Abstract
Objectives
Measurement of lipoprotein(a) [Lp(a)] is used in risk assessment of atherosclerotic cardiovascular disease (ASCVD). The aim of the current study was to evaluate performance characteristic of five different Lp(a) assays using the cobas c501 (Roche Diagnostics) analyzer.
Design and methods
Lp(a) was measured using five Lp(a) assays (Diazyme, Kamiya, MedTest, Randox, and Roche) configured to mg/dL units. Assays from Diazyme and Kamiya were also configured using nmol/L units in separate experiments. Studies included sensitivity, imprecision, linearity, method comparison, and evaluation of healthy subjects. Imprecision (intra-day, 20 replicates; inter-day, duplicates twice daily for five days) and linearity were evaluated using patient pools. Linearity assessed a minimum of five patient splits spanning the analytical measurement range (AMR). Method comparison used 80 residual serum samples. Specimens from 120 self-reported healthy subjects (61 females / 59 males) were also tested. Method comparison for two assays in nmol/L units was conducted using 96 residual serum samples.
Results
Assay sensitivities met all manufacturer claims. Imprecision studies demonstrated %CVs ranging from 2.5 to 5.2% for the low pool (average concentration from 7.3 to 12.4 mg/dL); high pool %CVs ranged from 0.8 to 3.0% (average concentrations from 31.5–50.2 mg/dL). Linearity was confirmed for all assays. Variation in accuracy was observed when comparing results to an all method average. Lp(a) results were higher in females versus males in self-reported healthy subjects.
Conclusions
All assays performed according to manufacturer described performance characteristics, although differences were observed across Lp(a) assays tested when compared to an all method average.
Keywords: Lipoprotein, Lipoprotein(a), Atherosclerotic cardiovascular disease, Lipids, Harmonization, Standardization
Abbreviations: AMR, analytical measurement range; apo(a), apolipoprotein(a); apoB-100, apolipoprotein B-100; ASCVD, antherosclerotic cardiovascular disease; CV, coefficient of variation; ELISA, enzyme linked immunosorbent assay; IFCC, International Federation of Clinical Chemistry; IFE, immunofixation electrophoresis; KIV2, kringle-4 type 2; LDL, low density lipoprotein; Lp(a), lipoprotein(a); NLMDRL, Northwest Lipid Metabolism and Diabetes Research Laboratories; R, correlation coefficient; VNTR, variable number of tandem repeat
Highlights
-
•
Five automated assays for Lp(a) measurement (mg/dL units) were compared.
-
•
Differences in accuracy were observed across the methods investigated.
-
•
Two assays were also compared using nmol/L units.
-
•
More Lp(a) assay traceability to the international reference material is needed.
1. Introduction
Lipoprotein(a) [Lp(a)] is a low-density lipoprotein (LDL)-like particle that contains the glycoprotein apolipoprotein(a) [apo(a)] covalently linked by a disulfide bond to apolipoprotein B-100 [apoB-100] [[1], [2], [3]]. Apo(a) shows homology to plasminogen, including a variable number of tandem repeat (VNTR) kringle-4 type 2 (KIV2) domains [4]. The varying number of KIV2 domains contributes to the heterogeneity of Lp(a) size between individuals. These different size Lp(a) particles are commonly referred to as Lp(a) “isoforms” [1].
Increased serum Lp(a) is considered an independent, inherited risk factor for atherosclerotic cardiovascular disease (ASCVD), including myocardial infarction and stroke [[5], [6], [7], [8]]. This risk is believed to be due to pro-atherogenic, pro-inflammatory, and pro-thrombotic properties of the Lp(a) particle [7,8]. While the apo(a) gene is a significant determinant of plasma Lp(a) concentration, variation in genetic and phenotypic associations have been observed across populations and ethnicities [[9], [10], [11], [12], [13]]. Genetic studies have also suggested that along with increased Lp(a) concentration, smaller isoform size may also be an independent risk factor for ASCVD [14].
Lp(a) can be measured using a variety of methods, including enzyme linked immunosorbent assays (ELISA) [15,16], latex-agglutination [15], immunoturbidimetry/immunonephelometry [17], electrophoresis [18], and immunofixation electrophoresis (IFE) [19]. Despite the development of a secondary reference material to assist in standardization of commercial Lp(a) assays [20,21], harmonization of Lp(a) remains elusive due to the size and composition heterogeneity of Lp(a), as well as the antibodies, methods, and calibration strategies used across kits [22,23]. A confounding factor is that Lp(a) can be reported in either mass units (mg/dL of the total Lp(a) particle) or molar concentration of particles (nmol/L) [24,25]. As each individual inherits two copies of the Lp(a) gene (with co-dominant expression), more than one Lp(a) isoform is often present in an individual.
To evaluate for differences in performance across automated clinical assays for Lp(a) measurement, the present study was conducted comparing immunoassays from five different vendors configured on a cobas c501 (Roche Diagnostics; Indianapolis, IN) chemistry instrument. Experiments included sensitivity, imprecision, linearity, method comparison, and an analysis of otherwise healthy donor specimens.
2. Materials and Methods
2.1. Assays
All testing was performed on a cobas c501 chemistry analyzer configured to Lp(a) assay manufacturer-provided instrument parameters. The Diazyme Lp(a) assay (La Jolla, CA) is a latex particle enhanced immunoturbidimetric assay using 5 level calibration (analytical measurement range (AMR), 5.4–100 mg/dL) [26]. The K-Assay Lp(a) assay (Kamiya Biomedical; Seattle, WA) is an immunoturbidimetric assay using multi-point calibration made with dilutions from a reconstituted high calibrator (AMR, 5–150 mg/dL) [27]. The Point Scientific Lp(a) assay (MedTest; Canton, MI) is a latex particle enhanced immunoturbidimetric assay using 5 level calibration (AMR, 5.4–100 mg/dL) [28]. The Randox Lp(a) assay (Crumlin, United Kingdom) is an immunoturbidimetric assay licensed from Denka Seiken using a 5 point calibration (AMR, 3–90 mg/dL). The Roche Lp(a) assay is a particle enhanced immunoturbidimetric assay using 5 level calibration (AMR, 6–80 mg/dl). The Diazyme and Randox assays were also configured using vendor-defined calibrators in nmol/L units for separate method comparison experiments. The Randox Lp(a) assay in nmol/L units is traceable to the WHO/IFCC SRM 2B Lp(a) reference material [29].
