Abstract
We functionally characterized the GH10 xylanase (SoXyn10A) and the GH11 xylanase (SoXyn11B) derived from the actinomycete Streptomyces olivaceoviridis E-86. Each enzyme exhibited differences in the produced reducing power upon degradation of xylan substrates. SoXyn10A produced higher reducing power than SoXyn11B. Gel filtration of the hydrolysates generated by both enzymes revealed that the original substrate was completely decomposed. Enzyme mixtures of SoXyn10A and SoXyn11B produced the same level of reducing power as SoXyn10A alone. These observations were in good agreement with the composition of the hydrolysis products. The hydrolysis products derived from the incubation of soluble birchwood xylan with a mixture of SoXyn10A and SoXyn11B produced the same products as SoXyn10A alone with similar compositions. Furthermore, the addition of SoXyn10A following SoXyn11B-mediated digestion of xylan produced the same products as SoXyn10A alone with similar compositions. Thus, it was hypothesized that SoXyn10A could degrade xylans to a smaller size than SoXyn11B. In contrast to the soluble xylans as the substrate, the produced reducing power generated by both enzymes was not significantly different when pretreated milled bagasses were used as substrates. Quantification of the pentose content in the milled bagasse residues after the enzyme digestions revealed that SoXyn11B hydrolyzed xylans in pretreated milled bagasses much more efficiently than SoXyn10A. These data suggested that the GH10 xylanases can degrade soluble xylans smaller than the GH11 xylanases. However, the GH11 xylanases may be more efficient at catalyzing xylan degradation in natural environments (e.g. biomass) where xylans interact with celluloses and lignins.
Keywords: β-xylanase, glycoside hydrolase family 10, glycoside hydrolase family 11, Streptomyces olivaceoviridis, 4-O-methyl glucuronoxylan, arabinoxylan, sugarcane bagasse
Abbreviations
BSA, bovine serum albumin; CAZy, Carbohydrate-Active enZymes; CBM, carbohydrate binding module; GH, glycoside hydrolase; GH10, glycoside hydrolase family 10; GH11, glycoside hydrolase family 11; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection; SoXyn10A, xylanase A from Streptomyces olivaceoviridis E-86 belonging glycoside hydrolase family 10; SoXyn11B, xylanase B from Streptomyces olivaceoviridis E-86 belonging glycoside hydrolase family 11.
INTRODUCTION
Plant cell walls are the most abundant renewable biomass in nature. Polysaccharides such as celluloses and xylans comprise the major portion of plant cell walls. Xylans are referred to as hemicelluloses, which are the second most abundant natural polysaccharide next to cellulose.1) The backbone of xylan, a β-1,4-linked xylopyranose polymer, is decorated at the O-2 and/or O-3 positions with arabinofuranose, acetyl, and 4-O-methyl-D-glucuronic acid and/or D-glucuronic acid side chains.2) Several types of enzymes such as endo-1,4-β-xylanases (E.C. 3.2.1.8), β-xylosidases, (E.C. 3.2.1.37), α-L-arabinofuranosidases (E.C. 3.2.1.55), α-glucuronidases (E.C. 3.2.1.139), and acetyl xylan esterases (E.C. 3.1.1.72) are necessary to completely degrade xylans.3) Previously, we have explored the substrate specificities of these enzymes.4),5),6),7),8)
Our studies suggested that there are variations in the substrate specificity of the enzymes that are dependent on the structure of the xylan branches. These differences in the enzymes greatly affect xylan degradation. However, only limited studies have explored xylan degradation based on the substrate specificities of the enzymes for xylan branches.
Endo-β-1,4-xylanases hydrolyze the xylan backbone, and are members of the glycoside hydrolase (GH) families 10 and 11 (CAZy website available at http://www.cazy.org/).9) To date, more than 600 xylanases (352 for GH10 and 272 for GH11) have been characterized, however many questions remain unanswered regarding the xylanolytic enzyme systems that decompose biomass. For example, it has been known that many organisms possess both GH10 and GH11 enzymes10) that differ in structure and produce distinct hydrolysis products.11),12) Several extensive reviews covering characteristics of xylanases have been published, and some of them are focused on the structure-function of xylanases belonging to individual GH families.12),13),14) However it is not clear why these microorganisms possess two types of xylanases, and how differences in substrate specificities between GH10 and GH11 enzymes affect xylan degradation.
