Abstract
Sampling impediments and paucity of suitable material for molecular analyses have precluded the study of speciation and radiation of deep-sea species in Antarctica. We analyzed barcodes together with genome-wide single nucleotide polymorphisms obtained from double digestion restriction site-associated DNA sequencing (ddRADseq) for species in the family Antarctophilinidae. We also reevaluated the fossil record associated with this taxon to provide further insights into the origin of the group. Novel approaches to identify distinctive genetic lineages, including unsupervised machine learning variational autoencoder plots, were used to establish species hypothesis frameworks. In this sense, three undescribed species and a complex of cryptic species were identified, suggesting allopatric speciation connected to geographic or bathymetric isolation. We further observed that the shallow waters around the Scotia Arc and on the continental shelf in the Weddell Sea present high endemism and diversity. In contrast, likely due to the glacial pressure during the Cenozoic, a deep-sea group with fewer species emerged expanding over great areas in the South-Atlantic Antarctic Ridge. Our study agrees on how diachronic paleoclimatic and current environmental factors shaped Antarctic communities both at the shallow and deep-sea levels, promoting Antarctica as the center of origin for numerous taxa such as gastropod mollusks.
Subject terms: Genomics, Zoology, Phylogenetics, Taxonomy
Introduction
Traditionally, deep-sea species are regarded as occupying a wide depth range (i.e. eurybathy) and as distributed across large biogeographical areas due to the supposed homogeneity of the deep-sea habitat1. However, unlike their shallow-water counterparts, little is known about the evolution and radiation of deep-sea species at a global scale2–4. Elucidating the factors that drive diversification in the deep is of profound importance for understanding how deep-sea taxa originated and diversified. Nonetheless, the paucity of taxonomic surveys of deep-sea invertebrates precludes us from having a sound assessment of the diversity and distributional patterns of deep-sea organisms5,6. Fossil evidence suggests that many post-Paleozoic taxa first appeared onshore even if they are now exclusive in the deep-sea7, and might have been displaced into deeper waters as a result of pressure from predation and/or competition5 or due to physical disturbances8. There is evidence suggesting that both shallow and deep-sea organisms share a common period of diversification around the Oligocene and Miocene, undoubtedly, due to major tectonic events during these epochs9–11.
Antarctica represents an interesting system for comparatively studying shallow versus deep water speciation processes. Firstly, low physical disturbance (below the influence of iceberg scour), cold temperatures, and intermittent food availability are shared characteristics for both the Southern Ocean (SO) shelf and deep-sea ecosystems as a whole12. Secondly, during the Eocene–Oligocene transition (~ 34 Mya13) and due to the effect of the glacial cycles, Antarctica acquired a considerably deeper shelf and slope than other ocean basins14. This fact led the Antarctic fauna into the tendency to eurybathy and widespread—often around the continent, i.e. circumpolar—distributions, but reaching the upper and more accessible continental shelf, compared to more typical deep-sea species15,16. Concurrently, the onset of the Antarctic Circumpolar Current (ACC) also occurred, and tectonic events leading to the opening of both the Drake and Tasmanian Passages17. The ACC connected shallow-water Antarctic fauna with deep water in the Atlantic, Indian, and Pacific Oceans contributing to the Cenozoic diversification in the SO18,19. This provides evidence that Antarctica may have acted as a center of origin for deep-sea taxa20–22, with its shelf taxa dispersing into deep water using the northward movement of the Antarctic Bottom Water (ABW, 20–5 Mya18), as a result of most of the Antarctic continental shelf being covered by grounded ice sheets during glacial periods17–19. Nonetheless, the potential ecological or historical mechanisms affecting patterns of spatial and temporal differentiation in Antarctic deep-sea fauna remains largely unexplored.
Notably, the Antarctic deep-sea floor appears to be rich in mollusk gastropod species compared to other ocean plains23. Ample evidence has been proposed for the Antarctic origin of two lineages of mollusks, Cephalaspidea and Nudipleura (Gastropoda: Heterobranchia)24–27, and dispersal through the deep sea by the ABW19,22,28. Thus, a suitable model for studying the origin, diversification, and biogeography of deep-sea organisms and their evolutionary links with shallow water faunas at the SO are cephalaspidean gastropod mollusks, particularly Philinoidea. Recent work on the systematics of philinoid snails from around the world resulted in the division of Philinidae sensu lato into five families29,30, including the SO endemic Antarctophilinidae. This family is composed of two genera, the monospecific Waegelea with the species W. antarctica and Antarctophiline with six known species, namely A. alata, A. apertissima, A. easmithi, A. gibba, and A. falklandica from shelf waters and A. amundseni from deeper plains29. Although some Antarctophilinidae species are supposed to be circumpolar, encompassing depth ranges from shallow down to 500 m, others appear to have more restricted distributions29,31. Antarctophilinid species are endemic to the SO with only rare or dubious records of some species from the adjacent Polar Front boundaries. Both sympatric and allopatric speciation events were described as shaping the family’s diversity with evidence for cryptic or hidden speciation also potentially occurring in this system29. Moreover, species records encompass all the Antarctic plateau from shallow to abyssal waters. Thereby, this paper aims to shed light on the origin and diversification in Antarctica by using the radiation of Antarctophilinidae as a case study to ascertain the patterns of diversification across shallow- and deep-water species from the SO and adjacent areas. For that purpose, we used high-throughput genomic data and novel machine learning approaches for unraveling genetic speciation processes in extant species. Moreover, we reevaluate the only known related fossil to further dig into the origin of the family.
Results
Phylogenetic reconstruction
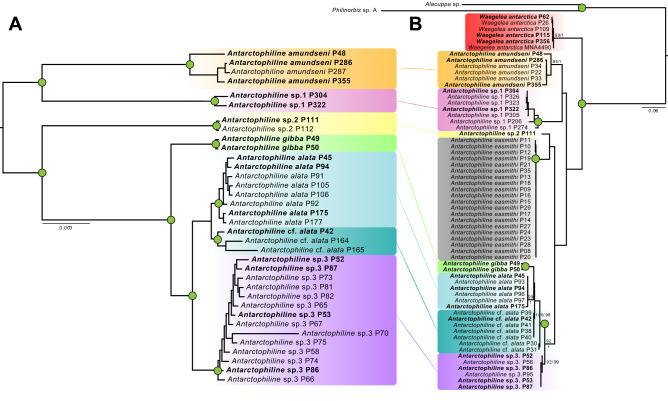
From a total of 142 extracted samples for molecular analyses, only 61 were successfully COI-barcoded and 40 were used in ddRADseq experiments (Table 1, Fig. 1A,B). The sequenced fragment of the COI gene included ca. 658 bp. All newly sequenced samples belong to Antarctophilinidae; the closely related cephalaspideans Alacuppa sp. and Philinorbis sp. were obtained from GenBank and used as outgroups. The maximum likelihood topology (Fig. 1B) showed the sister group to all Antarctophiline species was Waegelea antarctica (E. A. Smith, 1902), with samples ranging across the Drake Passage, Ross Sea, Scotia Arc, and Weddell Sea (65–500 m depth). Also, six clades corresponding to Antarctophiline species were identified, including A. easmithi (E Weddell Sea, 170–460 m depth) not present in the ddRADseq datasets, and depicted in grey in Fig. 1B, and a complex of species with affinity to A. alata.
Table 1.
Samples obtained from the SIO-BIC, MCZ, MNA, WAM, ZMBN (University Museum of Bergen), and ZSM, including voucher numbers, collecting site and date, geographical and bathymetric distribution, and COI barcode, when present.
