Abstract
Glaucoma, a disease characterized by progressive optic nerve degeneration, can be prevented through timely diagnosis and treatment. We characterized optic nerve photographs of 67,040 UK Biobank participants and used a multitrait genetic model to identify risk loci for glaucoma. A novel glaucoma polygenic risk score (PRS) enables effective risk stratification in unselected glaucoma cases, and modifies penetrance of MYOC p.Gln368Ter, the most common glaucoma-associated myocilin variant. In the unselected glaucoma population, individuals in the top PRS decile reach an absolute risk for glaucoma 10 years earlier than the bottom decile, and are at 15-fold increased risk of developing advanced glaucoma (top 10% vs. remaining 90% OR = 4.20). The PRS predicts glaucoma progression in prospectively monitored early manifest glaucoma cases (P = 0.004), and surgical intervention in advanced disease (P = 3.6 × 10−6). This glaucoma PRS will facilitate the development of a personalized approach for earlier treatment of high-risk individuals, with less intensive monitoring and treatment possible for lower-risk groups.
Glaucoma refers to a group of ocular conditions united by a clinically characteristic optic neuropathy associated with, but not dependent on, elevated intraocular pressure1. It is the leading cause of irreversible blindness worldwide and is predicted to affect 76 million by 20202,3. There is no single definitive biomarker for glaucoma, and diagnosis involves assessing clinical features, with characterization of the optic nerve head carrying the strongest evidential weight. Primary open-angle glaucoma (POAG) is the most prevalent subtype of glaucoma in people of European and African ancestry2,4. POAG is asymptomatic in the early stages, and currently approximately half of all cases in the community are undiagnosed even in developed countries5. Early detection is paramount as existing treatments are unable to restore vision that has been lost, and late presentation is a major risk factor for blindness6. Thus, better strategies to identify high-risk individuals are urgently needed7, and more refined approaches can capitalize on the fact that POAG is one of the most heritable of all common human diseases8–10. The lack of a currently cost-effective screening strategy for glaucoma7, coupled with very high heritability, make glaucoma an ideal candidate disease for the development and application of a polygenic risk score to facilitate risk stratification.
Overlap of features shared by healthy optic nerves with those in early stages of glaucoma makes it a difficult disease to diagnose early, necessitating costly ongoing monitoring of patients for progressive optic nerve degeneration1. Once a glaucoma diagnosis is established, rates of progression vary widely between individuals, and considerable time can elapse before surveillance techniques adequately differentiate slow from more rapidly progressing cases1. Progressive vision loss from glaucoma can be slowed, or in some cases halted, by timely intervention to reduce intraocular pressure using medical therapy, laser trabeculoplasty or incisional surgery1. The ability to predict progression is currently crude, with delays in treatment escalation for high-risk individuals an important and inevitable consequence, as well as substantial cost and morbidity associated with overtreatment of lower risk cases.
The chronicity, heritability, clinical heterogeneity and treatability of POAG make it an ideal candidate for genetic risk profiling11,12. In this study, we evaluated the optic nerve head in 67,040 UK Biobank participants (UKBB), enabling the largest genome-wide association study (GWAS) on optic nerve morphology to date, using vertical cup-disc ratio (VCDR) as an endophenotype for glaucoma. We then incorporated additional genetic data from a second well established glaucoma endophenotype, intraocular pressure (IOP), and combined this with glaucoma disease status using a recently developed multiple trait analysis of GWAS (MTAG)13 approach to first identify new risk loci for glaucoma, and then generate a comprehensive glaucoma polygenic risk score (PRS). We examined the impact of newly implicated glaucoma genes in independent case-control cohorts from Australia, the United States, and the United Kingdom, and then evaluated the utility of the PRS for predicting glaucoma risk, and important clinical outcomes in well-characterized cases across a range of disease severities.
Results
Study design.
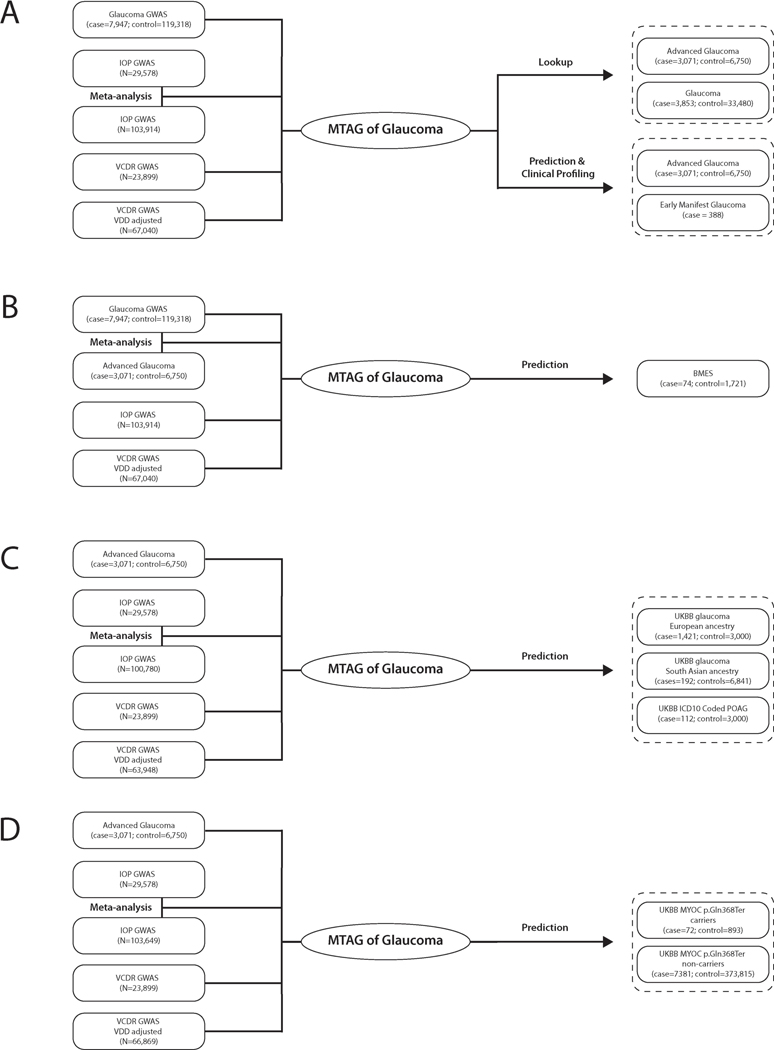
Our overall study design is illustrated in Extended Data Figure 1a. We first conducted a GWAS on glaucoma (7,947 cases and 119,318 controls) and on the key endophenotypes for glaucoma: VCDR (including new data on 67,040 UKBB participants, and International Glaucoma Genetics Consortium, IGGC, n = 23,899) and intraocular pressure (including data on 103,914 UKBB participants and GWAS summary statistics from IGGC, n = 29,578; Supplementary Table 1). These data were then combined using MTAG13 to identify new glaucoma risk loci and to construct a PRS. The clinical significance of the PRS was investigated in advanced glaucoma cases in two populations, and a separate prospectively monitored clinical cohort with early manifest glaucoma. The predictive ability of the PRS was also explored in other datasets; however, to ensure our results generalize to further cohorts, we selected mutually exclusive samples for inclusion in the discovery and testing datasets to ensure no sample overlap. When required, we re-derived the PRS to ensure no sample overlap (Extended Data Fig. 1b-d and Supplementary Note).
