Abstract
Genetic testing for BRCA1 and BRCA2 is crucial in diagnosing hereditary breast and ovarian cancer syndromes and has increased with the development of multigene panel tests. However, results classified as variants of uncertain significance (VUS) present challenges to clinicians in attempting to choose an appropriate management plans. We reviewed a total of 676 breast cancer patients included in the Korean Hereditary Breast Cancer (KOHBRA) study with a VUS on BRCA mutation tests between November 2007 and April 2013. These results were compared to the ClinVar database. We calculated the incidence and odds ratios for these variants using the Korean Reference Genome Database. A total of 58 and 91 distinct VUS in BRCA1 and BRCA2 were identified in the KOHBRA study (comprising 278 and 453 patients, respectively). A total of 27 variants in the KOHBRA study were not registered in the Single Nucleotide Polymorphism database. Among BRCA1 VUSs, 20 were reclassified as benign or likely benign, four were reclassified as pathogenic or likely pathogenic, and eight remained as VUSs according to the ClinVar database. Of the BRCA2 VUSs, 25 were reclassified as benign or likely benign, two were reclassified as pathogenic or likely pathogenic, and 33 remained as VUS according to the ClinVar database. There were 12 variants with conflicting interpretations of pathogenicity for BRCA1 and 18 for BRCA2. Among them, p.Leu1780Pro showed a particularly high odds ratio. Six pathogenic variants and one conflicting variant identified using ClinVar could be reclassified as pathogenic variants in this study. Using updated ClinVar information and calculating odds ratios can be helpful when reclassifying VUSs in BRCA1/2.
Subject terms: Cancer, Genetics, Medical research
Introduction
Hereditary breast and ovarian cancer syndrome (HBOC) has been shown to be associated with germline mutations in BRCA1 and BRCA21, spurring demands for genetic testing to identify pathogenic variations in these genes2. The identification of a pathogenic BRCA mutation in a patient diagnosed with breast cancer not only affects their treatment and prognosis, but also enables the prevention of other cancers3. Guidelines for the management of pathogenic variants in BRCA1 and BRCA2 recommend consideration of risk-reducing medications or surgeries4,5.
A genetic test for BRCA has four possible results: no mutation detected, pathogenic mutation, benign mutation, or variant of uncertain significance (VUS). A VUS is an alteration in the gene sequence that has an unknown effect on the function of the gene product. This leaves patients and their physicians with uncertainty due to the inability to interpret the result in a clinical context and a lack of specific guidelines regarding genetic counseling or prophylactic management in mutation carriers and their relatives6.
While an overall VUS rate of 7–15% in women who have received BRCA testing has been reported7, the frequency of VUS varies worldwide depending on the testing prevalence and population ancestry7,8. Researchers reported a frequency of VUS of 21% in African-Americans, 5–6% in people of European ancestry in the United States, and 15% in European laboratories9,10. Myriad Genetic Inc. (Salt Lake City, UT, USA) reported that they decreased the proportion of VUSs to 2.1% using accumulated data11. However, these databases are not public or accessible.
In this study, we aimed to explore the prevalence of VUS in the Korean population and to reclassify these variants using the ClinVar database and the Korean Reference Genome Database (KRGDB).
Results
Baseline characteristics
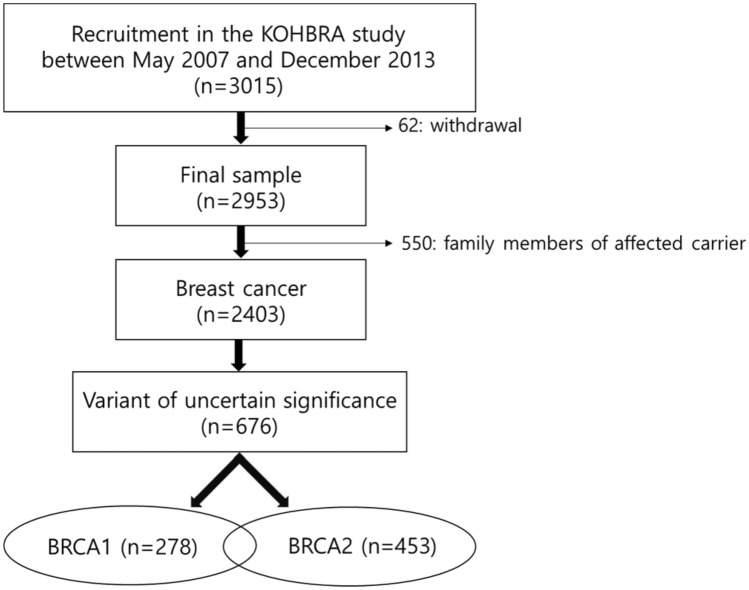
Among 2,403 breast cancer patients in the Korean Hereditary Breast Cancer (KOHBRA) study, more than a quarter, 676 (28.13%) patients, had mutations that were classified as VUS. Simultaneous mutations of BRCA1 and BRCA2 were observed in 55 (55/676, 8.14%) patients. Of the 676 subjects, 278 had a VUS in BRCA1, and 453 patients had a VUS in BRCA2 (Fig. 1). refSNP (RS) numbers were reviewed for 262 and 440 subjects with BRCA1 and BRCA2 mutations, respectively.
