Abstract
Late infantile Batten disease (CLN2 disease) is a rare, autosomal recessive, neurodegenerative lysosomal storage disease caused by mutations in the CLN2 gene encoding tripeptidyl peptidase 1 (TPP1). We tested intraparenchymal delivery of AAVrh.10hCLN2, a nonhuman serotype rh.10 adeno-associated virus vector encoding human CLN2, in a non-randomized trial consisting of 2 arms assessed over 18 months: an AAVrh.10hCLN2-treated cohort of 8 children with mild to moderate disease, and an untreated, Weill Cornell natural history cohort consisting of 12 children. The treated cohort was also compared to an untreated European natural history cohort of CLN2 disease. The vector was administered through 6 burr holes directly to 12 sites in the brain without immunosuppression. In an additional safety assessment under a separate protocol, 5 children with severe CLN2 disease were treated with AAVrh.10hCLN2. The therapy was associated with a variety of expected adverse events, none causing long-term disability. Induction of systemic anti-AAVrh.10 immunity was mild. Post-therapy, the treated cohort had a 1.3- to 2.6-fold increase in cerebral spinal fluid TPP1. There was a slower loss of grey matter volume in 4 of 7 children by MRI; and a 42.4% and a 47.5% reduction in the rate of decline of motor and language function, compared to the Weill Cornell natural history cohort (p<0.04), and European natural history cohort (p<0.0001), respectively. Intraparenchymal brain administration of AAVrh.10hCLN2 slowed the progression of disease in children with CLN2 disease. However, improvements in vector design and delivery strategies will be necessary to halt disease progression using gene therapy.
One sentence summary:
Administration of an adeno-associated virus coding for the CLN2 gene into the brain parenchyma slows the progression of CLN2 disease in children.
Editor’s summary:
Gene therapy for CLN2 disease
The current treatment for CLN2 disease, caused by mutations in the CLN2 gene, is infusion of human recombinant tripeptidyl peptidase 1 (TPP1) into the cerebrospinal fluid every other week, which slows but does not halt progression of the disease. Sondhi and colleagues sought an alternative treatment through gene therapy. They injected an adeno-associated virus vector expressing the normal human CLN2 coding sequence directly into the brain parenchyma of children with the disease. Progression of the disease was slowed in treated children, but not to the same degree as recombinant TPP1. Further improvements in gene therapy are needed before progression of CLN2 disease can be halted.
Introduction
CLN2 disease (also referred to as late infantile neuronal ceroid lipofuscinosis (LINCL), late infantile Batten disease, Janksy-Bielschowsky disease, and tripeptidyl peptidase 1 deficiency), is a uniformly fatal childhood autosomal recessive neurodegenerative lysosomal storage disorder caused by mutations in the CLN2 gene (1–6). The disease affects the central nervous system (CNS) and retina, with typical onset between ages 2 to 4 years old. The clinical course is characterized by progressive neurologic decline with cognitive impairment, visual failure, seizures, deterioration of motor and language skills, and death by ages 10 to 12 (2, 5, 7, 8). The disease is caused by mutations in the CLN2 gene, which encodes lysosomal tripeptidyl peptidase 1 (TPP1), an enzyme that cleaves tripeptides from the N-terminus of polypeptides imported into the lysosome (1, 9). The loss of TPP1 activity leads to accumulation of storage material in lysosomes, characterized as autofluorescent intracellular deposits by light microscopy (2, 10). There is allelic heterogeneity, but two CLN2 variants, G3556C (c.509–1G>C; intron 7 splice defect) and C3670T (c.622C>T; nonsense Arg208 to stop), are responsible for the majority of cases in Caucasian populations (http://www.ucl.ac.uk/ncl/CLN2mutationtable.htm) (1, 10, 11).
