Abstract
Net blotch, induced by the ascomycete Pyrenophora teres, has become among the most important disease of barley (Hordeum vulgare L.). Easily recognizable by brown reticulated stripes on the sensitive barley leaves, net blotch reduces the yield by up to 40% and decreases seed quality. The life cycle, the mode of dispersion and the development of the pathogen, allow a quick contamination of the host. Crop residues, seeds, and wild grass species are the inoculum sources to spread the disease. The interaction between the barley plant and the fungus is complex and involves physiological changes with the emergence of symptoms on barley and genetic changes including the modulation of different genes involved in the defense pathways. The genes of net blotch resistance have been identified and their localizations are distributed on seven barley chromosomes. Considering the importance of this disease, several management approaches have been performed to control net blotch. One of them is the use of beneficial bacteria colonizing the rhizosphere, collectively referred to as Plant Growth Promoting Rhizobacteria. Several studies have reported the protective role of these bacteria and their metabolites against potential pathogens. Based on the available data, we expose a comprehensive review of Pyrenophora teres including its morphology, interaction with the host plant and means of control.
Keywords: barley, Hordeum vulgare L., Pyrenophora teres, net blotch, plant growth promoting rhizobacteria
Introduction
Worldwide, net blotch caused by Pyrenophora teres Drechsler [anamorph Drechslera teres (Sacc.) Shoem] is a major foliar disease of barley (Hordeum vulgare L.) causing economic losses by reducing the grain quantity and quality. According to Smedegard-Petersen (1971), net blotch exists in two different forms: the spot and net form of net blotch (SFNB and NFNB), caused by P. teres f. maculata (Ptm) and P. teres f. teres (Ptt) respectively (Smedegård-Petersen, 1976). These two forms have been identified as similar morphologically, however, different at the genetic and pathophysiological levels (Campbell et al., 1999; Liu et al., 2011; Akhavan et al., 2016). Ptt forms dark-brown and longitudinal necrotic lesions, which can turn chlorotic (Lightfoot and Able, 2010), while Ptm is responsible for dark brown circular or elliptical spots with chlorosis on the surrounding leaf tissues (Gupta and Loughman, 2001; Jayasena et al., 2004). The differentiation of these forms has been reported in Sweden (Jonsson et al., 1997), France (Arabi et al., 1992), Western Australia (Gupta and Loughman, 2001), South Africa (Louw et al., 1996), and Western Canada (Akhavan et al., 2016).
The distinction between these two forms is also due to differences in fungal growth and in symptoms’ development (Lightfoot and Able, 2010). Indeed, compared to Ptt, Ptm germinates slowly. Additionally, Ptm forms more intracellular vesicles compared to Ptt and is responsible of the leaf cell death within the fungal penetration area. The hyphal growth of Ptt is more extensive than Ptm before the formation of appressoria on the leaf surface. The time required for Ptm infection is also shorter than Ptt (Liu et al., 2011). Therefore, Ptt feeds and infects as a necrotroph during the infection period and grows only intercellularly. Contrary, Ptm initially develops haustorial-like intracellular vesicles, feeding similarly to a biotroph, and then switches quickly to a necrotrophic growth. Thus, Ptt behaves as a necrotroph, while Ptm acts as a hemibiotroph (Lightfoot and Able, 2010). A recent study has demonstrated that Ptm has significantly higher necrotrophic and saprotrophic growth rates than Ptt (Ronen et al., 2019). Several toxins are produced by both forms of P. teres (Bach et al., 1979; Nukina et al., 1980; Barrault et al., 1982; Friis et al., 1991; Weiergang et al., 2002), namely proteinaceous toxins and low molecular weight aspergillomarasmine-derived toxins contributing to the necrosis and chlorosis (Sarpeleh et al., 2007, 2008). Ptt produces greater quantities of toxins in the culture medium (Lightfoot and Able, 2010). In addition, the toxin composition and amount are different and remain to be established in planta.
Pyrenophora teres f. teres and Pyrenophora teres f. maculata are morphologically very similar, while the disease symptoms are different. Recent studies have shown that they are two phylogenetically distinct species, which are considered to be genetically autonomous populations (Akhavan et al., 2016). A period of evolutionary separation has been suggested thanks to a study of intergenic regions (Ellwood et al., 2012). Primers were developed on the basis of Internal Transcripted Spacer (ITS) regions and they allow to identify and distinguish the two forms of P. teres (Leisova et al., 2006; Mclean et al., 2009). In several parts of the world, the genetic diversity and population of P. teres were explored by using random amplified polymorphic DNA (RAPD; Peever and Milgroom, 1994; Campbell et al., 1999, 2002), amplified fragment length polymorphism (AFLP; Rau et al., 2003; Leisova et al., 2005; Serenius et al., 2007), and simple sequence repeat (SSR) analysis (Keiper et al., 2008; Bogacki et al., 2010; Leišová-Svobodová et al., 2014). In addition, both forms of P. teres have cycles of sexual reproduction occurring on overwintering crop residues followed by multiple cycles of asexual reproduction during the vegetative season (Piening, 1968; Duczek et al., 1999). Therefore, Ptt and Ptm have a mixed breeding and an outcrossing mating system. Because of these characteristics, Ptt and Ptm fall into the category of pathogens having a high capacity to adapt to resistance genes of the plant host as well as to fungicides. The recombination between P. teres isolates can lead to multiple resistances, for example, towards several triazoles (Jalli, 2011; Poudel et al., 2018). Studies have reported that the sexual reproduction between Ptt and Ptm is inducible under laboratory conditions (Campbell et al., 1999; Jalli, 2011). However, other studies have indicated that, under field conditions, hybridizations between Ptt and Ptm are unusual or even absent (Rau et al., 2003, 2007; Serenius et al., 2007; Poudel et al., 2018).
The first genome assembly of Ptt was obtained using the Illumina Solexa sequencing platform leading to a 41.95 Mbp of total assembly size (Ellwood et al., 2010). There have since been additional genomes sequenced and deposited in publicly available repositories including 11 P. teres f. teres genomes and five P. teres f. maculata genomes (Wyatt et al., 2018; Wyatt and Friesen, 2020). The genome assemblies of both forms of P. teres were constructed from long DNA reads, optical and genetic maps. These genomes are highly collinear and each one is composed of 12 chromosomes. The Ptt genome is larger and more repetitive than the Ptm genome (Syme et al., 2018).
In 1973, Shipton and collaborators published the first review on barley net blotch. This review compiled available information concerning the repartition and importance of the disease (Shipton et al., 1973). From 1973 to 2011, other reviews have been published describing the disease epidemiology and the host resistance toward Ptm (Mclean et al., 2009; Liu et al., 2011). In 2020, Clare and collaborators reported the brief consensus maps for all loci published for both barley and P. teres (Clare et al., 2020). To the best of our knowledge, no review has yet described the pathology of the fungus in relation with biological means to increase the resistance of the host. Beneficial bacteria are attracting attention in light of their potential use in agriculture (Babalola, 2010; Dutta and Podile, 2010; Vejan et al., 2016, 2019; Ferreira et al., 2019; Kumari et al., 2019; Prasad et al., 2019; Kumar et al., 2020). The present review provides an overview of the existing knowledge on the interaction between P. teres and barley and summarizes the current and ongoing research on P. teres. It also presents the morphological description, development of the fungus and the interaction with barley, and synthesizes the knowledge on current means used to manage net blotch.
Taxonomy History and Focus on Pyrenophora Species
Established in 1809, the genus Helminthosporium became the repository for a large number of described taxa (Alcorn, 1988). Helminthosporium species attack the graminaceous plants in temperate regions (Sampson and Western, 1940). For instance, in 1943, Helminthosporium oryzae destroyed 90% of the rice in India, leading to a famine situation (Padmanabhan, 1973). This virulence was due to an increase of (i) the mean temperature, (ii) the rainfall, and (iii) the relative humidity. Nowadays, the major climate-change factors, including the increasing CO2, lead to an increase in humidity and therefore to optimal conditions for fungal development (Luck et al., 2011; Mikkelsen et al., 2015).
In 1923, Drechsler distinguished Helminthosporium species with cylindrical conidia growing over the whole cell surface and species with fusoid conidia germinating only at their ends (Drechsler, 1923). In 1929, Nisikado proposed two subgenera based on the description of Drechsler: Cylindro-Helminthosporium and Eu-Helminthosporium (Nisikado, 1929; Shoemaker, 1959; Alcorn, 1988). Later, Luttrell (1963) highlighted the differences between Helminthosporium sensu stricto and Graminicolous species in the development of conidia and conidiophores. Depending on the sexual states, Graminicolous species were separated into three genera, Bipolaris, Drechslera, and Exserohilum or Cochliobolus, Pyrenophora and Setosphaeria, respectively (Ibrahim et al., 1966; Ariyawansa, 2014). The genus Bipolaris contains Eu-Helminthosporium of Nisikado and Helminthosporium for lignicolous species (Shoemaker, 1959; Alcorn, 1988). Leonard and Suggs (1974) established the Exserohilum genus for species having strongly protuberant conidial hilum (Alcorn, 1988). In 1930, Cylindro-Helminthosporium became a genus under the name of Pyrenophora by Ito (1930).
Pyrenophora accounts for 135 species listed in Index Fungorum (2014), most of which are not pathogenic to humans (Ariyawansa, 2014). Gramineae and, more specifically, Hordeum vulgare, are considered as the main hosts for Pyrenophora. Depending on the barley’s symptoms, different species of Pyrenophora can be distinguished. Table 1 summarizes the main species of Pyrenophora involved in barley diseases.
Table 1.
Summary of the different species of Pyrenophora, pathogen of barley.
| Teleomorph stage | Anamorph stage | Host | Symptoms | Bibliographic references |
|---|---|---|---|---|
| Pyrenophora graminea | Drechslera graminea | barley | leaf stripe | (Tekauz, 1983; Porta-Puglia et al., 1986) |
| Pyrenophora japonica | Drechslera tuberosa | barley | leaf spot | (Campbell et al., 1999) |
| Pyrenophora teres f. teres | Drechslera teres f. teres | barley | net form of net blotch | (Brown et al., 1993; Jalli and Robinson, 2000; Afanasenko et al., 2007) |
| Pyrenophora teres f. maculata | Drechslera teres f. maculata | barley | spot form of net blotch | (Campbell et al., 1999; Jayasena et al., 2004; Mclean et al., 2009; Lartey et al., 2012) |
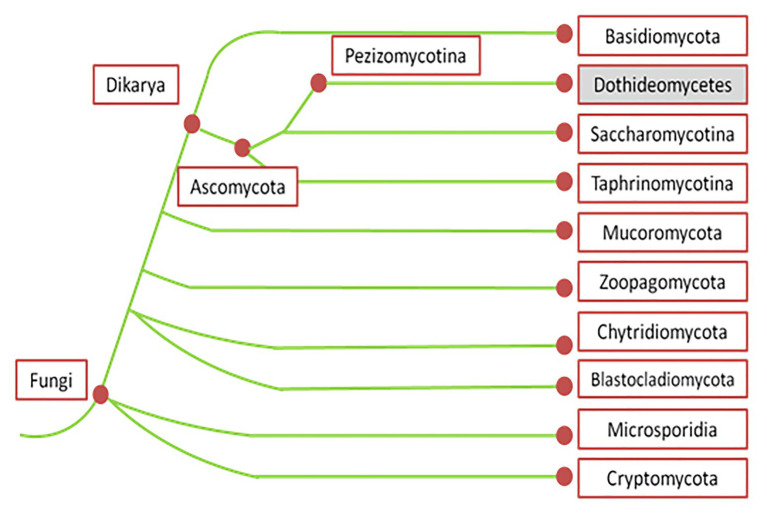
Pyrenophora graminea [Ito & Kuribayashi; anamorph stage: Drechslera graminea (Rabenhorst ex Schlechtendal Shoemaker], is a seed-borne pathogen of barley causing leaf stripe (Porta-Puglia et al., 1986). Pyrenophora japonica S. Ito & Kurib. (anamorph stage: Drechslera tuberosa Shoemaker) is identified as the agent causing leaf spot symptoms on barley (Campbell et al., 1999). Net blotch caused by P. teres [anamorph stage: D. teres (Sacc.) Shoemaker Drechsler] belongs to the kingdom Fungi, phylum Ascomycota, subphylum Pezizomycotina, and class Dothideomycetes (Figure 1; Liu et al., 2011).
Figure 1.
Phylogenetic tree of fungi according to MycoCosm portal (adapted from Grigoriev et al., 2014). According to this phylogenetic tree, Pyrenophora teres belongs to the kingdom Fungi, phylum Ascomycota, subphylum Pezizomucotina, and class Dothideomycetes.
Distribution and Economic Impact of Net Blotch
The ascomycete P. teres is the causal agent of net blotch on spring and winter barley. During the last decades, P. teres has spread throughout the world and ravaged crops in many countries: Australia (Gupta and Loughman, 2001; Mclean et al., 2010), Canada (Turkington et al., 2002; Akhavan et al., 2016), Europe (Dennis and Foister, 1942; Arabi et al., 1992; Plessl et al., 2005), South Africa (Smith and Rattray, 1930; Louw et al., 1996; Campbell et al., 1999), and the United States (Lartey et al., 2012). Net blotch causes important economic problems by reducing barley seed’s quality (Shipton, 1966; Plessl et al., 2005; Jayasena et al., 2007). For instance, in Australia, the economic losses are estimated to be $ 117 × 106 per year (Murray and Brennan, 2010). In addition, yield losses might reach 40% in years with extensive rainfall in Germany (Plessl et al., 2005).
