Abstract
Xyloglucan is a major hemicellulosic component in plant cell walls. Phytopathogenic fungi secrete cell wall-degrading enzymes on their infection to hosts, while the nature of the cell wall-lytic enzymes of such fungi are yet to be fully understood. Verticillium dahliae is a soil-borne fungus that causes vascular wilt diseases in a variety of commercially important crops worldwide. We purified two types of xyloglucanases, XEG12A and XEG74B, from the culture of naturally isolated Verticillium dahliae strain 2148. XEG12A showed a molecular size of 23 kDa with its maximal activity at pH 7.5. XEG12A specifically hydrolyzed xyloglucan with no activity on other β-glucans. XEG74B had a molecular size of 110 kDa with its optimum pH at 6.0. XEG74B primarily hydrolyzed xyloglucan, with a slight activity on β-1,3-1,4-glucan. Analysis of hydrolytic products of xyloglucanooligasaccharide (XXXGXXXG) by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) revealed that the both enzymes cleaved β-1,4-glucosidic linkage at the position of unbranched chain, while XEG74B showed a little fluctuation with the cleavage site. Both enzymes did not hydrolyzed xyloglucanoheptasaccharide (XXXG) at all. N-Terminal and internal amino acid sequencing of the enzymes revealed that XEG12A and XEG74B belonged to Glycoside Hydrolase (GH) Families 12 and 74, respectively. Based on these results we concluded that V. dahliae XEG12A and XEG74B were xyloglucan-specific endo-β-1,4-glucanases (EC 3.2.1.151).
Keywords: xyloglucan, xyloglucanase, Verticillium, fungi
Abbreviations
CMC, carboxymethyl cellulose; GH, glycoside hydrolase; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MS, mass spectrometry; SDS-PAGE, sodium dodecyl sulfate-denatured polyacryl amide gel electrophoresis; XXXG, xyloglucanoheptasaccharide or (Xylα1-6)Glcβ1-4(Xylα1-6)Glcβ1-4(Xylα1-6)Glcβ1-4Glc; XLXG, (Xylα1-6)Glcβ1-4(Galβ1-2Xylα1-6)Glcβ1-4(Xylα1-6)Glcβ1-4Glc; XXLG, (Xylα1-6)Glcβ1-4(Xylα1-6)Glcβ1-4(Galβ1-2Xylα1-6)Glcβ1-4Glc.
INTRODUCTION
Protection of vegetables from infectious disease is a worldwide issue in the field of agriculture. Genomic analysis of phytopathogenic fungi suggested the presence of several enzymes capable of degrading β-glucans of the plant cell wall.1) 2) Such enzymes are supposed to be involved in the infection to the plant hosts. Plant cell wall is primarily consisted from three kinds of polysaccharides, cellulose, hemicellulose, and pectinic substances. Xyloglucan is a major component of hemicellulose, that has β-1,4 glucan backborn with branches of α-1,6-Xyl. Occasional β-1-,2-Gal, Ara, or Fuc can be added to the branch of Xyl.3) Xyloglucan and xyloglucan oligosaccharides are thought to have roles in the control of differentiation and growth of plant cells.4) 5) 6) So far three types of fungal enzymes have been reported to be involved in the degradation of xyloglucan. Xyloglucan-specific endo-β-1,4-glucanase (EC 3.2.1.151) acts specifically on xyloglucan and has been classified according to Carbohydrate-Active enZymes (CAZy) Database into Glycoside Hydrolase (GH) families 12 and 74.7) 8) 9) Oligoxyloglucan reducing-end specific cellobiohydrolase (EC 3.2.1.150) acts on xyloglucan oligosaccharides in an exo-type manner and is known to belong to GH 74.10)
Oligoxyloglucan β-glycosidase (EC 3.2.1.120) also acts on xyloglucan oligosaccharides but Xylα1-6Glc is the hydrolytic product in this case.11)
In our screening of fungal strains for productivity of xyloglucan-degrading enzyme, a fungus Verticillium dahliae was chosen for its high xyloglucanolytic activity. V. dahliae is one of the major plant pathogenic fungi which causes wilt or vascular disease to many vegetables including tomato and egg plant, and to woody plants including linden and mountain maple.12) In this study we purified and characterized two types of xyloglucanase from V. dahliae.
MATERIALS AND METHODS
Reagents.