CLN2 disease has several features making it a good target for gene therapy using an adeno-associated virus (AAV) vector expressing the normal human CLN2 coding sequence (12–19). AAV vectors are efficient in transferring genes to the CNS, mediating persistent expression (20–25). Genotype/phenotype comparisons suggest that the severe phenotype should be ameliorated with an increase of CNS TPP1 amount to 5 to 10% of normal (12, 26). TPP1 is a secreted protein capable of cross-correcting neighboring cells via uptake by the mannose-6-phosphate receptor (27–29). Therefore, it is not necessary to transfer the normal CLN2 cDNA to all of the cells in the CNS, since the corrected cells will secrete TPP1 protein which will be taken up to correct neighboring cells. The concept that delivery of TPP1 to the CNS can be effective in treating CLN2 CNS disease is supported by the success of cerliponase alfa, a recombinant human TPP1 protein therapy administered biweekly to cerebral spinal fluid via a CNS reservoir, in slowing the progression of the CNS disease (30–32). If AAV-mediated CNS gene therapy with the CLN2 coding sequence could provide sufficient amounts of TPP1 throughout the CNS, it could provide a one-time therapy to treat the disease.
Based on efficacy studies in CLN2−/− mice and CNS biodistribution and safety studies in nonhuman primates (33–35), we chose the AAV serotype rh.10 expressing the normal human CLN2 coding sequence (AAVrh.10hCLN2) to treat children with CLN2 disease. The hypothesis of the study was that direct CNS administration of AAVrh.10hCLN2 was safe and would slow down the progression of the neurologic disease.
Results
We tested intraparenchymal delivery of AAVrh.10hCLN2, a nonhuman serotype rh.10 adeno-associated virus vector coding for human CLN2 (fig. S1). The vector, AAVrh.10hCLN2 (total dose of 2.85–9.0 ×1011 genome copies (gc)) was delivered directly via catheter into the CNS via 6 burr holes (3 bilaterally), with equal doses to 2 sites/burr hole (2.4–7.5×1010 gc in 150 μl per sire(fig. S2). The study was designed as a non-randomized trial comparing a treatment group that received AAVrh.10hCLN2 (cohort 1; n=8) and a non-treated natural history control cohort (“Weill Cornell natural history control cohort,” cohort 2; n=12). The study population was limited to children with CLN2 disease with specific genotypes and severity criteria, limiting the inclusion to those with mild to moderate disease as assessed by a clinical neurologic rating scale (table S1) (4, 36).
As the trial progressed, we participated in a collaborative publication of the natural history of untreated CLN2 disease, comparing disease progression of the Weill Cornell natural history control cohort to that of the “European DEM Child Natural History” cohort (5). Since the DEM child cohort is the untreated cohort from which natural history controls were used for the regulatory approval of cerliponase alfa therapy, although not part of our original design, we have used the European DEM Child Natural History cohort data as a replication control cohort (n=41, “DEM Child Natural History Replication Cohort,” cohort 3).
Finally, there were 5 children who had severe disease and did not meet the “mild to moderate” entry criteria (Weill Cornell LINCL scale <6) (4) and/or did not fit the genotype entry criteria. Under a separate protocol, these 5 children were treated and assessed for additional safety data (“therapy/safety only cohort,” cohort 4).
Description of the primary treatment and control cohorts
Cohort 1 (the therapy cohort, n=8, V1 to V8) included 4 females and 4 males (Table 1). Of the 8 subjects, 3 subjects were homozygous for either g.C3670T or g.G3556C and 2 were compound heterozygous for both genotypes. The remaining 3 were heterozygous for either g.C3670T or g.G3556C and a different mutation. The average age of first reported symptoms was 29 months (range 16–48 months). In 7 of 8 subjects in this group, the first reported symptom was speech delay, accompanied in some subjects by motor, balance, cognition and behavioral abnormalities. Six of the eight had the age of first seizure between 30 and 50 months of age, and 2 between 18 and 25 months. As described above, subject V8 was not included in the analysis.