Figure 1.
Schematic diagram of patient selection: BRCA1 and BRCA 2 (n = 55).
Reclassification using public databases
Table 1 shows the reclassification results for VUS according to the ClinVar database. We classified the results into four groups: benign/likely benign, VUS, conflicting interpretations of pathogenicity, and pathogenic/likely pathogenic. Benign/likely benign was the most common reclassification for both BRCA1 and BRCA2 VUSs. Table 2 shows the rank of VUS genes based on the number of patients.
Table 1.
Reclassification of patients diagnosed with variants of uncertain significance based on ClinVar data.
| BRCA1 | 58 mutations (n = 278) |
|---|---|
| Benign/ likely benign | 20 mutations (n = 193) |
| VUS | 8 mutations (n = 8) |
| Conflicting in interpretations of pathogenicity | 12 mutations (n = 55) |
| Pathogenic/ likely pathogenic | 4 mutations (n = 6) |
| Variants not registered in SNP database | 14 mutations (n = 16) |
| BRCA2 | 91 mutations (n = 453) |
|---|---|
| Benign/ likely benign | 25 mutations (n = 328) |
| VUS | 33 mutations (n = 58) |
| Conflicting in interpretations of pathogenicity | 18 mutations (n = 51) |
| Pathogenic/ likely pathogenic | 2 mutations (n = 3) |
| Variants not registered in SNP database | 13 mutations (n = 13) |
Table 2.
Top 10 high-frequency mutations based on the number of patients.
| BRCA1 | RS_number | ClinVar | No. of patients | OR | CI |
|---|---|---|---|---|---|
| c.4883T>C | rs4986854 | Benign | 57 | 0.8649 | 0.5865–1.2754 |
| c.4484 + 14A>G | rs80358022 | Benign | 41 | 0.5823 | 0.3845–0.8818 |
| c.2566T>C | rs80356892 | Benign | 40 | 0.3659 | 0.2491–0.5374 |
| c.3113A>G | rs16941 | Benign | 17 | 0.0079 | 0.0049–0.0128 |
| c.154C>T | rs80357084 | Conflicting interpretations of pathogenicity | 15 | 0.4646 | 0.2421–0.8914 |
| c.5339T>C | rs80357474 | Conflicting interpretations of pathogenicity | 12 | 8.5181 | 1.1192–64.8277 |
| c.4729T>C | rs80356909 | Conflicting interpretations of pathogenicity | 9 | 1.2874 | 0.4314–3.8417 |
| c.3448C>T | rs80357272 | Benign | 8 | 0.6349 | 0.2448–1.6464 |
| c.3548A>G | rs16942 | Benign | 8 | 0.0046 | 0.0025–0.0086 |
| c.547 + 14delG | rs273902771 | Conflicting interpretations of pathogenicity | 6 |
| BRCA2 | RS_number | ClinVar | No. of patients | OR | CI |
|---|---|---|---|---|---|
| c.10234A>G | rs1801426 | Benign | 67 | 0.6443 | 0.4627–0.8972 |
| c.8187G>T | rs80359065 | Benign | 57 | 0.7962 | 0.5445–1.1643 |
| c.9649-19G>A | rs11571830 | Benign | 40 | 0.5462 | 0.3609–0.8266 |
| c.5785A>G | rs79538375 | Benign | 33 | 0.8118 | 0.4922–1.339 |
| c.2971A>G | rs1799944 | Benign | 29 | 0.0432 | 0.0299–0.0623 |
| c.2350A>G | rs11571653 | Benign | 28 | 0.4624 | 0.2868–0.7456 |
| c.1744A>C | rs80358457 | Benign | 24 | 0.7135 | 0.4046–1.2581 |
| c.7052C>G | rs80358932 | Benign/likely benign | 12 | 0.6115 | 0.2826–1.3231 |
| c.6325G>A | rs79456940 | Conflicting interpretations of pathogenicity | 10 | 0.6488 | 0.2754–1.5285 |
| c.623T>G | rs80358865 | Uncertain significance | 9 | 1.2867 | 0.4313–3.8389 |
Of the 278 patients with a BRCA1 VUS, 58 VUSs were identified, and 44 had RS numbers. Twenty of these variants, found in 193 patients, were classified as benign/likely benign. The least common VUSs were classified as pathogenic/likely pathogenic and comprised four variants in six patients (Table 1).