We characterized two xylanases from Streptomyces olivaceoviridis E-86 because this organism is a robust producer of xylanases. One of the xylanases produced by S. olivaceoviridis E-86 is SoXyn10A (formerly FXYN), which is one of the most well-characterized xylanases.15),16),17) The following functional properties of SoXyn10A have been studied in detail: sugar-binding structures with various kinds of xylooligosacchrides,18),19) mechanism of enzyme catalysis,20),21),22) topology of the substrate binding cleft,23),24) protein engineering25),26),27) and production of xylooligosaccharides.28),29),30),31),32) In contrast, the other xylanase produced by S. olivaceoviridis E-86 designated as SoXyn11B (formerly GXYN) has not been well-characterized.33) Therefore, in this study, we functionally characterized SoXyn11B in comparison to SoXyn10A. Differences in the xylan and pretreated biomass hydrolytic activities between SoXyn10A and SoXyn11B were detected. We also demonstrated the effects of xylan structure and xylan substitutions on the degradation by xylanases. These studies revealed novel effective applications of substituted xylans for bioeconomy.
MATERIALS AND METHODS
Substrates.
Birchwood xylan, a kind of glucuronoxylan, was obtained from Sigma Chemical Company (St. Louis, MO, USA). Insoluble materials were removed from birchwood xylan by centrifugation and the soluble fraction was freeze-dried. The obtained powder was used as soluble birchwood xylan. Arabinoxylan from wheat flour (low viscosity) was purchased from Megazyme Co. Ltd (Wiscow, Ireland). Sugarcane bagasse was kindly gifted from Mitsui Sugar Co. Ltd. (Tokyo, Japan) and was milled to a length of 2 mm with a Force Mill FM-1 (ASONE Corp., Osaka, Japan). Pretreatment of milled bagasse with sodium chlorite and acetic acid (Wise method34)) was performed three times with milled bagasses, the insoluble material was desiccated in a dry oven at 60 °C. Pretreatment of milled bagasse with sodium hydroxide was conducted with the following protocol: 2 g of milled bagasse was placed in a 100 ml screw-top bottle and the inside of the bottle was replaced with nitrogen; next, 40 mL of a 180 mM NaOH solution was added to the bottle and the mixture was heated at 85 °C for 24 h; subsequently, the insoluble material was washed with distilled water until the pH reached 7.0; lastly, the insoluble material was desiccated in a dry oven at 60 °C.
Preparation of SoXyn10A and SoXyn11B.
SoXyn10A was prepared as described previously.25) The pET expression system (Novagen, Madison, WI, USA) was employed for the expression of SoXyn11B in Escherichia coli followed by purification of SoXyn11B. Recombinant SoXyn11B was prepared with the following protocol: the gene encoding the mature SoXyn11B was amplified from S. olivaceoviridis E-86 genomic DNA by PCR using Phusion DNA polymerase (Finnzymes, Espoo, Finland) with the primers forward, 5′-CAT ATG GCC ACG GTC ATC ACC ACC AAC CAG ACC-3′ (a NdeI site is underlined) and reverse, 5′-AAG CTT GCC GCT CAC CGT GAG GTT GGA GGA GCC GCT-3′ (a HindIII site is underlined); the amplified DNA was then cloned into pET30(+) (Novagen, Darmstadt, Germany) at the NdeI and HindIII restriction enzyme sites; next, recombinant enzymes were expressed using the T7 expression system in E. coli BL21gold cells (DE3; Novagen) and purified with a C-terminal histidine tag as described in a previous report.7) The purified protein showed a single band on SDS-PAGE with the expected molecular weight of 23,000, and displayed same optimal pH and temperature as those for the native enzyme.
Xylan degradation.
The enzyme assay mixture contained 300 μL of 50 mM acetate buffer (pH 6.0), 500 μL of 1 % (w/v) soluble birchwood xylan or wheat arabinoxylan, 100 μL of 1 % (w/v) bovine serum albumin (BSA) and 100 μL of the enzyme preparation (1 μM). The reactions were carried out at 30 °C for up to 72 h, and were terminated by heating at 100 °C for 20 min. The amount of reducing sugars was determined by the Somogyi-Nelson method with D-xylose as the standard.35) The protein concentration was calculated as 1 mg/mL when the absorbance at 280 nm was 1 and then determined using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA) with BSA as the standard. The milled bagasse degradation assay mixture contained 10 mg of milled bagasse, 250 μL of distilled water, 200 μL of McIlvaine buffer (pH 7.0), and 50 μL of 0.5 mg/mL enzyme solution. The reactions were incubated at 40 °C for 72 h. Insoluble materials were collected by centrifugation, washed three times with distilled water, then dried in the dry oven. The pentose content remaining in the residue of the enzyme reaction was quantified by the orcin-hydrochloride method36) after hydrolysis with 72 % sulfuric acid as described previously.37)
Chromatography of the hydrolysis products of soluble birchwood xylan.
