 (2008–2018 models). This is likely because of the importance of PM2.5 concentration data from a chemical transport model in the machine learning models (Supplementary Table 5). Although chemical transport models provide spatiotemporal estimates of PM2.5, they do not always predict accurately and many have shown that empirical models like the ones presented here can improve predictive performance of chemical transport models. However, the algorithms we used perform well even when one important predictor is missing. When CMAQ PM2.5 is not included, MAIAC AOD rises in variable importance (Supplementary Table 5). Although collinearity between variables does not matter for prediction with random forest, it most likely reduces the variable importance calculations via permutation54.
We have better predictive performance at lower levels of PM2.5. This is likely partially because a much larger number of observations at lower values allowed the model to be better trained at those values; it is well-known that models tend to have greater trouble predicting extremes. In the spatiotemporal subsets, we observed higher RMSE for the years 2012 and 2015–2018, which have some of the highest PM2.5 values due to high numbers of wildfires in our study domain during those years. The patterning of results by state is less clear, although it is notable that the RMSE values for California are lower than might be expected given the state’s higher-than-average PM2.5 levels. This is likely because over 40% of our monitoring locations and thus observations are from California. Finally, the RMSE values for Spring are lower than those from the other seasons, which is likely due to the predominance of lower PM2.5 values in the spring (Fig. 4). One thing to note related to the performance metrics in Table 3 is that sometimes the R2 values are quite low for the testing data for a given year or state because there were few observations within that state or year within the pre-selected 10% testing hold-out done across all of the data before the modelling; this is not a 10% of observations from each state or year.
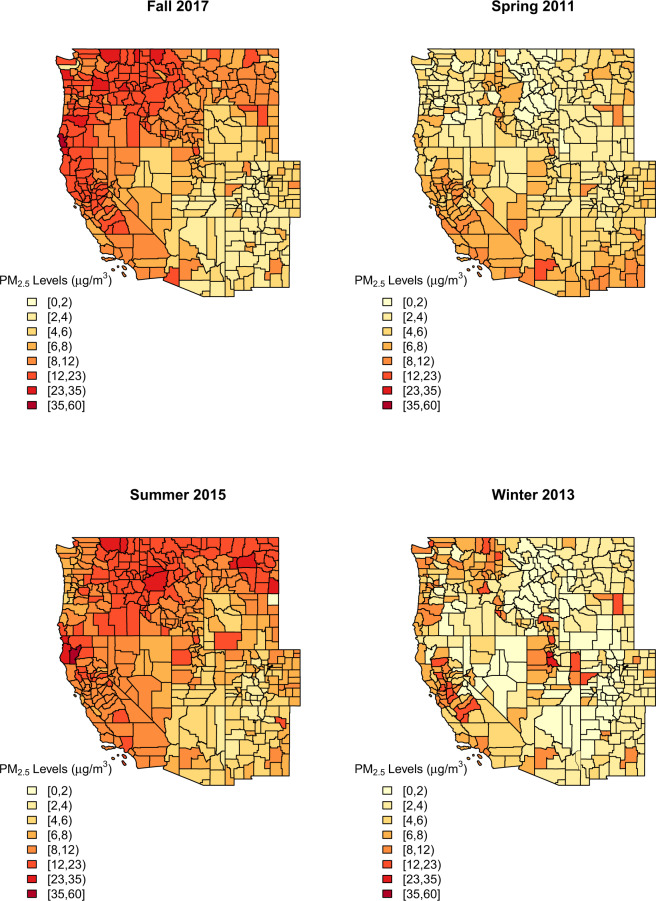
Fig. 4.
Map of seasonal average predicted PM2.5 by county for selected seasons using the 2008–2018 model. Winter = December, January, February; Spring = March, April, May; Summer = June, July, August; Fall = September, October, November.
We also provide RMSE values normalized by the mean of each corresponding dataset in Supplementary Tables 3 and 4 (corresponding to the spatial and random folds results respectively). These tables provide a sense of the predictive performance relative to the observed values. We observe that the model has larger error relative to the mean of the observed data for years and seasons with more wildfires. This is unsurprising given that squaring the errors in the RMSE calculation tends to weight large differences more heavily than the simple sum in the mean calculation. There is less of a clear pattern for the normalized RMSE by state, though the fact that our model does relatively well in California is likely due to having more air quality monitors and thus training data there. The spatial folds and random folds results follow the same overall pattern, though the random folds results have smaller errors compared to their mean values, likely because there is training data from all locations in all folds yet use of these metrics would lead to overconfident predictions in locations without monitoring data.
