Abstract
Temperature stress restricts plant growth and development. Antifreeze protein (AFP) can improve plants antifreeze ability. In our previous study, the AnAFP gene cloned from Ammopiptanthus nanus was confirmed to be an excellent candidate enhancing plant cold resistance. But, AnAFP protein shared similar structures with KnS type dehydrins including K, N and S domains except ice crystal binding domain A. Here, we generated AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS, and transformed them into ordinary and cold sensitive strains of E. coli, and Arabidopsis KS type dehydrin mutant to evaluate their function. Expression of AnAFPΔA decreases cold and heat tolerance in E. coli, meanwhile, AnAFP enhances heat tolerance in Arabidopsis, suggesting that domain A is a thermal stable functional domain. AnAFP, AnAFPΔA and AnAFPΔS localize in whole cell, but AnAFPΔK and AnAFPΔN only localizes in nucleus and cytoplasm, respectively, exhibiting that K and N domains control localization of AnAFP. Likewise, K domain blocks interaction between AnAFP and AnICE1. The result of RT-qPCR showed that expression of AnAFP, AnICE1 and AnCBF genes was significantly induced by high-temperature, indicating that the AnAFP is likely regulated by ICE1-CBF-COR signal pathway. Taken together, the study provides insights into understanding the mechanism of AnAFP in response to temperature stress and gene resource to improve heat or cold tolerance of plants in transgenic engineering.
Subject terms: Plant sciences, Plant biotechnology
Introduction
Temperature stress including low- and high-temperature stress restricts plant growth and development. Low temperature inhibits enzyme activity and destroys membrane permeability resulting in physiological disorder, metabolic obstruction and even cell death1,2. Similarly, high temperature leads to wilting, accumulation of reactive oxygen species (ROS), and destruction of membrane system3–5. After perceiving temperature stress, plants approach genes expression change, physiological and biochemical response via signal transduction5–8. The ICE1-CBF-COR pathway is the most studied signaling pathway under temperature stress in plants. Under low temperature conditions, a MYC-like basic helix loop helix (bHLH) transcription factor ICE1 (Inducer of CBF expression 1) induces expression of the CBF gene. The CBF transcription factor binds to the CRT/DRE (C-repeats/dehydration responsive) element (CCGAC) and activates the expression of cold-responsive genes (COR)9,10. In response to high temperature stress, the energy releasing from cell physiological disorder such as membrane fluidity increasing, DNA unwinding and protein subunit dissociation triggers transcriptional changes to restore homeostasis, promoting cell survival, and elaborating longer-term responses for adaptation, growth, and development11.
Antifreeze proteins (AFPs) were firstly found in polar fishes, as well as insects living in freeze-zone. AFPs prevent water from freezing by adsorbing to the ice surface and stopping the growth of minute ice crystals to large crystals in a non-colligative manner, and help the organisms survive in subzero temperature environments. The distribution of AFPs in different species appears to be the outcome of a combination of independent evolutionary events, which is probably the convergent evolution or horizontal gene transfer12. These AFPs genes were introduced into crops to improve their tolerance to low temperature stress. Expression of antifreeze protein of winter flounder or insect (Microdera puntipennis dzungarica) confers the cold tolerance to transgenic spring wheat at subzero temperatures, tobacco or tomato, respectively13–15. Meanwhile, the transgenic plants did not demonstrate significant tolerance improvement when compared to wild-types13–15. The non-colligative manner of the heterologous animal AFPs, as well as their expression rate, localization, and stability, might not be suitable for cellular environments in transgenic plants16. However, the AFPs from overwintering plants including Lolium perenne, Loliurn perenne and Ammopiptanthus nanus showed higher inhibitory effect on ice growth and recrystallization than that of AFPs of fishes and insects17–20.
Ammopiptanthus nanus (A. nanus) is a tertiary relict plant and evergreen broad-leaved shrub distrusted in deserts in Central Asia, exhibits excellent tolerance to abiotic stress including drought, low and high temperature. In our previous study, the AnAFP gene was cloned from xerophyte A. nanus, and evaluated its cold tolerance function by ectopic expression in Escherichia coli (E. coli), tobacco and maize21,22. Bioinformatics analysis showed that AnAFP shared high similarity with some members of KnS type dehydrins22. In addition to the ice crystal binding domain (A domain) of AFPs, AnAFP also has three conserved domains of dehydrins including K, S and N (Nuclear localization sequence) domains22,23. In order to evaluate the function of these domains, four mutants of AnAFP deleting A (AnAFPΔA), K (AnAFPΔK), S (AnAFPΔS) and N (AnAFPΔN) domain were generated by overlapping PCR, respectively. In this study, these four mutants and AnAFP were introduced into cold-sensitive and ordinary strains of E. coli, as well as Arabidopsis mutant of KnS type dehydrin gene AtHIRD11 to identify thermal stability of each domain under low- and high-temperature stress, respectively. Together with their subcellular localization, interacting proteins and induced endogenous expression, the regulation mechanism of the AnAFP protein in response to temperature stress was elucidated.
Results
Sequence of mutant genes
Through overlap PCR, the sequences of AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS were amplified from AnAFP (Figure S1a). Their encoding putative proteins were deleted of crystal binding domain A, and K, NLS and S domains of dehydrin, respectively (Figure S1b).