The hydrolysis products of soluble birchwood xylan formed in the reaction mixtures described above were subjected to high-performance anion-exchange chromatography and identified using pulsed amperometric detection (herein referred to as the HPAEC-PAD system). The samples were analyzed using a CarboPacTM PA1 column (4 × 250 mm, Dionex Corp., Sunnyvale, CA, USA) and eluted with 0.1 M NaOH (0–5 min), followed by a linear gradient (5–35 min) of sodium acetate (0–0.4 M) at a flow rate of 1 mL/min. In contrast, for the analysis of the size variation of the hydrolysis products, the mixture was applied to a Superdex 75 HR 10/30 column (GE Healthcare UK Ltd., Buckinghamshire, England), which had been equilibrated with deionized water at a flow rate of 0.5 ml/min and identified using a Shodex RI-71 detector (Showa Denko K.K., Tokyo, Japan).
RESULTS AND DISCUSSION
Effect of substitutions on the complete hydrolysis of substituted xylans.
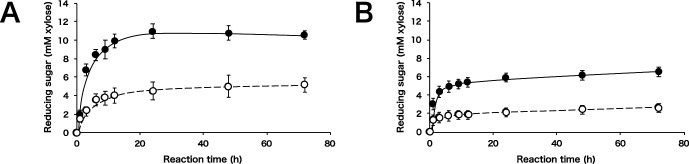
The hydrolysis time course of soluble birchwood xylan and wheat arabinoxylan by SoXyn10A and SoXyn11B are shown in Fig. 1. SoXyn10A produced more than twice as much reducing power from both substrates. In order to confirm whether this result was due to the difference in the decomposition rate of xylans, soluble birchwood xylan and its hydrolysates generated by SoXyn10A and SoXyn11B were subjected to gel filtration chromatography (Fig. 2). The peak corresponding to the original substrate disappeared completely and low molecular size products were observed in both SoXyn10A and SoXyn11B hydrolysates. Therefore, it was concluded that the difference in the produced reducing power is not mediated by the differences in the decomposition rate of xylans by both enzymes.
Fig. 1. Time course of xylan hydrolysis by SoXyn10A and SoXyn11B.
A: soluble birchwood xylan (glucuronoxylan), B: wheat arabinoxylan, closed circle; SoXyn10A, open circle; SoXyn11B. The experiments were performed in triplicate, and the value of average was plotted in the figure.
Fig. 2. Gel filtration chromatography of xylan hydrolysates generated by SoXyn10A and SoXyn11B.
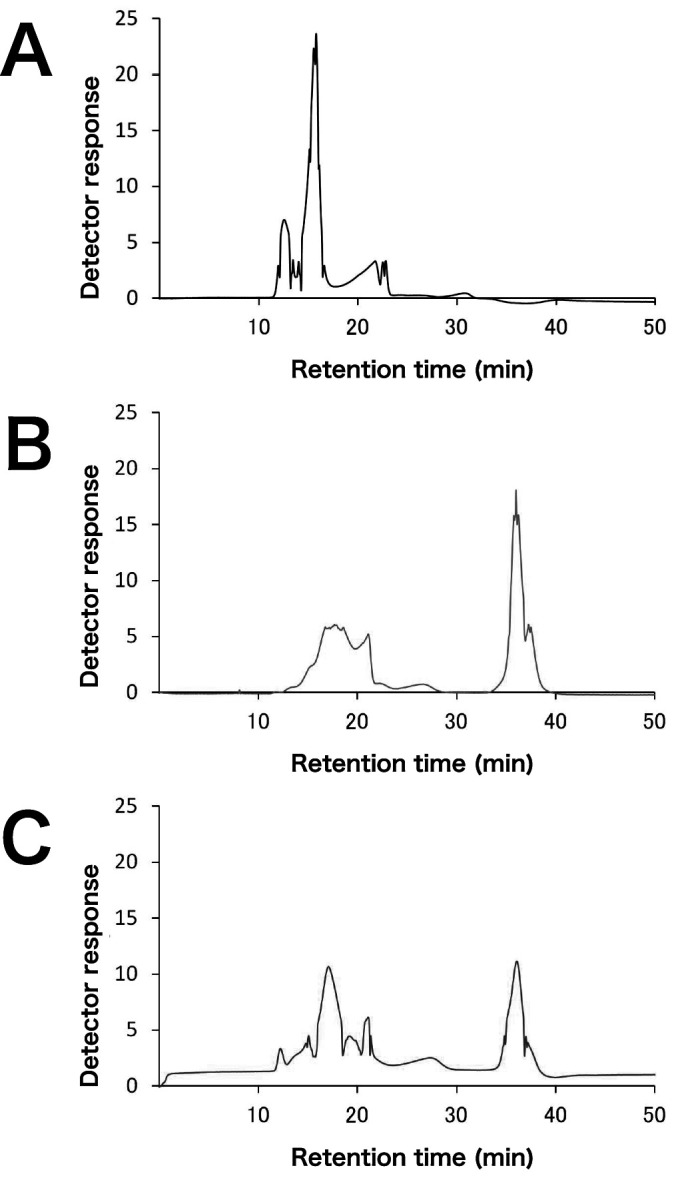
A: soluble birchwood xylan, B: hydrolysates of soluble birchwood xylan generated by SoXyn10A, C: hydrolysates of soluble birchwood xylan generated by SoXyn11B.
