How lipid droplets are formed is largely unclear. Chen et al. show that FIT2 recruits ER tubule-forming proteins and septin cytoskeletons to facilitate nascent LD formation by providing membrane curvature and bulging scaffold.
Abstract
Lipid droplets (LDs) are critical for lipid storage and energy metabolism. LDs form in the endoplasmic reticulum (ER). However, the molecular basis for LD biogenesis remains elusive. Here, we show that fat storage–inducing transmembrane protein 2 (FIT2) interacts with ER tubule-forming proteins Rtn4 and REEP5. The association is mainly transmembrane domain based and stimulated by oleic acid. Depletion of ER tubule-forming proteins decreases the number and size of LDs in cells and Caenorhabditis elegans, mimicking loss of FIT2. Through cytosolic loops, FIT2 binds to cytoskeletal protein septin 7, an interaction that is also required for normal LD biogenesis. Depletion of ER tubule-forming proteins or septins delays nascent LD formation. In addition, FIT2-interacting proteins are up-regulated during adipocyte differentiation, and ER tubule-forming proteins, septin 7, and FIT2 are transiently enriched at LD formation sites. Thus, FIT2-mediated nascent LD biogenesis is facilitated by ER tubule-forming proteins and septins.
Introduction
Lipid droplets (LDs) are spherical organelles covered with a monolayer of phospholipids (Martin and Parton, 2006; Murphy, 2001; Walther et al., 2017). Inside LDs, neutral lipids, triacylglycerols, and sterol esters are packed into an oil-like core (Martin and Parton, 2006; Murphy, 2001; Walther et al., 2017). LDs are generally used for intracellular fat storage, as a defense against lipotoxicity, and as a source for energy and lipid metabolism (Listenberger et al., 2003; Martin and Parton, 2006; Murphy, 2001). In addition to an excess of free fatty acids, LD formation can also be triggered by a variety of stresses and viral infection (Fei et al., 2009; Miyanari et al., 2007). In some bacteria, LDs can bind to DNA and serve as a survival strategy (Zhang et al., 2017). In humans, dysregulation of LD biogenesis and metabolism has been linked to obesity-related diseases, particularly fatty liver disease (Gross and Silver, 2014; Onal et al., 2017).
Eukaryotic LDs originate in the ER, the major site of lipid synthesis (Choudhary et al., 2015; Jacquier et al., 2011; Kassan et al., 2013). The accumulation of neutral lipids between the leaflets of ER membranes results in a lens-like structure (Choudhary et al., 2015; Gross and Silver, 2014; Walther et al., 2017). Further growth of the lens results in the budding of nascent LDs into the cytosol. These initial LDs often return to tether and fuse with the ER. The continuity between the cytosolic leaflet of the ER and the LD monolayer allows translocation of proteins and lipids from the ER to the LDs (Salo et al., 2016; Wilfling et al., 2014; Wilfling et al., 2013), and LDs can expand by attaching to the ER or merging with other LDs (Murphy et al., 2010; Wilfling et al., 2013).
The LD surface is decorated with proteins (Na et al., 2013), many of which contribute to LD function. Given the monolayer coverage and hydrophobic nature of the core, LD proteins can either associate as a peripheral membrane protein or insert shallowly into the surface layer (Hristova et al., 1999; Wilfling et al., 2013). Thus, a transmembrane (TM) hairpin that does not traverse the entire lipid bilayer or an amphipathic helix that can dip into membranes using its hydrophobic side are frequently found in LD-targeted proteins, and multi-spanning membrane proteins are usually excluded. Membrane curvature has consistently been shown to be preferred during LD formation (Santinho et al., 2020).
Several key proteins involved in LD biogenesis have been identified. The fat storage–inducing transmembrane protein (FIT) family, a conserved ER-resident protein family, plays an important role in LD biogenesis. FIT1 is expressed mostly in skeletal muscle, whereas FIT2 is expressed ubiquitously, with the highest levels found in adipose tissue (Kadereit et al., 2008). When FITs are depleted or deleted postnatally, LD accumulation is blocked and lethal enteropathy triggered (Goh et al., 2015; Miranda et al., 2014). Conversely, LD formation can be induced when FITs are overexpressed (Kadereit et al., 2008). FIT2 is capable of binding to DAG and triacylglycerol (Gross et al., 2011), regulates lipid homeostasis in the ER (Becuwe et al., 2020; Yap et al., 2020), and has been implicated in the regulation of nascent LD budding (Choudhary et al., 2015). A second key protein is the ER-localized integral membrane protein seipin. Deletion of seipin results in defects in LD biogenesis (Cartwright et al., 2015; Wang et al., 2016). Seipin has been suggested to be involved in ER-LD contact and to be critical for LD maturation (Binns et al., 2010; Grippa et al., 2015; Salo et al., 2016; Sui et al., 2018; Szymanski et al., 2007; Wang et al., 2016; Yan et al., 2018). The cytosolic phosphatidate phosphatase lipin also contributes to LD biogenesis. Mutations in lipin result in a decreased number of LDs (Adeyo et al., 2011). In a mouse model, deletion of lipin was shown to cause massive adipose tissue loss (Péterfy et al., 2001; Reue et al., 2000). Localization of lipin is phosphorylation dependent (O’Hara et al., 2006; Santos-Rosa et al., 2005). Furthermore, lipin has been suggested to regulate adipogenesis and phospholipid biosynthesis transcriptionally via nuclear import (Reue and Zhang, 2008). The detailed actions of these proteins are not clear; they may cooperate at different steps of LD biogenesis. Alternatively, they may work on different populations of LDs.
ER shapes are tightly linked to LD biogenesis. Two morphological domains, tubules and sheets, exist in the ER (Shibata et al., 2006). The tubular ER network is generated mainly by two classes of integral membrane proteins: reticulons/REEPs that stabilize curvature in tubules (Hu et al., 2008; Shibata et al., 2008; Voeltz et al., 2006) and the dynamin-like GTPase atlastin (ATL), which connects tubules through membrane fusion (Bian et al., 2011; Hu et al., 2009). Proteomic analysis of ER tubules has revealed that they are mainly involved in membrane trafficking, lipid metabolism, and organelle contact (Wang et al., 2017), many functions of which are directly related to LD biogenesis. ER sheets can be generated and stabilized by Climp-63, kinectin, and p180 (Shibata et al., 2010). Sheet-like membranes often expand when lipids accumulate in the ER or when ER stress is induced (Ron and Walter, 2007; Sriburi et al., 2004).
The involvement of ER-shaping protein in LD biogenesis has been hinted at. Many of these proteins, including ATL (Klemm et al., 2013), REEP1 (Falk et al., 2014; Renvoisé et al., 2016), and the three-way junction stabilizer lunapark (LNP; Krahmer et al., 2013), are found in LD-enriched fractions. ATL has been proposed to control LD size in Caenorhabditis elegans (Klemm et al., 2013). In addition, REEP1-knockout (KO) mice exhibit LD defects and severe lipoatrophy (Renvoisé et al., 2016). However, the precise mechanism underlying the regulation of LD biogenesis by these proteins is still largely unknown.
Through a screen of ER morphology regulators, we identified FIT2 in C. elegans. Further analysis revealed that FIT2 interacts with ER tubule-forming proteins and cytoskeletal septin. Similar LD defects are observed when FIT2 or these interacting proteins are depleted. Live cell imaging of nascent LD formation suggests that tubule-forming proteins and septin contribute to the early FIT2-mediated steps of LD formation.
Results
FIT2 regulates ER morphology
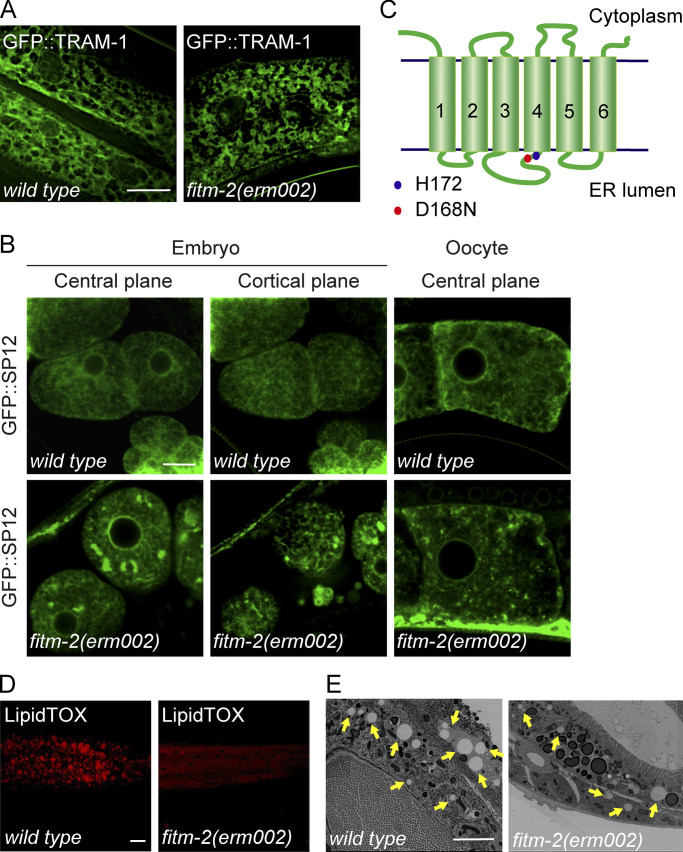
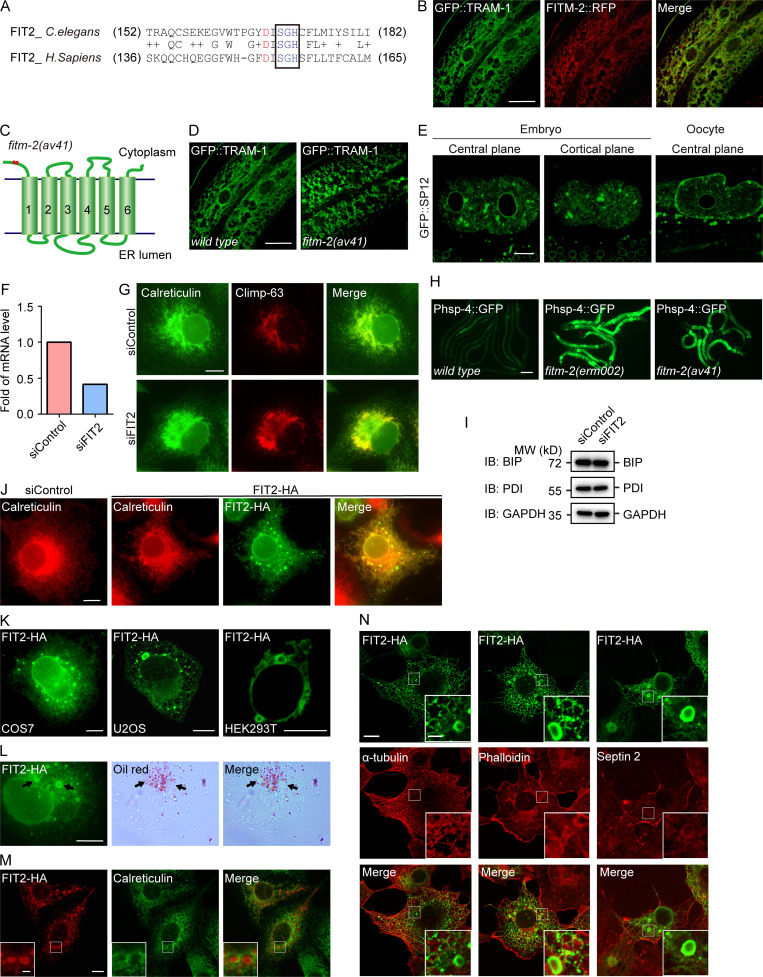
In an effort to identify new regulators of ER morphology in C. elegans, we performed a genetic screen in worms expressing GFP::TRAM-1 in the hypodermis. One mutant allele, erm002, exhibited condensed ER patches resembling ER sheet expansion (Fig. 1 A). When embryos and oocytes were monitored, patches of ER were also observed with another ER marker, GFP::SP12 (Fig. 1 B). Mapping and sequencing revealed that erm002 possessed an aspartic acid to asparagine mutation at conserved position 168 in FITM-2, a worm homologue of FIT2 (Fig. 1 C and Fig. S1 A). RFP-fused FITM-2 colocalized with ER marker TRAM-1 (Fig. S1 B), confirming that FITM-2 is an ER-resident protein. The same ER morphology was seen in another FITM-2 mutant, av41, which has a 5′-end stop codon and represents a null allele (Fig. S1, C–E).
Figure 1.
ER and LD morphology in fitm-2 mutant. (A) Representative 3D-SIM images of wild-type and fitm-2(dt002) animals carrying the qxIs439 (Phyp7::gfp::tram-1) transgene at the young adult stage, showing ER morphology in the hypodermis. Scale bar, 10 µm. (B) ER morphology of early embryos and oocytes, shown as single confocal planes, in wild-type and fitm-2(erm002) animals carrying the ojIs23 (Ppie-1::gfp::sp12) transgene. Scale bar, 10 µm. (C) Topology of FITM-2. The blue dot indicates the predicted enzyme active site His-172 in C. elegans. The red dot indicates the mutation site in fitm-2(erm002). (D) LipidTOX staining of larval stage L4 animals. Images are 3D projections of 7.5-µm confocal z-stacks that covered the posterior body forward from the vent. Scale bar, 10 µm. (E) Electron micrographs of the middle gut of the indicated young adult animals. Yellow arrows indicate LDs. Scale bar, 4 µm.
Figure S1.