Prediction data set validation
Descriptive statistics for the predicted PM2.5 values by geographic scale (county, ZIP code, and census tract) are provided in Supplementary Table 6 (for the 2008–2016 model) and Supplementary Table 7 (for the 2008–2018 model). Our metrics for comparing our model predictions to nearby observed monitors are shown in Table 4. When we consider only stable monitors (excluding monitors with less than 31 days of observations, 1% of the training data), the 2008–2018 model yields an RMSE of 5.6 μg/m3 and an R2 of 0.581; the 2008–2016 model yields an RMSE of 4.2 μg/m3 and an R2 of 0.696. When we include all observations, the 2008–2018 model yields an RMSE of 6.2 μg/m3 and an R2 of 0.643; the 2008–2016 model yields an RMSE of 5.1 μg/m3 and an R2 of 0.647.
Table 4.
Performance metrics (RMSE and R2) comparing model predictions with the closest nearby PM2.5 monitoring observations.
| Model | Compared to all monitors RMSE (μg/m3) (and R2) | Compared to stable monitors (excluding monitors with less than 31 days of observations, 1% of the training data) RMSE (μg/m3) (and R2) |
|---|---|---|
| 2008–2016 model | 5.1 (0.647) | 4.2 (0.696) |
| 2008–2018 model | 6.2 (0.643) | 5.6 (0.581) |
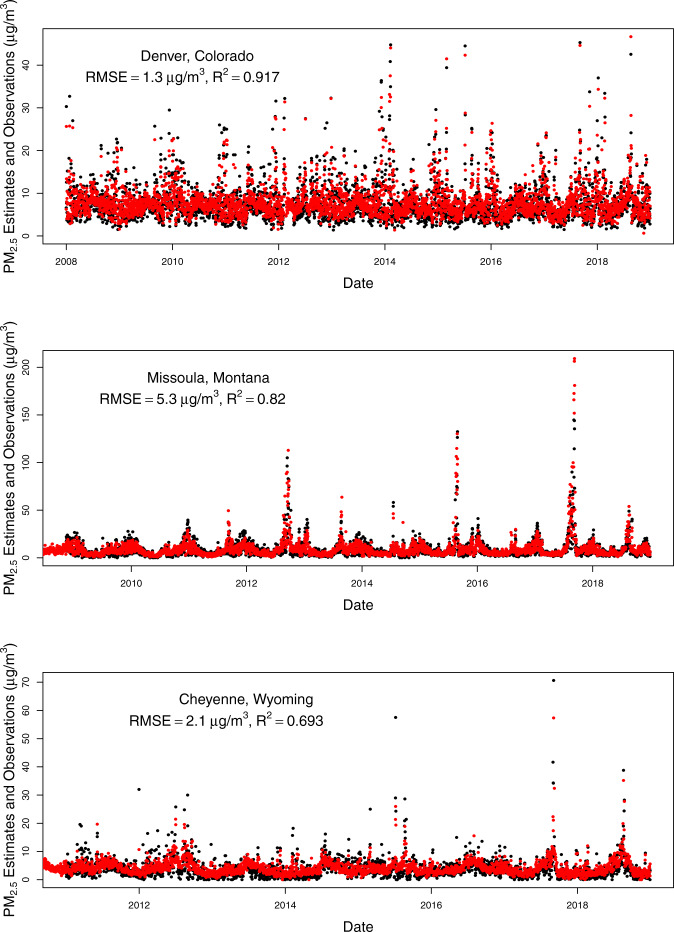
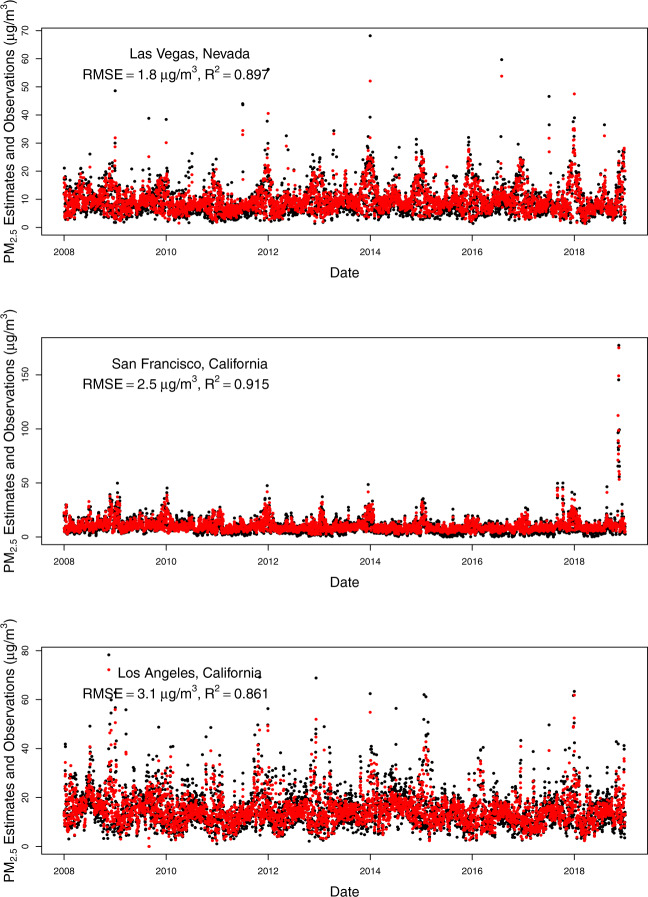
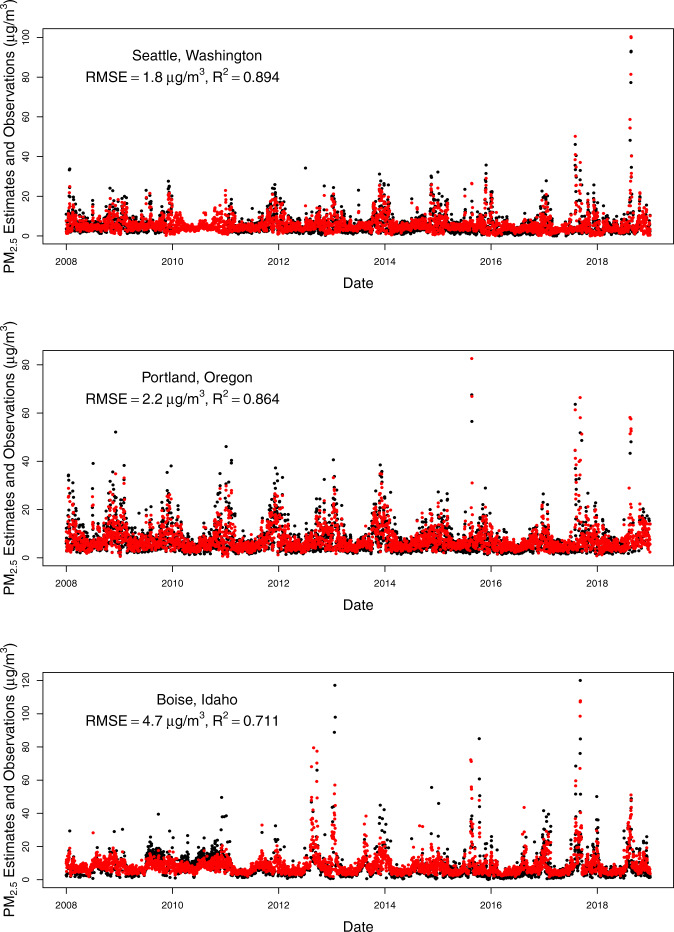
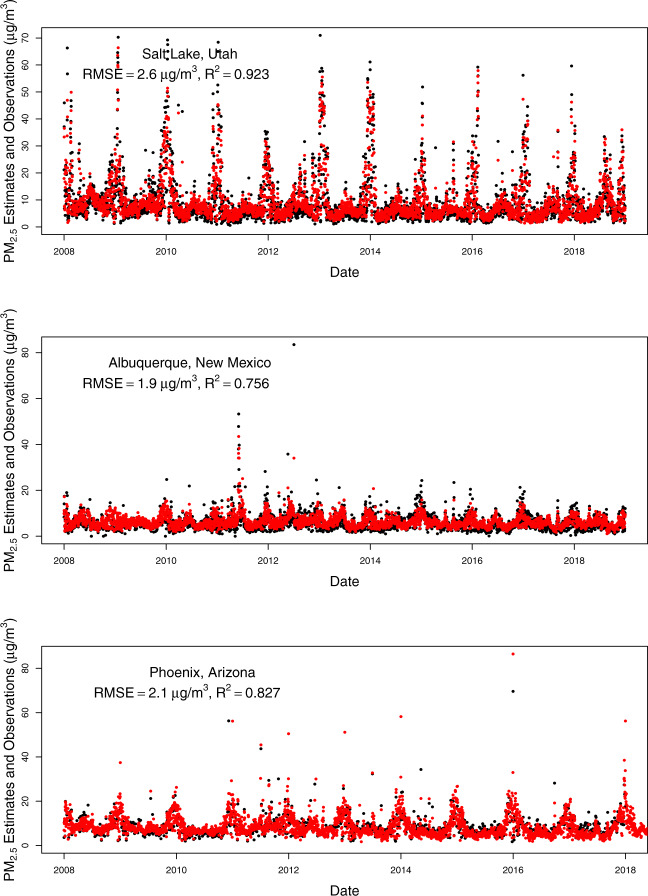
To assess the temporal alignment of our predicted values with observed values, we plotted time series for predicted daily PM2.5 for a monitoring station within a major city in each state of our study domain compared to observations from the closest monitors (Figs. 5–8). The monitoring stations were selected for having the most continuous monitoring observations in a large city within each state, but we included two large cities for California because of its large population and geographic variability with regards to air pollution. From these plots, you can see that our model predictions depict the temporal variability in