Xyloglucan was generously provided by Dainippon Sumitomo Pharma Co. (Tokyo/Osaka, Japan). Xyloglucan heptasaccharide (XXXG) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Xyloglucan tetradecasaccharide (XXXGXXXG) and isoprimeverose were purchased from Megazyme International Ireland Limited (Wicklow, Ireland). Barley β-glucan and β-1,3-1,6-glucan (Aureobasidium extracellular polysaccharide) were kindly provided by ADEKA Co. (Tokyo, Japan). Carboxymethyl cellulose was purchased from ICN Biomedicals Inc. (Tokyo, Japan). Cardran (polysaccharide of Alacaligenes facecalis) was from Kirin Food-Tech Company (Tokyo, Japan). Laminarin was purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). Konjac glucomannan were purchased from ORIHIRO Co. (Takasaki, Japan). Other reagents were purchased from Nacalai Tesque Inc. (Kyoto, Japan).
Reagents.
Xyloglucan was generously provided by Dainippon Sumitomo Pharma Co. (Tokyo/Osaka, Japan). Xyloglucan heptasaccharide (XXXG) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Xyloglucan tetradecasaccharide (XXXGXXXG) and isoprimeverose were purchased from Megazyme International Ireland Limited (Wicklow, Ireland). Barley β-glucan and β-1,3-1,6-glucan (Aureobasidium extracellular polysaccharide) were kindly provided by ADEKA Co. (Tokyo, Japan). Carboxymethyl cellulose was purchased from ICN Biomedicals Inc. (Tokyo, Japan). Cardran (polysaccharide of Alacaligenes facecalis) was from Kirin Food-Tech Company (Tokyo, Japan). Laminarin was purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). Konjac glucomannan were purchased from ORIHIRO Co. (Takasaki, Japan). Other reagents were purchased from Nacalai Tesque Inc. (Kyoto, Japan).
Fungal strain.
The fungus V. dahliae strain 2148 was obtained from the fungal culture stock library of Hirosaki University and was kept grown on PS agar (potato extract, 2 % sucrose and 1.8 % agar).
Purification of xyloglucan-specific endo-β-1,4-glucanases (XEGs).
V. dahliae strain 2148 was cultured in Czapek-Dox medium containing xyloglucan (1 % xyloglucan, 2 % NaNO3, 1 % K2HPO4, 0.5 % MgSO4・7H2O, 0.5 % KCl, 0.01 % FeSO4・7H2O, 0.01 % ZnSO4・7H2O, 0.005 % CuSO4・5H2O) at 30 °C for 16 days. Cells were removed by filtration and proteins in the culture filtrate were precipitated by 80 % saturated ammonium sulfate, then collected by centrifugation at 10,000 × G, 4 °C for 20 min. The resultant pellets were dissolved in 20 mM phosphate buffer (pH 7.0) containing 0.8 M ammonium sulfate. The insoluble substances were removed by centrifugation and the supernatants were applied to a column of TOYOPEARL Butyl-650 M (25 × 100 mm, Tosoh Co., Tokyo, Japan) that had been equilibrated with 20 mM phosphate buffer (pH 7.0) containing 0.8 M ammonium sulfate. Elution was performed with a decreasing gradient of ammonium sulfate (0.8-0 M) in 20 mM phosphate buffer (pH 7.0). The fractions containing xyloglucanase were pooled and concentrated by Macrosep Advance Centrifugal Devices (Pall Co., Washington, USA). Protein concentration was determined by the method of Lowry using a BSA standard.13) SDS-PAGE was performed according to the method described by Laemmli.14) Deglycosylation of the enzyme was performed by incubating the heat denatured enzyme (20 μg) with glycopeptidase F (1 mU, Takara Bio Inc., Kusatsu, Japan) at 37 °C for 16 h according to the manufacturers protocol. Gels were stained with Coomassie Brilliant Blue R250.
Enzyme assay.
Reaction mixture containing 40 mM sodium phosphate, pH 7.0, 0.4 % xyloglucan, and enzyme in a total 0.5 mL solution was incubated at 40 °C for a period of time. The reaction was stopped by heating for 5 min, then reducing sugars were determined by the method of Somogyi and Nelson.15) One unit of the enzyme was defined as an amount of the activity that released one micromole glucose per minute under the given conditions.
Internal amino acid sequencing.
To determine internal amino acid sequences, the purified protein was digested with V8 Protease (Takara Bio Inc.). The purified protein and peptide fragments were separated by SDS-PAGE and transferred from the gel onto a polyvinylidene difluoride membrane filter (Sequi-Blot PVDF membrane membranes, Bio-Rad Laboratories, Inc., Hercules, USA) in transfer buffer (10 mM CAPS (pH 10.0) and 10 % methanol). The amino acid sequence was determined by automatic sequential Edman degradation using a PPSQ-31A protein sequencer Model (Shimazu Co., Kyoto, Japan).
MALDI-TOF MS.
Molocular masses of the enzyme and xyloglucanoligosaccharides were analyzed by MALDI-TOF MS using Autoflex III Smartbeam mass spectrometer (Bruker Corp., Billerica, USA). Sinapinic acid (saturated solution) in 33 % CH3CN-0.066 % TFA or 2,5-dihydroxy-benzonic acid (5 mg/mL) in 30 % ethanol was used as a matrix. Bovine serum albumin and myoglobin were used as an external calibration standard.