Of the 453 patients with BRCA2 VUSs, 91 VUSs were identified, and 78 had RS numbers. The most common VUSs were benign/likely benign, comprising 25 variants in 328 patients. Meanwhile, pathogenic/likely pathogenic variants were the least common and included two variants in three patients.
Calculating minor allele frequency and plotting graphs thereof, we noted that minor allele frequencies for all variants, except for BRCA1 c.4987-92A>G(rs8176233), BRCA1 c.3113A>G(rs16941), BRCA1 c.3548A>G(rs16942), and BRCA2 c.2971A>G(rs1799944), had P values of 0.05 or less: all four of the variants lacking statistical significance were classified as benign (Supplementary Fig. S1).
In this study, six gene mutations previously classified as VUS were reclassified as likely pathogenic based on ClinVar review. The variants were BRCA1 c.5089T>C (p.Cys1697Arg), BRCA1 c.5509T>C (p. Trp1837Arg), BRCA1 c.5516T>C (p.Leu1839Ser), BRCA1 c.81-9C>G, BRCA2 c.8023A>G (p.Ile2675Val), and BRCA2 c.9004G>A (p.Glu3002Lys).
Odds ratio estimation
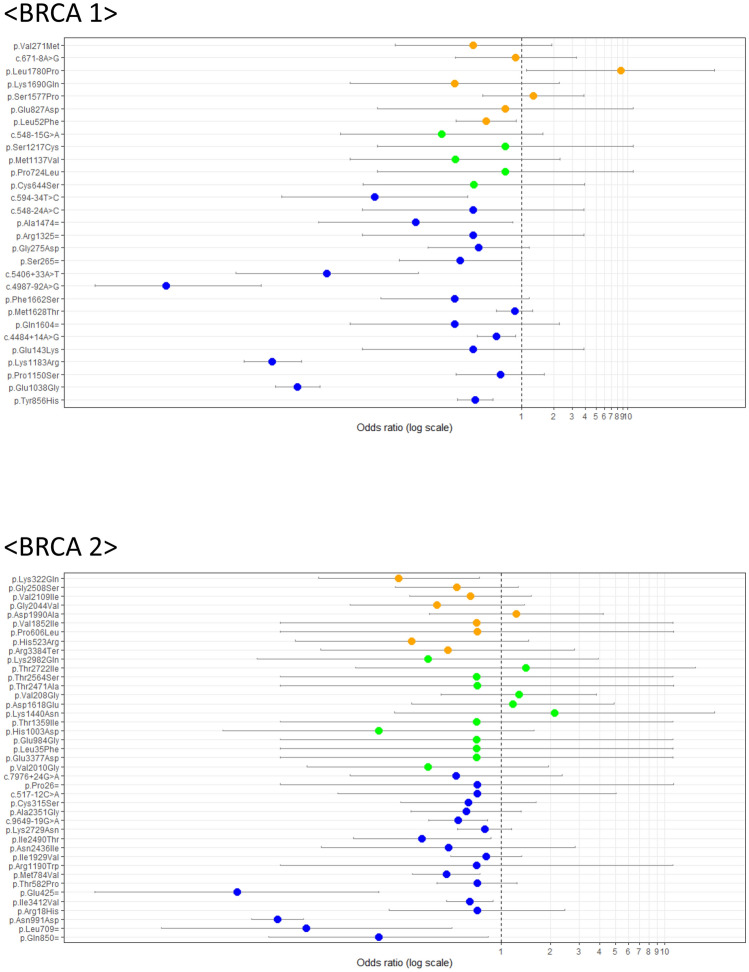
We calculated the odds ratios (ORs) for each variant using the KRGDB, as shown in Fig. 2. The OR of BRCA1 c.5339T>C (p.Leu1780Pro) was significantly high in analysis with the Wald method with 95% confidence intervals (CI). The corresponding P value was 0.0127 before and 0.889 after Bonferroni correction.
Figure 2.
Odds ratios (ORs) using Korean population data from the Korean Reference Genome Database (KRGDB). Variants that could be identified in the KRGDB were classified according to the ClinVar database (vertical axis). Round dots indicate ORs, and the continuous line through each dot indicates a 95% confidence interval. Variants with orange, green, and blue dots indicate conflicting interpretations of pathogenicity, uncertain significance, and benign/likely benign variants according to the ClinVar database, respectively.