2.2. Sensitivity
Sensitivity was assessed by running ten replicates of zero material (de-ionized water) and three replicates of non-zero material (e.g. respective manufacturer low calibrator or a low patient serum pool) using the sensitivity module in EP Evaluator 12 (Data Innovations; South Burlington, VT).
2.3. Imprecision
Intra-assay imprecision was assessed by testing low and high-concentration Lp(a) patient serum pools for 20 replicates in a single day. Inter-assay imprecision was assessed by testing low and high-concentration Lp(a) patient serum pools in duplicate twice daily, with a minimum of 2 hours between each run, for five days. Inter-assay imprecision was assessed using the complex precision module in EP Evaluator to determine for each pool the total, within run, between run, and between day imprecision. Acceptable imprecision criteria was defined as a percent coefficient of variation (%CV) ≤ 10%.
2.4. Linearity
Linearity was assessed by testing a minimum of five concentrations of high patient pool diluted with a low patient pool and tested in duplicate (or triplicate where additional reagent was available). Slopes and intercepts were determined using linear regression in SigmaPlot 14 (Systat; San Jose, CA).
2.5. Method comparison
Method comparisons were performed using residual adult serum samples stored at −20 °C (n = 80; 40 males, 40 females; 57.5 ± 17.9 years old) obtained in accordance with an IRB-approved protocol for use of de-identified clinical specimens. Prior to testing in singlicate, specimens were thawed, mixed, centrifuged, and checked for clots. Specimens were excluded (n = 17) from data analysis if they were below (n = 16) or above (n = 1) the AMR for any of the 5 assays. In the absence of a gold standard method, the average Lp(a) result of all methods was used as the comparator result for accuracy assessment [30,31], which was assessed using the alternate (quantitative) method comparison module in EP Evaluator. Deming regression was performed and slope, intercept, correlation coefficient (R), bias, and percent bias (%) were calculated. Method comparisons were plotted for each individual assay pair, as presented in the Supplementary Figures. Residual adult serum samples (n = 96; 49 males, 47 females; 57.4 ± 15.0 years old) were also tested on the Randox and Diazyme assays, configured with nmol/L reporting units. These specimens were concurrently tested using the Roche assay (configured with mg/dL reporting units) for comparison purposes. A frequency distribution of Randox (nmol/L) results divided by Roche (mg/dL) results [i.e. Lp(a) ratios] was generated using 0.05 bin sizes.
2.6. Self-declared healthy volunteers
Analysis of serum Lp(a) concentrations in self-declared healthy volunteers was conducted for all assays using specimens obtained from 120 individuals – 59 males (age, 40.2 ± 10.4 years old) and 61 females (age, 38.5 ± 11.4 years old) – according to an IRB-approved protocol for freshly collected specimens. These specimens had been obtained with a primary objective of establishing a new reference interval for chromogranin A. As such, exclusion criteria included any history of cancer or impaired renal function, as well as use of any proton pump inhibitors in the prior two weeks. Lp(a) results for healthy volunteers (mg/dL units) is presented in histograms by gender, and in tabular format as mean ± SD, median, 80th percentile, and 95th percentile.
2.7. Data analysis
Data analysis was conducted in EP Evaluator 12, Excel Office 365 (Microsoft; Redmond, WA), and SigmaPlot 14. Method comparison was assessed using Deming regression. Graphs were prepared in SigmaPlot. Individual (direct) method comparison graphs were generated in EP Evaluator. Data are presented as mean ± standard deviation (SD), unless otherwise indicated. Statistical comparison of healthy volunteer Lp(a) results was conducted using the Mann-Whitney rank sum test in SigmaPlot due to nonparametric data distributions. Statistical significance was defined as p < 0.05.
3. Results
All methods met manufacturer claims regarding sensitivity: observed (manufacturer claim): Diazyme, 0.7 mg/dL (1.3 mg/dL); Kamiya, 1.2 mg/dL (5.0 mg/dL); MedTest, 0.2 mg/dL (1.3 mg/dL); Randox, 0.7 mg/dL (3.0 mg/dL); Roche, 0.3 mg/dL (4.0 mg/dL). Lp(a) assays also demonstrated acceptable imprecision and met manufacturers’ claims, with CVs less than 6% in all cases (Table 1).
Table 1.
