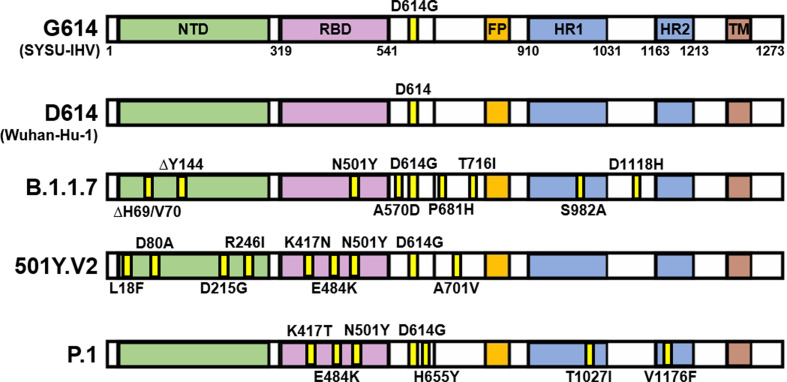
Since the outbreak of the pandemic, waves of epidemics caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants that harbor novel mutations have never paused. Globally, it undergoes rapid mutations that involve single-nucleotide polymorphism (SNP) dominantly, whereas ORF1ab and spike genes contain the most of more than 20,000 mutation sites reported within a year (Fang et al., 2021). Mutations inside spike protein are highly concerned for their potential impact on viral transmissibility and immune evasion, as spike protein is responsible for the interaction with the viral receptor angiotensin-converting enzyme 2 (ACE2) to mediate viral entry to the target cells. D614G identified in early 2020 is a globally dominant mutation (Korber et al., 2020). In late 2020, several variants were reported, which had caused continental and eventually worldwide epidemics. These notable variants include B.1.1.7 lineage (501Y.V1, Variant of Concern [VOC] 202012/01), 501Y.V2 variant (known as B.1.351 lineage), and P.1 lineage (also named 501Y.V3). In comparison with the D614G and D614 lineages identified in early 2020, they contain a large number of mutations within spike protein (Fig. 1 ).
Fig. 1.
Mutations in several notable SARS-CoV-2 variants. Schematic of the full-length spike protein of SARS-CoV-2 variants, including G614, D614, B.1.1.7, 501Y.V2, and P.1 lineage. Mutations in the spike protein of these different variants are shown in yellow labeled with the precise position. NTD, N terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain.
Characteristics of B.1.1.7, 501Y.V2, and P.1 variants
B.1.1.7 was first identified in London during September 2020, and within a month, this lineage emerged in other areas in the United Kingdom and many European countries. Before late January 2021, B.1.1.7 had been detected in more than 70 countries around the world (Galloway SE et al., 2020). Recently, it has been frequently reported in the United States. Compared with the origin lineage, B.1.1.7 exhibited higher transmissibility with 50–75% enhancement, and its R0 was estimated to be 1.75-fold higher. Moreover, juveniles were probably more sensitive to this variant and more prone to develop symptoms (Volz et al., 2021). Based on structural analysis for the recognition of receptor-binding domain (RBD) to ACE2, the increased transmissibility of B.1.1.7 could be due to the mutation (N501Y) on a significant site Asn501 that participates in the interaction between RBD and ACE2 and the additional deletion at positions 69 and 70 that could increase the capability (Leung et al., 2020). As for its impact on pathogenicity, B.1.1.7 infection was correlated with a higher risk of lethality (Horby et al., 2021). Referring to the results of neutralization assays tittering the neutralizing antibodies (NAbs) against B.1.1.7 lineage from different reports, the mutations in the spike caused no significant reduction of neutralizing activity of plasma from convalescents who suffered coronavirus disease 2019 (COVID-19) in early 2020, suggesting that NAbs induced by the infection of original viral lineages could still maintain activity against B.1.1.7 variant (Haynes et al., 2021; Li et al., 2021; Rees-Spear et al., 2021). Furthermore, most of the subsequent studies claimed that the mutations inside the spike protein of B.1.1.7 have minor effects on immune evasion, although N501Y mutation could eliminate the binding affinity of class 3 NAbs that target the N terminal domain (NTD) of the spike protein (Chen et al., 2021).