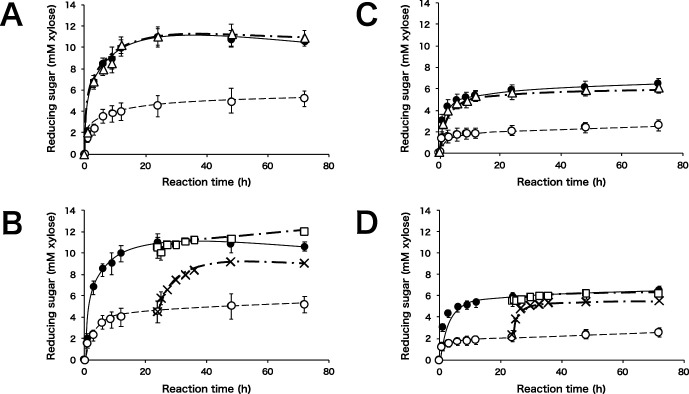
Next, we monitored the effects of a combination of SoXyn10A and SoXyn11B on soluble birchwood xylan hydrolysis. When soluble birchwood xylan was hydrolyzed by a mixture of SoXyn10A and SoXyn11B, the rate of hydrolysis was almost the same as when hydrolyzed by SoXyn10A alone (Fig. 3A). When SoXyn10A was added to the SoXyn11B hydrolysate, a significant increase in reducing power was observed. In contrast, SoXyn11B addition to the SoXyn10A hydrolysate did not result in an increase in reducing power (Fig. 3B).
Fig. 3. Time course of xylan hydrolysis with a combination of SoXyn10A and SoXyn11B.
A: soluble birchwood xylan, closed circle; SoXyn10A, open circle; SoXyn11B, open triangle; SoXyn10A+SoXyn11B, B: soluble birchwood xylan, closed circle; SoXyn10A, open circle; SoXyn11B, open square; SoXyn10A addition to SoXyn10A after 24 h, cross; SoXyn10A addition to SoXyn11B after 24 h, C: wheat arabinoxylan, closed circle; SoXyn10A, open circle; SoXyn11B, open triangle; SoXyn10A+SoXyn11B, D: wheat arabinoxylan, closed circle; SoXyn10A, open circle; SoXyn11B, open square; SoXyn10A addition to SoXyn10A after 24 h, cross; SoXyn10A addition to SoXyn11B after 24 h. In case of B and C, each 100 μL of SoXyn10A or SoXyn11B was added to the 1 mL of corresponding hydrolysate. The plotted reducing power was adjusted by account of volume of reaction mixture. The experiments were performed in triplicate, and the value of average was plotted in the figure.
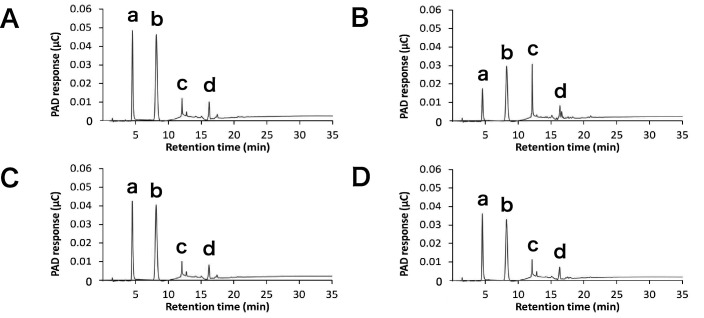
Based on the gel filtration results shown in Fig. 2, which showed that SoXyn11B completely hydrolyzed original substrate and the fact that the product of SoXyn11B was hydrolyzed by SoXyn10A, we hypothesized that SoXyn10A and SoXyn11B generated distinct products. Therefore, we analyzed the soluble birchwood xylan hydrolysis products formed by SoXyn10A and SoXyn11B by the HPAEC-PAD system (Fig. 4). SoXyn10A mainly produced xylose and xylobiose and produced minor amounts of xylotriose and substituted oligosaccharides (Fig. 4A). In contrast, SoXyn11B produced larger amounts of xylotriose than SoXyn10A and two peaks of substituted oligosaccharides were observed (Fig. 4B). The products generated by a combined mixture of SoXyn10A and SoXyn11B enzymes were similar to those obtained with SoXyn10A alone (Fig. 4C). When the soluble birchwood xylan degradation products from SoXyn11B were hydrolyzed by SoXyn10A, one of the two peaks of substituted oligosaccharides observed in the SoXyn11B HPAEC-PAD product profile disappeared. Moreover, the addition of SoXyn10A resulted in a decrease in xylotriose and an increase in both xylose and xylobiose (Fig. 4D). Since the birchwood xylan is a glucuronoxylan, the substitution is at the O-2 position of the xylan backbone harboring 4-O-methyl-D-glucuronic acid side chains. Similar results were obtained utilizing arabinoxylan as the substrate (Figs. 3C and D), which suggested that O-2 4-O-methyl-D-glucuronic acid and O-3 L-arabinofuranose substitutions in xylans similarly affected the attack of SoXyn10A and SoXyn11B on the hydrolysis of wheat arabinoxylan. Thus, the appearance that SoXyn10A produced much amounts of reducing power than SoXyn11B was not greatly influenced by the type of side chain.