Symptomatology
Pyrenophora teres causes disease and can infect leaves, stems, and kernels of barley (Liu et al., 2011). Like other plant diseases, symptoms’ appearance is dependent on the pathogen virulence, host genotype, and environment. Damages are different on resistant and susceptible varieties. Only few dot-like lesions are present with no development of a net-like pattern on highly resistant barley (Mclean et al., 2009; Liu et al., 2011).
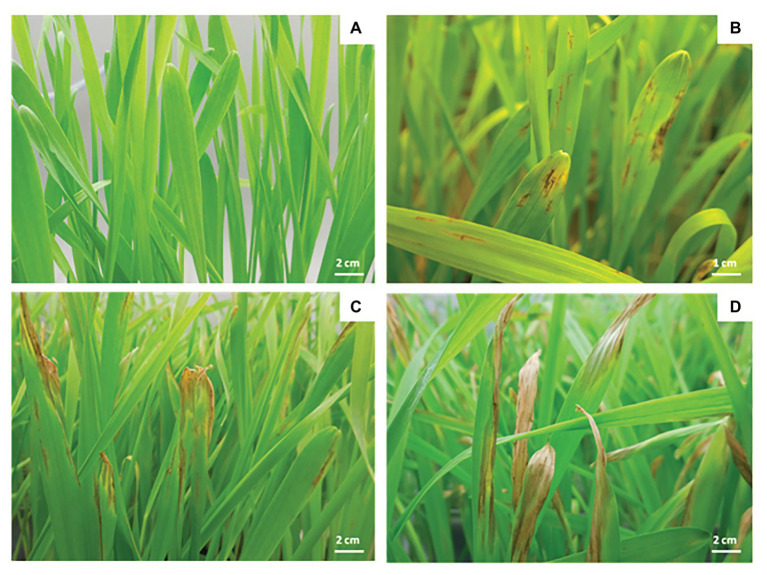
On susceptible varieties, the disease evolves quickly as shown in Figure 2. Twenty-two hours post infection on a six row winter barley named Siberia, the first symptoms appear as brown necrotic spots on infected tissues increasing in size to form elliptical or fusiform lesions to 3 by 6 mm (Figure 2B; Keon and Hargreaves, 1983; Mclean et al., 2009). These necrotic lesions may be accompanied by chlorotic lesions of varying width (Figure 2C). Upon further fungal development, these chlorotic lesions might lead to the entire leaf’s death (Figure 2D). The first wilts appear 40 h after infection (Barrault et al., 1982; Arabi et al., 2003). Then, the oldest leaves start to wither, followed by the youngest.
Figure 2.
Symptoms caused by P. teres on Siberia barley leaves. Control (A), 4 days after infection (B), 7 days after infection (C), and 10 days after infection (D).
These symptoms closely resemble those caused by P. japonica (Scott et al., 1991) and Cochliobolus sativus (Van den Berg, 1988), therefore examination of the conidia is often necessary to distinguish the pathogen agent (Mclean et al., 2009).
Toxins’ Production
The symptoms caused by P. teres are partially induced by various toxins (Weiergang et al., 2002). The produced phytotoxic compounds include pyrenolides, pyrenolines, and three peptide alkaloids, aspergilomarasmine A and its derivatives (Muria-Gonzalez et al., 2020). Pyrenolines (A and B) and pyrenolides (A, B, C, and D) constitute general toxins affecting different plants (Nukina and Hirota, 1992; Sarpeleh et al., 2007) and induced only brown necrotic spots or lesions similar to those induced by the pathogen 72 h after inoculation (Sarpeleh et al., 2007). Pyrenolines A and B represent a class of bioactive metabolites produced by P. teres (Coval et al., 1990). While tested on both monocots and dicots, pyrenoline A shows no host specificity. In addition, pyrenoline B is also active on several plant species but at higher concentrations compared to pyrenoline A. Pyrenolides A, B, and C produced by P. teres, exhibit growth inhibiting and morphogenic activities towards other fungi. For instance, the application of pyrenolide C allows hyphal growth inhibition and the formation of many irregularly swollen hyphae in Cochliobolous lunata (Nukina et al., 1980).
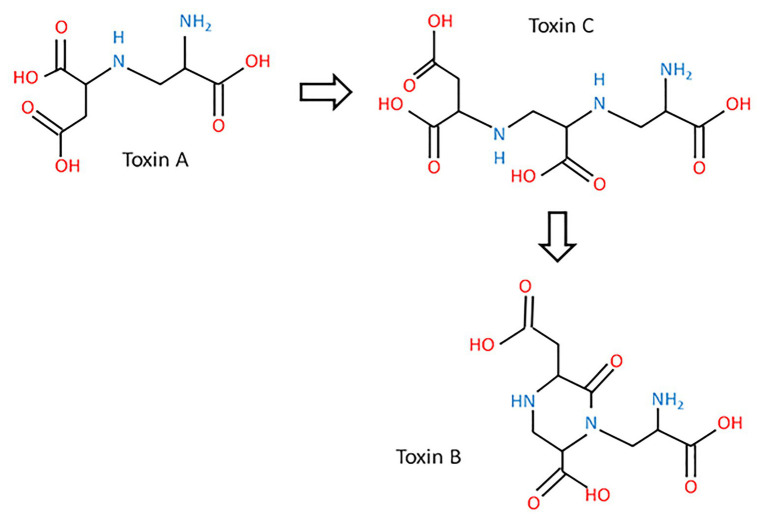
In addition to pyrenolines and pyrenolides, P. teres also produces three other toxins, designated as N-(2-amino-2-carboxyethyl) aspartic acid, anhydrospergillomarasmine A, and aspergilomarasmine A, symbolized by A, B, and C, respectively (Barrault et al., 1982; Friis et al., 1991). These peptides are thermostable with a low molecular weight and cause chlorosis, rather than the classical net blotch necrosis, after infiltration of the pure compound (Muria-Gonzalez et al., 2020). The aspergilomarasmines were first described by Haenni et al. (1965), as metabolites of Aspergillus oryzae and Aspergillus flavus (Haenni et al., 1965). According to their chemical structures, toxin A might be a precursor of toxin C (Figure 3; Friis et al., 1991).
Figure 3.
The biosynthetic pathway of aspergilomarasmine A (toxin C) and its derivatives (toxin A and toxin B) and their chemical structures (inspired from Friis et al., 1991).
These toxins are involved in the development of the net blotch symptoms (Mikhailova et al., 2010). Toxin A is responsible for necrosis, while toxin B and C cause chlorosis (Weiergang et al., 2002; Mikhailova et al., 2010). Moreover, aspergilomarasmines A and B disturb the water balance of the plant cell. Their activities are enhanced by the presence of metal ions, especially ferric ions (Friis et al., 1991). Separated by electrophoresis, toxin C is the first detectable toxin, which is accumulated between 10 and 16 days after inoculation (Friis et al., 1991). The quantity of toxins produced by the fungus impacts the severity of symptoms (Sarpeleh et al., 2007; Mclean et al., 2009). Aspergilomarasmine A (toxin C) and its derivative (toxin A) are toxics at a concentration of 0.25 mmol/L, while the other derivative of aspergilomarasmine A, toxin B, is toxic at 1 mmol/L (Bach et al., 1979; Weiergang et al., 2002). These three peptides belong to host-specific toxins (HST; Stergiopoulos et al., 2013). Their activities are dependent on the age of plants with a higher level on young leaves (Sarpeleh et al., 2008). Toxins of P. teres are considered as virulence factors defined as the degree of damage caused to a host and not as pathogenicity factors representing the qualitative capacity of a pathogen to infect and cause disease on a host (Sacristán and García-Arenal, 2008; Jalli, 2011).
Origin of Contamination
Characterized as a hemibiotroph, P. teres survives saprophytically between cropping seasons. The pathogen is present as mycelium on host crop residues, on seeds before sowing or wild grass species, forming a source of primary inoculum (Jordan and Allen, 1984; Brown et al., 1993). Pyrenophora teres is able to contaminate young shoots and the coleoptile (Jordan, 1981). The pathogenic factor and the quantity of primary inoculum from infected residues depend on several factors. Firstly, the environmental conditions and, more specifically, long periods of wet, increase the primary inoculum levels (Mclean et al., 2009). The amount of residues also directly impacts the disease intensity since the inoculum survives on infected residues. Secondly, the disease levels vary greatly depending on the cultural practice applied. Crop rotation, avoiding barley monoculture and eliminating or reducing primary inoculum in the field are means preventing the pathogen’s development (Liu et al., 2011). For instance, a minimum of 2 years between barley crops is required to prevent net blotch disease (Duczek et al., 1999). Concerning wild grass species, P. teres can infect Agropyron, Bromus, Elymus, Hordelymus, Stipa, and other Hordeum spp. (Mclean et al., 2009). These wild grass species can impact the epidemiology of net blotch, but they do not provide a significant additional inoculum to the next year’s crop (Brown et al., 1993).
Life Cycle of Pyrenophora teres
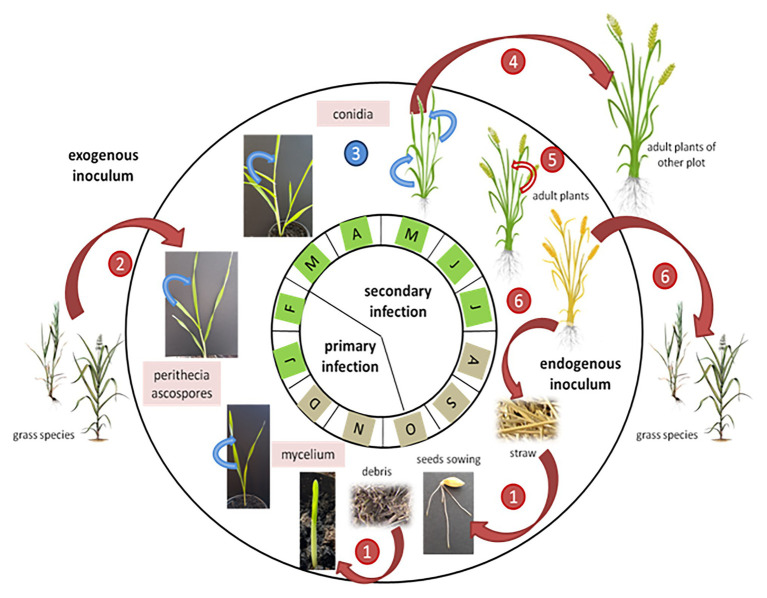
The P. teres lifecycle implies both an asexual and a sexual stage (Figure 4; Jalli, 2011). Conidia are produced during the asexual stage, whereas the sexual stage involves reproduction between isolates of compatible mating types and genetic recombination to produce ascospores (Fowler et al., 2017). The asexual stage occurs during the summer period on residues of the previous barley crop, and triggers the infection in autumn. At the end of the growing season, the fungus produces dark, globosely shaped pseudothecia, 1–2 mm in diameter (Figure 5A; Mclean et al., 2009). As sexual organs, pseudothecia represent the teleomorph or perfect state of P. teres. Once mature, pseudothecia produce asci containing three to eight ascospores measuring 18–28 μm × 43–61 μm, light brown and helical in shape (Liu et al., 2011). The ascospores production is temperature-dependent with an optimum between 15 and 20°C (Jordan, 1981; Mironenko et al., 2005). The spores have three or four transverse septa (Figure 5B). These ascospores are released on infected residues (Figure 5C) until spring constituting the primary inoculum (Duczek et al., 1999). These ascospores are dispersed into the air by the wind, or are splash-dispersed by the rain (Figure 4; Deadman and Cooke, 1989).
Figure 4.
Dynamics of net blotch epidemics adapted from Suffert et al. (2011). Red arrows indicate P. teres wind-dispersed infections and blue arrows indicate splash-dispersed infections. The months with a brown color indicate that the source of inoculum comes from swarming debris on the soil, while the months with a green color indicate a source of inoculum mainly from the aerial parts of the plants. The numbers indicate the pathogen’s infection stages: 1: infection by P. teres ascospores present on infected barley debris; 2: mycelium present on grass species infects barley young plants; 3: the net blotch disease progresses from the bottom to the top of the barley plant; 4: disseminated by the wind, conidia contaminate other barley plants; 5: heavily infected crops show abortion of the ear; and 6: Pyrenophora teres colonizes the senescent tissues and produces perithecia on straw and grass species.
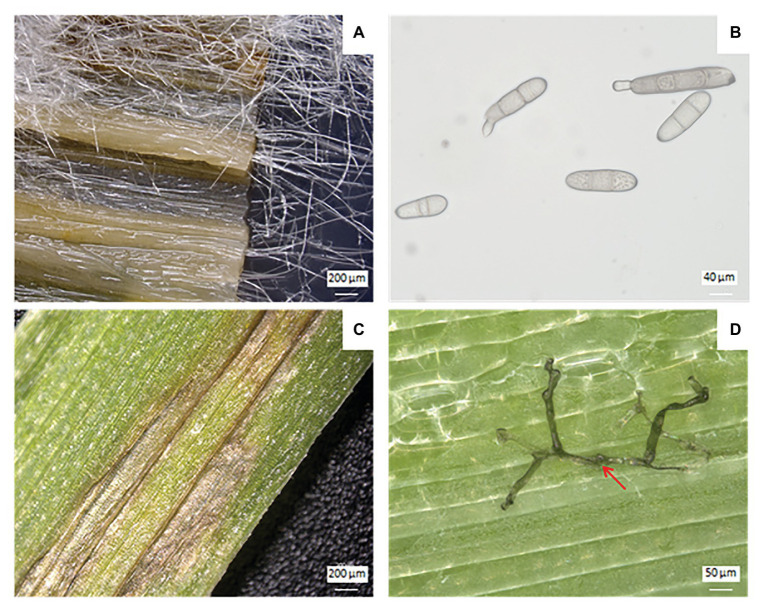
Figure 5.
