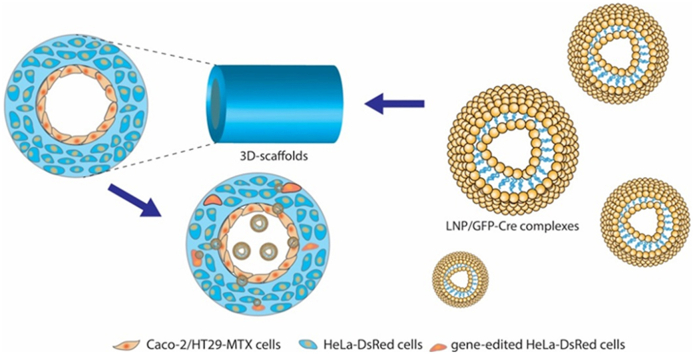
Abstract
Lipid nanoparticles are promising carriers for oral drug delivery. For bioactive cargos with intracellular targets, e.g. gene-editing proteins, it is essential for the cargo and carrier to remain complexed after crossing the epithelial layer of intestine in order for the delivery system to transport the cargos inside targeted cells. However, limited studies have been conducted to verify the integrity of cargo/carrier nanocomplexes and their capability in facilitating cargo delivery intracellularly after the nanocomplex crossing the epithelial barrier. Herein, we used a traditional 2D transwell system and a recently developed 3D tissue engineered intestine model and demonstrated the synthetic lipid nanoparticle (carrier) and protein (cargo) nanocomplexes are able to cross the epithelial layer and deliver the protein cargo inside the underneath cells. We found that the EC16-63 LNP efficiently encapsulated the GFP-Cre recombinase, penetrated the intestinal monolayer cells in both the 2D cell culture and 3D tissue models through temporarily interrupting the tight junctions between epithelial layer. After transporting across the intestinal epithelia, the EC16-63 and GFP-Cre recombinase nanocomplexes can enter the underneath cells to induce gene recombination. These results suggest that the in vitro 3D intestinal tissue model is useful for identifying effective lipid nanoparticles for potential oral drug delivery.
Keywords: Lipidoid nanoparticle, Protein delivery, Oral drug delivery, Genome engineering, 3D tissue model
Graphical abstract
Highlights
-
•
Employed a 3D intestine model for nanodrug screening.
-
•
Developed lipidoid nanoparticles for genome engineering protein delivery.
-
•
Used 3D tissue model to test lipidoid nanoparticles for potential oral drug delivery.
1. Introduction
The employment of most current FDA approved protein and nucleic acid based drugs involve invasive administration methods like injection and infusion. Oral drug delivery can avoid the discomfort and pain associated with needle-involved administrations, resulting in better patient acceptance [1,2]. However, in the gastrointestinal (GI) tract, various physiochemical and enzymatic factors greatly limit the rate and extent of drug absorption process. Many oral delivery systems have been developed to enhance the drug delivery efficiency. In particular, lipid-based carriers such as liposomes and solid lipid nanoparticles have received increasing attention for oral drug delivery due to their superior physicochemical and biological properties [3]. Nevertheless, insight into the in vivo biological fate of lipid nanoparticles after crossing the intestinal epithelial layer remains limited. Several studies showed that the lipid nanoparticles can be taken up or transported across the GI tract as intact nanoparticles after oral administration [4,5]. However, it is unclear whether the lipid nanoparticle/cargo nanocomplexes remain intact after crossing the epithelial layer. For the delivery of nucleic acids or gene-editing proteins, maintaining the intact structure of the nanocomplexes after intestinal absorption is crucial in order to deliver the cargo into the targeted tissues and cells. Thus, it is essential to develop a screening approach to investigate whether carrier/cargo nanocomplexes maintain its integrity after crossing the intestinal epithelial layer for intracellular cargo delivery.
Animal models have been extensively utilized to assess in vivo oral drug delivery using nanoparticles [[6], [7], [8], [9]]. However, these models often lack relevance to human physiological conditions, thus hindering the accurate prediction of the behavior of nanoparticles in human intestines. Many drugs have been tested successfully in animal studies but unfortunately failed in human trials [10]. Tissue-engineered human intestinal tissue models offer an alternative option to animal models by culturing human intestinal epithelial cells on 2D substrates, such as transwell inserts, or in 3D pre-fabricated scaffolds or matrices. These cell culture systems have been invaluable in improving our understanding of transport mechanisms of nanoparticle systems [11]. 2D transwell cell cultures are relatively simple to use and enable transport studies of drugs and chemicals as well as other metabolic activities in vitro [12]. 3D cell cultures in vitro, while more complex to establish and utilize, resemble the complex microenvironment found in vivo more closely and thus can more accurately reflect human outcomes [13].