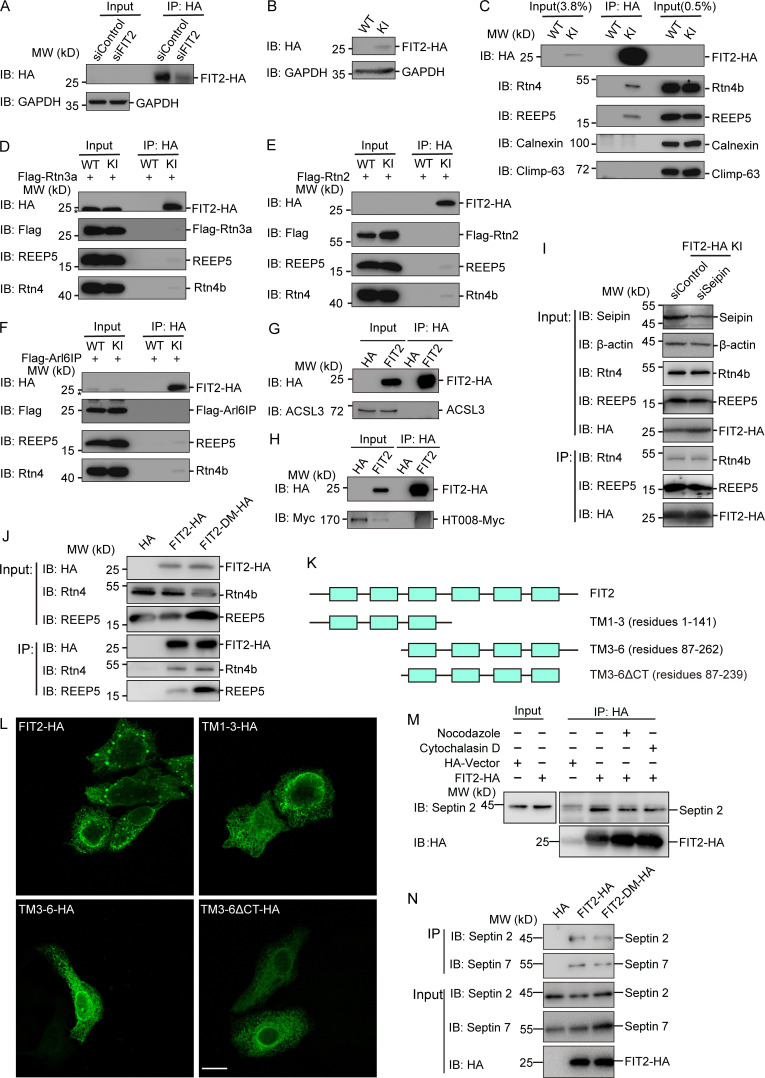
FIT2 is required in the maintenance of ER homeostasis. (A) Sequence alignment of FIT2. The mutation site in fitm-2(erm002) is highlighted in red. The black box outlines the conserved SGH sequence containing the predicted active residue His. (B) Colocalization of FITM-2::RFP and ER marker GFP::TRAM-1 in the hypodermis. 3D-SIM images. Scale bar, 10 µm. (C) Depiction of fitm-2(av41). Red dots show two stop codons on the N-terminus of FITM-2. (D) Representative images of fitm-2(av41) animals at the young adult stage carrying the qxIs439 (Phyp7::gfp::tram-1) transgene. The ER of WT animals shown in B was used for comparison. 3D-SIM images. Scale bar, 10 µm. (E) ER morphology of early embryos and oocytes in fitm-2(av41) animals carrying the ojIs23 (Ppie-1::gfp::sp12) transgene. Single confocal planes. Scale bar, 10 µm. (F) Relative mRNA level of FIT2 in control and FIT2-depleted COS-7 cells, indicating the knockdown efficiency. (G) ER morphology in control and FIT2-depleted COS-7 cells. Cells were transfected with control or FIT2 siRNA for 48 h, fixed, and stained with anti-calreticulin and anti–Climp-63 antibodies. Scale bar, 10 µm. (H) Representative images of wild-type and fitm-2 mutants at the young adult stage carrying the zcls4 (Phsp-4::gfp) transgene. Images were acquired under the same excitation light intensity. Scale bar, 100 µm. (I) Expression level of PDI and BIP in control and FIT2-knockdown COS-7 cells. (J) ER morphology in control and FIT2-HA–overexpressing COS-7 cells. Scale bar, 10 µm. (K) Bubble-like structures in COS-7, U2OS, and HEK293T cells overexpressing FIT2-HA. Single confocal planes. Scale bars, 10 µm. (L) COS-7 cells were overexpressed with FIT2-HA and stained with anti-HA antibody, as well as Oil Red. Arrows indicate bubble-like structures stained by Oil Red. Scale bar, 10 µm. (M) COS-7 cells were overexpressed with FIT2-HA and stained with anti-HA and anti-calreticulin antibodies. The insets show high-magnification details in the white squares. Scale bars: 10 µm (main panel) or 2 µm (inset). (N) Immunofluorescence assay using phalloidin (Dylight 405; Thermo Fisher Scientific) and antibodies targeting HA, septin 2, and α-tubulin in COS-7 cells transfected with FIT2-HA. Scale bar, 10 µm; 2 µm (inset). BIP, binding immunoglobulin protein; H. sapiens, Homo sapiens; PDI, protein disulfide isomerase; siControl, small interfering control; siFIT2, small interfering FIT2.
Next, we tested whether mammalian homologue FIT2 plays a role in ER morphogenesis. When FIT2 was depleted using siRNA in COS-7 cells (Fig. S1 F), ER sheets expanded, as indicated by sheet-specific marker Climp-63 (Fig. S1 G). These results suggest that FIT2 activity is linked to ER morphology and erm002 is a loss-of-function mutant of FITM-2.
Using mutant allele av41, FITM-2 deletion has been shown to result in small LDs with decreased numbers (Choudhary et al., 2015). The same phenotype was seen when LDs were visualized by LipidTOX staining in erm002 (Fig. 1 D). LD defects were further confirmed by EM (Fig. 1 E). As dysregulation of LD biogenesis often leads to altered lipid metabolism and subsequent ER stress, we tested the levels of ER stress in FITM-2 mutants using the Phsp-4::GFP reporter. Both erm002 and av41 exhibited substantial ER stress (Fig. S1 H). Interestingly, when mammalian FIT2 was depleted in COS-7 cells, no ER stress was detected (Fig. S1 I). Taken together, these results confirm that FITM-2 is involved in LD biogenesis, perhaps via regulation of ER morphology, and different FIT2 members may affect ER homeostasis to different extents.
To further investigate whether FIT2 activity is associated with ER shaping in higher eukaryotes, we overexpressed FIT2 in COS-7 cells. The ER morphology exhibited some changes, including redistribution of ER sheets toward the cell periphery (Fig. S1 J). In addition, puncta, or even larger bubble-like structures, were frequently observed. These structures were FIT2-positive (Fig. 2 A and Fig. S1 J) and were observed in a variety of cell lines, including U2OS and HEK293T cells (Fig. S1 K). To test whether these structures are LD related, we treated the cells with LipidTOX. The bubble-like structures contained a very low density of neutral lipids (Fig. 2 A). Therefore, these bubbles are likely byproducts of abnormal LD formation but not actual LDs. The same conclusion was drawn when another neutral lipid dye, Oil Red, was used (Fig. S1 L). Even though FIT2 has been proposed to be a regulator of LD budding, we did not observe an increased number of LDs when FIT2 was overexpressed. These results further suggest that FIT2 plays a role in LD formation and that FIT2 alone is not sufficient for LD formation.
Figure 2.
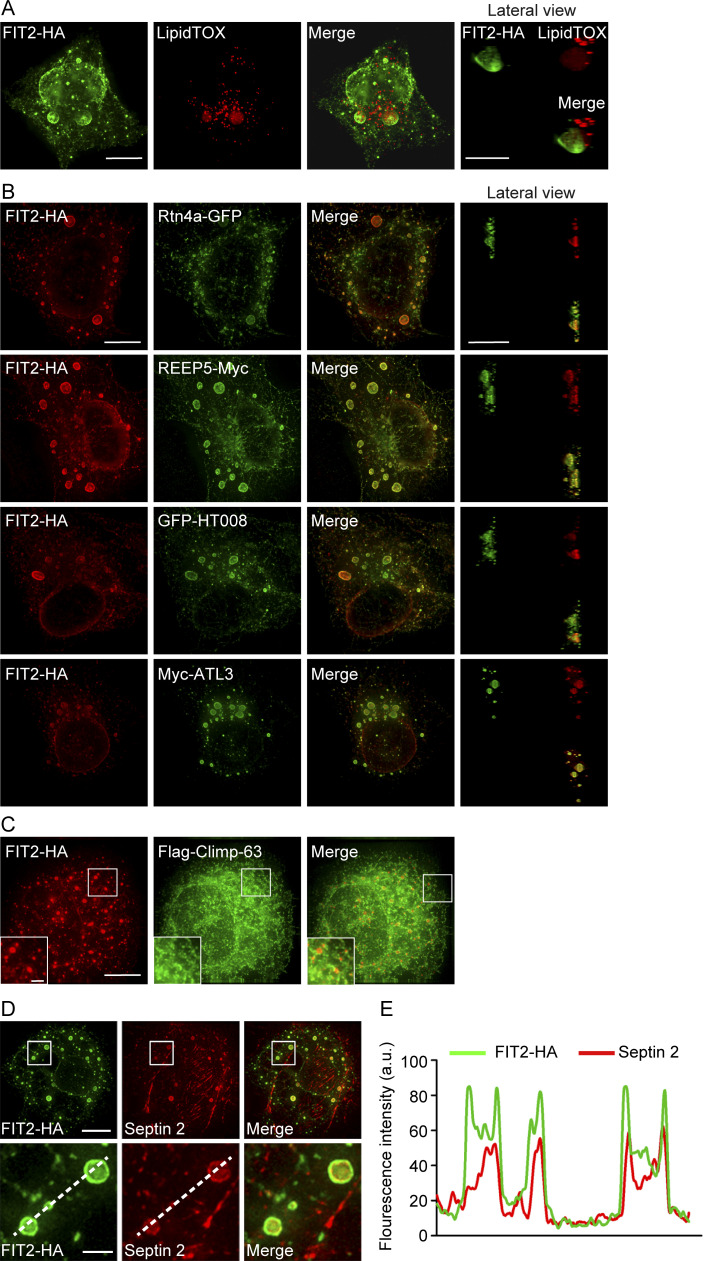
Proteins enriched in FIT2-induced ER bubbles. (A) COS-7 cells were transfected with FIT2-HA and stained with anti-HA antibodies and LipidTOX. Representative images were captured by 3D-SIM. A lateral view of a 3D bubble-like structure is shown on the right. Scale bars, 10 µm. (B) COS-7 cells were cotransfected with FIT2-HA and the indicated ER tubule proteins and stained with anti-HA antibodies alone or together with anti-Myc antibodies. Representative images were captured by 3D-SIM. Lateral views of 3D bubble-like structures are shown on the right. Scale bars, 10 µm. (C) As in B, but with COS-7 cells coexpressing FIT2-HA and Flag–Climp-63. The insets show magnified details in white squares. Scale bars: 10 µm; 2 µm (inset). (D and E) Distribution of endogenous septin 2 filaments (red) visualized in COS-7 cells expressing FIT2-HA (green). The bottom images show enlargements of the boxed regions on FIT2-induced ER bubbles. The line scans of the red and green fluorescence intensities indicate the colocalization of septin 2 filaments and FIT2-induced ER bubbles. Scale bar, 10 µm; 2 µm (inset).
Next, we tested whether FIT2-induced bubbles contain a specific set of ER proteins. When ER tubule-forming protein Rtn4a or REEP5 was cotransfected into COS-7 cells, it decorated the bubbles (Fig. 2 B). Similarly, when other known ER tubule-enriched proteins, such as HT008 and ATL3, were cotransfected, they localized to the rim of the bubbles (Fig. 2 B). In contrast, when Climp-63, an ER sheet-specific marker, was coexpressed with FIT2, it expanded the ER sheets but was not found in FIT2-positive bubbles (Fig. 2 C). Another commonly used luminal ER marker, calreticulin, also did not merge with the bubble signals (Fig. S1 M). These results suggest that FIT2 is concentrated in ER tubule-enriched regions.
FIT2 interacts with ER tubule-forming proteins
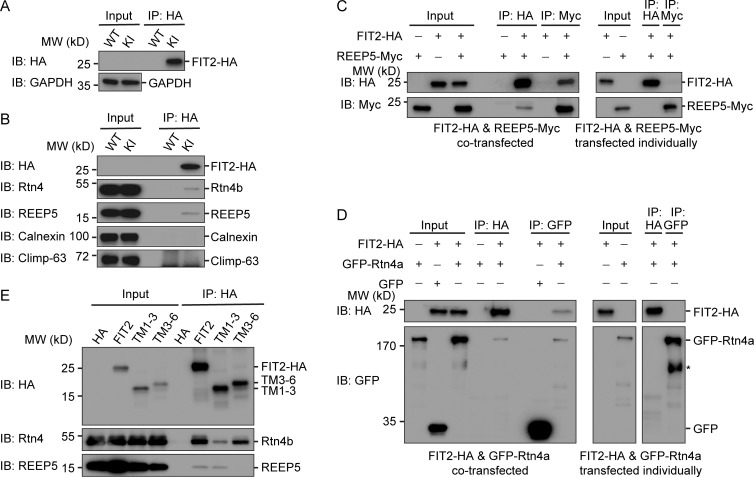
To test whether FIT2 interacts with ER tubule-enriched proteins, we performed CRISPR-based knock-in (KI) of two tandem HA-tags on the C-terminus of endogenous FIT2 in HepG2 and U2OS cells. The endogenous FIT2-HA could be detected using anti-HA antibodies (Fig. 3 A; and Fig. S2, A and B). When FIT2 was precipitated by anti-HA antibodies, both endogenous Rtn4 and REEP5 coprecipitated (Fig. 3 B and Fig. S2 C). As expected, unrelated ER resident proteins Climp-63 and calnexin did not interact with FIT2 under these conditions (Fig. 3 B and Fig. S2 C).