Vegetative and reproductive forms of P. teres and its symptoms on barley leaves. Mycelium of P. teres in barley leaf deposited on PDA medium (A), conidia of P. teres (B), chlorosis and necrosis symptoms on barley leaf caused by P. teres (C), and penetration of mycelium (red arrow) through barley leaf (D).
Following the primary infection, P. teres produces conidia measuring 30–174 μm × 15–23 μm constituting the asexual cycle (Liu et al., 2011). Generally, conidia are elongated with rounded ends. This secondary infection sets in 14–20 days after the primary infection and increases the severity of the disease (Mclean et al., 2009). The conidia, considered as the anamorph stage or imperfect stage of P. teres are found on conidiophores, solitary or grouped by two or three and swollen at the base (Mclean et al., 2009). Conidiophores need light to develop whereas conidia appear in the darkness (Wright and Sutton, 1990). Conidia of P. teres are recognizable from other pathogens due to their inside septum (Alcorn, 1988). The conidia are qualified as viable when they present more than two segments, otherwise they will not germinate and cannot penetrate plant tissues (Figure 5D). Between April and August, conidia are disseminated by the wind and/or rainfall to surrounding barley plants, or carried for long distances to reach new barley fields initiating, thus, the secondary infection cycle (Figure 4; Duczek et al., 1999). The infection process begins with the germination of ascospores or conidia on leaves. After penetrating the outer epidermal cell wall of barley, P. teres develops within a large intracellular vesicle called the primary vesicle (Liu et al., 2011). Then, vesicles are formed inside the sub-stomatal chamber leading to haustoria, the secondary hyphae (Keon and Hargreaves, 1983).
Thus, the pathogen contaminates the leaf, particularly the epidermal cells, within 48 h post infection (Jørgensen et al., 1998). The disease progresses from the bottom to the top of the plant (Figure 4). Further, the severity of damages is lower in older plants. Indeed, the older plants have a thicker cuticle limiting the penetration of the pathogen and have a greater capacity for producing antifungal substances (Khan and Boyd, 1969; Douiyssi et al., 1998). Once secondary infection is completed, P. teres colonizes the senescent tissues and produces pseudothecia on straw or weed residues (Figure 4; Liu et al., 2011).
Methods for the Disease Assessment
Field trials can be tested, known as “hill summer,” to follow the development of the disease and to test resistance to net blotch in different barley cultivars. For hill summer trials, barley seeds are sown in hill plots at a distance between hills of 50 cm at the beginning of August. Pyrenophora teres inoculation is conducted by distributing naturally infested straw debris before sowing (early August; König et al., 2013; Vatter et al., 2017).
The evaluation of the disease in the laboratory and under controlled conditions is as follows: the barley plants are grown in controlled enclosure (temperature, humidity, and light) following a cycle of 23°C day/22°C night, 80% relative humidity, 14/10 h day/night photoperiod. For example, the P. teres pathogen can be sprayed with a sprayer on the leaves at different stages of growth after sowing in order to follow the disease development. For the control condition, the barley plants are sprayed with a sterile water solution. As soon as the pathogen is inoculated, the barley plants are placed in a hood to increase the humidity level, allowing the pathogen to improve its development. This same experiment can be carried out in a greenhouse under less controlled conditions, i.e., with a natural photoperiod.
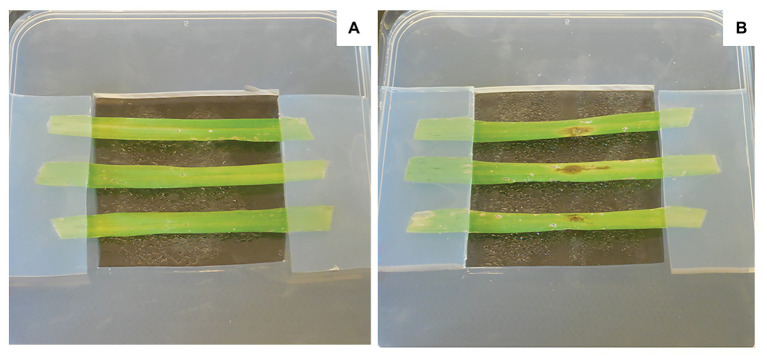
The disease can also be followed on detached leaves (Figure 6). Indeed, the detached plant leaf assay is a rapid technique of assessment under controlled conditions. For this experiment, barley leaves are cut to equal lengths and then placed on filter paper or on an agar-agar medium deposited or poured into a sterile plastic Petri dish. The barley leaves can be kept taut by means of agar–agar bands. Then, the inoculation is carried out by depositing 5–10 μl of a suspension of P. teres spores on previously injured barley leaves (Deadman and Cooke, 1986; El-Mor et al., 2017).
Figure 6.
Assessment on detached leaves. The barley leaves were disinfected in an ethanol bath at 70°C followed by rinsing in three successive sterile water baths. All the leaves were cut to the same length then placed on agar-agar medium. These leaves were then wounded with a wooden pick. A volume of 10 μl was deposited at the level of the wound containing sterile water for a control condition (A) or P. teres spores at a concentration of 105 spores.ml−1 for the infected condition (B).
The infection response based on the measurement of individual lesion sizes (dimension; mm) for each second leaf was assessed 15 days after inoculation of P. teres. The 0–5 scale was used in this methods, where the scores 0–5 indicate resistant and increasingly susceptible barley phenotypes (Arabi and Jawhar, 2010). The disease severity for net blotch was scored on a dozen barley plants using modified Saari and Prescott’s double-digit scale (D1D2, 00–99) scoring method, which was based on the severity scale to assess foliar diseases in cereals. The first digit (D1) represents the relative height of the disease on the plant and corresponds to the vertical disease progression. The second digit (D2) refers to the severity measured as diseased leaf area (Saari and Prescott, 1975; Jalata et al., 2020).
Genetics of the Interaction Between Barley and Pyrenophora Teres
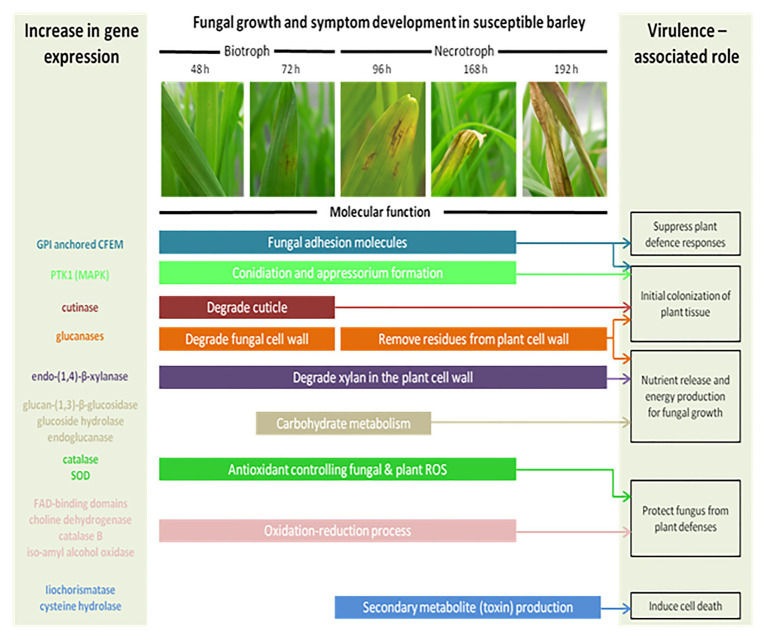
At the molecular level, several mechanisms are involved during the interaction between P. teres and barley. In many fungal diseases, the infection cycle begins with the development of penetration structures triggered by the perception of chemical and/or physical signals from the plant surface. During the initial interaction, the mitogen-activated protein kinase (MAPK) signal transduction pathway plays crucial roles in the pathogenesis process. In P. teres, the MAPK PTK1 gene allows the conidiation, appressorium formation, and pathogenicity on barley (Figure 7; Ruiz-Roldán et al., 2001). Other proteins containing a common domain in several fungal extracellular membrane proteins (CFEM), an eight cysteine-containing domain, are also required for appressorium development. CFEM-containing proteins could act as signal transducers or cell-surface receptors during host-pathogen interactions (Kulkarni et al., 2003; Ismail et al., 2014a).
Figure 7.
The schematic model of the fungal growth and symptoms’ development in a susceptible barley leaf. Each enzyme occurs at a specific time of infection. At 24 h, the spores germinate and form an appressorium through the plant epidermis supposing a biotrophic stage. By 96 h, P. teres is growing inside the barley leaf and develops chlorosis and necrosis suggesting a necrotroph stage. Colored bars represent the genes expressed during the plant/pathogen interaction and describe their molecular function(s) (Inspired from Ismail and Able, 2017).
Pyrenophora teres is able to modulate its cell wall in function of the vegetative or reproductive state and of the culture media used. Indeed, genes involved in the synthesis and remodeling of cell wall polysaccharides, namely chitin, β-(1,3)-glucan, mixed-linkage glucan, as well as endo/exoglucanases and a MAPK, varied in expression in P. teres spores and mycelium after cultivation on several media (Backes et al., 2020).
The cell wall of barley constitutes the first barrier to P. teres. To infect the plant, P. teres produces proteins degrading the plant cell wall (Muria-Gonzalez et al., 2020). These enzymes belong to cell wall degrading enzymes (CWDE; Figure 7). Each of these enzymes plays a role at several time points during the plant-pathogen interaction. Pyrenophora teres is characterized as a hemibiotrophic pathogen appearing as a biotroph during the first 24–48 h in contact with its host before switching to the necrotrophic stage (Ismail and Able, 2017). So, cutinase degrades the plant cuticle and participates to the initial colonization of plant tissues. Some of these proteins have been identified such as endo-(1,4)-β-xylanase and glucan-(1,3)-β-glucosidase precursors (Ismail et al., 2014b; Ismail and Able, 2016). Endo-(1,4)-β-xylanase is the key enzyme responsible for degradation of xylan, predominant hemicellulose in the plant cell (Beliën et al., 2006). Glucan-(1,3)-β-glucosidase precursor catalyzes the liberation of α-glucose from β-(1,3)-glucan, other principal component of plant cell wall (Boonvitthya et al., 2012). According to Ismail and Able (2017), genes coding for glycoside hydrolase family 43 protein, glycoside hydrolase family 105 protein, and endoglucanase-5 are highly expressed during the barley – P. teres interaction (Figure 7).
After the pathogen attack, the carbohydrate metabolism is impacted in plants. In barley, the sugar transporter genes are induced after pathogen attack (Williams et al., 2000; Bogacki et al., 2008). In the same way, the gene coding for a putative invertase allows to satisfy the increased metabolic demand on barley leaf tissue. An invertase-encoding gene, which cleaves sucrose to glucose and fructose, is induced in plants infected by fungal pathogens (Fotopoulos et al., 2003). The energy normally used for the primary metabolism is intended for defense mechanisms activated by the host plant against pathogens (Rojas et al., 2014). Consequently, these energy requirements for plant defense responses decrease the yield in barley (Shipton, 1966; Rojas et al., 2014).
Filamentous fungi may also produce effectors or proteinaceous toxins to facilitate the susceptible host colonization and notably cereal hosts (Gardiner et al., 2012; Ismail et al., 2014b; Muria-Gonzalez et al., 2020). Three proteins have been identified in P. teres: a cysteine hydrolase family protein, an endo-1,4- β-xylanase A, and an unknown secreted protein. The cysteine hydrolase family protein shows homology to an isochorismatase known as an enzyme suppressing plant defense (Ismail et al., 2014a).
During the interaction between P. teres and barley, a series of complex molecular and physiological processes activate and often lead to plant cell death, referred to as the hypersensitive reaction (HR; Goodman and Novacky, 1994; Greenberg, 1997; Heath, 2000; Lam et al., 2001; Greenberg and Yao, 2004; Mur et al., 2008; Künstler et al., 2016; Balint-Kurti, 2019). In the initiation of the HR, the oxidative burst triggers the production of reactive oxygen species (ROS). During the interaction between barley and pathogens, great quantities of ROS are produced resulting in an increase of antioxidant enzymes’ expression such as catalase and superoxide dismutase (SOD; Able, 2003). For instance, HvCSD1, a cytosolic SOD isolated from barley, is upregulated in barley tissues infected by P. teres (Lightfoot et al., 2017). The barley resistance to P. teres is correlated to this SOD activity increase and the HvCSD1 seems to be important in the cytosolic redox status maintenance. Other genes involved in oxidation-reduction processes and defense mechanisms in plants including FAD-binding domains proteins, a choline dehydrogenase, or an iso-amyl alcohol oxidase were upregulated during the infection of barley by P. teres (Ismail and Able, 2017). Identified in 16 P. teres proteins, FAD-binding domain proteins are associated with multiple secondary metabolite pathways (Ismail and Able, 2016; Muria-Gonzalez et al., 2020). During infection of Septoria nodorum in wheat, the activity of catalase was also increased with the virulence of the pathogen and with the time post inoculation (Maksimov et al., 2013; Ismail and Able, 2017). Contributing to amine and polyamine biosynthesis, the choline dehydrogenase plays a role in barley infection by P. teres. The isoamyl alcohol oxidase produces aspergilomarasmine-derivatives, a toxin responsible for the chlorotic symptoms observed and described above.