Expression of AnAFPΔA increases cold sensitivity of BX04
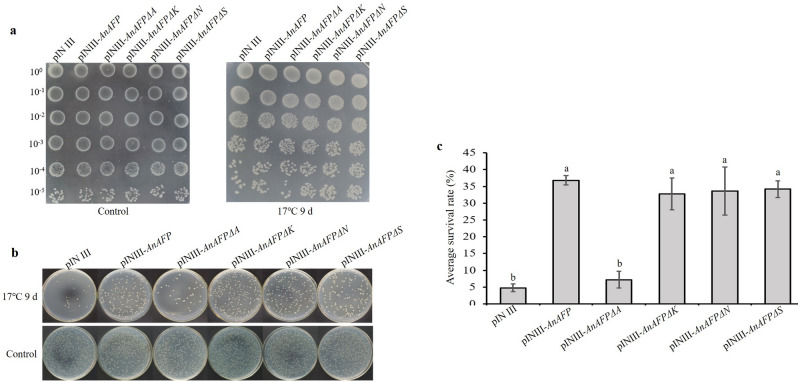
After low temperature treatment at 17 °C for 9 days, the cold-sensitive BX04 containing pINIII-AnAFPΔA showed growth defect and failed to form colonies with an average survival rate < 10%, which was similar to BX04 cells contain pINIII. However, the BX04 containing pINIII-AnAFPΔK, pINIII-AnAFPΔS and pINIII-AnAFPΔN grew vigorously and formed more colonies with an average survival rate > 30%, respectively, which was similar to BX04 cells with pINIII-AnAFP (Fig. 1). These results suggest that domain A is a functional domain of AnAFP protein related to cold tolerance.
Figure 1.
Phenotype of cold sensitive BX40 strain of E. coli with AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS gene under low temperature stress (17 °C) for 9 days. (a) Colony growth of different dilution times. (b) Monoclone of transformed strains. (c) Average survival rate of transformed strains. Lower case letter indicates the significant difference at p < 0.05 level in student’s t-test. The experiment was performed with three replicates. The data were presented as the mean values ± SD.
Expression of AnAFPΔA increases heat sensitivity of BL21
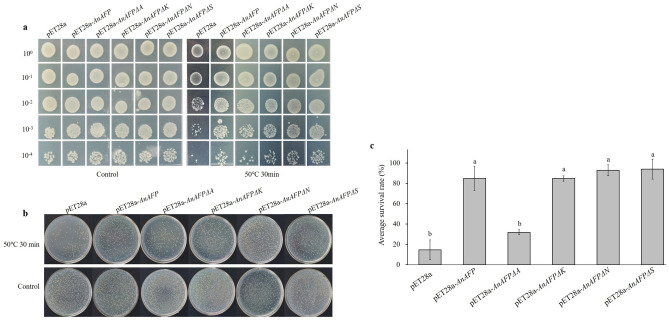
After IPTG induction, the expression of candidate genes in BL21 was confirmed and analyzed by SDS-PAGE (Figure S2). After 30 min of heat treatment at 50 °C, the colony growth of each dilution time of the E. coli BL21 transformed by pET28a was inhibited, and the average survival rate was only 14.46%. Whereas the BL21 cells with pET28a-AnAFP grew better, with an average survival rate of 85.29%. Notably, the BL21 cells with pET28a-AnAFPΔA was similar to BL21 with pET28a, the average survival was 32.41%. However, the BL21 cells with pET28a-AnAFPΔK, pET28a-AnAFPΔN, and pET28a-AnAFPΔS was similar to that transformed by pET28a-AnAFP, and average survival rates were 84.85%, 95.75% and 95.85%, respectively (Figs. 2 and S3), suggesting that A domain as ice crystal binding domain was also related to thermostable function of AnAFP protein.
Figure 2.
Thermotolerant phenotype of E. coli BL21strains transformed by AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS gene under heat stress (50 °C) for 30 min. (a) Colony growth of different dilution times. (b) Monoclone of transformed strains. (c) Average survival rate. Lower case letter indicates the significant difference at p < 0.05 level in student’s t-test. The experiment was performed with three replicates. The data were presented as the mean values ± SD.
Expression of mutant genes increase heat sensitivity of Arabidopsis
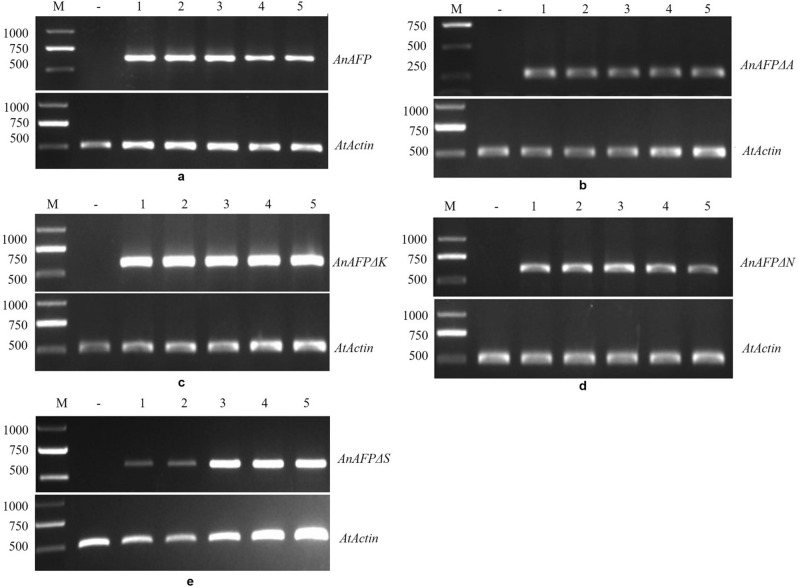
After screening with 50 mg/L kanamycin, the five homozygous lines for every gene were identified by PCR amplification of AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS, respectively (Figure S4), indicating that these genes were integrated into the genome of Arabidopsis. Subsequently, the ORF of AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS were amplified by RT-PCR from the T3 lines of transgenic lines, but not in untransformed mutant. Likewise, the specific fragment of AtActin was amplified both in transgenic lines and untransformed mutant (Figs. 3 and S5), indicating the ectopic expression of candidate genes in transgenic plants.