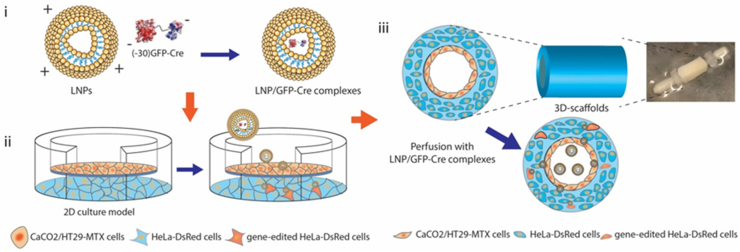
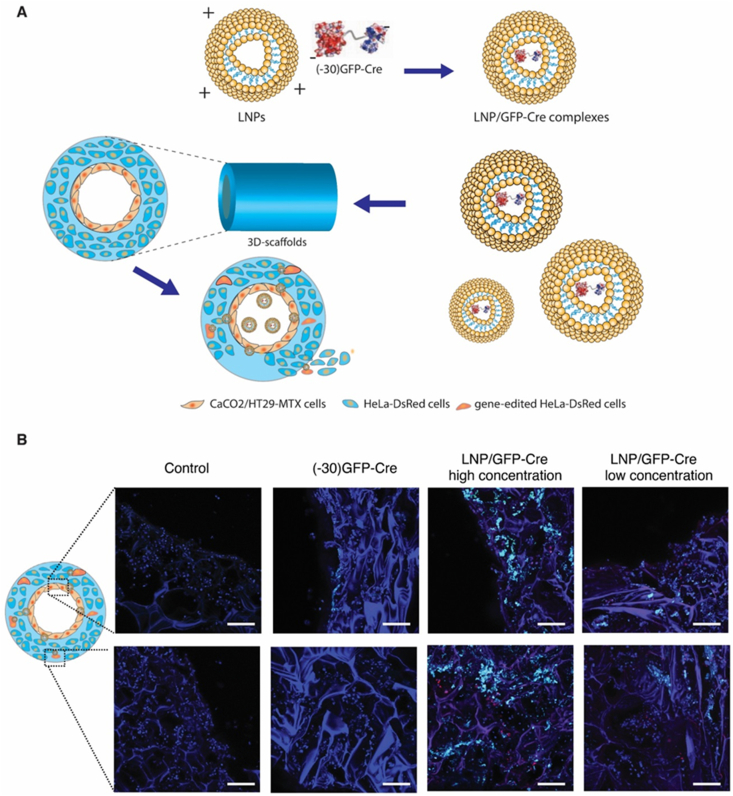
In the present study, we synthesized a series of lipidoid nanoparticles (LNPs) and demonstrated that cationic LNPs formed stable complexes with the gene-editing Cre recombinase ((-30)GFP-Cre) through electrostatic interactions. We first used a 2D transwell system to screen a library of LNPs and then futher validate the resulting LNPs in a 3D tissue engineered intestinal model we previously established [13]. In 2D Transwell systems, Caco-2/HT29-MTX cells were seeded on the transwell membrane forming a compact cell monolayer, and HeLa-DsRed cells were plated in the bottom chamber, to serve as a model for determining whether the escaped LNPs formed intact nanoparticles to exert their gene-editing function.
We found that the LNPs efficiently delivered the (−30)GFP-Cre protein, penetrating the cell monolayer seeded on the Transwell membranes in the 2D culture model and activating the HeLa-DsRed cells by endocytosis. We then incorporated LNP loaded (−30)GFP-Cre protein (LNP/GFP-Cre) into the 3D tissue engineered system, in which Caco-2/HT29-MTX cells were seeded on the surface of the lumen and the bulk space was incubated with the HeLa-DsRed cells. After delivering the LNP/GFP-Cre complexes into the 3D cell culture system, we validated that the complexes successfully penetrated the Caco-2/HT29-MTX monolayer and reached the scaffold bulk space to realize gene-editing functions (Scheme 1). In this way, series LNPs with encapsulated gene-editing proteins were screened with a combination of 2D Transwell and 3D scaffold tissue models. Some of the nanoparticles were found to maintain stable structures to penetrate these in vitro intestinal lumen mimics, suggestive towards successful screening of designs with potential to reach the circulatory system in vivo to generate gene-editing functions at other sites.
Scheme 1.
The illustration of LNPs-mediated (−30)GFP-Cre proteins delivery in 2D Transwell model and 3D tissue systems. (i) The self-assembly of the LNPs and (−30)GFP-Cre protein forming stable complexes through electronic interactions. (ii) The delivery of LNP/GFP-Cre complexes in 2D Transwell culture model. (iii) The delivery of LNP/GFP-Cre complexes in 3D tissue system.