RESULTS
Purification of two types of XEGs from V. dahliae.
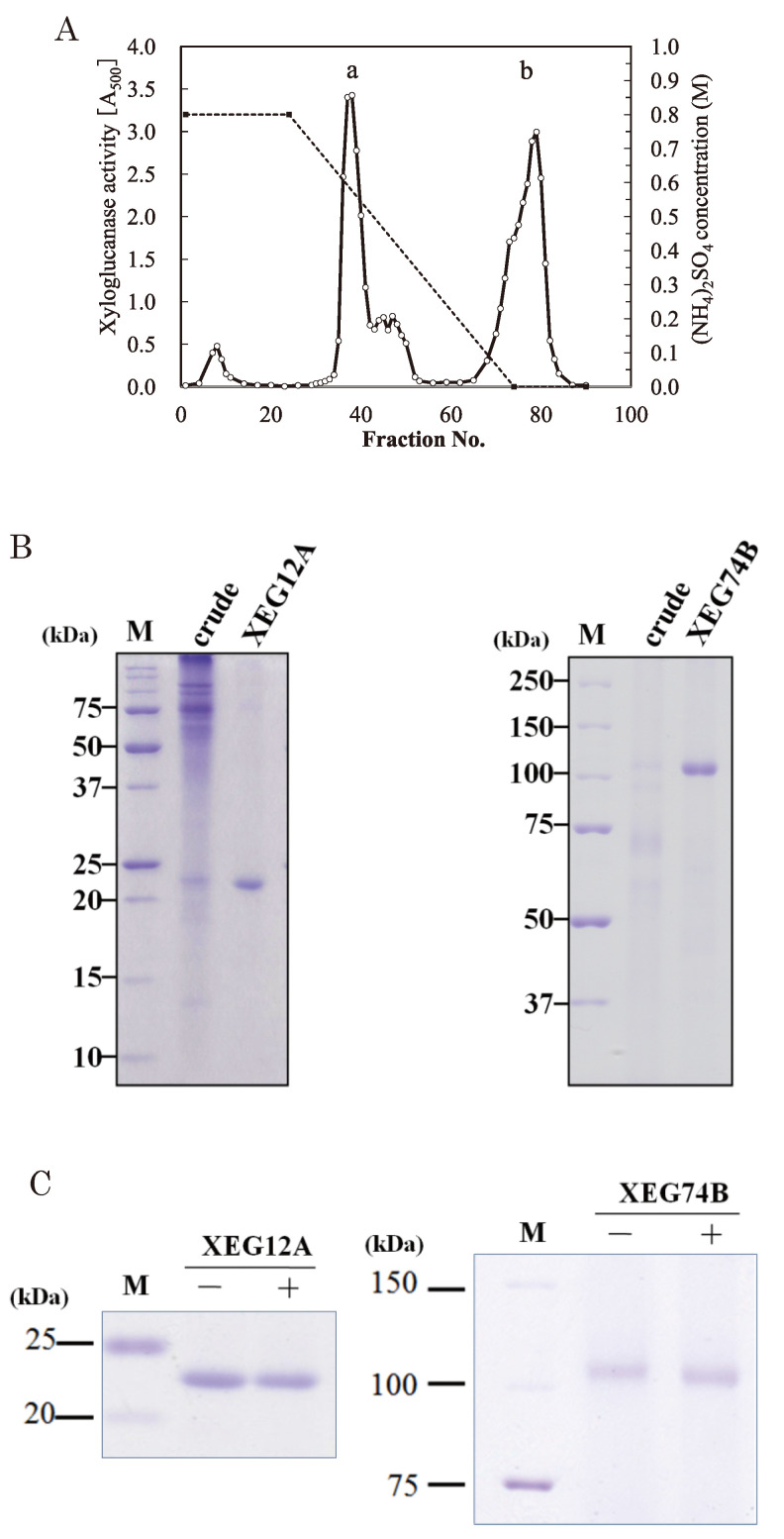
For the xyloglucanolytic activity we screened 335 strains of the plant pathogenic fungi those were isolated from the north-eastern area of Japan and were kept as the culture stock of the library. Among the fungal strains tested a group of Vericillium dahliae showed a high level of xyloglucanolytic activity with the strain 2148 as the best. Xyloglucanase was secreted by V. dahliae strain 2148 when the culture medium contained xyloglucan as the carbon source. When CMC was the sole carbon source xyloglucanase activity was not observed indicating that xyloglucan was an indispensable inducer for the production of the enzyme. The culture supernatants of the fungus also contained CMC hydrolytic activity but it was separated by a column chromatographly in high salts. Homogeneous preparation of XEG12A was obtained in a middle of the decreasing salt gradient in Butyl Toyopearl column chromatography (Table 1 and Fig. 1). XEG12A showed the molecular size of 23 kDa on SDS-PAGE (Fig. 1). Glycopeptidase F treatment of the enzyme did not cause a change of the molecular size (Fig. 1C, left), indication that N-glycosylation was negative. N-Terminal sequencing of the intact XEG12A protein was not successful provably due to an unknown blocking. Two large peptides (11 and 9 kDa) were obtained after protease digestion and 30 amino acid residues of the 9 kDa peptide was determined (Table 2). On searching with the Verticillium genomic database (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiHome.html. Access date; Oct 2014), the enzyme was identified as V. dahliae putative endoglucanase 1 (locus VDAG04017, accession EGY22579). Predicted protein had 246 amino acid residues with a calculated molecular mass 25,662 Da, and the enzyme belonged to GH family 12 (Fig. 2). N-Glycosylation motif was not found in its amino acid sequence. On MALDI-TOF MS analysis of XEG12A, the protein showed a signal of m/z 21,148 (data not shown). Thus XEG12A was supposed to have a secretory signal peptide of 25 amino acid residues at N-terminal of the mature protein of 221 amino acid residues.