The 501Y.V2 variant was first reported in Nelson Mandela Bay, South Africa, in early October 2020. Similar to the B.1.1.7, the 501Y.V2 variant was found to have robust transmissibility and rapidly became the dominant lineage over South Africa (> 90%) before December 2020 (Tegally et al., 2020). It emerged in other countries such as the United States as well. Specific mutations including K417N, E484K, and N501Y within RBD of 501Y.V2 increased its binding affinity to ACE2, whereas K417 and E484 of the dominant epitope within RBD, receptor-binding motif (RBM), were not conserved and had plasticity to mutate to escape the interaction with related NAbs (Thomson et al., 2021; Wang et al., 2021). These mutants were associated significantly with immune evasion. Especially, E484K was considered as one of the most important mutations that could lead to the insensitivity of NAbs (Liu et al., 2021a, 2021b). The combination of K417N, E484K, and 242–244 deletion mutations could robustly impact three classes of NAbs, which might consequently reduce the neutralizing activity of sera from more than 90% early convalescents (Li et al., 2021; Wibmer et al., 2021), while some of the samples showed a nearly 90% neutralization reduction (Greaney et al., 2021). Although the enhancement of pathogenicity of 501Y.V2 was not verified with conclusive evidence, it has been shown that a mouse-adapted SARS-CoV-2 strain with similar mutation sites (N501Y, Q493H, and K417N) could induce severe organ damages similar to COVID-19 in mice, which showed higher lethality in the infected aged male mice (Sun et al., 2020). More studies are needed to estimate the clinical risks of the 501Y.V2 variant.
The genomic sequence of P.1 lineage was first identified in Manaus, Brazil, which shared several mutation sites with 501Y.V2, including K417T, E484K, and N501Y. In the late December of 2020, P.1 lineage became dominant, as nearly 40% of cases in Manaus confirmed this variant (Resende et al., 2021). Surprisingly, more than 75% of residents in this area were infected with SARS-CoV-2 previously (Buss et al., 2021). These possible reinfection cases related to P.1 lineage alarm that the mutations within this variant may confer potent ability to escape the anti-SARS-CoV-2 immune memory established in the previous infection.
Besides these mutants listed above, several emerging variants with unclear risk were recently identified. The variant emerging in Southern California of the United States harbored a L452R mutation within RBM, which has been reported to be resistant to NAbs and was defined as B.1.429 lineage (Li et al., 2020; Zhang et al., 2021). Another variant, B.1.525 detected first in the United Kingdom in December 2020, now becomes dominant in Nigeria (Hodcroft et al., 2021; WHO, 2021). Mutations in the B.1.525 variant are partially similar to 501Y.V2 and B.1.1.7, respectively, which also include E484K and deletions within NTD. In addition, B.1.525 variant contains two other biologically significant mutations, Q677H and F888L. The risk of these new emerging mutants of concern still needs to be further estimated with more epidemiological and biological data.
Efficacy of SARS-CoV-2 vaccines against new variants
Till February 7, 2021, analyses on vaccine protection against novel SARS-CoV-2 variants had been reported, including the analyses of RNA vaccines from Moderna and Pfizer (BNT162b2), protein subunit vaccines from Novavax and Anhui Zhifei Longcom Biologic Pharmacy Co, Ltd, China, two adenovirus vaccines from Johnson & Johnson (JNJ) and AstraZeneca (AZN), and an inactivated virus vaccine from Beijing Institute of Biological Products (Beijing, China).
RNA vaccines
In the neutralization assays for the sera from participants vaccinated with BNT162b2, N501Y mutation within RBD, the most important region targeted by NAbs, had a slight impact on NAbs efficacy induced after vaccination. This result suggests that B.1.1.7 may not decrease the protective effect of BNT162b2, which could induce 7-fold higher NAb titers in comparison with the sera of COVID-19 convalescents (Muik et al., 2021; Tada et al., 2021). Nevertheless, another study claimed that 144–145 deletion in the spike protein of B.1.1.7 may eliminate the efficacy of a potent class of NAbs that binds to the NTD of the spike, consequently reducing NAb titers for nearly 3.85-fold in some volunteers (Shen et al., 2021; Collier et al., 2021). The same slight reduction of neutralizing activity against live B.1.1.7 variant (and also N501Y single-mutated variant) occurred in the volunteers acquiring mRNA-1273 from Moderna company, which decreased one to three-fold compared with the early stage lineage (Edara et al., 2021). Until the middle of March 2021, there was no detailed analysis of RNA vaccine efficacy reduction relating to B.1.1.7 or 501Y.V2 variants in clinical phase III trial, although in vitro studies showed that the neutralizing activity of plasma from RNA vaccinees may decrease against mutant variants, especially 501Y.V2, as the NAb titers of these plasmas were reduced from 2.74- to 7.6-fold against 501Y.V2 and 1.22-fold against P.1 strain (Liu et al., 2021a; Zhou et al., 2021). The E484K mutation may contribute mainly to this reduction since the neutralization assays against E484K single-mutated pseudotyped viruses showed a nearly 4.3-fold decrease in neutralizing activity (Jangra et al., 2021; Tada et al., 2021). A similar reduction of NAb titers in the plasma of BNT162b2 and mRNA-1273 vaccine recipients was also detected against B.1.429 variant, which contains E484K as well (Garcia-Beltran et al., 2021). A recent study found that in some convalescents who had recovered from COVID-19 for 4–8 months and received a single dose of RNA vaccine (from Pfizer or Moderna), immune memory was recalled rapidly. Although pre-vaccinated sera could barely neutralize 501Y.V2, the NAb titers were boosted for an approximate 1,000-fold increase in the post-vaccinated sera, which is still lower than that against the wild-type early stage strain (median titers, 5,156 versus 14,100) (Stamatatos et al., 2021).