Discovery of novel optic nerve morphology loci.
GWAS of VCDR (adjusted for vertical disc diameter) identified 76 statistically independent, genome-wide significant SNPs (66 loci), of which 49 SNPs (43 loci) had not previously been associated with VCDR (Supplementary Figs. 1 and 2, and Supplementary Table 2). Using LD score regression, we found no evidence for genomic inflation (intercept = 1.04, s.e. = 0.01, Supplementary Fig. 3). The genetic correlation between VCDR (adjusted for vertical disc diameter) and glaucoma in UKBB was 0.50 (s.e. = 0.05); the correlation in effect size estimates at the 76 SNPs was 0.60 (P = 9.0 × 10−9, Supplementary Fig. 4). We further combined UKBB VCDR (adjusted for vertical disc diameter) GWAS and IGGC VCDR GWAS summary statistics using MTAG, and identified 107 independent genome-wide significant SNPs (across 90 loci, Supplementary Table 3) for VCDR (adjusted for vertical disc diameter). As previously reported, the genetic correlation between intraocular pressure and glaucoma was high (0.71)15, but as expected the genetic correlation between VCDR (adjusted for vertical disc diameter) and intraocular pressure was substantially lower (0.22, s.e. = 0.03).
Discovery of novel glaucoma loci via multivariate analysis.
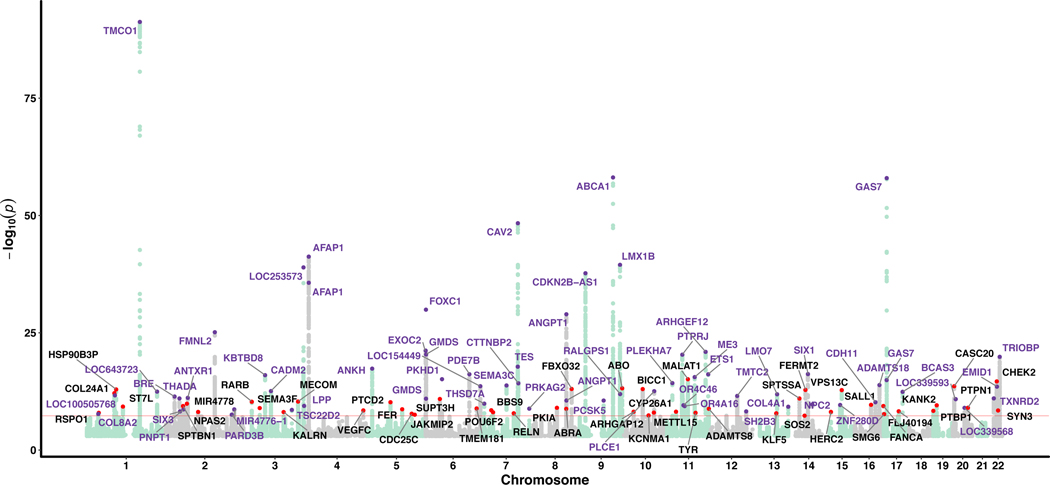
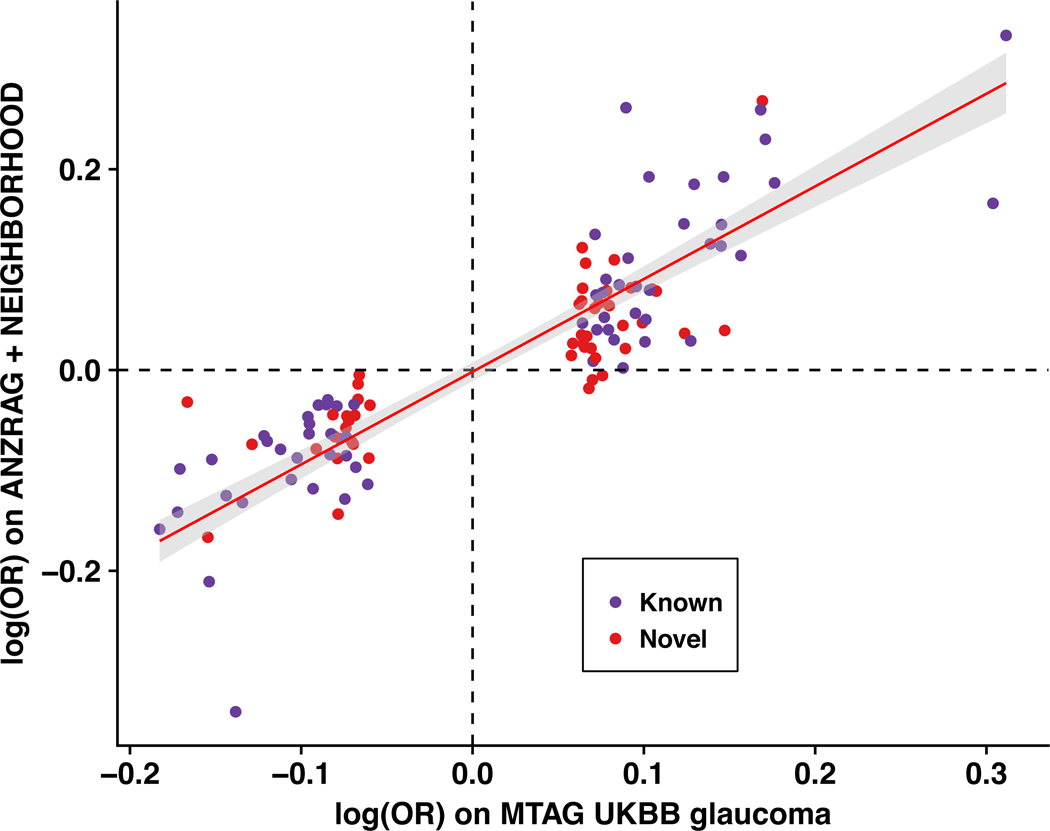
Given the high correlation between glaucoma and its endophenotypes, we then conducted a multivariate GWAS (with 8,002,429 SNPs after quality control) to identify 114 statistically independent SNPs (107 loci, P < 5 × 10−8) associated with glaucoma; this includes all previously published glaucoma loci as well as 49 novel loci (Fig. 1, Supplementary Figs. 5 and 6, and Supplementary Table 4). At the more stringent multiple testing threshold (P < 1 × 10−8) suggested by a simulation study16, 95 loci reach significance, 39 of which are novel (Supplementary Table 4); 27 of the 49 top SNPs at these novel loci were not associated individually with any of the individual input traits at the genome-wide significance level (P = 5 × 10−8) and were only found to reach this threshold for glaucoma due to the MTAG method leveraging the strong correlation between the input traits. We then attempted to replicate the 49 novel SNPs in two independent glaucoma cohorts (ANZRAG and NEIGHBORHOOD). Given the much smaller effective sample size of these replication cohorts (versus the discovery datasets from the MTAG analysis), we did not expect all of the SNPs to be strongly associated; rather, if they were genuine associations, we would expect the ORs to be highly concordant, with some of the smaller ORs being individually non-significant. The concordance between the discovery cohort and our replication cohorts log ORs was excellent (correlation 0.88, P = 1.6 × 10−36), indicating that our multivariate model was successful in identifying genuine glaucoma risk loci (Fig. 2 and Supplementary Fig. 7). Of the 49 novel SNPs, nine were replicated after Bonferroni correction (P < 0.05/49 = 0.001, one-sided test, bold text in Supplementary Table 4), 26 were associated at a nominal significance level (P < 0.05, one-sided test, italic text in Supplementary Table 4), and 46 (94%) were in the expected direction. While the concordance between the multivariate and the glaucoma replication sample log ORs was high, only nine of the 49 loci were significant for glaucoma after correction for multiple comparisons, and further studies are required to replicate the remaining 40 loci for glaucoma.