Figure 3.
Interactions between tubule-forming proteins and FIT2. (A) IP of FIT2-HA in WT and FIT2-HA KI HepG2 cells. WT and FIT2-HA KI cells were lysed in 1% digitonin-containing buffer. IP was performed with anti-HA antibodies. The samples were analyzed by immunoblotting (IB) with the indicated antibodies. (B) coIP of FIT2 and ER tubule-forming proteins in HepG2 cells. IP was performed as in A. Samples were analyzed by IB with antibodies of different ER membrane proteins. (C) FIT2-HA and REEP5-Myc were cotransfected into HEK293T cells and solubilized in triton-containing buffer or transfected individually into cells in different dishes, followed by mixing of the Triton-solubilized cell extracts. IP was performed with anti-HA or anti-Myc antibodies. (D) As in C, but with cells expressing FIT2-HA and/or GFP-Rtn4a. The asterisk (*) indicates degraded GFP-Rtn4a. (E) coIP of FIT2 truncations and Rtn4/REEP5. HEK293T cells expressing FIT2-HA, FIT2 truncations, or empty vector were lysed in 1% Triton-containing buffer and cell lysates immunoprecipitated with anti-HA antibodies.
Figure S2.
FIT2 interacts with tubule-forming proteins and septins. (A) IP of FIT2-HA in FIT2-HA KI HepG2 cells transfected with control or siFIT2 RNA. Cells were lysed in 1% digitonin-containing buffer. IP was performed with anti-HA antibodies. The samples were analyzed by immunoblotting (IB) with the indicated antibodies. (B) Verification of FIT2-HA KI U2OS cells by Western blotting. (C) coIP of FIT2 and ER tubule-forming proteins in U2OS cells. IP was performed as in A. Samples were analyzed by immunoblotting with antibodies of different ER membrane proteins. (D) coIP of FIT2 and Flag-Rtn3a. WT and FIT2-HA KI HepG2 cells were transfected with Flag-Rtn3a. IP was performed as in A with anti-HA antibodies. The asterisk indicates nonspecific bands. (E) coIP of FIT2 and Flag-Rtn2. (F) coIP of FIT2 and Flag-Arl6IP. The asterisk indicates nonspecific bands. (G) coIP of FIT2-HA and ACSL3. HEK293T cells were transfected with FIT2-HA or empty vector and lysed in 1% Triton-containing IP buffer. IP was performed with anti-HA antibody in 0.5% Triton-containing IP buffer. Samples were analyzed by Western blotting with anti-HA and anti-ACSL3 antibodies. (H) coIP of FIT2-HA and HT008-Myc. As in G, except with cells coexpressing FIT-HA and HT008-Myc. (I) coIP of endogenous FIT2-HA and tubule-forming proteins in FIT2-HA KI U2OS cells transfected with siControl or siSeipin. (J) coIP of overexpressed FIT-HA and FIT2 H155A/H214A mutant (FIT2-DM-HA) with ER tubule-forming proteins in HEK293T cells. (K) Schematic diagram of the FIT2 constructs used. (L) Immunofluorescence assay detected the subcellular location of the indicated FIT2 constructs. Scale bar, 20 µm. (M) 293T cells were transfected with FIT2-HA or HA-vector and then treated with nocodazole or cytochalasin D. The coIP assay was performed as in Fig. 4 A. (N) As in M, but coIP of exogenous FIT2-HA or FIT2-DM-HA with endogenous septin 2 and 7. siControl, small interfering Control; siFIT2, small interfering FIT2; siSeipin, small interfering seipin.
We also observed coprecipitation of Rtn3a with FIT2 (Fig. S2 D). However, when Arl6IP1 or Rtn2, other reticulon homology domain (RHD)–containing proteins, were tested, nearly no interaction was seen with FIT2, presumably because of RHD sequence variation (Fig. S2, E and F). In addition, two unrelated ER-resident and TM hairpin–containing proteins, ACSL3 and HT008, did not coprecipitate with FIT2-HA (Fig. S2, G and H). Taken together, these results show that FIT2 interacts specifically with ER tubule-forming proteins.
FIT2 is a six-TM–containing membrane protein with a few loops facing the cytosol (Fig. 1 B). We investigated the specific regions to which tubule-forming proteins bind. FIT2-HA and Flag-REEP5 were transfected into COS-7 cells individually or together. As described above, when cotransfected, FIT2-HA was able to precipitate Flag-REEP5 and vice versa (Fig. 3 C). However, when they were transfected individually and the extracts mixed, no coprecipitation was observed (Fig. 3 C). Similarly, FIT2-HA and Rtn4a-GFP (Fig. 3 D) only coimmunoprecipitated when they resided in the same membranes. These results suggest that FIT2 likely interacts with tubule-forming proteins through TM domains.
To further dissect the RHD-interacting regions in FIT2, we engineered truncation mutants of FIT2 (Fig. S2 K). When the first three TM domains of FIT2 (TM1-3, residues 1–141) were expressed and precipitated using anti-HA antibodies, endogenous REEP5 and Rtn4 were still pulled down, with reduced binding to Rtn4 (Fig. 3 E). In contrast, when the last four TM domains of FIT2 (TM3-6, residues 87–262) were used, Rtn4 retained its interactions with FIT2 but REEP5 was no longer bound (Fig. 3 E). These results suggest that tubule-forming proteins interact with different regions of the FIT2 TM domains. Notably, neither FIT2 truncated mutant was able to induce bubble-like structures in the ER (Fig. S2 L), confirming that all FIT2 TM domains are functionally important.
FIT2 interacts with cytoskeletal septins
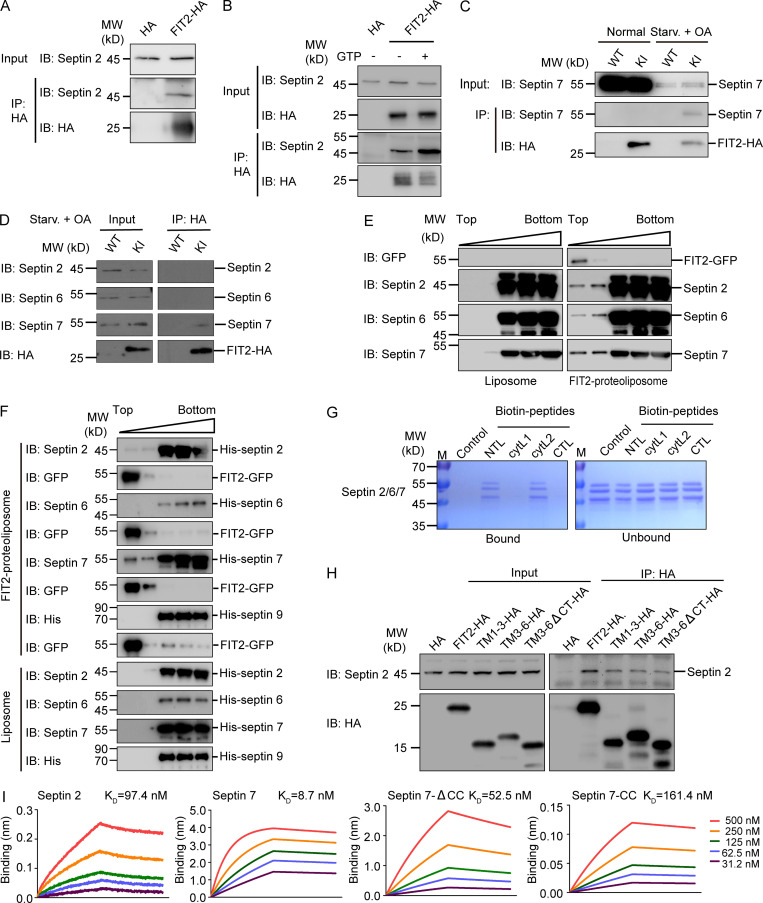
Cytoskeleton structures, particularly microtubules and septins, are suspected to play a role in LD growth and transportation (Akil et al., 2016; Boström et al., 2005; Moreno-Castellanos et al., 2017; Shubeita et al., 2008). When we attempted to identify additional FIT2-interacting proteins, we found that the septin cytoskeleton, but not microtubules or F-actins, colocalizes with FIT2-induced ER bubbles (Fig. 2, D and E; and Fig. S1 N). Septins form GTP-dependent oligomers in the form of a filament (Kim et al., 2011; Sirajuddin et al., 2007). In mammals, the core complex of septin filaments is composed of a septin 2 dimer flanked by two heterotypic dimers of septin 6 and septin 7 (Fig. S3 A). In cells expressing exogenous FIT2-HA, when the extracts were precipitated by anti-HA antibodies, septin 2, the core component of the septin skeleton, was found in the precipitate (Fig. 4 A). The interaction between FIT2 and endogenous septin 2 was enhanced when the extracts were treated with GTP (Fig. 4 B), presumably promoting septin assembly.
Figure S3.
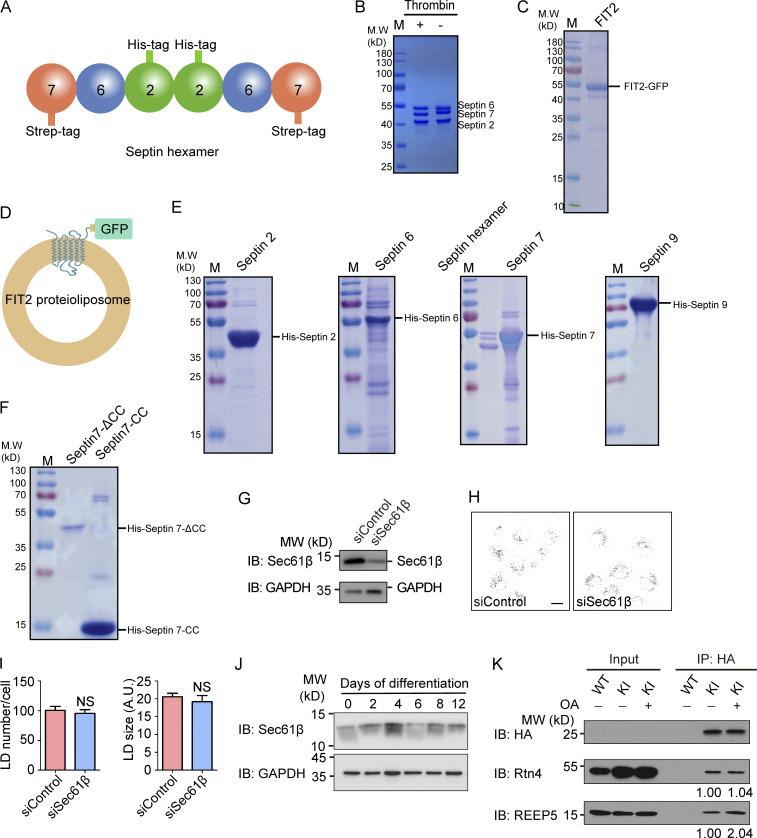
Purification of septins and FIT2. (A) Representation of the septin2/6/7 hexamer. (B) Purified septin2/6/7 hexamer was detected by SDS-PAGE and Coomassie blue staining. The strep tag on the C-terminus of septin 7 was cleaved by thrombin. (C) The purified FIT2-strep-His8-GFP fusion protein was detected by SDS-PAGE and Coomassie blue staining. (D) Representation of FIT2 proteoliposomes. (E) Purified His-tagged septins 2, 6, 7, and 9 were detected by SDS-PAGE and Coomassie blue staining. (F) Purified His-tagged septin 7 CC and septin 7ΔCC were detected by SDS-PAGE and Coomassie blue staining. (G–I) As in Fig. 5 A but with COS-7 cells depleted of Sec61β. n = 51–88 cells/group. Mann–Whitney test; NS, P > 0.05. Scale bar, 10 µm. Error bars represent SEM. (J) As in Fig. 6 D, Sec61β and GAPDH were detected by immunoblotting (IB). (K) coIP of FIT2 and Rtn4/REEP5 before and after OA treatment. WT and FIT2-HA KI HepG2 cells were delipidated by starving for 60 h or treating with OA for 15 min and lysed in 1% digitonin buffer. IP was performed with anti-HA antibodies, and samples were analyzed by immunoblotting. The relative amount of Rtn4 (coIP) or REEP5 (coIP) compared with FIT2-HA (IP) was quantified by Gel-Pro analyzer software. siSec61β, small interfering Sec61β.
Figure 4.
Interactions between septins and FIT2. (A) HEK293T cells transfected with FIT2-HA or empty HA vector were lysed, and coIP assay was performed by incubating cell lysates with anti-HA agarose beads. Immunoblotting (IB) was performed using the indicated antibodies. (B) As in A, but with or without 5 mM GTP in the lysates. (C) FIT2 KI U2OS cells were cultured under normal conditions or LD biogenesis conditions (fatty acid starvation [Starv.] for 60 h, OA treatment for 15 min). After DSP-mediated protein cross-linking, coIP of FIT2-HA with endogenous septin 7 was performed using anti-HA agarose beads. (D) As in C, coIP of FIT2-HA and endogenous septin 2, 6, and 7 in the LD biogenesis condition. (E and F) Coflotation assay using FIT2-reconstituted proteoliposomes and purified septin2/6/7 hexamer (E) or individual septins (septin 2, 6, 7, and 9; F). Proteins in each fraction were detected by IB using the indicated antibodies. (G) In vitro peptide pull-down assays were performed using the indicated biotinylated peptides and purified septin2/6/7 hexamer, followed by SDS-PAGE and Coomassie blue staining. (H) HEK293T cells were transfected with the indicated FIT2-HA truncations, coIP and IB assays performed as in A. (I) BLI assays using biotinylated FIT2 NTL peptide-coated sensor and individually purified septin 2, septin 7, septin 7-ΔCC, and septin 7-CC at the gradient concentration of 31.2–500 nM. The dissociation constant (Kd) is shown. CTL, C-terminal loop; ctyL1, cytosolic loop 1; ctyL2, cytosolic loop 2.