Assay imprecision.
| Method | Patient Pools | Mean Concentration (mg/dL) | Total Imprecision (SD) | Total Imprecision (%CV) | Within Run (%CV) | Between Run (%CV) | Between Day (%CV) |
|---|---|---|---|---|---|---|---|
| Diazyme | Low | 12.4 | 0.31 | 2.5 | 2.1 | 1.2 | 0.8 |
| High | 50.2 | 0.98 | 2.0 | 1.4 | 0.0 | 1.4 | |
| Kamiya | Low | 8.2 | 0.43 | 5.2 | 4.7 | 0.0 | 2.3 |
| High | 28.6 | 0.86 | 3.0 | 2.9 | 0.0 | 0.8 | |
| MedTest | Low | 12.3 | 0.48 | 3.9 | 2.8 | 2.8 | 0.0 |
| High | 47.5 | 0.55 | 1.1 | 0.8 | 0.9 | 0.0 | |
| Randox | Low | 9.5 | 0.32 | 3.3 | 3.2 | 0.7 | 0.0 |
| High | 35.7 | 0.29 | 0.8 | 0.5 | 0.5 | 0.4 | |
| Roche | Low | 7.3 | 0.31 | 4.3 | 4.0 | 0.0 | 1.6 |
| High | 31.5 | 0.36 | 1.1 | 0.8 | 0.8 | 0.0 |
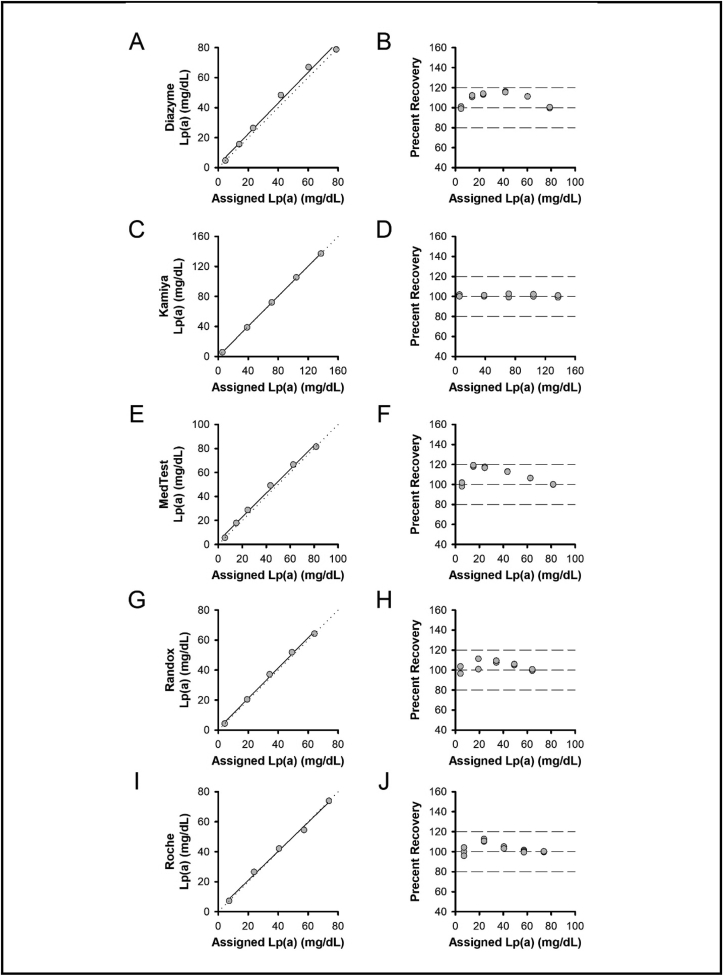
As shown in Fig. 1, all assays were linear (slope, intercept): Diazyme (Fig. 1A,B), 1.03, 2.05 mg/dL; Kamiya (Fig. 1C,D), 1.01, 0.25 mg/dL; MedTest (Fig. 1E,F), 1.00, 2.82 mg/dL; Randox (Fig. 1G,H), 1.01, 1.00 mg/dL; Roche (Fig. 1I,J), 0.99, 1.60 mg/dL.
Fig. 1.
Linearity. Linear regression (left panels) and percent recovery (right panels) for Lp(a) assays from Diazyme (A, B), Kamiya (C, D), MedTest (E, F), Randox (G, H), and Roche (I, J). Left panels, solid line shows linear regression and dotted line represents unity. Right panels, dashed lines represent 80%, 100%, and 120% recovery.
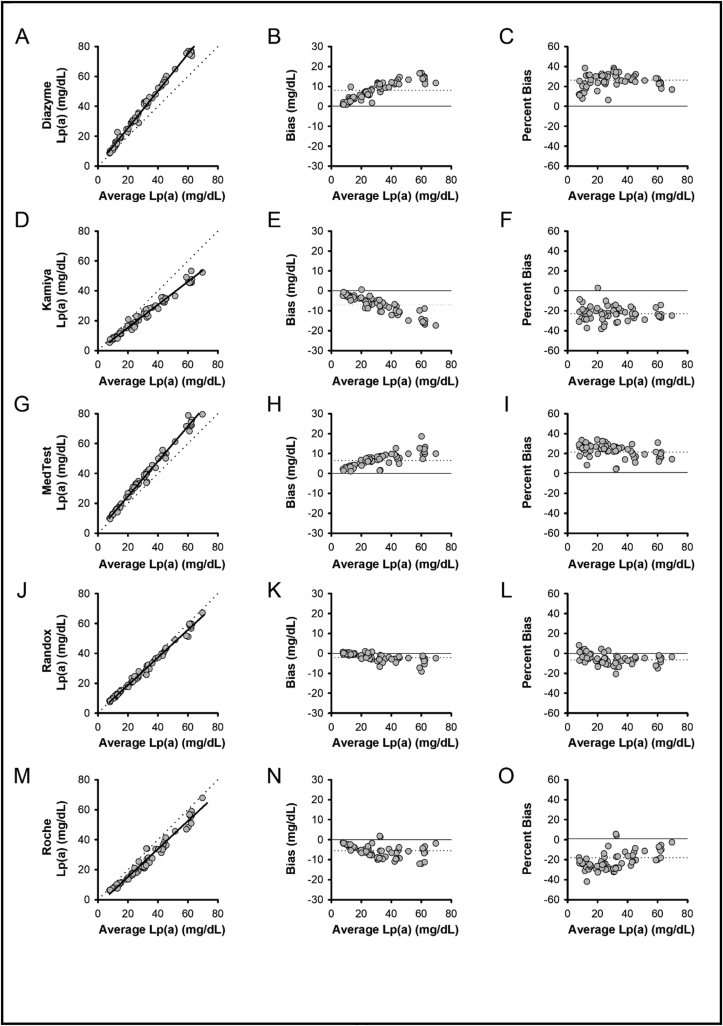
Accuracy assessment was performed by comparing assays to the average Lp(a) result across all methods (Fig. 2). As compared to all method averages, positive bias was observed for Diazyme (26.3%; Fig. 2A-C) and MedTest (21.4%; Fig. 2G-I) assays, whereas negative bias was observed for Kamiya (−23.0%; Fig. 2D-F) and Roche (−18.1%; Fig. 2M-O) assays. Results from the Randox assay most closely matched the all method average (−6.7%; Fig. 2J-L).