Figure 1 |. Manhattan plot displaying glaucoma-specific P values from the multi-trait GWAS (MTAG) analysis.
The samples used in multi-trait analysis is presented in Extended Data Figure1a. Novel SNPs are highlighted in red dots, with the nearest gene names in black text. Known SNPs are highlighted in purple dots, with the nearest gene names in purple text. The red line is the genome-wide significance level at 5 × 10-8.
Figure 2 |. Comparison of the effect sizes (log odds ratio) for 114 genome-wide significant independent SNPs identified from the glaucoma multiple trait analysis of GWAS in the UKBB versus those in independent glaucoma cohorts (meta-analysis of ANZRAG and NEIGHBORHOOD).
Pearson’s correlation coefficient is 0.88 (P = 1.6 × 10−36). The red line is the best fit line, with the 95% confidence interval region in grey. Novel glaucoma SNPs are highlighted in red and known SNPs in purple.
We conducted a genome-wide gene-based association analysis and a gene set enrichment analysis to assess which predefined biological pathways were enriched in our multitrait glaucoma GWAS; we found 196 genes and 14 gene sets, respectively, that were significant after Bonferroni correction (Supplementary Tables 5 and 6). The most significant pathways were also previously implicated (i.e. extracellular matrix, collagen, and circulatory system development)15,17. Further studies are warranted to investigate the role of these pathways in the risk of glaucoma.
Optimizing prediction of glaucoma risk by combining correlated traits.
We derived our PRS based on the MTAG of GWAS data from glaucoma and its endophenotypes. As well as increasing the number of SNPs that reach genome-wide significance (mean chi-squared statistic increased from 1.12 to 1.30, implying our effective sample size was 2.59 times larger than if we had used UKBB glaucoma cases and controls alone), our multivariate model improved the power of risk prediction by reducing the error in the estimate of the effect size for every SNP (assuming the MTAG homogeneity assumption is true, see Discussion)13. We first tested the discriminatory power of the MTAG-derived PRS in the ANZRAG cohort of advanced glaucoma. We found SNPs with MTAG P values ≤ 0.001 (corresponding to 2,673 uncorrelated SNPs after LD-clumping at r2 = 0.1 and P-value threshold at 0.001) had the highest Nagelkerke R2 (13.2%) and AUC (0.68, 95%CI: 0.67–0.70) (Supplementary Table 7). The MTAG PRS has better prediction ability than any of the input traits alone (Supplementary Table 8). Based on this, we set the P-value threshold at 0.001 for all the remaining prediction target sets (PROGRESSA, BMES, UKBB).
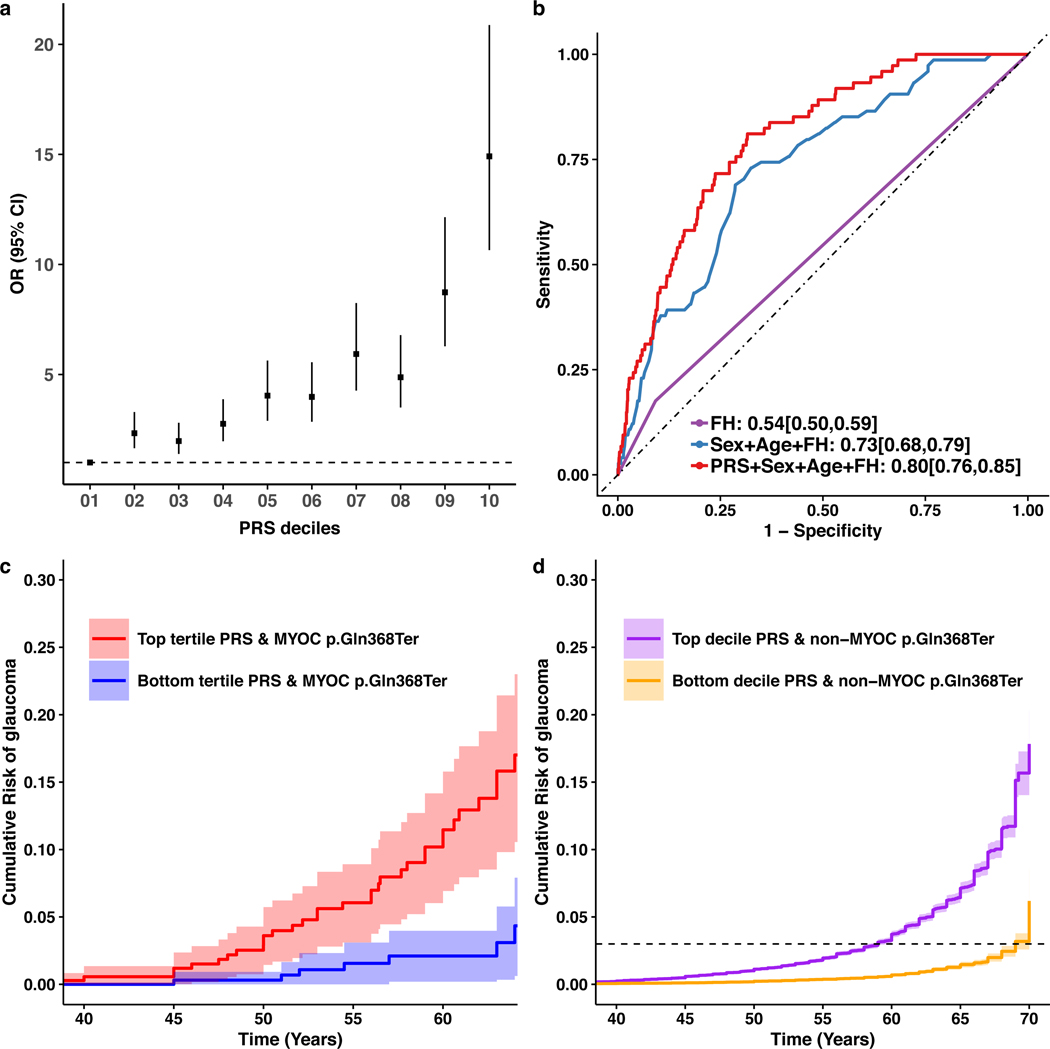
The MTAG-derived PRS was effective at separating advanced glaucoma individuals in terms of risk, with a clear dose-response over deciles (Fig. 3a and Supplementary Fig. 8). In ANZRAG, individuals in the top decile of the PRS had 14.9-fold higher risk (95%CI: 10.7–20.9) relative to the bottom decile, with even better discrimination for the more common high-tension glaucoma (OR = 21.5, 95%CI: 12.5–37.0) than normal-tension glaucoma (Supplementary Fig. 9). We replicated the dose-response of the PRS in a smaller UK advanced glaucoma dataset (Southampton and Liverpool); the top versus bottom PRS decile had OR = 11.6 (95%CI: 6.0–25.3), with again better discrimination for high-tension glaucoma (OR = 12.9, 95%CI: 6.2–31.3). While comparing the top and bottom deciles shows the dose-response across deciles, one can also consider the risk in the high PRS individuals versus all others; when this is done in ANZRAG, the OR is 4.2 and 8.5 in the top 10% and 1%, respectively, of individuals versus all remaining individuals (Supplementary Table 9).