Net Blotch Disease Management
Chemical Control
Causing 70% of cereal diseases, fungi are commonly controlled by fungicides. These products aim to ensure yield and to secure the quality of the harvest (Bartlett et al., 2002). Fungicides of the quinone outside inhibitors (QoI), the succinate dehydrogenase inhibitor (SDHI), and azole or demethylase inhibitor (DMI) classes are used as site-specific systemic fungicides (Mair et al., 2016). The foliar fungicide application effectiveness to control net blotch has been largely carried out (Sutton and Steele, 1983; Jayasena et al., 2002; Mclean et al., 2009). First studies have shown that triazole-based fungicides by pulverization allowed to control net blotch (Sutton and Steele, 1983; Van Den Berg and Rossnagel, 1990; Jayasena et al., 2002). Triazoles, known as DMI (propiconazole and prothioconazole), inhibit dimethylation between substrates that are necessary for the biosynthesis of ergosterol in fungi. In addition, SDHIs are also used to reduce the disease severity. In 1969, carboxin was the first SDHI fungicide launched, followed by several other SDHIs with a narrow spectrum of activity. The target of SDHI fungicide is the succinate dehydrogenase (SDH) enzyme or succinate ubiquinone oxidoreductases, which play an essential role in the tricarboxylic cycle and the mitochondrial electron transfer chain (Rehfus et al., 2016). The strobilurins, a new class of broad-spectrum fungicides, have been adopted these last years for net blotch control (Bartlett et al., 2002). Strobilurin fungicides were inspired by natural fungicidal derivatives of β-methoxyacrylic acid (Bartlett et al., 2002). Belonging to QoI (pyraclostrobin and picoxystrobin), strobilurins are natural substances isolated mainly from fungi and more specifically, Basidiomycetes. The strobilurin name is derived from the fungi genera Strobilurus (Balba, 2007). First introduced to the market in 1996, strobilurins inhibit mitochondrial respiration by blocking electron transfer at the level of cytochromes b and c (Bartlett et al., 2001; Gisi et al., 2002; Balba, 2007).
The antifungal efficacies depend also on the period of their application and how they are applied, as well as on the plant growth stage (Van Den Berg and Rossnagel, 1990). Seed treatments were successful if applied early in the season corresponding at Zadoks growth stage 23–24, but less at later growth stages (Martin, 1985). Barley seeds are considered as a source of inoculum for the ascomycete P. teres. The severity of the barley net blotch is reduced when a fungicide seed treatment is applied (Martin, 1985). Seed treatment effectiveness depends on fungal sensitivity, chemical fungitoxicity, and seed coverage quality. Iprodione is the fungicide providing the best control of dematiaceous fungi (Bipolaris and Drechslera) on seeds (Reis et al., 2012). Another study demonstrates the efficiency of one application of propiconazole at spike emergence for the management of net blotch (Sutton and Steele, 1983). A correct application of fungicides before the emergence of the flag leaf and the ear aims to protect the photosynthetic potential of the top four leaves, which contribute to 72% of the total yield (Mclean et al., 2009). A single application of propiconazole is not enough when the pathogen progresses quickly. A recent study demonstrates that two applications with the combination pyraclostrobin and epoxiconazole improved net blotch control and increased the yield in both experimental years (Stepanovic et al., 2016). Belonging to QoI, metyltetraprole is a new fungicide, which is effective against important cereal diseases, including net blotch (Suemoto et al., 2019). Further, the metyltetraprole suppresses succinate-cytochrome c reductase activity in QoI susceptible P. teres.
With time, resistant strains to these products have emerged (Jørgensen and Olsen, 2007). In Europe and in Australia, P. teres developed a resistance to DMI fungicides (Peever and Milgroom, 1993; Mair et al., 2016; Rehfus et al., 2016). Shortly after the first QoIs uses, resistant isolates to these antifungal products were detected in field populations (Gisi et al., 2002). More specifically, in 2003, resistance to QoI fungicides in P. teres was detected in France, Sweden, and Denmark. The resistance mechanism to QoIs has been identified as mutations in the mitochondrial target gene, cytochromes b (Sierotzki et al., 2007). In P. teres, this mutation has been described as a substitution of phenylalanine to leucine at amino acid position 129 (Sierotzki et al., 2007). To conclude, the fungicide exerts a selection pressure, which leads to the selection of isolates, which have a mutation providing fungicide resistance, while susceptible isolates will be eliminated. There is a subsequent increase in the number of resistant individuals in the population. Successive rounds of fungicide use repeat the selection of resistant isolates, which leads to the increase of the resistance mutation in the population each time the fungicide is used. Eventually, the resistant isolates will dominate the population and the effectiveness of the fungicide will be reduced (Gisi et al., 2000).
In addition to the resistance among several fungal species, azoles use has also been affected by a restriction with a wide range of significant toxicities, including hallucinations, hepatotoxicity, and QTc prolongation (Thompson et al., 2009; Gintjee et al., 2020). Faced with these problems, varietal selection, preventive agronomic measures, and biocontrol agents might be considered as alternative solutions to fungicides products.
Host Plant Resistance
Cereals are selected according to their disease resistance (Jonsson et al., 1998). By definition, a resistant genotype is characterized by having the least and smallest visible foliar lesions, a fungal restricted growth on the infected leaf tissue and an increased production of antifungal products by barley leaves (Graner et al., 1996). Several studies have demonstrated the existence of resistance genes and loci to net blotch depending on the different forms of P. teres.
Geschele (1928) has first demonstrated the resistance to P. teres f. teres to be quantitatively inherited (Clare et al., 2020). The genetic control of resistance to P. teres in barley was first conducted in United States in 1955 (Afanasenko et al., 2007). In 1955, the first gene Pt1 conferring the barley resistance to P. teres was found by Schaller. Later, two additional loci, designated Pt2 and Pt3 were identified by Mode and Schaller in 1958 (Mode and Schaller, 1958; Graner et al., 1996). Using different molecular techniques, several studies have identified net blotch resistance genes or quantitative trait loci (QTL) on all seven barley chromosomes (Mode and Schaller, 1958; Steffenson et al., 1996; Manninen et al., 2000; Gupta et al., 2011; Clare et al., 2020). Major QTL have been identified on barley chromosomes 1H (Bockelman et al., 1977; Manninen et al., 2000, 2006), 2H (Bockelman et al., 1977; Williams et al., 1999; Ma et al., 2004; Adawy et al., 2013; Tamang et al., 2019), 3H (Bockelman et al., 1977; Graner et al., 1996), 4H (Friesen et al., 2006; Grewal et al., 2008; Adawy et al., 2013; Islamovic et al., 2017), 5H (Manninen et al., 2006; Adawy et al., 2013), 6H (Ma et al., 2004; Friesen et al., 2006; Abu Qamar et al., 2008; Grewal et al., 2008; Gupta et al., 2011; Adawy et al., 2013), and 7H (Williams et al., 1999; Grewal et al., 2008; Mclean et al., 2009; Tamang et al., 2019). Localized on chromosome 6H, the Rpt5 locus has been reported by several studies and is considered to be essential in the P. teres f. teres – barley interaction (Clare et al., 2020). According to several studies, the majority of the markers significantly associated with NFNB resistance localize to the centromeric region of chromosome 6H (Abu Qamar et al., 2008; Adawy et al., 2013; Richards et al., 2016). In the same way, the high-resolution mapping of a dominant susceptibility locus located in the centromeric region of barley chromosome 6H has been described using markers (Richards et al., 2016). Therefore, these results indicate the importance of this region. In addition, the Rpt7 locus confers resistance to P. teres f. teres in barley on the chromosome 4H. Recently, 449 barley accessions were phenotyped for P. teres f. teres resistance in greenhouse trials. Using genome-wide association, the results identified 254 marker-trait associations corresponding to 15 QTLs. Four of these regions were new QTL not described in previous studies and are located on chromosome 3H at 233–350 Mpb, 5H at 579 Mbp, 6H at 406–410 Mpb and 7H at 5 Mbp, respectively (Novakazi et al., 2019).
Initially, the genetics conferring resistance to P. teres f. maculata contained three major designated loci and therefore has been considered less complex to compare the P. teres f. teres – barley interaction (Clare et al., 2020). Designated as Rpt4, Rpt6, and Rpt8, these three major loci confer in barley a resistance to P. teres f. maculata. The Rpt4, Rpt6, and Rpt8 loci are localized on chromosome 7H, 5H, and 4H, respectively. Burlakoti et al. (2017) revealed the effect of two- and six-row barley, and concluded that the two-row barley (13%) resistant to P. teres f. maculata was less than the six-row barley (43%) tested.
The use of resistant varieties is proving to be one of the most effective methods with the least environmental impact. Nevertheless, this selection has a high financial cost for farmers (Mclean et al., 2009).
Preventive Agronomic Measures
Based on the life cycle described earlier in this review, three sources can form the primary inoculum of which infected seeds, crop debris, and straw residue. Therefore, the first step to control net blotch is the deletion of the primary inoculum of P. teres by sowing healthy seeds (Jalli, 2011). After harvesting barley kernels, debris and straw residues are other sources of primary inoculum. A study has demonstrated that amounts of residue infested can increase disease intensity and thus reduce the yield (Adee, 1989). To limit the primary inoculum and reduce straw debris, straw may be baled and removed from fields leaving standing stubble after the harvest (Jordan and Allen, 1984). In addition, some farmers practiced open field burning a few years ago. However, due to the pollution (smoke and smuts) generated the loss of organic matter and damage to wildlife and hedges, this practice of burning is now prohibited. Today, new means exist to eliminate the quantity of inoculum present on the straw residues such as chopping and burying (Jordan and Allen, 1984).
In addition to these measures to limit the sources of inoculum, preventive agronomic measures play an essential role in the management of net blotch. Crop rotation is beneficial to reduce the severity of the pathogen. A minimum of 2 years between barley crops is required to prevent net blotch (Duczek et al., 1999). The type of seedling has also an influence on the severity of net blotch. Today, agricultural trends aim to reduce field trips with direct seeding for example. Consisting of sowing the seeds directly in undisturbed soil, direct sowing increases the quantity of P. teres present on crop debris and straw residues (Jordan and Allen, 1984; Mclean et al., 2009). Therefore, direct sowing reduces the cost of production but increases P. teres severity (Jørgensen and Olsen, 2007).
Biological Control
The development of environmentally friendly methods based on biological agents is of great interest in the context of the establishment of a sustainable agriculture (Cazorla et al., 2007). The seeking and the selection of biological agents targeting the pathogen, plays an essential role in the success of biological control strategies (Cazorla et al., 2007). Biological control brings together a set of biocontrol agents including microorganisms (bacteria, fungus, and viruses), macroorganisms (birds, insects, and nematodes), as well as molecules derived from these organisms (natural substances and chemical mediators; Pérez-García et al., 2011). Biological control results from a combination of mechanisms, including competition for nutrients and space, production of antibiotics, and induced systemic resistance to pathogens.
To effectively limit pathogens, biocontrol agents need to rely on plant colonization strategies and maintain a high population density (Ghorbanpour et al., 2018). Competition for nutrients and the space between biological control agents and pathogens is considered the primary mode of action (Sharma et al., 2009). The antibiotics production and antibiosis phenomenon constitute the second mechanism, which controls the development and spread of pathogens (Sharma et al., 2009). The last means for biological control agents to limit the development of pathogens is by inducing systemic resistance. Following recognition with a beneficial organism, the plant can respond systemically and rapidly to the perception of the pathogen. The phenomenon called “priming” allows the plant host to set up more quickly and massively the various defense mechanisms against pathogens (Conrath et al., 2002).
In this review, we only highlight the bacteria defined as plant growth promoting rhizobacteria (PGPR) and used in biological control thanks to their beneficial effects.
The term “PGPR” was first coined in 1978 and these bacteria are classified into two groups. The first group includes bacterial strains able to colonize roots, enhance emergence, stimulate growth either directly, by the capability of synthesizing plant growth-promoting substance, or indirectly, by changing the microbial composition in the rhizosphere in favor of the beneficial micro-organisms (Ait Barka et al., 2000, 2002; Hardoim et al., 2008; Turan et al., 2012). They may induce a systemic resistance to pathogens and modulate the plant regulatory mechanisms through the production of hormones such as auxin, cytokinins, and gibberellins (phytostimulators; Van Loon et al., 1998; Ait Barka et al., 2000, 2006; Bloemberg and Lugtenberg, 2001). In addition, some beneficial bacteria fix atmospheric nitrogen, solubilize inorganic nutrients limiting plant growth, stimulate nutrient delivery and uptake by plant roots and improve nutrient and water management (biofertilizers; Babalola, 2010; Afzal et al., 2019). A second group prevents or decreases the deleterious effects of pathogens (Turan et al., 2012). These bacteria are used in agricultural practices against diseases in light of the improved plant performance under environmental stress and, consequently, of yield enhancement (Ait Barka et al., 2000). The biocontrol agents that are best-characterized belong to the genus Pseudomonas, Streptomyces Paraburkholderia, and Bacillus (Bloemberg and Lugtenberg, 2001; Beneduzi et al., 2012; Esmaeel et al., 2016, 2018; Shafi et al., 2017; Newitt et al., 2019). For instance, a study shows that Bacillus substilis has antagonist activity against the causal agent of Fusarium head blight in wheat under in vitro and greenhouse assays (Palazzini et al., 2016).