PM2.5 concentrations at monitoring stations nicely including the high values when they occur (most often during wildfire seasons) even for cities in which the air pollution during the most recent wildfire seasons have dwarfed the PM2.5 levels in previous wildfire seasons (e.g., San Francisco, Seattle). There is quite good agreement between the predictions (which are from the nearest prediction location (centroid of census tract, ZIP code, or county) to each selected monitoring location (R2 values range from 0.693 in Cheyenne, WY to 0.923 in Salt Lake City, UT, with all but three above 0.80). The RMSE values also vary, but these are more impacted by the range of PM2.5 values observed at a given location. The models predict worse at higher values, thus locations with more PM2.5 variability (mostly driven by high PM2.5 values during wildfire seasons) tend to have higher RMSE values (e.g., Missoula, Montana). It is important to note that these are predictions in our prediction data set (either census tract, ZIP code, or county centroid) that are nearest to the monitor and not at that exact location. In particular, the distances from the nearest prediction to the monitor in Santa Fe, New Mexico and Boise, Idaho were nearly an order of magnitude larger than the distances for each of the other cities, which contributed to their lower R2 and higher RMSE values.
Fig. 6.
Time series of daily PM2.5 estimates (red) at the nearest prediction locations (centroid of census tract, ZIP code, or county) near daily PM2.5 monitoring observations (black) for Denver, CO, Missoula, MT, and Cheyenne, WY using the 2008–2018 models (with RMSE and R2 values for that location).
Fig. 7.
Time series of daily PM2.5 estimates (red) at the nearest prediction locations (centroid of census tract, ZIP code, or county) near daily PM2.5 monitoring observations (black) for Las Vegas, NV, San Francisco, CA, and Los Angeles, CA using the 2008–2018 models (with RMSE and R2 values for that location).
Fig. 5.