Figure 3 |. Multiple trait analysis of GWAS PRS prediction.
a, Odds ratio (OR) of developing advanced glaucoma in the ANZRAG cohort (with 1,734 advanced glaucoma cases and 2,938 controls) for each PRS decile. The square dots are the OR values (adjusted for sex and the first four principal components) and the error bars are 95% confidence interval. The dashed line is the reference at the bottom PRS decile (OR = 1). b, AUCs of PRS in BMES. The MTAG-derived PRS provided additional predictive ability on top of traditional risk factors (age, sex, and self-reported family history (FH), DeLong’s test P = 0.002). The AUC is based on a logistic regression model with the coefficients for age, sex, FH and PRS estimated from the BMES data (Supplementary Table 10). c, Cumulative risk of glaucoma in UKBB MYOC p.Gln368Ter carriers stratifying by the PRS (adjusted for sex and first six genetic principal components). Here the cumulative risk of tertiles (with 95% confidence intervals) of PRS are displayed given the relatively small number of MYOC p.Gln368Ter carriers (n = 965). d, Cumulative risk of glaucoma for people in the top and bottom decile (with 95% confidence intervals) of PRS of the UKBB who do not have the MYOC p.Gln368Ter variant (adjusted for sex and first six genetic principal components). The dashed line is the reference line of cumulative risk at 3%.
Glaucoma risk score performance in individuals carrying high penetrance variants.
Previous studies indicated that PRS modifies the penetrance of rare BRCA1/2 mutation carriers for breast, ovarian, and prostate cancers18,19. Although the MTAG-derived PRS only contains common variants, given it indexes general glaucoma risk, we hypothesized that it could stratify individuals carrying known high-penetrance glaucoma variants. Pathogenic MYOC (Myocilin) gene variants account for 2–4% of POAG cases among most populations, the most common disease-causing variant being p.Gln368Ter (rs74315329)20. Penetrance is age-related and is lower in population-based than family-based studies20,21. We speculated that this difference in penetrance could be due to enrichment of common glaucoma-associated variants in families modifying age-related penetrance. Within UKBB, we identified 965 MYOC p.Gln368Ter carriers based on imputation (Supplementary Note)22. Figure 3c shows the cumulative risk of glaucoma in p.Gln368Ter carriers, stratifying by PRS tertiles. For p.Gln368Ter carriers in the lowest tertile PRS, glaucoma risk remained very low (2%) up to age 60. In contrast, the highest tertile PRS group had substantially increased risk of early diagnosis, reaching a 6-fold increase in absolute risk of glaucoma by age 60, relative to the lowest PRS tertile (considering whole age range, hazard ratio = 3.4, 95%CI: 1.7–6.6). This supports the utility of PRS in optimizing risk stratification and prediction, and early screening for patients carrying high penetrance MYOC variants in the presence of high PRS scores.
Potential for glaucoma risk score in screening in the general population.
We considered a general population screening scenario using UKBB (PRS was re-derived to ensure no sample overlap; Extended Data Fig. 1d), where we excluded the 965 MYOC p.Gln368Ter carriers. Over the 40–69 year old age range for individuals sampled in UKBB, glaucoma prevalence increases from 0.1% at age 40, reaching 3% (95%CI: 2.9–3.1%) by age 64. The MTAG-derived PRS stratifies UKBB participants very effectively; for those in the top PRS decile, 3% prevalence (prevalence in general population) is reached by age 59, while it takes an additional 10 years for this disease prevalence to be reached for people in the bottom PRS decile. Alternatively, the prevalence can be well stratified by PRS deciles (Fig. 3d).
To benchmark the performance of the MTAG-derived PRS with traditional risk factors, we computed the AUC in datasets for which this was possible: BMES, UKBB glaucoma (broad glaucoma definition), and UKBB POAG (ICD-10 definition) (Fig. 3b, Supplementary Table 11 (PRS was rederived to ensure no sample overlap), and Extended Data Fig. 1). In the BMES, our PRS provided additional predictive ability beyond that imparted by traditional risk factors (age, sex, and self-reported family history (FH)), with a significant change in the AUC (from 0.73 to 0.80, P = 0.002, Fig. 3b). Clear improvement in prediction using this PRS is also observed in people of South Asian ancestry (Supplementary Table 11), though we were underpowered to explore this further across other groups.
A previous study examined the cost-effectiveness requirements for glaucoma screening and highlighted the key age 50–60 bracket7. In the BMES data (Extended Data Fig. 1b), screening only those with a top decile PRS identified 40% of all early onset cases in age 50–60 bracket (40% of the 10 cases, P = 0.013). Such individuals represent a set of individuals likely to benefit from referral for immediate clinical assessment—with skilled clinical examination, retinal imaging, and visual fields. We replicated this result in the UKBB POAG cohort (ICD10 cases in Extended Data Fig. 1c, top 10% PRS screening finds 29% of 24 cases aged 50–60, P = 0.0075). In this way, PRS-based screening would satisfy the cost-effectiveness requirements of Burr et al.7, identify a meaningful proportion of cases, and capture those cases most at risk of severe disease.
Clinical implications of the glaucoma risk score.