In the rhizosphere, a community of several strains is often more stable, suppresses a broader range of pathogens and sets up different mechanisms of biological control (Jetiyanon and Kloepper, 2002). The great potential for using bacteria has been shown as an alternative to fungicides in the management of plant diseases (Ait Barka et al., 2002).
The microorganisms are able to produce a wide range of antimicrobial peptides including small bacteriocins and fungal defensins (Waghu and Idicula-Thomas, 2020). In addition, these microorganisms are also able to produce secondary metabolites such as peptaibols, cyclopeptides, and pseudopeptides by non-ribosomal synthesis (Montesinos et al., 2012). The beneficial bacteria also produce antifungal antibiotics, named cyclic lipopeptides, which allow acting as antagonistic agents against pathogens. These molecules with low-molecular-weight are deleterious to the growth of other organisms (Beneduzi et al., 2012). Synthesized in a non-ribosomal manner, lipopeptides exhibit surfactant and antimicrobial activities due to amphiphilic features that have drawn attention (Cazorla et al., 2007). These antibiotic compounds are mostly produced by Bacillus species and Burkholderia and more specifically, Paraburkholderia phytofirmans species (Ongena et al., 2007; Pérez-García et al., 2011; Esmaeel et al., 2016, 2018).
Use of Biocontrol Toward Pyrenophora teres
Concerning P. teres, many species of Trichoderma produce secondary metabolites like volatile organic compounds (VOCs) in the presence of P. teres. These VOCs have been identified as sesquiterpenes, diterpenes, terpenoids, and eight-carbon compounds. In addition, VOCs inhibit mycelium growth and lead to unpigmented mycelium with vacuolization (Moya et al., 2018). In the same way, Trichoderma isolates significantly decrease the severity of P. teres, up to 55% on barley seedlings, 70% on leaves and 77% on stems (Moya et al., 2020). Finally, the fungus Clonostachys rosea isolate IK726 reduces the infection caused by P. teres under controlled conditions (Jensen et al., 2016).
The use of biological agents is considered as one of the most promising methods for more rational and safer crop-management practices (Ongena and Jacques, 2008) since they reduce the inputs use and increase the plant vigor. Future experiments will confirm the biological control use to limit net blotch.
Conclusion and Future Perspectives
This review provides a reference point on net blotch of barley by highlighting P. teres severity, the changing complexity during the interaction between barley and P. teres and the management of net blotch control. Net blotch has become a major foliar disease of barley in many countries of the world. Caused by the ascomycete P. teres, this disease causes significant grain yield loss and reduces grain quality. Net blotch develops quickly when the environmental conditions are optimal including long periods of wet and cultural practice used. Additionally, P. teres produces a large quantity of toxins improving its virulence factor. The variability of the fungal pathogenicity leads to the conclusion that symptoms’ occurrence is dependent on the host genotype, the pathogen virulence, and the environment. The net blotch control provides a significant challenge now and in the future. Chemical control, host plant resistance, and preventive agronomic measures are used for net blotch management. Among them, as an environmental-friendly means, the biocontrol agents appear as a promising tool towards a sustainable agriculture.
Author Contributions
AB wrote the paper. All authors have proofread, amended, and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge BAYER SAS Lyon for providing the P. teres, and more specifically Marie-Pascale Latorse, Stéphane Brunet, and Catherine Wantier for their participation in this study.
Footnotes
Funding. This work was supported by grand-Reims, grand-Est Region, and the European Union funding through the INTERREG France-Wallonie-Vlaanderen a SmartBioControl project (BioScreen project).
References
- Able A. J. (2003). Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma 221, 137–143. 10.1007/s00709-002-0064-1, PMID: [DOI] [PubMed] [Google Scholar]
- Abu Qamar M., Liu Z. H., Faris J. D., Chao S., Edwards M. C., Lai Z., et al. (2008). A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor. Appl. Genet. 117, 1261–1270. 10.1007/s00122-008-0860-x, PMID: [DOI] [PubMed] [Google Scholar]
- Adawy S. S., Diab A. A., Sayed A. H. I., Ibrahim S. D., El-Morsy S. I., Saker M. M. (2013). Construction of genetic linkage map and QTL analysis of net blotch resistance in barley. Int. J. Adv. Biotechnol. Res. 4, 348–363. [Google Scholar]
- Adee E. A. (1989). The effect of primary inoculum level of Pyrenophora tritici-repentis on tan spot epidemic development in wheat. Phytopathology 79, 873–877. 10.1094/Phyto-79-873 [DOI] [Google Scholar]
- Afanasenko O., Mironenko N., Filatova O., Kopahnke D., Krämer I., Ordon F. (2007). Genetics of host-pathogen interactions in the Pyrenophora teres f. teres (net form)—barley (Hordeum vulgare) pathosystem. Eur. J. Plant Pathol. 117, 267–280. 10.1007/s10658-006-9093-5 [DOI] [Google Scholar]
- Afzal I., Shinwari Z. K., Sikandar S., Shahzad S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. 10.1016/j.micres.2019.02.001, PMID: [DOI] [PubMed] [Google Scholar]
- Ait Barka E., Belarbi A., Hachet C., Nowak J., Audran J. C. (2000). Enhancement of in vitro growth and resistance to gray mould of Vitis vinifera co-cultured with plant growth-promoting rhizobacteria. FEMS Microbiol. Lett. 186, 91–95. 10.1111/j.1574-6968.2000.tb09087.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ait Barka E., Gognies S., Nowak J., Audran J. C., Belarbi A. (2002). Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote the grapevine growth. Biol. Control 24, 135–142. 10.1016/S1049-9644(02)00034-8 [DOI] [Google Scholar]
- Ait Barka E., Nowak J., Clément C. (2006). Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl. Environ. Microbiol. 72, 7246–7252. 10.1128/AEM.01047-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan A., Turkington T., Askarian H., Tekauz A., Xi K., Tucker J. R., et al. (2016). Virulence of Pyrenophora teres populations in western Canada. Can. J. Plant Pathol. 38, 183–196. 10.1080/07060661.2016.1159617 [DOI] [Google Scholar]
- Alcorn J. L. (1988). The taxonomy of "Helminthosporium" species. Annu. Rev. Phytopathol. 26, 37–56. 10.1146/annurev.py.26.090188.000345 [DOI] [Google Scholar]
- Arabi M. I. E., Al-Safadi B., Charbaji T. (2003). Pathogenic variation among isolates of Pyrenophora teres, the causal agent of barley net blotch. J. Phytopathol. 151, 376–382. 10.1046/j.1439-0434.2003.00734.x [DOI] [Google Scholar]
- Arabi M. I., Barrault G., Sarrafi A., Albertini L. (1992). Variation in the resistance of barley cultivars and in the pathogenicity of Drechslera teres f. sp. maculata and D. teres f. sp. teres isolates from France. Plant Pathol. 41, 180–186. 10.1111/j.1365-3059.1992.tb02336.x [DOI] [Google Scholar]
- Arabi M., Jawhar M. (2010). Greenhouse method for assessing spot blotch resistance in barley. Plant Pathol. J. 26, 421–423. 10.5423/PPJ.2010.26.4.421 [DOI] [Google Scholar]
- Ariyawansa H. (2014). Pyrenophora. Mycosphere 5, 351–362. 10.5943/mycosphere/5/2/9 [DOI] [Google Scholar]
- Babalola O. O. (2010). Beneficial bacteria of agricultural importance. Biotechnol. Lett. 32, 1559–1570. 10.1007/s10529-010-0347-0, PMID: [DOI] [PubMed] [Google Scholar]
- Bach E., Christensen S., Dalgaard L., Larsen P. O., Olsen C. E., Smedegård-Petersen V. (1979). Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres. Physiol. Plant Pathol. 14, 41–46. 10.1016/0048-4059(79)90023-7 [DOI] [Google Scholar]
- Backes A., Hausman J. F., Renaut J., Ait Barka E., Jacquard C., Guerriero G. (2020). Expression analysis of cell wall-related genes in the plant pathogenic fungus Drechslera teres. Genes 11:300. 10.3390/genes11030300, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balba H. (2007). Review of strobilurin fungicide chemicals. J. Environ. Sci. Health 42, 441–451. 10.1080/03601230701316465, PMID: [DOI] [PubMed] [Google Scholar]
- Balint-Kurti P. (2019). The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178. 10.1111/mpp.12821, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrault G., Al-Ali B., Petitprez M., Albertini L. (1982). Contribution à l’étude de l’activité toxique de l’Helminthosporium teres, parasite de l’orge (Hordeum vulgare). Can. J. Bot. 60, 330–339. 10.1139/b82-046 [DOI] [Google Scholar]
- Bartlett D. W., Clough J. M., Godfrey C. R. A., Godwin J. R., Hall A. A., Heaney S. P., et al. (2001). Understanding the strobilurin fungicides. Pesticide Outlook 12, 143–148. 10.1039/b106300f [DOI] [Google Scholar]
- Bartlett D. W., Clough J. M., Godwin J. R., Hall A. A., Hamer M., Parr-Dobrzanski B. (2002). The strobilurin fungicides. Pest Manag. Sci. 58, 649–662. 10.1002/ps.520, PMID: [DOI] [PubMed] [Google Scholar]
- Beliën T., Van Campenhout S., Robben J., Volckaert G. (2006). Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 19, 1072–1081. 10.1094/MPMI-19-1072, PMID: [DOI] [PubMed] [Google Scholar]
- Beneduzi A., Ambrosini A., Passaglia L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. 10.1590/S1415-47572012000600020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350. 10.1016/S1369-5266(00)00183-7, PMID: [DOI] [PubMed] [Google Scholar]
- Bockelman H. E., Sharp E. L., Eslick R. F. (1977). Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can. J. Bot. 55, 2142–2148. 10.1139/b77-242 [DOI] [Google Scholar]
- Bogacki P., Keiper F. J., Oldach K. H. (2010). Genetic structure of south Australian Pyrenophora teres populations as revealed by microsatellite analyses. Fungal Biol. 114, 834–841. 10.1016/j.funbio.2010.08.002, PMID: [DOI] [PubMed] [Google Scholar]
- Bogacki P., Oldach K. H., Williams K. J. (2008). Expression profiling and mapping of defence response genes associated with the barley-Pyrenophora teres incompatible interaction. Mol. Plant Pathol. 9, 645–660. 10.1111/j.1364-3703.2008.00485.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonvitthya N., Tanapong P., Kanngan P., Burapatana V., Chulalaksananukul W. (2012). Cloning and expression of the Aspergillus oryzae glucan 1,3-beta-glucosidase A (exgA) in Pichia pastoris. Biotechnol. Lett. 34, 1937–1943. 10.1007/s10529-012-1001-9, PMID: [DOI] [PubMed] [Google Scholar]
- Brown M. P., Steffenson B. J., Webster R. K. (1993). Host range of Pyrenophora teres f. teres isolates from California. Plant Dis. 77, 942–947. 10.1094/PD-77-0942 [DOI] [Google Scholar]
- Burlakoti R. R., Gyawali S., Chao S., Smith K. P., Horsley R. D., Cooper B., et al. (2017). Genome-wide association study of spot form of net blotch resistance in the upper midwest barley breeding programs. Phytopathology 107, 100–108. 10.1094/PHYTO-03-16-0136-R [DOI] [PubMed] [Google Scholar]
- Campbell G. F., Crous P. W., Lucas J. A. (1999). Pyrenophora teres f. maculata, the cause of Pyrenophora leaf spot of barley in South Africa. Mycol. Res. 103, 257–267. 10.1017/S0953756298007114 [DOI] [Google Scholar]
- Campbell G. F., Lucas J. A., Crous P. W. (2002). Evidence of recombination between net- and spot-type populations of Pyrenophora teres as determined by RAPD analysis. Mycol. Res. 106, 602–608. 10.1017/S0953756202005853 [DOI] [Google Scholar]
- Cazorla F. M., Romero D., Pérez-García A., Lugtenberg B. J. J., De Vicente A., Bloemberg G. (2007). Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 103, 1950–1959. 10.1111/j.1365-2672.2007.03433.x, PMID: [DOI] [PubMed] [Google Scholar]
- Clare S. J., Wyatt N. A., Brueggeman R. S., Friesen T. L. (2020). Research advances in the Pyrenophora teres–barley interaction. Mol. Plant Pathol. 21, 272–288. 10.1111/mpp.12896, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U., Pieterse C. M. J., Mauch-Mani B. (2002). Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. 10.1016/S1360-1385(02)02244-6, PMID: [DOI] [PubMed] [Google Scholar]
