Abstract
Background
SARS-CoV-2 predisposes patients to secondary infections; however, a better understanding of the impact of coinfections on the outcome of hospitalized COVID-19 patients is still necessary.
Aim
To analyse death risk due to coinfections in COVID-19 patients.
Methods
The odds of death of 212 severely ill COVID-19 patients were evaluated, with detailed focus on the risks for each pathogen, site of infection, comorbidities and length of hospitalization.
Findings
The mortality rate was 50.47%. Fungal and/or bacterial isolation occurred in 89 patients, of whom 83.14% died. Coinfected patients stayed hospitalized longer and had an increased odds of dying (odds ratio (OR): 13.45; R2 = 0.31). The risk of death was increased by bacterial (OR: 11.28) and fungal (OR: 5.97) coinfections, with increased levels of creatinine, leucocytes, urea and C-reactive protein. Coinfections increased the risk of death if patients suffered from cardiovascular disease (OR: 11.53), diabetes (OR: 6.00) or obesity (OR: 5.60) in comparison with patients with these comorbidities but without pathogen isolation. The increased risk of death was detected for coagulase-negative Staphylococcus (OR: 25.39), Candida non-albicans (OR: 11.12), S. aureus (OR: 10.72), Acinetobacter spp. (OR: 6.88), Pseudomonas spp. (OR: 4.77), and C. albicans (OR: 3.97). The high-risk sites of infection were blood, tracheal aspirate, and urine. Patients with coinfection undergoing invasive mechanical ventilation were 3.8 times more likely to die than those without positive cultures.
Conclusion
Severe COVID-19 patients with secondary coinfections required longer hospitalization and had higher risk of death. The early diagnosis of coinfections is essential to identify high-risk patients and to determine the right interventions to reduce mortality.
Keywords: Coinfections, COVID-19, Fungal, Bacterial, Mortality
Introduction
Since December 2019, SARS-CoV-2 has led to more than 100 million COVID-19 cases around the world, resulting in almost three million deaths [1]. SARS-CoV-2 infects primarily the lungs and there is no effective treatment available [2]. Most COVID-19 patients present with mild or moderate disease; but patients with comorbidities may require mechanical ventilation and intensive care, which predispose to secondary and opportunistic infections [3].
Considering previous data from other respiratory virus diseases such as severe acute respiratory syndrome (SARS-CoV-1), Middle East respiratory syndrome (MERS), some authors have pointed out that COVID-19 patients may also be more susceptible to bacterial/fungal coinfection [3,4]. These coinfections represent a severe risk of morbidity and mortality as observed during influenza A (H1N1) pandemic in 2009 and may be present in 8% (62/806) of COVID-19 patients [5,6]. Goyal et al. reported a 6% rate of bacteraemia during hospital admission, whereas Wang et al. reported that 29 out of 69 patients had bacterial/fungal coinfection [7,8]. Acinetobacter, Klebsiella, Entero-bacter, Aspergillus, and Candida are among the main genera that cause secondary infection in COVID-19 patients [[8], [9], [10]].
Although some studies report coinfections and pathogens isolated, the risk imposed by these infections on patients' morbidity and mortality is still unclear. This study aimed to analyse the association between fungal/bacterial coinfections and mortality of patients with severe COVID-19 admitted to a public tertiary hospital in Belo Horizonte, Brazil [11]. The primary endpoint was the risk of death, comparing patients with and without fungal and bacterial secondary infection; secondary endpoints included: (i) patients' risk of death in the presence of both coinfection and comorbidities; (ii) risks associated with the opportunistic pathogen; (iii) site of isolation; and (iv) implications on length of hospitalization. This knowledge will help to improve the diagnostic approaches to identify high-risk patients, and to determine interventions that reduce mortality.
Methods
Study design and participants
In this cohort we evaluated data from 212 severely ill COVID-19 patients (from May to November 2020) admitted to the Eduardo de Menezes hospital (Fundação Hospitalar do Estado de Minas Gerais), Belo Horizonte, Brazil, which is exclusively dedicated to attend COVID-19 patients during the pandemic. All patients received laboratory diagnosis of COVID-19 (SARS-CoV-2 reverse transcription–polymerase chain reaction positive in nasopharyngeal samples) and were followed from admission until the outcome (discharge or death). Most of the patients required critical care unit admission. Either written or oral informed consent from patients or their legal representatives was obtained before the enrolment. Both the National Ethics Committee (Comissão Nacional de Ética em Pesquisa – CONEP) and the hospital's Ethics Committee approved the study (CAAE: 30627320.6.0000.0008).
Data collection
Upon enrolment, information about age, sex, comorbidities and length of hospital stay were obtained from medical records. Other data such as signs and symptoms, use of mechanical ventilation and results of blood tests performed at the time of suspected secondary infection were collected.
Bacterial and fungal strains were isolated from clinical samples and identified according to the hospital standards procedures. The techniques used to identify pathogens are based on biochemical and morphological methods. Sometimes the available culture media and reagents are not the ideal ones for pathogen identification at the species level.
Definitions
COVID-19 cases were classified as severe according to the Clinical Spectrum of SARS-CoV-2 Infection established by the National Institutes of Health, which includes SpO2 <94% on room air at sea level, a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) <300 mmHg, respiratory frequency >30 breaths/min, or lung infiltrates >50% [12].