Purification of xyloglucanases from V. dahiae.
The reactions were carried out in 40 mM phosphate buffer, pH 7.0, at 40 °C with 0.4 % xyloglucan as the substrate.
| Purification step (mg) | Protein | Total activity (U) | Specific activity (U/mg) | Yield (%) | Purification (fold) |
| Crude | 595.9 | 220.6 | 0.37 | 100 | 1.0 |
| Ammonium sulfate precipitation | 72.7 | 67.9 | 0.93 | 30.8 | 2.5 |
| Butyl Toyopearl 650M | |||||
| Xgl12A | 7.1 | 33.2 | 4.70 | 15.0 | 12.7 |
| Xgl74B | 4.9 | 16.9 | 3.42 | 7.7 | 9.2 |
The reactions were carried out in 40 mM phosphate buffer, pH 7.0, at 40°C with 0.4% xyloglucan as the substrate.
Fig. 1.
Purification of xyloglucanases from V. dahliae.
(A) Elution profile of Butyl Toyopearl column chromatography. Thick bars a and b corresponded to the activities of XEG12A and XEG74B, respectively. Every 10 mL of the effluents were collected per a tube. (B) SDS-PAGE of purified XEG12A and XEG74B (right lane of each gel). Total 20 μg of the purified enzymes were applied. M; molecular markers. (C) Deglycosylation of XEG12A and XEG74B. The symbols minus and plus are without and with glycopeptidase F treatment, respectively. Gels were stained with Coomassie Blue.
Table 2.
Amino acid sequences of V. dahliae xyloglucanases.
| 9K peptide (12A) | IMIWLGSLGGAGPISATGSTIANPTIAGTTXRLYQ |
| N-terminus (74B) | AFEWQNARMGG |
| 20K peptide (74B) | AFEWQNARMGGGGGFVPGISFHPXVXGVAY |
| 18K peptide (74B) | TSAVSGGATSRIFVGTAXXVTXS |
| 10K peptide (74B) | KALYLSYSXGTGPYXGTSXSV |
| 12K peptide (74B) | FYYSISTPXAP |
| 26K peptide (74B) | VKSAPGGTELFSAV |
| 25K peptide (74B) | LFSAVGXXSGFT |
The letters 12A or 74B meant that the sequence was from XEG12A or XEG74B, respectively. X; unidentified residue.
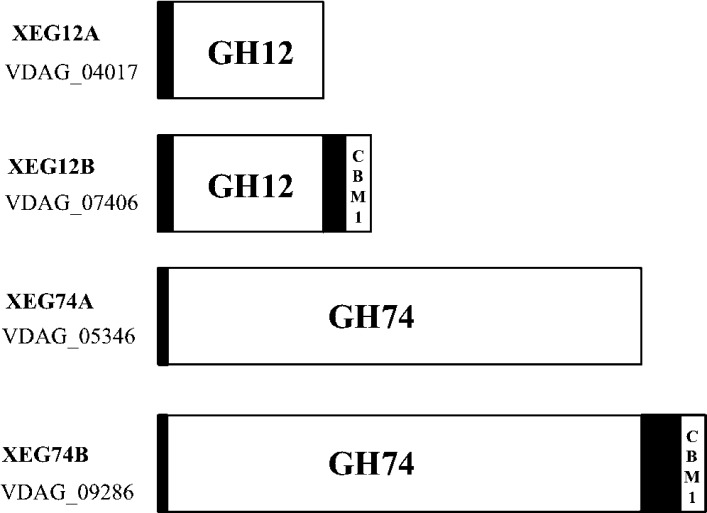
Fig. 2.