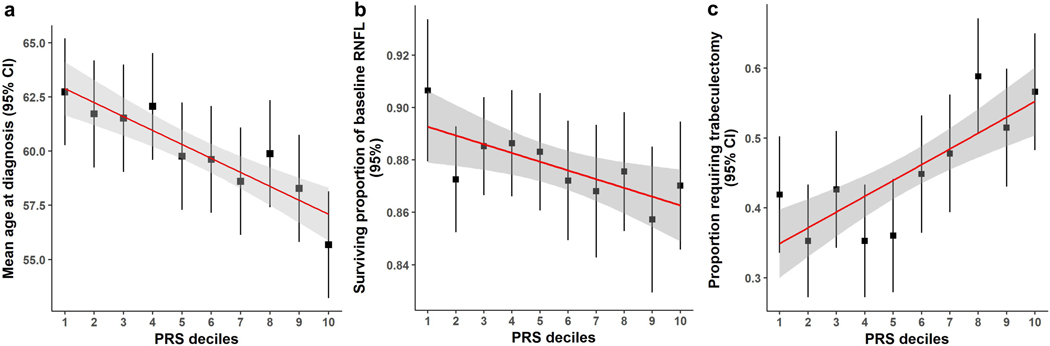
We evaluated the predictive power of the PRS in advanced glaucoma; in 1,336 ANZRAG advanced POAG cases with accurate age at diagnosis information available (Supplementary Table 12), the PRS was significantly associated with age at diagnosis of POAG (P = 1.8 × 10−5). Individuals in the top 10% of the PRS distribution were on average diagnosed 7 years younger than people in the bottom 10% (Fig. 4a). We also found ANZRAG individuals with higher PRS had more family members affected by glaucoma (P = 3.5 × 10−9 ), with the highest decile having twice as many members affected (Supplementary Fig. 10).
Figure 4 |. Clinical implications of the glaucoma PRS.
a, Mean age at diagnosis (years) for each decile of PRS in the ANZRAG cohort (linear regression P = 1.8 × 10−5). A total of 1,336 cases had accurate age at diagnosis information. We calculated the mean age at diagnosis for each decile of PRS, adjusted for sex and the first four principal components in a linear regression model. The square dots are the regression-based mean age at diagnosis, with error bars for 95% confidence intervals. The red line is the line of best fit, with 95% confidence intervals in grey. b, Proportion of preserved baseline retinal nerve fibre layer for PROGRESSA participants with early manifest glaucoma plotted against PRS decile (n = 388; linear regression P = 0.004). The square dots are the retinal nerve fibre layer proportions, with error bars showing 95% confidence intervals. The remaining retinal nerve fibre layer proportion is calculated for the most affected quadrant of the most affected eye of each patient — as determined on optical coherence tomography scans at baseline and latest follow-up scan. c, Proportion of patients requiring trabeculectomy in either eye in the ANZRAG POAG cohort (linear regression P = 3.6 × 10−6). There were 1,360 cases with records of surgical treatment status. The square dots represent the observed average proportion of cases in each decile of PRS who required trabeculectomy, with 95% confidence interval bars. The line of best fit is shown in red, with 95% confidence interval shaded in grey.
Retinal nerve fibre layer thinning is a major structural change evident in early stage glaucoma23. In the early manifest glaucoma (PROGRESSA) cohort, the PRS predicted both the proportion lost and rate of loss of peripapillary retinal nerve fibre layer. Given that glaucomatous loss of retinal ganglion cells generally progresses unequally between eyes, with some quadrants of the retina damaged more rapidly than others, we analyzed the most affected quadrant of the most affected eye in individuals with early manifest glaucoma and greater than two years of longitudinal optical coherence tomography data. The PRS was significantly associated with the proportion of retinal nerve fibre layer lost from baseline to most recent review, even after adjustment for known risk factors: age, intraocular pressure and retinal nerve fibre layer thickness at presentation (P = 0.004; Fig. 4b and Supplementary Table 13). Expressed in terms of rate of loss, each decile change in PRS was associated with an accelerated progression rate of 0.05 μm/year, which was twice the rate of thinning per mmHg (approximately 1 decile change for intraocular pressure) of baseline intraocular pressure (0.022 μm/year).
Incisional surgery for glaucoma (trabeculectomy) is highly effective at reducing intraocular pressure, but has significant complications which can adversely impact vision1. Trabeculectomy is performed either when intraocular pressure is unable to be controlled with medical or laser therapy, or when there is progressive visual field loss despite well controlled intraocular pressure. Patients with a high PRS were more likely to have undergone surgery for glaucoma (Fig. 4c and Supplementary Figure 11). In the ANZRAG cohort of POAG cases, a higher PRS was associated with requiring trabeculectomy, even after adjustment for maximum recorded intraocular pressure and age (P = 3.6 × 10−6), the OR of requiring trabeculectomy in either eye for people in the top PRS decile was 1.78 (95%CI: 1.07–3.00) compared to the bottom decile. We observed a very similar trend in our UK replication (Southampton/Liverpool) samples (Supplementary Fig. 11).
Discussion
Through a large-scale multivariate GWAS we identified novel genes for glaucoma, the leading cause of irreversible blindness worldwide2. Despite a smaller replication cohort, many of these novel hits were replicated, and all but three SNPs showed a consistent direction of effect. We then expanded this analysis to derive a PRS and interrogated its utility across a wide spectrum of clinically relevant glaucoma outcomes.
From the multivariate GWAS, we identified 49 novel loci associated with glaucoma (nine of which replicated after correction for multiple comparisons in independent glaucoma case-control cohorts; 26 were replicated with P < 0.05). Interestingly, most of the loci replicated at P < 0.001 are at genes previously associated with glaucoma risk factors (myopia, CCT, IOP, VCDR). Specifically, RSPO1 is associated with ocular axial length24. BICC1 is associated with myopia and corneal astigmatism25,26,27. POU6F2 modulates corneal thickness and increases glaucoma risk in animal experiments28. FBXO32, PTPN1, and VPS13C are associated with IOP15,29,30, while CASC20 was identified in our VCDR (adjusted for vertical disc diameter) GWAS. These findings show that our multivariate GWAS improves power to identify novel glaucoma genes and advance our understanding of the causes of glaucoma risk.
The MTAG-derived PRS was validated in independent samples, confirming its high predictive ability. Individuals in the top PRS decile were at 15-fold increased risk of advanced glaucoma, and at 21.5-fold increased risk of advanced high tension glaucoma, relative to the bottom decile, which represents a substantial improvement on previously reported genetic profiling strategies (where, based on SNPs that were genome-wide significantly associated with intraocular pressure and SNPs previously associated with VCDR and glaucoma, top decile individuals had a 5.6-fold increased risk)15. This new glaucoma PRS also outperforms those derived from other well-studied conditions; for example, our OR comparing the top 1% PRS individuals versus the remaining individuals was 8.5, which is higher than that seen in a recent study which surveyed coronary artery, atrial fibrillation, type 2 diabetes, inflammatory bowel disease and breast cancer31. The etiology of complex diseases depends on both environmental and genetic factors; thus, PRS alone will never achieve the very high predictive power (e.g. AUC > 0.99) required for accurate population screening32. Our glaucoma PRS will be primarily useful for stratifying individuals into risk groups; for example in the BMES data, screening the top decile of the PRS in individuals between 50–60 years old identifies 40% of cases. Moreover, as argued by Khera et al.31, individuals with a high PRS for glaucoma are likely to be at a similar risk to individuals carrying rare “high penetrance” MYOC mutations21. Finally, the PRS performance for glaucoma is particularly noteworthy given the clinical implications of identifying at-risk individuals and the prevention of irreversible blindness with readily available treatment proven to be effective at preventing visual loss.