Time series of daily PM2.5 estimates (red) at the nearest prediction locations (centroid of census tract, ZIP code, or county) near daily PM2.5 monitoring observations (black) for Seattle, WA, Portland, OR, and Boise, ID using the 2008–2018 models (with RMSE and R2 values for that location).
Fig. 8.
Time series of daily PM2.5 estimates (red) at the nearest prediction locations (centroid of census tract, ZIP code, or county) near daily PM2.5 monitoring observations (black) for Salt Lake City, UT, Albuquerque, NM, and Phoenix, AZ using the 2008–2018 models (with RMSE and R2 values for that location).
We also mapped seasonal averages of our daily PM2.5 predictions by county to assess the spatial patterning in the predictions and whether they aligned with the spatial pattern in the monitored data (Fig. 4). The year picked for each season was to demonstrate our model’s ability to predict the spatial pattern of PM2.5 during specific times when PM2.5 was particularly high in certain regions such as the large wildfires in Arizona in the spring of 2011, in the North in summer 2015, in the Pacific Northwest and California in the Fall of 2017, and to show the PM2.5 from inversions in Utah and California in the winter of 2013. Not all counties had monitoring data, so it was not possible to plot the monitoring data in the same way. Overall, these maps show lower PM2.5 levels in the spring; Spring 2011 is just one example of this. In the winter, in the western US counties, many areas experience quite low levels of PM2.5, but there are some regions that have higher levels due to wintertime inversions (e.g., Salt Lake City, California’s Central Valley). We also chose to highlight two seasons with high wildfire activity: the summer of 2015 when there were many wildfires burning in the Pacific northwest and the fall of 2017 when California had a large number of wildfires. We note that, as these are seasonal averages, the PM2.5 levels are not as high as the maximum daily predicted levels for each county.