| Species | Code | Voucher number | Barcode | Latitude | Longitude | Gear | Depth (m) | Location | Date | Cruise number | Station number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| W. antarctica (E. A. Smith, 1902) | P26 | ZMBN 121313 | MK015702 | 71° 7.3′ S | 11° 28.4′ W | TVG | 65 | E Weddell Sea | 18-Feb-98 | ANT XV/3 | 48/209 |
| W. antarctica | P62 | SIO-BIC M12655 | MN486297 | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 5-Oct-11 | NBP11-05 | SS2a/36 |
| W. antarctica | P109 | SIO-BIC M13658 | MN486298 | 61° 13′ 3.7″ S | 54° 15′ 17.1″ W | BLT | 202–223 | Elephant Island | 22-Oct-11 | NBP11-05 | EI1/81 |
| W. antarctica | P115 | SIO-BIC M17788 | MN486299 | 62° 52′ 20.7″ S | 57° 11′ 32.5″ W | BLT | 150–247 | Bransfield Strait | 24-Oct-11 | NBP11-05 | BS1/86 |
| W. antarctica | P117 | SIO-BIC M17789 | – | 62° 52′ 20.7″ S | 57° 11′ 32.5″ W | BLT | 150–247 | Bransfield Strait | 24-Oct-11 | NBP11-05 | BS1/86 |
| W. antarctica | P118 | SIO-BIC M17790 | – | 62° 52′ 20.7″ S | 57° 11′ 32.5″ W | BLT | 150–247 | Bransfield Strait | 24-Oct-11 | NBP11-05 | BS1/86 |
| W. antarctica | P356 | MNA11027 | MN486300 | 62° 55.99′ S | 58° 40.67′ W | AGT | 547 | Bransfield Strait | 3-May-13 | ANT XXIX/3 | 227-2 |
| W. antarctica | MNA 04490 | MN486301 | 74° 45′ 52.2″ S | 164° 4′ 55.3″ E | D | 100 | Adélie Cove, Ross Sea | 8-Jan-10 | PNRA XXV Exp 09/10 | DR4 | |
| A. amundseni Moles, Avila & Malaquias, 2019 | P22 | ZMBN 121347 | MK015698 | 73° 36.6′ S | 22° 24.7′ W | BT | 736 | E Weddell Sea | 5-Feb-98 | ANT XV/3 | 48/097 |
| A. amundseni | P33 | ZMBN 121314 | MK015708 | 71° 18.61′ S | 13° 56.12′ W | EBS | 910 | E Weddell Sea | 21-Dec-03 | ANT XXI/2 | PS65/232-1 |
| A. amundseni | P34 | ZMBN 121348 | MK015709 | 71° 18.61′ S | 13° 56.12′ W | EBS | 910 | E Weddell Sea | 21-Dec-03 | ANT XXI/2 | PS65/232-1 |
| A. amundseni | P48 | SIO-BIC M13655 | MN486278 | 55° 4′ 51.8″ S | 35° 10′ 21.4″ W | BLT | 196–253 | South Georgia | 29-Sep-11 | NBP11-05 | SG3a/23 |
| A. amundseni | P286 | ZSM 27239 | MN486279 | 71° 18′ 25.2″ S | 13° 58′ 13.2″ W | EBS | 1048 | E Weddell Sea | 20-Feb-05 | ANT XXII/3 | PS67/074-6-E |
| A. amundseni | P287 | ZSM 27239 | – | 71° 18′ 25.2″ S | 13° 58′ 13.2″ W | EBS | 1048 | E Weddell Sea | 20-Feb-05 | ANT XXII/3 | PS67/074-6-E |
| A. amundseni | P355 | MNA 11026 | MN486280 | 62° 55.99′ S | 58° 40.67′ W | AGT | 547 | Bransfield Strait | 3-May-13 | ANT XXIX/3 | 227-2 |
| Antarctophiline sp. 1 | P206 | ZSM 20854 | MN486283 | 62° 57′ 48″ S | 27° 52′ 8.4″ W | AGT | 4548 | Weddell Sea—S South Sandwich Islands | 16-Mar-02 | ANT XIX/4 | PS61/138-4 |
| Antarctophiline sp. 1 | P274 | ZSM 21093 | MN486284 | 60° 39′ 11.4″ S | 53° 56′ 51″ W | EBS | 2893 | N Elephant Island | 30-Jan-02 | ANT XIX | PS61/046-7 |
| Antarctophiline sp. 1 | P304 | ZSM 34346 | MN486285 | 52° 2′ 31.8″ S | 0° 0′ 36″ E | AGT | 2996 | NW Bouvet Island, S Atlantic Ocean | 6-Dec-07 | ANT XXIV/2 | PS71/013-15 |
| Antarctophiline sp. 1 | P305 | ZSM 34346 | MN486286 | 52° 2′ 31.8″ S | 0° 0′ 36″ E | AGT | 2996 | NW Bouvet Island, S Atlantic Ocean | 6-Dec-07 | ANT XXIV/2 | PS71/013-15 |
| Antarctophiline sp. 1 | P322 | MN486287 | 52° 0.36′ S | 10° 1.47′ E | EBS | 3705–3757 | NW Bouvet Island, S Atlantic Ocean | 20-Jan-12 | ANT XXVIII/3 | PS79/081-18 | |
| Antarctophiline sp. 1 | P323 | MN486288 | 52° 0.18′ S | 10° 0.72′ E | EBS | 3743–3763 | NE Bouvet Island, S Atlantic Ocean | 20-Jan-12 | ANT XXVIII/3 | PS79/081-17 | |
| Antarctophiline sp. 1 | P326 | MN486289 | 52° 0.18′ S | 10° 0.72′ E | EBS | 3743–3763 | NE Bouvet Island, S Atlantic Ocean | 20-Jan-12 | ANT XXVIII/3 | PS79/081-17 | |
| Antarctophiline sp. 2 | P111 | SIO-BIC M17786 | MN486290 | 62° 52′ 20.7″ S | 57° 11′ 32.5″ W | BLT | 150–247 | Bransfield Strait | 24-Oct-11 | NBP11-05 | BS1/86 |
| Antarctophiline sp. 2 | P112 | SIO-BIC M17787 | – | 62° 52′ 20.7″ S | 57° 11′ 32.5″ W | BLT | 150–247 | Bransfield Strait | 24-Oct-11 | NBP11-05 | BS1/86 |
| A. easmithi Moles, Avila & Malaquias, 2019 | P08 | ZMBN 121327 | MK015684 | 71° 18.6′ S | 12° 18.1′ W | AGT | 173 | E Weddell Sea | 25-Jan-98 | ANT XV/3 | PS48/006 |
| A. easmithi | P09 | ZMBN 121328 | MK015685 | 71° 18.6′ S | 12° 18.1′ W | AGT | 173 | E Weddell Sea | 25-Jan-98 | ANT XV/3 | PS48/006 |
| A. easmithi | P10 | ZMBN 121329 | MK015686 | 71° 19.3′ S | 12° 24.7′ W | TVG | 182 | E Weddell Sea | 28-Jan-98 | ANT XV/3 | PS48/027 |
| A. easmithi | P11 | ZMBN 121330 | MK015687 | 70° 52.7′ S | 10° 34.8′ W | AGT | 230 | E Weddell Sea | 30-Jan-98 | ANT XV/3 | PS48/044 |
| A. easmithi | P12 | ZMBN 121331 | MK015688 | 70° 52.7′ S | 10° 34.8′ W | AGT | 230 | E Weddell Sea | 30-Jan-98 | ANT XV/3 | PS48/044 |
| A. easmithi | P13 | ZMBN 121332 | MK015689 | 70° 52.7′ S | 10° 34.8′ W | AGT | 230 | E Weddell Sea | 30-Jan-98 | ANT XV/3 | PS48/044 |
| A. easmithi | P14 | ZMBN 121333 | MK015690 | 70° 52.7′ S | 10° 34.8′ W | AGT | 230 | E Weddell Sea | 30-Jan-98 | ANT XV/3 | PS48/044 |
| A. easmithi | P15 | ZMBN 121334 | MK015691 | 70° 54′ S | 10° 28.2′ W | AGT | 232 | E Weddell Sea | 31-Jan-98 | ANT XV/3 | PS48/062 |
| A. easmithi | P16 | ZMBN 121335 | MK015692 | 70° 54′ S | 10° 28.2′ W | AGT | 232 | E Weddell Sea | 31-Jan-98 | ANT XV/3 | PS48/062 |
| A. easmithi | P17 | ZMBN 121336 | MK015693 | 70° 54′ S | 10° 28.2′ W | AGT | 232 | E Weddell Sea | 31-Jan-98 | ANT XV/3 | PS48/062 |
| A. easmithi | P18 | ZMBN 121337 | MK015694 | 72° 51.7′ S | 19° 7.9′ W | BT | 439 | E Weddell Sea | 3-Feb-98 | ANT XV/3 | PS48/078 |
| A. easmithi | P19 | ZMBN 121338 | MK015695 | 72° 50.5′ S | 19° 28′ W | BT | 463 | E Weddell Sea | 3-Feb-98 | ANT XV/3 | PS48/082 |
| A. easmithi | P20 | ZMBN 121339 | MK015696 | 72° 50.5′ S | 19° 28′ W | BT | 463 | E Weddell Sea | 3-Feb-98 | ANT XV/3 | PS48/082 |
| A. easmithi | P21 | ZMBN 121340 | MK015697 | 72° 50.5′ S | 19° 28′ W | BT | 463 | E Weddell Sea | 3-Feb-98 | ANT XV/3 | PS48/082 |
| A. easmithi | P23 | ZMBN 121341 | MK015699 | 73° 39.1′ S | 20° 59.6′ W | D | 211 | E Weddell Sea | 8-Feb-98 | ANT XV/3 | PS48/128 |
| A. easmithi | P24 | ZMBN 121342 | MK015700 | 73° 39.1′ S | 20° 59.6′ W | D | 211 | E Weddell Sea | 8-Feb-98 | ANT XV/3 | PS48/128 |
| A. easmithi | P27 | ZMBN 121343 | MK015703 | 70° 50.5′ S | 10° 41.8′ W | BT | 307 | E Weddell Sea | 19-Feb-98 | ANT XV/3 | PS48/222 |
| A. easmithi | P28 | ZMBN 121344 | MK015704 | 71° 18′ S | 12° 15′ W | AGT | 184 | E Weddell Sea | 27-Feb-98 | ANT XV/3 | PS48/277 |
| A. easmithi | P29 | ZMBN 121345 | MK015705 | 71° 18′ S | 12° 15′ W | AGT | 184 | E Weddell Sea | 27-Feb-98 | ANT XV/3 | PS48/277 |
| A. easmithi | P35 | ZMBN 121346 | MW509525 | 71° 04.30′ S | 11° 33.92′ W | BT | 309 | E Weddell Sea | 23-Dec-03 | ANT XXI/2 | PS65/253-1 |
| A. gibba (Strebel, 1908) | P49 | SIO-BIC M12896 | MN486281 | 55° 2′ S | 35° 26′ W | BLT | 125 | South Georgia | 29-Sep-11 | NBP11-05 | SG3/22 |
| A. gibba | P50 | SIO-BIC M12896 | MN486282 | 55° 2′ S | 35° 26′ W | BLT | 125 | South Georgia | 29-Sep-11 | NBP11-05 | SG3/22 |
| A. alata (Thiele, 1912) | P45 | WAMS101214 | MN486272 | 59° 28′ 11.3″ S | 27° 16′ 44.8″ W | AGT | 230 | Southern Thule, South Sandwich Islands | 8-Mar-17 | ACE2016-17 | 90/2590 |
| A. alata | P91 | SIO-BIC M17793 | – | 59° 23′ 40.8″ S | 27° 18′ 41.7″ W | BLT | 103–221 | S South Sandwich Islands | 7-Oct-11 | NBP11-05 | SS3/42 |
| A. alata | P92 | SIO-BIC M17794 | – | 59° 23′ 40.8″ S | 27° 18′ 41.7″ W | BLT | 103–221 | S South Sandwich Islands | 7-Oct-11 | NBP11-05 | SS3/42 |
| A. alata | P93 | SIO-BIC M17795 | MN486273 | 59° 23′ 40.8″ S | 27° 18′ 41.7″ W | BLT | 103–221 | S South Sandwich Islands | 7-Oct-11 | NBP11-05 | SS3/42 |