Figure 3.
Ectopic expression of AnAFP (a), AnAFPΔA (b), AnAFPΔK (c), AnAFPΔN (d), AnAFPΔS (e) genes in T3 transgenic Arabidopsis by RT-PCR. M: DNA molecular weight marker DL2000; –: Untransformed mutant; 1, 2, 3, 4, 5 indicates independent transgenic line.
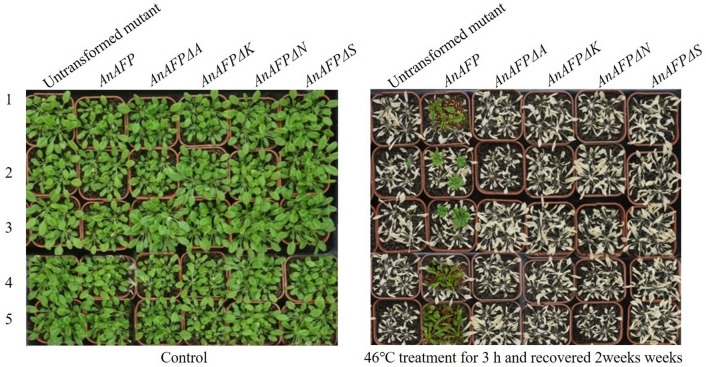
As shown in Fig. 4, before heat treatment, the transgenic lines and untransformed mutants grew vigorously. After 46 °C treatment for 3 h and then recovered 2 weeks, the transgenic lines transformed by AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS and untransformed mutants all died. However, a few plants of transgenic line transformed by AnAFP still survived. This result suggests that ectopic expression of AnAFP improves heat tolerance of heat sensitive Arabidopsis mutant. However, deletion of any one of A, K, N and S domains will lead to loss of heat resistance function of AnAFP protein.
Figure 4.
Phenotype of T3 transgenic lines and untransformed mutant under heat treatment. 1, 2, 3, 4, 5 indicates independent transgenic line. Five T3 lines were planted in pots, and grown in green house at 22 ℃ and 60–70% relative humidity under a 10 h light/14 h dark photoperiod. One-month-old seedlings were used for heat-shock treatment at 46 °C for 3 h, and recovered for 2 weeks at 22 ℃, and investigated for phenotype.
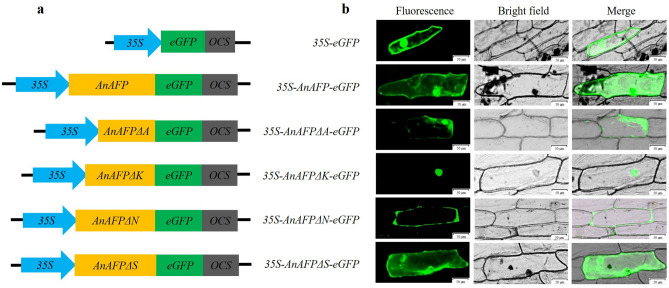
Deletion of domains K and N alters subcellular localization
Subcellular localization results showed that green fluorescence signals were observed both in cytoplasm and nucleus of onion epidermal cells transformed by 35S-eGFP, 35S-AnAFP-eGFP, 35S-AnAFPΔA-eGFP, and 35S-AnAFPΔS-eGFP, respectively. However, green fluorescence was observed only in the nucleus of onion epidermal cells with 35S-AnAFPΔK-eGFP, and only in the cytoplasm of onion epidermal cells with 35S-AnAFPΔN-eGFP (Fig. 5). These results suggest that the deletion of domains K and N changes the subcellular localization of AnAFP protein.
Figure 5.
Subcellular localization of AnAFP and its deletion mutants in onion epidermal cells. (a) The diagram of vector for transient expression. (b) Green fluorescence observed by confocal microscopy. Scale bars = 50 µm.