Fig. 2.
Method Comparisons. Results from Lp(a) assays versus the average of all methods showing method comparison (A, D, G, J, M), absolute bias (B, E, H, K, N), and percent bias (C, F, I, L, O) for Diazyme (A–C), Kamiya (D–F), MedTest (G–I), Randox (J–L), and Roche (M-O). Left panel method comparisons: solid lines show Deming regression, dotted lines represent unity. Middle panel absolute bias plots: solid line represents zero bias, dotted lines represent absolute bias. Right panel percent bias plots: solid line represents zero bias, dotted lines represent percent bias.
Summary of method comparison results is presented in Table 2. All methods demonstrated correlation coefficients of greater than 0.98.
Table 2.
Method comparison summary results.
| Method | Slope | Intercept (mg/dL) | R | Bias (mg/dL) | % Bias |
|---|---|---|---|---|---|
| Diazyme | 1.238 | 0.763 | 0.9954 | 8.03 | 26.4 |
| Kamiya | 0.770 | 0.009 | 0.9899 | −6.99 | −23.0 |
| MedTest | 1.170 | 1.327 | 0.9949 | 6.51 | 21.4 |
| Randox | 0.926 | 0.213 | 0.9956 | −2.03 | −6.7 |
| Roche | 0.931 | −3.419 | 0.9849 | −5.51 | −18.1 |
Individualized (direct) method comparisons across all assays are included in the Supplementary Figures.
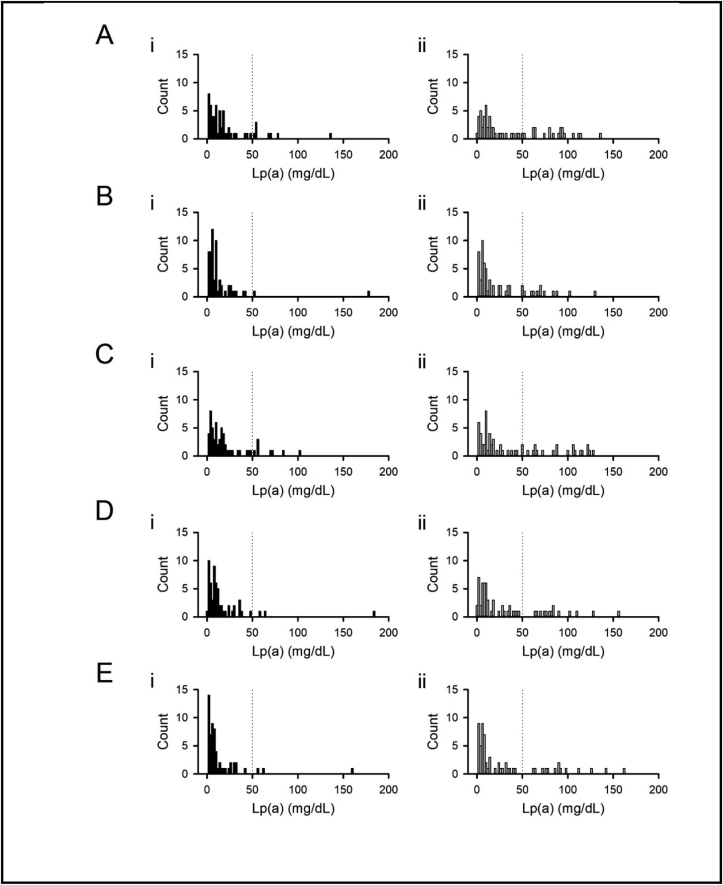
Lp(a) results from 120 self-disclosed healthy donors (61 females and 59 males) are presented by gender in Fig. 3 with summary data included in Table 3. Female population Lp(a) results were higher than male Lp(a) results across all assays (Diazyme, p = 0.032; Kamiya, p = 0.035; MedTest, p = 0.040; Randox, p = 0.041; Roche, p = 0.025).
Fig. 3.
– Lp(a) Results from Self-Disclosed Volunteers. Lp(a) results from male (i, left) and female (ii, right) volunteers when tested using Diazyme (A), Kamiya (B), MedTest (C), Randox (D), and Roche (E) assays. Dotted line, 50 mg/dL cutoff.
Table 3.