While current treatments are effective in preventing or reducing POAG progression12, many patients are not diagnosed before irreversible damage to visual function has already occurred. Earlier diagnosis of glaucoma can reduce glaucoma blindness, and our work demonstrates that people with a higher PRS require earlier clinical assessment. In the UKBB, individuals in the top PRS decile reach an equivalent absolute risk for glaucoma 10 years earlier than people in the bottom decile. In advanced glaucoma cases, individuals in the top decile were diagnosed 7 years earlier than those in the bottom decile. Similarly, the MTAG-derived PRS was associated with significantly earlier disease onset in UK Biobank MYOC p.Gln368Ter carriers who are at high disease risk. The MTAG-derived PRS can also identify people with early manifest glaucoma who are at higher probability of disease progression, as well as the likelihood of requiring surgical intervention, which is highly effective at reducing intraocular pressure, but carries substantial treatment morbidity meaning it should always be targeted specifically to those at higher risk of disease progression and blindness.
A concern with the MTAG method is the homogeneous assumption, which could be violated for some SNPs that have no effect on one trait but are non-null for other traits (i.e. it is possible that a small number of the variants may be more specific for IOP or VCDR rather than glaucoma). The homogeneity assumption has been studied in detail by Turley et al.13 We have evaluated the possible inflation using max False Discovery Rate (maxFDR) as recommended13. The baseline maxFDR for MTAG glaucoma-specific input GWAS summary statistics is 0.049, and the maxFDR for MTAG glaucoma-specific output summary statistics is 0.03. As these are similar, there is no evidence of inflation due to violation of the homogeneity assumption. As recommended by the MTAG authors, we also performed replication analysis to assess the credibility of novel SNPs in two independent data sets (an Australasian cohort of advanced glaucoma (ANZRAG) and a consortium of cohorts from the United States (NEIGHBORHOOD)); this analysis shows there is very good concordance between the MTAG-based effect sizes and those from the glaucoma cohorts. Furthermore, using MTAG output instead of the individual input traits improves the predictions in independent cohorts (Supplementary Table 8), providing additional evidence that we are not merely identifying IOP- or VCDR-specific loci that have no effect on glaucoma. Further research needs to be undertaken to investigate the biological mechanisms of these novel genes on glaucoma risk.
A limitation of this work, is that in our 7,947 UKBB glaucoma cases, only a small proportion had documented disease subtype; however, since the proportion of UK glaucoma cases that have POAG is high (87% in a recent study4), this is unlikely to have a large influence on our results. A further limitation is that it is not yet clear how applicable our findings are to other populations. We showed that the PRS improved prediction accuracy over and above traditional risk factors in homogeneous groups (as defined by genetic principal components) of either European or South Asian ancestry. The performance of the PRS in other populations should be tested to investigate the generalizability of our findings. The performance of the PRS in aiding clinical decision making and guiding earlier treatment could be evaluated prospectively in a longitudinal intervention study, with participants randomized to have their PRS provided or withheld from their treating specialist.
In summary, we have applied a multivariate approach using weighted data on glaucoma, and endophenotypes intraocular pressure and VCDR, to identify novel glaucoma loci, and develop a polygenic risk score. This PRS was shown to be predictive of: 1) increasing risk of advanced glaucoma; 2) glaucoma status significantly beyond traditional risk factors; 3) earlier age of glaucoma diagnosis; 4) high levels of absolute risk in persons carrying high penetrance glaucoma variants; 5) increasing likelihood of disease progression in early stage disease, and 6) increasing likelihood of incisional glaucoma surgery in advanced disease. This glaucoma PRS has good predictive power across a range of clinical cohorts and its application will facilitate the rational allocation of resources through clinical screening and timely treatment in high-risk patients, with reduced clinical monitoring costs in lower risk groups.
Methods
Study design and overview.
Our overall study design is illustrated in Extended Data Figure 1. We first conducted a GWAS on glaucoma and on the key endophenotypes for glaucoma: VCDR and intraocular pressure. These data were then combined using MTAG13, a method for combining multiple genetically correlated traits to maximize power for identifying new loci and improving genetic risk prediction. Specifically, our MTAG analysis outputs glaucoma-specific effect size estimates and P-values for single nucleotide polymorphisms (SNPs) across the genome. Newly associated loci (P < 5 × 10−8) were validated in two independent cohorts with well-characterised POAG. We created a PRS based on the MTAG GWAS summary statistics. The clinical significance of the PRS was investigated in advanced glaucoma cases in two populations, and a separate prospectively monitored clinical cohort with early manifest glaucoma. The predictive ability of the PRS was also explored in other datasets; however, to ensure our results generalize to further cohorts, we selected mutually exclusive samples for inclusion in the discovery and testing datasets to ensure no sample overlap. When required, we re-derived the PRS to avoid any sample overlap (Extended Data Fig. 1). Study procedures were performed in accordance with the World Medical Association Declaration of Helsinki ethical principles for medical research.
Study populations.
Detailed information of individual studies, phenotypic definitions, and genetic quality control procedures are provided in the Supplementary Note.
The UK Biobank (UKBB) is a population-based study of half a million people living in the United Kingdom33. We measured VCDR and vertical disc diameter in all subjects with gradable retinal images (67,040 participants following exclusions, detailed in Supplementary Note) and undertook a GWAS to identify SNPs influencing optic nerve head morphology. Vertical disc diameter adjustment of the VCDR was used to account for optic cup and disc size covariation14,34. To improve power in the multi-trait analysis, we combined the VCDR data with data on corneal-compensated intraocular pressure (103,914 participants) and glaucoma (7,947 cases, 119,318 controls) in the MTAG analysis15. We also used publicly available VCDR and intraocular pressure GWAS summary results for individuals of European descent from the International Glaucoma Genetics Consortium (IGGC; nVCDR = 23,899, nintraocular pressure = 29,578)35.
The Australian & New Zealand Registry of Advanced Glaucoma (ANZRAG) comprises 3,071 POAG cases of European descent, who were compared to 6,750 controls36,37. For sub-analyses restricted to advanced POAG, there were 1,734 advanced POAG cases and 2,938 controls, and of these cases 1,336 participants had accurate age at diagnosis information available. Replication of the ANZRAG findings was performed using 332 advanced glaucoma cases from Southampton and Liverpool in the United Kingdom; for case-control analysis, cases were matched to 3,000 randomly selected European ancestry individuals from the QSkin Sun and Health study38. The National Eye Institute Glaucoma Human Genetics Collaboration Heritable Overall Operational Database (NEIGHBORHOOD) GWAS results were generated through meta-analyzing summary data from eight independent datasets (3,853 POAG cases, 33,480 controls) of European ancestry from the United States39.