Patients with signs and symptoms of coinfection during COVID-19 hospitalization and with further positive cultures, as defined by CDC, were also included in this study [13]. Only healthcare-associated infections were included. Data on the growth of the micro-organisms were analysed in relation to the risk of death. Microbiological findings not related to a higher risk of death were considered as colonization. The presence of positive cultures related to increased risk of death was considered as an infection. The clinical specimens analysed included blood, tracheal aspirate, urine, catheter tip, mini-bronchoalveolar lavage (mini-BAL), sputum, and refluid (blood collected at the access to the central venous catheter).
Data on antimicrobials were collected, whose prescription was carried out according to the Guidelines Assistance for COVID-19 [14].
Statistical analyses
Upon the determination of the main factors influencing patients' deaths, patients were compared according to whether they had been with or without fungal and/or bacterial isolation. Multiple regression and stratified univariate analysis were used to assess the risk of death for patients with comorbidities in the presence and absence of positive cultures. We calculated the death risk associated with each isolated pathogen, site of infection, and the implications of coinfection on length of hospitalization. The odds ratios (OR) were presented with 95% confidence intervals and P < 0.05 was considered statistically significant. Data were expressed as absolute values, mean ± SD, or as percentages. Categorical variables were compared using the χ2-test. Continuous variables were compared using Student's t-test or Mann–Whitney test. EpiInfo 7.2, GraphPad Prism 5.0 and Microsoft Excel 2007 software were used in the analyses.
Results
Clinical characteristics
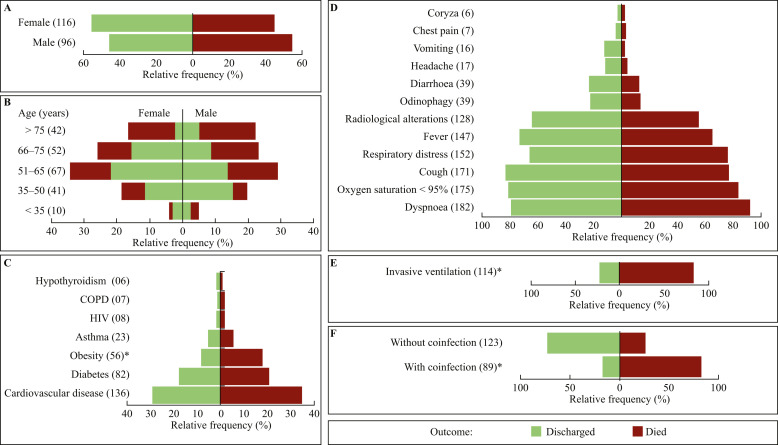
The patients' demographic and clinical characteristics are shown in Figure 1 . From the 212 patients, 96 (45.3%) were female and 116 (54.7%) were male. Overall mortality was 50.5% (107), being 54.3% among men and 45.8% among women (Figure 1A), with no significant difference between them (OR: 0.72; P = 0.2202). Moreover, 44.2% of patients were aged >65 years (Figure 1B). The average age for those who were discharged (56.8 ± 14.3 years) was significantly lower than that of those who died (66.7 ± 13.8 years) (P < 0.0001). Mortality was higher among older patients: 81.0% in the group aged >75 years and 51.9% in the group aged 66–75 years. Regarding the comorbidities, cardiovascular diseases (46.0%) and diabetes mellitus (38.5%) were the most frequent (Figure 1C), and obesity increased the death risk (OR: 2.66; P = 0.0029). Dyspnoea (85.9%), oxygen saturation <95% (82.6%), cough (80.3%), respiratory distress (71.3%), and fever (69.3%) were common, whereas chest pain (3.8%) and coryza (2.8%) were uncommon. None of the symptoms was related to a higher risk of death (P > 0.05) (Figure 1D). A total of 114 (53.8%) patients underwent invasive mechanical ventilation; of whom 79.0% (90) died (Figure 1E), indicating that this procedure increased risk of death (OR: 17.86; R 2 = 0.38; P < 0.001). Fungal and/or bacterial positive cultures occurred in 89 (41.8%) patients (Figure 1F), of whom 83.1% died. Coinfections increased the death risk (OR: 13.45; R 2 = 0.31; P < 0.0001).
Figure 1.
Demographics and clinical characteristics of hospitalized COVID-19 patients. Comparative analyses of COVID-19 case fatality rates by sex (A), age range (B), comorbidities and lifestyle (C), symptoms (D), invasive ventilation (E), and presence or absence of fungal and/or bacterial coinfections (F). The values in parentheses indicate the number of occurrences in the respective category. All relative frequency values are expressed as a percentage of the study cohort (N = 212). ∗P < 0.05 according to the logistic regression analysis. COPD, chronic obstructive pulmonary disease; HIV, human immunodeficiency virus.
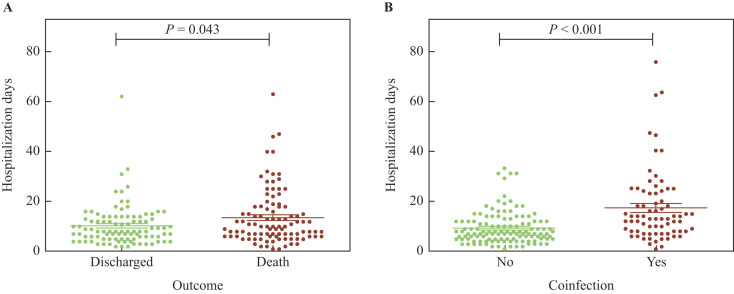
The mean hospitalization length was significantly longer (P = 0.043) among patients who died (13.6 days) compared to those who were discharged (10.3 days) (Figure 2 A). Patients with positive cultures stayed hospitalized (17.3 days) longer than those without them (9.3 days) (P = 0.001) (Figure 2B).