Next, we tested whether endogenous FIT2 binds to the septin cytoskeleton. FIT2-HA precipitated from FIT2 KI cells, but none of the endogenous septin components, coprecipitated (Fig. 4 C). We reasoned that FIT2-septin interactions were transient during nascent LD formation; therefore, we performed coimmunoprecipitation (coIP) upon oleic acid (OA)–induced LD biogenesis after starvation and clearance of existing LDs. In addition, we added dithiobis(succinimidyl propionate) (DSP) to stabilize transient interactions via chemical cross-linking. Under these conditions, endogenous septin 7 readily coprecipitated with endogenous FIT2-HA (Fig. 4 C), despite the levels of septin 7 being greatly reduced due to starvation (Fig. 4 C). Interestingly, endogenous FIT2 only exhibited detectable interactions with septin 7, not septin 2 and 6 (Fig. 4 D). These results confirm that FIT2 and cytoskeletal septin interact endogenously, likely through septin 7.
We also examined whether FIT2 and septins interact directly using purified component. GFP-tagged full-length FIT2 was expressed in SF9 insect cells, purified, and reconstituted into proteoliposomes (Fig. S3, C and D). Septin hexamer, which was formed by septin 2, 6, 7 (two of each), was either coexpressed and copurified or individually isolated (Fig. S3, B and E) and subsequently incubated with FIT2-containing proteoliposomes. When FIT2 vesicles were floated in a density gradient, we observed coflotation of septin 2/6/7 (Fig. 4 E). When individual septins were tested, only septin 7 had prominent coflotation; septin 2 interacted weakly with FIT2, and septin 6 showed no detectable binding (Fig. 4 F). We also tested septin 9, another common septin cytoskeleton component, and found no appreciable interactions with FIT2 (Fig. 4 F and Fig. S3 E). Taken together, these results indicate that FIT2 interacts directly with septin 7 of the cytoskeletal septins.
The cytosolic localization of septin suggests that it may engage FIT2 through its cytosolic loops. Thus, we performed pull-down assays using purified septin hexamer and synthesized biotinylated peptides according to the sequences of the FIT2 loops (N-terminal loop, residues 1–23, [NTL]; cytosolic loop 1, residues 79–93, [cytL1]; cytosolic loop 2, residues 167–190, [cytL2]; C-terminal loop, residues 240–262, [CTL]; Table S1). When peptides were precipitated by streptavidin beads, coprecipitated septins were analyzed. We found that the NTL and cytL2 interacted with septin (Fig. 4 G). These binding sites were confirmed in cells using truncated FIT2 (Fig. 4 H). We also analyzed interactions between purified septin 2 or 7 and the NTL peptide using bio-layer interferometry (BLI). Biotinylated peptide was immobilized to streptavidin sensor and incubated with increasing concentrations of septin 2 or 7. The association and disassociation of septins with the sensor, which influenced the thickness of the layer on the sensor tip, were measured as an interference wavelength shift. Consistently, we observed stronger binding by septin 7 than septin 2 (Fig. 4 I). To narrow down the regions of septin 7 that interact with FIT2, we purified the C-terminal coiled coil (CC) domain of septin 7 (residues 332–437) and a truncated septin 7 that lacks the CC domain (SEPT7-ΔCC, residues 1–331; Fig. S3 F). SEPT7-ΔCC, which consisted of mostly the GTPase domain, had similar interactions with FIT2-NTL similar to those of full-length septin 7, whereas SEPT7 CC had a much weaker interaction with the peptide (Fig. 4 I). Finally, we disassembled other cytoskeleton components, including microtubules and actin filaments, and found no impact on septin association with FIT2 (Fig. S2 M). Collectively, these results indicate that septin 7, particularly its GTPase domain, physically interacts with FIT2.
FIT2-interacting proteins regulate LD biogenesis
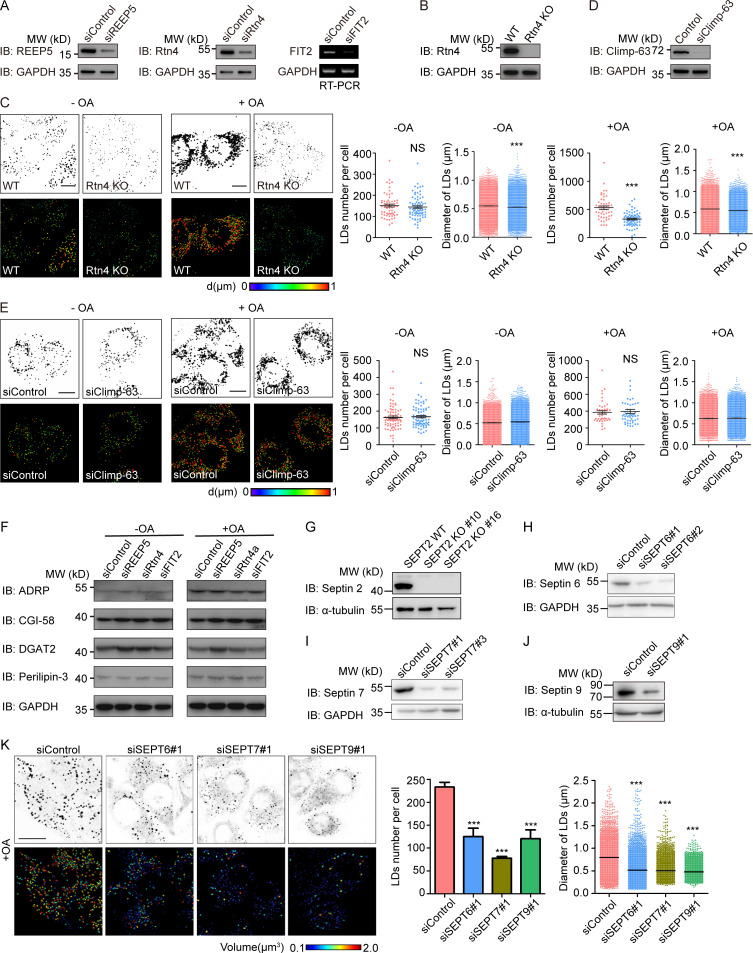
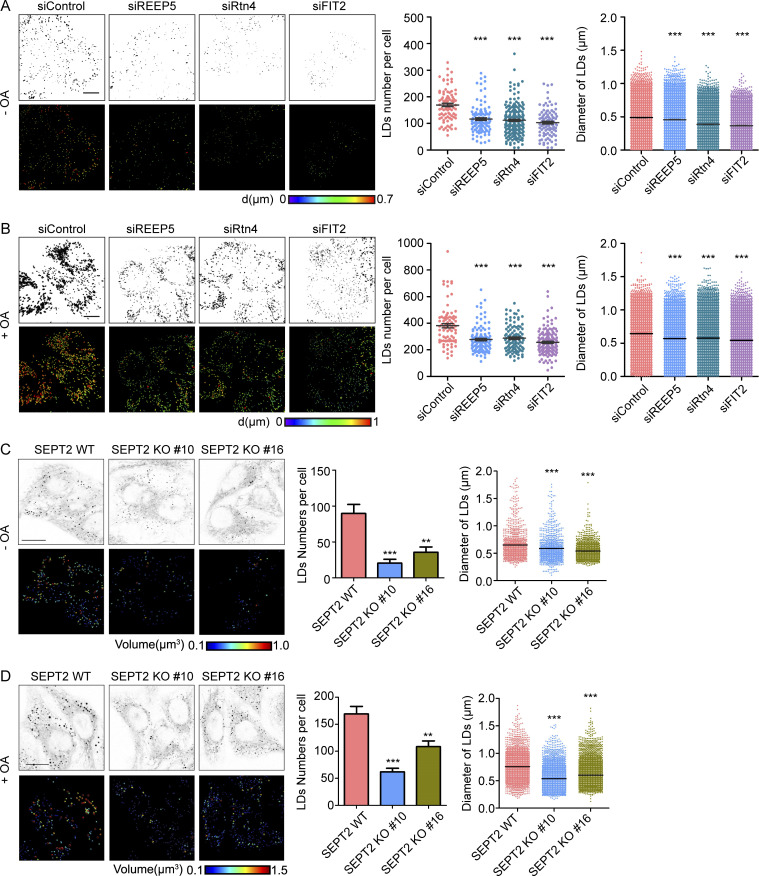
To test whether newly identified FIT2-interacting proteins participate in LD biogenesis, we depleted these proteins individually in HepG2 cells (Fig. S4, A and B), which have a known tendency to store neutral lipids in LDs, and monitored LD number and size by LipidTOX staining. As in FIT2-depleted cells, both LD number and size were decreased in cells depleted of REEP5 or Rtn4 (Fig. 5 A and Fig. S4 C). Similar results were obtained when cells were treated with OA to stimulate LD formation (Fig. 5 B and Fig. S4 C), but depletion of an unrelated ER protein, Sec61β, had no effect (Fig. S3, G–I). When tubule-forming proteins are eliminated, the morphological balance in the ER shifts toward sheets (Voeltz et al., 2006). However, depletion of Climp-63, an ER-shaping protein for sheet formation, did not affect LD biogenesis (Fig. S4, D and E). In addition, depletion of REEP5 or Rtn4 did not alter the levels of a known regulator of LD biogenesis (Fig. S4 F). These results suggest that tubule-forming proteins contribute specifically to LD biogenesis.
Figure S4.
Tubule-forming proteins are required in the maintenance of LD homeostasis in HepG2 cells. (A) The levels of FIT2 and FIT2-related tubule-forming proteins were determined by Western blotting or real-time PCR as indicated after HepG2 cells were transfected with siRNAs for 48 h. (B) Western blotting of Rtn4 in WT and Rtn4-KO HepG2 cells. (C) LipidTOX staining of LDs and quantification of number of LDs per cell, as well as LD size, in WT and Rtn4 KO HepG2 cells without (−OA) or with (+OA) OA treatment. Colorized pictures were acquired using Imaris software. Different colors indicate different LD diameters, as labeled. n = 50–76 cells/group, total LD numbers >2,000. Mann–Whitney test; ***, P < 0.001. Scale bar, 10 µm. (D) The level of Climp-63 was determined by Western blotting after HepG2 cells were transfected with the indicated siRNAs for 48 h. (E) As in C, except in Control and Climp-63–depleted HepG2 cells. For groups without OA treatment, n = 76 and 74 cells, total LD numbers >2,000. For groups with OA treatment, n = 45 and 51 cells, total LD numbers >2,000. Mann–Whitney test; NS, P > 0.05. Scale bar, 10 µm. (F) Levels of LD-related proteins in HepG2 cells transfected with the indicated siRNAs. (G) Septin 2 KO (SEPT2 KO) HepG2 cells were generated using CRISPR/Cas9, two single clones (#10 and #16) picked, and the protein level of septin 2 in WT and SEPT2 KO cells measured by immunoblotting (IB). (H–J) The knockdown efficiencies of siRNAs targeting septin 6, 7, and 9 were tested by Western blotting. (K) As in Fig. 5 D, LipidTOX staining and Imaris 3D images of OA-treated HepG2 cells transfected with the indicated siRNAs. The numbers and sizes of the LDs in each group were determined by Imaris surface analysis. n = 50–76 cells/group, n = 74–101 cells/group, total LD numbers >2,000. Mann–Whitney test; ***, P < 0.001. Scale bar, 10 µm. siClimp-63, small interfering Climp-63; siControl, small interfering Control; si FIT2, small interfering siFIT2; siREEP5, small interfering REEP5; siRtn4, small interfering Rtn4; siSEPT, small interfering SEPT. Error bars represent SEM.
Figure 5.
FIT2-interacting proteins in LD biogenesis. (A) LipidTOX staining of LDs and quantification of LDs per cell, as well as LD size, in control and tubule-forming protein-depleted HepG2 cells without OA treatment. 3D pictures of LDs were acquired under a confocal microscope by serial layer scanning along the z axis. Colorized pictures were acquired using Imaris software. Different colors indicate different LD diameters, as labeled. n = 86–200 cells/group. All graphs are representative of three repetitions, total LDs numbers >2,000. Mann–Whitney test; ***, P < 0.001. Scale bar, 10 µm. (B) As in A, except each group of cells was treated with 0.2 mM OA for 6 h before LipidTOX staining. n = 82–143 cells/group, total LDs numbers >2,000. Mann–Whitney test; ***, P < 0.001. Scale bar, 10 µm. (C and D) As in A and B, LDs were stained in WT (SEPT2-WT) and septin 2 KO (SEPT2-KO) HepG2 cells with (C) or without (D) OA treatment. The numbers and sizes (diameter) of LDs were measured. For groups without OA treatment, n = 46–76 cells/group; for groups with OA treatment, n = 53–78 cells/group, total LD numbers >2,000. Mann–Whitney test; **, P < 0.01; ***, P < 0.001. Scale bar, 10 µm. siControl, small interfering Control; siFIT2, small interfering FIT2; siREEP5, small interfering siREEP5; siRtn4, small interfering Rtn4. Error bars represent SEM.