Discussion
We re-evaluated genetic results in patients with VUSs in BRCA1 and BRCA2 using the ClinVar database. Since genetic characteristics can vary by ethnicity7,9,10, this study aimed to identify the prevalence of VUS in the Korean population and to re-classify the results of initial genetic tests. Although initial genetic testing results in the KOHBRA study revealed a VUS in 676 patients (278 patients for BRCA1 and 453 patients for BRCA2) out a total of 2,403 breast cancer patients (28.13%, 676/2,403), re-evaluation revealed a lower frequency of VUSs (8.03%, 193/2,403). About a third of the variants that were originally classified as VUS in the KOHBRA study were reclassified as benign/likely benign or pathogenic/likely pathogenic, accounting for two-thirds of all VUS patients (71.45%, 483/676). This result suggests that a re-classification approach using the ClinVar database can reduce the frequency of VUSs in the Korean population.
About two-thirds of the VUSs in the KOHBRA study were reclassified as benign or likely benign [193/278 (69.42%) of BRCA1 patients, 328/453 (72.41%) of BRCA2 patients, and 471/676 (69.67%) of all patients]. A third of the mutation types classified as VUS in the KOHBRA study were downgraded to benign or likely benign (20/58 mutations in BRCA1 and 25/91 in BRCA2). These results were consistent with previous studies1,12,13: So et al. reported that 30/75 (40%) of VUS patients were reclassified as benign or likely benign14. In our study, six patients with BRCA1 VUS (four mutations) and three patients with BRCA2 VUS (two mutations) were reclassified as pathogenic or likely pathogenic.
In this study, six mutations in nine patients were reclassified from VUSs to pathogenic or likely pathogenic variants. BRCA1 c.5089T>C (p.Cys1697Arg), BRCA1 c.5509T>C (p. Trp1837Arg), BRCA1 c.5516T>C (p.Leu1839Ser), and BRCA1 c.81-9C>G were interpreted as likely pathogenic among variants in BRCA1. BRCA2 c.8023A>G (p.Ile2675Val) and BRCA2 c.9004G>A (p.Glu3002Lys) were interpreted as pathogenic/likely pathogenic among variants in BRCA2 based on the ClinVar database. Since all six mutations are supported by sufficient evidence in functional studies (PS3)15–17, have not been reported in a genomic database for the general Korean population (PM2), are classified as pathogenic in ClinVar (PP5), and are deleterious mutations (PP3) according to an in silico study18, they should be reclassified as likely pathogenic or pathogenic mutations. For patients with these mutations, additional genetic counseling and proper management, such as familial genetic testing, risk-reducing medications, or risk-reducing surgery, are needed for the prevention of cancer.
Interestingly, we identified several cases with conflicting interpretations of pathogenicity, for which reviewed the current status of available evidence in the literature (Table 3). In such instances, calculating ORs can help to reclassify them. The KRGDB, which is a large-scale, single-race database collected from 1,722 Koreans, is built for precision medicine research. We analyzed the KRGDB data using the Wald method to obtain ORs and 95% CIs for all VUS variants (Fig. 2). Several mutations (7 BRCA1 and 9 BRCA2 mutations) were evaluated. Most of them showed no significance. However, BRCA1 c.5339T>C (p.Leu1780Pro) showed possible pathogenicity, while BRCA1 c.154C>T (p.Leu52Phe) and BRCA2 c.964A>C (p.Lys322Gln) were potentially benign. However, with the most conservative multiple test correction, Bonferroni correction, the statistically significance of c.5339T>C disappeared. This non-significant level should be carefully interpreted, because several previous studies indicated that this c.5339.T>C variant is pathogenic or likely pathogenic according to clinicopathologic features and American College of Medical Genetics (ACMG) guidelines13,19–21. Therefore, even though the P value after multiple correction indicated a lack of statistical significance, c.5339T>C should be interpreted as a pathogenic or likely pathogenic variant in light of other functional evaluations, including co-segregation, in vitro, and in silico analyses. In verifying additional pathogenic mutation candidates, we merely referred to OR and CI values, but do not insist that OR values alone should be applied. With further accumulation of data in the future, we may be able to recalculate these ORs of P values. Meanwhile, variants BRCA 1 c.5014_5016delCAC, BRCA 1 c.5332G>A, BRCA2 c.182T>C, BRCA 2 c.1909 + 22delT, BRCA 2 c.8486A>G, and BRCA2 c.8954-5A>G have not been reported in the general population and should remain classified as VUS, since there are no reports on their deleterious function.
Table 3.