| A. alata | P94 | SIO-BIC M13654 | MN486274 | 59° 23′ 40.8″ S | 27° 18′ 41.7″ W | BLT | 103–221 | S South Sandwich Islands | 7-Oct-11 | NBP11-05 | SS3/42 |
| A. alata | P96 | SIO-BIC M17796 | MN486275 | 59° 23′ 11.4″ S | 27° 18′ 49.2″ W | BLT | 403–501 | S South Sandwich Islands | 8-Oct-11 | NBP11-05 | SS3/44 |
| A. alata | P97 | SIO-BIC M17797 | MN486276 | 59° 23′ 11.4″ S | 27° 18′ 49.2″ W | BLT | 403–501 | S South Sandwich Islands | 8-Oct-11 | NBP11-05 | SS3/44 |
| A. alata | P105 | SIO-BIC M17798 | – | 59° 23′ 11.4″ S | 27° 18′ 49.2″ W | BLT | 403–501 | S South Sandwich Islands | 8-Oct-11 | NBP11-05 | SS3/44 |
| A. alata | P106 | SIO-BIC M17799 | – | 59° 23′ 11.4″ S | 27° 18′ 49.2″ W | BLT | 403–501 | S South Sandwich Islands | 8-Oct-11 | NBP11-05 | SS3/44 |
| A. alata | P175 | ZSM 15955 | MN486277 | 70° 50′ 12″ S | 10° 35′ 24″ W | BT | 271 | E Weddell Sea | 4-Oct-00 | ANT XVII/3 | 136-1 |
| A. alata | P177 | ZSM 15955 | – | 70° 50′ 12″ S | 10° 35′ 24″ W | BT | 271 | E Weddell Sea | 4-Oct-00 | ANT XVII/3 | 136-1 |
| A. cf. alata | P30 | ZMBN 121350 | MK015706 | 54° 30.01′ S | 3° 13.97′ E | AGT | 260 | Bouvet Island | 24-Nov-03 | ANT XXI/2 | PS65/019-1 |
| A. cf. alata | P31 | ZMBN 121351 | MK015707 | 54° 22.49′ S | 3° 17.58′ E | AGT | 134 | Bouvet Island | 24-Nov-03 | ANT XXI/2 | PS65/028-1 |
| A. cf. alata | P38 | ZMBN 121323 | MK015710 | 62° 58.18S | 60° 42.23 W | SD | 10 | Fumarole Bay, Deception Island | 25-Jan-13 | ACTIQUIM-4 | A4-453 |
| A. cf. alata | P39 | ZMBN 121324 | MK015711 | 62° 58.18S | 60° 42.23 W | SD | 10 | Fumarole Bay, Deception Island | 25-Jan-13 | ACTIQUIM-4 | A4-453 |
| A. cf. alata | P40 | ZMBN 121315 | MK015712 | 62° 58.18S | 60° 42.23 W | SD | 10 | Fumarole Bay, Deception Island | 25-Jan-13 | ACTIQUIM-4 | A4-453 |
| A. cf. alata | P41 | ZMBN 121325 | MK015713 | 62° 58.18S | 60° 42.23 W | SD | 10 | Fumarole Bay, Deception Island | 25-Jan-13 | ACTIQUIM-4 | A4-453 |
| A. cf. alata | P42 | ZMBN 121326 | MK015714 | 62° 58.18S | 60° 42.23 W | SD | 10 | Fumarole Bay, Deception Island | 25-Jan-13 | ACTIQUIM-4 | A4-453 |
| A. cf. alata | P164 | MCZ 393955 | – | 62° 40′ S | 60° 38′ W | RD | 216 | S Livingston Island, South Shetlands Islands | 19-Feb-94 | BENTART-95 | 100R |
| A. cf. alata | P165 | MCZ 393955 | – | 62° 40′ S | 60° 38′ W | RD | 216 | S Livingston Island, South Shetlands Islands | 19-Feb-94 | BENTART-95 | 100R |
| Antarctophiline sp. 3 | P52 | SIO-BIC M13656 | MN486291 | 56° 42′ 50.6″ S | 27° 1′ 35.8″ W | BLT | 134–142 | N South Sandwich Islands | 3-Sep-11 | NBP11-05 | SS1A/30 |
| Antarctophiline sp. 3 | P53 | SIO-BIC M12975 | MN486292 | 58° 28.1′ S | 26° 13.1′ W | BLT | 164–172 | N South Sandwich Islands | 5-Oct-11 | NBP11-05 | SS2/34 |
| Antarctophiline sp. 3 | P56 | SIO-BIC M17800 | MN486293 | 58° 22′ S | 26° 16′ W | BLT | 153–420 | N South Sandwich Islands | 5-Oct-11 | NBP11-05 | SS2a/36 |
| Antarctophiline sp. 3 | P58 | SIO-BIC M17801 | – | 58° 22′ S | 26° 16′ W | BLT | 153–420 | N South Sandwich Islands | 5-Oct-11 | NBP11-05 | SS2a/36 |
| Antarctophiline sp. 3 | P65 | SIO-BIC M17802 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P66 | SIO-BIC M17803 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P67 | SIO-BIC M17804 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P70 | SIO-BIC M17805 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P73 | SIO-BIC M17806 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P74 | SIO-BIC M17807 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P75 | SIO-BIC M17808 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P81 | SIO-BIC M17809 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P82 | SIO-BIC M17810 | – | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P86 | SIO-BIC M17811 | MN486294 | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P87 | SIO-BIC M17812 | MN486295 | 58° 22.71′ S | 26° 17′ W | BLT | 134–260 | N South Sandwich Islands | 6-Oct-11 | NBP11-05 | SS2a/38 |
| Antarctophiline sp. 3 | P95 | SIO-BIC M13093 | MN486296 | 59° 23.19′ S | 27° 18.82′ W | BLT | 403–501 | N South Sandwich Islands | 8-Oct-11 | NBP11-05 | SS3/44 |
Specimens in bold were included in the ddRADseq phylogenetic analysis. Gear types: AGT Agassiz trawl, BLT Blake trawl, BT bottom trawl, D rock dredge, ES epibenthic sled, RD Rauschert dredge, SD SCUBA diving, TVG TV grab.
Figure 1.
Phylogenetic relationships of antarctophilinids based on maximum likelihood (ML), identical topology was recovered through Bayesian inference (BI), colored boxes illustrating species hypotheses. (A) Tree based on ddRADseq data of Matrix 2 (depicted in Figure S1). (B) Tree based on COI sequences showing similar clades, but also including the sister group Waegelea antarctica in red and Antarctophiline easmithi in grey. Green dots denote full support for both bootstrap (ML) and posterior probability values (BI). Samples in bold are present in both trees.
The initial ddRADseq Matrix 1 including both antarctophilinid genera (i.e. Antarctophiline and Waegelea) included 40 individuals, 3893 loci, and 38.5% missing data (not depicted; see “Methods” section). The outgroup included five individuals of W. antarctica, a species with a circumpolar distribution. Matrix 2 was built to increase the number of loci and resolution of the tree, including only the 35 individuals in the genus Antarctophiline. The final Matrix 2 dataset contained 5411 loci and 41.6% missing data (see summary statistics in Supplementary Table S1 and plotted matrix visualization in Fig. S1). A particularly long branch was found in sample P70 (Fig. 1A), most likely due to cross-contamination and/or high levels of missing data (see the section below). The subset to specifically address the A. gibba/A. alata species complex includes 27 individuals (Matrix 3), 5411 loci, and 41.6% missing data. Although several clustering thresholds were tested, only a 75% threshold of similarity produced an output to construct a matrix. This is due to the high genetic divergences among species, and thus, the high amount of singleton reads that makes the clustering within and across samples computationally too demanding. The ML tree of Matrix 2 was rooted with A. amundseni Moles, Avila & Malaquias, 2019 plus the abyssal Antarctophiline sp. 1 (Fig. 1A) as the sister group to the rest of Antarctophiline species. This was also found in the tree of Matrix 1 (Fig. S2), for which an identical topology and maximum bootstrap support (BS = 100) and posterior probability (PP = 1) values were recovered for most nodes, but sample P70 due to potential contamination.
Distinctiveness in genetic lineages
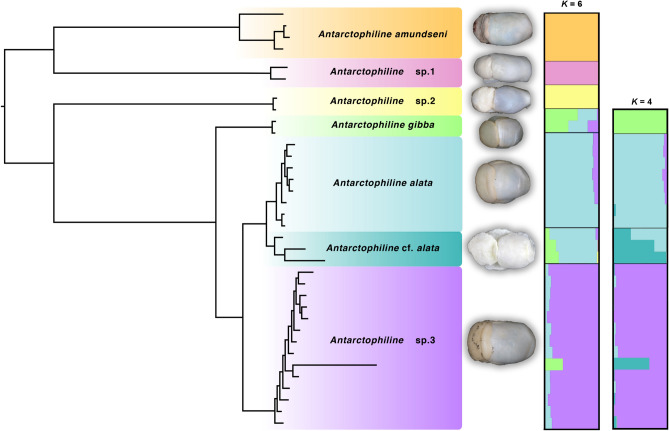
For Matrix 2, STRUCTURE (Evanno method32) favored an optimal K = 6 (Fig. 2), recovering all six a priori barcoded Antarctophiline species as distinct clusters, including the eurybathic A. amundseni (South Georgia, 200 m depth; Bransfield Strait, 550 m depth; E Weddell Sea, 740–1050 m depth), the abyssal Antarctophiline sp. 1 (Scotia Sea, Weddell Sea, NW Bouvet Island, 2900–4500 m depth), a shallow-water Antarctophiline sp. 2 (Bransfield Strait, 200 m depth), and distinct clades of the A. gibba/A. alata species complex not recovered in the barcode phylogeny (including Antarctophiline sp. 3). For Matrix 3, STRUCTURE retrieved an optimal K = 4 (Fig. 2), clearly splitting A. gibba (South Georgia, 125 m depth) from three potentially cryptic shallow-water species with affinity to A. alata tentatively named: A. alata (S of the South Sandwich Islands plus E Weddell Sea, 100–500 m depth), A. cf. alata (South Shetland Islands, 10–200 m depth), and Antarctophiline sp. 3 (N of the South Sandwich Islands, 130–500 m depth).