Self-disclosed healthy volunteer Lp(a) results.
|
I. Males (n = 59) – Lp(a), mg/dL | ||||
|---|---|---|---|---|
| Assay | Mean ± SD | Median | 80th Percentile | 95th Percentile |
| Diazyme | 20.0 ± 24.5 | 12.2 | 29.5 | 67.3 |
| Kamiya | 13.5 ± 24.3 | 6.9 | 16.4 | 39.6 |
| MedTest | 20.7 ± 22.5 | 13.0 | 33.2 | 70.1 |
| Randox | 15.7 ± 26.2 | 8.3 | 22.3 | 47.5 |
| Roche |
13.2 ± 23.2 |
6.0 |
18.4 |
41.9 |
| II. Females (n = 61) – Lp(a), mg/dL | ||||
|
Assay |
Mean ± SD |
Median |
80th Percentile |
95th Percentile |
| Diazyme | 36.4 ± 37.2 | 16.2 | 79.9 | 105.5 |
| Kamiya | 25.0 ± 29.4 | 9.9 | 49.8 | 83.2 |
| MedTest | 39.2 ± 40.6 | 17.2 | 83.3 | 121.6 |
| Randox | 30.6 ± 36.5 | 11.3 | 64.8 | 101.2 |
| Roche | 30.4 ± 39.6 | 8.2 | 63.6 | 110.4 |
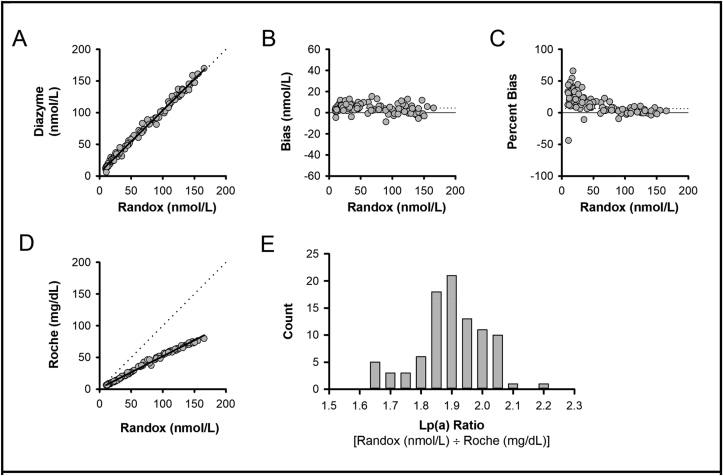
The Randox and Diazyme assays were then configured and calibrated per vendor specifications to report in nmol/L reporting units. A total of 47 residual adult female specimens and 49 residual adult male specimens were tested on the Randox and Diazyme assays (configured with nmol/L reporting units; Fig. 4A-C; slope 0.983, intercept 5.45 nmol/L, R 0.9958, bias 4.30 nmol/L, percent bias 6.5%). These specimens were concurrently tested using the Roche assay (configured with mg/dL reporting units), for comparison purposes [Fig. 4D; Randox (nmol/L) versus Roche (mg/dL); slope 0.505, R 0.9963, bias not shown due to differing units]. Using this comparison for assays with different reporting units, a frequency distribution of Lp(a) ratios [Randox (nmol/L)/Roche (mg/dL)] was generated (Fig. 4E).
Fig. 4.
– Comparison of Lp(a) Results Using Diazyme (nmol/L), Randox (nmol/L), and Roche (mg/dL) Assay Configurations. Results from the Randox Lp(a) assay versus the Diazyme Lp(a) assay showing method comparison (A), absolute bias (B), and percent bias (C). (A) Solid line shows Deming regression, dotted line represents unity. (B) Solid line represents zero bias, dotted line represents absolute bias. (C) Solid line represents zero bias, dotted line represents percent bias. (D) Method comparison of Randox (x-axis; nmol/L) versus Roche (y-axis; mg/dL). Solid line shows Deming regression, dotted line represent unity. (E) Frequency distribution of Lp(a) ratios [Randox Lp(a) (nmol/L)/Roche Lp(a) (mg/dL)].
4. Discussion
The present study provides a comparative analysis of five automated immunoturbidimetric assays for Lp(a) measurement. While sensitivity, linearity, and precision performed per manufacturer specifications, differences in accuracy were observed across methods. Lp(a) size heterogeneity and lack of assay standardization continue to impact Lp(a) assay comparability [23]. Differences in patient Lp(a) results across assays are multifactorial and likely due to lack of assay harmonization, inability to harmonize assays in mg/dL reporting units due to Lp(a) isoform size heterogeneity, co-dominant expression of the Lp(a) gene, vendor-specific calibrators and calibration set point strategies to compensate for expected Lp(a) size differences observed in patient specimens, and vendor-specific assay configurations. Global availability of assays harmonized to the international reference material using nmol/L reporting units will help to minimize differences seen across assays.
In order to improve harmonization in Lp(a) assays, the Working Group for the Standardization of Lipoprotein(a) Assays was formed by the International Federation of Clinical Chemistry (IFCC) [21]. After extensive evaluation, a secondary reference material for Lp(a) was proposed (IFCC SRM 2B) [20], and it was subsequently accepted by the WHO Expert Committee on Biological Standardization [i.e. Lp(a) SRM 2B] [32]. It is important to emphasize that due to Lp(a) isoform heterogeneity, this secondary reference material can only be used to standardize assays in nmol/L and not mg/dL units. Size heterogeneity remains a critical limitation in achieving assay harmonization [33]. Many vendors have participated in the harmonization activities described above, as well as initiatives from the Northwest Lipid Metabolism and Diabetes Research Laboratories (NLMDRL) [15,22,23]. Other approaches include use of common calibrator materials or antibodies from previously characterized assays (e.g. Denka Seiken), or use of previously characterized methods as predicate devices to establish substantial equivalence in regulatory approvals.
While all assays passed acceptability criteria for linearity experiments, deviations from 100% recovery were observed in several assays (Fig. 1; percent recovery panels). It should be noted, however, that Lp(a) isoform size is typically inversely correlated to Lp(a) concentration (i.e. small isoforms with less KIV2 repeats are typically associated with higher serum Lp(a) concentrations, whereas larger isoforms with more KIV2 repeats are typically associated with lower serum Lp(a) concentrations) [34]. Because high Lp(a) concentration specimens are often used as source materials for calibrator development, smaller isoform sizes may be over-represented. This can lead to under-quantifying high Lp(a) concentrations and over-quantifying low Lp(a) concentrations when using assays that are not isoform independent [24]. Assay manufacturers may also set or adjust calibration points to compensate for isoform size-dependent differences expected in varying Lp(a) concentrations [3,22,24]. In this context, the observed differences in percent recovery (Fig. 1B,F,H,J) does not necessarily represent non-linearity, but rather they could also reflect compensation in calibrator set points to more accurately measure serum Lp(a) concentrations in actual (non-diluted) clinical specimens.