Domain structures of cryptic xyloglucanases annotated in V. dahliae genome.
VDAG number means the gene locus in V. dahliae genome. XEG12A and XEG74B are identified in this work. N-Terminal black boxes are cryptic signal sequences.
XEG74B was also purified by the same column chromatography, in this case the enzyme was eluted in the latter part of the salt gradient (Table 1 and Fig. 1). XEG74B showed a molecular size of 110 kDa on SDS-PAGE (Fig. 1). Deglycosylation by glycopeptidase F caused a slight decrease of the molecular size (Fig. 1C, right), suggesting that the enzyme had a little N-glycosylation. N-Terminal and internal amino acid sequences of XEG74B were obtained with and V8-proteolytic peptides, respectively (Table 2). On searching with the Verticillium genomics database the enzyme was identified as V. dahliae putative Cel74A (locus VDAG 09286, accession EGY18952). Predicted protein had 816 amino acid residues with a calculated molecular mass 85, 362 Da, and the enzyme belonged to GH family 74 (Fig. 2). Two of N-glycosylation motifs were found in the amino acid sequence. Amino acid sequencing of the Peptide 1 indicated that the N-terminal 16 residues were deleted from the encoded protein.
Optimal conditions for XEG12A and XEG74B.
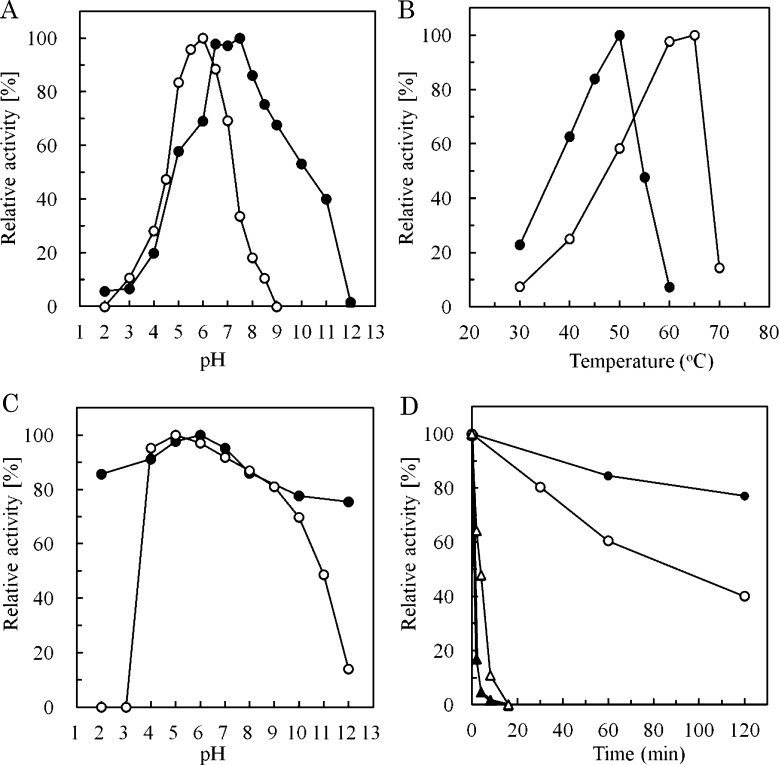
Highest activity of XEG12A was obtained at pH 8.0 with over 85 % of the activity observed between pH 6.5 and 8.5 (Fig. 3A). It should be noted that XEG12A showed 50 % of the activity at pH 10.0, for the other fungal XEGs did not show activity in alkaline condition over pH 9.0.16) This nature was also observed with the alkaline stability of XEG12A (Fig. 3C). XEG74B showed maximal activity at pH 6.0 and did not show alkaline tolerance over pH 9.0 (Fig. 3A). As to thermal performance XEG12A showed maximal activity at 50 °C while XEG74B preferred a higher temperature than XEG12A between 60 to 65 °C (Fig. 3B). XEG12A showed more stability than XEG74B after pre-heating at 50 °C for 120 min (Fig. 3D) but the both were almost inactive after pre-heating at 60 °C (Fig. 3D, triangles).
Fig. 3. Optimal conditions for XEG12A and XEG74B.
The optimum pH was measured by incubating the reaction mixture at various pH for 20 min at 40 °C (A). For the determination of pH stability the enzymes were pre-warmed in various pH of buffers at 30 °C for 120 min, then remaining activities were measured with xyloglucan as the substrate at pH 7.0 (B). Universal buffer solutions (pH 2.0‒12.0) those were prepared from 0.4 M boric acid / 0.1 M citric acid and 0.2 M trisodium phosphate were used. The optimum temperature was determined by incubating the reaction mixture at various temperature in 40 mM sodium phosphate buffer (pH 7.0) (C). To determine thermal stability the enzymes were pre-heated at 50 °C for various period, then remaining activities were measured with the substrate at 40 °C for 20 min (D, circles). Residual activities after the pre-heating at 60 °C are also shown (triangles). Closed symbols are for XEG12A and open symbols for XEG74B, respectively.