Fig. 4. HPAEC-PAD analysis of degradation products.
A: degradation products of birchwood xylan generated by SoXyn10A after 24 h, B: degradation products of birchwood xylan generated by SoXyn11B after 24 h, C: degradation products of birchwood xylan generated with a mixture of SoXyn10A and SoXyn11B after 24 h, D: addition of SoXyn10A to degradation products of birchwood xylan generated by SoXyn11B after 48 h. a: xylose, b: xylobiose, c: xylotriose, and d: aldotetrauronic acid were determined from the retention time of standards.
Biely et al. analyzed the mode of action of xylanases belonging to GH10 (XlnA) and GH11 (XlnB, XlnC) families derived from Streptomyces lividans for various polysaccharide and oligosaccharide substrates.38) Their results are in good agreement with our results. In their experiments, GH10 also showed higher catalytic efficiency compared to GH11, indicating the superiority of GH10 for xylan hydrolysis. Bond-cleavage frequencies of these three xylanases suggested that GH10 releases shorter oligosaccharides than GH11. They concluded that the differences in the catalytic efficiency of the enzymes are due to the difference in the mode of xylan degradation.
From these observations, it is apparent that the substrate specificity of GH10 and GH11 is different and GH10 is more efficient at catalyzing xylan degradation. In addition, these studies suggested that only GH10 is sufficient for xylan hydrolysis. Indeed, genomewide analysis of polysaccharide-degrading enzymes of wood-decay fungi demonstrated that GH10 is conserved in all strains but GH11 is not.10) However, since many microorganisms have both GH10 and GH11 xylanases, GH11 xylanase must have a significant role in the natural environment. Therefore, the biomass degradation activities of SoXyn10A and SoXyn11B were compared.
Comparison of the hydrolytic activities of GH10 and GH11 xylanases for biomass xylans.
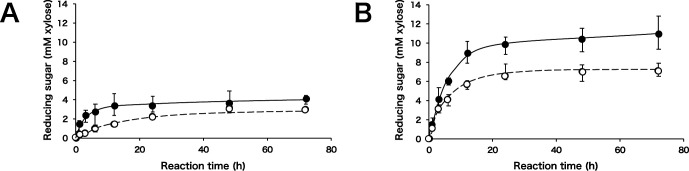
The time course of pretreated milled bagasse hydrolysis by SoXyn10A and SoXyn11B is shown in Fig. 5. The activities of both enzymes for chlorite treated milled bagasse are lower than that for NaOH treated milled bagasse. In addition, SoXyn10A produced larger amounts of reducing power from both substrates compared to SoXyn11B. Interestingly, the produced reducing powers from SoXyn10A and SoXyn11B hydrolysates of pretreated milled bagasses were not significantly different compared to the reducing powers from SoXyn10A and SoXyn11B hydrolysates from soluble xylans.
Fig. 5. Degradation of pretreated milled bagasse.
A: milled bagasse treated three times with sodium chlorite, B: milled bagasse treated with 180 mM NaOH. closed circle; SoXyn10A, open circle; SoXyn11B. The experiments were performed in triplicate, and the value of average was plotted in the figure.