Usage Notes
All of our data files are available in the RData format. An RData file can be opened in R using the “load” command. Note that the state data frames range in size from about 2 million rows to about 34 million rows. A computer with lots of memory and/or cloud computing may be necessary to analyse these data.
Because the predictive model is continuous and not confined to positive numbers, low predictions (close to 0) will sometimes go negative. Because this is physically unrealistic, we recommend replacing the few negative PM2.5 predictions with 0, as we have done to generate the plots and performance metrics shown in the Technical Validation section.
Then, our PM2.5 estimates may be merged with health data or other data at the county, ZIP code, and census tract levels based on FIPS codes (for county and census tract) and ZCTA5 codes (for ZIP code). If a research team is concerned about our imputation technique, then they may wish to exclude any rows that have one or more missing variables.
Supplementary information
Acknowledgements
This work was supported by Earth Lab through the University of Colorado Boulder’s Grand Challenge Initiative. We also thank the following individuals and groups who helped us obtain additional monitoring data: Denise Odenwalder, Charles Pearson, and Joseph McCormack at the California Air Resources Board; Seth Lyman at Utah State University for the Uintah Basin PM2.5 data; Geoff Silcox at the University of Utah for the PM2.5 data from the Persistent Cold Air Pool Study (PCAPS). Publication of this article was funded by the University of Colorado Boulder Libraries Open Access Fund.
Online-only Table
Author contributions
C.E.R. conception and design, project supervision, writing and editing. E.M.C. design, acquisition, analysis, and interpretation of the data, writing and editing. M.M.M. design, data acquisition and analysis, writing and editing. G.L. data acquisition. All authors approve the submitted version of this paper.
Code availability
All code used for downloading and processing the data used in this project, including the machine learning and technical validation code, may be accessed at 10.5281/zenodo.449926455.
To ensure that our work is reproducible, all code is written in open-source languages. Some scripts are in R and others are in Python. Python scripts need Python 3; R versions beyond 3.5.1 should suffice.
The scripts on Zenodo are a copy of our GitHub repository of scripts and contains the following files and directories:
• General_Project_Functions: Scripts to obtain the prediction set locations and tools that are generally useful during the data processing, such as making buffers around points and reprojecting point coordinates.
• Get_PM25_Observations: Scripts to process PM2.5 observations from across the western U.S. These observations are used to train our machine learning models.
• Get_Earth_Observations: Scripts to download and process observations from data sets that are used both as inputs for our machine learning models during training and as inputs for our models in the prediction stage. The file Overall_steps provides all necessary directions. Individual README files (in each folder) provide more details if there are any.
• Merge_Data: Scripts to merge all the data together and derive some spatio-temporal variables.
• Machine_Learning: Scripts to run and evaluate our machine learning models. The folder Final_scripts contains all code used for our final analysis. The code in the Exploring_models folder was all preliminary testing.
• Estimate_PM25: Scripts to use our machine learning models to make final predictions and to explore the prediction data sets over time and space.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-021-00891-1.