Conflicting interpretations of pathogenicity.
| BRCA1 | RS_number | (Likely) Pathogenic | Uncertain significance | (Likely) Benign | No. of patients | OR | CI |
|---|---|---|---|---|---|---|---|
|
c.1357G>C, p.Glu453Gln |
rs768054411 | 3 | 1 | 1 | |||
|
c.154C>T p.Leu52Phe |
rs80357084 | 5 | 3 | 15 | 0.4646 | 0.2421–0.8914 | |
|
c.2481A>C p.Glu827Asp |
rs397508970 | 2 | 1 | 1 | 0.7082 | 0.0446–11.2359 | |
|
c.2726A>T p.Asn909Ile |
rs80357127 | 8 | 1 | 2 | |||
|
c.4729T>C p.Ser1577Pro |
rs80356909 | 1 | 7 | 9 | 1.2874 | 0.4314–3.8417 | |
|
c.5014_5016delCAC p.His1673del |
rs80358343 | 3 | 4 | 1 | |||
|
c.5068A>C p.Lys1690Gln |
rs397507239 | 8 | 1 | 1 | 0.2358 | 0.0246–2.2601 | |
|
c.5332G>A p.Asp1778Asn |
rs80357112 | 2 | 3 | 1 | |||
|
c.5339T>C p.Leu1780Pro |
rs80357474 | 4 | 1 | 12 | 8.5181 | 1.1192–64.8277 | |
|
c.547 + 14delG c.547 + 14delG |
rs273902771 | 2 | 4 | 6 | |||
|
c.671-8A>G c.671-8A>G |
rs80358144 | 1 | 4 | 5 | 0.8852 | 0.2385–3.2849 | |
|
c.811G>A p.Val271Met |
rs80357244 | 1 | 11 | 2 | 0.3537 | 0.065–1.9254 |
| BRCA2 | RS_number | (Likely) Pathogenic | Uncertain significance | (Likely) Benign | No. of patients | OR | CI |
|---|---|---|---|---|---|---|---|
|
c.10150C>T p.Arg3384Ter |
rs397507568 | 1 | 5 | 2 | 0.4717 | 0.0791–2.8122 | |
|
c.1568A>G p.His523Arg |
rs80358443 | 2 | 9 | 2 | 0.2855 | 0.0554–1.4714 | |
|
c.1817C>T p.Pro606Leu |
rs80358469 | 5 | 1 | 1 | 0.7145 | 0.0448–11.4046 | |
|
c.182T>C p.Leu61Pro |
rs1555280374 | 2 | 1 | 1 | |||
| c.1909 + 22delT | rs276174816 | 1 | 7 | 1 | |||
| c.317-10A>G | rs81002824 | 1 | 1 | 1 | |||
|
c.3256A>G p.Ile1086Val |
rs80358571 | 5 | 2 | 3 | |||
|
c.4599A>C p.Lys1533Asn |
rs80358694 | 3 | 5 | 1 | |||
|
c.5554G>A p.Val1852Ile |
rs80358777 | 4 | 2 | 1 | 0.7078 | 0.0446–11.225 | |
|
c.5969A>C p.Asp1990Ala |
rs148618542 | 6 | 1 | 7 | 1.2391 | 0.3642–4.216 | |
|
c.6101G>A p.Arg2034His |
rs80358849 | 2 | 1 | 1 | |||
|
c.6131G>T p.Gly2044Val |
rs56191579 | 1 | 8 | 4 | 0.4078 | 0.1194–1.3933 | |
|
c.6325G>A p.Val2109Ile |
rs79456940 | 2 | 10 | 10 | 0.6488 | 0.2754–1.5285 | |
|
c.7522G>A p.Gly2508Ser |
rs80358978 | 7 | 6 | 9 | 0.5353 | 0.2254–1.2713 | |
|
c.8092G>A p.Ala2698Thr |
rs80359052 | 6 | 4 | 1 | |||
|
c.8486A>G p.Gln2829Arg |
rs80359100 | 2 | 1 | 1 | |||
| c.8954-5A>G | rs886040949 | 5 | 1 | 1 | |||
|
c.964A>C p.Lys322Gln |
rs11571640 | 3 | 9 | 4 | 0.2375 | 0.0766–0.7367 |
Interpretations of VUSs are complex. Functional studies for reclassifying VUS could be a promising approach. Traditionally, VUS interpretation has depended on inductive conclusions based on information of individual patients22. However, many potential variants in BRCA are present at low variant allele frequencies, with phenotypes that are incompletely penetrant. Findlay et al. reported the application of saturation genome editing (SGE) to measure the functional outcomes of all possible single nucleotide variants (SNVs) in key areas of BRCA115. Functional effects were almost concordant with the established assessments of pathogenicity. Function scores using SGE could help with interpreting the significance of VUSs by providing functional classification and assessment of ambiguous or newly-discovered variants.