Next, we generated septin 2–deleted HepG2 cells using CRISPR/Cas9 (Fig. S4 G) and analyzed LD number and size. In both the presence and absence of OA treatment, the LDs in these cells became fewer and smaller (Fig. 5, C and D). We also depleted FIT2-interacting septin 7 and observed similar changes (Fig. S4, I and K). Because septins usually act as hetero-oligomers, we found that depletion of septin 6 and 9 caused similar defects (Fig. S4, H and K). These data confirm that septin polymers are involved in LD biogenesis.
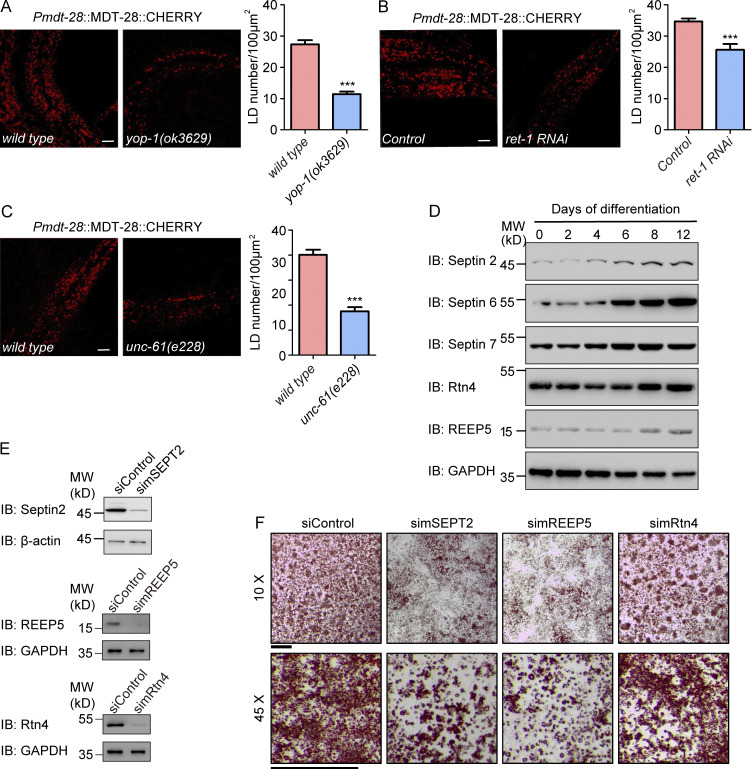
To further confirm the LD phenotype in a physiological setting, we tested FIT2-interacting protein mutants in C. elegans. Worms possess one reticulon, RET-1, and one REEP5 homologue, YOP-1. The yop-1 mutant strain ok3629 contained a reduced number of LDs (Fig. 6 A). Similarly, treatment with ret-1 RNAi resulted in fewer LDs (Fig. 6 B), and the mutant of septin homologue unc-61 (e228) had fewer LDs (Fig. 6 C). These results suggest that tubule-forming proteins and septins have a conserved role in LD biogenesis.
Figure 6.
Functional tests of FIT2-interacting proteins. (A) Representative confocal images of wild-type and yop-1(ok3629) larval stage L4 animals carrying the ldrIs2 (Pmdt28::mdt28::cherry) transgene. Images are 3D projections of 2.62-µm confocal z-stacks that covered the posterior body forward from the vent. LD quantification is shown on the right. n = 16 (wild-type) and 18 (yop-1) animals. Unpaired t test, ***, P < 0.001. Scale bar, 10 µm. (B) As in A, but with RET-1–depleted animals generated by RNAi feeding. n = 7 (wild-type) and 11 (ret-1) animals. Unpaired t test, ***, P < 0.001. Scale bar, 10 µm. (C) As in A, but with unc-61(e228) mutant. n = 10 (wild-type) and 13 (unc-61) animals. Unpaired t test, ***, P < 0.001. Scale bar, 10 µm. Error bars represent SEM. (D) Differentiated 3T3-L1 cells were collected at the indicated times, and protein levels were detected by immunoblotting (IB) with the indicated antibodies. (E) The knockdown efficiency of the indicated siRNAs in 3T3-L1 cells was measured by IB. (F) Representative images of 3T3-L1 adipocytes. On day 10, differentiated 3T3-L1 cells were fixed and stained with Oil Red. Images were captured using a stereomicroscope under the indicated magnifications. Scale bar, 2 mm. siControl, small interfering Control; simSEPT2: small interfering mouse SEPT2; simREEP5: small interfering mouse REEP5; simRtn4, small interfering mouse Rtn4.
To strengthen the linkage between FIT2-associated proteins and LD biogenesis, we measured the levels of these proteins during adipocyte differentiation, a process that demands elevated levels of LD formation. Consistently, the expression of septins (septin 2, 6, and 7) and REEP5 was induced in insulin-treated 3T3-L1 cells (Fig. 6 D). The levels of Rtn4 also increased, but to a lesser extent, whereas those of an unrelated ER-resident protein, Sec61β, did not change much (Fig. S3 J). Depletion of FIT2-associated proteins blocked insulin-induced 3T3-L1 preadipocyte differentiation (Fig. 6, E and F). In addition, interactions between FIT2 and REEP5 were slightly enhanced when LD formation was stimulated by OA treatment (Fig. S3 K), but they were not affected upon seipin depletion (Fig. S2 I) or by mutating the enzymatic activity residues (H155 and H214) of FIT2 (Fig. S2 J). Similarly, the FIT2 H155A/H214A mutant interacted with septin 2 or septin7 similar to WT FIT2 (Fig. S2 N).
FIT2 and FIT2-interacting proteins mark peripheral LD formation sites
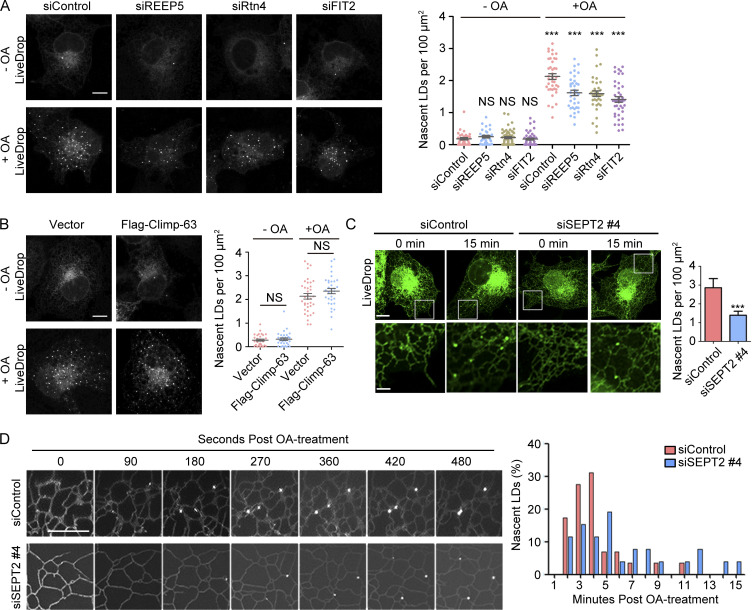
FIT2 has been proposed to be critical in the budding of nascent LDs from the ER (Choudhary et al., 2015). The LD defects observed with mutants of FIT2-interacting proteins coincide with mutants of FIT2, suggesting that they cooperate with FIT2 during LD formation. To analyze their precise roles in LD biogenesis, we monitored nascent LD formation using the recently developed early stage LD marker LiveDrop, which is composed of the TM domain of GPAT4 fused with GFP (Wang et al., 2016). Initially, cells were starved to consume most of the existing LDs, and subsequent formation of new LDs was followed with LiveDrop upon the addition of OA (Fig. S5 A). Comparing control COS-7 cells with cells depleted of FIT2 or FIT2-interacting tubule-forming proteins (REEP5 and Rtn4), fewer nascent LDs were seen in mutant cells (Fig. 7 A). As expected, septin 2 knockdown, but not overexpression of unrelated protein Climp-63 (Fig. 7 B), resulted in a decreased number of LDs (Fig. 7 C) and a delay in nascent LD formation (Fig. 7 D).
Figure S5.
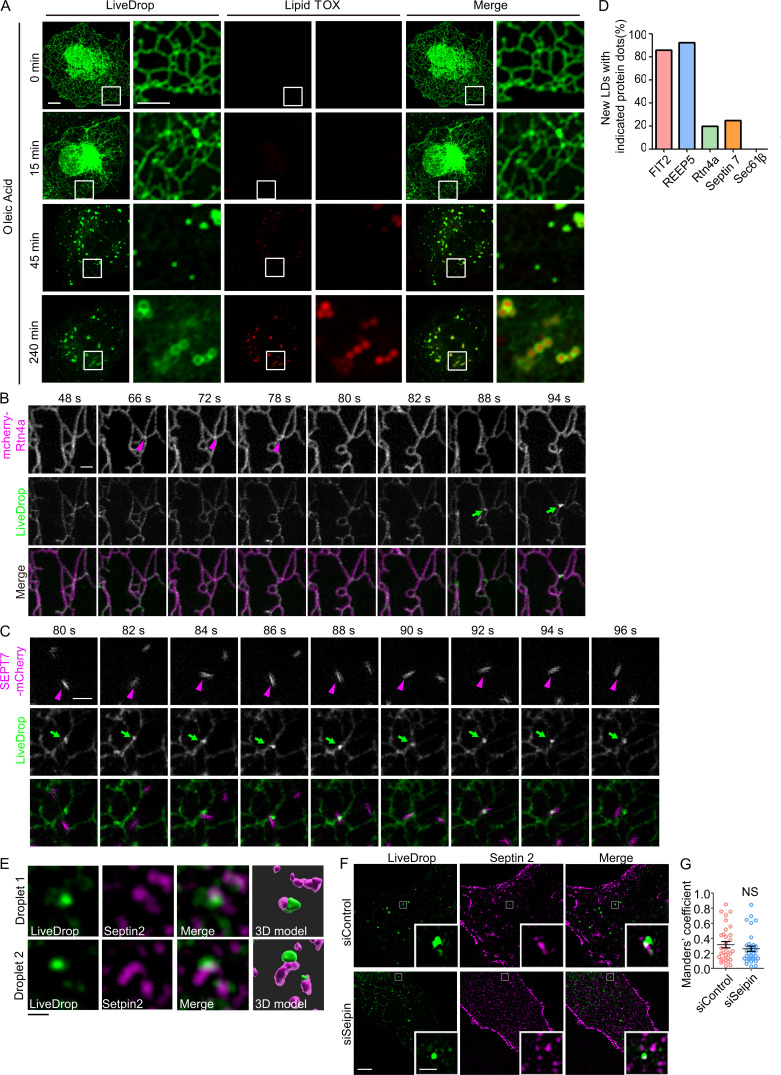
Tubule-forming proteins and septins participate in the early steps of LD biogenesis. (A) As in Fig. 7 A, COS-7 cells transfected with GFP-LiveDrop (green) were fixed at the indicated time points and stained with LipidTOX (red). The boxed regions are magnified on the right. Scale bar, 10 µm; 5 µm (inset). (B) Rtn4a appeared at LD biogenesis sites before LiveDrop accumulation. As in Fig. 8 A, but with cells expressing mcherry-Rtn4a and LiveDrop. Magenta arrowheads indicate the accumulation of Rtn4a, and green arrows indicate the nascent LDs. Scale bar, 1 µm. (C) Living cell images show the colocalization between septin 7 and nascent LDs. As in Fig. 8 A, but with cells expressing SEPT7-mCherry and LiveDrop. Magenta arrowheads indicate the accumulation of septin 7, and green arrows indicate the nascent LDs. Scale bar, 1 µm. (D) Percentage of nascent LDs with the indicated protein dots that appeared before LiveDrop accumulation in Fig. 8, A and B and Fig. S5, B and C. n = 10–14 nascent LDs in three cells for each group. (E) Representative images of the colocalization of nascent LDs (LiveDrop) and endogenous septin 2 in Fig. 8 C. 3D models were computed by Imaris software. Scale bar, 1 µm. (F and G) As in Fig. 8, E and D, but COS-7 cells were transfected with siControl or siSeipin. The Mander’s coefficient was measured by ImageJ. In each group, 30–40 ROIs in 8–12 cells were measured. Unpaired t test, NS, P > 0.05. Scale bar, 5 µm; 1 µm (inset). ROI, region of interest; siControl, small interfering Control; siSeipin, small interfering seipin. Error bars represent SEM.
Figure 7.
FIT2-interacting proteins in early steps of LD formation. (A) Representative images of remaining LDs after delipidation and nascent LDs formed after OA treatment in control and ER tubule-forming protein–depleted COS-7 cells. Each group of cells expressing LiveDrop was delipidated for 60 h and then treated with 0.2 mM OA for 15 min to induce nascent LD formation. Quantification of LDs is shown on the right. n = 36–42 cells/group. Unpaired t test; NS, P > 0.05. Scale bar, 10 µm. (B) As in A, but with cells overexpressing Flag–Climp-63. n = 33–37 cells/group. One-way ANOVA, ***, P < 0.001. Scale bar, 10 µm. (C) As in A, but nascent LDs in septin 2 knockdown COS-7 cells were counted. The bottom images show enlargements of the boxed regions over the tubular ER network. n = 13–16 cells/group. Unpaired t test, ***, P < 0.001. Scale bar, 10 µm; 3 µm (inset). Error bars represent SEM. (D) Time-lapse stills of nascent LD formation in control or septin 2 knockdown COS-7 cells. The histogram on the right shows the quantification of the frequency of nascent LDs in the indicated time frame. n = 3 or 4 cells/group. Scale bar, 5 µm. siControl, small interfering Control; siFIT2, small interfering FIT2; siREEP5, small interfering siREEP5; siRtn4, small interfering Rtn4; siSEPT, small interfering SEPT.