Figure 2.
Length of hospitalization period. Hospitalization (days) for patients who were discharged or who died (A) and for the presence or absence of fungal and/or bacterial coinfections (B). P-values calculated by Mann–Whitney U-test.
Pathogen isolation and mortality risk considering COVID-19 risk factors
Stratified analyses were performed to assess whether the pathogen isolation increased the risk of death in patients with comorbidities (Table I ).
Table I.
Influence of coinfection on the risk of death for severe COVID-19 patients with comorbidities
| Comorbidity/risk factor | Total (negative/positive culture) |
OR (95% CI) | P-value | R2 | |
|---|---|---|---|---|---|
| Discharged | Death | ||||
| Cardiovascular disease (N = 136) | 62 (52/10) | 74 (23/51) | 11.53 (4.99–26.63) | <0.0001 | 0.28 |
| Diabetes (N = 82) | 38 (28/10) | 44 (14/30) | 6.00 (2.29–15.69) | 0.0002 | 0.17 |
| Obesity (N = 56) | 18 (12/06) | 38 (10/28) | 5.60 (1.66–18.92) | 0.0042 | 0.15 |
| Invasive ventilation (N = 114) | 24 (14/10) | 90 (25/65) | 3.63 (1.14–11.49) | 0.0005 | 0.07 |
| Asthma (N = 23) | 11 (8/03) | 12 (5/7) | 3.73 (0.65–21.58) | 0.1500 | 0.10 |
| COPD (N = 7) | 03 (1/02) | 04 (2/2) | 0.5 (0.02–11.09) | 0.6830 | 0.03 |
| HIV (N = 8) | 04 (2/2) | 04 (0/4) | Undefineda | 0.1266 | 0.33 |
| Hypothyroidism (N = 6) | 04 (3/1) | 02 (0/2) | Undefineda | 0.1138 | 0.50 |
COPD, chronic obstructive pulmonary disease; HIV, human immunodeficiency virus.
Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using stratified univariate analysis.
R2 was calculated using regression model.
Undefined because OR could not be calculated with a zero cell.
Coinfections increased the risk of death in patients with cardiovascular disease (OR: 11.53; R 2 = 0.28; P < 0.001), diabetes (OR: 6.00; R 2 = 0.17; P < 0.001), and obesity (OR: 5.60; R 2 = 0.15; P < 0.001) comparing with patients with these comorbidities but without micro-organism isolation. Due to the absence of positive cultures among patients with HIV and hypothyroidism, ORs were not calculated in these cases. Patients with positive cultures undergoing invasive mechanical ventilation were 3.8 times more likely to die than those who were undergoing the same procedure but without micro-organism isolation (P = 0.005).
Influence of secondary pathogen isolation on biochemical and haematological parameters
Next, biochemical and haematological markers were verified by comparing COVID-19 patients with and without positive cultures (Table II ). Creatinine, total leucocyte count, urea, and C-reactive protein were increased in patients with positive cultures, whereas the levels of haemoglobin were decreased. Platelet numbers and lactate concentrations were not different between the two groups.
Table II.
Biochemical and haematological parametersa of COVID-19 patients with and without positive cultures
| Parameter | Positive culture |
P-value | |
|---|---|---|---|
| No (N = 123) | Yes (N = 89) | ||
| Serum creatinine concentration (mg/dL) | 1.53 ± 1.85 | 2.40 ± 1.85 | <0.0001 |
| Leucocyte count (/mm³) | 10,705 ± 8434 | 14,429 ± 7357 | <0.0001 |
| Urea (mg/dL) | 60.66 ± 49.28 | 113.7 ± 52.25 | <0.0001 |
| C-reactive protein (CRP) (mg/L) | 113.9 ± 98.76 | 159.6 ± 99.18 | 0.0002 |
| Haemoglobin concentration (g/dL) | 12.53 ± 2.01 | 11.23 ± 2.31 | <0.0001 |
| Platelet count (/mm³) | 268,233 ± 109,669 | 248,012 ± 118,474 | 0.1112 |
| Lactate concentration (mmol/L) | 1.53 ± 1.13 | 1.59 ± 1.05 | 0.4979 |
Data are presented as mean ± SD.
P-values calculated by Mann–Whitney U-test.
The values refer to tests carried out at the same time of secondary pathogen isolation.
Aetiological agents of coinfections
Data related to the identified micro-organisms, their anatomical sites of origin, as well as administered antimicrobials are described in Table III .
Table III.