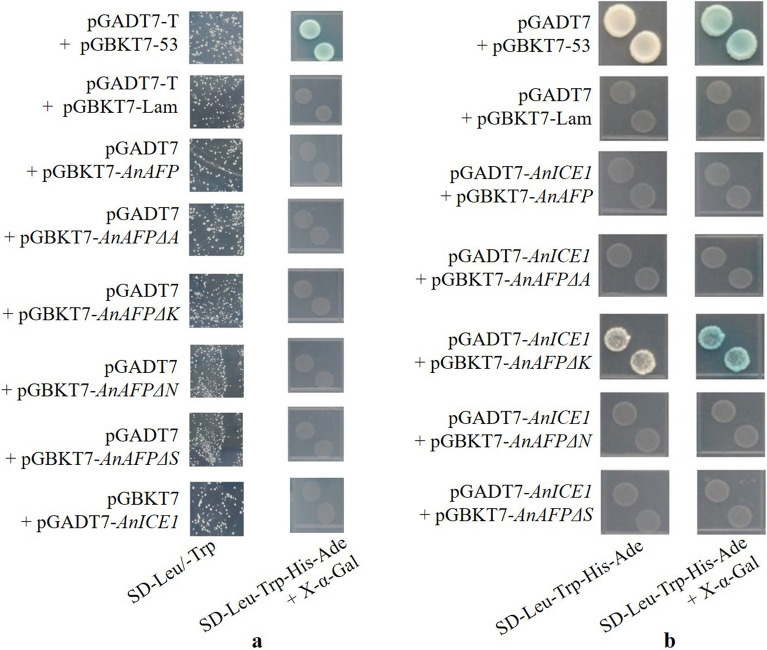
Domain K blocks interaction of AnAFP with AnICE1
As shown in Fig. 6, the yeast Y2H Gold co-transformed by bait vector and trap vector could grow normally on SD/-Leu-Trp plates, but not grow and not turn blue on SD/-Leu-Trp-His-Ade with X-α-Gal plates except positive control (pGADT7-T + pGBKT7-53), showing that these proteins have no toxicity and no autoactivation in yeast cell. Likewise, the yeast cells with pGBKT7-AnAFPΔK and pGADT7-AnICE1 could grow and be stained to blue on the auxotroph SD/-Leu-Trp-His-Ade with X-α-gal plates, while the cells co-transformed by pGBKT7-AnAFP, pGBKT7-AnAFPΔA, pGBKT7-AnAFPΔN, pGBKT7-AnAFPΔS, and pGADT7-AnICE1 did not grow, suggesting that AnAFPΔK interacts with AnICE1. This result indicates that domain K of AnAFP protein blocks its interaction with AnICE1 protein.
Figure 6.
Protein interaction by Y2H. (a) The toxicity and autoactivation assay. (b) Y2H between AnICE1 and AnAFP, as well as its deletion mutants.
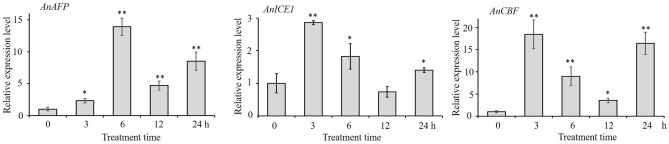
Endogenous expression of AnAFP, AnICE1 and AnCBF genes in response to high temperature stress
The results of real-time quantitative PCR (RT-qPCR) showed that the expression of AnAFP, AnICE1 and AnCBF genes were significantly upregulated by high temperature stress. The expression of AnAFP reached to 13.91 times of control at 6 h of treatment. Meanwhile, at 3 h of treatment, the expression of AnICE1 and AnCBF increased to 2.86 and 18.50 times of control (0 h), respectively (Fig. 7). This result suggests that the upregulated expression of AnAFP gene in response to high temperature stress maybe regulated by the signaling pathway of ICE1-CBF-COR.
Figure 7.
Relative expression levels of AnAFP, AnICE1 and AnCBF genes under high temperature (45 °C) treatment. The AnGAPDH gene was used as internal reference. The 2−ΔΔCT method of the CFX Manger™ software version 2.0 (Bio-Rad, USA) was used to normalize the expression differentiation between reference gene and investigated genes. The experiment was performed with three replicates. The data were presented as the mean values ± SD.
Discussion
All AFPs are conserved for their ice crystal binding sites, although difference in their structural characteristics due to their different evolution24,25. Under low temperature stress (17 °C for 9 days), the colony growth and average survival rates of the cold-sensitive E. coli BX04 with AnAFPΔA were significantly poorer and lower than the strains transformed with AnAFP, as well as AnAFPΔK, AnAFPΔN and AnAFPΔS mutants (Fig. 1). Under high temperature stress (50 °C for 30 min), the colony growth and average survival rate of E. coli BL21 (DE3) transformed by AnAFPΔA were also significantly poorer and lower than the strains transformed by AnAFP, as well as AnAFPΔK, AnAFPΔN and AnAFPΔS (Fig. 2). These results confirmed the conserved thermostable function of ice crystal binding domain (domain A) in AnAFP proteins. However, the function of each domain of AnAFP is not indispensable. All T3 plants of Arabidopsis mutant of KnS type dehydrin athird11 transformed by AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS mutants died after high temperature stress (46 °C for for 3 h), while a few T3 plants of the same mutant transformed by the AnAFP gene survived (Fig. 4). Subcellular localization showed that the mutants without domains A and S were localized in the cytoplasm and nucleus, which was same as AnAFP protein. However, the mutant without domain K was only localized in the nucleus, while the mutant without domain N was localized only in the cytoplasm (Fig. 5). This result suggests that domains K and N determine the subcellular localization of AnAFP protein and affect its function. Similar phenomenon was observed for KnS-type dehydrin ZmDHN13 in maize26.