References
- 1.Achilleos S, et al. Acute effects of fine particulate matter constituents on mortality: A systematic review and meta-regression analysis. Environ. Int. 2017;109:89–100. doi: 10.1016/j.envint.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing Y-F, Xu Y-H, Shi M-H, Lian Y-X. The impact of PM2.5 on the human respiratory system. J. Thorac. Dis. 2016;8:E69–74. doi: 10.21037/jtd.2016.06.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajagopalan S, Al-Kindi SG, Brook RD. Air Pollution and Cardiovascular Disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018;72:2054–2070. doi: 10.1016/j.jacc.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 4.Klepac P, Locatelli I, Korošec S, Künzli N, Kukec A. Ambient air pollution and pregnancy outcomes: A comprehensive review and identification of environmental public health challenges. Environ. Res. 2018;167:144–159. doi: 10.1016/j.envres.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Hamra GB, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ. Health Perspect. 2014;122:906–911. doi: 10.1289/ehp/1408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fann N, Kim S-Y, Olives C, Sheppard L. Estimated Changes in Life Expectancy and Adult Mortality Resulting from Declining PM2.5 Exposures in the Contiguous United States: 1980–2010. Environ. Health Perspect. 2017;125:097003. doi: 10.1289/EHP507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClure CD, Jaffe DA. US particulate matter air quality improves except in wildfire-prone areas. Proc. Natl. Acad. Sci. USA. 2018;115:7901–7906. doi: 10.1073/pnas.1804353115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Dell K, Ford B, Fischer EV, Pierce JR. Contribution of Wildland-Fire Smoke to US PM2.5 and Its Influence on Recent Trends. Environ. Sci. Technol. 2019;53:1797–1804. doi: 10.1021/acs.est.8b05430. [DOI] [PubMed] [Google Scholar]
- 9.Reid CE, et al. Associations between respiratory health and ozone and fine particulate matter during a wildfire event. Environ. Int. 2019;129:291–298. doi: 10.1016/j.envint.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 10.US EPA. PM 2.5 Policy and Guidance | Ambient Monitoring Technology Information Center | US EPA. https://www3.epa.gov/ttnamti1/pmpolgud.html (2016).
- 11.Brokamp, C., Brandt, E. B. & Ryan, P. H. Assessing Exposure to Outdoor Air Pollution for Epidemiological Studies: Model-based and Personal Sampling Strategies. J. Allergy Clin. Immunol. 10.1016/j.jaci.2019.04.019 (2019). [DOI] [PubMed]
- 12.Liu JC, Pereira G, Uhl SA, Bravo MA, Bell ML. A systematic review of the physical health impacts from non-occupational exposure to wildfire smoke. Environ. Res. 2015;136:120–132. doi: 10.1016/j.envres.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, et al. Satellite-based high-resolution mapping of ground-level PM2.5 concentrations over East China using a spatiotemporal regression kriging model. Sci. Total Environ. 2019;672:479–490. doi: 10.1016/j.scitotenv.2019.03.480. [DOI] [PubMed] [Google Scholar]
- 14.Lassman W, et al. Spatial and temporal estimates of population exposure to wildfire smoke during the Washington state 2012 wildfire season using blended model, satellite, and in situ data. GeoHealth. 2017;1:106–121. doi: 10.1002/2017GH000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid CE, et al. Spatiotemporal prediction of fine particulate matter during the 2008 northern California wildfires using machine learning. Env. Sci Technol. 2015;49:3887–96. doi: 10.1021/es505846r. [DOI] [PubMed] [Google Scholar]
- 16.Di Q, et al. Assessing PM2.5 Exposures with High Spatiotemporal Resolution across the Continental United States. Env. Sci Technol. 2016;50:4712–21. doi: 10.1021/acs.est.5b06121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Q, et al. An ensemble-based model of PM2.5 concentration across the contiguous United States with high spatiotemporal resolution. Environ. Int. 2019;130:104909. doi: 10.1016/j.envint.2019.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, et al. Estimating PM2.5 Concentrations in the Conterminous United States Using the Random Forest Approach. Environ. Sci. Technol. 2017;51:6936–6944. doi: 10.1021/acs.est.7b01210. [DOI] [PubMed] [Google Scholar]
- 19.Park Y, et al. Estimating PM2.5 concentration of the conterminous United States via interpretable convolutional neural networks. Environ. Pollut. Barking Essex 1987. 2020;256:113395. doi: 10.1016/j.envpol.2019.113395. [DOI] [PubMed] [Google Scholar]
- 20.Bellinger C, Jabbar MSM, Zaiane O, Osornio-Vargas A. A systematic review of data mining and machine learning for air pollution epidemiology. Bmc Public Health. 2017;17:907. doi: 10.1186/s12889-017-4914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. EPA. AirData website File Download page. https://aqs.epa.gov/aqsweb/airdata/download_files.html#Daily (2020).
- 22.US EPA. PM 2.5 Visibility (IMPROVE) | Ambient Monitoring Technology Information Center | US EPA. https://www3.epa.gov/ttnamti1/visdata.html (2016).
- 23.California Air Resources Board. AQMIS 2 - Air Quality and Meteorological Information System. https://www.arb.ca.gov/aqmis2/aqmis2.php (2020).