2. Results and discussion
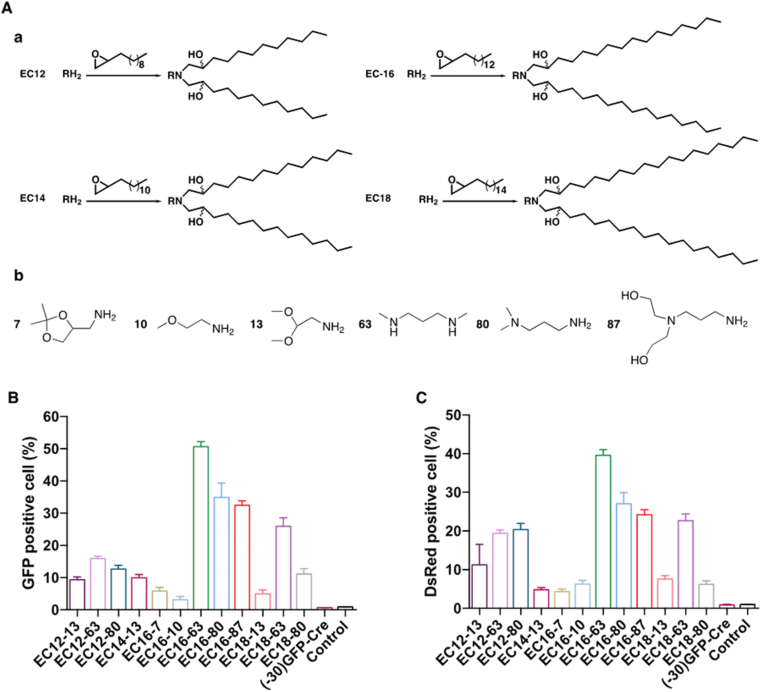
Previously, we reported a series of cationic lipidoids that were successfully employed to intracellularly deliver therapeutic proteins for cancer treatment [[14], [15], [16]]. The properties of proteins for delivery, such as charge intensity, molecular weight, and hydrophobicity impacted delivery efficiency [17,18]. In addition, different proteins for delivery required distinct lipidoids formulations. Thus, to verify effective LNPs for (−30)GFP-Cre delivery, we first screened a small library of lipidoids which has been narrowed to 12 lipidoids based on a preliminary study. The library of the lipidoids was synthesized through the ring-opening reaction between the lipophilic tails and various aliphatic amine heads, where the lipophilic tails were 1,2-epoxydodecane (EC12), 1,2-epoxytetradecane (EC14), 1,2-epoxyhexadecane (EC-16), and 1,2-epoxyoctadecane (EC18), respectively. The lipidoids were named by the lipophilic tail and the amine head number (Fig. 1A). The LNPs were subsequently obtained by formulation with cholesterol, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE)-PEG2K. Next, to evaluate the intracellular protein delivery efficiency, the Cre recombinase, which has been fused to a negatively charged GFP variant to generate green fluorescence spontaneously, was used as a model cargo. The HeLa-DsRed cell line expressing the red fluorescence DsRed upon Cre-mediated gene recombination was used to determine the (−30)GFP-Cre mediated gene recombination efficiency. Following LNPs-mediated (−30)GFP-Cre delivery into the HeLa-DsRed cells for 6 h, the percentage of GFP-positive cells was determined to evaluate the uptake efficiency of LNP/GFP-Cre complexes by flow cytometry. As shown in Fig. 1B, negligible GFP-positive cells could be detected in the free (−30)GFP-Cre group, indicating that free GFP-Cre protein could not be taken up by the cells without the LNPs. In contrast, a significantly higher proportion of GFP-positive cells was obtained after the LNP/Cre-GFP treatments, where the EC16-63, EC16-80, EC16-87 and EC18-63 LNPs exhibited more than 30% GFP-positive cells among the 12 LNPs tested. Particularly, EC16-63 mediated (−30)GFP-Cre delivery achieved the highest delivery efficiency, with more than 50% of GFP-positive cells observed among the four LNPs. We further tested the gene recombination efficiency of Cre recombinase by detecting the red fluorescence DsRed generated from the HeLa-DsRed cells after 24 h incubation with the LNP/GFP-Cre complexes. As shown in Fig. 1C, limited red fluorescence signal was detected after the free (−30)GFP-Cre proteins treatment, demonstrating that the (−30)GFP-Cre alone did not induce the expression of DsRed in the cells. Compared with cells treated with free (−30)GFP-Cre without LNPs, those receiving LNP/GFP-Cre complexes expressed a more obvious red fluorescence signal, revealing that the LNPs efficiently delivered the (−30)GFP-Cre into the HeLa-DsRed cells and then induced gene recombination functions. Among all of the lipidoids tested, the performance of EC16-63, EC16-80, EC16-87 and EC18-63-mediated (−30)GFP-Cre delivery exhibited the most enhanced genetic functionality, consistent with the intracellular efficiency shown in Fig. 1B. Compared with EC16-87, EC18-63 mediated (−30)GFP-Cre delivery exhibited a slightly lower proportion of DsRed-positive cells. Taken together, three formulations of lipidoids, EC16-63, EC16-80, EC16-87 were considered as useful LNPs from the initial library screening, which were then adopted in follow on experiments.
Fig. 1.
(A) The synthesis of cationic lipidoids conducted through the ring-opening reaction between the lipophilic tails (a) and various aliphatic amine heads (b). The cellular uptake (B) and DsRed expression level (C) of HeLa-DsRed cells treated with (−30)GFP-Cre alone or different LNP/GFP-Cre complexes. Untreated cells served as controls.
The hydrodynamic size and polydispersity index (PDI) of three LNP/GFP-Cre complexes was examined through dynamic light scattering (DLS) (Figure S1, Supporting information). The three LNPs exhibited comparable hydrodynamic sizes, approximately 57.2 nm in diameter, while the size dramatically increased to over 200 nm with the addition of the (−30)GFP-Cre protein. In particular, after the co-incubation with (−30)GFP-Cre protein, the size of EC16-80 increased to about 400 nm. The increased size demonstrated that the LNPs could form stable complexes with the (−30)GFP-Cre protein.