Micro-organisms, site of micro-organism isolation, and antimicrobials used
| Patient | Sex | Pathogen, site of isolation | Antibacterial | Antifungal |
|---|---|---|---|---|
| 1 | M | Pseudomonas aeruginosa, TA | CEF, VAN, POL, AMO, CLA, DOX | |
| 2 | M | Candida tropicalis, TA; Klebsiella pneumoniae, blood, mini-BAL; Staphylococcus aureus, CT | CEF, AZI, MER, VAN, POL, AMI, TIG, SUL | MIC |
| 3 | M | Candida albicans, TA | CEF, MER, VAN, POL | |
| 4 | F | Candida albicans, urine; Pseudomonas fluorescens, TA; Candida glabrata, urine | MER, VAN, POL | FLU |
| 5 | M | Candida albicans, TA | MER, VAN, TAZ | |
| 6 | F | Candida albicans, urine; Pseudomonas fluorescens, TA | CEF, MER, VAN, POL, AMI | MIC |
| 7 | F | Candida glabrata, TA, urine | MER, VAN, POL, TAZ | |
| 8 | F | Stenotrophomonas maltophilia, TA; Staphylococcus aureus, blood, TA; Pseudomonas aeruginosa, TA; Staphylococcus NC, blood | CEF, AZI, VAN, POL, AMO | |
| 9 | F | Pseudomonas spp., TA; Candida tropicalis, blood | CEF, MER, VAN, POL, CLA | |
| 10 | F | Candida tropicalis, urine | CEF, AZI, TAZ, CLA, TET | |
| 11 | F | Candida albicans, urine | CEF, AZI, VAN, COL, CEP, | |
| 12 | M | Enterobacter spp., TA | CEF, AZI, MER, VAN, CLA, COL | |
| 13 | M | Klebsiella pneumoniae, blood, TA | CEF, AZI, MER, VAN, POL, COL | |
| 14 | M | Aspergillus spp., sputum | CEF, AZI, MER, CLI, SUL | |
| 15 | M | Candida albicans, urine; Acinetobacter baumannii, TA; Klebsiella pneumoniae, CT; Staphylococcus aureus, blood; Staphylococcus NC, CT | CEF, MER, VAN, POL, CLA, LEV, | MIC |
| 16 | M | Staphylococcus NC, blood; Candida albicans, TA; Aspergillus spp., TA; Candida parapsilosis, TA | CEF, AZI, VAN, POL | |
| 17 | F | Staphylococcus NC, blood, CT | CEF, MER, VAN, POL, CLA | AMB |
| 18 | F | Candida albicans, TA | CEF, AZI, MER, VAN, POL, LEV | |
| 19 | F | Acinetobacter baumannii, TA, CT; Candida glabrata, urine; Candida tropicalis, TA; Staphylococcus NC, blood, CT; Stenotrophomonas maltophilia, TA; Enterococcus spp., blood | CEF, AZI, MER, VAN, POL, TAZ, | MIC |
| 20 | M | Candida tropicalis, TA | CEF, AZI | |
| 21 | M | Pseudomonas aeruginosa, TA, blood | CEF, AZI, MER, VAN, POL, AMO, | |
| 22 | F | Stenotrophomonas maltophilia, TA; Acinetobacter spp., TA | CEF, AZI, MER, VAN, POL, LEV | |
| 23 | M | Pseudomonas aeruginosa, TA | CEF, AZI, MER, VAN, POL | |
| 24 | F | Acinetobacter baumannii, TA, CT; Candida albicans, TA; Candida glabrata, urine; Pseudomonas aeruginosa, urine; Candida tropicalis, urine | CEF, AZI, MER, VAN, POL, CEP | |
| 25 | F | Pseudomonas aeruginosa, urine | CEF, AZI, MER, VAN, TAZ | |
| 26 | F | Acinetobacter baumannii, TA | CEF, AZI, MER, VAN, LEV, COL, SUL | FLU |
| 27 | F | Staphylococcus NC, CT; Candida albicans, TA | CEF, AZI, MER, VAN, POL | MIC |
| 28 | F | Acinetobacter baumannii, TA, CT | CEF, AZI, MER | FLU |
| 29 | M | Acinetobacter baumannii, TA; Klebsiella pneumoniae, blood, TA | CEF, MER, VAN, POL, CLA, AMI, | FLU |
| 30 | F | Pseudomonas aeruginosa, TA; Candida tropicalis, TA | CEF, AZI, VAN, AMI | |
| 31 | M | Pseudomonas aeruginosa, TA; Enterobacter sakasakii, TA; Stenotrophomonas maltophilia, TA | CEF, MER, VAN, POL, CLA, COL, | |
| 32 | F | Candida albicans, TA; Staphylococcus NC, blood | MER, VAN, POL, TAZ | |
| 33 | F | Acinetobacter spp., TA; Staphylococcus aureus, TA | CEF, AZI, MER, VAN, POL, COL, | |
| 34 | M | Candida albicans, sputum | CEF, AZI | |
| 35 | F | Candida albicans, TA; Acinetobacter baumannii, TA; Staphylococcus aureus, TA; Enterobacter aerogenes, TA | AZI, VAN, POL, TAZ | |
| 36 | M | Staphylococcus NC, blood, CT; Stenotrophomonas maltophilia, blood; Acinetobacter baumannii, TA | CEF, AZI, VAN, CLA, COL | |
| 37 | M | Pseudomonas aeruginosa, TA | CEF, VAN, POL, CLA | |
| 38 | M | Candida albicans, TA; Pseudomonas spp., TA; Candida tropicalis, TA | AZI, MER, CLA, SUL | |
| 39 | M | Staphylococcus aureus, TA; Stenotrophomonas maltophilia, TA | CEF, AZI, MER, VAN, POL | |
| 40 | M | Candida tropicalis, TA | CEF, AZI, MER, VAN, POL, | MIC |
| 41 | M | Candida albicans, sputum; Aspergillus spp., sputum | AZI, AMO | |
| 42 | F | Candida albicans, TA; Acinetobacter baumannii, TA | CEF, AZI, MER, VAN, POL, | |
| 43 | M | Pseudomonas aeruginosa, TA; Acinetobacter baumannii, CT | CEF, AZI, VAN, POL, | |
| 44 | F | Acinetobacter baumannii, TA; Candida albicans, urine | CEF, AZI, MER, VAN, POL, | MIC |