The four BRCA1 mutations (c.5089T>C (p.Cys1697Arg), c.5509T>C (p.Trp1837Arg), c.5516T>C (p.Leu1839Ser), and c.81-9C>G) identified as likely pathogenic using the ClinVar database were identified as non-functional in Findlay’s study15. The c.5339T>C (p.Leu1780Pro) variant identified as having conflicting interpretations of pathogenicity in the ClinVar database was also identified as “non-functional” in the functional study results, suggesting this variant as pathogenic15. On the other hand, other variants with conflicting interpretations of pathogenicity in the ClinVar database (including c.154C>T (p.Leu52Phe), c.5068A>C (p.Lys1690Gln), and c.5332G>A (p.Asp1778Asn)) have been categorized as functional or intermediate15. The other BRCA1 variants with conflicting interpretations of pathogenicity in the ClinVar database could not be evaluated according to Findlay’s study, as it analyzed only RING and BRCT domains as targets15.
Cosegregation analysis may also be helpful in re-evaluating VUSs. Zuntini et al. performed cosegregation analysis for 13 VUSs in 11 kindreds to improve VUS evaluation, and two variants were found to have additional supporting evidence of pathogenicity23. Among the variants that were reclassified as pathogenic variants in our study, BRCA1 c.5509T>C (p. Trp1837Arg) was discussed in Zuntini's study as well; however, due to the distinct nature of our data, cosegregation analysis could not be conducted. Cosegregation analysis may help improve understanding of VUSs and provide genetic counseling for specific families, sufficient for pedigree analysis.
When ORs were calculated using the KRGDB for all KOHBRA data, the OR of BRCA1 c.5339T>C (p.Leu1780Pro) was found to be significantly elevated (Fig. 2). This variant was also identified as non-functional in Findlay’s study. In addition, several studies have suggested that this mutation is pathogenic based on other evidence, including a strong family history of breast and ovarian cancer, absence in general population data, impaired function demonstrated by in silico studies, and triple negativity in clinicopathologic features13,14,21. Previous studies have used a similar approach to reclassify some variants24,25.
One limitation of the study is that we reviewed the VUSs by assigning them to a database based on a mostly Caucasian population. Researchers contributing to the Single Nucleotide Polymorphism (SNP) database or ClinVar tend to be concentrated in Western countries, leading to a lack of registration of major variants in Asian populations or a lack of interpretation of variants such as L1780P. Nevertheless, this study was meaningful in that it confirmed VUS status in Koreans using a prospective study and lays the groundwork for broadening our understanding of VUSs and conducting further research. Another limitation of the study is that there is missing information in the KOHBRA data, which are necessary to reclassify VUSs (e.g., 27 variants were not submitted to the SNP database). However, the lacking data comprised only 4.29% of the cohort and would unlikely weaken the power of the current study.
Taken together, most of the mutations that were classified as VUS in the KOHBRA study were reclassified as benign. Four VUSs in BRCA1 and two in BRCA2 VUS were reclassified as pathogenic or likely pathogenic. When ORs were calculated using the KRGDB for all KOHBRA data, the OR of BRCA1 c.5339T>C (p.Leu1780Pro) was significantly high, although ClinVar considered BRCA1 c.5339T>C to have conflicting interpretations of pathogenicity. These seven mutations could be reclassified as likely pathogenic or pathogenic mutations, according to ACMG guidelines. Since the mutations classified as benign in ClinVar have a high normal frequency, it is desirable to judge them as benign.
However, some VUSs remained as having conflicting interpretations of pathogenicity, rather than being re-assessed as benign or pathogenic. Their characteristics will likely be more discernable with the accumulation of more information. When a VUS is reclassified as pathogenic/likely pathogenic, appropriate management, including risk-reducing medication and surgery, should be discussed with patients and their families. In addition to collecting individual data, functional studies using genetic techniques, such as SGE, could help contribute to the functional classification and assessment of VUSs.
Methods
Subjects
The study population was obtained from the Korean Hereditary Breast Cancer (KOHBRA) study26. The KOHBRA study is a multicenter prospective cohort study designed to investigate the prevalence and causes of hereditary breast cancer in the Korean population. Through the study, 3,015 subjects were recruited between May 2007 and December 2013 from 36 institutions27. The eligibility criteria were as follows: (1) breast cancer patients with a family history of breast or ovarian cancer; (2) breast cancer patients without a family history of breast or ovarian cancer (non-familial) who were aged 40 years or younger at diagnosis and were diagnosed with bilateral breast cancer or another primary malignancy; (3) male breast cancer patients; and (4) family members of BRCA 1/2 mutation carriers. After excluding several subjects, a total of 2953 subjects (1228 familial breast cancer patients, 1175 non-familial breast cancer patients, and 550 family members of affected carriers) were evaluated. We identified 676 breast cancer patients with VUS on BRCA mutation tests.