Figure 2.
Phylogram of antarctophilinids based on Matrix 2 (left) and STRUCTURE plots (right), showing the posterior probability for individual assignments of samples to different genetic clusters. Both plots show the result for the most likely number of genetic clusters for Matrix 2 (K = 6) and Matrix 3 (K = 4). Dorsal pics of preserved type specimens for each cluster are also depicted.
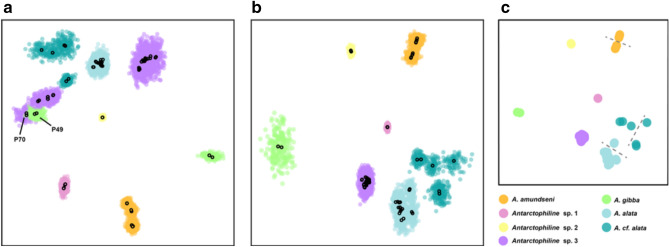
Clustering with the VAE output of the full dataset resulted in 8–10 clusters (Fig. 3A). However, examining the VAE plot, it is apparent that samples P70 and P49 were misplaced and likely confounded an accurate representation of the data, for example, showing two widely divergent Antarctophiline sp. 3 clusters. As previously mentioned, P70 likely contains contamination and was removed from subsequent analyses. Additionally, the placement of P49 with Antarctophiline sp. 3 (specifically with P70) instead of the other A. gibba (P50) is perhaps driven by a combination of admixture (Fig. 1A) and high levels of missing data (up to 80%, see Fig. S1) in this sample, and was also removed. VAE output of the dataset with these two samples removed (Fig. 3B) recovers a single cluster for all Antarctophiline sp. 3 samples, more in line with the results from COI-barcoding and ddRADseq analyses. “Partition around medoids” (PAM) and hierarchical clustering on this VAE output recover seven clusters (Fig. 3C), while the gap statistic favors 10 clusters, splitting A. alata, A. cf. alata, and A. amundseni into two clusters each. The gap statistic is thus probably over-splitting these taxa as overlapping VAE standard deviations indicate one cluster for A. amundseni and at most two clusters for A. cf. alata (Fig. 3B). Given the concordance between genetic clustering across multiple approaches, we favor seven species, as shown in the VAE clustering (Fig. 3B) results that match those in the phylogeny (Fig. 1) and the sum of the STRUCTURE analyses (Fig. 2).
Figure 3.
Variational autoencoder (VAE) showing two haplotypes (circles) per sample and clustering results. (A) VAE output on the full dataset with a mean (black outlined circle) and standard deviation (colored circles) for each sample. (B) VAE output on the dataset with P49 and P70 removed. (C) Results of PAM and hierarchical clustering analyses on the VAE output of the dataset with P49 and P70 removed, favoring seven clusters. Dashed lines indicate further split clusters recovered with the gap statistic.
Fossil systematic reassessment
Class Gastropoda Cuvier, 1795
Order Cephalaspidea Fischer, 1883
Superfamily Philinoidea Gray, 1850 (1815)
Family Antarctophilinidae Moles, Avila & Malaquias, 2019
Material examined (Fig. 4). King George Island, Melville Peninsula (Crab Creek locality, I), Cape Melville Formation, Lower Miocene: 1 specimen, ZPAL Ga. IV/26, length = 11 mm, width = 5 mm.
Figure 4.

Images of the single fossil of Philinoidea found in Antarctica (King George Island) dated from the Lower Miocene and originally attributed to the Japanese Scaphander yonabaruensis Mac Neil, 1960 (Karczewski, 1987), but here redesignated to Antarctophilinidae. (a) Ventral view. (b) Dorsal view. (c) Lateral view. (d) Apical view.
Diagnosis Shell ovate-subquadrate, slightly flattened dorsoventrally; aperture wide; apex obtuse, slightly sunken; outer lip convex; posterior edge of outer lip obtuse, not protruding beyond apex; columellar wall concave; growth lines visible.
Remarks Although originally attributed to Scaphander yonabaruensis Mac Neil, 1960 known from the Miocene of Japan, in Okinawa33, Beu34 suggested its similarity to the genus Philine. Indeed, we believe the overall shell morphology matches the recently erected family Antarctophilinidae which epitomizes most Philinoidea diversity known from Antarctic waters.
Discussion
This study, grounded on a large collecting effort in remote and abyssal areas in the SO aided by phylogenetic analyses using ddRADseq-derived SNP data, increases our understanding of Antarctic benthic gastropod species distributions and diversity. Our barcode and STRUCTURE analyses served as a starting point for discerning among genetic lineages35. Since incongruences in the number of distinct genetic groups recovered in both Matrix 2 and 3 were found using STRUCTURE, we used novel unsupervised machine learning methods36. VAEs proved to be powerful and resolutive when using genomic data for recovering congruent genetic lineages across methods, some of which correspond to species hypothesis. Our results corroborated the latest systematic assessment by Moles et al.29 on species diversity and support the likelihood of further cryptic diversity in Antarctophilinidae. Out of the eight delimited species three are considered undescribed, the abyssal Antarctophiline sp. 1, the shallow-water species Antarctophiline sp. 2 from Bransfield Strait, and Antarctophiline sp. 3 from the South Sandwich Islands. Regarding A. cf. alata and, to a certain extent, Antarctophiline sp. 3, these are considered to be at the early stages of speciation (potential cryptic species). Taxonomic descriptions of the undescribed species, as well as the validity of certain synonymized taxa within the A. alata species complex (Philine gouldi Doello-Jurado, 1918, and P. amoena Thiele, 1925), will follow in a separate manuscript. Ecological restrictions to bottom-dwelling habitats may be a driver for the morphological convergence in species of Philinoidea29,30,37. Here, genomic data have enhanced the phylogenetic resolution of Antarctophilinidae species obtained through Sanger analysis, particularly for less diverged species, an approach that has been previously applied only to a handful of Antarctic samples (e.g., Ref. 38).
A total of seven species of Antarctophiline are found in Antarctic shallow waters, the five species analyzed here: A. alata, A. easmithi, A. gibba, Antarctophiline sp. 2, and Antarctophiline sp. 3 from the vicinities of the Scotia Arc and western Antarctic Peninsula plus the species from the Ross Sea A. apertissima (E. A. Smith, 1902) and A. falklandica (Powell, 1951) (also from the Falkland Islands39,40). Contrary, the deep-sea fauna is less diverse, with only two species known to date. Independent colonization of the continental shelf from the slope during interglacial cycles, < 23 Mya14,41, and the presence of habitat refugia in the Antarctic Peninsula tip and adjacent islands42,43 may explain the high species endemism and richness found in this study—but this remains to be tested. This phenomenon has been referred to as the Antarctic Biodiversity Pump21,44 and sustains habitat fragmentation during glacial maxima as the driving force towards allopatric speciation. Secondly, the present island patchiness across the Scotia Arc (from South Georgia to the South Shetland Islands) may have allowed for rapid radiation and speciation processes due to the availability of different ecological niches38,45,46. This could explain the relatively restricted distributions found for A. gibba, Antarctophiline sp. 2, and Antarctophiline sp. 3. Additionally, high species richness is expected due to the elevated productivity of these shallow waters during warm seasons41, a pivotal influence controlling Antarctic benthic diversity47. In fact, A. gibba is endemic to South Georgia, an island considered a hotspot for gastropod diversity, with more than 50 endemic species recorded48. Nonetheless, the high degree of single species found at each collecting site underlines the difficulty of gathering data on Antarctic ecosystems, and thus, conclusions should be made with caution, especially concerning deep-sea samples49. Overall, our study provides evidence for high diversity in a group of species previously considered to be rather low, which may be the result of fluctuating paleoclimatic history and current habitat heterogeneity.
Antarctica has long been considered a center of radiation of marine benthic taxa19–21, and heterobranch gastropods in particular24,25,27. Here, the single fossil record from the Oligocene–Early Miocene at King George Island is attributed to Antarctophilinidae (Fig. 4), proposing that these snails were present in shallow waters of the South Shetland Islands at least 20 Mya. Although limited water transport is hypothesized during the Eocene 50–34 Mya, and probably until the mid-Miocene 15 Mya28,50, Trans-Antarctic migrations through a Ross-Weddell seaway through the Amundsen Sea could partially explain the disjunct distributions of several shallow-water gastropods41,51. In our study, molecular data for W. antarctica supports this disjunct distribution and, the specimens found in Peter I Island—an intermediate locality in the Bellingshausen Sea—further reinforce this hypothesis (reexamined material from Aldea & Troncoso52). This has been suggested for other marine benthic taxa50,53, for which a circumpolar distribution seems unlikely, but instead, a disjunct distribution has been documented. Our compiling evidence suggests that the Scotia Arc and the Weddell Sea have played a pivotal role in the evolution and radiation of many molluscan species from the Miocene forward. However, until comprehensive sampling has been carried out—something difficult to accomplish in Antarctica—our understanding of species distributions remains somewhat limited.