| 45 | M | Enterococcus spp., CT; Stenotrophomonas maltophilia, TA; Candida albicans, CT, sputum, urine; Staphylococcus NC, CT; Pseudomonas aeruginosa, CT, TA; Acinetobacter baumannii, TA | MER, VAN, POL, COL, AMP, AMI, SUL | MIC |
| 46 | M | Enterobacter spp., TA | CEF, VAN, CLA | |
| 47 | M | Candida albicans, TA | CEF, VAN, POL, CLA | |
| 48 | M | Candida albicans, TA | MER, VAN, POL | |
| 49 | M | Staphylococcus NC, CT | CEF, MER, VAN, POL | |
| 50 | M | Candida glabrata, TA; Acinetobacter baumannii, TA | MER | MIC |
| 51 | M | Pseudomonas aeruginosa, TA | CEF, AZI, VAN, POL | |
| 52 | M | Aspergillus spp., TA; Candida albicans, TA | CEF, AZI, VAN, POL | |
| 53 | M | Enterococcus spp., CT, TA; Candida albicans, TA; Klebsiella pneumoniae, blood, TA; Enterobacter spp., TA | CEF, AZI, MER, VAN, POL, COL, AMI, | |
| 54 | M | Staphylococcus NC, CT | CEF, AZI, MER, VAN, POL, COL | |
| 55 | M | Acinetobacter baumannii, TA; Candida albicans, urine; Pseudomonas aeruginosa, TA | CEF, AZI, MER, VAN, POL, | |
| 56 | M | Staphylococcus aureus, blood, TA; Candida albicans, urine; Staphylococcus NC, CT; Enterococcus spp., urine | CEF, AZI, POL, TEI | |
| 57 | M | Pseudomonas aeruginosa, TA | CEF, AZI, VAN, POL, AMI, TIG | |
| 58 | F | Staphylococcus aureus, CT; Stenotrophomonas maltophilia, TA; Staphylococcus NC, CT | CEF, AZI, MER, VAN, POL, SUL | MIC |
| 59 | F | Candida albicans, urine | AZI, AMO, | |
| 60 | F | Candida albicans, TA; Staphylococcus NC, blood | CEF, AZI, AMO | FLU |
| 61 | M | Staphylococcus NC, CT; Stenotrophomonas maltophilia, TA; Candida tropicalis, urine | CEF, AZI, MER, VAN, POL, AMI, OXA, SUL | |
| 62 | F | Candida albicans, TA | CEF, AZI, VAN | |
| 63 | F | Candida albicans, TA | CEF, AZI, MER, VAN, POL, COL | |
| 64 | F | Candida kefir, urine; Acinetobacter baumannii, TA | CEF, AZI, MER, VAN, POL, AMO | FLU |
| 65 | F | Candida tropicalis, TA; Pseudomonas fluorescens, TA | CEF, AZI, VAN, POL | |
| 66 | F | Staphylococcus NC, CT; Klebsiella aerogenes, TA; Candida albicans, TA; Candida glabrata, TA | CEF, AZI, MER, COL | |
| 67 | F | Acinetobacter baumannii, blood, TA, CT; Candida tropicalis, urine | ||
| 68 | F | Candida albicans, urine | AZI, AMO | |
| 69 | F | Staphylococcus aureus, blood, TA; Klebsiella spp., TA | CEF, AZI, VAN, POL | |
| 70 | F | Acinetobacter baumannii, TA | CEF, AZI, VAN, POL | |
| 71 | M | Staphylococcus NC, blood; Klebsiella oxytoca, TA; Klebsiella aerogenes, TA; Escherichia coli, TA | CEF, AZI, VAN, POL | FLU |
| 72 | F | Acinetobacter baumannii, CT, TA; Staphylococcus NC, CT | CEF, AZI, VAN, POL, CEP | |
| 73 | F | Klebsiella pneumoniae, TA | CEF, AZI, MER, VAN, POL | |
| 74 | M | Enterococcus sp., CT, TA; Staphylococcus NC, CT | CEF, AZI, MER, VAN, POL | MIC |
| 75 | M | Candida albicans, TA | CEF, AZI, MER, VAN, POL | |
| 76 | F | Acinetobacter baumannii, TA; Candida albicans, TA | CEF, AZI, MER, VAN, POL, TAZ, COL | |
| 77 | F | Candida albicans, TA | AZI, AMP | |
| 78 | M | Staphylococcus aureus, blood, TA; Acinetobacter baumannii, TA, urine; Klebsiella aerogenes, TA; Staphylococcus, NC, CT; Enterobacter gergoviae, TA | CEF, AZI, MER, VAN, | MIC |
| 79 | M | Klebsiella pneumoniae, TA | MER, VAN, POL, TAZ, AMI, TET | |
| 80 | F | Candida glabrata, blood, CT, refluid, urine; Candida tropicalis, TA | CEF, MER, VAN, POL, CLA, LEV | FLU |
| 81 | F | Staphylococcus aureus, TA | CEF, VAN, POL, CLA | |
| 82 | M | Candida glabrata, urine; Candida tropicalis, urine | MER, POL, TAZ, CLA | FLU |
| 83 | M | Candida albicans, urine, CT; Enterococcus faecalis, CT | CEF, AZI, VAN, POL, CLI | |
| 84 | M | Pseudomonas aeruginosa, TA | CEF, AZI, VAN, POL | |
| 85 | M | Candida albicans, CT | MER, VAN, POL | FLU |
| 86 | M | Staphylococcus aureus, TA; Candida albicans, blood; Pseudomonas aeruginosa, TA | CEF, VAN, POL, CLA | |
| 87 | M | Klebsiella aerogenes, urine | CEF, CLA | |
| 88 | M | Candida albicans, TA; Staphylococcus NC, CT | CEF, AZI, MER, VAN, POL, CEP | MIC |
| 89 | F | Candida tropicalis, urine; Staphylococcus NC, CT | CEF, AZI, MER, VAN, POL, TAZ, AMI, TIG, SUL | MIC |
NC, negative coagulase; TA, tracheal aspirate; BAL, bronchoalveolar lavage; CT, catheter tip; CEF, ceftriaxone; AZI, azithromycin; MER, meropenem; VAN, vancomycin; POL, polymyxin B; CLA, clarithromycin; AMO, amoxicillin; CEP, cefepime; SUL, sulfamethoxazole; DOX, doxycycline; TEI, teicoplanin; AMP, ampicillin; COL, sodium colistimethate; CLI, clindamycin; TAZ, Tazocin; OXA, oxacillin; LEV, levofloxacin; TIG, tigecycline; AMI, amikacin; TET, tetracycline; MIC, micafungin; FLU, fluconazole; AMB, amphotericin B.