These results were reclassified using the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) based on refSNP (RS) numbers. Odds ratios (ORs) for each variant were calculated using Korean population data from the KRGDB, which was established by conducting whole genome sequencing of 1,722 Koreans28. Variants that were not registered in the Single Nucleotide Polymorphism (SNP) database are shown in Table 1. In this study, variations without RS numbers were also included in the denominator when checking the overall frequency of VUS.
BRCA 1/2 mutation analysis
BRCA 1/2 genetic testing was performed using genomic DNA from the peripheral blood. Of the 2403 patients, clinicians used fluorescence confirmation sensitive capillary (gel) electrophoresis on 1183 patients, direct sequencing on 1101 patients, and denaturing high-performance liquid chromatography on 119 patients27. Each testing method was selected according to the laboratory linked to the institution. All BRCA 1/2 mutations were classified as pathogenic, VUS, or polymorphic.
Statistical analysis
Chi-square or Fisher’s exact tests were used for categorical variables. ORs and 95% confidence intervals were obtained using the Wald method. We retrieved allele frequencies from 1722 Koreans (KRGDB). For each variant, an OR was calculated based on its occurrence in 2403 patient cases and in the KRGDB. All analyses were performed using SPSS version 23 (SPSS; Chicago, IL, USA), and statistical significance was defined as P < 0.05.
Mutation nomenclature
All sequence variations are described according to the HUGO-approved systematic nomenclature (http://www.hgvs.org/mutnomen/) using GenBank reference sequences (NM_007294.2 for BRCA 1 and NM_000059.3 for BRCA 2). The breast cancer information core nomenclature is also presented for convenience.
This study was approved by the Institutional Review Board of Severance Hospital, Yonsei University. (IRB# 4-2017-0255).
Supplementary information
Author contributions
Conceptualization: H.S.P. and J.H.K. Methodology and software: S.P., S.-T.L. and J.S.P. Formal analysis and investigation: J.H.K., S.P., S.-T.L. and H.S.P. Writing—original draft preparation: J.H.K. and H.S.P. Co-first authors: J.H.K. and S.P. Writing—review and editing: J.H.K., S.P., S.-T.L., S-W.K., J.S.P., and H.S.P. Resources: J.W.L., M.H.L., S.K.P., W.-C.N., D.H.C., W.H., and S.H.J. Supervision: S.-T.L., S.-W.K., J.S.P., and H.S.P.
Funding
The present research has been supported by Korea Breast Cancer Foundation (KBCF-2016U002).
Data availability
The genotype and clinical phenotype data that support the findings of this study are not publicly available due to ethical and patient consent constraints. However, genotype and basic clinical phenotype data are available upon reasonable request from the corresponding author [H.S.P.] under a collaboration and data usage agreement.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The Funding section in the original version of this Article was omitted. The Funding section now reads: “The present research has been supported by Korea Breast Cancer Foundation (KBCF-2016U002)”.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joo Heung Kim and Sunggyun Park.
Change history
3/8/2023
A Correction to this paper has been published: 10.1038/s41598-023-31093-x
Contributor Information
Hyung Seok Park, Email: imgenius@yuhs.ac.
Ji Soo Park, Email: PMJISU@yuhs.ac.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87792-w.
References
- 1.Park KS, Cho EY, Nam SJ, Ki CS, Kim JW. Comparative analysis of BRCA1 and BRCA2 variants of uncertain significance in patients with breast cancer: a multifactorial probability-based model versus ACMG standards and guidelines for interpreting sequence variants. Genet. Med. Off. J. Am. Coll. Med. Genet. 2016;18:1250–1257. doi: 10.1038/gim.2016.39. [DOI] [PubMed] [Google Scholar]
- 2.Grindedal EM, et al. Current guidelines for BRCA testing of breast cancer patients are insufficient to detect all mutation carriers. BMC Cancer. 2017;17:438. doi: 10.1186/s12885-017-3422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotsopoulos J. BRCA mutations and breast cancer prevention. Cancers. 2018;10:524. doi: 10.3390/cancers10120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Practice Bulletin No 182: hereditary breast and ovarian cancer syndrome. Obstet. Gynecol. 130, e110–e126 (2017). [DOI] [PubMed]
- 5.Krontiras H, Farmer M, Whatley J. Breast cancer genetics and indications for prophylactic mastectomy. Surg. Clin. N. Am. 2018;98:677–685. doi: 10.1016/j.suc.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Richter S, et al. Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013;24(Suppl 8):viii69–viii74. doi: 10.1093/annonc/mdt312. [DOI] [PubMed] [Google Scholar]