The Y2H result indicates that domain K of AnAFP protein blocks its interaction with AnICE1 proteins (Fig. 6). This result can be explained by the molecular shield model of dehydrin to protect functional proteins27,28. The internal disordered structure of dehydrin protein occupies the space between the target proteins and reduces their collision. This kind of shield forms a loose structure around the target protein but not interact with it like the classical molecular chaperone.
In our previous study, the AnAFP was found to be a KnS type dehydrin and the AnAFP gene was identified as a member of COR genes22. Dehydrin and COR genes can be activated by CBF participating ICE1-CBF-COR pathway29–32 In the present study, the RT-qPCR result showed that the endogenous expression of AnAFP, AnICE1 and AnCBF genes was significantly upregulated by high temperature stress (Fig. 7). It suggests that high temperature stress also induces the expression of COR genes. In response to low and high temperature stress, the endogenous expression of AnAFP is probably induced by signaling pathway of ICE1-CBF-COR. In Arabidopsis, the expression of heat shock proteins was also found to be extensively overlapped with non-heat stress response pathways33. Therefore, the regulation of the endogenous expression of AnAFP in A. nanus can be concluded as signaling pathway sketched in Fig. 8.
Figure 8.
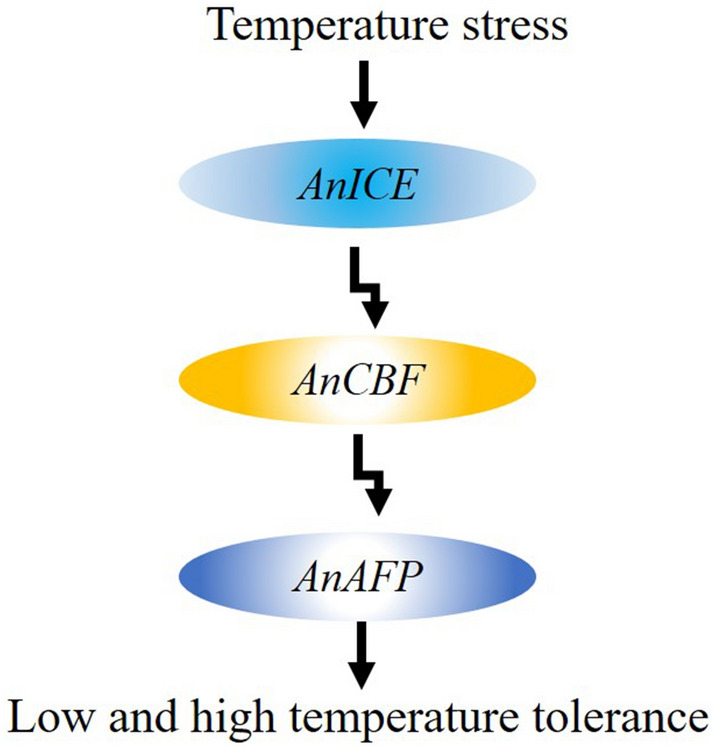
Signaling model of AnAFP in A. nanus in response to temperature stress.
Conclusion
In the presents study, we generated AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS, and transformed them into ordinary and cold sensitive strains of E. coli, and Arabidopsis KS type dehydrin mutant to evaluate their function. It’s confirmed that domain A is a thermal stable functional domain, K and N domains control localization of AnAFP. Likewise, K domain blocks interaction between AnAFP and AnICE1. The expression of AnAFP, AnICE1 and AnCBF genes was significantly induced by high-temperature, indicating that the AnAFP is likely regulated by ICE1-CBF-COR signal pathway.
Materials and methods
Evaluation of AnAFP domains in cold resistance of E. coli
Two pairs of specific primers (Table S1) with restriction sites of EcoRI/BamHI were designed by using CE Design V1.04 (http://www.downcc.com/soft/281907.html), and used to amplify open reading frame (ORF) of AnAFP and four mutants deleting domain A (AnAFPΔA), K (AnAFPΔK), S (AnAFPΔS) and N (AnAFPΔN) created in our previous study, respectively. The products were cloned into the EcoRI/BamHI site of prokaryotic expression vector pINIII to generate pINIII-AnAFP, pINIII-AnAFPΔA, pINIII-AnAFPΔK, pINIII-AnAFPΔS and pINIII-AnAFPΔN, and confirmed by sequencing, respectively.
The re-constructed plasmids were transformed into competent cells of E. coli cold-sensitive strain BX04. After confirming by screening with 50 mg/L ampicillin, PCR amplification and sequencing, the positive colonies were transferred into LB liquid medium and incubated at 37 °C until OD600 = 0.5–0.6. According to the methods of Yang et al.34 and Deng et al.21 with minor modification, the cultured cells were treated at 17 °C for 9 days with three replicates. All samples were diluted by 100 to 107 times, respectively. The 5 μL and 100 μL of them were plated onto LB plates with 50 mg/L ampicillin, incubated at 37 °C for 12 h and photographed, respectively. Before treatment, 5 μL of them were plated onto LB plates with 50 mg/L ampicillin and incubated at 37 °C for 12 h for control. The colonies number with spraying 100 μL cells were counted and used to calculate average survival rates under cold stress.