Of the 212 patients, 34 (16.0%) had only bacterial positive cultures, 25 (11.8%) had only fungal positive culture, and 30 (14.2%) had positive cultures for both. The risk of death was increased in patients who had positive cultures for bacteria (OR: 11.28; P < 0.00005), fungi (OR: 5.97; P < 0.00005), and mixed (bacteria and fungi) (OR: 6.10; P = 0.001) compared to those with negative cultures. More than one micro-organism was isolated in most patients undergoing mechanical ventilation. The isolation of only one micro-organism occurred frequently from tracheal aspirate samples. Among patients who did not undergo invasive mechanical ventilation, the isolation of only one micro-organism predominated, mainly from urine samples.
Of the 64 patients with bacterial positive cultures, the isolates were: Staphylococcus spp. 45.3% (29), Acinetobacter spp. 32.8% (21), Pseudomonas spp. 32.8% (21), Stenotrophomonas spp. 14.06% (9), Klebsiella spp. 12.5% (8), Enterobacter spp. 9.4% (6), Enterococcus spp. 9.4% (6) and Escherichia coli 6% (1). The mortality rates of COVID-19 patients suffering from these coinfections were: coagulase-negative Staphylococcus (NC) 95.5%, Staphylococcus aureus 90.9%, Acinetobacter spp. 85.7%, Pseudomonas spp. 81.0%, Enterobacter spp. 83.3%, Klebsiella spp. 75%, Enterococcus spp. 100%, Stenotrophomonas spp. 100%, and Escherichia spp. 100%. Regarding fungi, isolates were Candida spp. 98.2% (54) and Aspergillus spp. 5.5% (3). The mortality rates were: Candida non-albicans 90.5%, Candida albicans 76.3%, and Aspergillus spp. 66.7%.
Increased risk of death (comparing with patients without coinfection) was verified when Staphylococcus NC (OR: 25.40), Staphylococcus aureus (OR: 10.72), Acinetobacter spp. (OR: 6.88), Pseudomonas spp. (OR: 4.77), Candida non-albicans (OR: 11.12), and Candida albicans (OR: 3.97) were isolated (Table IV ).
Table IV.
Risk of death associated with isolation of bacteria and fungi from COVID-19 patients
| Genus | OR (95% CI) | P-value |
|---|---|---|
| Staphylococcus spp. | ||
| Staphylococcus NC | 25.40 (3.35–192.66) | <0.0001 |
| Staphylococcus aureus | 10.72 (1.35–85.32) | 0.0060 |
| Acinetobacter spp. | 6.88 (1.96–24.11) | 0.0007 |
| Pseudomonas spp. | 4.77 (1.55–14.70) | 0.0033 |
| Enterobacter spp. | 5.10 (0.58–44.40) | 0.1032 |
| Klebsiella spp. | 3.06 (0.60–15.52) | 0.1582 |
| Enterococcus spp. | Undefineda | 0.0141 |
| Stenotrophomonas spp. | Undefineda | 0.0024 |
| Escherichia spp. | Undefineda | 0.3219 |
| Candida spp. | ||
| Candida non-albicans | 11.12 (2.52–49.07) | 0.0001 |
| Candida albicans | 3.97 (1.77–8.87) | 0.0004 |
| Aspergillus spp. | 1.98 (0.18–22.18) | 0.5729 |
NC, negative coagulase.
Undefined because odds ratio (OR) could not be calculated with a zero cell. OR and 95% confidence interval (CI) were calculated using univariate logistic regression model. Data were calculated considering the isolated micro-organisms individually.
Site of pathogen isolation and risk of death
Bacteria were frequently isolated from tracheal aspirate (N = 45), followed by catheter tip (N = 21), blood (N = 19), urine (N = 4), and mini-BAL (N = 1) (Table V ). Bacteria in the blood (OR: 21.03) and tracheal aspirate (OR: 11.94) were associated with an increased risk of death (P ≤ 0.001). On the other hand, their isolation from the urine did not increase risk of death (P = 0.9848).
Table V.