Table 1 –
Study Cohorts
| Group | Number of subjects | Sex | Age at report of first symptom (months) | Age of first reported seizure (months) | Most common first symptom |
|---|---|---|---|---|---|
| Cohort 1 - Treatment1 | 8, 7 with follow-up | 4M/4F | 16 – 48 | 18 – 50 | Speech delay |
| Cohort 2 - No treatment control2 | 12 | 4M/8F | 24 – 36 | 30 – 54 | Speech delay |
| Cohort 3 - DEM child replication control3 | 41 | 24M/17F | 12 – 53 | 0 – 106 | Seizures |
| Cohort 4 - Treatment/safety only4 | 5 | 1M/4F | 24 – 42 | 30 – 36 | Speech delay |
Mild to moderate disease, of the n=8, 7 had follow-up during the 18-month study period.
The pre-therapy data from subject S5 (cohort 4) over the 5 months prior to therapy was used as part of the Weill Cornell natural history data (referred to as subject C3 in cohort 2). Prior to receiving treatment subject C3 had 2 assessments that were 5 months apart.
Cohort 3 – the untreated European DEM child cohort was used as a replication control group (5).
Cohort 4 – treated children with severe disease, used for additional safety data.
M: male; F: female; DEM child: A consortium that studies natural history of neuronal ceroid lipofuscinoses disorders.
Cohort 2 (the Weill Cornell natural history cohort, n=12, C1 to C12) included 8 females and 4 males (Table 1). 92% (11/12) were homozygous or heterozygous for either g.C3670T or g.G3556C, with 5 (42%) heterozygous for g.C3670T and g.G3556C and 2 (17%) homozygous for g.C3670T. The remaining 4 were heterozygous for either g.C3670T or g.G3556C and a different mutation (see Table 2 for genotypes). 55% manifested symptoms at about 24 months and 45% had symptoms by 36 months. In 10 of 12, the first reported symptom was speech delay, accompanied in some by balance, motor, cognition and behavioral issues. Almost all had the age of first seizure between 30 and 50 months of age, with the latest at 54 months.
Table 2.
Assessment of Cohort 1 with the Motor + Language Parameters1
| Subject2/genotype | Study visit3 | Age at assessment (months) | Time before or after vector administration (months) | Motor score4 | Language score5 | Total score6 |
|---|---|---|---|---|---|---|
| V17 | 1 | 83.0 | − 4.6 | 1.0 | 1.7 | 2.7 |
| G3556C/G3556C | 2 | 87.4 | − 0.2 | 1.3 | 2.0 | 3.3 |
| Vector | 87.6 | 0 | - | - | - | |
| 88.8 | + 1.2 | 1.0 | 1.0 | 2.0 | ||
| 4 | 93.9 | + 6.3 | 1.0 | 1.3 | 2.3 | |
| 5 | 99.9 | + 12.3 | 1.0 | 1.0 | 2.0 | |
| 6 | 106.6 | + 19.0 | 0.0 | 1.0 | 1.0 | |
| V28 | 1 | 39.6 | − 2.5 | 2.7 | 2.0 | 4.7 |
| C3670T/G3556C | 2 | 41.0 | -1.1 | 3.0 | 2.0 | 5.0 |
| 3 | 42.0 | − 0.1 | 2.7 | 2.0 | 4.7 | |
| Vector | 42.1 | 0 | - | - | - | |
| 4 | 43.1 | + 1.0 | 3.0 | 2.0 | 5.0 | |
| 5 | 48.4 | + 6.3 | 3.0 | 2.0 | 5.0 | |
| 6 | 54.6 | + 12.5 | 2.3 | 2.0 | 4.3 | |
| 7 | 60.1 | + 18.0 | 2.3 | 2.0 | 4.3 | |