- Coval S. J., Hradil C. M., Lu H. S. M., Clardy J., Satouri S., Strobel G. A. (1990). Pyrenoline-A and -B, two new phytotoxins from Pyrenophora teres. Tetrahedron Lett. 31, 2117–2120. 10.1016/0040-4039(90)80086-2 [DOI] [Google Scholar]
- Deadman M. L., Cooke B. M. (1986). A comparison of detached leaf, greenhouse and field experiments for screening barley cultivars to Drechslera teres. Ir. J. Agric. Res. 25, 63–70. [Google Scholar]
- Deadman M. L., Cooke B. M. (1989). An analysis of rain-mediated dispersal of Drechslera teres conidia in field plots of spring barley. Ann. Appl. Biol. 115, 209–214. 10.1111/j.1744-7348.1989.tb03378.x [DOI] [Google Scholar]
- Dennis R. W. G., Foister C. E. (1942). List of diseases of economic plants recorded in Scotland. Trans. Br. Mycol. Soc. 25, 266–306. 10.1016/S0007-1536(42)80018-2 [DOI] [Google Scholar]
- Douiyssi A., Rasmusson D. C., Roelfs A. P. (1998). Responses of barley cultivars and lines to isolates of Pyrenophora teres. Plant Dis. 82, 316–321. 10.1094/PDIS.1998.82.3.316, PMID: [DOI] [PubMed] [Google Scholar]
- Drechsler C. (1923). Some graminicolous species of Helminthosporium. J. Agric. Res. 24, 641–740. [Google Scholar]
- Duczek L. J., Sutherland K. A., Reed S. L., Bailey K. L., Lafond G. P. (1999). Survival of leaf spot pathogens on crop residues of wheat and barley in Saskatchewan. Can. J. Plant Pathol. 21, 165–173. 10.1080/07060669909501208 [DOI] [Google Scholar]
- Dutta S., Podile A. R. (2010). Plant growth promoting rhizobacteria (PGPR): the bugs to debug the root zone. Crit. Rev. Microbiol. 36, 232–244. 10.3109/10408411003766806, PMID: [DOI] [PubMed] [Google Scholar]
- Ellwood S. R., Liu Z., Syme R. A., Lai Z., Hane J. K., Keiper F., et al. (2010). A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres. Genome Biol. 11:R109. 10.1186/gb-2010-11-11-r109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood S. R., Syme R. A., Moffat C. S., Oliver R. P. (2012). Evolution of three Pyrenophora cereal pathogens: recent divergence, speciation and evolution of non-coding DNA. Fungal Genet. Biol. 49, 825–829. 10.1016/j.fgb.2012.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- El-Mor I. M., Fowler R. A., Platz G. J., Sutherland M. W., Martin A. (2017). An improved detached-leaf assay for phenotyping net blotch of barley caused by Pyrenophora teres. Plant Dis. 102, 760–763. 10.1094/PDIS-07-17-0980-RE, PMID: [DOI] [PubMed] [Google Scholar]
- Esmaeel Q., Miotto L., Rondeau M., Leclère V., Clément C., Jacquard C., et al. (2018). Paraburkholderia phytofirmans PsJN-plants interaction: from perception to the induced mechanisms. Front. Microbiol. 9:2093. 10.3389/fmicb.2018.02093, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeel Q., Pupin M., Kieu N. P., Chataigné G., Béchet M., Deravel J., et al. (2016). Burkholderia genome mining for nonribosomal peptide synthetases reveals a great potential for novel siderophores and lipopeptides synthesis. Microbiologyopen 5, 512–526. 10.1002/mbo3.347, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C. M. H., Soares H. M. V. M., Soares E. V. (2019). Promising bacterial genera for agricultural practices: an insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 682, 779–799. 10.1016/j.scitotenv.2019.04.225, PMID: [DOI] [PubMed] [Google Scholar]
- Fotopoulos V., Gilbert M. J., Pittman J. K., Marvier A. C., Buchanan A. J., Sauer N., et al. (2003). The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atβfruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol. 132, 821–829. 10.1104/pp.103.021428, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. A., Platz G. J., Bell K. L., Fletcher S. E. H., Franckowiak J. D., Hickey L. T. (2017). Pathogenic variation of Pyrenophora teres f. teres in Australia. Australas. Plant Pathol. 46, 115–128. 10.1007/s13313-017-0468-1 [DOI] [Google Scholar]
- Friesen T. L., Faris J. D., Lai Z., Steffenson B. J. (2006). Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled-haploid barley population. Genome 49, 855–859. 10.1139/g06-024, PMID: [DOI] [PubMed] [Google Scholar]
- Friis P., Olsen C. E., Møller B. L. (1991). Toxin production in Pyrenophora teres, the ascomycete causing the net-spot blotch disease of barley (Hordeum vulgare L.). J. Biol. Chem. 266, 13329–13335. 10.1016/S0021-9258(18)98843-5, PMID: [DOI] [PubMed] [Google Scholar]
- Gardiner D. M., Mcdonald M. C., Covarelli L., Solomon P. S., Rusu A. G., Marshall M., et al. (2012). Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8:e1002952. 10.1371/journal.ppat.1002952, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschele E. E. (1928). The response of barley to parasitic fungi Helminthosporium teres Sacc. Bull. Appl. Bot. Genet. Plant Breed. 19, 371–384. [Google Scholar]
- Ghorbanpour M., Omidvari M., Abbaszadeh-Dahaji P., Omidvar R., Kariman K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 117, 147–157. 10.1016/j.biocontrol.2017.11.006 [DOI] [Google Scholar]
- Gintjee T. J., Donnelley M. A., Thompson G. R. (2020). Aspiring antifungals: review of current antifungal pipeline developments. J. Fungi 6:28. 10.3390/jof6010028, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisi U., Chin K. M., Knapova G., Färber R. K., Mohr U., Parisi S., et al. (2000). Recent developments in elucidating modes of resistance to phenylamide, DMI and strobilurin fungicides. Crop Prot. 19, 863–872. 10.1016/S0261-2194(00)00114-9 [DOI] [Google Scholar]
- Gisi U., Sierotzki H., Cook A., Mccaffery A. (2002). Mechanisms influencing the evolution of resistance to Qo inhibitor fungicides. Pest Manag. Sci. 58, 859–867. 10.1002/ps.565, PMID: [DOI] [PubMed] [Google Scholar]
- Goodman R. N., Novacky A. J. (1994). The hypersensitive reaction in plants to pathogens: A resistance phenomenon. St. Paul, MN (USA): American Phytopathological Society. [Google Scholar]
- Graner A., Foroughi-Wehr B., Tekauz A. (1996). RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica 91, 229–234. [Google Scholar]
- Greenberg J. T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. 10.1146/annurev.arplant.48.1.525, PMID: [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Yao N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6, 201–211. 10.1111/j.1462-5822.2004.00361.x, PMID: [DOI] [PubMed] [Google Scholar]
- Grewal T. S., Rossnagel B. G., Pozniak C. J., Scoles G. J. (2008). Mapping quantitative trait loci associated with barley net blotch resistance. Theor. Appl. Genet. 116, 529–539. 10.1007/s00122-007-0688-9, PMID: [DOI] [PubMed] [Google Scholar]
- Grigoriev I. V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., et al. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704. 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Li C., Loughman R., Cakir M., Westcott S., Lance R. (2011). Identifying genetic complexity of 6H locus in barley conferring resistance to Pyrenophora teres f. teres. Plant Breed. 130, 423–429. 10.1111/j.1439-0523.2011.01854.x [DOI] [Google Scholar]
- Gupta S., Loughman R. (2001). Current virulence of Pyrenophora teres on barley in Western Australia. Plant Dis. 85, 960–966. 10.1094/PDIS.2001.85.9.960, PMID: [DOI] [PubMed] [Google Scholar]
- Haenni A. L., Robert M., Vetter W., Roux L., Barbier M., Lederer E. (1965). Structure chimique des aspergillomarasmines A et B. Helv. Chim. Acta 48, 729–750. 10.1002/hlca.19650480409, PMID: [DOI] [PubMed] [Google Scholar]
- Hardoim P. R., Van Overbeek L. S., Van Elsas J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. 10.1016/j.tim.2008.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Heath M. C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. 10.1023/A:1026592509060, PMID: [DOI] [PubMed] [Google Scholar]
- Ibrahim F. M., Threleall R. J., Gregory P. H. (1966). The application of numerical taxonomy to some graminicolous species of Helminthosporium. Proc. R. Soc. Lond. 165, 362–388. 10.1098/rspb.1966.0072 [DOI] [Google Scholar]
- Islamovic E., Bregitzer P., Friesen T. L. (2017). Barley 4H QTL confers NFNB resistance to a global set of P. teres f. teres isolates. Mol. Breed. 37, 1–12. 10.1007/s11032-017-0621-028127252 [DOI] [Google Scholar]
- Ismail I. A., Able A. J. (2016). Secretome analysis of virulent Pyrenophora teres f. teres isolates. Proteomics 16, 2625–2636. 10.1002/pmic.201500498, PMID: [DOI] [PubMed] [Google Scholar]
- Ismail I. A., Able A. J. (2017). Gene expression profiling of virulence-associated proteins in planta during net blotch disease of barley. Physiol. Mol. Plant Pathol. 98, 69–79. 10.1016/j.pmpp.2017.03.007 [DOI] [Google Scholar]
- Ismail I. A., Godfrey D., Able A. J. (2014a). Fungal growth, proteinaceous toxins and virulence of Pyrenophora teres f. teres on barley. Australas. Plant Pathol. 43, 535–546. 10.1007/s13313-014-0295-6 [DOI] [Google Scholar]
- Ismail I., Godfrey D., Able A. J. (2014b). Proteomic analysis reveals the potential involvement of xylanase from Pyrenophora teres f. teres. in net form net blotch disease of barley. Australas. Plant Pathol. 43, 715–726. 10.1007/s13313-014-0314-7 [DOI] [Google Scholar]
- Ito S. (1930). On some new ascigerous stages of the species of Helminthosporium parasitic on cereals. Proc. Imper. Acad. 6, 352–355. 10.2183/pjab1912.6.352 [DOI] [Google Scholar]
- Jalata Z., Mekbib F., Lakew B. (2020). Generation mean analysis of net blotch and scald diseases on barley. World J. Agric. Res. 4, 142–149. 10.12691/wjar-8-4-6 [DOI] [Google Scholar]
- Jalli M. (2011). Sexual reproduction and soil tillage effects on virulence of Pyrenophora teres in Finland. Ann. Appl. Biol. 158, 95–105. 10.1111/j.1744-7348.2010.00445.x [DOI] [Google Scholar]
- Jalli M., Robinson J. (2000). Stable resistance in barley to Pyrenophora teres f. teres isolates from the Nordic-Baltic region after increase on standard host genotypes. Euphytica 113, 71–77. 10.1023/A:1003912825455 [DOI] [Google Scholar]
- Jayasena K. W., George E., Loughman R., Hardy G. (2004). First record of the teleomorph stage of Drechslera teres f. maculata in Australia. Australas. Plant Pathol. 33, 455–456. 10.1071/AP04032 [DOI] [Google Scholar]
- Jayasena K. W., Loughman R., Majewski J. (2002). Evaluation of fungicides in control of spot-type net blotch on barley. Crop Prot. 21, 63–69. 10.1016/S0261-2194(01)00118-1 [DOI] [Google Scholar]
- Jayasena K. W., Van Burgel A., Tanaka K., Majewski J., Loughman R. (2007). Yield reduction in barley in relation to spot-type net blotch. Australas. Plant Pathol. 36, 429–433. 10.1071/AP07046 [DOI] [Google Scholar]
- Jensen B., Lübeck P. S., Jørgensen H. J. L. (2016). Clonostachys rosea reduces spot blotch in barley by inhibiting prepenetration growth and sporulation of Bipolaris sorokiniana without inducing resistance. Pest Manag. Sci. 72, 2231–2239. 10.1002/ps.4260, PMID: [DOI] [PubMed] [Google Scholar]
- Jetiyanon K., Kloepper J. W. (2002). Mixtures of plant growth-promoting rhizobacteria for induction of systemic resistance against multiple plant diseases. Biol. Control 24, 285–291. 10.1016/S1049-9644(02)00022-1 [DOI] [Google Scholar]
- Jonsson R., Bryngelsson T., Gustafsson M. (1997). Virulence studies of Swedish net blotch isolates (Drechslera teres) and identification of resistant barley lines. Euphytica 94, 209–218. 10.1023/A:1002924424200 [DOI] [Google Scholar]
- Jonsson R., Bryngelsson T., Jalli M., Gustafsson M. (1998). Effect of growth stage on resistance to Drechslera teres f. teres in barley. J. Phytopathol. 146, 261–265. 10.1111/j.1439-0434.1998.tb04688.x [DOI] [Google Scholar]
- Jordan V. W. L. (1981). Aetiology of barley net blotch caused by Pyrenophora teres and some effects on yield. Plant Pathol. 30, 77–87. 10.1111/j.1365-3059.1981.tb01232.x [DOI] [Google Scholar]
- Jordan V. W. L., Allen E. C. (1984). Barley net blotch: influence of straw disposal and cultivation methods on inoculum potential, and on incidence and severity of autumn disease. Plant Pathol. 33, 547–559. 10.1111/j.1365-3059.1984.tb02879.x [DOI] [Google Scholar]
- Jørgensen H. J. L., Lübeck P. S., Thordal-Christensen H., De Neergaard E., Smedegaard-Petersen V. (1998). Mechanisms of induced resistance in barley against Drechslera teres. Phytopathology 88, 698–707. 10.1094/PHYTO.1998.88.7.698, PMID: [DOI] [PubMed] [Google Scholar]