Evaluation of AnAFP domains in heat resistance of E. coli
Two pairs of specific primers (Table S2) with restriction sites of NdeI/HindIII were designed, and used to amplify ORF of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔS and AnAFPΔN using the above plasmids as template, respectively. The products were cloned into Nde I/Hind III site of prokaryotic expression vector pET28a to generate pET28a-AnAFP, pET28a-AnAFPΔA, pET28a-AnAFPΔK, pET28a-AnAFPΔS and pET28a-AnAFPΔN, and confirmed by sequencing, respectively.
The recombined plasmids were transferred into competent cells of E. coli BL21 (DE3). After confirming by screening with 50 mg/L kanamycin, and PCR amplification and sequencing, the positive colonies were transferred into LB liquid medium and incubated at 37 °C until OD600 = 0.5–0.6. The ectopic expression of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔS and AnAFPΔN was induced by 0.5 mmol/L IPTG, detected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). As described by Li et al.35 with minor modification, the induced cells were splinted into six centrifuge tubes, three of them were treated at 50 °C for 30 min, while others were incubated at 37 °C for 30 min, respectively. All samples were diluted by 100 to 104 times, respectively. The 5 μL and 100 μL of them were plated onto LB plates with 50 mg/L kanamycin, incubated at 37 °C for 12 h, and photographed. The colonies number with spraying 100 μL cells were counted and used to calculate average survival rate under heat stress.
Plasmids reconstruction and Arabidopsis transformation
Two pairs of specific primers (Table S3) with restriction sites of BspI/PstI were designed, and used to amplify ORF of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔS and AnAFPΔN, respectively. The products were cloned into Bsp I/Pst I site of plants expression vector pCAMBIA2300-35S-eGFP to generate 35S-AnAFP-eGFP, 35S-AnAFPΔA-eGFP, 35S-AnAFPΔK-eGFP, 35S-AnAFPΔS-eGFP and 35S-AnAFPΔN-eGFP, and confirmed by sequencing, respectively. These plasmids were transferred into competent cells of Agrobacterium tumefaciens GV3101 by freeze–thaw method.
After confirming by screening with 50 mg/L rifampicin and 50 mg/L kanamycin on YEB plates, PCR amplification and sequencing, the positive colonies were transferred into liquid medium YEB and incubated at 28 °C until OD600 = 1.0–1.5. The cells were collected by centrifugation at 4 °C and 4000 r/min for 10 min, resuspended and adjusted to OD600 = 1.0 with 5% (wt/vol) fresh sucrose solution, added surfactant Silwet L-77 to a concentration of 0.02% (vol/vol), and used to transform Arabidopsis mutant of the KnS type dehydrin gene AtHIRD11 (AT1G54410) by floral-dip method.
Heat tolerance of transgenic Arabidopsis
As described by Sun et al.36, T1 seeds were surface-sterilized with 75% ethanol for 1 min and 10% NaClO for 10 min, and plated onto 1/2 MS plates with 50 mg/L kanamycin (Sigma, USA) for screening of transgenic plants, which were used to produce T2 generation. The T2 plants with 3:1 segregating-ratio to resistance/susceptibility of kanamycin were self-pollinated to generate T3. The homozygous lines without segregation were collected from T3, and were identified by PCR amplification using the primers (Table S4) for the specific fragments of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS. The total RNA of every line was extracted by RNA extractor kit (Sangon, China), reacted with RNase-free DNase I, and used to reverse transcribed into cDNA using PrimeScript RT Reagent Kit (TaKaRa, Dalian). The ectopic expression of the transformed genes was identified by reverse transcription PCR (RT-PCR) using the above primers. The AtActin gene was amplified and used as reference. Five T3 lines were planted in pots, and grown in green house at 22 ℃ and 60–70% relative humidity under a 10 h light/14 h dark photoperiod. One-month-old seedlings were used for heat-shock treatment at 46 °C for 3 h, and recovered for 2 weeks at 22 ℃, and investigated for phenotype.
Subcellular localization
The 35S-AnAFP-eGFP, 35S-AnAFPΔA-eGFP, 35S-AnAFPΔK-eGFP, 35S-AnAFPΔS-eGFP and 35S-AnAFPΔN-eGFP plasmids were precipitated onto 50 mg of 0.6 µm gold particles by 2.5 mol/L CaCl2 and 0.1 mol/L spermidine, respectively, and used for transformation of intraepidermal cells of onion bulbs by microprojectile bombardment on DuPont PDS 1000/He (Bio-Rad, USA). The green fluorescence signal was observed using laser confocal microscope LSM 800 (Carl Zeiss, Germany).
Yeast two-hybrid
Two pairs of specific primers (Table S5) with restriction sites of NdeI/BamHI were designed, and used to amplify ORF of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔS and AnAFPΔN, respectively. The products were cloned into NdeI/BamHI sites of yeast two hybrid (Y2H) bait vector pGBKT7 to generate pGBKT7-AnAFP, pGBKT7-AnAFPΔA, pGBKT7-AnAFPΔK, pGBKT7-AnAFPΔS and pGBKT7-AnAFPΔN, and confirmed by sequencing, respectively. Another pair of specific primers (Table S6) with restriction sites of NdeI/BamHI were designed and used to amplify ORF of AnICE1 from the cDNA of A. nanus seedlings. The products were cloned into Y2H trap vector pGADT7 to generate pGADT7-AnICE1 and confirmed by sequencing.