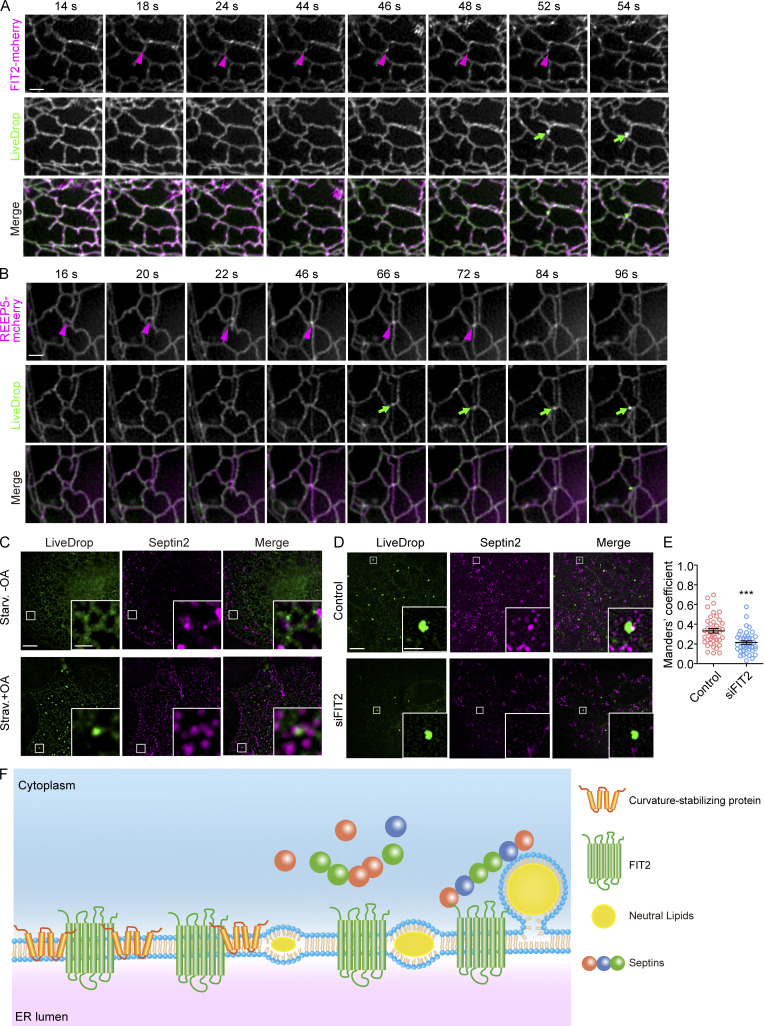
Finally, we followed nascent LD formation in the cell periphery using LiveDrop and live cell super-resolution microscopy. When fluorescently labeled FIT2 was cotransfected with LiveDrop, FIT2 formed puncta along the tubule first, and a few seconds later LiveDrop puncta became visible in close proximity. The maturation of LiveDrop puncta was accompanied by the disappearance of FIT2 puncta (Fig. 8 A and Video 1). These events accounted for ∼85% of newly formed LDs in the cell periphery (Fig. S5 D). A similar pattern was observed with REEP5 and Rtn4a (Fig. 8 B, Fig. S5 B, Video 2, and Video 3), but only in the case of Rtn4a, its puncta forming at ∼20% of nascent LDs in the cell periphery (Fig. S5 D). When septin 7–mCherry was cotransfected with LiveDrop, septin 7–positive puncta were recruited to some LiveDrop puncta, even though these events occurred transiently (Fig. S5 C and Video 4). We also analyzed the overlap between LiveDrop puncta and endogenous septin 2 during nascent LD formation in fixed COS-7 cells. The septin 2 puncta had a significant overlap with LiveDrop puncta, and the degree of overlap decreased when FIT2, but not seipin, was depleted in these cells (Fig. 8, C–E; and Fig. S5, E–G). Taken together, these results indicate that FIT2-interacting proteins, particularly REEP5, Rtn4a, and septins, contribute to the early steps of LD formation, likely in concert with FIT2.
Figure 8.
FIT2-interacting proteins at the nascent LD formation site. (A) COS-7 cells expressing FIT2-mCherry and LiveDrop were delipidated for 72 h and then treated with 0.2 mM OA (final concentration) to induce nascent LD formation. Live cell images were acquired by GI-SIM after OA addition at t = 0. Magenta arrowheads indicate the accumulation of FIT2, and green arrows indicate the nascent LDs. Scale bar, 1 µm. (B) As in A, but with cells expressing REEP5-mCherry and LiveDrop. Magenta arrows indicate the accumulation of REEP5, and green arrows indicate the nascent LDs. Scale bar, 1 µm. (C) Representative 3D-SIM images of colocalization between LiveDrop and endogenous septin 2 in delipidated COS-7 cells with or without OA treatment. Enlargements of the boxed regions are shown. Scale bar, 5 µm; 1 µm (inset). (D and E) As in C, but in COS-7 cells transfected with siControl and siSeipin. The Mander’s coefficient was measured by ImageJ. In each group, 45–50 ROIs in 10–15 cells were measured. Unpaired t test, ***, P < 0.001. Scale bar, 5 µm; 1 µm (inset). Error bars represent SEM. (F) A model of FIT2-mediated nascent LD formation. ROI, region of interest; siControl, small interfering Control; siFIT2, small interfering FIT2; siSeipin, small interfering seipin; Starv., starvation.
Video 1.
The accumulation of FIT2 at LD formation site. Associated with Fig. 8 A. GI-SIM images of OA-treated COS-7 cells expressing mCherry-FIT2 (magenta) and LiveDrop (green) were taken with a time interval of 2 s. The playback speed is at 5.7× real time. The circle highlights nascent LD formation site.
Video 2.
The accumulation of REEP5 at LD formation site. Associated with Fig. 8 B. GI-SIM images of OA-treated COS-7 cells expressing mCherry-REEP5 (magenta) and LiveDrop (green) were taken with a time interval of 2 s. The playback speed is at 5.7× real time. The circle highlights nascent LD formation site.
Video 3.
The accumulation of Rtn4 at LD formation site. Associated with Fig. S5 B. GI-SIM images of OA-treated COS-7 cells expressing mCherry-Rtn4a (magenta) and LiveDrop (green) were taken with a time interval of 2 s. The playback speed is at 5.7× real time. The circle highlights nascent LD formation site.
Video 4.
The accumulation of Septin 7 at LD formation site. Associated with Fig. S5 C. GI-SIM images of OA-treated COS-7 cells expressing mCherry-Septin 7 (magenta) and LiveDrop (green) were taken with a time interval of 2 s. The playback speed is at 5.7× real time. The circle highlights nascent LD formation site.
Discussion
Our results reveal previously unidentified interactions between FIT2, the known regulator of LD budding, and ER tubule-forming proteins and the cytoskeletal septin. Mutation of FIT2 causes abnormal budding of newly formed LDs; thus, fewer and smaller LDs are seen (Choudhary et al., 2015; Goh and Silver, 2013; Kadereit et al., 2008). FIT2 can bind to neutral lipids through its TM segments (Gross et al., 2011), is predicted to be a potential lipid phosphatase or phosphotransferase (Hayes et al., 2018), and subsequently was shown to be an acyl–coenzyme A diphosphatase (Becuwe et al., 2020), suggesting a converging role in orchestrating lipids for LD formation. Similar defects of fewer and/or smaller LDs are affiliated with the loss or mutation of tubule-forming proteins and septins consistent with a cooperative role with FIT2. We noted that FIT2 clusters often appear at sites of LD formation and precede the appearance of LiveDrop puncta, implying that localized activity is needed at an early stage of nascent LD formation. Coincidently, REEP5 or Rtn4 behaves the same (Fig. 8 F). These findings could explain why overexpression of FIT2 alone does not boost LD formation.
The interactions between FIT2 and tubule-forming proteins can be categorized into two types: REEP5 mainly engages the N-terminal half of FIT2, and Rtn4 binds to both halves. These findings support that the interactions are specific and may have different functions. One benefit of tubule-forming proteins is membrane curvature. The biophysical property of the oil lens and the budding and fission processes could all be related to curvature generation (Santinho et al., 2020). The intrinsic curvature of phospholipids in the ER is critical for the architecture of LDs (Choudhary et al., 2018). Thus, budding site–concentrated FIT2 may introduce yet another layer of curvature generation, in this case by integral membrane proteins, to the birth of LDs. However, LD defects caused by depletion of tubule-forming proteins could not be easily recapitulated by the domination of ER sheets, structures with less curvature than tubules. Therefore, the presence of ER tubules per se is not sufficient for efficient LD formation. As FIT2 is an acyl–coenzyme A diphosphatase, it is reasonable to speculate that the association of tubule-forming proteins regulates the enzymatic activity of FIT2. The requirement of this activity of FIT2 is also supported by the loss-of-function mutation we identified in worms. The equivalent mutation in humans in FIT2, D151N, is very close to the potential active site, H155, in the luminal face of FIT2 between TM3 and TM4.
The colocalization of FIT2, ER tubule-forming proteins, and the nascent LD marker LiveDrop implies spatial control of LD formation in subdomains of the ER. Almost all nascent LDs observed in our live cell tracing experiments formed in ER tubules. However, nascent LDs can also be generated in the cell center, where tubules and sheets are interconnected in a dense network; thus, currently available tools will likely not be able to resolve the spatial regulation of LD formation. Notably, ER sheets have high membrane curvature similar to tubules at sheet edges, and ER tubule-forming proteins localize to the edges of sheets (Shibata et al., 2010). In theory, nascent LDs can bud from ER sheets, even if membrane curvature and ER tubule-forming proteins are needed.
The involvement of septin in LD biogenesis has been proposed, but as indirect regulation through microtubules (Akil et al., 2016). The physical contacts between the GTPase domain of septin 7 and two cytosolic loops of FIT2 indicate that the role of septins can be direct. Other septins are known to interact with phospholipids through a C-terminal amphipathic helix. Thus, it is likely that FIT2-anchored septins hold onto the surface phospholipids of newly formed LDs and serve as a handrail (Fig. 8 F). Such scaffolds would not only stabilize the budding LDs but would also ensure directional budding of the neutral lipid–enriched membrane compartment. Notably, each septin hexamer is ∼25 nm in length, and nascent LDs are 100–200 nm in diameter (Sirajuddin et al., 2007; Walther et al., 2017). Therefore, the “handrail” may not need heavy bundling of the septin units.
Our results, together with previous findings, led to a refined model of early stage LD formation (Fig. 8 F). Neutral lipids within the two leaflets of ER membranes may accumulate with FIT2, which recruits ER tubule-forming proteins to stabilize the curvature of the growing oil lens or promote lipid preparation. The outward bulging of the oil lens may be supported further by FIT2-interacting septins. The actual actions of tubule-forming proteins on FIT2 remain to be determined. Given the diversity and redundancy of these proteins, it is very likely that FIT2 is differentially regulated in different cell types. In addition, FIT2 likely acts independently from other LD biogenesis regulators, such as seipin. The FIT2-organized interacting complex facilitates the generation of the complicated, but fine-tuned, landscape of the fat-storage organelle.
Materials and methods
Strains
The following strains were used in this work: N2 Bristol (WT), fitm-2(erm002), fitm-2(av41), qxIs439(Phyp7::gfp::tram-1), ojIs2 (Ppie-1::gfp::sp12), ldrIs2 (Pmdt28::mdt28::cherry), and zcls4(Phsp-4::gfp).
Identification, mapping, and cloning of fitm-2
The fitm-2(erm002) allele was identified in screens for mutations with altered ER morphology in qxIs439(Phyp7::gfp::tram-1) animals, which expressed the GFP::TRAM-1 fusion protein in hypodermal cells. Mapping single nucleotide polymorphism markers placed erm002 between 2 and 2.95 on linkage group I. The molecular lesion in erm002 was then determined by whole-genome sequencing. A transgene containing fosmid WRM0637bE08 or the single gene fitm-2 restored ER morphology in erm002 mutants.
Generation of fitm-2(av41) C. elegans
We generated the early nonsense mutation in fitm-2 using CRISPR/Cas9 technology as described previously (Choudhary et al., 2015). Briefly, we designed two gRNAs (Table S1) that target two overlapping fitm-2 Cas9 sites and cloned the designer gRNAs into plasmid pDD162. A repair template was synthesized containing two stop codons on both targeted PAM sites (codons 17 and 20) and a HindIII restriction site for screening purposes (Table S1). The gRNA containing pDD162 plasmids and the repair template were injected into C. elegans germlines. The F2 progeny were picked. The fitm-2(av41) sequence was confirmed and balanced with hT2 [bli-4(e937) let-?(q782) qIs48] (I;III). Non-GFP animals were picked and observed.
RNAi inactivation in C. elegans
For RNAi feeding assays, L1 larvae expressing MDT-28::CHERRY were plated onto Nematode Growth Medium plates containing 1 mM IPTG and seeded with ret-1 double-stranded RNA bacterial clones. The animals were grown at 20°C, and the F1 progeny was examined at L4 stage.
Worm LipidTOX staining
Animals at L4 stage were suspended in 1 ml of water before adding 100 µl of freshly prepared 4% paraformaldehyde solution and mixing. The mixture was frozen and (not completely) thawed three times. The worms were allowed to settle and washed three times with PBS. Next, 1 ml of LipidTOX dye diluted in PBS (1:1,000) was added, and the worms incubated for 30 min at room temperature (RT) with occasional gentle agitation. The worms were then washed three times with PBS. After most of the staining solution was removed, the fixed worms were mounted onto 2% agarose pads for microscopic observation and photography.