Substrate specificity.
Hydrolytic activity of XEG12A was highly restricted to xyloglucan (Table 3). None of other β-glycans including CMC (carboxymethylated β-1,4-glucan), konjac glucomannan (hybrid of β-1,4-Glc and β-1,4-Man), barley β-glucan (β-1,3- and β-1,4-glucan), yeast β-glucan (β-1,3- and β-1,6-glucan, laminarin (β-1,3-glucan), and curdlan (β-1,3-glucan) were the substrate. On the other hand XEG74B showed a partial activity toward barley β-glucan besides the major preference to xyloglucan (Table 3).
Table 3.
Substrate specificity of V. dahliae xyloglucanases.
| Substrate | (linkage) | Relative activity (%) | |
| XEG12A | XEG74B | ||
| Xyloglucan | (β1,4) | 100 | 100 |
| CM celulose | (β1,4) | N.D. | N.D. |
| Glucomannan | (β1,4) | N.D. | N.D. |
| Barley glucan | (β1,3-1,4) | N.D. | 10 |
| Yeast glucan | (β1,3) | N.D. | N.D. |
| Laminarin | (β1,3) | N.D. | N.D. |
The reactions were carried out at 40 °C for 20 min with 0.4 % of each glycans as the substrate. N.D., not detected.
Analysis of xyloglucanolytic products.
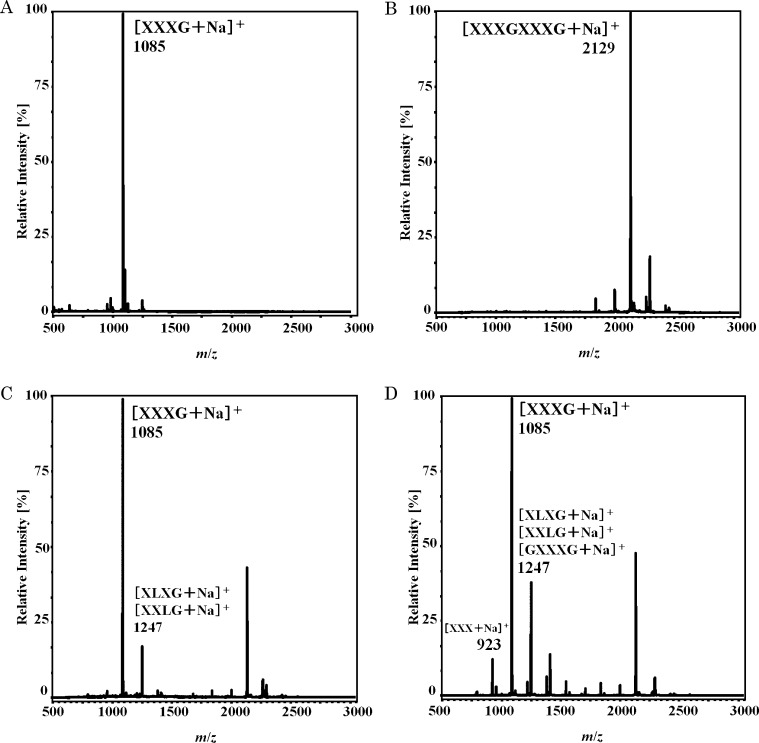
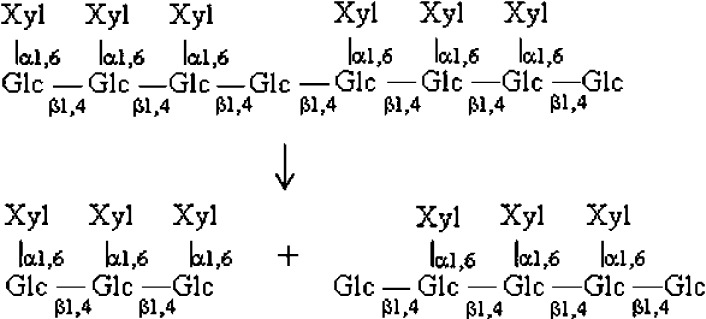
When XXXGXXXG (XXXGXXXG + Na+, m/z 2147, Fig. 4B) was digested with XEG12A, a signal of XXXG + Na+ (m/z 1085) was exclusively observed on MALDI-TOF MS (Fig. 4C). When XXXG was the substrate not a signal of oligosaccharide smaller than XXXG was detected (data not shown). This meant that the minimal product of the enzyme should be XXXG, and that XEG12A specifically cleaved the linkage between the heptasaccharide units.