| V39 | 1 | 52.2 | − 1.5 | 1.0 | 1.0 | 2.0 |
| G3556C/G4655A | 2 | 53.6 | − 0.1 | 1.0 | 1.7 | 2.7 |
| Vector | 53.7 | 0 | - | - | - | |
| 3 | 54.6 | + 0.9 | 1.0 | 0.0 | 1.0 | |
| 4 | 60.5 | + 6.7 | 1.0 | 0.0 | 1.0 | |
| 5 | 65.8 | + 12.0 | 0.3 | 0.0 | 0.3 | |
| 6 | 72.4 | + 18.7 | 1.0 | 0.0 | 1.0 | |
| V410 | 1 | 53.9 | − 2.2 | 2.0 | 1.7 | 3.7 |
| G3556C/G3556C | 2 | 55.9 | − 0.2 | 2.0 | 2.0 | 4.0 |
| Vector | 56.1 | 0 | - | - | - | |
| 3 | 57.0 | + 0.9 | 1.3 | 1.0 | 2.3 | |
| 4 | 61.9 | + 5.8 | 1.0 | 1.0 | 2.0 | |
| 5 | 68.4 | + 12.3 | 1.0 | 1.3 | 2.3 | |
| 6 | 74.1 | + 18.0 | 0.0 | 0.0 | 0.0 | |
| V511 | 1 | 32.2 | − 1.8 | 2.0 | 1.0 | 3.0 |
| G3556C/C3084T | 2 | 33.8 | − 0.2 | 3.0 | 2.0 | 5.0 |
| Vector | 34.0 | 0 | - | - | - | |
| 3 | 34.8 | + 0.8 | 3.0 | 1.7 | 4.7 | |
| 4 | 40.0 | + 6.0 | 2.0 | 2.0 | 4.0 | |
| 5 | 46.0 | + 12.0 | 3.0 | 2.0 | 5.0 | |
| 6 | 54.3 | + 20.3 | 2.0 | 2.0 | 4.0 | |
| V612 | 1 | 63.1 | − 0.8 | 1.0 | 1.0 | 2.0 |
| G3556C/G4013T | 2 | 63.7 | − 0.2 | 1.0 | 1.0 | 2.0 |
| Vector | 63.9 | 0 | - | - | - | |
| 3 | 65.4 | + 1.4 | 1.0 | 1.0 | 2.0 | |
| 4 | 69.5 | + 5.6 | 0.7 | 1.0 | 1.7 | |
| 5 | 95.0 | + 31.1 | 0.0 | 0.0 | 0.0 | |
| V713 | 1 | 59.5 | − 1.1 | 2.0 | 1.7 | 3.7 |
| C3670T/G3556C | 2 | 60.4 | − 0.2 | 2.0 | 1.7 | 3.7 |
| Vector | 60.6 | 0 | - | - | - | |
| 3 | 61.7 | + 1.1 | 1.0 | 2.0 | 3.0 | |
| 4 | 67.3 | + 6.7 | 1.0 | 1.0 | 2.0 | |
| 5 | 73.3 | +12.7 | 0.0 | 1.0 | 1.0 | |
| 6 | 77.9 | + 17.3 | 0.0 | 1.3 | 1.3 | |
| V814 | 1 | 57.3 | − 0.4 | 2.0 | 1.7 | 3.7 |
| C3670T/C3670T | Vector | 57.7 | 0 | - | - | - |
| 2 | 83.9 | + 26.2 | 0.0 | 0.0 | 0.0 | |
The motor and language data are provided for all subjects in cohort 1. The rows highlighted in light grey are pre-administration, dark grey is vector administration and the unshaded rows are post-administration visits. The clinical assessment of motor + language was performed prospectively using defined standard operating procedures (SOPs) based on 3 to 4 observers, with specific rules on how the data was evaluated. The primary, on-site assessor was a pediatric neurologist who had been trained on implementing the scale. The assessment of each child was videotaped by a trained technician following a SOP for recording the assessment and editing for review by 2 to 3 other pediatric neurologists who were trained on implementing the scale. All were blinded to the subjects’ treatment status. In the event of discrepancy of more than 1 point between the 2 blinded scorers, a 3rd pediatric neurologist, also blinded, scored the video in order to act as a tie-breaker. The final score was an average of the assessment of 3 to 4 reviewers (primary + 2 to 3 additional reviewers), minimizing bias and subjective interpretation. The data provided here is the final score.
