Abstract
There is growing interest in the characterization of protein complexes and their interactions with ligands using native ion mobility mass spectrometry. A particular challenge, especially for membrane proteins, is preserving noncovalent interactions and maintaining native-like structures. Different approaches have been developed to minimize activation of protein complexes by manipulating charge on protein complexes in solution and the gas-phase. Here, we report the utility of polyamines that have exceptionally high charge-reducing potencies with some molecules requiring 5-fold less than trimethylamine oxide to elicit the same effect. The charge-reducing molecules do not adduct to membrane protein complexes and are also compatible with ion-mobility mass spectrometry, paving the way for improved methods of charge reduction.
Graphical Abstract
Native mass spectrometry (MS) is an emerging biophysical approach for characterizing the structure and function of biomolecular assemblies, especially for membrane protein complexes.1–4 Under tuned conditions that minimize perturbation to protein structure, native MS can maintain noncovalent interactions in the gas phase that enables quantitative studies of ligand binding,5–7 subunits stoichiometry8,9 and lipid–protein interactions.6,10–12 Ion-mobility (IM) spectrometry in conjunction with MS provides means for the separation of ions by their size and charge along with determination of the rotationally averaged collision cross section (CCS). IM-MS approaches have also been developed to characterize the resistance of membrane proteins and their complexes with ligands to unfolding in the gas-phase.7,11,13 Of late, native MS has uncovered that specific protein–lipid interactions can allosterically modulate other interactions with protein,6,12 lipid,10 and drug14–16 molecules.
Membrane proteins are typically encapsulated in detergent micelles for native MS studies. The detergent solubilized complexes are ionized using nanoelectrospray ionization followed by application of minimal collisional activation to release the protein complex from the detergent micelle.17,18 In general, the mass spectrometer is tuned to preserve the structure and noncovalent interactions of protein complexes.19 However, even under carefully tuned instrument conditions, protein structure can inadvertently be disrupted, resulting in measurements that deviate from those obtained by other biophysical techniques. An attractive approach to preserve the noncovalent interactions is to reduce the charges carried by protein molecules to reduce their internal energy and minimize Coulombic repulsion thereby lowering the propensity for unfolding or activation.20–22 It has been well-established that detergents play an important role in the number of charge states acquired by membrane proteins.11,17 N-Dodecyl-β-d-maltopyranoside (DDM), a commonly used detergent for purifying and solubilizing membrane proteins, results in charges states that are difficult to minimize activation energy resulting in partial unfolding of membrane proteins.11,14,17 The discovery of charge-reducing detergents, such as tetraethylene glycol monooctyl ether (C8E4) and n-dodecyl-N,N-dimethylamine-N-oxide (LDAO), has revolutionized the native MS field by enabling IM measurements of compact, native-like complexes with collision cross section (CCS) values that are in direct agreement with those calculated from atomic coordinates.11 In addition to aiding the preservation of native-like states of protein complexes and increased spacing between bound adducts, which has been shown to be beneficial for observing more lipid bound states of membrane proteins.23
Numerous studies have reported methods to achieve lower charge states of proteins for MS measurements.21,23–27 Adding charge-reducing chemicals to protein samples, such as the abundant naturally occurring osmolyte28–30 trimethylamine N-oxide (TMAO), has been shown to significantly lower the average charge state (Zavg) of protein complexes.23,31,32 Imidazole and derivatives thereof have shown marginal charge reduction for membrane protein complexes (by about ~2–3 charge states), especially in cases when noncharge-reducing detergents are used.21,33 Besides the addition of small molecules, synthetic oligoglycerol detergents have been developed for membrane protein complexes that exhibit varying charge-reducing properties.24 Despite recent advances in manipulation of charge, these charge-reducing modalities have a number of limitations. Some charge-reducing molecules require high concentration to be effective or an undesirable tendency to adduct to protein complexes leading to broadened mass spectral peaks. There is a need to further investigate charge-reducing approaches that are not only effective but also applicable to a wide range of proteins.
EXPERIMENTAL SECTION
Protein Expression and Purification.
AmtB was expressed and purified as previously described11 with the exception of a modified expression plasmid was used. The AmtB gene from Escherichia coli with N-terminal TEV protease cleavable His10 and MBP (described in previous study11) was amplified by polymerase chain reaction (PCR) and cloned into pCOLA backbone, which also coexpresses GlnK with Y51F mutation (removes possible heterogeneity arising from uridylation34) as a TEV protease cleavable N-terminal fusion to Strep-tag II as described before.5 The AmtB used in this study contains the C312T mutation that retains the ability to couple to GlnK (manuscript in preparation). For soluble proteins, streptavidin was purchased from Sigma, and egg white lysozyme was purchased from Amresco. GlnK was expressed and purified as previously described.5
Preparation of Charge-Reducing Reagents.
Trimethylamine N-oxide was purchased from Cayman Chemical. Spermidine was purchased from Beantown Chemical. Spermine was purchased from Alfa Aesar. Histamine, bis(2-aminoethyl)amine, bis(3-aminopropyl)amine, tris(2-aminoethyl)amine, 3,3′-diamino-N-methyldipropylamine, tris-(3-aminopropyl)amine, piperazine, 1-(2-aminoethyl)-piperazine, and 1,4-bis(3-aminopropyl)piperazine were purchased from Sigma-Aldrich. Protein was buffer exchanged into 200 mM ammonium acetate (supplemented with 0.5% C8E4 for membrane protein). Charge-reducing reagents stocks were made by dissolving into the same buffer as the protein and were mixed with protein to obtain desired final concentrations of charge-reducing reagent.
Native Mass Spectrometry (MS) Analysis of AmtB Native.
MS was performed on a Q Exactive Ultra-High Mass Range (UHMR) from ThermoFisher Scientific, modified with a custom reverse entry ion source (REIS) and 1.5 m drift tube. A Synapt G1 HDMS instrument with a 32-k RF generator from Waters Corporation was also used for membrane and soluble protein measurements. Nanoelectrospray ionization was performed as previously described.19 UHMR parameters applied for AmtB included 250 °C capillary temperature, 1.40 kV spray voltage, 200 maximum inject time, resolution set to 6250 without averaging, 50 eV in-source CID, 40 CE, trapping gas pressure of 5. Ion transfer was set to high m/z mode, with in-source desolvation voltage set to −180 V.
Native MS Analysis of Soluble Proteins.