Not all assays evaluated in the present report support nmol/L reporting per the manufacturers. For example, MedTest [28] and Kamiya [27] only provide calibrator set points in mg/dL units, Diazyme [26] and Randox [35] provide calibrator set points in either mg/dL or nmol/L units, whereas Roche provides nmol/L calibrator set points internationally [36] but only mg/dL calibrator set points in the United States [37]. Conversion factors are discouraged due to risk of introducing inaccuracies caused by variation in Lp(a) isoform distribution, mass, and composition [22,25]. The Randox (nmol/L)/Roche (mg/dL) Lp(a) ratio distributions shown in Fig. 4E, along with those previously described elsewhere [24], emphasize this important point. An important step toward greater harmonization in Lp(a) reporting will be availability of nmol/L calibration set points by all assay manufacturers and to all clinical laboratory settings. The method comparison experiments in the present report emphasize the importance of moving toward concentration-based (nmol/L) reporting.
Population Lp(a) distributions vary based on ethnicity as does association with ASCVD risk [6,13]. The exact Lp(a) concentrations used as clinical cutoffs associated with ASCVD risk have varied across studies [38]. As examples, 30 mg/dL and more frequently 50 mg/dL cutoffs – the latter corresponding to either 100 nmol/L [38] or 125 nmol/L [39], depending on the 80th percentile population estimates – are commonly described and vary based on study design and ethnicity of population under evaluation [40,41]. Summaries of guidelines and consensus statements on Lp(a) cutoffs, testing, and therapeutic recommendations have previously been reported [38,40].
A limitation of the present report is that we do not have ethnicity or clinical history information available for self-reported healthy volunteers evaluated. We therefore cannot determine why larger Lp(a) results were observed in the population of females tested. Previously published reports have provided conflicting results on factors that may influence Lp(a) concentrations in females (e.g. pregnancy, menopause, or hormone replacement therapy) [13,[42], [43], [44], [45], [46]]. We also do not have data related to the Lp(a) isoform or genotype for these specimens. Finally, as most assays were configured using mg/dL calibrator set points, results are not directly traceable to the Lp(a) SRM 2B reference material.
CRediT authorship contribution statement
Sara P. Wyness: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Jonathan R. Genzen: Conceptualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing, Supervision.
Declaration of competing interest
JRG, contract research support to ARUP Laboratories from Fujirebio Diagnostics. For the present study, Lp(a) reagents were supplied by Diazyme and Roche for their respective assays in the five assay comparison studies. This work was supported by the ARUP Institute for Clinical and Experimental Pathology.
Acknowledgements
The authors would like to acknowledge Lp(a) reagent support from Diazyme and Roche.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.plabm.2021.e00218.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Utermann G. The mysteries of lipoprotein(a) Science. 1989;246(4932):904–910. doi: 10.1126/science.2530631. [DOI] [PubMed] [Google Scholar]
- 2.Berg K. Lp(a) lipoprotein: an overview. Chem. Phys. Lipids. 1994;67–68:9–16. doi: 10.1016/0009-3084(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 3.Wieringa G. Lipoprotein(a): what’s in a measure? Ann. Clin. Biochem. 2000;37(Pt 5):571–580. doi: 10.1258/0004563001899799. [DOI] [PubMed] [Google Scholar]
- 4.Eaton D.L., Fless G.M., Kohr W.J., McLean J.W., Xu Q.T., Miller C.G., Lawn R.M., Scanu A.M. Partial amino acid sequence of apolipoprotein(a) shows that it is homologous to plasminogen. Proc. Natl. Acad. Sci. U. S. A. 1987;84(10):3224–3228. doi: 10.1073/pnas.84.10.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta A., Virani S.S., Ayers C.R., Sun W., Hoogeveen R.C., Rohatgi A., Berry J.D., Joshi P.H., Ballantyne C.M., Khera A. Lipoprotein(a) and family history predict cardiovascular disease risk. J. Am. Coll. Cardiol. 2020;76(7):781–793. doi: 10.1016/j.jacc.2020.06.040. [DOI] [PubMed] [Google Scholar]
- 6.Pearson K., Rodriguez F. Lipoprotein(a) and cardiovascular disease prevention across diverse populations. Cardiol. Ther. 2020;9:275–292. doi: 10.1007/s40119-020-00177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsimikas S. A test in context: lipoprotein(a): diagnosis, prognosis, controversies, and emerging therapies. J. Am. Coll. Cardiol. 2017;69(6):692–711. doi: 10.1016/j.jacc.2016.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Shah N.P., Pajidipati N.J., McGarrah R.W., Navar A.M., Vemulapalli S., Blazing M.A., Shah S.H., Hernandez A.F., Patel M.R. Lipoprotein (a): an update on a marker of residual risk and associated clinical manifestations. Am. J. Cardiol. 2020;126:94–102. doi: 10.1016/j.amjcard.2020.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boerwinkle E., Leffert C.C., Lin J., Lackner C., Chiesa G., Hobbs H.H. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 1992;90(1):52–60. doi: 10.1172/JCI115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Utermann G., Duba C., Menzel H.J. Genetics of the quantitative Lp(a) lipoprotein trait. II. Inheritance of Lp(a) glycoprotein phenotypes. Hum. Genet. 1988;78(1):47–50. doi: 10.1007/BF00291233. [DOI] [PubMed] [Google Scholar]