Recombinant protein vaccine
Novavax released the phase III trial results of NVX-CoV2373 on January 29, 2021, which were conducted in the United Kingdom and South Africa. In the United Kingdom, NVX-CoV2373 had a protective efficacy of 85.6% against B.1.1.7 lineage, which was slightly lower in comparison with the 95.6% efficacy against the SARS-CoV-2 early stage strain. However, the results of the South Africa phase IIb trial indicated that only 60% efficacy was observed in the HIV-negative population, whereas the overall efficacy fell to 49.4% if the statistics included HIV-positive subjects (Shinde et al., 2021). In addition, another study also found 1.3- to 2.5-fold and 1.1- to 3-fold decreases of neutralizing activity among NVX-CoV2373-vaccinated individuals against N501Y-containing and three mutations-containing (K417N, E484K, and N501Y) pseudotyped viruses, respectively (Wang et al., 2021). Another recombinant vaccine, ZF2001, a dimeric RBD vaccine on phase 3 trial, could encouragingly induced robust neutralizing activity against live 501Y.V2 variant, while the titers were declined for only 1.6-fold compared with nonmutated strains (Huang et al., 2021).
Adenovirus-vectored vaccine
The first adenovirus-vectored vaccine in clinical phase III trial that reported their results of efficacy against 501Y.V2 variant was JNJ-manufactured Ad26.COV2.S vaccine on January 29, 2021 (Johnson & Johnson, 2021). After a single-dose vaccination, the overall 66% protective efficacy in preventing moderate to severe COVID-19 has been reached. The trials were performed on different continents, and the efficacy was 72%, 66%, and 57% in the United States (cases included early stage strain and B.1.1.7 variant), Latin America, and South Africa, respectively, which suggested that different efficacy may be correlated with immune evasion variants. Furthermore, it has been reported that 95% of COVID-19 cases from Ad26.COV2.S-receiving volunteers in South Africa were infected with the 501Y.V2 variant. Another phase II/III trial for the adenovirus-vectored vaccine, ChAdOx1 nCOV-19 (AZD1222) from AstraZeneca and the University of Oxford, had released a result of 66.7% overall efficacy. The in vitro results of sera neutralizing activity against live B.1.1.7 variant indicated that the vaccine-induced NAb titers were ninefold lower than that against a canonical non-B.1.1.7 lineage. Nevertheless, the efficacy that prevented symptomatic COVID-19 was slightly lower for B.1.1.7 compared with the non-B.1.1.7 lineage, which provided a potential correlation between in vitro neutralizing activity and overall efficacy (Voysey et al., 2021). However, 501Y.V2 still showed robust resistance to vaccine-induced humoral immunity, against which 50% titers of focus reduction neutralization test (FRNT) decreased for nearly 9-fold among plasma from AZD1222 vaccinees (Zhou et al., 2021).
Inactivated virus vaccine
Lately, an inactivated virus vaccine (BBIBP-CorV) was reported to have efficacy against the 501Y.V2 variant. In live virus neutralization assays, serum samples from BBIBP-CorV vaccine recipients were tested and found to neutralize HB02 (the early stage 614D strain), 614G strain, and 501Y.V2 variant with the mean titers of 110.9, 107.2, and 70.9, respectively, suggesting that the titers only decreased by 1.6-fold against 501Y.V2 variant (Huang et al., 2021).