Measurements for soluble proteins were collected on a Synapt G1 HDMS from Waters. For GlnK, source temperature was lowered to 80 °C, and trap and transfer collision energy was set to 40 and 25, respectively. Trap wave velocity and height set to 300 m/s and 1 V, respectively. IMS wave velocity and height set to 300 m/s and 18 V, respectively. For streptavidin, source temperature was set to 80 °C, and trap and transfer collision energy was set to 20 and 10, respectively. Trap wave and velocity and height set to 300 m/s and 1 V, respectively. IMS wave velocity and height set to 300 m/s and 18 V, respectively. For lysozyme, capillary voltage was set to 1.5 kV. Source temperature was set to 50 °C, with 50 and 2 V on sampling and extraction cone, respectively. Trap and transfer CE were set to 30 and 15, respectively. Trap gas flow rate was set to 5 mL/min. Trap wave and velocity and height set to 100 m/s and 0.4 V, respectively. IMS wave velocity and height set to 100 m/s and 4 V, respectively. Soluble protein spectra were collected from 1000 to 10000 m/z. All mass spectra were deconvoluted using Unidec.35
Ion-Mobility Mass Spectrometry.
Ion-Mobility Mass Spectrometry (IM-MS) was performed on a home-built periodic focusing drift-tube (PF-DT) Fourier-transform IM (FT-IM) orbitrap platform, as described previously.36–39 Briefly, ions are generated via a nano-ESI source before entering a heated capillary transfer tube at 155 °C, which are then focused into a RF-ion funnel (~3 Torr) before being differentially pumped into a DC only periodic focusing funnel with a field strength of 50 V/cm. Ions are then gated using a modified Bradbury–Nelson gate that pulses ions into the 1.5 m PF-DT for mobility separation, with a second gate post-IM to allow for implementation of FT-IM. Two MKS calibrated manometers where placed on both ends of the PF-DT to ensure steady-state of He (99.999% purity). Ions are then transferred via a multipole into the HCD cell of the Thermo Scientific Exactive Series platform. FT-IM-PF-DT data is processed into ATDs and subsequently to CCS using custom Python 3.7 scripts utilizing Scipy40 and multiplierz.41
Determination of Half Maximal Effective Concentration (EC50).
Native mass spectra of AmtB solubilized in 2× CMC C8E4 were processed with UniDec35 to determine the weighted average charge state (Zavg). The half maximal effective concentration (EC50) or potency of charge-reducing compounds was calculated using a modified form of the Hill equation:42
where nH is the Hill coefficient, b is the value of the upper asymptote, and X is the concentration of charge-reducing molecule. Zavg data was essentially inverted to enable the equation above to be fitted to the experimental data as follows:
where Zavg,max is the maximum average charge state measured without any charge-reducing molecule added. The equation was fitted to the inverted data using Python (version 3.7) scripts making use of NumPy and SciPy modules.43,44 The resulting fits had an R2 value of 0.98 or better. Z′ and b were converted back to units of charge:
Plots were generated using Python and the Matplotlib module.45
RESULTS AND DISCUSSION
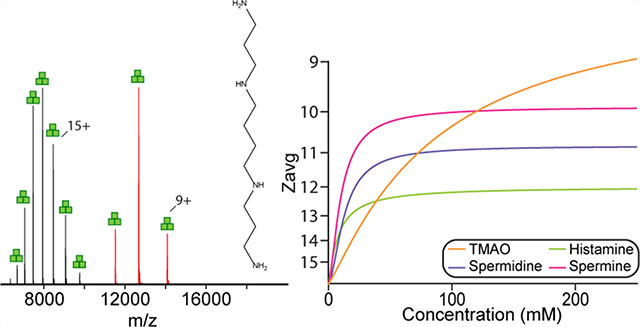
Inspired by recent reports of TMAO and imidazole derivatives’ effectiveness for charge-reducing protein complexes,21,23,31,32 we explored the potential of other small molecules found in biology and characterized their impact on the trimeric ammonia channel (AmtB), a model integral membrane protein (Figure 1). The first molecule we investigated was histamine, an amine-functionalized imidazole involved in the immune response and plays important roles in the nervous system.46 The addition of 100 mM histamine, which the charge-reducing properties have not been studied before, to AmtB solubilized in C8E4 resulted in a reduction of the Zavg by four compared to the mass spectrum acquired in the absence of histamine (Zavg of 16.2) (Figures 1B and S1). For reference, the addition of 100 mM TMAO results in a broad charge state distribution with a Zavg of 10.9 (Figure 1A). We next explored spermidine and spermine, natural polyamines found in cells and involved in many cellular functions, such as promoting cellular proliferation and required for replication of many viruses.47–50 Compared to histamine, spermidine, and spermine at 100 mM displayed pronounced charge-reducing properties with Zavg of 10.9 and 10.0, respectively (Figure 1C). Unlike TMAO, histamine, spermidine, and spermine resulted in narrow charge state distributions (Figure 1D). More specifically, the total number of charge states (Ztot) for 10 and 250 mM TMAO was 13 and 6, respectively. The Ztot for histamine, spermidine, and spermine at the highest concentration investigated are 3, 4, and 3, respectively (Figure 1D). Moreover, the quality of the mass spectra for these molecules did not diminish, unlike previous reports showing the various degree of adduction to soluble proteins.51
Figure 1.

Natural compounds are potent charge-reducing molecules. (A–C) Representative mass spectra of AmtB in C8E4 doped with 100 mM of (A) TMAO, (B) histamine, or (C) spermidine. Data was acquired on an ultrahigh mass range (UHMR) Orbitrap mass spectrometer under gentle instrument conditions. Structures of molecules are shown in the inset. (D) Total number of charge states for AmtB in the presence of compounds at different concentrations. (E) Plot of average charge states (Zavg, dots) as a function of concentration of charge-reducing molecules. A modified form of Hill equation was fit to the data (solid lines) to determine the half maximal effective concentration (EC50). (F) Mass spectra of AmtB doped with different concentrations of spermine ranging from 0 to 250 mM.
We next titrated AmtB with different charge-reducing molecules to better understand the potency of these compounds. For example, AmtB in the presence of increasing concentrations of spermine progressively reduced charge on the protein complex (Figure 1F). At low concentrations of spermine the charge state distribution was broad and asymmetric. Interestingly, at higher spermine concentrations the charge-reducing effect was diminished and Zavg reached a plateau around 10 (Figure 1E). In a similar fashion to spermine, histamine and spermidine displayed a rapid decrease in Zavg at lower concentrations and reached a plateau at higher concentrations, over 100 mM (Figure 1E). The trend for TMAO differed from other compounds where Zavg gradually decrease with increasing concentrations (Figure 1E). To quantify the potency of these compounds, a modified form of the Hill equation was fit to the Zavg data to determine the half maximal effective concentration (EC50) and the minimum Zavg value (or upper asymptote). With the exception of TMAO, the charge-reducing molecules had an EC50 ~ 10 Mm whereas the value for minimum Zavg (Zavg,min) was the lowest for spermine (Table S1). In contrast, TMAO with an EC50 of 87 mM for the range of concentrations screened is nearly 9-fold greater than the other molecules. Lastly, it is worth noting that 10 mM spermine achieved the same degree of charge reduction for AmtB as 250 mM histamine, underscoring the potency of this class of compounds.