Subjects V1-V8, Cohort 1, received vector administration as outlined in footnotes 7–14. The genotypes for each subject are provided; following are the alternate nomenclatures for each mutation: G3556C (c.509–1G>C; intron 7 splice); C3670T (c.622C>T; nonsense Arg208 to stop); G4665A (1094 G>A; Cys365Tyr); C3084T (379 C>T; Arg127X); and G4013T (851G>T; Gly248Val).
Each subject typically underwent 2 motor + language assessments prior to vector administration and 4 assessments (scheduled for months 1, 6, 12 and 18) post administration.
Motor score – Scale of 0–3, 3 is normal, 2 is abnormal, but independent, 1 is abnormal, requires assistance and 0 is non-ambulatory
Language – Scale of 0–3, 3 is normal, 2 is abnormal, 1 is barely understandable, requires assistance and 0 is unintelligible or no speech
Composite of motor + language
Subject V1 received vector at age 87.6 months
Subject V2 received vector at age 42.1 months
Subject V3 received vector at age 53.7 months
Subject V4 received vector at age 56.1 months
Subject V5 received vector at age 34.0 months
Subject V6 received vector at age 63.9 months
Subject V7 received vector (lower dose – 2.85×1011 gc) at age 60.6 months
Subject V8 received vector (lower dose – 2.85×1011 gc) at age 57.7 months. V8 did not return for any of the interim follow up visits, the study team was able to get one measurement on subject at their home location, 2.2 year after vector administration. Due to no data points in the 18±1-month study period, a rate of decline was not calculated for this subject.
Description of the ancillary cohorts
The European DEM Child natural history replication control cohort (cohort 3) consisted of 41 CLN2 genotype-confirmed subjects (Table 1) (5). Twenty-four were males and 17 females. The mutations included 71% heterozygous or homozygous for g.C3670T or g.G3556C. Of those, 42% were homozygous at both alleles for g.C3760T and 7% homozygous for g.G3556C. Of the remainder, 39% were either heterozygous for both mutations or heterozygous with a different mutation; 12% did not have either of the two common mutations. The average age of first observed clinical symptom was 33 months (range 12 to 53 months). For the majority (88%) the age of first seizure was 30 to 50 months, 2 had the first seizure at 0 to 30 months, 2 at 50 to 70 months and 1 at 106 months.
The safety-only treated cohort (cohort 4) comprised of 5 children (n=4 females, n=1 male, S1 to S5; Table 1). The average age of first reported symptoms was 36 months (range 24–42 months). In 4 of 5, the first reported symptom was speech delay, accompanied in some subjects by gait, balance, motor, cognition and behavioral issues. All had the first seizure between 30 and 36 months of age.
Safety of CNS administration of AAVrh.10hCLN2
Treatment with AAVrh.10hCLN2 was well tolerated, with minimal serious adverse events in the acute/post-operative period (0–14 days) and over the 18-month study period (14 days – 18 months).
Vector infusion time, surgery time and duration of anesthesia was similar for all subjects treated with CNS administration of the AAVrh.10hCLN2 vector (tables S2 and S3). Vector administration was well tolerated in both cohorts 1 and 4. The children were discharged from the hospital an average of 3.0 ± 1.0 days for cohort 1 and 5.0 ± 1.4 days for cohort 4. For analysis of safety, the data from cohorts 1 and 4 were combined. In the acute period, a severe adverse event (SAE) occurred in 6 of 13 children, including seizures, abnormal movements and emesis (Table 3). For the seizures and abnormal movements (3 of 13), it was not possible to determine whether these were related to the administration procedure or study drug, and thus they were ascribed to both. Other acute SAE included MRI identification of hematoma (1 of 13) and hemorrhagic contusion (1 of 13).