Accordingly, we determined solubilized pentose from pretreated bagasses and remaining pentose in each hydrolysis residues. We found that the hydrolysis rates estimated from solubilized pentoses overestimate the recovery of sugars, because total amounts of pentoses in every hydrolysates calculated from solubilized and remaining pentoses were far more than 100 % despite the significant amount of pentoses were detected in the each hydrolysis residues. Thus, we estimated the hydrolysis rates from the remaining pentose content in each hydorolysis residues. The remaining pentose content in milled bagasse residues after 72 h of hydrolysis by SoXyn10A and SoXyn11B suggested that SoXyn11B released larger amounts of xylans from pretreated milled bagasse than SoXyn10A (Table 1). Since chlorite treatment removes lignins from biomass but maintains xylan modifications (e.g. acetylation),39) the xylans in chlorite treated milled bagasse are more similar to native xylan than NaOH treated xylans. The fact that SoXyn11B showed significantly higher activity for chlorite treated milled bagasse than SoXyn10A may suggest that the GH11 xylanases play a major role in biomass degradation. It is known that the GH10 xylanase has a TIM-barrel structure and GH11 has a jellyroll structure,11) and the molecular weights of the catalytic domains of SoXyn10A and SoXyn11B are approximately 30,000 and 20,000, respectively. The compact wide shallow structure of the substrate binding cleft of GH11 may facilitate more efficient substrate binding compared to GH10. This difference in the efficiency of substrate binding is crucial because of the complex structure of lignin, cellulose and hemicellulose networks, which are present in native plant cell walls. These observations are also supported by the genomes of Arabidopsis thaliana and Oryza sativa. These plants possess GH10 xylanases, but there are no GH11 xylanase genes found in these plant genomes (http://www.cazy.org/e1.html).40) Even though many microorganisms that actively decompose biomass possess both GH10 and GH11 xylanases, plants may not need both xylanases because they do not induce drastic changes in their plant cell walls to oppose turgor pressure and to protect against external attacks from microorganisms and insects.
Table 1.
Enzymatic hydrolysis rate of xylan in pretreated milled bagasse
| NaClO2 treated (%) | NaOH treated (%) | |
|---|---|---|
| SoXyn10A | 33.6 ± 4.63 | 54.0 ± 4.10 |
| SoXyn11B | 53.9 ± 4.39 | 63.4 ± 1.73 |
The hydrolysis rate was estimated by quantifying the amount of pentose remaining in the each enzymatic hydrolysis residue. The experiments were performed in triplicate, and the value of average was indicated.
In this study, we functionally characterized GH10 and GH11 xylanases from S. olivaceoviridis E-86. We found that the GH11 xylanase may be more efficient at catalyzing biomass degradation than the GH10 xylanase. It is important to note that there are many biomass-degrading organisms that contain different types of GH10 and GH11 xylanases, which harbor different carbohydrate-binding modules (CBMs). This suggests that different organisms have distinct mechanisms to degrade biomass. Therefore, by comparing additional types of xylanases from various origins, the mechanism of biomass degradation by each microorganism will become clear and the strategy used to efficiently degrade biomass will be elucidated.
REFERENCES
- 1).Bhatia L., Johri S., and Ahmad R.: An economic and ecological perspective of ethanol production from renewable agro waste: a review. AMB Express, 2, 65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Scheller H.V. and Ulvskov P.: Hemicelluloses. Annu. Rev. Plant Biol., 61, 263–289 (2010). [DOI] [PubMed] [Google Scholar]
- 3).Biely P.: Xylanolytic enzymes. in Handbook of Food Enzymology, J.R. Whitaker, A. Voragen, and D. Wong, eds., Marcel Dekker Inc., New York, pp. 879–916 (2003). [Google Scholar]
- 4).Matsuo N., Kaneko S., Kuno A., Kobayashi H., and Kusakabe I.: Purification, characterization and gene cloning of two α-L-arabinofuranosidases from streptomyces chartreusis GS901. Biochem. J., 346, 9–15 (2000). [PMC free article] [PubMed] [Google Scholar]
- 5).Ichinose H., Nishikubo N., Demura T., and Kaneko S.: Characterization of α-L-arabinofuranosidase related to the secondary cell walls formation in Arabidopsis thaliana. Plant Biotech., 27, 259–266 (2010). [Google Scholar]