Given the results for amine-functionalized linear molecules, cyclic amines such as piperazine and piperazine derivatives were investigated for their ability to charge-reduce AmtB in C8E4 (Figure 2 and Tables S1 and S2). Titration of piperazine revealed a shallow change in Zavg with increasing concentration and reaching a Zavg,min of 14. Piperazine derivatives with one aminoethyl (AEP, 1-(2-aminoethyl)piperazine) or two aminopropyl (APP, 1,4-bis(3-aminopropyl)piperazine) substituents increased the amount charge-reduction with a Zavg,min of 13 and 10, respectively (Figure 2B–C). Piperazine and derivatives have a range of EC50 values (7–12 mM), and like other amine containing molecules, the charge-reducing effect reached a plateau at higher concentrations. Interestingly, substitution of piperazine with amine containing substituents not only increased the ability to charge-reduce but also resulted in narrowing of the charge state distribution where Ztot decreased from 7 to 5 at the highest concentration studied (Table S3). However, APP, the best piperazine derivative, was not as potent in regards to charge-reduction compared to spermine. Taken together, these results suggest that aliphatic polyamines represent promising candidates as charge-reducing agents.
Figure 2.
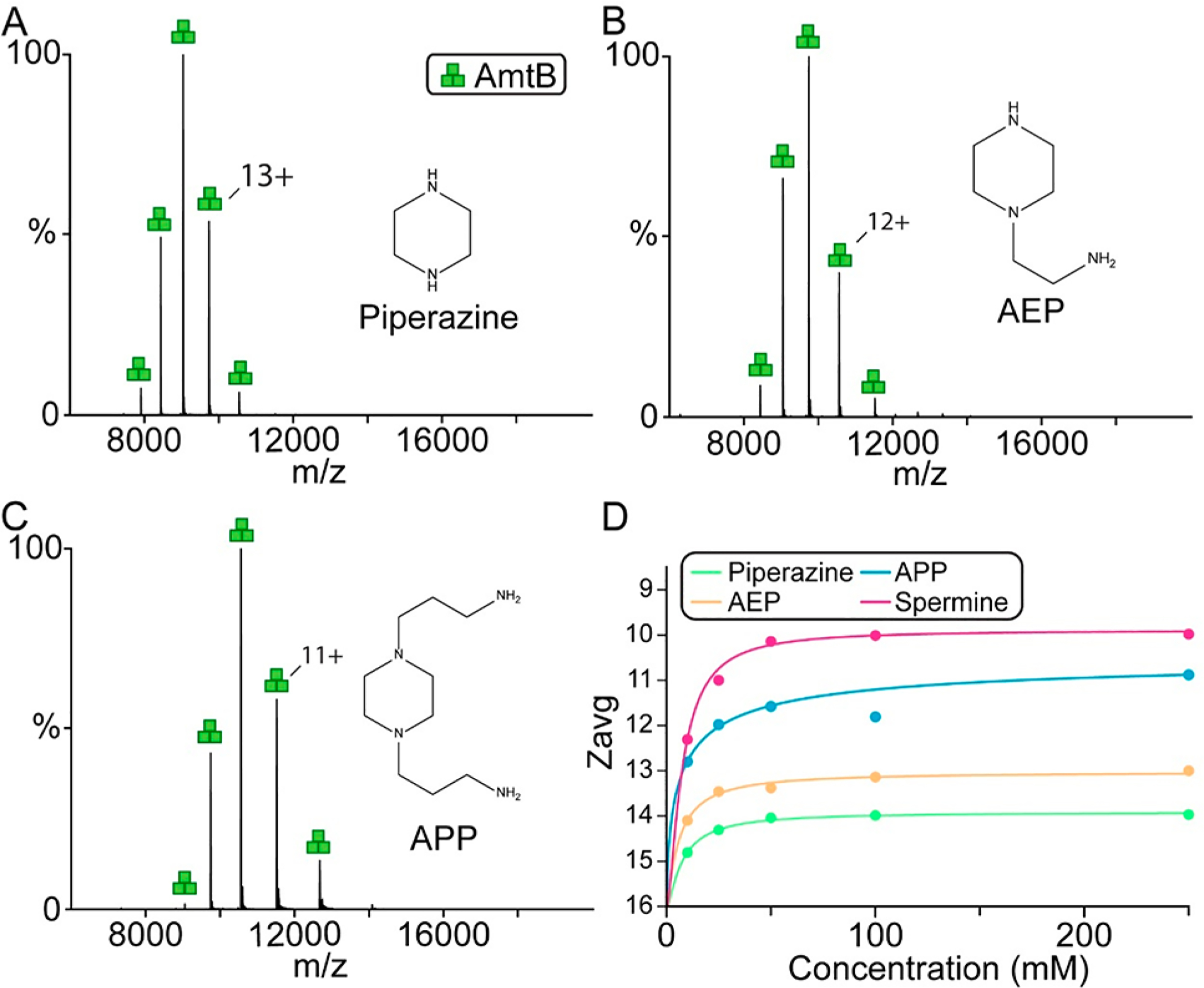
Charge reduction of AmtB by piperazine and amine derivatives thereof. (A–C) Representative mass spectra of AmtB in C8E4 doped with 100 mM of (A) piperazine, (B) AEP, and (C) APP. Structures of molecules are shown in the inset. (D) Plot of Zavg (dots) as a function of concentration of charge-reducing molecules and regression of a modified Hill equation (solid lines).