EM of C. elegans
In the high-pressure freeze (HPF) procedure, several living worms were picked into a type A specimen carrier (200-µm well) containing 2% agarose to avoid voids and overfilling. Next, they were placed in a treated type B carrier (flat) with hexadecane and closed before freezing in the HPF device (Leica; HPM100). Following HPF, the fast-frozen samples were immersed in a freezing tube containing 2% osmium tetroxide in 98% acetone/2% water and placed into the freeze substitution device (Leica; EM AFS) set to the following parameters: T1 = −90°C for 72 h, S1 = 5°C/h, T2 = −60°C for 12 h, S2 = 5°C/h, T3 = −30°C for 10 h, then slowly warmed to 10°C (5°C/h). Following freeze substitution, samples were rinsed four times in 100% acetone (15 min each) at RT. Next, they were stained using 0.5% uranyl acetate dissolved in 90% acetone/10% methanol (filtered before use) and incubated for 2 h in the dark at RT. After staining, samples were rinsed four times in 100% acetone (15 min each) at RT, and the samples were transferred into new 2-ml Eppendorf tubes. Next, they were infiltrated in graded mixtures (1:5, 1:3, 1:1, 1:3) of resin (EMS; Resin Mixture: 16.2 ml SPI-PON812, 10 ml Dodecenylsuccinic anhydride, 8.9 ml Methyl-5-Norbornene-2,3-Dicarboxylic Anhydride, and 1.5% Benzyldimethylamine) and acetone, then changed to 100% resin, which was replaced four times over the next 3 d on a rotator. Finally, worms were embedded and polymerized for 12 h at 45°C and 48 h at 60°C. The ultrathin sections (70 nm sethick) were sectioned with a microtome (Leica; EM UC7) and examined by a scanning electron microscope (FEI Helios; Nanolab 600i SEM) in immersion high-magnification mode (Concentric higher energy electron detector, 2 kV, 0.34 nA).
Constructs
GFP-HT008, Flag–Climp-63, GFP-Rtn4a, Myc-ATL3, and RFP-sec61β constructs were described previously (Hu et al., 2009; Shibata et al., 2008; Wang et al., 2017). FIT2 and REEP5 were amplified from the human cDNA library. Full-length FIT2 and truncated FIT2 (TM1-3, TM3-6, TM3-6ΔC) were subcloned into pCI-neo-2HA, and REEP5 was ligated into pcDNATM4/TO/myc-His B as indicated. For mcherry-Rtn4a, mCherry was amplified and replaced the GFP tag in GFP-Rtn4a. For REEP5-mCherry, REEP5 was amplified from REEP5-Myc and inserted into the pCI-neo vector with mCherry on the C-terminus, and Septin 7 was inserted into the pmCherry-N1 vector with mCherry on the C-terminus. For C. elegans expression, fitm-2 was amplified from the C. elegans cDNA library and subcloned into the pPD49.26 vector with the hyp7 promoter on the N-terminus and RFP on the C-terminus. The Flag-Lipin1b plasmid was a gift from Pingsheng Liu’s laboratory (Institute of Biophysics, CAS, Beijing, China). Septin2/6/7 hexamer dual vectors were obtained from Dr. Thomas Lecuit (Aix-Marseille University, Marseille, France), and individual septins (septin 2, 6, 7, and 9) and septin 7 truncations (SEPT7 CC and SEPT7ΔCC) were inserted into the pET28a vectors with a His-tag on the N-terminus.
BLI
Purified septins used for BLI were exchanged into assay buffer (50 mM Tris-Cl, 150 mM KCl, and 0.02% Tween 20, pH 8.0) and diluted to a gradient concentration of 32.5–500 nM. Biotinylated FIT2 NTL peptides were diluted in assay buffer to the final concentration of 100 nM and then loaded onto the streptavidin sensors for 300 s. Association and dissociation were measured for 600 s by dipping the sensors into gradient protein solutions or blank assay buffers. The BLI assays were performed using Octer96 and data analysis performed in FortéBio Data Analysis 7.0 software.
Cell culture, transfection, and immunofluorescence
HepG2 cells (a gift from Pingsheng Liu’s laboratory), COS-7 cells, U2OS cells, and HEK293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS at 37°C in 5% CO2. Transfections were performed using Lipofectamine 3000 (Invitrogen) for DNA plasmids, except for transfection of plasmids after delipidation, which was performed using X-tremeGENE HP DNA Transfection Reagent (Roche) and Lipofectamine RNAi MAX (Invitrogen) for siRNAs according to the manufacturer’s instructions. Cell samples were harvested 24 h after plasmid transfection or 48 h after siRNA transfection unless otherwise indicated. Immunofluorescence experiments were performed as described previously. Transfected cells were grown on coverslips and immunostained with primary antibodies and various Alexa Fluor–conjugated secondary antibodies (Invitrogen). Images were captured at RT on a 3D–structured illumination microscopy (3D-SIM) or Zeiss LSM700 confocal microscope as indicated. The primary antibodies used are mouse–anti-HA (ORIGENE; TA180128), rabbit–anti-HA (Abcam), rabbit–anti-Myc (Sigma; C3956), mouse–anti-Flag (Sigma), rabbit–anti-calreticulin (Abcam; ab2907), mouse–anti–Climp-63 (Enzo; ENZ-ABS669-0100), rabbit–anti–septin 2 (Abcam; ab187654), and mouse–anti–α-tubulin (Abcam; ab7291).
coIP
For ER tubule-forming protein immunoprecipitation (IP), cells were lysed in 1% digitonin containing IP buffer (25 mM Hepes, 150 mM KAc, 2 mM Mg(Ac)2, and protease inhibitor cocktails). For septin IP, cells were lysed in 1% Triton X-100 IP buffer (50 mM Tris-Cl, 150 mM KCl, 2 mM MgCl2, and protease inhibitor cocktails). Cell lysates were incubated with anti-HA magnetic beads (Bimake; B26202) or anti-HA agarose (Sigma; A2095) for 2 h at 4°C. Washed precipitates were separated by SDS-PAGE and immunoblotted with various antibodies. For DSP-mediated cross-linking coIP, FIT2-HA KI U2OS cells were cultured in fatty acid–free medium (1% fatty acid–free BSA in DMEM) for 60 h and then treated with 20 mM OA for 15 min. The culture medium was then removed, and 1 mM DSP in PBS was added. The dishes were incubated at RT for 30 min. The cross-linking reaction was stopped by the addition of 20 mM Tris-Cl (final concentration, pH 7.8) for 15 min at RT. Cells were washed with PBS three times and then lysed. coIP and immunoblotting were then performed. The primary antibodies used for immunoblotting are mouse–anti-GAPDH (Proteintech; 60004–1), rabbit–anti-NOGO (Rtn4; Novus; NB100-5681), rabbit–anti-REEP5 (Proteintech; 14643–1-AP), rabbit–anti-calnexin (Proteintech; 10427–2-AP), rabbit–anti-GFP (Proteintech; 50430–2-AP), rabbit–anti–septin 7 (Abcam; ab175229), rabbit–anti–septin 6 (Immunoway; YT4205), rabbit–anti-MSF (septin 9; Abcam; ab114099), mouse–anti-His (MBL; D291-3), mouse–anti–β-actin (abgent; AM1021B), rabbit–anti-BIP (Abcam; ab21685), mouse–anti-PDI (Abcam; ab2792), mouse–anti-ACSL3 (Abnova; H00002181-B01P), mouse–anti-BSCL2 (seipin; Abnova; H00026580-A02), and rabbit–anti-sec61β (Abcam; ab78276).
CRISPR-mediated gene targeting
FIT2-HA KI HepG2 cells and U2OS cells were generated following a previously described procedure (Ran et al., 2013). Briefly, the gRNA sequences of human FIT2 were ligated into the BbsI sites of px260a-m2 vector. The template plasmids were constructed using pUC57 vector as the backbone, with the insertion of a left homologous arm (∼600-bp genomic DNA sequence upstream of the TAA region of FIT2), two tandem HA sequences (5′-TACCCATACGACGTCCCAGACTACGCT-3′), a 2A sequence (5′-GGCTCCGGAGAGGGCAGGGGAAGTCTTCTAACGTGCGGGGACGTGGAGGAAAATCCCGGCCCC-3′), a neo sequence, and a right homologous arm (∼800-bp genomic DNA sequence downstream of TAA region of FIT2). The two plasmids were cotransfected into cells, and FIT2-HA KI cells were screened using G418 and then verified by PCR sequencing and Western blotting.
Rtn4 KO and septin 2 KO HepG2 cells were generated using the gRNA sequences in Table S1. Single cells were sorted 48 h after transfection and verified by PCR sequencing and Western blotting.
LipidTOX staining of mammalian cells
HepG2, COS-7, and U2OS cells were grown on 60-cm confocal dishes. Cells were fixed in 4% paraformaldehyde for 30 min, washed with PBS buffer, and stained with LipidTOX dye diluted in PBS (1:3,000) for 30 min. LipidTOX was removed by washing with PBS three times. Images were captured on a Zeiss LSM700 confocal microscope.
Statistical analysis
3D pictures of LDs in HepG2 cells were acquired under a confocal microscope by serial layer scanning along the z axis. Statistical analysis of LD number and size was performed using Imaris software by creating the surface of each LD using the same parameters in the control and experimental groups for each experiment. Statistical analysis of LD number and size in single confocal plane pictures was performed in ImageJ using the same threshold value in the control and experimental groups for each experiment. LD size was visualized in the form of a cross-sectional area in pixels. The gray values of the Western blot bands were analyzed in Gel-Pro. The results are presented as mean ± SEM. Student’s t test, Mann-Whitney test, and one-way ANOVA were performed for statistical analyses. Data distribution was assumed to be normal, but this was not formally tested.
RNA extraction and RT-PCR
Total RNA was extracted using an RNA isolation kit (Magen) according to the manufacturer’s instructions. cDNA reverse transcribed from poly-A mRNA was used as the template for PCR. RT-PCR was performed in QuantStudio 7 Flex (Applied Biosystems). The primers used for RT-PCR are in Table S1.
Delipidation and LD induction
Delipidation and LD induction were performed as described previously (Salo et al., 2016). Cells were delipidated by culturing in serum-free medium supplemented with 5% lipoprotein-deficient serum (Sigma) for 60 h. Nascent LD formation was induced by adding 0.2 mM OA (final concentration) for the indicated times.
3D-SIM super-resolution microscopy and image analysis
3D-SIM images of fixed cells were acquired at RT on the DeltaVision OMX V3 imaging system (Applied Precision; GE) with a ×100 1.4 oil objective (Olympus UPlanSApo), solid-state multimode lasers (488 nm, 405 nm, and 561 nm), and electron-multiplying charge-coupled device cameras (Evolve; 512 × 512; Photometrics). Serial z-stack sectioning was performed at 250-nm intervals in conventional mode. The microscope is routinely calibrated with 100-nm fluorescent spheres to calculate both the lateral and axial limits of image resolution. Conventional image stacks were processed by deconvolution methods using softWoRx 5.0 (Applied Precision; GE Healthcare) with the following settings: Wiener filter enhancement = 0.900 and winner filter smoothing = 0.800. Pixel registration was corrected to be <1 pixel for all channels using 100 nm Tetraspeck beads.
Live cell imaging by gazing incidence structured illumination microscopy (GI-SIM)
Transfected COS-7 cells were plated on 25-mm coverslips coated with polylysine and delipidated for 60 h. Before imaging, OA was added to the culture medium at a final concentration of 0.2 mM at time point 0. Pictures were acquired every 2 s. All images were acquired by GI-SIM system (Li et al., 2015) using the Olympus 1.49-NA objective under physiological conditions (37°C and 5% CO2). At each time point, we acquired three raw images at successive phase steps of 0, 1/3, and 2/3 of the illumination period. We then repeated this process with the standing wave excitation pattern rotated ±120° with respect to the first orientation, for a total of nine raw images. The film speed for the rebuild image was one frame per second. The excitation intensity was 40 W/cm2.
Protein purification
The purification of septin2/6/7 hexamer was performed as described previously (Mavrakis et al., 2014; Mavrakis et al., 2016). The septin2/6/7 hexamer with a His-tag and thrombin-cleaved Strep-tag were coexpressed in Escherichia coli. The proteins were isolated by Ni-NTA and StrepTactin chromatography (GE Healthcare). The individual septins (septin 2, 6, 7, and 9) with N-terminal His-tags were expressed in E. coli and isolated by Ni-NTA chromatography.
The FIT2 protein was expressed using the Bac-to-Bac Baculovirus Expression System (Invitrogen). Full-length FIT2 was inserted into the pFastbac1-C-tag vector, which encodes a C-terminal Strep-His8-GFP-tag. Recombinant Bacmid and baculovirus were generated according to the user manual. The baculovirus-infected Sf9 cells were harvested and resuspended in lysis buffer containing 50 mM Tris-Cl, 150 mM NaCl, and 1 mM EDTA and lysed by sonication. The lysate was centrifuged at 32,000g for 1 h, and the pellet was collected and homogenized in lysis buffer containing 1% Fos-choline 12 using a Dounce glass tissue grinder. The FIT2-Strep-His8-GFP fusion protein was isolated by StrepTactin chromatography.
Biotin-peptide pull-down assay
Biotinylated cytosolic FIT2 peptides (Table S1) were synthesized by GL Biochem and dissolved in DMSO. Strep-tag cleaved septin hexamer (1 µM) and peptides (20 µM) were used for pull-down assays in a buffer containing 50 mM Tris-Cl, 150 mM KCl, 2 mM MgCl2, and 2 mM GTP. The protein-peptide mixture was incubated with streptavidin resin (Thermo Fisher Scientific; Pierce) for 1 h. The precipitates were washed and analyzed by SDS-PAGE and Coomassie blue staining.