- 24.Colorado State University. Federal Land Manager Environmental Database. http://views.cira.colostate.edu/fed/DataWizard/Default.aspx (2020).
- 25.Desert Research Institute. Fire Cache Smoke Monitor Archive. https://wrcc.dri.edu/cgi-bin/smoke.pl (2020).
- 26.Lyman, S., Mansfield, M. & Tran, H. UBAQR_2018_AnnualReport.pdf. 110, https://usu.app.box.com/s/rigadr7yt7ipir4gzj75vfaazoe8u8mt (2018).
- 27.Utah DEQ. DAQ Home> AMP> Data Archive> Particulate PM2.5. http://www.airmonitoring.utah.gov/dataarchive/archpm25.htm (2020).
- 28.Silcox GD, Kelly KE, Crosman ET, Whiteman CD, Allen BL. Wintertime PM2.5 concentrations during persistent, multi-day cold-air pools in a mountain valley. Atmos. Environ. 2012;46:17–24. [Google Scholar]
- 29.Lyaspustin, A. & Wang, Y. MODIS/Terra and Aqua MAIAC Land Aerosol Optical Depth Daily L2G 1 km SIN Grid V006 – MCD19A2.006 [Data set]. NASA EOSDIS Land Processes DAAC. 10.5067/MODIS/MCD19A2.006 (2018).
- 30.Chudnovsky AA, Kostinski A, Lyapustin A, Koutrakis P. Spatial scales of pollution from variable resolution satellite imaging. Env. Pollut. 2012;172C:131–138. doi: 10.1016/j.envpol.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Geng G, et al. Satellite-Based Daily PM2.5 Estimates During Fire Seasons in Colorado. J. Geophys. Res.-Atmospheres. 2018;123:8159–8171. doi: 10.1029/2018JC014327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Ma T, Xu Q, Song X. Using MAIAC AOD to verify the PM2.5 spatial patterns of a land use regression model. Environ. Pollut. Barking Essex 1987. 2018;243:501–509. doi: 10.1016/j.envpol.2018.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Lee HJ. Benefits of High Resolution PM2.5 Prediction using Satellite MAIAC AOD and Land Use Regression for Exposure Assessment: California Examples. Environ. Sci. Technol. 2019;53:12774–12783. doi: 10.1021/acs.est.9b03799. [DOI] [PubMed] [Google Scholar]
- 34.National Centers for Environmental Information (NOAA). North American Mesoscale Forecast System (NAM). https://www.ncdc.noaa.gov/data-access/model-data/model-datasets/north-american-mesoscale-forecast-system-nam (2020).
- 35.Liu Y, Sarnat JA, Kilaru V, Jacob DJ, Koutrakis P. Estimating ground-level PM2.5 in the eastern United States using satellite remote sensing. Env. Sci Technol. 2005;39:3269–78. doi: 10.1021/es049352m. [DOI] [PubMed] [Google Scholar]
- 36.Bowman DC, Lees JM. Near real time weather and ocean model data access with rNOMADS. Comput. Geosci. 2015;78:88–95. doi: 10.1016/j.cageo.2015.02.013. [DOI] [Google Scholar]
- 37.Koman PD, et al. Mapping Modeled Exposure of Wildland Fire Smoke for Human Health Studies in California. Atmosphere. 2019;10:308. doi: 10.3390/atmos10060308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giglio L, Csiszar I, Justice CO. Global distribution and seasonality of active fires as observed with the Terra and Aqua Moderate Resolution Imaging Spectroradiometer (MODIS) sensors. J. Geophys. Res. Biogeosciences. 2006;111:G02016. doi: 10.1029/2005JG000142. [DOI] [Google Scholar]
- 39.Whiteman CD, Hoch SW, Horel JD, Charland A. Relationship between particulate air pollution and meteorological variables in Utah’s Salt Lake Valley. Atmos. Environ. 2014;94:742–753. doi: 10.1016/j.atmosenv.2014.06.012. [DOI] [Google Scholar]
- 40.USGS. About 3DEP Products & Services. https://www.usgs.gov/core-science-systems/ngp/3dep/about-3dep-products-services (2015).
- 41.Homer C, et al. Completion of the 2011 National Land Cover Database for the Conterminous United States - Representing a Decade of Land Cover Change Information. Photogramm. Eng. Remote Sens. 2015;81:345–354. [Google Scholar]
- 42.Social Explorer. T2. Population Density (per sq. mile) [3] - Social Explorer Tables (SE) - Census 2010. Social Explorerhttps://www.socialexplorer.com/data/C2010/metadata/?ds=SE&table=T002 (2010).