- 11.Utermann G., Kraft H.G., Menzel H.J., Hopferwieser T., Seitz C. Genetics of the quantitative Lp(a) lipoprotein trait. I. Relation of LP(a) glycoprotein phenotypes to Lp(a) lipoprotein concentrations in plasma. Hum. Genet. 1988;78(1):41–46. doi: 10.1007/BF00291232. [DOI] [PubMed] [Google Scholar]
- 12.Mooser V., Scheer D., Marcovina S.M., Wang J., Guerra R., Cohen J., Hobbs H.H. The Apo(a) gene is the major determinant of variation in plasma Lp(a) levels in African Americans. Am. J. Hum. Genet. 1997;61(2):402–417. doi: 10.1086/514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enkhmaa B., Anuurad E., Berglund L. Lipoprotein (a): impact by ethnicity and environmental and medical conditions. J. Lipid Res. 2016;57(7):1111–1125. doi: 10.1194/jlr.R051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleheen D., Haycock P.C., Zhao W., Rasheed A., Taleb A., Imran A., Abbas S., Majeed F., Akhtar S., Qamar N., Zaman K.S., Yaqoob Z., Saghir T., Rizvi S.N.H., Memon A., Mallick N.H., Ishaq M., Rasheed S.Z., Memon F.U., Mahmood K., Ahmed N., Frossard P., Tsimikas S., Witztum J.L., Marcovina S., Sandhu M., Rader D.J., Danesh J. Apolipoprotein(a) isoform size, lipoprotein(a) concentration, and coronary artery disease: a mendelian randomisation analysis. Lancet Diabetes Endocrinol. 2017;5(7):524–533. doi: 10.1016/S2213-8587(17)30088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcovina S.M., Lippi G., Guidi G. Lipoprotein(a) immunoassays: comparison of a semi-quantitative latex method and two monoclonal enzyme immunoassays. Int. J. Clin. Lab. Res. 1995;25(4):201–204. doi: 10.1007/BF02592698. [DOI] [PubMed] [Google Scholar]
- 16.Stroop D.M., Glueck C.J., McCray C., Speirs J., Schumacher H.R. Measurement of lipoprotein (a): comparison of Macra and Imubind methods. Ann. Clin. Lab. Sci. 1996;26(4):329–339. [PubMed] [Google Scholar]
- 17.Scharnagl H., Stojakovic T., Dieplinger B., Dieplinger H., Erhart G., Kostner G.M., Herrmann M., Marz W., Grammer T.B. Comparison of lipoprotein (a) serum concentrations measured by six commercially available immunoassays. Atherosclerosis. 2019;289:206–213. doi: 10.1016/j.atherosclerosis.2019.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Baudhuin L.M., Hartman S.J., O’Brien J.F., Meissner I., Galen R.S., Ward J.N., Hogen S.M., Branum E.L., McConnell J.P. Electrophoretic measurement of lipoprotein(a) cholesterol in plasma with and without ultracentrifugation: comparison with an immunoturbidimetric lipoprotein(a) method. Clin. Biochem. 2004;37(6):481–488. doi: 10.1016/j.clinbiochem.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Guadagno P.A., Summers Bellin E.G., Harris W.S., Dayspring T.D., Hoefner D.M., Thiselton D.L., Stanovick B., Warnick G.R., McConnell J.P. Validation of a lipoprotein(a) particle concentration assay by quantitative lipoprotein immunofixation electrophoresis. Clin. Chim. Acta. 2015;439:219–224. doi: 10.1016/j.cca.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Tate J.R., Berg K., Couderc R., Dati F., Kostner G.M., Marcovina S.M., Rifai N., Sakurabayashi I., Steinmetz A. International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) standardization project for the measurement of lipoprotein(a). Phase 2: selection and properties of a proposed secondary reference material for lipoprotein(a) Clin. Chem. Lab. Med. 1999;37(10):949–958. doi: 10.1515/CCLM.1999.140. [DOI] [PubMed] [Google Scholar]
- 21.Tate J.R., Rifai N., Berg K., Couderc R., Dati F., Kostner G.M., Sakurabayashi I., Steinmetz A. International Federation of Clinical Chemistry standardization project for the measurement of lipoprotein(a). Phase I. Evaluation of the analytical performance of lipoprotein(a) assay systems and commercial calibrators. Clin. Chem. 1998;44(8 Pt 1):1629–1640. [PubMed] [Google Scholar]
- 22.Marcovina S.M., Albers J.J. Lipoprotein (a) measurements for clinical application. J. Lipid Res. 2016;57(4):526–537. doi: 10.1194/jlr.R061648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcovina S.M., Albers J.J., Scanu A.M., Kennedy H., Giaculli F., Berg K., Couderc R., Dati F., Rifai N., Sakurabayashi I., Tate J.R., Steinmetz A. Use of a reference material proposed by the International Federation of Clinical Chemistry and Laboratory Medicine to evaluate analytical methods for the determination of plasma lipoprotein(a) Clin. Chem. 2000;46(12):1956–1967. [PubMed] [Google Scholar]
- 24.Tsimikas S., Fazio S., Viney N.J., Xia S., Witztum J.L., Marcovina S.M. Relationship of lipoprotein(a) molar concentrations and mass according to lipoprotein(a) thresholds and apolipoprotein(a) isoform size. J Clin Lipidol. 2018;12(5):1313–1323. doi: 10.1016/j.jacl.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 25.McConnell J.P., Guadagno P.A., Dayspring T.D., Hoefner D.M., Thiselton D.L., Warnick G.R., Harris W.S. Lipoprotein(a) mass: a massively misunderstood metric. J Clin Lipidol. 2014;8(6):550–553. doi: 10.1016/j.jacl.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Diazyme Lipoprotein (a) Assay Package Insert. 70980 Rev. C. Diazyme Laboratories: Poway, CA.