The yeast strain Y2H Gold was transformed with every plasmid using Yeastmaker Yeast Transformation System 2 (Clotech, Japan), and used to test of self-activation and toxicity of AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔN, AnAFPΔS, and AnICE1 proteins. Subsequently, the yeast strain Y2H Gold was co-transformed by each pair of bait vector pGBKT7-AnAFP, pGBKT7-AnAFPΔA, pGBKT7-AnAFPΔK, pGBKT7-AnAFPΔN, pGBKT7-AnAFPΔS, and trap vector pGADT7-AnICE1, respectively. The transformants were screened on the auxotroph SD/-Trp-Leu plates at 30 °C for 3–5 days. The mono-colonies were transferred onto the auxotroph SD/-Leu-Trp-His-Ade plates, incubated at 30 °C for 3–5 days, and strained with X-α-Gal.
Real-time quantitative PCR
The seeds of A. nanus were surface-sterilized with 75% ethanol for 10 min, and planted in soil and grown in green house at 25 °C 12 h light/ 20 °C 12 h dark and 60–70% relative humidity. At the six-leaf stage, the seedlings with same size were treated at high temperature of 45 °C for 0 (negative control), 3, 6, 12, and 24 h with three biological replicates, as described by Yu et al.37. The total RNA was extracted, and reverse transcribed into cDNA as above.
For pairs of specific primers (Table S7) were designed, and used to amplify a 150–250 bp fragment for AnAFP, AnAFPΔA, AnAFPΔK, AnAFPΔN and AnAFPΔS, as well as the internal reference gene AnGAPDH. The RT-qPCR was performed using SYBR Green Super Mix (Bio-Rad, USA) by two step real-time PCR cycles (95 °C 30 s; 95 °C 5 s, 50–65 °C 30 s, 39 cycles) in CFX96 Real-Time System (Bio-Rad, USA). The 2−ΔΔCT method was used to normalize the expression differentiation between reference gene and investigated genes38.
Statistical analysis
All experiments were conducted with three replicates. The data are presented as the mean values ± standard deviation (SD). Statistical difference was analyzed using Student’s t tests.
Supplementary Information
Acknowledgements
This research was funded by the National Key Science and Technology Special Project (2016ZX08003004-005), and the Applied Basic Project of Science and Technology Department of Sichuan Province (2018JY0470). We thank Professor Wei Chi from Institute of Botany, Chinese Academy of Sciences for presenting the pINIII vector and E. coli BX04 strain. The present study complies with all relevant institutional, national, and international guidelines and legislation.
Author contributions
H.Y. administrated the project. H.Z. carried out the experiments. Y.Z. analyzed the data. H.Y. and W.L. wrote the manuscript. Y.L., and Q.Y. took part in the experiments. F.F provided technical support. Y.Z. and F.F. designed the research. All the authors approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
YuanYuan Zhang, Email: 199774@mnu.cn.
FengLing Fu, Email: ffl@sicau.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88021-0.
References
- 1.Guy CL. Freezing tolerance of plants: Current understanding and selected emerging conceopts. Can. J. Bot. 2003;81:1216–1223. doi: 10.1139/b03-130. [DOI] [Google Scholar]
- 2.Hong JH, Savina M, Du J, Devendran A, Ramakanth KK, Tian X, Sim WS, Mironova VV, Xu J. A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell. 2017;170:102–113. doi: 10.1016/j.cell.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 4.Farmer EE, Mueller MJ. ROS-mediated lipid peroxidation and RES-activated signaling. Ann. Rev. Plant Biol. 2013;1:429–450. doi: 10.1146/annurev-arplant-050312-120132. [DOI] [PubMed] [Google Scholar]
- 5.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, Shi Y, Yang S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019;222:1690–1704. doi: 10.1111/nph.15696. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Dai XY, Xu YY. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Wang WH, He EM, Guo Y, Tong Q, Zheng H. Chloroplast calcium and ROS signaling networks potentially facilitate the primed state for stomatal closure under multiple stresses. Environ. Exp. Bot. 2016;122:85–93. doi: 10.1016/j.envexpbot.2015.09.008. [DOI] [Google Scholar]
- 9.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Nat. Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnusamy V, Ohta M, Kanrar S. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene. Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baena-Gonzalez E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung RCF, Ng TB, Wong JH. Antifreeze proteins from diverse organisms and their applications: An overview. Curr. Protein. Pept. Sc. 2017;18:262–283. doi: 10.2174/1389203717666161013095027. [DOI] [PubMed] [Google Scholar]
- 13.Hightower R, Baden C, Penzes E, Lund P, Dunsmuir P. Expression of antifreeze proteins in transgenic plants. Plant. Mol. Biol. 1991;17:1013–1021. doi: 10.1007/BF00037141. [DOI] [PubMed] [Google Scholar]
- 14.Khanna HK, Daggard GE. Targeted expression of redesigned and codon optimized synthetic gene leads to recrystallisation inhibition and reduced electrolyte leakage in spring wheat at subzero temperatures. Plant. Cell. Rep. 2006;25:1336–1346. doi: 10.1007/s00299-006-0191-9. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Qiu L, Dai C, Wang J, Luo J, Zhang F, Ma J. Expression of insect (Microdera puntipennis dzungarica) antifreeze protein MpAFP149 confers the cold tolerance to transgenic tobacco. Plant Cell Rep. 2008;27:1349–1358. doi: 10.1007/s00299-008-0562-5. [DOI] [PubMed] [Google Scholar]