Risk of death associated with the presence of bacteria and fungi by type of clinical specimen
| Site of pathogen isolation | Death (%) | OR (95% CI) | P-value |
|---|---|---|---|
| Bacteria | |||
| Blood (N = 19) | 94.74 | 21.03 (2.75–160.71) | 0.0001 |
| Tracheal aspirate (N = 45) | 88.89 | 11.94 (4.49–31.81) | 0.0000 |
| Urine (N = 4) | 50.00 | 0.98 (0.14–7.10) | 0.9848 |
| Catheter tip (N = 21) | 100 | Undefineda | 0.0000 |
| Mini-BAL (N = 1) | 100 | Undefineda | 0.3219 |
| Fungi | |||
| Tracheal aspirate (N = 33) | 84.85 | 7.09 (2.62–19.20) | 0.0000 |
| Urine (N = 23) | 82.61 | 5.45 (1.78–16.63) | 0.0011 |
| Sputum (N = 3) | 33.33 | 0.48 (0.04–5.44) | 0.5508 |
| Catheter tip (N = 4) | 100 | Undefineda | 0.0460 |
| Blood (N = 4) | 100 | Undefineda | 0.0460 |
| Refluid (N = 1) | 100 | Undefineda | 0.3218 |
Undefined because odds ratio (OR) could not be calculated with a zero cell. ORs and 95% confidence interval (CIs) were calculated using univariate logistic regression model.
Fungi were frequently isolated from tracheal aspirate (N = 33), followed by urine (N = 25), blood (N = 4), catheter tip (N = 4), sputum (N = 3), and mini-BAL (N = 1). The isolation of fungi in the tracheal aspirate (OR: 7.09) and urine (OR: 5.45) was associated with an increased risk of death (P ≤ 0.002), which did not occur with isolation from sputum (OR: 0.49) (Table V).
Discussion
In all, 212 COVID-19 hospitalized patients were analysed, 54.7% of whom were male and 75.9% were aged >50 years, which is consistent with previous studies on COVID-19 in China and Brazil [10,15]. The mortality data from our study corroborate a Brazilian study, in which 59% of the patients admitted to the intensive care unit and 80% of those who were mechanically ventilated died [15]. Unlike one previous report, we did not find higher mortality among male patients [16]. The average age of deceased patients was higher than that of those discharged, which is in line with previous studies showing that 65% of patients aged >50 years were more likely to develop severe COVID-19 [10,15,17]. Dyspnoea, oxygen saturation <95%, and cough were the main symptoms reported at admission and are compatible with those previously described [18]. Cardiovascular disease, diabetes, and obesity, the main comorbidities found in our study, are also cited by other authors as related to severe COVID-19. Organ damage in these patients may contribute to multiple organ failure and consequent mortality [19].
Patients with COVID-19 may present with reduction in the lymphocyte count, which affects cell-mediated immune response by decreasing CD4 and CD8 T cells, suggesting that SARS-CoV-2 consumes many immune cells and inhibits the cellular immune function [20,21]. Therefore, the immune dysregulation increases susceptibility to coinfection, which can be diagnosed in 50% (27/54) of patients with COVID-19 who died [22]. In our analysis, the presence of coinfections significantly increased the length of hospitalization and death risk, which has an impact on public health costs and the scarcity of beds available for new patients.
Coinfections also cause haematological and biochemical disbalance, worsening the general clinical condition [23]. The present sutdy found significant changes in the levels of creatinine, haemoglobin, leucocyte count, and urea in patients with positive cultures compared to those without them.
In our cohort, the secondary pathogen isolation increased the risk of death in patients with cardiovascular disease, diabetes, obesity, and when invasive mechanical ventilation was used. Interestingly, non-aureus Staphylococcus was associated with a higher risk of death. Despite being often considered as a commensal micro-organism, it is one of the most common causes of catheter-related infections, and its treatment may be complicated by the presence of biofilms and resistance to antibiotics [24]. In most cases of isolation of NC Staphylococcus from catheter, other micro-organisms were also isolated from this clinical sample, pointing out that the catheter may act as a reservoir for colonization by multiple micro-organisms, possibly in biofilms. In this study, 97.2% of the patients received antibacterial drugs, a higher rate than that described by other authors [25]. As previously discussed, the possibility of coinfections should not be ignored, but it is essential that the need for antibacterial treatments is carefully evaluated, so the selection of resistant micro-organisms is not accelerated [26].
Although bacterial infections are more common in critically ill patients, fungal infections should not be underestimated, as they are also associated with increased mortality, longer hospital stays and increased hospital costs [27]. We detected a high prevalence of Candida spp., which is the main fungus causing infections in critically ill patients [28]. It may be part of the human microbiota, making it difficult to distinguish between colonization and infection.
The increased risk of death (depending on the site of isolation and species) may indicate that they are acting as pathogens. The higher risk of death from infections by Candida non-albicans may be related to the lower sensitivity of some species to antifungals. It is worth mentioning that patients with severe COVID-19 undergo many interventions that favour opportunistic infections (mechanical ventilation, broad-spectrum antibacterial, corticosteroids, parenteral nutrition, central venous catheter). If a patient's condition shows strong evidence of a fungal infection, this possibility should not be ignored [29].
Overall, this study has some limitations. First, because of its retrospective nature, data availability was limited to the medical records at the hospital. Second, the total population in the analysed period was larger than those presented in this study, since data from patients who were transferred to other hospitals and those who were discharged from the hospital were excluded. Third, we must consider the small sample size related to some analysed data. In these cases, studies with a larger sample size should be performed in order to offer more data. Finally, our analyses were limited to the available laboratory data. However, we believe that our data lead to the better understanding of fungal and bacterial coinfections in patients with severe COVID-19.