The results for spermine and other natural polyamines prompted us to investigate analogues of polyamines. Bis(2-aminoethyl)amine (B2A) and bis(3-aminopropyl)amine (B3A) are symmetric variants of spermidine (contains aminopropyl and aminobutyl groups) and addition of these compounds resulted in charge-reduction of AmtB (Figure 3A and B). However, B2A and B3A were not as potent compared to spermine with a Zavg of 11. A marginal increase in charge-reducing properties (Zavg decrease by ~1) was observed for 3,3-diamino-N-methyldipropylamine (DMP), which differs from B3A by a methyl group (Figure 3C). Tris(2-aminoethyl)-amine (T2A) and tris(3-aminopropyl)amine (T3A) are molecules with four amines and showed increase charge-reducing properties compared to their counterparts B2A and B3A, respectively. T3A at the highest concentration reduced charge at comparable levels to that of spermine and spermidine. Notably, the addition of an amine resulted in a reduction of both Ztot and Zavg,min. No correlation is observed between EC50 and Zavg,min for these molecules.
Figure 3.
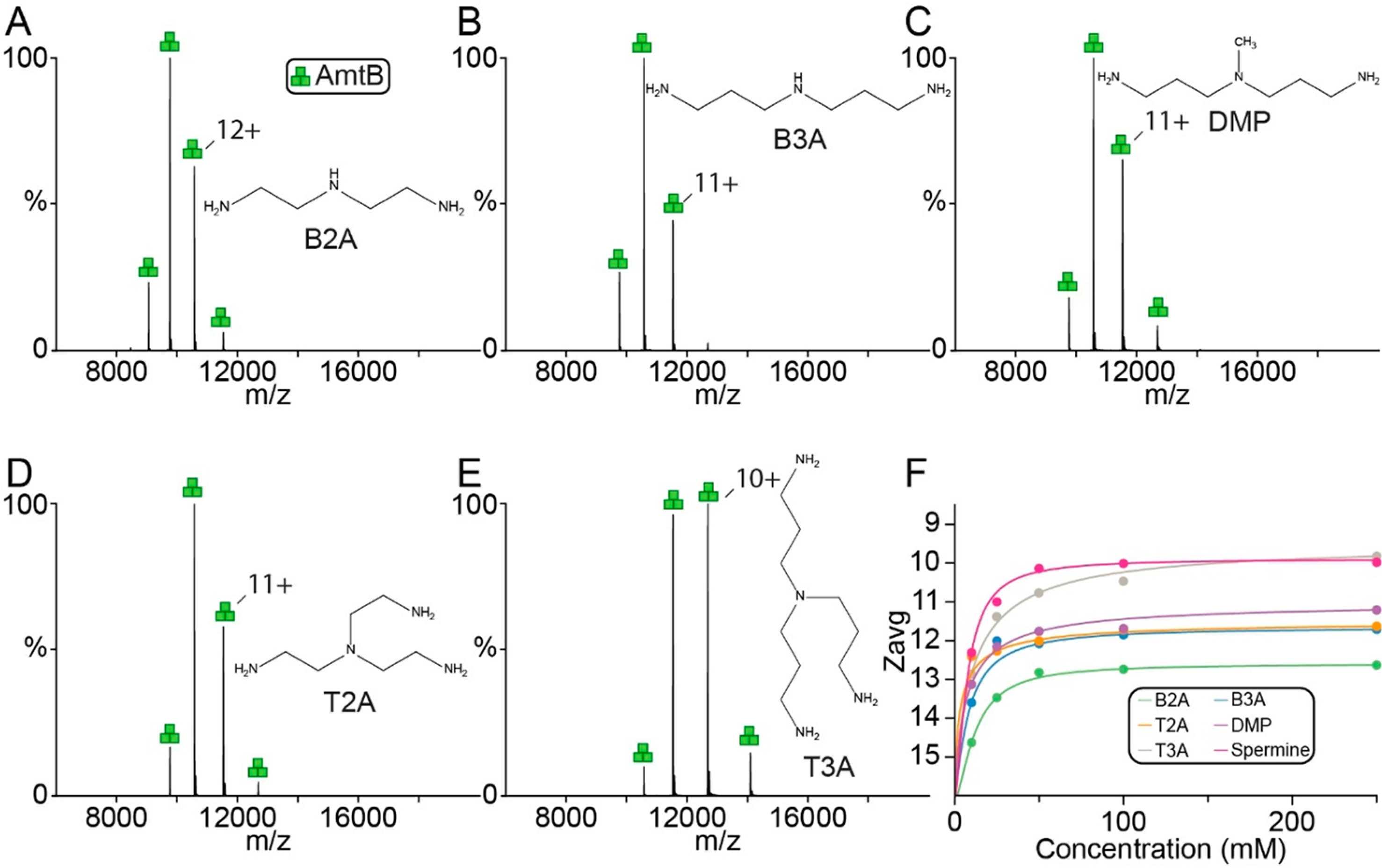
Charge-reduction of AmtB by synthetic polyamines. (A–E) Representative mass spectra of AmtB in C8E4 doped with 100 mM of (A) B2A, (B) B3A, (C) DMP, (D) T2A, and (E) T3A. Structures of molecules are shown in the inset. (F) Plot of Zavg (dots) as a function of concentration of charge-reducing molecules and regression of a modified Hill equation (solid lines).
We next evaluated the performance of charge-reducing molecules for native ion mobility mass spectrometry (IM-MS) studies. Native IM-MS data was collected for a subset of molecules at 50 mM concentration on a high-resolution Fourier-transform ion-mobility (FT-IM) PF-DT Orbitrap platform36 (Figure 4). The mass spectrum of AmtB in C8E4 acquired on the FT-IM-PF-DT Orbitrap is comparable to that obtained using the commercial ionization source (Figure 4 and S1). However, lower charge states for AmtB in the presence of spermine were observed on the FT-IM-PF-DT, consistent with previous results.36,52 The collision cross section (CCS) profiles for the lowest (C8E4) and most charge-reducing (addition of spermine) show a 0.4% difference with a slightly decreased in the ion mobility resolving power (RIM) of spermine (Figure 4C). Here, CCS values for AmtB in the absence and presence of different charge-reducing molecules show, at most, a 3% difference (Figure 4D).
Figure 4.