- 16.Griffith M, Yaish MWF. Antifreeze proteins in overwintering plants: A tale of two activities. Trends Plant Sci. 2004;9:399–405. doi: 10.1016/j.tplants.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Bredow M, Vanderbeld B, Walker VK. Ice-binding proteins confer freezing tolerance in transgenic Arabidopsis thaliana. Plant. Biotechnol. J. 2017;15:68–81. doi: 10.1111/pbi.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pudney PD, Buckley SL, Sidebottom CM. The physicochemical characterization of a boiling stable antifreeze protein from a perennial grass (Loliurn perenne) Arch. Biochem. Biophys. 2003;410:238–245. doi: 10.1016/S0003-9861(02)00697-5. [DOI] [PubMed] [Google Scholar]
- 19.Kuiper MJ, Davies PL, Walker VK. A theoretical model of a plant antifreeze protein from Lolium perenne. Biophys. J. 2001;81:3560–3565. doi: 10.1016/S0006-3495(01)75986-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S, Yin L, Mu S. Discovery of an antifreeze protein in the leaves of Ammopiptanthus nanus. Can. J. Plant Sci. 2010;90:35–40. doi: 10.4141/CJPS09060. [DOI] [Google Scholar]
- 21.Deng LQ, Yu HQ, Liu YP, Jiao PP, Zhou SF, Zhang SZ, Li WC, Fu FL. Heterologous expression of antifreeze protein gene AnAFP from Ammopiptanthus nanus enhances cold tolerance in Escherichia coli and tobacco. Gene. 2014;539:132–140. doi: 10.1016/j.gene.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Cao Y, Zheng H, Feng W, Qu J, Fu F, Li W, Yu H. Chilling tolerance identification of transgenic maize with antifreeze protein gene (AnAFP) of Ammopiptanthus nanus and chilling tolerance mechanism. Crop J. 2020 doi: 10.1016/j.cj.2020.08.011. [DOI] [Google Scholar]
- 23.Koag M, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009;150:1503–1514. doi: 10.1104/pp.109.136697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley ME, Sykes BD. The role of side chain conformational flexibility in surface recognition by Tenebrio molitor antifreeze protein. Protein Sci. 2003;12:1323–1331. doi: 10.1110/ps.0369503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya BD, Ido B, Peter LD. Ice-binding proteins and their function. Ann. Rev. Biochem. 2016;85:515–542. doi: 10.1146/annurev-biochem-060815-014546. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Wang L, Zhang T, Yang X, Li D. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 2017;7:7361. doi: 10.1038/s41598-017-07852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabortee S, Tripathi R, Watson M, Schierle GS, Kurniawan DP, Kaminski CF, Wise MJ, Tunnacliffe A. Intrinsically disordered proteins as molecular shields. Mole. Biosyst. 2012;8:210–219. doi: 10.1039/C1MB05263B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephanie LH, Verena S, Janet M, Kaley AH, David MM, Erik TB, Shruti NP, Steffen PG. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013;163:1376–1386. doi: 10.1104/pp.113.226803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lissarre M, Ohta M, Sato A, Miura K. Cold-responsive gene regulation during cold acclimation in plants. Plant. Signal Behav. 2010;5:948–952. doi: 10.4161/psb.5.8.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez-Hernandez M, Romero I, Escribano MI, Merodio C, Sanchez-Ballesta MT. Deciphering the role of CBF/DREB transcription factors and dehydrins in maintaining the quality of table grapes cv. autumn royal treated with high CO2 levels and stored at 0 ℃. Front. Plant Sci. 2017;8:1591. doi: 10.3389/fpls.2017.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 32.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 33.Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C, Wang L, Siva VS, Shi X, Jiang Q, Wang J, Zhang H, Song L. A novel cold-regulated cold shock domain containing protein from scallop Chlamys farreri with nucleic acid-binding activity. PLoS ONE. 2012;7:e32012. doi: 10.1371/journal.pone.0032012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Yang F, Lu B, Chen D, Yang W. Thermotolerance and molecular chaperone function of the small heat shock protein HSP20 from hyperthermophilic archaeon, Sulfolobus solfataricus P2. Cell Stress Chaperon. 2012;17:103–108. doi: 10.1007/s12192-011-0289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun FA, Yu HQ, Qu JT, Cao Y, Ding L, Feng WQ, Muhammad HBK, Li WC, Fu FL. Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int. J. Mol. Sci. 2020;21:996. doi: 10.3390/ijms21030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Han N, Zhang Y, Tao Y, Chen L, Liu Y, Zhou S, Fu F, Li W. Cloning and characterization of vacuolar H+-pyrophosphatase gene (AnVP1) from Ammopiptanthus nanus and its heterologous expression enhances osmotic tolerance in yeast and Arabidopsis thaliana. Plant Growth Regul. 2017;81:385–397. doi: 10.1007/s10725-016-0215-6. [DOI] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.