The Blue Mountains Eye Study (BMES) is a population-based cohort study investigating the etiology of common ocular diseases among suburban residents aged 49 years or older in Australia5. Data from 74 POAG cases and 1,721 controls of European descent with genotype information were included.
The Progression Risk Of Glaucoma: RElevant SNPs with Significant Association (PROGRESSA) study is a prospective longitudinal study of the clinical and genetic risk factors, and course of early-stage glaucoma (n = 388). Patients with confirmed early manifest POAG on sequential automated perimetry testing were consecutively recruited from ophthalmology clinics in South Australia (detailed criteria in Supplementary Note). Individuals underwent six-monthly evaluation of intraocular pressure, optic disc assessment, retinal nerve fibre layer analysis by optical coherence tomography, and achromatic Humphrey visual field perimetry. Longitudinal data were used from all visits since baseline presentation; participants were followed for one to eight years. The change in retinal nerve fibre layer was measured between the baseline optical coherence tomography and the most recent scan in the most-affected quadrant of the most-affected eye. Treating clinicians and graders were unaware of the patient’s genetic risk for glaucoma or any PRS data.
POAG in the ANZRAG, NEIGHBORHOOD, BMES, and PROGRESSA cohorts was defined as outlined previously40, and in accordance with the consensus statement from the World Glaucoma Association41. Intraocular pressure was not used in the clinical case definition of POAG41.
Statistical analysis.
Detailed information on the statistical analysis is provided in the Supplementary Note.
For the VCDR (adjusted for vertical disc diameter) and intraocular pressure GWAS in UKBB, we used linear mixed models (BOLT-LMM software) to account for cryptic relatedness and population stratification adjusting for sex, age and the first ten principal components42. We meta-analyzed UKBB intraocular pressure GWAS results with those from the IGGC using the inverse variance weighted method (METAL software) 43. For the UKBB glaucoma GWAS, we removed relatives (pi-hat > 0.2 calculated using identity by descent determined based on autosomal markers) and used PLINK software for association analysis44.
We then conducted a multitrait GWAS using the MTAG (version 1.0.7) software to combine the European descent GWAS summary statistics from UKBB glaucoma, UKBB VCDR (adjusted for vertical disc diameter), IGGC VCDR and the intraocular pressure meta-analysis (Extended Data Fig. 1)13. MTAG performs joint analysis of GWAS summary results from related traits to improve statistical power to identify new genes and to maximize the predictive ability of our polygenic risk scores13. In MTAG, GWAS summary results from related traits are used to construct the variance–covariance matrix of their SNP effects and estimation error; MTAG improves the accuracy of effect estimates by incorporating information from other genetic correlated traits. The MTAG method explicitly models sample overlap in the input studies and provides valid estimates even when sample overlap is present13. To benchmark the increase in effective sample size relative to just using UKBB glaucoma, we calculated , where and are the mean chi-squared statistics from MTAG and the UKBB glaucoma analyses, respectively13.
We used a stepwise model selection procedure in the GCTA-COJO software to identify independent genome-wide significant SNPs45. Gene-based and pathway analysis were conducted in MAGMA (v1.06), as implemented in FUMA (version 1.3.1)46,47.
Prediction was based on the estimated glaucoma odds ratios (OR) from the MTAG analysis. To derive a PRS, we considered a range of P-value thresholds (5 × 10−8, 1 × 10−5, 0.001, 0.05, 1) with LD-clumping r2 = 0.1 for inclusion of SNPs in the prediction model, applying each to our first prediction cohort (advanced glaucoma from ANZRAG). To avoid falsely inflating prediction accuracy, we applied the threshold with greatest predictive value in ANZRAG (P ≤ 0.001) for the subsequent predictions into other target sets (rather than repeatedly taking the best P-value threshold for each of the datasets). We tested the LDpred48 approach for PRS construction although the predictions were no better than those from the thresholding approach described above. There was no sample overlap between any of the training and target datasets (Extended Data Fig. 1).
Bivariate LD score regression was used to estimate the genetic correlation between pairs of traits49. The “pROC” package was used to calculate the area under the curve (AUC)50. Analyses were performed with R software51.
Data availability.
UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. GWAS summary statistics from the glaucoma MTAG analysis are available for research uses at URL (https://doi.org/10.6084/m9.figshare.10635854) after publication. We will return the derived data fields following the UK biobank policy and in due course they will be available through the UK Biobank Access Management System.
Extended Data
Extended Data Fig. 1. Study design.
We applied the multi-trait analysis of GWAS (MTAG) algorithm to datasets of European descent (unless otherwise specified). a, We applied MTAG to four datasets (glaucoma case-control GWAS from the UKBB; GWAS meta-analysis of intraocular pressure (IOP) from the International Glaucoma Genetics Consortium (IGGC) and the UKBB; Vertical cup-disc ratio (VCDR) GWAS data that was either adjusted for vertical disc diameter (VDD) in the UKBB dataset; or not adjusted for VDD in the IGGC). Novel variants identified through this analysis were then confirmed in two independent data sets: an Australasian cohort of advanced glaucoma (ANZRAG) and a consortium of cohorts from the United States (NEIGHBORHOOD). The clinical significance of the PRS derived from the MTAG analysis was validated in independent samples: first, in advanced glaucoma cases (ANZRAG and samples from Southampton/Liverpool in the UK), and second, in a prospectively monitored clinical cohort with early manifest glaucoma (PROGRESSA). b, Prediction in BMES, where we removed the IGGC VCDR and IGGC IOP GWAS from the training datasets, given that they contain BMES data. c, Prediction in the UKBB glaucoma and ICD-10 POAG cases. Here we removed all glaucoma cases and 3,000 controls with IOP/VCDR measurements as well as their relatives from UKBB VCDR/IOP GWAS. We also evaluated the performance of PRS in non-European ancestry (192 cases and 6,841 controls of South Asian ancestry in UKBB). d, Cumulative risk of glaucoma in UKBB. For the analysis of MYOC p.Gln368Ter carriers (n = 965; cases = 72; controls = 893), participants were stratified into tertiles of PRS. We also examined cumulative risk of glaucoma in the general population (i.e. in MYOC p.Gln368Ter non-carriers, n = 381,196; cases = 7,381; controls = 373,815) stratifying by deciles of the PRS. The discovery and testing datasets were designed to derive the PRS with no sample overlap (Supplementary Note).