- 27.K-Assay Lp(a) Package Insert. Rev. 2009-09-16. Kamiya Biomedical: Seattle, WA.
- 28.Pointe Scientific Lipoprotein (a) Assay Package Insert. Rev. 10/18. MedTest: Canton, MI.
- 29.Randox lipoprotein(a) assay. https://www.randox.com/lipoproteina/ Accessed 19 February 2021.
- 30.Reitsma J.B., Rutjes A.W., Khan K.S., Coomarasamy A., Bossuyt P.M. A review of solutions for diagnostic accuracy studies with an imperfect or missing reference standard. J. Clin. Epidemiol. 2009;62(8):797–806. doi: 10.1016/j.jclinepi.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Rutjes A.W., Reitsma J.B., Coomarasamy A., Khan K.S., Bossuyt P.M. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol. Assess. 2007;11(50) doi: 10.3310/hta11500. iii, ix-51. [DOI] [PubMed] [Google Scholar]
- 32.Dati F., Tate J.R., Marcovina S.M., Steinmetz A., International Federation of Clinical Chemistry and Laboratory Medicine. IFCC Working Group for Lipoprotein(a) Assay Standardization. First WHO/IFCC international reference reagent for lipoprotein(a) for immunoassay--Lp(a) SRM 2B. Clin. Chem. Lab. Med. 2004;42(6):670–676. doi: 10.1515/CCLM.2004.114. [DOI] [PubMed] [Google Scholar]
- 33.Dati F., Tate J.R. Reference materials for the standardization of the apolipoproteins A-1 and B, and lipoprotein(a) EJIFCC. 2001;13(3):73–79. [PMC free article] [PubMed] [Google Scholar]
- 34.Utermann G., Menzel H.J., Kraft H.G., Duba H.C., Kemmler H.G., Seitz C. Lp(a) glycoprotein phenotypes. Inheritance and relation to Lp(a)-lipoprotein concentrations in plasma. J. Clin. Invest. 1987;80(2):458–465. doi: 10.1172/JCI113093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipoprotein(a) (Lp(a)) Package Insert. 06 May 16, Rev. 002. Randox: Crumlin, UK.
- 36.Tina-quant Lipoprotein (a) Gen.2 For Accurate and Reliable Assessment of Cardiovascular Risk . Risch-Rotkreuz; Switzerland: 2019. Roche Diagnostics International. [Google Scholar]
- 37.LPA2 Tina-quant Lipoprotein (a) Gen. 2 Package Insert. 2015-01, V 2.0. Roche Diagnostics: Indianapolis, IN.
- 38.Wilson D.P., Jacobson T.A., Jones P.H., Koschinsky M.L., McNeal C.J., Nordestgaard B.G., Orringer C.E. Use of Lipoprotein(a) in clinical practice: a biomarker whose time has come. A scientific statement from the National Lipid Association. J Clin Lipidol. 2019;13(3):374–392. doi: 10.1016/j.jacl.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Grundy S.M., Stone N.J., Bailey A.L., Beam C., Birtcher K.K., Blumenthal R.S., Braun L.T., de Ferranti S., Faiella-Tommasino J., Forman D.E., Goldberg R., Heidenreich P.A., Hlatky M.A., Jones D.W., Lloyd-Jones D., Lopez-Pajares N., Ndumele C.E., Orringer C.E., Peralta C.A., Saseen J.J., Smith S.C., Jr., Sperling L., Virani S.S., Yeboah J. AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines. J. Am. Coll. Cardiol. 2018;73(24):3168–3209. doi: 10.1016/j.jacc.2018.11.002. 2019. [DOI] [PubMed] [Google Scholar]
- 40.Tsimikas S., Stroes E.S.G. The dedicated "Lp(a) clinic": a concept whose time has arrived? Atherosclerosis. 2020;300:1–9. doi: 10.1016/j.atherosclerosis.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Guan W., Cao J., Steffen B.T., Post W.S., Stein J.H., Tattersall M.C., Kaufman J.D., McConnell J.P., Hoefner D.M., Warnick R., Tsai M.Y. Race is a key variable in assigning lipoprotein(a) cutoff values for coronary heart disease risk assessment: the Multi-Ethnic Study of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2015;35(4):996–1001. doi: 10.1161/ATVBAHA.114.304785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costello B.T., Silverman E.R., Doukky R., Braun L.T., Aggarwal N.T., Deng Y., Li Y., Lundberg G., Williams K.A., Sr, Volgman A.S. Lipoprotein(a) and increased cardiovascular risk in women. Clin. Cardiol. 2016;39(2):96–102. doi: 10.1002/clc.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frohlich J., Dobiasova M., Adler L., Francis M. Gender differences in plasma levels of lipoprotein (a) in patients with angiographically proven coronary artery disease. Physiol. Res. 2004;53(5):481–486. [PubMed] [Google Scholar]
- 44.Cook N.R., Mora S., Ridker P.M. Lipoprotein(a) and cardiovascular risk prediction among women. J. Am. Coll. Cardiol. 2018;72(3):287–296. doi: 10.1016/j.jacc.2018.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fanshawe A.E., Ibrahim M. The current status of lipoprotein (a) in pregnancy: a literature review. J. Cardiol. 2013;61(2):99–106. doi: 10.1016/j.jjcc.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Forbang N.I., Criqui M.H., Allison M.A., Ix J.H., Steffen B.T., Cushman M., Tsai M.Y. Sex and ethnic differences in the associations between lipoprotein(a) and peripheral arterial disease in the Multi-Ethnic Study of Atherosclerosis. J. Vasc. Surg. 2016;63(2):453–458. doi: 10.1016/j.jvs.2015.08.114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.