Possible strategies for vaccine development against new variants
Given that in the United Kingdom, the primary B.1.1.7 lineage was recently identified to carry E484K mutation in a fraction of viral sequences (Wise, 2021), it is rational to infer that E484K, which arose in 501Y.V2 and P1 variants independently, may appear as a similar tendency of immune evasion under immune pressure (Eguia et al., 2020). Considering some studies reported that the NAb titers against viruses harboring this mutation have reduced and the efficacy of various vaccines had declined in epidemic areas of these variants, strategies for developing vaccines against immune evasion variants are urgently needed to prevent new waves of pandemics due to the rapid emergence of viral mutants. This requirement is similar to the vaccine development strategy of influenza viruses, which frequently undergo mutation so that the vaccines targeting the viral variants are routinely generated.
The risk of the E484K mutation warns us to design and estimate new vaccine strategies that could rapidly respond to the dynamically evolving viruses. Alongside E484K, other mutation hotspots, such as N501Y, K417N, and K417T, should be included in the immunogen design. Multivalent immunogens containing different variants may confer potent and broader protective efficacy against frequently mutated viruses (Quan et al., 2008; Ma et al., 2020).
In addition to updating the design of immunogen, the optimization of inoculation strategy would be another solution to deal with variants. In the mouse model, receiving heterologous two-dose vaccination, including RNA and adenovirus-vectored vaccines, induced higher titers of NAbs than either single-dose vaccination or two doses of adenovirus-vectored vaccine, along with superior cell immune response (Spencer et al., 2021).
Importantly, if vaccines carrying mutations are demanded in case of spreading of immune evasive variants, the additional boosting dose, so-called ‘third dose’, of updated vaccine could be inoculated to provide adequate protection, whether with a heterologous dose or homologous vaccine. Considering the plasma of convalescents exhibited poor neutralization ability against new variants, the populations who recovered from COVID-19 or asymptomatic infection should also be included in vaccine recipients, in which a more potent immune response would be induced due to the immune memory (Saadat et al., 2021). Moreover, for those who are naïve to vaccination, a multivalent vaccine could be considered to improve efficacy and cover a wider range of variants.
Summary
COVID-19 pandemic emerged at the end of 2019 had caused more than 2.6 million death and billions of economic losses. Productive vaccine programs under accelerated studies and clinical trials indeed relieve the anxiety for possible upcoming of another ‘Spanish influenza’ in the 21st century. However, the problem that whether the evolution of the virus would run faster than the vaccine development still remains to be resolved. More studies, especially on structural analysis of immunogen, antibody-epitope interaction prediction, and the underlying mechanisms for both humoral and cellular immunity against SAR-CoV-2 infection, are crucial for vaccine design and vaccination strategy, which will be an important aspect in the next period of vaccine development.
Conflict of interest
The authors have no conflicts of interest.
Acknowledgements
This work was supported by the National Special Research Program of China for Important Infectious Diseases (2018ZX10302103 and 2017ZX10202102), the Special 2019-nCoV Project of the National Key Research and Development Program of China (2020YFC0841400), the First Panel of 2021 Emergency Program of Guangzhou Laboratory (EKPG21-24), the Special 2019-nCoV Program of the Natural Science Foundation of China (NSFC) (82041002), the Special Research and Development Program of Guangzhou (202008070010), the Important Key Program of NSFC (81730060), the Joint-Innovation Program in Healthcare for Special Scientific Research Projects of Guangzhou (201803040002) to H. Zhang.