- Jørgensen L. N., Olsen L. V. (2007). Control of tan spot (Drechslera tritici-repentis) using cultivar resistance, tillage methods and fungicides. Crop Prot. 26, 1606–1616. 10.1016/j.cropro.2007.01.009 [DOI] [Google Scholar]
- Keiper F. J., Grcic M., Capio E., Wallwork H. (2008). Diagnostic microsatellite markers for the barley net blotch pathogens, Pyrenophora teres f. maculata and Pyrenophora teres f. teres. Australas. Plant Pathol. 37, 428–430. 10.1071/AP08035 [DOI] [Google Scholar]
- Keon J. P. R., Hargreaves J. A. (1983). A cytological study of the net blotch disease of barley caused by Pyrenophora teres. Physiol. Plant Pathol. 22, 321–329. 10.1016/S0048-4059(83)81019-4 [DOI] [Google Scholar]
- Khan T. N., Boyd W. J. R. (1969). Environmentally induced variability in the host reaction of barley to net blotch. Aust. J. Biol. Sci. 22, 1237–1244. 10.1071/BI9691237 [DOI] [Google Scholar]
- König J., Perovic D., Kopahnke D., Ordon F. (2013). Development of an efficient method for assessing resistance to the net type of net blotch Pyrenophora teres f. teres in winter barley and mapping of quantitative trait loci for resistance. Mol. Breed. 32, 641–650. 10.1007/s11032-013-9897-x [DOI] [Google Scholar]
- Kulkarni R. D., Kelkar H. S., Dean R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. 10.1016/S0968-0004(03)00025-2, PMID: [DOI] [PubMed] [Google Scholar]
- Kumar A., Kumar R., Kumari M., Goldar S. (2020). Enhancement of plant growth by using PGPR for a sustainable agriculture: a review. Int. J. Curr. Microbiol. App. Sci. 9, 152–165. 10.20546/ijcmas.2020.902.019 [DOI] [Google Scholar]
- Kumari B., Mallick M. A., Solanki M. K., Solanki A. C., Hora A., Guo W. (2019). “Plant growth promoting rhizobacteria (PGPR): modern prospects for sustainable agriculture” in Plant health under biotic stress. 2nd Edn. eds. Sayyed R. Z., Reddy M. S., Antonius S. (Singapore: Springer; ), 109–127. [Google Scholar]
- Künstler A., Bacsó R., Gullner G., Hafez Y. M., Király L. (2016). Staying alive—is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 93, 75–84. 10.1016/j.pmpp.2016.01.003 [DOI] [Google Scholar]
- Index Fungorum (2014). http://www.indexfungorum.org/names/IndexFungorumPublicationsListing.asp
- Lam E., Kato N., Lawton M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. 10.1038/35081184, PMID: [DOI] [PubMed] [Google Scholar]
- Lartey R. T., Caesar-Tonthat T. C., Caesar A. J., Sainju U. M., Evans R. G. (2012). First report of spot form net blotch caused by Pyrenophora teres f. maculata on barley in the Mon-dak area of the United States. Plant Dis. 97:143. 10.1094/PDIS-07-12-0657-PDN, PMID: [DOI] [PubMed] [Google Scholar]
- Leisova L., Kucera L., Minarikova V. (2005). AFLP-based PCR markers that differentiate spot and net forms of Pyrenophora teres. Plant Pathol. 54, 66–73. 10.1111/j.1365-3059.2005.01117.x [DOI] [Google Scholar]
- Leisova L., Minarikova V., Kucera L., Ovesna J. (2006). Quantification of Pyrenophora teres in infected barley leaves using real-time PCR. J. Microbiol. Methods 67, 446–455. 10.1016/j.mimet.2006.04.018, PMID: [DOI] [PubMed] [Google Scholar]
- Leišová-Svobodová L., Minaříková V., Matušinsky P., Hudcovicová M., Ondreičková K., Gubiš J. (2014). Genetic structure of Pyrenophora teres net and spot populations as revealed by microsatellite analysis. Fungal Biol. 118, 180–192. 10.1016/j.funbio.2013.11.008, PMID: [DOI] [PubMed] [Google Scholar]
- Leonard K. J., Suggs E. G. (1974). Setosphaeria prolata, the ascigerous state of Exserohilum prolatum. Mycologia 66, 281–297. 10.2307/3758363 [DOI] [Google Scholar]
- Lightfoot D. J., Able A. J. (2010). Growth of Pyrenophora teres in planta during barley net blotch disease. Australas. Plant Pathol. 39, 499–507. 10.1071/AP10121 [DOI] [Google Scholar]
- Lightfoot D. J., Mcgrann G. R. D., Able A. J. (2017). The role of a cytosolic superoxide dismutase in barley–pathogen interactions. Mol. Plant Pathol. 18, 323–335. 10.1111/mpp.12399, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Ellwood S. R., Oliver R. P., Friesen T. L. (2011). Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 12, 1–19. 10.1111/j.1364-3703.2010.00649.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw J. P. J., Crous P. W., Holz G. (1996). Relative importance of the barley net blotch pathogens Pyrenophora teres f. teres (net-type) and P. teres f. maculata (spot-type) in South Africa. African. Afr. Plant Prot. 2, 1–7. 10.10520/AJA10233121_225 [DOI] [Google Scholar]
- Luck J., Spackman M., Freeman A., Bicki P. T., Griffiths W., Finlay K., et al. (2011). Climate change and diseases of food crops. Plant Pathol. 60, 113–121. 10.1111/j.1365-3059.2010.02414.x [DOI] [Google Scholar]
- Luttrell E. S. (1963). Taxonomic criteria in Helminthosporium. Mycologia 55, 643–674. 10.2307/3756441 [DOI] [Google Scholar]
- Ma Z. Q., Lapitan N. L. V., Steffenson B. (2004). QTL mapping of net blotch resistance genes in a doubled-haploid population of six-rowed barley. Euphytica 137, 291–296. 10.1023/B:EUPH.0000040441.36990.58 [DOI] [Google Scholar]
- Mair W. J., Deng W., Mullins J. G. L., West S., Wang P., Besharat N., et al. (2016). Demethylase inhibitor fungicide resistance in Pyrenophora teres f. sp. teres associated with target site modification and inducible overexpression of Cyp51. Front. Microbiol. 7:1279. 10.3389/fmicb.2016.01279, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov I. V., Yarullina L. G., Burkhanova G. F., Zaikina E. A. (2013). Relationship between the aggressiveness and catalase activity of Septoria nodorum berk. in wheat. Biol. Bull. 40, 441–446. 10.1134/S1062359013050099 [DOI] [PubMed] [Google Scholar]
- Manninen O. M., Jalli M., Kalendar R., Schulman A., Afanasenko O., Robinson J. (2006). Mapping of major spot-type and net-type net-blotch resistance genes in the Ethiopian barley line CI 9819. Genome 49, 1564–1571. 10.1139/g06-119, PMID: [DOI] [PubMed] [Google Scholar]
- Manninen O., Kalendar R., Robinson J., Schulman A. H. (2000). Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol. Gen. Genet. 264, 325–334. 10.1007/s004380000326, PMID: [DOI] [PubMed] [Google Scholar]
- Martin R. A. (1985). Disease progression and yield loss in barley associated with net blotch, as influenced by fungicide seed treatment. Can. J. Plant Pathol. 7, 83–90. [Google Scholar]
- Mclean M. S., Howlett B. J., Hollaway G. J. (2009). Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley: a review. Crop Past. Sci. 60, 303–315. 10.1071/CP08173 [DOI] [Google Scholar]
- Mclean M. S., Howlett B. J., Hollaway G. J. (2010). Spot form of net blotch, caused by Pyrenophora teres f. maculata, is the most prevalent foliar disease of barley in Victoria, Australia. Australas. Plant Pathol. 39, 46–49. 10.1071/AP09054 [DOI] [Google Scholar]
- Mikhailova L. A., Ternyuk I. G., Mironenko N. (2010). Pyrenophora teres, an agent causing wheat leaf spot. Microbiology 79, 561–565. 10.1134/S0026261710040223 [DOI] [Google Scholar]
- Mikkelsen B. L., Jørgensen R. B., Lyngkjær M. F. (2015). Complex interplay of future climate levels of CO2, ozone and temperature on susceptibility to fungal diseases in barley. Plant Pathol. 64, 319–327. 10.1111/ppa.12272 [DOI] [Google Scholar]
- Mironenko N. V., Afanasenko O. S., Filatova O. A., Kopahnke D. (2005). Genetic control of virulence of Pyrenophora teres drechs, the causative agent of net blotch in barley. Russ. J. Genet. 41, 1389–1394. 10.1007/s11177-006-0011-x, PMID: [DOI] [PubMed] [Google Scholar]
- Mode C. J., Schaller C. W. (1958). Two additional factors for host resistance to net blotch in barley. Agron. J. 50, 15–18. 10.2134/agronj1958.00021962005000010005x [DOI] [Google Scholar]
- Montesinos E., Badosa E., Cabrefiga J., Planas M., Feliu L., Bardají E. (2012). “Antimicrobial peptides for plant disease control. From discovery to application” in Small wonders: Peptides for disease control. eds. Rajasekaran K., Cary J. W., Jaynes J. M., Montesinos E. (Washington, DC: ACS Symposium Series, American Chemical Society; ), 235–262. [Google Scholar]
- Moya P., Barrera V., Cipollone J., Bedoya C., Kohan L., Toledo A., et al. (2020). New isolates of Trichoderma spp. as biocontrol and plant growth–promoting agents in the pathosystem Pyrenophora teres-barley in Argentina. Biol. Control 141, 1–8. 10.1016/j.biocontrol.2019.104152 [DOI] [Google Scholar]
- Moya P. A., Girotti J. R., Toledo A. V., Sisterna M. N. (2018). Antifungal activity of Trichoderma VOCs against Pyrenophora teres, the causal agent of barley net blotch. J. Plant Prot. Res. 58, 45–53. 10.24425/119115 [DOI] [Google Scholar]
- Mur L. A. J., Kenton P., Lloyd A. J., Ougham H., Prats E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. 10.1093/jxb/erm239, PMID: [DOI] [PubMed] [Google Scholar]
- Muria-Gonzalez M. J., Zulak K. G., Allegaert E., Oliver R. P., Ellwood S. R. (2020). Profile of the in vitro secretome of the barley net blotch fungus, Pyrenophora teres f. teres. Physiol. Mol. Plant Pathol. 109:101451. 10.1016/j.pmpp.2019.101451 [DOI] [Google Scholar]
- Murray G. M., Brennan J. P. (2010). Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 39, 85–96. 10.1071/AP09064 [DOI] [Google Scholar]
- Newitt J. T., Prudence S. M. M., Hutchings M. I., Worsley S. F. (2019). Biocontrol of cereal crop diseases using Streptomycetes. Pathogens 8:78. 10.3390/pathogens8020078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisikado Y. (1929). Studies on the Helminthosporium diseases of Gramineae in Japan. Ber. Ohara Inst. 4, 111–126. [Google Scholar]
- Novakazi F., Afanasenko O., Anisimova A., Platz G. J., Snowdon R., Kovaleva O., et al. (2019). Genetic analysis of a worldwide barley collection for resistance to net form of net blotch disease (Pyrenophora teres f. teres). Theor. Appl. Genet. 132, 2633–2650. 10.1007/s00122-019-03378-1, PMID: [DOI] [PubMed] [Google Scholar]
- Nukina M., Hirota H. (1992). Pyrenolide D, a new cytotoxic fungal metabolite from Pyrenophora teres. Biosci. Biotechnol. Biochem. 56, 1158–1159. 10.1271/bbb.56.1158, PMID: [DOI] [PubMed] [Google Scholar]
- Nukina M., Ikeda M., Sassa T. (1980). Two new pyrenolides, fungal morphogenic substances produced by Pyrenophora teres (Diedicke) Drechsler. Agric. Biol. Chem. 44, 2761–2762. [Google Scholar]
- Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. 10.1016/j.tim.2007.12.009, PMID: [DOI] [PubMed] [Google Scholar]
- Ongena M., Jourdan E., Adam A., Paquot M., Brans A., Joris B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. 10.1111/j.1462-2920.2006.01202.x, PMID: [DOI] [PubMed] [Google Scholar]
- Padmanabhan S. Y. (1973). The great Bengal famine. Annu. Rev. Phytopathol. 11, 11–24. 10.1146/annurev.py.11.090173.000303 [DOI] [Google Scholar]
- Palazzini J. M., Dunlap C. A., Bowman M. J., Chulze S. N. (2016). Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: genome sequencing and secondary metabolite cluster profiles. Microbiol. Res. 192, 30–36. 10.1016/j.micres.2016.06.002, PMID: [DOI] [PubMed] [Google Scholar]
- Peever T. L., Milgroom M. G. (1993). Genetic correlations in resistance to sterol biosynthesis-inhibiting fungicides in Pyrenophora teres. Phytopathology 83, 1076–1082. 10.1094/Phyto-83-1076 [DOI] [Google Scholar]
- Peever T. L., Milgroom M. G. (1994). Genetic structure of Pyrenophora teres populations determined with random amplified polymorphic DNA markers. Can. J. Bot. 72, 915–923. 10.1139/b94-116 [DOI] [Google Scholar]
- Pérez-García A., Romero D., De Vicente A. (2011). Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 22, 187–193. 10.1016/j.copbio.2010.12.003, PMID: [DOI] [PubMed] [Google Scholar]
- Piening L. (1968). Development of barley net blotch from infested straw and seed. Can. J. Plant Sci. 48, 623–625. 10.4141/cjps68-118 [DOI] [Google Scholar]
- Plessl M., Heller W., Payer H. D., Elstner E. F., Habermeyer J., Heiser I. (2005). Growth parameters and resistance against Drechslera teres of spring barley (Hordeum vulgare L. cv. Scarlett) grown at elevated ozone and carbon dioxide concentrations. Plant Biol. 7, 694–705. 10.1055/s-2005-873002, PMID: [DOI] [PubMed] [Google Scholar]