Liposome reconstitution and coflotation assay
Liposome preparation, protein reconstitution, and proteoliposome flotation assay were performed as described previously (Bian et al., 2011; Yan et al., 2015). Briefly, the FIT2-GFP fusion protein was reconstituted at a protein/lipid molar ratio of 1:2,000. Approximately 20 mM of liposomes or proteoliposomes was mixed with 1 µM septin2/6/7 hexamer or individual septins in 100 µl of 1.9 M sucrose. The sucrose gradient (1.9 M, 1.25 M, and 0.25 M sucrose) was centrifuged at 174,000g for 1 h at 4°C. The gradient fractions were collected and analyzed by SDS-PAGE and immunoblotting.
Insulin-induced 3T3-L1 preadipocyte differentiations
3T3-L1 cells were maintained in DMEM supplemented with 10% FBS. Before differentiation, 5 × 104 cells were seeded in a 6-well plate and cultured until 90% confluence. To initiate differentiation, the culture medium was removed on day 0 and Differentiation Medium I (500 µM 3-isobutyl-1-methylxanthine, 1 µM dexamethasone, 2 µM rosiglitazone, 10 µg/ml insulin, and 10% FBS in DMEM) was added. On day 3, Differentiation Medium I was removed and Differentiation Medium II (10 µg/ml insulin and 10% FBS in DMEM) was added. On day 7, Differentiation Medium II was removed, and fresh culture medium (10% FBS in DMEM) was added. The differentiated cells were cultured until the adipocyte-like cells were obtained on day 10.
Online supplemental material
Fig. S1 shows that FIT2 is required in the maintenance of ER homeostasis. Fig. S2 shows that FIT2 interacts with tubule-forming proteins and septins. Fig. S3 shows the purification of septins and FIT2. Fig. S4 shows that tubule-forming proteins are required in the maintenance of LD homeostasis in HepG2 cells. Fig. S5 shows that tubule-forming proteins and septins participate in the early steps of LD biogenesis. Table S1 lists sequences for primers, siRNAs, gRNAs, and peptides. Video 1, Video 2, Video 3, and Video 4 show colocalization of LiveDrop with FIT2, REEP5, Rtn4a, and septin 7, respectively.
Supplementary Material
lists sequences for primers, siRNAs, gRNAs, and peptides used in this study.
Acknowledgments
We thank Drs. William Prinz and Pedro Carvalho for critically reading the manuscript; Ms. Shuoguo Li, Ms. Yun Feng, and Dr. Xixia Li (Center for Biological Imaging, Institute of Biophysics) for the 3D–structured illumination microscopy, 3D data processing, and EM work; and Dr. Thomas Lecuit (Aix-Marseille University) for providing the septin 2/6/7 dual vectors.
J. Hu is supported by the National Science Foundation of China (grant nos. 91854202 and 31630020), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB39000000), and the National Key Research and Development Program (grant no. 2016YFA0500201). B. Yan is supported by the National Science Foundation of China (grant no. 31700729). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (grant no. P40 OD010440).
The authors declare no competing financial interests.
Author contributions: F. Chen, B. Yan, and J. Hu conceived the project. J. Hu supervised the project. J. Hu and B. Yan designed the experiments. F. Chen and B. Yan performed all the experiments with the help of J. Ren, R. Lyu., Y. Wu, and Y. Guo. D. Li and H. Zhang helped analyze the data. J. Hu and B. Yan wrote the manuscript with input from all authors.
References
- Adeyo, O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., and Goodman J.M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055. 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil, A., Peng J., Omrane M., Gondeau C., Desterke C., Marin M., Tronchère H., Taveneau C., Sar S., Briolotti P., et al. 2016. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat. Commun. 7:12203. 10.1038/ncomms12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe, M., Bond L.M., Pinto A.F.M., Boland S., Mejhert N., Elliott S.D., Cicconet M., Graham M.M., Liu X.N., Ilkayeva O., et al. 2020. FIT2 is an acyl-coenzyme A diphosphatase crucial for endoplasmic reticulum homeostasis. J. Cell Biol. 219:e202006111. 10.1083/jcb.202006111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., and Hu J.. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981. 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns, D., Lee S., Hilton C.L., Jiang Q.X., and Goodman J.M.. 2010. Seipin is a discrete homooligomer. Biochemistry. 49:10747–10755. 10.1021/bi1013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström, P., Rutberg M., Ericsson J., Holmdahl P., Andersson L., Frohman M.A., Borén J., and Olofsson S.O.. 2005. Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler. Thromb. Vasc. Biol. 25:1945–1951. 10.1161/01.ATV.0000179676.41064.d4 [DOI] [PubMed] [Google Scholar]
- Cartwright, B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, V., Ojha N., Golden A., and Prinz W.A.. 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211:261–271. 10.1083/jcb.201505067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, V., Golani G., Joshi A.S., Cottier S., Schneiter R., Prinz W.A., and Kozlov M.M.. 2018. Architecture of Lipid Droplets in Endoplasmic Reticulum Is Determined by Phospholipid Intrinsic Curvature. Curr. Biol. 28:915–926.e9. 10.1016/j.cub.2018.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, J., Rohde M., Bekhite M.M., Neugebauer S., Hemmerich P., Kiehntopf M., Deufel T., Hübner C.A., and Beetz C.. 2014. Functional mutation analysis provides evidence for a role of REEP1 in lipid droplet biology. Hum. Mutat. 35:497–504. 10.1002/humu.22521 [DOI] [PubMed] [Google Scholar]
- Fei, W., Wang H., Fu X., Bielby C., and Yang H.. 2009. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem. J. 424:61–67. 10.1042/BJ20090785 [DOI] [PubMed] [Google Scholar]
- Goh, V.J., and Silver D.L.. 2013. The lipid droplet as a potential therapeutic target in NAFLD. Semin. Liver Dis. 33:312–320. 10.1055/s-0033-1358521 [DOI] [PubMed] [Google Scholar]
- Goh, V.J., Tan J.S., Tan B.C., Seow C., Ong W.Y., Lim Y.C., Sun L., Ghosh S., and Silver D.L.. 2015. Postnatal Deletion of Fat Storage-inducing Transmembrane Protein 2 (FIT2/FITM2) Causes Lethal Enteropathy. J. Biol. Chem. 290:25686–25699. 10.1074/jbc.M115.676700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa, A., Buxó L., Mora G., Funaya C., Idrissi F.Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, D.A., and Silver D.L.. 2014. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Crit. Rev. Biochem. Mol. Biol. 49:304–326. 10.3109/10409238.2014.931337 [DOI] [PubMed] [Google Scholar]
- Gross, D.A., Zhan C., and Silver D.L.. 2011. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA. 108:19581–19586. 10.1073/pnas.1110817108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, M., Choudhary V., Ojha N., Shin J.J., Han G.S., Carman G.M., Loewen C.J., Prinz W.A., and Levine T.. 2018. Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes. Microb. Cell. 5:88–103. 10.15698/mic2018.02.614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova, K., Wimley W.C., Mishra V.K., Anantharamiah G.M., Segrest J.P., and White S.H.. 1999. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J. Mol. Biol. 290:99–117. 10.1006/jmbi.1999.2840 [DOI] [PubMed] [Google Scholar]
- Hu, J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., and Prinz W.A.. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250. 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Hu, J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561. 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Kadereit, B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., and Silver D.L.. 2008. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA. 105:94–99. 10.1073/pnas.0708579105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.S., Froese C.D., Estey M.P., and Trimble W.S.. 2011. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195:815–826. 10.1083/jcb.201106131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm, R.W., Norton J.P., Cole R.A., Li C.S., Park S.H., Crane M.M., Li L., Jin D., Boye-Doe A., Liu T.Y., et al. 2013. A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Rep. 3:1465–1475. 10.1016/j.celrep.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer, N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R.V. Jr., and Walther T.C.. 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics. 12:1115–1126. 10.1074/mcp.M112.020230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Shao L., Chen B.C., Zhang X., Zhang M., Moses B., Milkie D.E., Beach J.R., Hammer J.A. III, Pasham M., et al. 2015. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 349:aab3500. 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger, L.L., Han X., Lewis S.E., Cases S., Farese R.V. Jr., Ory D.S., and Schaffer J.E.. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100:3077–3082. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S., and Parton R.G.. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378. 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- Mavrakis, M., Azou-Gros Y., Tsai F.C., Alvarado J., Bertin A., Iv F., Kress A., Brasselet S., Koenderink G.H., and Lecuit T.. 2014. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat. Cell Biol. 16:322–334. 10.1038/ncb2921 [DOI] [PubMed] [Google Scholar]
- Mavrakis, M., Tsai F.C., and Koenderink G.H.. 2016. Purification of recombinant human and Drosophila septin hexamers for TIRF assays of actin-septin filament assembly. Methods Cell Biol. 136:199–220. 10.1016/bs.mcb.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Miranda, D.A., Kim J.H., Nguyen L.N., Cheng W., Tan B.C., Goh V.J., Tan J.S., Yaligar J., Kn B.P., Velan S.S., et al. 2014. Fat storage-inducing transmembrane protein 2 is required for normal fat storage in adipose tissue. J. Biol. Chem. 289:9560–9572. 10.1074/jbc.M114.547687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari, Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K.. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- Moreno-Castellanos, N., Rodríguez A., Rabanal-Ruiz Y., Fernández-Vega A., López-Miranda J., Vázquez-Martínez R., Frühbeck G., and Malagón M.M.. 2017. The cytoskeletal protein septin 11 is associated with human obesity and is involved in adipocyte lipid storage and metabolism. Diabetologia. 60:324–335. 10.1007/s00125-016-4155-5 [DOI] [PubMed] [Google Scholar]
- Murphy, D.J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325–438. 10.1016/S0163-7827(01)00013-3 [DOI] [PubMed] [Google Scholar]
- Murphy, S., Martin S., and Parton R.G.. 2010. Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One. 5:e15030. 10.1371/journal.pone.0015030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, H., Zhang P., Ding Y., Yang L., Wang Y., Zhang H., Xie Z., Yang F., Cichello S., and Liu P.. 2013. Proteomic studies of isolated lipid droplets from bacteria, C. elegans, and mammals. Methods Cell Biol. 116:1–14. 10.1016/B978-0-12-408051-5.00001-2 [DOI] [PubMed] [Google Scholar]
- O’Hara, L., Han G.S., Peak-Chew S., Grimsey N., Carman G.M., and Siniossoglou S.. 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281:34537–34548. 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onal, G., Kutlu O., Gozuacik D., and Dokmeci Emre S.. 2017. Lipid Droplets in Health and Disease. Lipids Health Dis. 16:128. 10.1186/s12944-017-0521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péterfy, M., Phan J., Xu P., and Reue K.. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27:121–124. 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoisé, B., Malone B., Falgairolle M., Munasinghe J., Stadler J., Sibilla C., Park S.H., and Blackstone C.. 2016. Reep1 null mice reveal a converging role for hereditary spastic paraplegia proteins in lipid droplet regulation. Hum. Mol. Genet. 25:5111–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue, K., and Zhang P.. 2008. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 582:90–96. 10.1016/j.febslet.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue, K., Xu P., Wang X.P., and Slavin B.G.. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41:1067–1076. 10.1016/S0022-2275(20)32011-3 [DOI] [PubMed] [Google Scholar]
- Ron, D., and Walter P.. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529. 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Salo, V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santinho, A., Salo V.T., Chorlay A., Li S., Zhou X., Omrane M., Ikonen E., and Thiam A.R.. 2020. Membrane Curvature Catalyzes Lipid Droplet Assembly. Curr. Biol. 30:2481–2494.e6. 10.1016/j.cub.2020.04.066 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941. 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., Voeltz G.K., and Rapoport T.A.. 2006. Rough sheets and smooth tubules. Cell. 126:435–439. 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Shibata, Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., and Voeltz G.K.. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904. 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., and Rapoport T.A.. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788. 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubeita, G.T., Tran S.L., Xu J., Vershinin M., Cermelli S., Cotton S.L., Welte M.A., and Gross S.P.. 2008. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 135:1098–1107. 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin, M., Farkasovsky M., Hauer F., Kühlmann D., Macara I.G., Weyand M., Stark H., and Wittinghofer A.. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315. 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Sriburi, R., Jackowski S., Mori K., and Brewer J.W.. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167:35–41. 10.1083/jcb.200406136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, X., Arlt H., Brock K.P., Lai Z.W., DiMaio F., Marks D.S., Liao M., Farese R.V. Jr., and Walther T.C.. 2018. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217:4080–4091. 10.1083/jcb.201809067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Chung J., and Farese R.V. Jr. 2017. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582. 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Li S., Wang H., Shui W., and Hu J.. 2017. Quantitative proteomics reveal proteins enriched in tubular endoplasmic reticulum of Saccharomyces cerevisiae. eLife. 6:e23816. 10.7554/eLife.23816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling, F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling, F., Thiam A.R., Olarte M.J., Wang J., Beck R., Gould T.J., Allgeyer E.S., Pincet F., Bewersdorf J., Farese R.V. Jr., and Walther T.C.. 2014. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 3:e01607. 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Sun S., Wang W., Shi J., Hu X., Wang S., Su D., Rao Z., Hu J., and Lou Z.. 2015. Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J. Cell Biol. 210:961–972. 10.1083/jcb.201502078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN Binds Anionic Phospholipids. Dev. Cell. 47:248–256.e4. 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Yap, W.S., Shyu P. Jr., Gaspar M.L., Jesch S.A., Marvalim C., Prinz W.A., Henry S.A., and Thibault G.. 2020. The yeast FIT2 homologs are necessary to maintain cellular proteostasis and membrane lipid homeostasis. J. Cell Sci. 133:jcs248526. 10.1242/jcs.248526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Yang L., Ding Y., Wang Y., Lan L., Ma Q., Chi X., Wei P., Zhao Y., Steinbüchel A., et al. 2017. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 8:15979. 10.1038/ncomms15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists sequences for primers, siRNAs, gRNAs, and peptides used in this study.