- 7.Calo V, et al. The clinical significance of unknown sequence variants in BRCA genes. Cancers. 2010;2:1644–1660. doi: 10.3390/cancers2031644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: distribution and clinical implications. Curr. Opin. Obstet. Gynecol. 2010;22:72–78. doi: 10.1097/GCO.0b013e328332dca3. [DOI] [PubMed] [Google Scholar]
- 9.Lindor NM, Goldgar DE, Tavtigian SV, Plon SE, Couch FJ. BRCA1/2 sequence variants of uncertain significance: a primer for providers to assist in discussions and in medical management. Oncologist. 2013;18:518–524. doi: 10.1634/theoncologist.2012-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ready K, et al. Cancer risk management decisions of women with BRCA1 or BRCA2 variants of uncertain significance. Breast J. 2011;17:210–212. doi: 10.1111/j.1524-4741.2010.01055.x. [DOI] [PubMed] [Google Scholar]
- 11.Eggington JM, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin. Genet. 2014;86:229–237. doi: 10.1111/cge.12315. [DOI] [PubMed] [Google Scholar]
- 12.Macklin S, Durand N, Atwal P, Hines S. Observed frequency and challenges of variant reclassification in a hereditary cancer clinic. Genet. Med. Off. J. Am. Coll. Med. Genet. 2018;20:346–350. doi: 10.1038/gim.2017.207. [DOI] [PubMed] [Google Scholar]
- 13.Park JS, et al. Identification of a novel BRCA1 pathogenic mutation in Korean patients following reclassification of BRCA1 and BRCA2 variants according to the ACMG standards and guidelines using relevant ethnic controls. Cancer Res. Treat. 2017;49:1012–1021. doi: 10.4143/crt.2016.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.So MK, et al. Reinterpretation of BRCA1 and BRCA2 variants of uncertain significance in patients with hereditary breast/ovarian cancer using the ACMG/AMP 2015 guidelines. Breast Cancer (Tokyo, Japan) 2019;26:510–519. doi: 10.1007/s12282-019-00951-w. [DOI] [PubMed] [Google Scholar]
- 15.Findlay GM, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas K, et al. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum. Mol. Genet. 2012;21:3993–4006. doi: 10.1093/hmg/dds222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet C, et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J. Med. Genet. 2008;45:438–446. doi: 10.1136/jmg.2007.056895. [DOI] [PubMed] [Google Scholar]
- 18.Lee JS, et al. Reclassification of BRCA1 and BRCA2 variants of uncertain significance: a multifactorial analysis of multicentre prospective cohort. J. Med. Genet. 2018;55:794–802. doi: 10.1136/jmedgenet-2018-105565. [DOI] [PubMed] [Google Scholar]
- 19.Park HS, et al. Clinicopathological features of patients with the BRCA1 c.5339T>C (p.Leu1780Pro) variant. Cancer Res. Treat. 2020;52:680–688. doi: 10.4143/crt.2019.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu JM, et al. Suggestion of BRCA1 c.5339T>C (p.L1780P) variant confer from ‘unknown significance’ to ‘likely pathogenic’ based on clinical evidence in Korea. Breast (Edinburgh, Scotland) 2017;33:109–116. doi: 10.1016/j.breast.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Yoon KA, et al. Clinically significant unclassified variants in BRCA1 and BRCA2 genes among Korean breast cancer patients. Cancer Res. Treat. 2017;49:627–634. doi: 10.4143/crt.2016.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borg A, et al. Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study. Hum. Mutat. 2010;31:E1200–1240. doi: 10.1002/humu.21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuntini R, et al. Dealing with BRCA1/2 unclassified variants in a cancer genetics clinic: Does cosegregation analysis help? Front. Genet. 2018;9:378. doi: 10.3389/fgene.2018.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minucci A, et al. Additional molecular and clinical evidence open the way to definitive IARC classification of the BRCA1 c.5332G>A (p.Asp1778Asn) variant. Clin. Biochem. 2019;63:54–58. doi: 10.1016/j.clinbiochem.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Zuntini R, et al. BRCA1 p.His1673del is a pathogenic mutation associated with a predominant ovarian cancer phenotype. Oncotarget. 2017;8:22640–22648. doi: 10.18632/oncotarget.15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han SA, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin. Oncol. (R. Coll. Radiol. (Great Br.)) 2011;23:434–441. doi: 10.1016/j.clon.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Kang E, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res. Treat. 2015;151:157–168. doi: 10.1007/s10549-015-3377-4. [DOI] [PubMed] [Google Scholar]
- 28.Jung, K. S. et al. KRGDB: the large-scale variant database of 1722 Koreans based on whole genome sequencing. Database J. Biol. Databases Curation2020, 1–7 (2020) 10.1093/database/baz146.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotype and clinical phenotype data that support the findings of this study are not publicly available due to ethical and patient consent constraints. However, genotype and basic clinical phenotype data are available upon reasonable request from the corresponding author [H.S.P.] under a collaboration and data usage agreement.