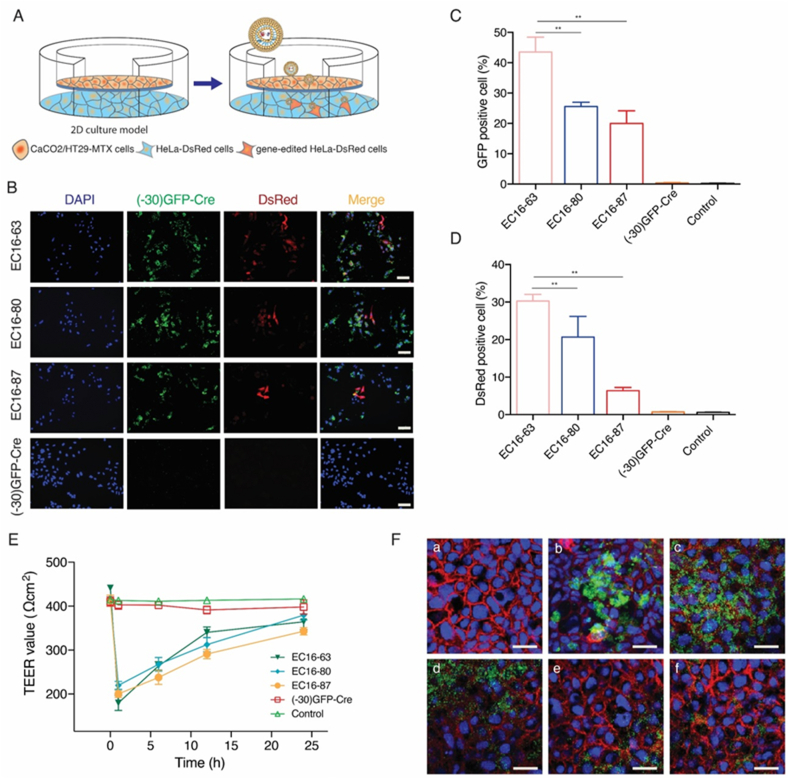
Having demonstrated that three lipidoids, EC16-63, EC16-80, and EC16-87 could efficiently deliver (−30)GFP-Cre into the cells to induce gene recombination, we further detected their deliver efficiency using 2D Transwell models. The Transwell membrane was seeded with human intestinal Caco-2/HT29-MTX cells forming a compact cell monolayer in the upper chamber with mucus-secreting features, since the mucus layer is significant in protecting the epithelial cells from damage from gut fluids (Fig. 2A). Meanwhile, we also plated HeLa-DsRed cells in the bottom of the chamber to evaluate delivery efficiency of the LNPs. After incubating the LNP/GFP-Cre complexes in the upper chamber for 48 h, GFP-positive cells in the lower chamber were observed using fluorescence microscopy and then analyzed through flow cytometry. No fluorescence signal was observed after the addition of (−30)GFP-Cre protein into the upper chamber (Fig. 2B), indicating that (−30)GFP-Cre alone did not penetrate the cell monolayer composed of the Caco-2/HT29-MTX cells, which could be attributed to protein absorbance by the mucus. In contrast, green fluorescence was observed in the three LNPs-mediated (−30)GFP-Cre delivery groups, demonstrating that the LNPs loaded with the proteins could travel across the mucus layer, subsequently penetrate the cell monolayer and eventually deliver the protein into the HeLa-DsRed cells seeded at the bottom of the chamber.
Fig. 2.
(A) Illustration of LNPs-mediated (−30)GFP-Cre protein delivery in 2D Transwell culture model. (B) The fluorescence images of the HeLa-DsRed cells seeded on the bottom of the chamber after (−30)GFP-Cre proteins alone or different LNP/GFP-Cre complexes were added into the upper chamber for 48 h. Scale bar: 200 μm. The cellular uptake (C) and DsRed expression level (D) of HeLa-DsRed cells seeded on the bottom of the chamber after (−30)GFP-Cre proteins alone or different LNP/GFP-Cre complexes were added into the upper chamber for 48 h **p < 0.01. (E) The TEER value change of the epithelial monolayer during the penetration of LNP/GFP-Cre complexes. (F) The CLSM images of immunostaining of the Transwell membrane after treated with different LNP/GFP-Cre complexes. (a) Control; (b) (−30)GFP-Cre proteins alone; LNP/GFP-Cre complexes treatment for 1 h (c), 6 h (d), 12 h (e), and 24 h (f), respectively. Red, cadherin; Blue, DAPI; Green, (−30)GFP-Cre proteins. Scale bar: 15 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The mucus layer consists of crosslinked and entangled mucin fibers secreted by goblet cells and submucosal glands, which exhibit negatively charged properties [19]. Upon penetrating the mucus layer, cationic lipidoids can neutralize the negatively charged density of mucin, leading to the disruption of the crosslinks in the mucus and thereby decreasing the adhesive interactions with the nanoparticles [20]. In addition, the addition of PEG2k in the formulation of the LNPs can enhance the stability of the nanoparticles in the mucus and accelerate the ability of the nanoparticles to cross the mucus layer [20]. The proportion of GFP-positive cells was further quantified through flow cytometer, where EC16-63 achieved highest delivery efficiency among the three lipidoids (Fig. 2C). Meanwhile, we also analyzed the gene recombination efficiency of complexes after (−30)GFP-Cre delivery. Once delivered into the HeLa-DsRed cells, the complexes released the (−30)GFP-Cre proteins and activated the gene-editing function to achieve the expression of intracellular red fluorescence. As shown in Fig. 2D, there was no DsRed signal in the (−30)GFP-Cre protein-treated group, since the free protein could not efficiently enter into the cells. Meanwhile, red signal was observed in the LNPs-mediated protein delivery, indicating successful gene recombination induced by Cre protein. DsRed-positive cells were harvested and counted by flow cytometry, where it was confirmed that EC16-63 provided the most efficient gene recombination, likely due to the relatively higher delivery efficiency among the three lipidoids.