- Porta-Puglia A., Delogu G., Vannacci G. (1986). Pyrenophora graminea on winter barley seed: effect on disease incidence and yield losses. J. Phytopathol. 117, 26–33. 10.1111/j.1439-0434.1986.tb04356.x [DOI] [Google Scholar]
- Poudel B., Mclean M. S., Platz G. J., Mcilroy J. A., Sutherland M. W., Martin A. (2018). Investigating hybridisation between the forms of Pyrenophora teres based on Australian barley field experiments and cultural collections. Eur. J. Plant Pathol. 2, 465–473. 10.1007/s10658-018-1574-9 [DOI] [Google Scholar]
- Prasad M., Srinivasan R., Chaudhary M., Choudhary M., Jat L. K. (2019). “Plant growth promoting rhizobacteria (PGPR) for sustainable agriculture: perspectives and challenges” in PGPR amelioration in sustainable agriculture. eds. Singh A. K., Kumar A., Singh P. K. (Duxford, UK: Woodhead Publishing; ), 129–157. [Google Scholar]
- Rau D., Attene G., Brown A. H. D., Nanni L., Maier F. J., Balmas V., et al. (2007). Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Curr. Genet. 51, 377–392. 10.1007/s00294-007-0126-1, PMID: [DOI] [PubMed] [Google Scholar]
- Rau D., Brown A. H. D., Brubaker C. L., Attene G., Balmas V., Saba E., et al. (2003). Population genetic structure of Pyrenophora teres Drechs. the causal agent of net blotch in Sardinian landraces of barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 947–959. 10.1007/s00122-002-1173-0, PMID: [DOI] [PubMed] [Google Scholar]
- Rehfus A., Miessner S., Achenbach J., Strobel D., Bryson R., Stammler G. (2016). Emergence of succinate dehydrogenase inhibitor resistance of Pyrenophora teres in Europe. Pest Manag. Sci. 72, 1977–1988. 10.1002/ps.4244, PMID: [DOI] [PubMed] [Google Scholar]
- Reis E. M., Danelli A., Casa R. T. (2012). Fungicides, seed dresser adjuvants and storage time in the control of Drechslera teres in barley seeds. Summa Phytopathol. 38, 187–191. 10.1590/S0100-54052012000300001 [DOI] [Google Scholar]
- Richards J., Chao S., Friesen T., Brueggeman R. (2016). Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 6, 1809–1818. 10.1534/g3.116.028902, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C. M., Senthil-Kumar M., Tzin V., Mysore K. S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5:17. 10.3389/fpls.2014.00017, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen M., Sela H., Fridman E., Perl-Treves R., Kopahnke D., Moreau A., et al. (2019). Characterization of the barley net blotch pathosystem at the center of origin of host and pathogen. Pathogens 8:275. 10.3390/pathogens8040275, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Roldán M. C., Maier F. J., Schäfer W. (2001). PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant Microbe Interact. 14, 116–125. 10.1094/MPMI.2001.14.2.116, PMID: [DOI] [PubMed] [Google Scholar]
- Saari E. E., Prescott J. M. (1975). Scale for appraising the foliar intensity of wheat diseases. Plant Dis. Rep. 59, 377–380. [Google Scholar]
- Sacristán S., García-Arenal F. (2008). The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. 10.1111/j.1364-3703.2007.00460.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson K., Western J. H. (1940). Two diseases of grasses caused by species of Helminthosporium not previously recorded in Britain. Trans. Br. Mycol. Soc. 24, 255–263. 10.1016/S0007-1536(40)80010-7 [DOI] [Google Scholar]
- Sarpeleh A., Wallwork H., Catcheside D. E. A., Tate M. E., Able A. J. (2007). Proteinaceous metabolites from Pyrenophora teres contribute to symptom development of barley net blotch. Phytopathology 97, 907–915. 10.1094/PHYTO-97-8-0907, PMID: [DOI] [PubMed] [Google Scholar]
- Sarpeleh A., Wallwork H., Tate M. E., Catcheside D. E. A., Able A. J. (2008). Initial characterisation of phytotoxic proteins isolated from Pyrenophora teres. Physiol. Mol. Plant Pathol. 72, 73–79. 10.1016/j.pmpp.2008.05.003 [DOI] [Google Scholar]
- Scott D. B. (1991). Identity of Pyrenophora isolates causing net-type and spot-type lesions on barley. Mycopathologia 116, 29–35. [Google Scholar]
- Serenius M., Manninen O., Wallwork H., Williams K. (2007). Genetic differentiation in Pyrenophora teres populations measured with AFLP markers. Mycol. Res. 111, 213–223. 10.1016/j.mycres.2006.11.009, PMID: [DOI] [PubMed] [Google Scholar]
- Shafi J., Tian H., Ji M. (2017). Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip. 31, 446–459. 10.1080/13102818.2017.1286950 [DOI] [Google Scholar]
- Sharma R. R., Singh D., Singh R. (2009). Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: a review. Biol. Control 50, 205–221. 10.1016/j.biocontrol.2009.05.001 [DOI] [Google Scholar]
- Shipton W. A. (1966). Effect of net blotch infection of barley on grain yield and quality. Aust. J. Exp. Agric. 6, 437–440. 10.1071/EA9660437 [DOI] [Google Scholar]
- Shipton W. A., Khan T. N., Boyd W. J. R. (1973). Net blotch of barley. Rev. Plant Pathol. 52, 269–290. [Google Scholar]
- Shoemaker R. A. (1959). Nomenclature of Drechslera and Bipolaris, grass parasites segregated from ‘Helminthosporium. Can. J. Plant Pathol. 28, S212–S220. 10.1080/07060660609507377 [DOI] [Google Scholar]
- Sierotzki H., Frey R., Wullschleger J., Palermo S., Karlin S., Godwin J., et al. (2007). Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 63, 225–233. 10.1002/ps.1330, PMID: [DOI] [PubMed] [Google Scholar]
- Smedegard-Petersen V. (1971). Pyrenophora teres f. teres and Pyrenophora teres f. maculata on barley in Denmark. Arskrift den kgle Vet-Landbohojsk 971, 124–144. [Google Scholar]
- Smedegård-Petersen V. (1976). Pathogenesis and genetics of net-spot blotch and leaf stripe of barley caused by Pyrenophora teres and Pyrenophora graminea, Available at: https://agris.fao.org/agris-search/search.do?recordID=XE7621191 (Accessed April 10, 2018).
- Smith N. J. G., Rattray J. M. (1930). Net blotch, spot blotch, and leaf-stripe diseases of barley in South Africa. S. Afr. J. Sci. 27, 341–351. [Google Scholar]
- Steffenson B. J., Hayes P. M., Kleinhofs A. (1996). Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor. Appl. Genet. 92, 552–558. 10.1007/BF00224557, PMID: [DOI] [PubMed] [Google Scholar]
- Stepanovic M., Rekanovic E., Milijašević-Marčić S., Potocnik I., Todorovic B., Stepanovic J. (2016). Field efficacy of different fungicide mixtures in control of net blotch on barley. Pestic. Fitomed. 31, 51–57. 10.2298/PIF1602051S [DOI] [Google Scholar]
- Stergiopoulos I., Collemare J., Mehrabi R., De Wit P. J. G. M. (2013). Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 37, 67–93. 10.1111/j.1574-6976.2012.00349.x, PMID: [DOI] [PubMed] [Google Scholar]
- Suemoto H., Matsuzaki Y., Iwahashi F. (2019). Metyltetraprole, a novel putative complex III inhibitor, targets known QoI-resistant strains of Zymoseptoria tritici and Pyrenophora teres. Pest Manag. Sci. 75, 1181–1189. 10.1002/ps.5288, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffert F., Sache I., Lannou C. (2011). Early stages of Septoria tritici blotch epidemics of winter wheat: build-up, overseasoning, and release of primary inoculum. Plant Pathol. 60, 166–177. 10.1111/j.1365-3059.2010.02369.x [DOI] [Google Scholar]
- Sutton J., Steele P. (1983). Effects of seed and foliar fungicides on progress of net blotch and yield of barley. Can. J. Plant Sci. 63, 631–639. 10.4141/cjps83-080 [DOI] [Google Scholar]
- Syme R. A., Martin A., Wyatt N. A., Lawrence J. A., Muria-Gonzalez M. J., Friesen T. L., et al. (2018). Transposable element genomic fissuring in Pyrenophora teres is associated with genome expansion and dynamics of host-pathogen genetic interactions. Front. Genet. 9:130. 10.3389/fgene.2018.00130, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang P., Richards J. K., Alhashal A., Poudel R. S., Horsley R. D., Friesen T. L., et al. (2019). Mapping of barley susceptibility/resistance QTL against spot form net blotch caused by Pyrenophora teres f. maculata using RIL populations. Theor. Appl. Genet. 132, 1953–1963. 10.1007/s00122-019-03328-x, PMID: [DOI] [PubMed] [Google Scholar]
- Tekauz A. (1983). Reaction of Canadian barley cultivars to Pyrenophora graminea, the incitant of leaf stripe. Can. J. Plant Pathol. 5, 294–301. 10.1080/07060668309501614 [DOI] [Google Scholar]
- Thompson G. R., Cadena J., Patterson T. F. (2009). Overview of antifungal agents. Clin. Chest Med. 30, 203–215. 10.1016/j.ccm.2009.02.001, PMID: [DOI] [PubMed] [Google Scholar]
- Turan M., Gulluce M., Şahin F. (2012). Effects of plant-growth-promoting rhizobacteria on yield, growth, and some physiological characteristics of wheat and barley plants. Commun. Soil Sci. Plant Anal. 43, 1658–1673. 10.1080/00103624.2012.681739 [DOI] [Google Scholar]
- Turkington T. K., Orr D. D., Clear R. M., Patrick S. K., Burnett P. A., Xi K. (2002). Fungal plant pathogens infecting barley and wheat seed from Alberta. Can. J. Plant Pathol. 24, 302–308. 10.1080/07060660209507013 [DOI] [Google Scholar]
- Van Den Berg C. G. J. (1988). Epidemiology of Pyrenophora teres and its effect on again yield of Hordeum vulgare. PhD thesis. Saskatoon, Canada: University of Saskatchewan.
- Van Den Berg C., Rossnagel B. G. (1990). Effect of tilt on severity of spot-type net blotch, grain yield and yield components in barley. Can. J. Plant Sci. 70, 473–480. 10.4141/cjps90-055 [DOI] [Google Scholar]
- Van Loon L. C., Bakker P. A., Pieterse C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. 10.1146/annurev.phyto.36.1.453, PMID: [DOI] [PubMed] [Google Scholar]
- Vatter T., Maurer A., Kopahnke D., Perovic D., Ordon F., Pillen K. (2017). A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS One 12:e0186803. 10.1371/journal.pone.0186803, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejan P., Abdullah R., Khadiran T., Ismail S., Boyce A. N. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability—a review. Molecules 21:573. 10.3390/molecules21050573, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejan P., Khadiran T., Abdullah R., Ismail S., Dadrasnia A. (2019). Encapsulation of plant growth promoting Rhizobacteria—prospects and potential in agricultural sector: a review. J. Plant Nutr. 42, 2600–2623. 10.1080/01904167.2019.1659330 [DOI] [Google Scholar]
- Waghu F. H., Idicula-Thomas S. (2020). Collection of antimicrobial peptides database and its derivatives: applications and beyond. Protein Sci. 29, 36–42. 10.1002/pro.3714, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiergang I., Jørgensen H. J. L., Møller I. M., Friis P., Smedegaard-Petersen V. (2002). Correlation between sensitivity of barley to Pyrenophora teres toxins and susceptibility to the fungus. Physiol. Mol. Plant Pathol. 60, 121–129. 10.1006/pmpp.2002.0384 [DOI] [Google Scholar]
- Williams L. E., Lemoine R., Sauer N. (2000). Sugar transporters in higher plants - a diversity of roles and complex regulation. Trends Plant Sci. 5, 283–290. 10.1016/S1360-1385(00)01681-2, PMID: [DOI] [PubMed] [Google Scholar]
- Williams K. J., Lichon A., Gianquitto P., Kretschmer J. M., Karakousis A., Manning S., et al. (1999). Identification and mapping of a gene conferring resistance to the spot form of net blotch (Pyrenophora teres f. maculata) in barley. Theor. Appl. Genet. 99, 323–327. 10.1007/s001220051239 [DOI] [Google Scholar]
- Wright K. H., Sutton J. C. (1990). Inoculum of Pyrenophora tritici-repentis in relation to epidemics of tan spot of winter wheat in Ontario. Can. J. Plant Pathol. 12, 149–157. 10.1080/07060669009501018 [DOI] [Google Scholar]
- Wyatt N. A., Friesen T. L. (2020). Four reference quality genome assemblies of Pyrenophora teres f. maculata: a resource for studying the barley spot form net blotch interaction. Mol. Plant Microbe Interact. 34, 135–139. 10.1094/MPMI-08-20-0228-A, PMID: [DOI] [PubMed] [Google Scholar]
- Wyatt N. A., Richards J. K., Brueggeman R. S., Friesen T. L. (2018). Reference assembly and annotation of the Pyrenophora teres f. teres isolate 0-1. G3-Genes Genom. Genet. 8, 11–18. 10.1534/g3.117.300196 [DOI] [PMC free article] [PubMed] [Google Scholar]