Table 3.
Serious Adverse Events with Direct CNS Administration of AAVrh.10hCLN2 to Cohorts 1 and 41
| Number of Events (% of total events) | ||
|---|---|---|
| Event | Acute/post-operative2 | Chronic3 |
| Any serious adverse event (SAE) | 6 (28.6) | 15 (71.4) |
| Any SAE related to study drug4 | 3 (14.3) | 3 (14.3) |
| Any SAE related to drug administration4 | 6 (28.6) | 7 (33.3) |
| Specific SAE | ||
| Episodes of increased seizures5 | 1 (4.8) | 6 (28.6) |
| Dystonia | 0 (0) | 1 (4.8) |
| Episodes of increased abnormal movements6 | 2 (9.5) | 0 (0) |
| Emesis | 1 (4.8) | 1 (4.8) |
| Hematoma, hemorrhagic contusion | 1 (4.8) | 1 (4.8) |
| Hygroma | 0 (0) | 1 (4.8) |
| Pneumocephalus | 1 (4.8) | 0 (0) |
| Bronchospasm | 0 (0) | 1 (4.8) |
| Aspiration | 0 (0) | 1 (4.8) |
| Pneumonia | 0 (0) | 1 (4.8) |
| Elevated hepatic enzymes | 0 (0) | 2 (9.5)7 |
Serious adverse events (SAE), as defined by 21 CFR 312 (a).
SAEs that occurred from day 0 (day of procedure/vector administration) through day 14 (14 days post-vector administration); reported as number of occurrences (% all occurrences).
SAEs that occurred from month 1 (starting 15 days after the vector administration) through month 18 (540 days post-vector administration); reported as number of occurrences (% all occurrences).
In most cases, in the actual/post-operative period, it is not possible to distinguish as to whether the SAE resulted from the study drug or the drug administration; whenever the SAE was reported as “likely” or “probably” related to the study drug it was listed as “SAE related to study drug”
General tonic-clonic seizures, myoclonic seizures.
Abnormal facial movements, facial twitches/dyskinesia
Transient, mild elevation of ALT, AST at 6 months in S5, cohort 4, resolved without therapy, this subject received the lower dose of 2.85×1011 gc
In the chronic period, there were 15 SAE, with 3 definitely or possibly related to the study drug, and 7 related to the drug administration (Table 3). Among the SAE observed after 14 days were rare cases of increased seizures, dyskinesia, emesis, hygroma, pneumocephalus, bronchospasm, aspiration, pneumonia and mild, transiently elevated hepatic enzymes. Elevated hepatic enzymes (ALT, AST) were observed in only 1 subject (S5, Cohort 4) at month 6 and spontaneously resolved without therapy. There was no evidence of a preexisting condition that would make this child more susceptible to liver damage from the vector. In the preclinical studies, we observed mild, intermittently elevated liver enzymes, but no evidence for consistent elevated liver abnormalities (34). Assessment of cerebral spinal fluid (CSF) at 6 to 12 months in cohort 1 and 4 showed no abnormal accumulation of inflammatory cells (table S4).