The mechanism of LNPs penetration of the 2D Transwell model was systemically investigated. First, we examined the cytotoxicity of the lipidoids by MTT (Figure S2, Supporting information). The Caco-2/HT29-MTX cells were treated for 48 h with the same dose of LNP/GFP-Cre complexes as used in the investigation of delivery efficiency in 2D Transwell models. The lipidoids did not induce significant cytotoxicity against the Caco-2/HT29-MTX cells. Since the LNPs caused limited cytotoxicity, the transepithelial electrical resistance (TEER, EVOM2™ Epithelial Voltohmmeter) was measured in real-time to evaluate the integrity of tight junction dynamics of the monolayer [21]. During the transepithelial procedure, the TEER value of (−30)GFP-Cre group was generally consistent with the benchmark value of control group (Fig. 2E), which indicated that negative charged (−30)GFP-Cre had no effect on the disruption of tight junction of epithelium. On the contrary, a significant decrease of TEER values was detected in LNP/GFP-Cre complexes in the first hour, suggesting that the tight junctions between the epithelial monolayer were opened and LNP/GFP-Cre complexes mainly penetrated the epithelium via paracellular approach. Subsequently, a progressive recovery of the TEER values was achieved in a time-dependent manner, indicating that the tight junctions of the cell monolayers was gradually recovered and LNP/GFP-Cre complexes could reversibly open the tight junction of epithelium. We also detected the tight junction changes of the cell monolayer through immunochemistry staining using EC16-63 lipidoid as a model (Fig. 2F). There was strong green fluorescence observed in LNP/GFP-Cre group in the first hour, indicating that there was an accumulation of LNPs at the surface of the epithelial cells. Moreover, the green fluorescence gradually decreased in a time-dependent manner, demonstrating that most of the LNP/GFP-Cre complexes had penetrated the cell monolayer seeded on the Transwell membranes and reached the bottom of the chamber. In contrast, after 24 h treatment with (−30)GFP-Cre alone, there remained an obvious green fluorescence at the surface, providing direct evidence that the (−30)GFP-Cre proteins were incapable of penetrating the cell monolayer. Meanwhile, a reduced red fluorescence could be seen in the first hour after incubation with the LNP/GFP-Cre complexes, indicating that the tight junctions were interpreted by the cationic lipidoids. Further, after treatment for 24 h, the tight junctions exhibited recovery to normal levels, which was probably related to the fact that the LNPs had penetrated the cell monolayer and could not interrupt the tight junctions. Taken together, we conclude that compared with (−30)GFP-Cre alone, the LNPs-mediated protein delivery prevented (−30)GFP-Cre from adhesive interactions with the mucin, facilitating accelerated crossing the mucus layer, and penetrated the cell monolayer seeded on the Transwell membrane through the interruption of stight junction without cell damage.
Finally, based on the results with the 2D Transwell model, we investigated the performance of the LNP/GFP-Cre complexes in the 3D tissue engineered intestinal system. Compared with the 2D Transwell model, the 3D system provides more accurate assessment of the behavior of nanoparticles, since the 3D systems geometrically mimic the architecture of the human intestine, while also providing physiological conditions that more closely mimic human conditions (e.g., formation of natural oxygen gradients, higher levels of mucous formation, and interactions with gut bacteria [13,22]). We utilized this tissue to evaluate the LNPs using EC16-63 lipidoid as models. First, the lumen surface of the 3D tissues was seeded with Caco-2/HT29-MTX cells and the bulk space was filled with HeLa-DsRed cells. Seconds, the 3D scaffold were perfused with LNP/GFP-Cre complexes, incubated for 48 h and then washed with PBS (Fig. 3A). As shown in Fig. 3B, green fluorescence generated from (−30)GFP-Cre was only observed at the surface of the lumen, revealing that the (−30)GFP-Cre accumulated on these surfaces but were incapable of reaching the bulk space. In contrast, after treatment with LNP/GFP-Cre complexes at different concentrations, an accumulation of green fluorescence was obtained in both the lumen and bulk space, demonstrating that the LNPs-mediated protein delivery penetrated the lumen and reach the deeper bulk space. More importantly, when (−30)GFP-Cre was delivered into the bulk space by LNPs, a the red fluorescence of DsRed was detected, indicating that the (−30)GFP-Cre was functioned as delivered based on intracellular gene recombination. The successful LNPs-mediated (−30)GFP-Cre protein delivery indicated that the 3D tissue models provided a useful platform for monitoring the behavior of the lipidoids nanoparticles. There are several key factors underlying the success of our LNP component to cross intestinal epithelium and intracellularly deliver genome editing proteins. First, our lipidiods exhibit superior advantages. Cationic lipidiods can complex with and protect the annionic charged GFP-Cre. Moreover, the positive charged molecule can open the negatively charged sites of tight junction proteins of cell monolayer, facilitating paracellular permeability of intestinal epithelium [23]. Cationic lipidiods can also contribute to the intracellular uptake and further endosomal escape of GFP-Cre. Second, the addition of PEG can uniquely endow nanoparticles with a mucoinert surface leading to the rapid mucus penetration [24]. Third, modifying our LNP with cholesterol could contribute to the stable structure of nanoparticle, thus benefiting the GFP-Cre delivery [25]. In addition, doping DOPE into LNP could further improve the cargo intracellular delivery and endosomal escape [26]. Therefore, based on the synergistic effect of lipidoid, PEG, cholesterol and DOPE, our LNP can successfully penetrate the intestinal epithelium and deliver genome editing proteins. To achieve the application in human, the LNP is designed to be ornamented with multifunction, e.g. anit-acid capacity, in the future.