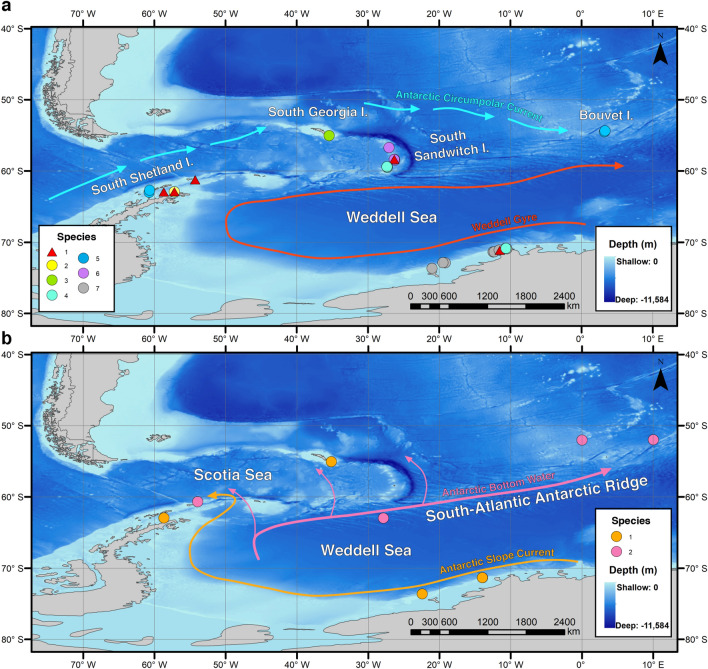
The onset of the ACC led to the isolation of the Antarctic continent and subsequent cool down with the likely extinction of shallow-water faunas17. During interglacial periods of shelf ice retreat, the unpopulated shelf could have been re-colonized by fauna from the slope54 or shelters on the continental shelf14,42,43. Species dispersal and gene flow at subtidal and shelf depths have been increasingly studied in SO areas with enough evidence of contrasting patterns related to the disparity in species life histories55,56, usually challenging the concept of well-connected, circumpolar distributions57–59 but see Moore et al.60. Extensive geographical distributions were found in species such as A. alata (including A. cf. alata) and W. antarctica, which occur at 10–500 m depth over the Eastern Weddell Sea, the South Shetland Islands, and the Southern South Sandwich Islands, even extending towards the more remote Bouvet Island. The Weddell Gyre is a clockwise current known for connecting shallow shelf waters along the Weddell Sea coast to the South Shetland Islands (through the tip of the Antarctic Peninsula), and towards the South Sandwich Islands, which might ultimately reach Bouvet Island61,62. Dispersal through this hydrographic jet could explain the current distribution among the studied species (see Fig. 5a). The Scotia Arc faunal gateway, helped by oceanic eddies, might ultimately be responsible for the distribution found across the Sub-Antarctic islands of South Sandwich and South Georgia63,64. Unfortunately, scarce is information on the life history of Antarctophilinidae. The studied species A. gibba lays egg masses at the superficial waters of South Georgia, these containing thousands of large eggs lacking a planktonic phase65. Just recently, the larval development of W. antarctica was studied in detail showing early juvenile stages hatching with a relict larval velum, being able to drift away in water currents66. We hypothesize this is a drifting mode that may explain our current knowledge of distribution in this family (see Ref. 67).
Figure 5.
Bathymetric maps showing the distribution of the antarctophilinid specimens from the Atlantic sector of the Southern Ocean including the Scotia Sea, Eastern Weddell Sea, and South Atlantic. (a) Shallow water species: (1) Waegelea antarctica; (2) Antarctophiline sp. 2; (3) A. gibba; (4) A. alata; (5) A. cf. alata; (6) Antarctophiline sp. 3; (7) A. easmithi. (b) Deep-sea species: (1) A. amundseni; (2) Antarctophiline sp. 1.
Cenozoic glacial-interglacial cycles may have also represented the environmental force that shaped the evolutionary trend toward eurybathy in many Antarctic benthic invertebrates14,54,68. During periods of extension of the continental ice sheet, an Antarctophiline lineage from the shelf may have been forced to go into deep slope refugia. In this sense, the sister group to the shallow water Antarctophiline clade is a deep-sea group composed of the recently described A. amundseni and an undescribed abyssal species, both displaying a bathymetrically separated distribution but with widespread geographical distributions. At the upper bathyal zone, which includes the Antarctic slope, we found populations of A. amundseni across the Eastern Weddell Sea and the Bransfield Strait. The Antarctic Slope Current circulating in a counterclockwise direction could have been responsible for such distributions69. Far below, very distant populations of Antarctophiline sp. 1 at 2900–4500 m depth were recorded west of the Antarctic Peninsula, over the Scotia Ridge, and towards the northern and eastern regions of Bouvet Island. The distribution expands through the South-Atlantic Antarctic Ridge (Fig. 5b), strikingly covering a linear distance of more than 3900 km. The eastern jet of the ABW (i.e., Weddell Sea Bottom Water) feeds the Atlantic abyssal waters all over the North Atlantic as part of the Global Thermohaline Circulation70–72 and indeed reflects the distribution patterns reported here. An expected longer life cycle in the deep sea73 may have driven these species into alternative modes of reproduction and dispersal, but the absence of detailed ecological information for Antarctic Philinoidea precludes any conclusions. Alternatively, geological events may explain the current distribution of disjunct species of sea snails across ocean basins74.
During the past two decades, an increasing number of molecular studies on different taxa have challenged the three central paradigms of Antarctic benthic lineages55, i.e., isolation64,75, circumpolarity15,76, and eurybathic distributions16,77. Our evidence, based on genomic data and novel machine learning approaches, also challenges these long-standing concepts of Antarctic benthic species. Habitat segregation either through shelf refugia or current ecosystem heterogeneity at Antarctic shelf depths may have favored species flocks59. Contrarily to the widespread longitudinal distribution of some species78, bathymetrically separated distributions are a common phenomenon found in Antarctophilinidae29, with higher species diversity and endemism found at shelf depths. Nonetheless, we have to bear in mind the potential biases of sampling efforts across depths, with the lower part of the slope and abyss, seldom explored compared to shallower depths49. Shallow-water and slope species are thought to have colonized abyssal depths during the late Mesozoic and Cenozoic epochs19. The resulting sinking of cold, saline water adjacent to the Antarctic continent and its subsequent movement northwards at abyssal depths has resulted in colonization from the Antarctic for many invertebrate families and genera. Deep-sea communities seem to harbor less species-level diversity, probably because of more homogeneity in their habitats, compared to the shallow water environments5. Strikingly, Antarctica has acted as a center of origin and radiation of certain benthic taxa19,20, including Antarctophilinidae mollusks.
Methods
Taxon sampling
Antarctic cruises by researchers from multiple institutions were conducted, including the Italian National Antarctic Museum (MNA, Section of Genoa, Italy), the Western Australian Museum (WAM, Perth, Australia), the Benthic Invertebrate Collection at Scripps Institution of Oceanography (SIO-BIC, La Jolla, CA, USA), and the Bavarian State Collection of Zoology (ZSM, Munich, Germany). Sampling stations covered a wide geographical and bathymetric range (10–4550 m) during several cruises (German, Spanish, and US Antarctic Programs) from 1994 to 2017 carried out in the Atlantic sector of the Southern Ocean (Table 1). Distribution maps for the 142 specimens collected, color-coded by species for both continental shelf (Fig. 5a) and slope (Fig. 5b) plains were designed in Arc-GIS 10.3 (Esri, Redlands, CA). Samples were mostly collected by dredging and trawling, but some were collected manually during SCUBA. When possible, specimens were photographed alive on board and preserved in either 70% or 95% EtOH for molecular purposes. Once back in the laboratory, all specimens were photographed dorsally and ventrally using a Keyence VHX-6000 Digital Microscope system at the Museum of Comparative Zoology (MCZ) before dissection.
Additionally, material deposited at the Institute of Paleobiology, Polish Academy of Sciences, of the single philinoid fossil, precisely from Antarctica and originally attributed to Scaphander (Scaphandridae)33, was morphologically reassessed.
DNA extraction, library preparation, and sequencing
DNA was extracted from a fragment of the left parapodial lobe using the AutoGenprep 965 Tissue Protocol (AutoGen Inc., Holliston, MA). Initial ‘DNA barcoding’ of all samples was carried out by sequencing a fragment of the mitochondrial cytochrome c oxidase subunit I (COI) using primer pair LCO1490 and HCO219879. PCR amplifications were completed in 25-µL reactions with Illustra PuReTaq Ready-To-Go PCR Beads (GE Healthcare, Chicago, IL) with initial denaturation for 5 min at 94 °C, 35 cycles (15 s at 94 °C, 5 s at 48 °C, 15 s at 68 °C), and a final extension step for 7 min at 72 °C. Amplifications were cleaned with incubation of 1 µL ExoSAP-IT (Affymetrix, Santa Clara, CA) and sequencing reactions were performed in 10-µL reactions using BigDye ver. 1 chain-termination chemistry on an ABI3730xl (Applied Biosystems Inc., Foster City, CA). Sequences were edited and aligned with MUSCLE80, as implemented in Geneious v. 11.0.381. All sequences were submitted to GenBank (see Table 1 for accession numbers).