- 6).Maehara T., Fujimoto Z., Ichinose H., Michikawa M., Harazono K., and Kaneko S.: Crystal structure and characterization of the glycoside hydrolase family 62 α-L-arabinofuranosidase from Streptomyces coelicolor. J. Biol. Chem., 289, 7962–7972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Yagi H., Maehara T., Tanaka T., Takehara R., Teramoto K., Yaoi K., and Kaneko S.: 4-O-Methyl modifications of glucuronic acids in xylans are indispensable for substrate discrimination by GH67 α-glucuronidase from Bacillus halodurans C-125. J. Appl. Glycosci., 64, 115–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Maehara T., Yagi H., Sato T., Ohnishi-Kameyama M., Fujimoto Z., Kamino K., Kitamura Y., John F. St., Yaoi K., and Kaneko S.: GH30 glucuronoxylan-specific xylanase from Streptomyces turgidiscabies C56. Appl. Environ. Microbiol., 84, e01850-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Lombard V., Ramulu H.G., Drula E., Coutinho P.M., and Henrissat B.: The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res., 42, 490–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Hori C., Gaskell J., Igarashi K., Samejima M., Hibbett D., Henrissat B., and Cullen D.: Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia, 105, 1412–1427 (2013). [DOI] [PubMed] [Google Scholar]
- 11).Chakdar H., Kumar M., Pandiyan K., Singh A., Nanjappan K., Kashyap P.L., Srivastava A.K.: Bacterial xylanases: biology to biotechnology. 3 Biotech, 6, 150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Pollet A., Delcour J.A., and Courtin C.M.: Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit. Rev. Biotechnol., 30, 176–191 (2010). [DOI] [PubMed] [Google Scholar]
- 13).Collins T., Gerday C., and Feller G.: Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev., 29, 3–23 (2005). [DOI] [PubMed] [Google Scholar]
- 14).Paës G., Berrin J.G., and Beaugrand J.: GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv., 30, 564–592 (2012). [DOI] [PubMed] [Google Scholar]
- 15).Kusakabe I., Kawaguchi M., Yasui T., and Kobayashi T.: Purification and some properties of extracellular xylanase from Streptomyces sp. E-86. Nippon Nogeikagaku Kaishi, 51, 429–437 (1977). (in Japanese) [Google Scholar]
- 16).Fujimoto Z., Kuno A., Kaneko S., Yoshida S., Kobayashi H., Kusakabe v, and Mizuno H.: Crystal structure of Streptomyces olivaceoviridis E-86 β-xylanase containing xylan-binding domain. J. Mol. Biol., 300, 575–585 (2000). [DOI] [PubMed] [Google Scholar]
- 17).Kaneko S., Ito S., Fujimoto Z., Kuno A., Ichinose H., Iwamatsu S., and Hasegawa T.: Importance of interactions of the α-helices in the catalytic domain N- and C-terminals of the family 10 xylanase from Streptomyces olivaceoviridis E-86 to the stability of the enzyme. J. Appl. Glycosci., 56, 165–171 (2009). [Google Scholar]
- 18).Fujimoto Z., Kuno A., Kaneko S., Kobayashi H., Kusakabe I., and Mizuno H.: Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module. J. Mol. Biol., 316, 65–78 (2002). [DOI] [PubMed] [Google Scholar]
- 19).Fujimoto Z., Kaneko S., Kuno A., Kobayashi H., Kusakabe I., and Mizuno H.: Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86. J. Biol. Chem., 279, 9606–9614 (2004). [DOI] [PubMed] [Google Scholar]
- 20).Suzuki R., Fujimoto Z., Ito S., Kawahara S., Kaneko S., Taira K., Hasegawa T., and Kuno A.: Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86. J. Biochem., 146, 61–70 (2009). [DOI] [PubMed] [Google Scholar]
- 21).Kuno A., Shimizu D., Kaneko S., Hasegawa T., Gama Y., Hayashi K., Kusakabe I., and Taira K.: Significant enhancement in the binding of p‐nitrophenyl‐β‐D‐xylobioside by the E128H mutant F/10 xylanase from Streptomyces olivaceoviridis E‐86. FEBS Lett., 450, 299–305 (1999). [DOI] [PubMed] [Google Scholar]
- 22).Suzuki R., Fujimoto Z., Kaneko S., Hasegawa T., and Kuno A.: Enhanced azidolysis by the formation of stable Ser–His catalytic dyad in a glycoside hydrolase family 10 xylanase mutant. J. Appl. Glycosci., 65, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Kaneko S., Ichinose H., Fujimoto Z., Kuno A., Yura K., Go M., Mizuno H., Kusakabe I., and Kobayashi H.: Structure and function of a family 10 β-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex. J. Biol. Chem., 279, 26619–26626 (2004). [DOI] [PubMed] [Google Scholar]