Fig. 3.
(A) Illustration of the LNP/GFP-Cre (EC16-63) complexes delivered in 3D tissue systems. (B) The CLSM images of the scaffolds after treatment with (−30)GFP-Cre alone or different concentrations of complexes. The images were captured at the surface of the lumen and the bulk space of the 3D scaffolds, respectively. Red, DsRed signal; Green, (−30)GFP-Cre protein; Blue, DAPI. Scale bar: 200 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In summary, we used a combinatorial library approach to synthesize a series of cationic lipidoids as a protein delivery platform. Using the (−30)GFP-Cre proteins as a model, we found that the cationic lipidoids could protect the proteins from adhesive interactions of the mucus layer, subsequently penetrate the cell monolayer in 2D Transwell systems through the temporary interruption of tight junctions, and finally facilitate protein delivery into HeLa-DsRed cells seeded on the bottom of the chambers, achieving efficient intracellular gene recombination. Based on these 2D cell culture results with transwells, EC16-63 was selected for the test in 3D tissue model. After prefusion with the LNP/GFP-Cre complexes, the LNPs efficiently penetrated the cell monolayers on the surface of the lumen and reached the deeper bulk space to induce the expression of DsRed in the HeLa-DsRed cells through genetic recombination, indicating that the 3D system was beneficial for evaluating the performance of the LNPs. In summary, the combination of 2D and 3D cell and tissue culture provided a convenient platform to screen and validate potential LNPs, which were able to condense and deliver gene-editing proteins into stable nanoparticles, penetrate the intestinal cell monolayer, and maintain integrity to realize function.
2.1. Experimental methods
Formulation of LNPs. The lipidoids were synthesized according to previously reported methods [15,16,27]. Briefly, the hydrophilic tails (1,2-epoxydodecane, 1,2-epoxytetradecane, 1,2-epoxyhexadecane, and 1,2-epoxyoctadecane) and individual amine head groups were mixed in a 5 ml Teflon-lined glass screw-top vial at a molar ratio of 2.4:1 (epoxide:amine), followed by a reaction at 80 °C without solvent for 48 h. The mixtures were then cooled to room temperature and purified through the flash chromatography on silica gel. The LNPs were formulated by the lipidoid, cholesterol, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and DSPE-PEG2K at a mass ratio of 16:4:1:4 in the ethanol and then added to the sodium acetate solution (pH 5.0, 25 mM). The mixture was dialyzed against the pure water for 4 h using Slide-A-Lyzer MINI Dialysis Device (Millipore, 3.5 K MWCO, 0.1 ml).
Expression and purification of (-30)GFP-Cre protein. The plasmid harboring (−30)GFP-Cre protein was expressed in E. coli BL21 STAR (DE3)-competent cells (Life Technologies). The E. coli was incubated in LB broth containing 100 mg/ml ampicillin at 37 °C overnight. Afterwards, the culture was added with isopropyl β-d-1-thio-galactopyranoside (IPTG) and incubated at 20 °C overnight. Then, the cells were collected through centrifugation at 8000 g and resuspended in the lysis buffer ((20 mM Tris, 1 M KCl, 20% glycerol, pH 8.0). The cells were lysed by sonication and then centrifuged at 8,000 g for 15 min. Next, the precipitate was incubated with nickel-NTA resin at 4 °C for 30 min to capture His-tagged (−30)GFP-Cre protein. The resin was then transferred to a 20 ml-gravity column (Bio-Rad) and washed with 25 mM wash buffer (20 mM imidazole, 20 mM Tris, 1 M KCl, 20% glycerol, pH 8.0) for three times. Then, the column was washed with 20 ml elution buffer (250 mM imidazole, 20 mM Tris, 1 M KCl, 20% glycerol, pH 8.0) for five times. Finally, the purified protein was dialyzed against lysis buffer and then concentrated by an Amicon ultracentrifugal filter (Millipore; 100 KDa MWCO). After expression and purification, the (−30)GFP-Cre content was quantified through BCA Kit (Invitrogen).
Intracellular delivery of LNP/GFP-Cre complexes. The HeLa-DsRed cells were plated at 48-well plate at a density of 40,000 cells/well and incubated at 37 °C overnight. The LNPs were incubated with (−30)GFP-Cre at a mass ratio of 10:8 at room temperature for 15 min, forming stable complexes. The complexes containing 0.8 μg (−30)GFP-Cre were then added into individual wells for 6 h and then harvest for intracellular green fluorescence through flow cytometer (BD FACS Calibur, BD Science, CA). Similarly, for analysis of the gene recombination efficiency, the cells were treated with complexes under the same condition for 24 h, and then harvest for analyzing intracellular DsRed fluorescence using flow cytometer.