Supplementary Material
Acknowledgements
This work was conducted using the UK Biobank Resource (application number 25331) and publicly available data from the International Glaucoma Genetics Consortium. The UK Biobank was established by the Wellcome Trust medical charity, Medical Research Council (UK), Department of Health (UK), Scottish Government, and Northwest Regional Development Agency. It also had funding from the Welsh Assembly Government, British Heart Foundation, and Diabetes UK. The eye and vision dataset has been developed with additional funding from The NIHR Biomedical Research Centre at Moorfields Eye Hospital and the UCL Institute of Ophthalmology, Fight for Sight charity (UK), Moorfields Eye Charity (UK), The Macula Society (UK), The International Glaucoma Association (UK) and Alcon Research Institute (USA). This work was also supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (#1107098; 1116360, 1116495, 1023911), the Ophthalmic Research Institute of Australia, the BrightFocus Foundation, UK and Eire Glaucoma Society and Charitable Funds from Royal Liverpool University Hospital. S.M., J.E.C., K.P.B., and A.W.H. are supported by NHMRC Fellowships. S.M. was supported by an Australian Research Council Future Fellowship. L.R.P. is supported by NIH R01 EY015473. X.H. is supported by the University of Queensland Research Training Scholarship and QIMR Berghofer PhD Top Up Scholarship. We thank David Whiteman, Rachel Neale and Catherine Olson for providing access to QSKIN samples for use as controls as part of NHMRC Grant 1063061. We thank Scott Wood, John Pearson and Scott Gordon from QIMR Berghofer for support. The NEIGHBORHOOD consortium is supported by NIH grants P30 EY014104, R01 EY015473 and R01 EY022305.
Footnotes
Competing interests
D.A.M. is consultant/advisor to Allergan, Inc. J.E.C., A.W.H. and S.M. are listed as co-inventors on a patent application for the use of genetic risk scores to determine risk and guide treatment.
References:
- 1.Weinreb RN & Khaw PT Primary open-angle glaucoma. Lancet 363, 1711–1720 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Tham Y-C et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Quigley HA & Broman AT The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol 90, 262–267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan MPY et al. Glaucoma and intraocular pressure in EPIC-Norfolk Eye Study: cross sectional study. BMJ 358, j3889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell P, Smith W, Attebo K. & Healey PR Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology 103, 1661–1669 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Fraser S, Bunce C. & Wormald R. Risk factors for late presentation in chronic glaucoma. Invest. Ophthalmol. Vis. Sci 40, 2251–2257 (1999). [PubMed] [Google Scholar]
- 7.Burr JM et al. The clinical effectiveness and cost-effectiveness of screening for open angle glaucoma: a systematic review and economic evaluation. Health Technol. Assess 11, iii–iv, ix–x, 1–190 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Wang K, Gaitsch H, Poon H, Cox NJ & Rzhetsky A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat. Genet 49, 1319–1325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanfilippo PG, Hewitt AW, Hammond CJ & Mackey DA The heritability of ocular traits. Surv. Ophthalmol 55, 561–583 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Choquet H. et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun 9, 2278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leske MC, Heijl A, Hyman L, Bengtsson B. & Komaroff E. Factors for progression and glaucoma treatment: the Early Manifest Glaucoma Trial. Curr. Opin. Ophthalmol 15, 102–106 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Garway-Heath DF et al. Latanoprost for open-angle glaucoma (UKGTS): a randomised, multicentre, placebo-controlled trial. Lancet 385, 1295–1304 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Turley P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bengtsson B. The variation and covariation of cup and disc diameters. Acta Ophthalmol. 54, 804–818 (1976). [DOI] [PubMed] [Google Scholar]
- 15.MacGregor S. et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat. Genet 50, 1067–1071 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Zheng Z, Visscher PM & Yang J. Quantifying the mapping precision of genome-wide association studies using whole-genome sequencing data. Genome Biol. 18, 86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L. et al. Genome-wide analysis identified 17 new loci influencing intraocular pressure in Chinese population. Sci. China Life Sci 62, 153–164 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Kuchenbaecker KB et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst 109, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecarpentier J. et al. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J. Clin. Oncol 35, 2240–2250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt AW, Mackey DA & Craig JE Myocilin allele-specific glaucoma phenotype database. Hum. Mutat 29, 207–211 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Han X. et al. Myocilin gene Gln368Ter variant penetrance and association with glaucoma in population-based and registry-based studies. JAMA Ophthalmol. 137, 28–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gharahkhani P. et al. Accurate imputation-based screening of Gln368Ter Myocilin variant in primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci 56, 5087–5093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na JH et al. Detection of glaucoma progression by assessment of segmented macular thickness data obtained using spectral domain optical coherence tomography. Invest. Ophthalmol. Vis. Sci 53, 3817–3826 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Cheng C-Y et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am. J. Hum. Genet 93, 264–277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickrell JK et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet 48, 709–717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhoeven VJM et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet 45, 314–318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes MC et al. Identification of a candidate gene for astigmatism. Invest. Ophthalmol. Vis. Sci 54, 1260–1267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King R. et al. Genomic locus modulating corneal thickness in the mouse identifies POU6F2 as a potential risk of developing glaucoma. PLoS Genet. 14, e1007145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khawaja AP et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet 50, 778–782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao XR, Huang H, Nannini DR, Fan F. & Kim H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum. Mol. Genet 27, 2205–2213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References:
- 33.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X. et al. Genome-wide association analysis of 95,549 individuals identifies novel loci and genes influencing optic disc morphology. Hum. Mol. Genet (2019). doi: 10.1093/hmg/ddz193 [DOI] [PubMed] [Google Scholar]
- 35.Springelkamp H. et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet 26, 438–453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souzeau E. et al. Australian and New Zealand Registry of Advanced Glaucoma: methodology and recruitment. Clin. Experiment. Ophthalmol 40, 569–575 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Gharahkhani P. et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet 46, 1120–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen CM et al. Cohort profile: the QSkin Sun and Health Study. Int. J. Epidemiol 41, 929–929i (2012). [DOI] [PubMed] [Google Scholar]
- 39.Wiggs JL et al. The NEIGHBOR consortium primary open-angle 655 glaucoma genome-wide association study: rationale, study design, and clinical variables. J. Glaucoma 22, 517–525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon YH, Fingert JH, Kuehn MH & Alward WLM Primary open-angle glaucoma. N. Engl. J. Med 360, 1113–1124 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinreb RN, Garway-Heath DF, Leung C, Medeiros FA & Liebmann J. Diagnosis of Primary Open Angle Glaucoma: WGA consensus series - 10. (Kugler Publications, 2017). [Google Scholar]
- 42.Loh P-R et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willer CJ, Li Y. & Abecasis GR METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–75, S1–3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Leeuw CA, Mooij JM, Heskes T. & Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe K, Taskesen E, van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilhjálmsson BJ et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet 97, 576–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulik-Sullivan B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robin X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team. R: A Language and Environment for Statistical Computing. (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. GWAS summary statistics from the glaucoma MTAG analysis are available for research uses at URL (https://doi.org/10.6084/m9.figshare.10635854) after publication. We will return the derived data fields following the UK biobank policy and in due course they will be available through the UK Biobank Access Management System.