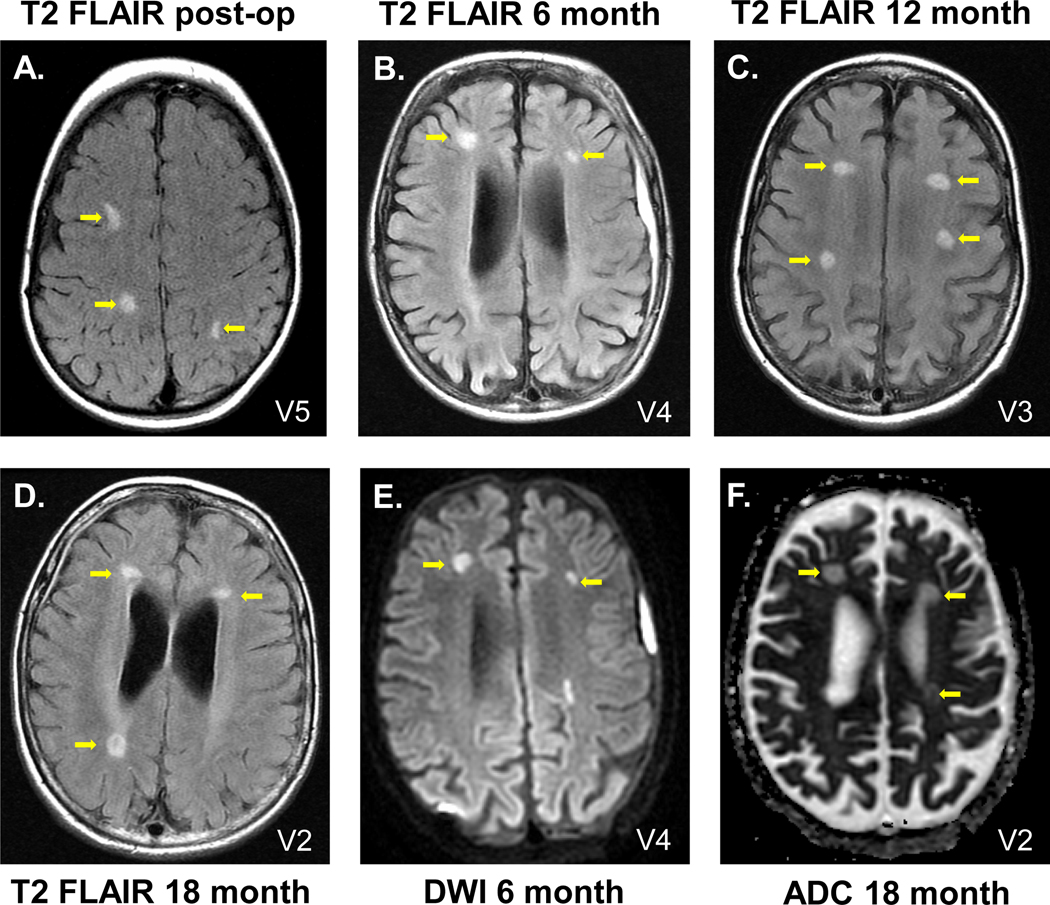
MRI assessed within 48 hr of vector administration demonstrated T2 hyperintensities (measured by T2 FLAIR), diffusion hyperintensity (measured by diffusion weighted imaging) and restriction of diffusion assessed by apparent diffusion coefficient localized to the sites of vector administration (Fig. 1, Table 4). During the course of the study, these localized abnormalities persisted 6 to 12 months post-therapy in most subjects, while in others these abnormalities resolved (fig. S3). Quantification of the extent of the hyperintense T2 signal in the MRI data demonstrated that the average volume of the hyperintense signal represented <0.3% of the total brain volume, and there was no increase or decrease of this volume with time (table S5). There were no clinical sequelae attributable to these MRI findings. Despite the lack of clinical correlate, our working hypothesis, based on our studies in nonhuman primates (34), was that the MRI findings localized to the vector administration sites represented mild persistent edema/inflammation in the areas at the tip of the catheter where the highest concentration of the vector was deposited. Based on this, we decided for subsequent subjects to reduce the dose by ½ log (from a total dose of 9 × 1011 gc divided into 12 sites) to a total dose of 2.85 × 1011 gc divided equally among 12 sites. Of the eight subjects that received the vector in cohort 1, 6 received the original dose and the last two (V7, V8) received the ½ log lower dose. Of the 5 subjects in cohort 4, 3 (S3, S4, S5) received the ½ log lower dose. Importantly, there was no association of T2 flair, ADC or DWI (Table 4), with dose for post-operative (p values: 1, 0.5, 1), month 6 (p values: 0.2, 0.5, 0.5), month 12 (p values: 1, 0.5, 1) and month 18 (p values: 0.08, 0.5, 1).