LNPs-mediated (-30)GFP-Cre protein delivery in 2D Transwell culture model. The Caco-2 and HT29-MTX cells (3:1) were planted on the Transwell membrane (Pore size: 0.4 μm; Costar Corp.) at a density of 2 × 105 cells/cm2 in the DEME culture medium containing 10% FBS and 10 μg/ml human transferrin until the Caco-2/HT29-MTX cells were cultured to 100% confluence where TEER values reached over 400 Ω/cm2. The HeLa-DsRed cells were seeded on the bottom of the 24-well plates at a density of 4 × 104 cells/well. Then the LNPs complexes containing 80 μg (−30)GFP-Cre protein were added into the upper chamber of the Transwell system for 48 h. The TEER value was measured using the Millicell ERS Voltohmmeter (Millpore) during the period of delivery. After 48 h, the HeLa-DsRed cells were collected and analyzed by the flow cytometer. The Transwell membrane was fixed with 4% paraformaldehyde (PFA, Santa Cruz), treated with 0.1% Triton X-100 in PBS solution, and then blocked with 5% BSA solution for 2 h. Afterwards, the membranes were stained with the anti-E-cadherin (2.5 μg/ml, Invitrogen) at 4 °C overnight and then treated with Alexa Fluo 594 goat-anti mouse secondary antibody for 1 h. Subsequently, the Transwell membranes were washed with PBS for three times and scanned using Leica SP2 confocal microscope (Leica Microsystems).
LNPs-mediated (-30)GFP-Cre protein delivery in 3D intestinal tissue models. The 3D in vitro tissue model adapted in the study was generated by incorporating human intestinal epithelial cells (Enterocyte-like Caco-2 and Goblet-like HT29-MTX cells) and HeLa-DsRed cells into 3D silk scaffolds. Briefly, 3D silk scaffolds with a hollow architecture were prepared using silk fibroin and PDMS molds which was reported in our previous studies [13,28]. Caco-2/HT29-MTX cells were seeded on the luminal surface of the scaffold; while HeLa-DsRed cells were resuspended in collagen gel and delivered into the bulk space of the 3D scaffolds for 10 days. Then the LNP/GFP-Cre complexes containing 100 μg (high concentration) or 60 μg (low concentration) (−30)GFP-Cre protein were added into individual 3D tissue systems and cultured for 48 h. Afterwards, the scaffolds were washed with PBS for 3 times and fixed with 4% PFA at 4 °C overnight. Subsequently, the scaffolds were cut into small pieces by a scissors and then scanned by Leica SP2 confocal microscopy (Leica Microsystems).
CRediT authorship contribution statement
Tao Yang: designed the experiments, performed experiments, Formal analysis, and organized data, and wrote the manuscript. Haobo Han: designed the experiments, performed experiments, Formal analysis, and organized data, and wrote the manuscript. Ying Chen: designed the experiments, performed experiments, Formal analysis, and organized data, and wrote the manuscript. Liu Yang: helped provide materials and conduct the experiments. Rachael Parker: helped provide materials and conduct the experiments. Yamin Li: helped provide materials and conduct the experiments, revised the manuscript. David L. Kaplan: generated the idea, provide the funding support, interpreted and, Formal analysis, and critically revised and finalized the manuscript. Qiaobing Xu: generated the idea, provide the funding support, interpreted and, Formal analysis, and critically revised and finalized the manuscript.
Declaration of competing interest
There are no conflicts of interest to declare.
Acknowledgement
Q. X. acknowledges the funding support by NIH Grant R01 EB027170-01. D. L. K. acknowledges the funding support by NIH grant 5U19AI131126-04.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.03.027.
Contributor Information
David L. Kaplan, Email: david.kaplan@tufts.edu.