Fig. 4.
MS analyses of reaction products of xyloglucan oligosaccharides by XEG12A and XEG74B.
XXXGXXXG were treated with XEG12A or XEG74 at 40 °C for 16 h in 40 mM sodium phosphate buffer (pH 7.0), then products were analyzed by MALDI-TOF MS. Panels A, B, C, and D are XXXG and XXXGXXXG as the standards, and the reaction products of XEG12A and of XEG74B, respectively.
DISCUSSION
In this study we characterized two xyloglucanases, XEG12A and XEG74B from phytopathogenic fungi, Verticillium dahliae. As to the reaction condition XEG12A preferred more alkaline condition than XEG74B, while the latter had more heat tolerance than XEG12A. The result of SDS-PAGE indicated the molecular size of XEG74B as 110 kDa, while calculated mass was 85 kDa. When N-glycosylation of XEG74B was examined by glycopeptidase F digestion, N-glycan was shown not to exceed 5 kDa (Fig. 1C). As the protein of XEG74B had threonin-rich region at the C-terminal part, we supposed that unknown O-glycosylation might have occurred.
On the degradation of β-glucans the both enzymes cleaved xyloglucan as their primary substrate. XEG12A did not act on other glucans and showed the strict specificity towards xyloglucan. A similar enzyme has been reported from Aspergillus niger.16) On the other hand XEG74B partially cleaved barley β-glucan, that suggested a slight looseness in its substrate specificity. Fungal xyloglucanases of GH74 so far have shown rather strict specificity toward xyloglucan9) 17) except that Cel74A of Trichoderma hydrolyzed β-glucans other than xyloglucan.18) When the degradation products of xyloglucan oligosaccharides were analyzed on MALDI-TOF MS, it was shown that the both Verticillium XEGs did not act on XXXG. This meant that the minimal substrate for the both enzymes should be over heptasaccharide. When XXXGXXXG was the substrate XEG12A made XXXG as a sole product. A cleavable site in XXXGXXXG was fixed for the enzyme, suggesting its stiff manner of action on xyloglucan. In the hydrolysis of the same substrate by XEG74B the major product was XXXG but hexasaccharide and octasaccharide were also made. In this case the cleavage should be explained as Fig. 5. The results indicated that XEG74B also cleaved β-1,4 glucosidic bond in Xylα1-6Glcβ1-4Glc other than the linkage between the heptasaccharide units. As to other fungal enzyme Trichoderma Cel74A was reported to make XX, XGXX, and XG on the hydrolysis of XXXGXXXG.18)
Fig. 5.
Presumed minor cleavage of a xyloglucano tetradecasaccharide by XEG74B.
Plant pathogenic fungi secrete plant cell wall degrading enzymes on invasion of host cells, while some plant hosts are known to produce proteins inhibitory to the enzymes.19) Xyloglucan-specific endo-β-1,4-glucanase inhibitor proteins (XEGIPs) is supposed to binds specifically to GH 12 xyloglucan-specific endo-β-1,4-glucanase (EC 3.2.1.151).20) However a direct interaction between XEGIPs and xyloglucanase of pathogenic fungi has yet to be verified. Whether Verticillium XEG12A binds such inhibitory phytoprotein should be a point of concern.
Besides the gene for XEG12A we found another gene that encoded a homologous polypeptide on the locus VDAG 07406 of V. dahliae genome. Predicted amino acid sequence of the gene did not match completely to the sequence we obtained, thus we called it as XEG12B. XEG12B was estimated to be a protein of 307 amino acid residues with a carbohydrate binding module (CBM I) at the C-terminus (Fig. 2). We also found a gene (VDAG 05346) encoding a protein homologous to XEG74B (Fig. 2). The protein of 718 amino acid residues was supposed to be a putative Cel74A without CBMI. Enzymatic nature is unknown but we termed it XEG74A. As to V. dahliae we detected a weak xyloglucanolytic activity other than XEG12A and XEG74B in the course of the Butyl Toyopearl column chromatography (Fig. 1A). The effluents containing the activity were not homogeneous but we did not observe proteins of the size close to XEG12 (data not shown). Presence of XEG74A in the culture broth was possibly expected but details will be revealed in our future work.
ACKNOWLEDGMENTS
We thank Dr. Kazuaki Tanaka of the Laboratory of Phytopathology, Hirosaki University, for generously providing the fungal stock culture library.