Successful DNA extractions were then quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA), and 100–700 ng of genomic DNA for each sample was used for double-digest restriction site-associated DNA (ddRAD). Libraries were prepared following Peterson et al.82 protocol with some modifications, using the enzymes EcoRI-HF and BfaI (New England Biolabs, Ipswich, MA) for digestion. Ca. 50–200 ng of fragmented DNA from each individual was later ligated using the customized P1 and P2 adapters with internal barcodes. Between 15–25 individual samples were then pooled together and size-selected to a range of 350–550 bp using a Blue Pippin (Sage Science Inc., Beverly, MA). Each size-selected pool was amplified through PCR and an Illumina P5 barcode was added using a Phusion High-Fidelity PCR Kit (New England Biolabs). PCRs were conducted with an initial denaturation for 30 s at 98 °C, 10–15 cycles (10 s at 98 °C, 30 s at 65 °C, 30 s at 72 °C), and a final extension for 10 min at 72 °C. Amplified libraries were checked and quantified with a TapeStation 2200 (Agilent Technologies, Carpinteria, CA). Amplified libraries were then cleaned using a ratio of 1:1.5 Agilent beads and quantified using the qPCR Kapa Quantification Kit (Kapa Biosystems, Wilmington, MA). Libraries were multiplexed and paired-end sequenced (150 bp) on an Illumina HiSeq 2500 (Illumina Inc., San Diego, CA) at the Bauer Core Facility, Harvard University (Cambridge, MA).
Matrix construction
Barcode demultiplexing, quality control, within-sample clustering, and between-pair variant calling were carried out using ipyrad v. 0.7.2883,84. Settings and data processing steps are default commonly used with ddRAD data. Only reads with unambiguous barcodes and Phred Q scores ≥ 33 were retained, and loci with more than five undetermined bases were additionally discarded. Maxdepth was set to 10,000 to avoid uneven sequencing coverage of amplified fragments and highly similar genomic repetitive regions. Thereafter, all individuals from the different libraries were analyzed together, first clustering per locus with a clustering threshold for de novo assembly of 75% (i.e., level of sequence similarity at which two sequences are identified as being homologous, and thus cluster together); additionally, clustering thresholds at 80, 85, and 90% were attempted. Adapters were trimmed and reads < 35 bp discarded. To avoid over-inflation of estimated heterozygosity, we required a minimum of six reads for each cluster during consensus base-calling and up to four shared polymorphic sites per called locus. For finding the consensus loci and clustering across samples we used a minimum number of samples per locus of 50% of total species and a maximum number of SNPs per locus of 20%, only allowing for a 0.05% of ambiguous positions per consensus locus, setting the maximum number of indels to eight (thus filtering out poor final alignments), and a 50% of polymorphic sites per locus. Full datasets were used for the phylogenetic analyses while unlinked SNPs (one randomly selected SNP per locus) were used for the STRUCTURE analysis and variational autoencoder (VAE) plots. Matrix condenser85 (available at https://bmedeiros.shinyapps.io/matrix_condenser) was used to visualize the matrix, discard samples with very low coverage (i.e., less than 20% of loci), and construct the final dataset for the downstream analyses. Three different matrices were then constructed for further analyses: Matrix 1, including both the genera Waegelea Moles, Avila & Malaquias, 2019 and Antarctophiline Chaban, 2016; Matrix 2, including all the species of the genus Antarctophiline and with an increased number of shared loci (Fig. S1); and Matrix 3, as a subset of Matrix 2 only including the A. gibba (Strebel, 1908) / A. alata (Thiele, 1912) species complex. Although depicted in the COI phylogenetic tree, all extractions of A. easmithi Moles, Avila & Malaquias, 2019 failed ddRAD library prep, thus they were not included in the matrix construction.
Phylogenetic and distinct genetic lineages analyses
A phylogenetic tree was inferred on ddRADseq-derived single nucleotide polymorphisms (SNPs) using the maximum likelihood (ML) criterion implemented in RAxML v. 8.2.1186 under the GTRGAMMA model. Nodal support was estimated via a rapid Bootstrap analysis (1500 replicates). Bayesian inference (BI) was conducted in MrBayes v. 3.2.687 with the GTR + I + G model88. We ran four independent Markov chain Monte Carlo (MCMC) chains for 2 million generations, sampling every 1000 generations and discarding 10% of the trees as burn-in for each MCMC run before convergence. Convergence was achieved when the potential scale reduction factor (PSRF) was close to 1.0 for all parameters. Trees were visualized in FigTree v. 1.4.489 and edited in Adobe Illustrator CC 2018.
Genetic structure and optimal clustering were analyzed in STRUCTURE v. 2.3.490 using matrices with unlinked SNPs for the Antarctophiline total dataset (Matrix 2) and the A. gibba and A. alata species complex (Matrix 3). SNP matrices were run for 1 million generations using an admixture model and 100,000 burn-in on K values ranging from 2 to 10 for Matrix 2 and 2–5 for Matrix 3, with eight replicates each. An optimal K value was calculated through the Evanno method32 in the Structure Harvester Web v. 0.6.9491 (http://taylor0.biology.ucla.edu/structureHarvester/). CLUMPAK92 (http://clumpak.tau.ac.il/) was used for graphical visualization that was later edited in Adobe Illustrator CC 2018.
To further visualize data and perform clustering on samples we use a VAE93 for dimensionality reduction of SNP data. VAEs are a type of machine learning algorithm rooted in Bayesian statistics that relies on neural networks and unsupervised learning to learn a reduced-dimension representation (latent space) of high dimensionality data. This approach allows for easy visualization of the mean and standard deviation of each sample in latent space. The use of VAEs in species delimitation and clustering with genetic data was recently demonstrated by Derkarabetian et al.36. The STRUCTURE formatted file was converted to “one-hot encoding” and the VAE was run using the “sp_deli” script (https://github.com/sokrypton/sp_deli) from Derkarabetian et al.36. An analysis was run on the full Antarctophiline dataset (Matrix 2). However, given potential contamination of sample P70 and issues with sample P49 (see “Results” section), a second analysis was run with these two samples removed. For both datasets, the VAE was run five times and the analysis with the lowest loss (a measure of the difference between input and reconstructed SNPs) was considered optimal. A single estimate of loss was calculated for each analysis by discarding the first 50% of generations as burn-in and calculating the average loss across the second 50% of generations. This average measure of loss for each analysis is akin to the likelihood estimate and burn-in associated with Bayesian phylogenetic analyses. Clustering was performed on the VAE output using only the mean of each sample; the two-dimensional representation (i.e., mean of each sample in latent space) was used as input for multiple clustering methods implemented in R94: “partition around medoids” (PAM) clustering using the cluster R package95, with the optimal K having the highest average silhouette width96; PAM clustering with the optimal K inferred via the gap statistic with k-means clustering using the factoextra R package97; and hierarchical clustering with the mclust R package98.
Supplementary Information
Acknowledgements
We are indebted to all collectors from the German, Spanish, and US cruises and campaigns: especially to G. W. Rouse, C. Avila, K. Linse, E. Schwabe, A. Brandt, C. Alvaro, M. Ballesteros, W. Brökeland, M. Choudhury, R. Cumming, A. Hickling, M. Malyutina, J. Naretto, and M. Raupach. Thanks are due to the Institute of Paleobiology, Polish Academy of Sciences, for kindly providing the images of the fossil specimen. We would like to thank T.J. da Cunha, B. de Medeiros, L.R. Benavides, and C.M. Baker (OEB, MCZ) for their help during the lab and bioinformatic work and to J. Giménez (Institut de Ciències del Mar–CSIC) for helping with map design. We are also grateful to the Bauer Core and the RC Computing team. Lab and analytical work were supported by internal funds from the Faculty of Arts and Sciences, Harvard University, to GG and by NSF Grant DEB-1754278, which provided the salary for SD. JM was supported by a Fundación Ramón Areces postdoctoral fellowship. NGW acknowledges the National Science Foundation (NSF ANT-1043749) and the ACE Foundation and Ferring Pharmaceuticals for funding cruises. Two anonymous reviewers provided comments that helped improve the manuscript.
Author contributions
J.M. conceived the original idea, experimental design, and conducted lab work. J.M. and S.D. conducted analyses. J.M., S.S., M.S., J.S.T. and N.G.W. contributed with samples. G.G. contributed with laboratory space, reagents and sequencing costs. J.M. wrote the initial manuscript. All authors contributed to the final manuscript.
Data availability
DNA sequences: GenBank accession numbers are listed in Table 1. Demultiplexed raw reads of ddRADseq-derived data: SRA BioProject PRJNA600882. Matrices and tree files are available in the Harvard Dataverse: https://doi.org/10.7910/DVN/HZJCCS.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87244-5.
References
- 1.Sanders HL. Marine benthic diversity: a comparative study. Am. Nat. 1968;102:234–282. doi: 10.1086/282541. [DOI] [Google Scholar]
- 2.Etter RJ, et al. Phylogeography of a pan-Atlantic abyssal protobranch bivalve: implications for evolution in the Deep Atlantic. Mol. Ecol. 2011;20:829–843. doi: 10.1111/j.1365-294X.2010.04978.x. [DOI] [PubMed] [Google Scholar]
- 3.Rex MA, et al. A source-sink hypothesis for abyssal biodiversity. Am. Nat. 2005;165:163–178. doi: 10.1086/427226. [DOI] [PubMed] [Google Scholar]
- 4.Thistle, D. The deep-sea floor: an overview. in Ecosystems of the World, Vol. 28 (ed. Tyler, P. A.), 5–39 (Elsevier, 2003).