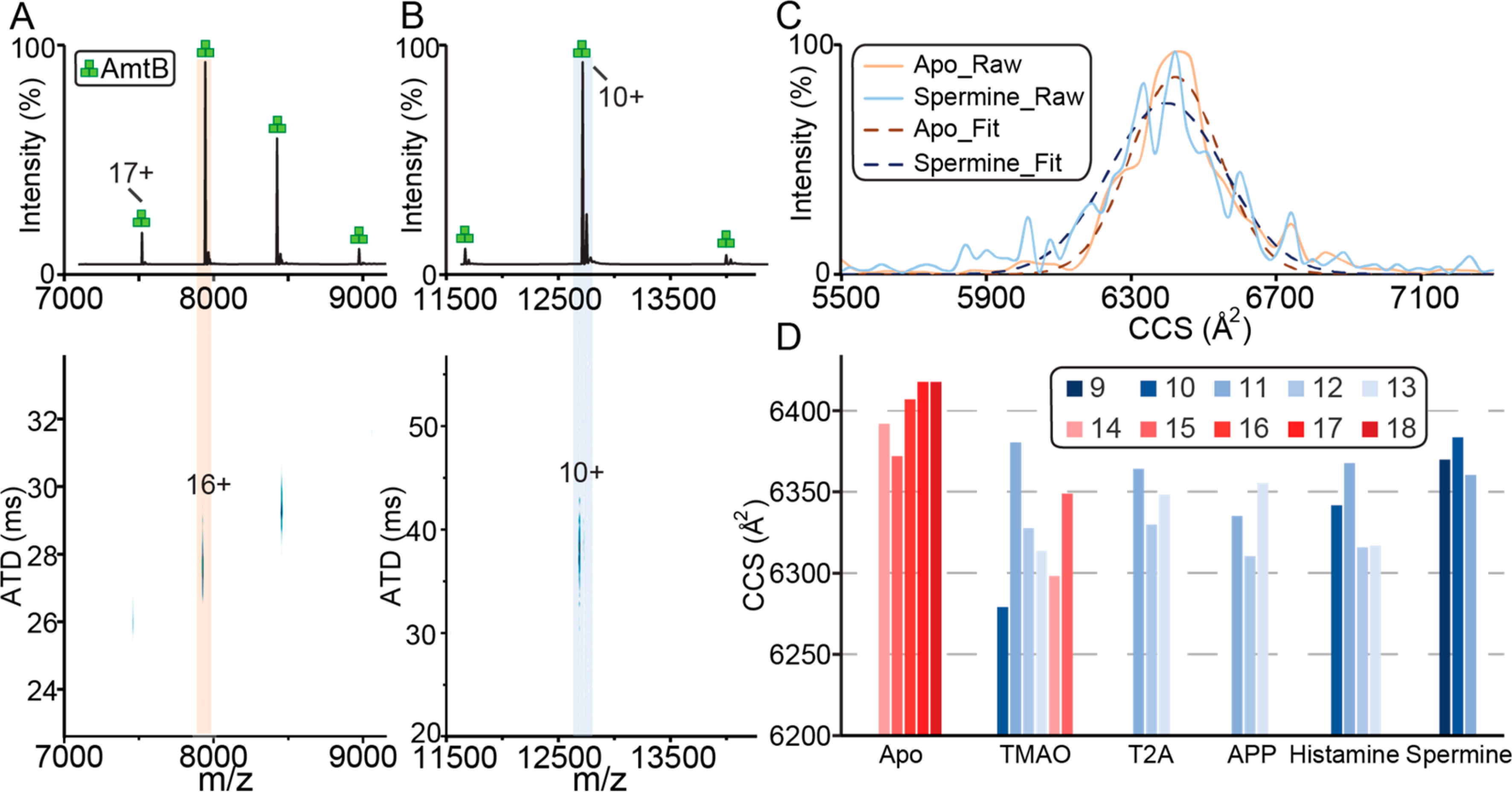
Native IM-MS of trimeric AmtB on a FT-IM-PF-DT Orbitrap platform. (A, B) Ion mobility mass spectra of AmtB in (A) C8E4 and in the presence of (B) 50 mM spermine. (C) Collision cross section (CCS) profiles for the most abundant charge states. A Gaussian function was fitted (solid lines) to the data (dashed lines) to determine the centroid CCS. (D) Plot of CCS for different charge states from AmtB in the presence of different charge-reducing molecules.
We also explored the charge-reducing effect of these reagents on AmtB solubilized in the non-charge-reducing detergent DDM. Mixtures of AmtB in DDM with different charge-reducing molecules and at various concentrations could not generate interpretable mass spectra (Figure S2). This finding is consistent with a recent study that reports charge-reduction of membrane proteins with alkali metal acetate is dependent on detergent.26 However, the mechanism behind this phenomenon is not clear but a plausible explanation is that DDM may directly compete with these molecules interacting with the membrane protein. Alternatively, the larger micelle size of DDM may prevent meaningful contacts between these molecules and the protein. Nevertheless, C8E4 exhibits charge-reducing properties that are amplified (mutually or inclusive) with charge-reducing molecules, such polyamines. These results highlight the fact that there is an urgent need for detergents engineered for native MS applications that have desirable attributes, such as charge-reducing properties, ease of release from the protein complex, and the ability to stabilize membrane proteins.
The ability of these molecules to charge-reduce soluble proteins was also investigated. The monomeric lysozyme, trimeric GlnK, and tetrameric streptavidin were mixed with a charge-reducing reagent at equimolar ratios (Figure S3). Some of the charge-reducing molecules adducted to the protein giving rise to an uninterpretable mass spectrum. This adduction behavior indicates that they interact strongly with proteins. Charge-reduced mass spectra for the three soluble proteins were observed for T3A, while the others were largely protein-dependent. For example, lysozyme in the presence of B2A was poorly resolved whereas spermine, APP and B3A resulted in resolved mass spectra. Data quality and charge reduction patterns differed for GlnK and streptavidin. It is interesting to note that TMAO’s ability to charge-reduce does not prevent the gas-phase dissociation of oligomeric proteins tested. Recently, charge reduction of soluble proteins by TMAO has been reported to be dependent on collisional activation.31 We next investigated the impact of increasing collision energy and cone voltage for soluble proteins. Activation of lysozyme in the presence of 50 mM spermine using either collision energy or cone voltage did not result enhanced charge reduction (Figures S6–7). In summary, polyamines can charge-reduce soluble proteins but their efficacy appears to be protein-dependent.
To better understand the mechanism of charge reduction for membrane proteins, we conducted studies of AmtB in C8E4 with either 100 mM spermine or TMAO at different solution or capillary temperatures. Native mass spectra of AmtB in the presence of spermine or TMAO acquired under different solution temperatures (32.0 to 37.0 °C) showed similar results (Figure S8). AmtB with either charge-reducing additive was introduced into the mass spectrometer with the capillary temperature ranging from 100 to 430 °C. For spermine, a capillary temperature below 200 °C resulted in severe adduction, presumably spermine molecules, and a resolved mass spectrum was not obtained. For both additives, increasing the capillary temperature resulted in reduction of Zavg (Figure 5). The Zavg of AmtB with spermine marginally decreased from 9.8 to 9.5 at 430 °C (Figure 5A). For TMAO, Zavg significantly decreased from 12.0 to 9.3 at the highest capillary temperature (Figure 5B). We also noticed that application of backing pressure to nanoESI emitter for AmtB with TMAO resulted in significant reduction in charge (Figure 5C and D). This observation can be rationalized by the production of larger droplets resulting in a higher concentration of TMAO in the final droplets formed in the electrospray process. Taken together, our findings agree with previous reports51,53 and indicate that charge reduction occurs in the final stages of the desolvation process.