Qiaobing Xu, Email: qiaobing.xu@tufts.edu.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Ensign L.M., Cone R., Hanes J. Oral drug delivery with polymeric nanoparticles: the gastrointestinal mucus barriers. Adv. Drug Deliv. Rev. 2012;64:557–570. doi: 10.1016/j.addr.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anselmo A.C., Gokarn Y., Mitragotri S. Non-invasive delivery strategies for biologics. Nat. Rev. Drug Discov. 2019;18:19–40. doi: 10.1038/nrd.2018.183. [DOI] [PubMed] [Google Scholar]
- 3.Niu Z., Conejos-Sánchez I., Griffin B.T., O'Driscoll C.M., Alonso M.J. Lipid-based nanocarriers for oral peptide delivery. Adv. Drug Deliv. Rev. 2016;106:337–354. doi: 10.1016/j.addr.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Chai G.H., Xu Y., Chen S.Q., Cheng B., Hu F.Q., You J., Du Y.Z., Yuan H. Transport mechanisms of solid lipid nanoparticles across Caco-2 cell monolayers and their related cytotoxicology. ACS Appl. Mater. Interfaces. 2016;8:5929–5940. doi: 10.1021/acsami.6b00821. [DOI] [PubMed] [Google Scholar]
- 5.Roger E., Lagarce F., Garcion E., Benoit J.-P. Lipid nanocarriers improve paclitaxel transport throughout human intestinal epithelial cells by using vesicle-mediated transcytosis. J. Contr. Release. 2009;140:174–181. doi: 10.1016/j.jconrel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Wong H.L., Bendayan R., Rauth A.M., Li Y., Wu X.Y. Chemotherapy with anticancer drugs encapsulated in solid lipid nanoparticles. Adv. Drug Deliv. Rev. 2007;59:491–504. doi: 10.1016/j.addr.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Qi J., Zhuang J., Lu Y., Dong X., Zhao W., Wu W. In vivo fate of lipid-based nanoparticles. Drug Discov. Today. 2017;22:166–172. doi: 10.1016/j.drudis.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Guimaraes P.P.G., Zhang R., Spektor R., Tan M., Chung A., Billingsley M.M., El-Mayta R., Riley R.S., Wang L., Wilson J.M., Mitchell M.J. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Contr. Release. 2019;28:404–417. doi: 10.1016/j.jconrel.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y., He H., Xia F., Li Y., Lu Y., Chen D., Qi J., Lu Y., Zhang W., Wu W. In vivo fate of lipid-silybin conjugate nanoparticles: implications on enhanced oral bioavailability. Nanomedicine. 2017;13:2643–2654. doi: 10.1016/j.nano.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Mak I.W., Evaniew N., Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- 11.Gamboa J.M., Leong K.W. In vitro and in vivo models for the study of oral delivery of nanoparticles. Adv. Drug Deliv. Rev. 2013;65:800–810. doi: 10.1016/j.addr.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimpel C., Teubl B., Absenger M., Meindl C., Fröhlich E., Leitinger G., Zimmer A., Roblegg E. Development of an advanced intestinal in vitro triple culture permeability model to study transport of nanoparticles. Mol. Pharm. 2014;11:808–818. doi: 10.1021/mp400507g. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y., Lin Y., Davis K.M., Wang Q., Rnjak-Kovacina J., Li C., Isberg R.R., Kumamoto C.A., Mecsas J., Kaplan D.L. Robust bioengineered 3D functional human intestinal epithelium. Sci. Rep. 2015;5:13708. doi: 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altınoğlu S.A., Wang M., Li K.Q., Li Y., Xu Q. Intracellular delivery of the PTEN protein using cationic lipidoids for cancer therapy. Biomater. Sci. 2016;4:1773–1780. doi: 10.1039/c6bm00580b. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Bolinger J., Yu Y., Glass Z., Shi N., Yang L., Wang M., Xu Q. Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles. Biomater. Sci. 2019;7:596–606. doi: 10.1039/c8bm00637g. [DOI] [PubMed] [Google Scholar]
- 16.Wang M., Alberti K., Sun S., Arellano C.L., Xu Q. Combinatorially designed lipid‐like nanoparticles for intracellular delivery of cytotoxic protein for cancer therapy. Angew Chem. Int. Ed. Engl. 2014;53:2893–2898. doi: 10.1002/anie.201311245. [DOI] [PubMed] [Google Scholar]
- 17.Yang T., Li C., Wang X., Zhao D., Huang Y. Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle. Bioactive Materials. 2020;5:1053–1061. doi: 10.1016/j.bioactmat.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Dsk D., Dong Samways H. Vol. 2. 2017. Fabrication of self-assembling nanofibers with optimal cell uptake and therapeutic delivery efficiancy; p. 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boegh M., Nielsen H.M. Mucus as a barrier to drug delivery–understanding and mimicking the barrier properties. Basic Clin. Pharmacol. Toxicol. 2015;116:179–186. doi: 10.1111/bcpt.12342. [DOI] [PubMed] [Google Scholar]
- 20.Lai S.K., Wang Y.Y., Hanes J. Mucus-penetrating nanoparticles for drug and gene delivery to mucosal tissues. Adv. Drug Deliv. Rev. 2009;61:158–171. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan B., Kolli A.R., Esch M.B., Abaci H.E., Shuler M.L., Hickman J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M., Lee J., Chen Y., Hoshino K. Studies of nanoparticle delivery with in vitro bio-engineered microtissues. Bioactive Materials. 2020;5:924–937. doi: 10.1016/j.bioactmat.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y., Mi F., Chen C., Chang W., Peng S., Liang H., Sung H. Preparation and characterization of nanoparticles shelled with chitosan for oral insulin delivery. Biomacromolecules. 2007;8(1):146–152. doi: 10.1021/bm0607776. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Lai K., Suk S., Pace A., Cone R., Hanes J. Addressing the PEG mucoadhesivity paradox to engineer nanoparticles that “slip” through the human mucus barrier. Angew. Chem. Int. Ed. 2008;47(50):9726–9729. doi: 10.1002/anie.200803526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Z., Fan J., Kim S., Bezouglaia O., Fartash A., Wu B., Aghaloo T., Lee M. Delivery of siRNA via cationic Sterosomes to enhance osteogenic differentiation of mesenchymal stem cells. J. Contr. Release. 2015;217:42–52. doi: 10.1016/j.jconrel.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun S., Wang M., Alberti K., Choy A., Xu Q. DOPE facilitates quaternized lipidoids (QLDs) for in vitro DNA delivery. Nanomed. Nanotechnol. Biol. Med. 2013;9(7):849–854. doi: 10.1016/j.nano.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q., Georgakoudi I., Liu D.R., Xu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Zhou W., Roh T., Estes M.K., Kaplan D.L. In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PloS One. 2017;12 doi: 10.1371/journal.pone.0187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.