- 5.Eilertsen MH, Malaquias MAE. Speciation in the dark: diversification and biogeography of the deep-sea gastropod genus Scaphander in the Atlantic Ocean. J. Biogeogr. 2015;42:843–855. doi: 10.1111/jbi.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubili C, et al. Species diversity in the cryptic abyssal holothurian Psychropotes longicauda (Echinodermata) Deep Res. Part II Top. Stud. Oceanogr. 2016 doi: 10.1016/j.dsr2.2016.04.003. [DOI] [Google Scholar]
- 7.Jablonski D. Evolutionary innovations in the fossil record: patterns in time and space. J. Exp. Zool. Mol. Dev. Evol. 2005;304B:504–519. doi: 10.1002/jez.b.21075. [DOI] [PubMed] [Google Scholar]
- 8.Barnes DKA, Conlan KE. Disturbance, colonization and development of Antarctic benthic communities. Philos. Trans. R. Soc. B Biol. Sci. 2007;362:11–38. doi: 10.1098/rstb.2006.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabezas P, Sanmartín I, Paulay G, Macpherson E, Machordom A. Deep under the sea: unraveling the evolutionary history of the deep-sea squat lobster paramunida (decapoda, munididae) Evolution (N. Y.) 2012;66:1878–1896. doi: 10.1111/j.1558-5646.2011.01560.x. [DOI] [PubMed] [Google Scholar]
- 10.Hunter RL, Halanych KM. Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the Drake Passage in the Southern Ocean. J. Hered. 2008;99:137–148. doi: 10.1093/jhered/esm119. [DOI] [PubMed] [Google Scholar]
- 11.Williams ST, et al. Cenozoic climate change and diversification on the continental shelf and slope: evolution of gastropod diversity in the family Solariellidae (Trochoidea) Ecol. Evol. 2013;3:887–917. doi: 10.1002/ece3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke A, Crame JA. Evolutionary dynamics at high latitudes: speciation and extinction in polar marine faunas. Philos. Trans. R. Soc. B Biol. Sci. 2010;365:3655–3666. doi: 10.1098/rstb.2010.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachos J, et al. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science (80-) 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 14.Thatje S, Hillenbrand C-D, Larter R. On the origin of Antarctic marine benthic community structure. TRENDS Ecol. Evol. 2005;20:534–540. doi: 10.1016/j.tree.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Beu AG, Griffin M, Maxwell PA. Opening of Drake Passage gateway and Late Miocene to Pleistocene cooling reflected in Southern Ocean molluscan dispersal: evidence from New Zealand and Argentina. Tectonophysics. 1997;281:83–97. doi: 10.1016/S0040-1951(97)00160-1. [DOI] [Google Scholar]
- 16.Brey T, et al. Do Antarctic benthic invertebrates show an extended level of eurybathy? Antarct. Sci. 1996;8:3–6. doi: 10.1017/S0954102096000028. [DOI] [Google Scholar]
- 17.Katz ME, et al. Impact of Antarctic circumpolar current development on late Paleogene ocean structure. Science (80-) 2011;332:1076–1079. doi: 10.1126/science.1202122. [DOI] [PubMed] [Google Scholar]
- 18.Brandt A, et al. First insights into the biodiversity and biogeography of the Southern Ocean deep sea. Nature. 2007;447:307–311. doi: 10.1038/nature05827. [DOI] [PubMed] [Google Scholar]
- 19.Strugnell JM, Rogers AD, Prodöhl PA, Collins MA, Allcock AL. The thermohaline expressway: the Southern Ocean as a centre of origin for deep-sea octopuses. Cladistics. 2008;24:853–860. doi: 10.1111/j.1096-0031.2008.00234.x. [DOI] [PubMed] [Google Scholar]
- 20.Briggs JC. Marine centres of origin as evolutionary engines. J. Biogeogr. 2003;30:1–18. doi: 10.1046/j.1365-2699.2003.00810.x. [DOI] [Google Scholar]
- 21.Rogers AD. Evolution and biodiversity of Antarctic organisms: a molecular perspective. Philos. Trans. R. Soc. B. 2007;362:2191–2214. doi: 10.1098/rstb.2006.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinogradova NG. Zoogeography of the abyssal and hadal zones. Adv. Mar. Biol. 1997;32:325–387. doi: 10.1016/S0065-2881(08)60019-X. [DOI] [Google Scholar]
- 23.Schrödl M, Bohn JM, Brenke N, Rolán E, Schwabe E. Abundance, diversity, and latitudinal gradients of southeastern Atlantic and Antarctic abyssal gastropods. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010;58:49–57. doi: 10.1016/j.dsr2.2010.10.009. [DOI] [Google Scholar]
- 24.Martynov AV, Schrödl M. The new Arctic side-gilled sea slug genus Boreoberthella (Gastropoda, Opisthobranchia): Pleurobranchoidean systematics and evolution revisited. Polar Biol. 2009;32:53–70. doi: 10.1007/s00300-008-0503-3. [DOI] [Google Scholar]
- 25.Moles J, Wägele H, Schrödl M, Avila C. A new Antarctic heterobranch clade is sister to all other Cephalaspidea (Mollusca: Gastropoda) Zool. Scr. 2017;46:127–137. doi: 10.1111/zsc.12199. [DOI] [Google Scholar]
- 26.Schrödl M. Sea Slugs of Southern South America. ConchBooks; 2003. [Google Scholar]
- 27.Wägele H, Klussmann-Kolb A, Vonnemann V, Medina M. Heterobranchia I. The Opisthobranchia. In: Ponder WF, Lindberg DR, editors. Phylogeny and Evolution of the Mollusca. University of California; 2008. pp. 385–408. [Google Scholar]
- 28.Lawver LA, Gahagan LM. Evolution of Cenozoic seaways in the circum-Antarctic region. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2003;198:11–37. doi: 10.1016/S0031-0182(03)00392-4. [DOI] [Google Scholar]
- 29.Moles J, Avila C, Malaquias MAE. Unmasking Antarctic mollusc lineages: novel evidence from philinoid snails (Gastropoda: Cephalaspidea) Cladistics. 2019;35:487–513. doi: 10.1111/cla.12364. [DOI] [PubMed] [Google Scholar]
- 30.Oskars TR, Bouchet P, Malaquias MAE. A new phylogeny of the Cephalaspidea (Gastropoda: Heterobranchia) based on expanded taxon sampling and gene markers. Mol. Phylogenet. Evol. 2015;89:130–150. doi: 10.1016/j.ympev.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Moles J, Avila C, Malaquias MAE. Systematic revision of the Antarctic gastropod family Newnesiidae (Heterobranchia: Cephalaspidea) with the description of a new genus and a new abyssal species. Zool. J. Linn. Soc. 2018;183:763–775. doi: 10.1093/zoolinnean/zlx089. [DOI] [Google Scholar]
- 32.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 33.Karczewski L. Gastropods from the Cape Melville Formation (Lower Miocene) of King George Island, West Antarctica. Palaeontol. Pol. 1987;49:127–146. [Google Scholar]
- 34.Beu AG. Before the ice: biogeography of Antarctic Paleogene molluscan faunas. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2009;284:191–226. doi: 10.1016/j.palaeo.2009.09.025. [DOI] [Google Scholar]
- 35.Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol. Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 36.Derkarabetian S, Castillo S, Koo PK, Ovchinnikov S, Hedin M. A demonstration of unsupervised machine learning in species delimitation. Mol. Phylogenet. Evol. 2019;139:106562. doi: 10.1016/j.ympev.2019.106562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malaquias MAE, Ohnheiser LT, Oskars TR, Willassen E. Diversity and systematics of philinid snails (Gastropoda: Cephalaspidea) in West Africa with remarks on the biogeography of the region. Zool. J. Linn. Soc. 2017;180:1–35. [Google Scholar]
- 38.Leiva C, et al. Population substructure and signals of divergent adaptive selection despite admixture in the sponge Dendrilla antarctica from shallow waters surrounding the Antarctic Peninsula. Mol. Ecol. 2019;28:1–20. doi: 10.1111/mec.15135. [DOI] [PubMed] [Google Scholar]
- 39.Powell AWB. Antarctic and subantarctic mollusca: Pelecypoda and Gastropoda. Discov. Rep. 1951;26:47–196. doi: 10.5962/bhl.part.16335. [DOI] [Google Scholar]
- 40.Rudman WB. The genus Philine (Opisthobranchia, Gastropoda) Proc. Malacol. Soc. Lond. 1972;40:171–187. [Google Scholar]
- 41.Linse K, Griffiths HJ, Barnes DKAA, Clarke A. Biodiversity and biogeography of Antarctic and sub-Antarctic mollusca. Deep Res. Part II Top. Stud. Oceanogr. 2006;53:985–1008. doi: 10.1016/j.dsr2.2006.05.003. [DOI] [Google Scholar]
- 42.Hemery LG, et al. Comprehensive sampling reveals circumpolarity and sympatry in seven mitochondrial lineages of the Southern Ocean crinoid species Promachocrinus kerguelensis (Echinodermata) Mol. Ecol. 2012;21:2502–2518. doi: 10.1111/j.1365-294X.2012.05512.x. [DOI] [PubMed] [Google Scholar]
- 43.Layton KKS, Rouse GW, Wilson NG. A newly discovered radiation of endoparasitic gastropods and their coevolution with asteroid hosts in Antarctica. BMC Evol. Biol. 2019;19:180. doi: 10.1186/s12862-019-1499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke, A. & Johnston, I. A. Evolution and adaptive radiation of Antarctic fishes. Trends Ecol. Evol. 11, 212–218 (1996). [DOI] [PubMed]
- 45.Chenuil A, et al. Understanding processes at the origin of species flocks with a focus on the marine Antarctic fauna. Biol. Rev. 2018;93:481–504. doi: 10.1111/brv.12354. [DOI] [PubMed] [Google Scholar]
- 46.Raguá-Gil JM, Gutt J, Clarke A, Arntz WE. Antarctic shallow-water mega-epibenthos: shaped by circumpolar dispersion or local conditions? Mar. Biol. 2004;144:829–839. doi: 10.1007/s00227-003-1269-3. [DOI] [Google Scholar]