Figure 5.
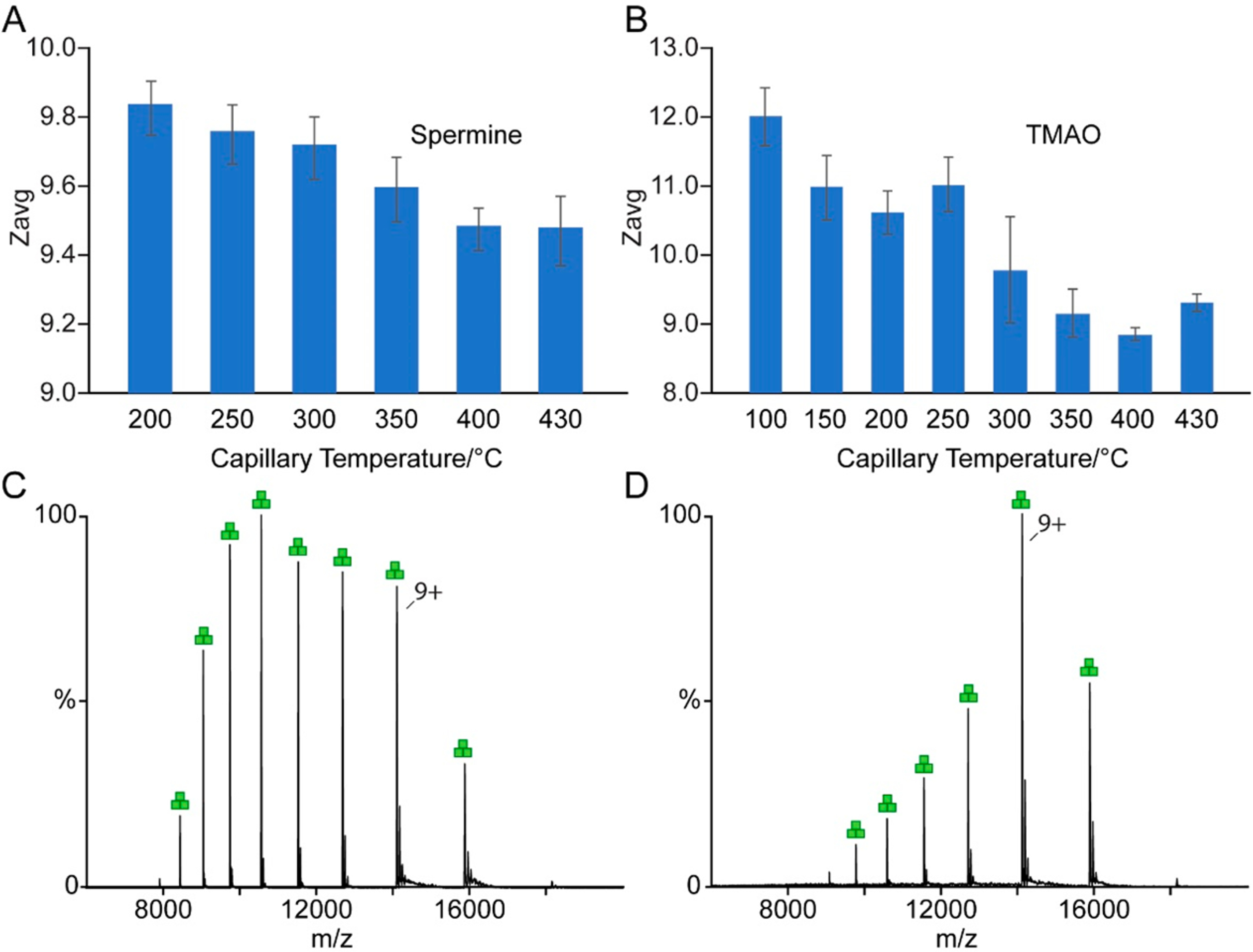
Effects of capillary temperature and backing pressure on charge-reduction of AmtB. (A, B) Zavg of AmtB in the presence of 100 mM (A) spermine or (B) TMAO acquired at different capillary temperatures. (C, D) Mass spectra of AmtB charge reduced by 100 mM TMAO (C) without and (D) with application of backing pressure to the nanoESI emitter.
By comparison of the chemical structures and their potency of charge reduction, a plausible explanation is that potency is correlated with their gas-phase basicity. Heck and co-workers have shown using soluble proteins a linear relationship between the additive gas-phase basicity (or proton affinity) and average charge state.53 For polyamines, it has been reported that increasing either the number of nitrogen or carbon atoms results in higher gas-phase basicity.54 Following on this generalization, the installation of a methyl group at the secondary amine of B3A makes it more basic that in turn improves its charge-reduction potency. Moreover, increasing the number of amino groups within the molecule, which increase gas-phase basicity, also leads to a better charge-reducing molecule as seen for piperazine and derivatives thereof. Our observation of the potency of these charge-reducing chemicals reveals there is a correlation with EC50 and empirically measured gas-phase basicity, which agrees with the previous reports.51,53–55 Another contributing feature of the charge-reducing molecule is the number of carbons, which would increase gas-phase basicity.54–57 Polyamines with more carbons generally show greater degree of charge reduction.54 Among these charge-reducing chemicals, TMAO appears to be an outlier due to the significant difference in the Hill plot compared to other polyamines, which could be explained by the difference of charge reduction mechanism of TMAO. It has been proposed that TMAO reduces charge of proteins by the proton transfer from gas-phase collision.31 However, it remains difficult to discern if this process is result of (i) dissociation of the additive ligand from the protein either taking a proton with it or leaves the proton on the protein,51,53,58 (ii) gas-phase collision of the additive with protein resulting in proton transfer to the additive,31 or (iii) a combination of both processes.