- 24).Kaneko S., Ichinose H., Fujimoto Z., Iwamatsu S., Kuno A., and Hasegawa T.: Substrate recognition of a family 10 xylanase from Streptomyces olivaceoviridis E-86: A study by site-directed mutagenesis to make an hindrance around the entrance toward the substrate-binding cleft. J. Appl. Glycosci., 56, 173–179 (2009). [Google Scholar]
- 25).Kaneko S., Kuno A., Fujimoto Z., Shimizu D., Machida S., Satoe Y., Yura K., Go M., Mizuno H., Taira K., Kusakabe I., and Hayashi K.: An investigation of the nature and function of module 10 in a family F/10 xylanase FXYN of Streptomyces olivaceoviridis E-86 by module shuffling with the Cex of Cellulomonas fimi and by site-directed mutagenesis. FEBS Lett., 460, 61–66 (1999). [DOI] [PubMed] [Google Scholar]
- 26).Kaneko S., Iwamatsu S., Kuno A., Fujimoto Z., Sato Y., Yura K., Go M., Mizuno H., Taira K., Hasegawa T., Kusakabe I., and Hayashi K.: Module shuffling of a family F/10 xylanase: replacement of modules M4 and M5 of the FXYN of Streptomyces olivaceoviridis E-86 with those of the Cex of Cellulomonas fimi. Protein Eng., 13, 873–879 (2000). [DOI] [PubMed] [Google Scholar]
- 27).Ichikawa T., Kuno A., Taki M., Hohsaka T., Sisido M., Kaneko S., Taira K., Kobayashi H., and Hasegawa T.: Site-directed incorporation of non-natural amino acid into Streptomyces xylanase. Nucleic Acids Symp. Ser., 48, 161–162 (2004). [DOI] [PubMed] [Google Scholar]
- 28).Kusakabe I., Ohgushi S., Yasui T., and Kobayashi T.: Structures of the arabinoxylo-oligosaccharides from the hydrolytic products of corncob arabinoxylan by a xylanase from Streptomyces. Agric. Biol. Chem., 47, 2713–2723 (1983). [Google Scholar]
- 29).Yoshida S., Kusakabe I., Matsuo N., Shimizu K., Yasui T., and Murakami K.: Structure of rice-straw arabinoglucuronoxylan and specificity of Streptomyces xylanase toward the xylan. Agric. Biol. Chem., 54, 449–457 (1990). [PubMed] [Google Scholar]
- 30).Matsuo N., Yoshida S., Kusakabe I., and Murakami K.: Chemical structure of xylan in cotton-seed cake. Agric. Biol. Chem., 55, 2905–2907 (1991). [PubMed] [Google Scholar]
- 31).Yoshida S., Satoh T., Shimokawa S., Oku T., Ito T., and Kusakabe I.: Substrate specificity of Streptomyces β-xylanase toward glucoxylan. Biosci. Biotechol. Biochem., 58, 1041–1044 (1994). [DOI] [PubMed] [Google Scholar]
- 32).Yoshida S., Ono T., Matsuo N., and Kusakabe I.: Structure of hardwood xylan and specificity of Streptomyces β-xylanase toward the xylan. Biosci. Biotechol. Biochem., 58, 2068–2070 (1994). [DOI] [PubMed] [Google Scholar]
- 33).Kaneko S., Kuno A., Muramatsu M., Iwamatsu S., Kusakabe I., and Hayashi K.: Purification and characterization of a family G/11 β-xylanase from Streptomyces olivaceoviridis E-86. Biosci. Biotechnol. Biochem., 64, 447–451 (2000). [DOI] [PubMed] [Google Scholar]
- 34).Wise L.E., Murphy M., and d'Addieco A.A.: Chlorite holocellulose, its fractionnation and bearing on summative wood analysis and on studies on the hemicelluloses. Paper Trade J., 122, 35–43 (1946). [Google Scholar]
- 35).Somogyi M.: Notes on sugar determination. J. Biol. Chem., 195, 19–23 (1952). [PubMed] [Google Scholar]
- 36).Fernell W.R. and King H.K.: The simultaneous determination of pentose and hexose in mixtures of sugars. Analyst, 78, 80–83 (1953). [Google Scholar]
- 37).Sluiter A., Hames B., Ruiz R., Scarlata C., Sluiter J., and Templeton D.: Determination of sugars, byproducts, and degradation products in liquid fraction process samples. National Renewable Energy Laboratory, USA : (2008). [Google Scholar]
- 38).Biely P., Kluepfel D., Morosoli R., and Shareck F.: Mode of action of three endo-β-1,4-xylanases of Streptomyces lividans. Biochim. Biophys. Acta., 1162, 246–254 (1993). [DOI] [PubMed] [Google Scholar]
- 39).de Carvalho D.M., Martínez-Abad A., Evtuguin D.V., Colodette J.L., Lindström M.E., Vilaplana F., and Sevastyanova O.: Isolation and characterization of acetylated glucuronoarabinoxylan from sugarcane bagasse and straw. Carbohydr. Polym., 156, 223–234 (2017). [DOI] [PubMed] [Google Scholar]
- 40).Lafond M., Guais O., Maestracci M., Bonnin E., and Giardina T.: Four GH11 xylanases from the xylanolytic fungus Talaromyces versatilis act differently on (arabino)xylans. Appl. Microbiol. Biotechnol., 98, 6339–6352 (2014). [DOI] [PubMed] [Google Scholar]