- 47.Arntz WE, Brey T, Gallardo A. Antarctic zoobenthos. Oceanogr. Mar. Biol. Annu. Rev. 1994;32:241–304. [Google Scholar]
- 48.Linse K, Griffiths HJ, Barnes DKA, Clarke A. Biodiversity and biogeography of Antarctic and sub-Antarctic Mollusca. Deep Sea Res. Part II Top. Stud. Oceanogr. 2006;53:985–1008. doi: 10.1016/j.dsr2.2006.05.003. [DOI] [Google Scholar]
- 49.Jörger KM, Schrödl M, Schwabe E, Würzberg L. A glimpse into the deep of the Antarctic polar front—diversity and abundance of abyssal molluscs. Deep Res. Part II Top. Stud. Oceanogr. 2014;108:93–100. doi: 10.1016/j.dsr2.2014.08.003. [DOI] [Google Scholar]
- 50.Barnes DKA, Hillenbrand C-DD. Faunal evidence for a late quaternary trans-Antarctic seaway. Glob. Change Biol. 2010;16:3297–3303. doi: 10.1111/j.1365-2486.2010.02198.x. [DOI] [Google Scholar]
- 51.Moles J, et al. The end of the cold loneliness: 3D comparison between Doto antarctica and a new sympatric species of Doto (Heterobranchia: Nudibranchia) PLoS ONE. 2016;11:e0157941. doi: 10.1371/journal.pone.0157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aldea C, Troncoso J. Systematics and distribution of shelled molluscs (Gastropoda, Bivalvia and Scaphopoda) from the South Shetland Islands to the Bellingshausen Sea, West Antarctica. Iberus. 2008;26:1–75. [Google Scholar]
- 53.Strugnell JM, Watts PC, Smith PJ, Allcock AL. Persistent genetic signatures of historic climatic events in an Antarctic octopus. Mol. Ecol. 2012;21:2775–2787. doi: 10.1111/j.1365-294X.2012.05572.x. [DOI] [PubMed] [Google Scholar]
- 54.Clarke A, Aronson RB, Crame JA, Gili J-M, Blake DB. Evolution and diversity of the benthic fauna of the Southern Ocean continental shelf. Antarct. Sci. 2004;16:559–568. doi: 10.1017/S0954102004002329. [DOI] [Google Scholar]
- 55.Allcock AL, Strugnell JM. Southern Ocean diversity: New paradigms from molecular ecology. TRENDS Ecol. Evol. 2012;27:520–528. doi: 10.1016/j.tree.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Riesgo A, Taboada S, Avila C. Evolutionary patterns in Antarctic marine invertebrates: an update on molecular studies. Mar. Genomics. 2015;23:1–13. doi: 10.1016/j.margen.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Allcock AL, et al. Cryptic speciation and the circumpolarity debate: a case study on endemic Southern Ocean octopuses using the COI barcode of life. Deep Res. Part II Top. Stud. Oceanogr. 2011;58:242–249. doi: 10.1016/j.dsr2.2010.05.016. [DOI] [Google Scholar]
- 58.Janosik AM, Halanych KM. Unrecognized Antarctic biodiversity: a case study of the genus Odontaster (Odontasteridae; Asteroidea) Integr. Comp. Biol. 2010;50:981–992. doi: 10.1093/icb/icq119. [DOI] [PubMed] [Google Scholar]
- 59.Wilson NG, Maschek JA, Baker BJ. A species flock driven by predation? Secondary metabolites support diversification of slugs in Antarctica. PLoS ONE. 2013;8:e80277. doi: 10.1371/journal.pone.0080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore JM, Carvajal JI, Rouse GW, Wilson NG. The Antarctic circumpolar current isolates and connects: structured circumpolarity in the sea star Glabraster antarctica. Ecol. Evol. 2018;8:10621–10633. doi: 10.1002/ece3.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orsi AH, Nowlin WD, Jr, Whitworth T., III On the circulation and stratification of the Weddell Gyre. Deep Sea Res. Part I Oceanogr. Res. Pap. 1993;40:169–203. doi: 10.1016/0967-0637(93)90060-G. [DOI] [Google Scholar]
- 62.Ryan S, Schröder M, Huhn O, Timmermann R. On the warm inflow at the eastern boundary of the Weddell Gyre. Deep Res. Part I Oceanogr. Res. Pap. 2016;107:70–81. doi: 10.1016/j.dsr.2015.11.002. [DOI] [Google Scholar]
- 63.Barnes DKA, Hodgson DA, Convey P, Allen CS, Clarke A. Incursion and excursion of Antarctic biota: past, present and future. Glob. Ecol. Biogeogr. 2006;15:121–142. doi: 10.1111/j.1466-822X.2006.00216.x. [DOI] [Google Scholar]
- 64.Clarke A, Barnes DKA, Hodgson DA. How isolated is Antarctica? TRENDS Ecol. Evol. 2005;20:1–3. doi: 10.1016/j.tree.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Seager JR. Reproductive biology of the Antarctic opistobranch Philine gibba Strebel. J. Exp. Mar. Biol. Ecol. 1979;41:51–74. doi: 10.1016/0022-0981(79)90080-7. [DOI] [Google Scholar]
- 66.Moran, A. L., Toh, M.-W. A., Lobert, G. T., Ely, T. & Marko, P. B. Egg masses and larval development of the Antarctic cephalaspidean slug Waegelea antarctica, with notes on egg masses of the related Antarctophiline alata. J. Molluscan Stud. (2021). (In press)
- 67.Wilson NG, Schrödl M, Halanych KM. Ocean barriers and glaciation: evidence for explosive radiation of mitochondrial lineages in the Antarctic sea slug Doris kerguelenensis (Mollusca, Nudibranchia) Mol. Ecol. 2009;18:965–984. doi: 10.1111/j.1365-294X.2008.04071.x. [DOI] [PubMed] [Google Scholar]
- 68.Smale DA, Brown KM, Barnes DKA, Fraser KPP, Clarke A. Ice scour disturbance in Antarctic waters. Science (80-) 2008;321:371. doi: 10.1126/science.1158647. [DOI] [PubMed] [Google Scholar]
- 69.Thompson AF, Stewart AL, Spence P, Heywood KJ. The Antarctic slope current in a changing climate. Rev. Geophys. 2018;56:741–770. doi: 10.1029/2018RG000624. [DOI] [Google Scholar]
- 70.Crame JA. Bipolar molluscs and their evolutionary implications. J. Biogeogr. 1993;20:145–161. doi: 10.2307/2845668. [DOI] [Google Scholar]
- 71.Orsi AH, Johnson GC, Bullister JL. Circulation, mixing, and production of Antarctic bottom water. Prog. Oceanogr. 1999;43:55–109. doi: 10.1016/S0079-6611(99)00004-X. [DOI] [Google Scholar]
- 72.Pawlowski J, et al. Bipolar gene flow in deep-sea benthic foraminifera. Mol. Ecol. 2007;16:4089–4096. doi: 10.1111/j.1365-294X.2007.03465.x. [DOI] [PubMed] [Google Scholar]
- 73.Moles J, et al. Giant embryos and hatchlings of Antarctic nudibranchs (Mollusca: Gastropoda: Heterobranchia) Mar. Biol. 2017;164:114. doi: 10.1007/s00227-017-3143-8. [DOI] [Google Scholar]
- 74.Almada F, Levy A, Robalo JI. Not so sluggish: the success of the Felimare picta complex (Gastropoda, Nudibranchia) crossing Atlantic biogeographic barriers. PeerJ. 2016;4:e1561. doi: 10.7717/peerj.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Page TJ, Linse K. More evidence of speciation and dispersal across the Antarctic Polar front through molecular systematics of Southern Ocean Limatula (Bivalvia: Limidae) Polar Biol. 2002;25:818–826. doi: 10.1007/s00300-002-0414-7. [DOI] [Google Scholar]
- 76.Fassio G, et al. An Antarctic flock under the Thorson’s rule: diversity and larval development of Antarctic Velutinidae (Mollusca: Gastropoda) Mol. Phylogenet. Evol. 2019;132:1–13. doi: 10.1016/j.ympev.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 77.Schwabe E, Bohn JM, Engl W, Linse K, Schrödl M. Rich and rare—first insights into species diversity and abundance of Antarctic abyssal Gastropoda (Mollusca) Deep Sea Res. Part II Top. Stud. Oceanogr. 2007;54:1831–1847. doi: 10.1016/j.dsr2.2007.07.010. [DOI] [Google Scholar]
- 78.Crame JA. Key stages in the evolution of the Antarctic marine fauna. J. Biogeogr. 2018;45:986–994. doi: 10.1111/jbi.13208. [DOI] [Google Scholar]
- 79.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 80.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eaton DAR. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics. 2014;30:1844–1849. doi: 10.1093/bioinformatics/btu121. [DOI] [PubMed] [Google Scholar]
- 84.Eaton, D. A. R. & Overcast, I. ipyrad v. 0.7.28. https://github.com/dereneaton/ipyrad (accessed 1 Feb 2018) (2018).
- 85.de Medeiros, B. A. S. Matrix Condenser v.1.0 (2019).
- 86.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2011;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 89.Rambaut, A. Figtree, a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree (2014).
- 90.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Earl DA, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 92.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kingma, D. P. & Welling, M. Auto-encoding variational Bayes. In Proceedings of the International Conference on Learning Representations (ICLR) (arXiv:1312.6114v10 [stat.ML], 2013).
- 94.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018).
- 95.Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M. & Hornik, K. cluster: Cluster Analysis Basics and Extensions. https://CRAN.R-project.org/package=cluster (2018).
- 96.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 97.Kassambara, A. & Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses (2017).
- 98.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J. 2017;8:205–233. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accession numbers are listed in Table 1. Demultiplexed raw reads of ddRADseq-derived data: SRA BioProject PRJNA600882. Matrices and tree files are available in the Harvard Dataverse: https://doi.org/10.7910/DVN/HZJCCS.