The results of this study also point to an additional feature that implicates the direct interaction of charge-reducing molecules and the protein. For example, there are marked differences when comparing spermidine and B3A, which differ by Zavg,tot by one charge. In addition, longer alkyl chains may interact more strongly with membrane proteins or detergent micelles. Different detergent environments may have different degrees of accessible protein surface area because of potential changes in droplet size or detergent micelle size. Taken together, the varied effects of polyamines on soluble proteins may be due to the hydrophobicity of the molecules in combination with their basicity. In short, the system composed of detergent encapsulated membrane proteins with charge-reducing molecules is complex and further studies are warranted to better understand the dependence of detergent on charge reduction.
CONCLUSION
In summary, this study reports a set of potent charge-reducing molecules for native IM-MS studies. The reported molecules do not adduct nor impede the analysis of membrane proteins for native MS studies. We also demonstrate improved charge-reduction potency; spermidine requires nearly 10-fold less in concentration compared to TMAO to achieve the same degree of charge reduction. Moreover, the polyamines are advantageous over TMAO as a narrower charge state distribution is observed improving the signal-to-noise ratio. Polyamines provide new approaches to manipulate charge in an effort to preserve noncovalent interactions and minimize perturbation and overactivation of protein complexes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) (DP2GM123486), the NIH (P41GM128577–01), and the Strategic Transformative Research Program (awarded to A.L. and L.F.).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c01826.
Contains tabulated names and abbreviations, EC50 values, and number of charge states (S2–S4), mass spectra for membrane protein in absence of charge reducing molecules and different detergents (S5–S6), mass spectra for soluble proteins (S7, S10–11), ion mobility mass spectrometry data for AmtB (S8–S9), and mass spectra acquired at different solution temperatures (S12) (PDF)
Contributor Information
Jixing Lyu, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Yang Liu, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Jacob W. McCabe, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Samantha Schrecke, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
Lei Fang, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
David H. Russell, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States
Arthur Laganowsky, Department of Chemistry, Texas A&M University, College Station, Texas 77843, United States.
REFERENCES
- (1).Calabrese AN; Radford SE Methods 2018, 147, 187–205. [DOI] [PubMed] [Google Scholar]
- (2).Hilton GR; Benesch JLJR Soc., Interface 2012, 9 (70), 801–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Loo JA Mass Spectrom. Rev. 1997, 16 (1), 1–23. [DOI] [PubMed] [Google Scholar]
- (4).Gupta K; Donlan JAC; Hopper JTS; Uzdavinys P; Landreh M; Struwe WB; Drew D; Baldwin AJ; Stansfeld PJ; Robinson CV Nature 2017, 541 (7637), 421–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cong X; Liu Y; Liu W; Liang X; Russell DH; Laganowsky AJ Am. Chem. Soc. 2016, 138 (13), 4346–9. [DOI] [PubMed] [Google Scholar]
- (6).Cong X; Liu Y; Liu W; Liang X; Laganowsky A Nat. Commun. 2017, 8 (1), 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fantin SM; Parson KF; Niu S; Liu J; Polasky DA; Dixit SM; Ferguson-Miller SM; Ruotolo BT Anal. Chem. 2019, 91 (24), 15469–15476. [DOI] [PubMed] [Google Scholar]
- (8).Wilson JW; Rolland AD; Klausen GM; Prell JS Anal. Chem. 2019, 91 (15), 10204–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Barrera NP; Isaacson SC; Zhou M; Bavro VN; Welch A; Schaedler TA; Seeger MA; Miguel RN; Korkhov VM; van Veen HW; Venter H; Walmsley AR; Tate CG; Robinson CV Nat. Methods 2009, 6 (8), 585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Patrick JW; Boone CD; Liu W; Conover GM; Liu Y; Cong X; Laganowsky A Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (12), 2976–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson CV Nature 2014, 510 (7503), 172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yen HY; Hoi KK; Liko I; Hedger G; Horrell MR; Song W; Wu D; Heine P; Warne T; Lee Y; Carpenter B; Pluckthun A; Tate CG; Sansom MSP; Robinson CV Nature 2018, 559 (7714), 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Allison TM; Reading E; Liko I; Baldwin AJ; Laganowsky A; Robinson CV Nat. Commun. 2015, 6, 8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Marcoux J; Wang SC; Politis A; Reading E; Ma J; Biggin PC; Zhou M; Tao H; Zhang Q; Chang G; Morgner N; Robinson CV Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (24), 9704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Bolla JR; Sauer JB; Wu D; Mehmood S; Allison TM; Robinson CV Nat. Chem. 2018, 10 (3), 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gault J; Donlan JA; Liko I; Hopper JT; Gupta K; Housden NG; Struwe WB; Marty MT; Mize T; Bechara C; Zhu Y; Wu B; Kleanthous C; Belov M; Damoc E; Makarov A; Robinson CV Nat. Methods 2016, 13 (4), 333–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Reading E; Liko I; Allison TM; Benesch JL; Laganowsky A; Robinson CV Angew. Chem., Int. Ed. 2015, 54 (15), 4577–81. [DOI] [PubMed] [Google Scholar]
- (18).Borysik AJ; Hewitt DJ; Robinson CV J. Am. Chem. Soc. 2013, 135 (16), 6078–83. [DOI] [PubMed] [Google Scholar]
- (19).Hopper JTS; Yu YT-C; Li D; Raymond A; Bostock M; Liko I; Mikhailov V; Laganowsky A; Benesch JLP; Caffrey M; Nietlispach D; Robinson CV Nat. Methods 2013, 10 (12), 1206–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hall Z; Politis A; Bush MF; Smith LJ; Robinson CV J. Am. Chem. Soc. 2012, 134 (7), 3429–38. [DOI] [PubMed] [Google Scholar]