REFERRENCES
- 1).Soanes D.M. and Talbot N.J.: Comparative genomic analysis of phytopathogenic fungi using expressed sequence tag (EST) collections. Mol. Plant. Pathol., 7, 61-70 (2006). [DOI] [PubMed] [Google Scholar]
- 2).Neumann M.J. and Dobinson K.F.: Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae. Fungal Genet. Biol., 38, 54-62 (2003). [DOI] [PubMed] [Google Scholar]
- 3).McNeil M., Darvill A.G., Fry S.C., and Albersheim P.: Structure and function of the primary cell walls of plants. Annu. Rev. Biochem., 53, 625-663 (1984). [DOI] [PubMed] [Google Scholar]
- 4).McDougall G.J. and Fry S.C.: Inhibition of auxin-stimulated growth of pea stem segments by a specific nonasaccharide of xyloglucan. Planta, 175, 412-416 (1998). [DOI] [PubMed] [Google Scholar]
- 5).McDougall G.J. and Fry S.C.: Xyloglucan oligosaccharides promote growth and activate cellulase : evidence for a role of cellulase in cell expansion. Plant Physiol., 93, 1042-1048 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Takeda T., Furuta, Y., Awano T., Mizuno K., Mitsuishi Y., and Hayashi T.: Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc. Natl. Acad. Sci. USA, 99, 9055-9060 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Lombard V., Ramulu H.G., Drula E., Coutinho, P.M., and Henrissat B.: The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic. Acids. Res., 42, D490-D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Pauly, M., Andersen, L.N., and Kauppinen, S., Kofod L.V., York W.S., Albersheim P., and Darvill A.: A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology, 9, 93-100 (1999). [DOI] [PubMed] [Google Scholar]
- 9).Yaoi K. and Mitsuishi Y.: Purification, characterization, cDNA cloning, and expression of a xyloglucan endoglucanase from Geotrichum sp. M128. FEBS Lett., 560, 45-50 (2004). [DOI] [PubMed] [Google Scholar]
- 10).Yaoi K. and Mitsuishi Y.: Purification, characterization, cloning, and expression of a novel xyloglucan-specific glycosidase, oligoxyloglucan reducing end-specific cellobiohydrolase. J. Biol. Chem., 277, 48276-48281 (2002). [DOI] [PubMed] [Google Scholar]
- 11).Kato Y., Matsushita J., Kubodera T., and Matsuda K.: A novel enzyme producing isoprimeverose from oligoxyloglucans of Aspergillus oryzae. J. Biochem., 97, 801-810 (1985). [DOI] [PubMed] [Google Scholar]
- 12).Harada Y., Furueda, T., and Murata K.: Verticillium Wilt of Tilia japonica and Acer palmatum, the First Report on the Occurrence of Verticillium dahliae on Trees in Japan. Ann. Phytopathol. Soc. Jpn., 63, 345-350 (1997). (in Japanese) [Google Scholar]
- 13).Lowry H., Rosebrough N.J., Farr A.L., and Randall R.J.: Protein measurement with the Folin phenol reagent. J. Biol. Chem., 193, 265-275 (1951). [PubMed] [Google Scholar]
- 14).Laemmli U.K.: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685 (1970). [DOI] [PubMed] [Google Scholar]
- 15).Somogyi M.: Notes on sugar determination. J. Biol. Chem., 195, 19-23 (1952). [PubMed] [Google Scholar]
- 16).Master E. R., Zheng Y., Storms R., Tsang A., and Powlowski J.: A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem. J., 411, 161-170 (2008). [DOI] [PubMed] [Google Scholar]
- 17).Ishida T., Yaoi K., Hiyoshi A., Igarashi, K., and Samejima M.: Substrate recognition by glycoside hydrolase family 74 xyloglucanase from the basidiomycete Phanerochaete chrysosporium. FEBS J., 274, 5727-5736 (2007). [DOI] [PubMed] [Google Scholar]
- 18).Desmet T., Cantaert T., Gualfetti P., Nerinckx W., Gross L., Mitchinson C., and Piens K.: An investigation of the substrate specificity of the xyloglucanase Cel74A from Hypocrea jecorina. FEBS J., 274, 356-363 (2007). [DOI] [PubMed] [Google Scholar]
- 19).Yoshizawa T., Shimizu T., Yamabe M., Taichi, M., Nishiuchi Y., Shichijo N., Unzai S., Hirano H., Sato M., and Hashimoto H.: Crystal structure of basic 7S globulin, a xyloglucan-specific endo-β-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-β-glucanase. FEBS J., 278, 1944-1954 (2011). [DOI] [PubMed] [Google Scholar]
- 20).Xie W., Hao L., and Goodwin P.H.: Role of a xyloglucan-specific endo-β-1,4-glucanase inhibitor in the interactions of Nicotiana benthamiana with Colletotrichum destructivum, C. orbiculare or Pseudomonas syringae pv. tabaci. Mol. Plant Pathol., 9, 191-202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]