- 43.Didan, K. MOD13A3 MODIS/Terra vegetation Indices Monthly L3 Global 1km SIN Grid V006 [Data set]. NASA EOSDIS Land Processes DAAC (2015).
- 44.U.S. Department of Transportation Federal Highway Administration. National Highway Planning Network - Tools - Processes - Planning - FHWA. https://www.fhwa.dot.gov/planning/processes/tools/nhpn/index.cfm (2014).
- 45.Katzfuss M. A multi-resolution approximation for massive spatial datasets. J. Am. Stat. Assoc. 2017;112:201–214. doi: 10.1080/01621459.2015.1123632. [DOI] [Google Scholar]
- 46.Xu Y, et al. Evaluation of machine learning techniques with multiple remote sensing datasets in estimating monthly concentrations of ground-level PM2.5. Environ. Pollut. 2018;242:1417–1426. doi: 10.1016/j.envpol.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 47.Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat. Surv. 2010;4:40–79. doi: 10.1214/09-SS054. [DOI] [Google Scholar]
- 48.Watson GL, Telesca D, Reid CE, Pfister GG, Jerrett M. Machine learning models accurately predict ozone exposure during wildfire events. Environ. Pollut. 2019;254:112792. doi: 10.1016/j.envpol.2019.06.088. [DOI] [PubMed] [Google Scholar]
- 49.R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2018).
- 50.Kuhn, M. & Contributions from Jed Wing, S. W., Andre Williams, Chris Keefer and Allan Engelhardt. caret: Classification and Regression Training. (2012).
- 51.Dean-Mayer, Z. A. & Knowles, J. E. caretEnsemble: Ensembles of Caret Models (2019).
- 52.Mayer, M. missRanger: Fast Imputation of Missing Values (2019).
- 53.Reid C, Maestas M, Considine E, Li G. 2021. Machine learning derived daily PM2.5 concentration estimates from by County, ZIP code, and census tract in 11 western states 2008–2018. figshare. [DOI] [PMC free article] [PubMed]
- 54.Gregorutti B, Michel B, Saint-Pierre P. Correlation and variable importance in random forests. Stat. Comput. 2017;27:659–678. doi: 10.1007/s11222-016-9646-1. [DOI] [Google Scholar]
- 55.Maestas MM, Considine E, Li G, colleenereid, Joseph M. 2021. earthlab/Western_states_daily_PM2.5: Release for SciData publication. Zenodo. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Reid C, Maestas M, Considine E, Li G. 2021. Machine learning derived daily PM2.5 concentration estimates from by County, ZIP code, and census tract in 11 western states 2008–2018. figshare. [DOI] [PMC free article] [PubMed]
- Maestas MM, Considine E, Li G, colleenereid, Joseph M. 2021. earthlab/Western_states_daily_PM2.5: Release for SciData publication. Zenodo. [DOI]
Supplementary Materials
Data Availability Statement
All code used for downloading and processing the data used in this project, including the machine learning and technical validation code, may be accessed at 10.5281/zenodo.449926455.
To ensure that our work is reproducible, all code is written in open-source languages. Some scripts are in R and others are in Python. Python scripts need Python 3; R versions beyond 3.5.1 should suffice.
The scripts on Zenodo are a copy of our GitHub repository of scripts and contains the following files and directories:
• General_Project_Functions: Scripts to obtain the prediction set locations and tools that are generally useful during the data processing, such as making buffers around points and reprojecting point coordinates.
• Get_PM25_Observations: Scripts to process PM2.5 observations from across the western U.S. These observations are used to train our machine learning models.
• Get_Earth_Observations: Scripts to download and process observations from data sets that are used both as inputs for our machine learning models during training and as inputs for our models in the prediction stage. The file Overall_steps provides all necessary directions. Individual README files (in each folder) provide more details if there are any.
• Merge_Data: Scripts to merge all the data together and derive some spatio-temporal variables.
• Machine_Learning: Scripts to run and evaluate our machine learning models. The folder Final_scripts contains all code used for our final analysis. The code in the Exploring_models folder was all preliminary testing.
• Estimate_PM25: Scripts to use our machine learning models to make final predictions and to explore the prediction data sets over time and space.