- (21).Townsend JA; Keener JE; Miller ZM; Prell JS; Marty MT Anal. Chem. 2019, 91 (22), 14765–14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lemaire D; Marie G; Serani L; Laprevote O Anal. Chem. 2001, 73 (8), 1699–706. [DOI] [PubMed] [Google Scholar]
- (23).Patrick JW; Laganowsky AJ Am. Soc. Mass Spectrom. 2019, 30, 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Urner LH; Liko I; Yen HY; Hoi KK; Bolla JR; Gault J; Almeida FG; Schweder MP; Shutin D; Ehrmann S; Haag R; Robinson CV; Pagel K Nat. Commun. 2020, 11 (1), 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Scalf M; Westphall MS; Smith LM Anal. Chem. 2000, 72 (1), 52–60. [DOI] [PubMed] [Google Scholar]
- (26).Petroff JT; Tong A; Chen LJ; Dekoster GT; Khan F; Abramson J; Frieden C; Cheng WWL Anal. Chem. 2020, 92, 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hopper JT; Sokratous K; Oldham NJ Anal. Biochem. 2012, 421 (2), 788–90. [DOI] [PubMed] [Google Scholar]
- (28).Lidbury I; Murrell JC; Chen Y Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (7), 2710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Velasquez MT; Ramezani A; Manal A; Raj DS Toxins 2016, 8 (11), 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yancey PH J. Exp. Biol. 2005, 208 (15), 2819–30. [DOI] [PubMed] [Google Scholar]
- (31).Kaldmäe M; Österlund N; Lianoudaki D; Sahin C; Bergman P; Nyman T; Kronqvist N; Ilag LL; Allison TM; Marklund EG; Landreh MJ Am. Soc. Mass Spectrom. 2019, 30 (8), 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gault J; Lianoudaki D; Kaldmäe M; Kronqvist N; Rising A; Johansson J; Lohkamp B; Lain S; Allison TM; Lane DP; Marklund EG; Landreh MJ Phys. Chem. Lett. 2018, 9 (14), 4082–4086. [DOI] [PubMed] [Google Scholar]
- (33).Mehmood S; Marcoux J; Hopper JT; Allison TM; Liko I; Borysik AJ; Robinson CV J. Am. Chem. Soc. 2014, 136 (49), 17010–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gruswitz F; O’Connell J 3rd; Stroud RM Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (1), 42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Marty MT; Baldwin AJ; Marklund EG; Hochberg GK; Benesch JL; Robinson CV Anal. Chem. 2015, 87 (8), 4370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Clowers BH; Russell DH Anal. Chem. 2018, 90, 10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Poltash ML; McCabe JW; Patrick JW; Laganowsky A; Russell DH J. Am. Soc. Mass Spectrom. 2019, 30 (1), 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Russell DH TrAC, Trends Anal. Chem. 2020, 124, 115533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Poltash ML; Shirzadeh M; McCabe JW; Moghadamchargari Z; Laganowsky A; Russell DH Chem. Commun. 2019, 55 (28), 4091–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Virtanen P; Gommers R; Oliphant TE; Haberland M; Reddy T; Cournapeau D; Burovski E; Peterson P; Weckesser W; Bright J; van der Walt SJ; Brett M; Wilson J; Millman KJ; Mayorov N; Nelson ARJ; Jones E; Kern R; Larson E; Carey CJ; Polat İ; Feng Y; Moore EW; VanderPlas J; Laxalde D; Perktold J; Cimrman R; Henriksen I; Quintero EA; Harris CR; Archibald AM; Ribeiro AH; Pedregosa F; van Mulbregt P; Vijaykumar A; Bardelli AP; Rothberg A; Hilboll A; Kloeckner A; Scopatz A; Lee A; Rokem A; Woods CN; Fulton C; Masson C; Häggström C; Fitzgerald C; Nicholson DA; Hagen DR; Pasechnik DV; Olivetti E; Martin E; Wieser E; Silva F; Lenders F; Wilhelm F; Young G; Price GA; Ingold G-L; Allen GE; Lee GR; Audren H; Probst I; Dietrich JP; Silterra J; Webber JT; Slavič J; Nothman J; Buchner J; Kulick J; Schönberger JL; de Miranda Cardoso JV; Reimer J; Harrington J; Rodríguez JLC; Nunez-Iglesias J; Kuczynski J; Tritz K; Thoma M; Newville M; Kümmerer M; Bolingbroke M; Tartre M; Pak M; Smith NJ; Nowaczyk N; Shebanov N; Pavlyk O; Brodtkorb PA; Lee P; McGibbon RT; Feldbauer R; Lewis S; Tygier S; Sievert S; Vigna S; Peterson S; More S; Pudlik T; Oshima T; Pingel TJ; Robitaille TP; Spura T; Jones TR; Cera T; Leslie T; Zito T; Krauss T; Upadhyay U; Halchenko YO; Vázquez-Baeza Y Nat. Methods 2020, 17 (3), 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Alexander WM; Ficarro SB; Adelmant G; Marto JA Proteomics 2017, 17 (15–16), 1700091. [DOI] [PubMed] [Google Scholar]
- (42).Hill AV Journal of Physiology 1910, 40, iv–vii. [Google Scholar]
- (43).van der Walt S; Colbert SC; Varoquaux G Comput. Sci. Eng. 2011, 13 (2), 22–30. [Google Scholar]
- (44).Millman KJ; Aivazis M Comput. Sci. Eng. 2011, 13 (2), 9–12. [Google Scholar]
- (45).Hunter JD Comput. Sci. Eng. 2007, 9 (3), 90–95. [Google Scholar]
- (46).Nieto-Alamilla G; Marquez-Gomez R; Garcia-Galvez AM; Morales-Figueroa GE; Arias-Montano JA Mol. Pharmacol. 2016, 90 (5), 649–673. [DOI] [PubMed] [Google Scholar]
- (47).Thomas T; Thomas TJ Biochemistry 1993, 32 (50), 14068–74. [DOI] [PubMed] [Google Scholar]
- (48).Li MM; MacDonald MR Cell Host Microbe 2016, 20 (2), 123–4. [DOI] [PubMed] [Google Scholar]
- (49).Mounce BC; Olsen ME; Vignuzzi M; Connor JH Microbiol. Mol. Biol. Rev. 2017, DOI: 10.1128/MMBR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Handa AK; Fatima T; Mattoo AK Front. Chem. 2018, 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Verkerk UH; Peschke M; Kebarle PJ Mass Spectrom. 2003, 38 (6), 618–31. [DOI] [PubMed] [Google Scholar]
- (52).Poltash ML; McCabe JW; Patrick JW; Laganowsky A; Russell DH J. Am. Soc. Mass Spectrom. 2019, 30, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Catalina MI; van den Heuvel RH; van Duijn E; Heck AJ Chem. - Eur. J. 2005, 11 (3), 960–8. [DOI] [PubMed] [Google Scholar]
- (54).Reyzer ML; Brodbelt JS J. Am. Soc. Mass Spectrom. 1998, 9 (10), 1043–1048. [Google Scholar]
- (55).Aue DH; Webb HM; Bowers MT J. Am. Chem. Soc. 1972, 94 (13), 4726–4728. [Google Scholar]
- (56).da Silva EF J. Phys. Chem. A 2005, 109 (8), 1603–7. [DOI] [PubMed] [Google Scholar]
- (57).Brauman JI; Riveros JM; Blair LK J. Am. Chem. Soc. 1971, 93 (16), 3914–3916. [Google Scholar]
- (58).Wagner ND; Kim D; Russell DH Anal. Chem. 2016, 88 (11), 5934–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