References
- Buss L.F., Prete C.A., Abrahim C.M.M., Mendrone A., Salomon T., de Almeida-Neto C., França R.F.O., Belotti M.C., Carvalho M.P.S.S., Costa A.G., et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A., Meng B., Datir R., Walls A.C., Kemp S.S.A., Bassi J., Pinto D.,, Fregni C.S., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Edara V.V., Floyd K., Lai L., Gardner M., Hudson W., Piantadosi A., Waggoner J.J., Babiker A., Ahmed R., Xie X., et al. Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant. medRxiv. 2021 doi: 10.1101/2021.02.02.21250799. [DOI] [Google Scholar]
- Eguia R., Crawford K.H.D., Stevens-Ayers T., Kelnhofer-Millevolte L., Greninger A.L., Englund J.A., Boeckh M.J., Bloom J.D. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17:e1009453. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Liu S., Shen J., Lu A.Z., Zhang Y., Li K., Liu J., Yang L., Hu C.-D., Wan J. 2021. Updated SARS-CoV-2 single nucleotide variants and mortality association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway SE P.P., MacCannell D.R. Emergence of SARS-CoV-2 B.1.1.7 lineage — United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2020;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv. 2021 doi: 10.1101/2021.02.14.21251704. [DOI] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes W.A., Kamath K., Lucas C., Shon J., Iwasaki A. Impact of B.1.1.7 variant mutations on antibody recognition of linear SARS-CoV-2 epitopes. medRxiv. 2021 doi: 10.1101/2021.01.06.20248960. [DOI] [Google Scholar]
- Hodcroft E.B., Domman D.B., Oguntuyo K., Snyder D.J., Diest M.V., Densmore K.H., Schwalm K.C., Femling J., Carroll J.L., Scott R.S., et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. MedRxiv. 2021 doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- Horby P., Huntley C., Davies N., Edmunds J., Ferguson N., Medley G., Semple C. Scientific Advisory Group for Emergencies, GOV; UK: 2021. NERVTAG: Note on B.1.1.7 Severity, 20 January 202.https://www.gov.uk/government/publications/nervtag-note-on-b117-severity-20-january-2021 [Google Scholar]
- Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Neutralization of SARS-CoV-2 VOC 501Y.V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. BioRxiv. 2021 doi: 10.1101/2021.02.01.429069. [DOI] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021 doi: 10.1101/2021.01.26.21250543. [DOI] [Google Scholar]
- Johnson, Johnson Announces single-shot Janssen COVID-19 vaccine candidate met primary endpoints in interim analysis of its Phase 3 ENSEMBLE trial. https://www.janssen.com/johnson-johnson-announces-single-shot-janssen-covid-19-vaccine-candidate-met-primary-endpoints
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Shum M.H.H., Leung G.M., Lam T.T.Y., Wu J.T. Early empirical assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 Spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Ma X., Deng J., Chen Q., Liu W., Peng Z., Qiao Y., Lin Y., He X., Zhang H. Differential efficiencies to neutralize the novel mutants B.1.1.7 and 501Y.V2 by collected sera from convalescent COVID-19 patients and RBD nanoparticle-vaccinated rhesus macaques. Cell. Mol. Immunol. 2021;18:1058–1060. doi: 10.1038/s41423-021-00641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2021 doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Sarkar R., Türeci Ö., Dormitzer P.R., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan F.S., Steinhauer D., Huang C., Ross T.M., Compans R.W., Kang S.-M. A bivalent influenza VLP vaccine confers complete inhibition of virus replication in lungs. Vaccine. 2008;26:3352–3361. doi: 10.1016/j.vaccine.2008.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. The impact of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Bezerra J.F., Vasconcelos R.H.T., Arantes I., Appolinario L., Mendonça A.C., Paixao A.C., Rodrigues A.C.D., Silva T., Rocha A.S., et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020, Virological. 2021. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584
- Saadat S., Tehrani Z.R., Logue J., Newman M., Frieman M.B., Harris A.D., Sajadi M.M. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS-CoV-2. J. Am. Med. Assoc. 2021;325:1467–1469. doi: 10.1001/jama.2021.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral Spike vaccines. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.03.002. 529–539. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., et al. Preliminary efficacy of the NVX-CoV2373 COVID-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A.J., McKay P.F., Belij-Rammerstorfer S., Ulaszewska M., Bissett C.D., Hu K., Samnuan K., Wright D., Sharpe H.R., Gilbride C., et al. Heterologous vaccination regimens with self-amplifying RNA and adenoviral COVID vaccines induce superior immune responses than single dose vaccine regimens in mice. Nat. Commun. 2021;12:2893. doi: 10.1038/s41467-021-23173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.-H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. A single mRNA immunization boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.02.05.21251182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., Yang G., Li R.-T., Fan H., Ye Q., Deng Y.-Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.11.10.377333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., Landau N.R. Neutralization of viruses with european, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv. 2021 doi: 10.1101/2021.02.05.430003. [DOI] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole Á., et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- Voysey M., Costa Clemens S.A., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . COVID-19 Weekly Epidemiological Update World Health Organization. World Health Organization; 2021. Covid-19 weekly epidemiological update, Data as received by who from national authorities, as of 21 February 2021, 10 am cet. [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wise J. Covid-19: the E484K mutation and the risks it poses. BMJ. 2021;372:n359. doi: 10.1136/bmj.n359. [DOI] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in southern California. J. Am. Med. Assoc. 2021;325 doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]