In conclusion, our results indicate that severely ill COVID-19 patients with bacterial and/or fungal coinfections require longer hospital stays and present a higher relative risk of death compared to those without coinfections. Furthermore, coinfections may increase the risk of death in subsets of patients with different comorbidities. Thus, we recommend the early identification of bacterial and fungal infections, since it will help to identify high-risk patients, and to determine the appropriate interventions to reduce mortality.
Conflict of interest statement
None declared.
Funding sources
This study was supported by Fundação de Amparo a Pesquisa do Estado de Minas Gerais – FAPEMIG (Grant APQ-00267-20) and Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq and Brazilian Ministry of Health (440010/2018-7). D.A.S. (303762/2020-9) and J.S.A. (302081/2018-6) are research fellows of the CNPq.
References
- 1.World Health Organization. WHO coronavirus disease (COVID-19) dashboard. Available at: https://covid19.who.int/[last accessed March 2021].
- 2.Stasi C., Fallani S., Voller V., Silvestri C. Treatment for COVID-19: an overview. Eur J Pharmacol. 2020;889:173644. doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lansbury L., Lim B., Baskaran V., Lim W.S. Coinfections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. https://doi:10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cauley L.S., Vella A.T. Why is coinfection with influenza virus and bacteria so difficult to control? Discov Med. 2015;19:33–40. [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson T.M., Moore L.S.P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., et al. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. https://doi:10.1093/cid/ciaa530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., et al. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. https://doi:10.1056/NEJMc2010419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. https://doi:10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alanio A., Dellière S., Sofiane F., Stéphane B., Mégarbane B. High prevalence of putative invasive pulmonary aspergillosis in critically ill COVID-19 patients. Resp Med. 2020;8:E48–E49. doi: 10.1016/S2213-2600(20)30237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-67362030211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contou D., Claudinon A., Pajot O., Micaëlo M., Flandre P.L., Dubert M., et al. Bacterial and viral coinfections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intens Care. 2020;10:119. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Institutes of Health . 2020. Clinical spectrum of SARS-CoV-2 infection.https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum Available at: [last accessed September 2020] [Google Scholar]
- 13.Centers for Disease Control and Prevention/NHSN. Surveillance definitions for specific types of infections 2020. Available at: https://portaldeboaspraticas.iff.fiocruz.br/wp-content/uploads/2020/10/17pscNosInfDef_current.pdf [last accessed November 2020].
- 14.Guidelines Assistance for COVID-19, FHEMIG 2021. Available at: http://www.fhemig.mg.gov.br/index.php?preview=1&option=com_dropfiles&format=&task=frontfile.download&catid=1440&id=14569&Itemid=1000000000000 [last accessed March 2021].
- 15.Ranzani O.T., Bastos L.S.L., Gelli J.G.M., Marchesi J.F., Baião F., Hamacher S., et al. Characterisation of the first 250 000 hospital admissions for COVID-19 in Brazil: a retrospective analysis of nationwide data. Lancet Respir Med. 2021;15:S2213–S2600. doi: 10.1016/S2213-2600(20)30560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark A., Jit M., Warren-Gash C., Guthrie B., Wang H.H.X., Mercer S.W., et al. Centre for the Mathematical Modelling of Infectious Diseases COVID-19 working group. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob Health. 2020;8:e1003–e1017. doi: 10.1016/S2214-109X(20)30264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah S.J., Barish P.N., Prasad P.A., Kistler A., Neff N., Kamm J., et al. Clinical features, diagnostics, and outcomes of patients presenting with acute respiratory illness: a retrospective cohort study of patients with and without COVID-19. EClinicalMedicine. 2020;27:100518. doi: 10.1016/j.eclinm.2020.100518. https://doi:10.1016/j.eclinm.2020.100518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and multiorgan response. Curr Probl Cardiol. 2020;45:100618. doi: 10.1016/j.cpcardiol.2020.100618. https://doi:10.1016/j.cpcardiol.2020.100618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P., Liu Z., Chen Y., Xiao Y., Huang X., Fan X.-G. Bacterial and fungal infections in COVID-19 patients: a matter of concern. Infect Control Hosp Epidemiol. 2020;41:1124–1125. doi: 10.1017/ice.2020.156. httpps://doi:10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frater J.L., Zini G., d’Onofrio G., Rogers H.J. COVID-19 and the clinical hematology laboratory. Int J LabHematol. 2020;42:11–18. doi: 10.1111/ijlh.13229. httpps://doi:10.1111/ijlh.13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otto M. Staphylococcus epidermidis – the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., et al. Bacterial coinfection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bengoechea J.A., Bamford C.G. SARS-CoV-2, bacterial coinfections, and AMR: the deadly trio in COVID-19? EMBO Mol Med. 2020;12 doi: 10.15252/emmm.202012560. https://doi:10.15252/emmm.202012560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verweij P.E., Alanio A. Fungal infections should be part of the core outcome set for COVID-19. Lancet Infect Dis. 2020;7:S1473–S3099. doi: 10.1016/S1473-3099(20)30591-0. https://doi:10.1016/S1473-3099(20)30591-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vincent J.L., Rello J., Marshall J., Silva E., Anzueto A., Martin C.D., et al. EPIC II Group of Investigators. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. https://doi:10.1001/jama.2009.1754 [DOI] [PubMed] [Google Scholar]
- 29.Schnabel R.M., Linssen C.F., Guion N., van Mook W.N., Bergmans D.C. Candida pneumonia in intensive care unit? Open Forum Infect Dis. 2014;1(1):ofu026. doi: 10.1093/ofid/ofu026. https://doi:10.1093/ofid/ofu026 [DOI] [PMC free article] [PubMed] [Google Scholar]