Figure 1. Axial T2 FLAIR (T2 FLAIR), diffusion weighted imaging (DWI) and apparent diffusion coefficient (ADC) MRI assessment of participants post-therapy.
MRI abnormalities were localized at the sites of the catheter tips, where there is the highest concentration of the administered vector. A-D. Examples of T2 FLAIR. A. Participant V5, 1 day post-administration; B. V4, 6 months; C. V3, 12 months; D. V2, 18 months. E. Example of DWI, participant V4, 6 months. F. Example of ADC, participant V2, 18 months. Yellow arrows identify the abnormalities. See Table 4 for the complete dataset of T2 FLAIR, DWI and ADC abnormalities observed.
Table 4.
| Pre-transfer | Post-operative | Month 6 | Month 12 | Month 18 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort | Subject | Total dose (gc) | T2 | DWI/ADC | T2 | DWI/ADC | T2 | DWI/ADC | T2 | DWI/ADC | T2 | DWI/ADC |
| 1 | V1 | 9.0×1011 | – | –/– | + | +/+ | + | +/+ | + | +/– | + | –/– |
| V2 | 9.0×1011 | – | –/– | + | –/– | + | +/+ | + | +/+ | + | +/+ | |
| V3 | 9.0×1011 | – | –/– | + | +/+ | + | –/– | + | –/– | + | –/– | |
| V4 | 9.0×1011 | – | –/– | + | –/– | + | +/+ | + | +/+ | + | –/– | |
| V5 | 9.0×1011 | – | –/– | + | –/– | + | –/– | + | +/– | + | +/– | |
| V6 | 9.0×1011 | – | –/– | + | +/+ | – | –/– | ND4 | ND | ND | ND | |
| V7 | 2.85×1011 | – | –/– | + | +/+ | + | –/– | ND | ND | ND | ND | |
| V8 | 2.85×1011 | – | –/– | + | +/+ | ND | ND | ND | ND | –5 | –/–5 | |
| 4 | S16 | 9.0×1011 | – | –/– | + | +/+ | ND | ND | ND | ND | ND | ND |
| S2 | 9.0×1011 | – | –/– | + | +/+ | + | –/– | – | –/– | + | –/– | |
| S3 | 2.85×1011 | – | –/– | + | +/+ | – | –/– | + | +/– | – | –/– | |
| S4 | 2.85×1011 | – | –/– | + | +/+ | – | –/– | + | –/– | + | –/– | |
| S5 | 2.85×1011 | – | –/– | + | +/+ | –5 | –/–5 | –5 | –/–5 | ND | ND | |
“T2” - T2 hyperintensities localized to the site of vector administration as assessed by T2 FLAIR; “DWI” - hyperintensities localized to the site of vector administration as assessed via diffusion weighted imaging; “ADC” – apparent diffusion coefficient, DWI data quantified
“–” indicates absence of T2 hyperintensities, “+” indicates presence of T2 hyperintensities
“–/–” indicates absence of DWI hyperintensities and of the associated diffusion restriction, “+/–” indicates presence of DWI hyperintensities but the absence of the associated diffusion restriction, “+/+”indicates presence of DWI hyperintensities and also of the associated diffusion restriction
ND - not done; some of the follow up MRIs were not done because of the difficulty of travel to Weill Cornell for the MRI; in these circumstances, whenever possible, the study team did the clinical assessment of the child at the subject’s home
For Subjects V8 and S5, some of the MRI scans were at their local hospitals
Subject S1 discontinued from further participation in the study and no follow-up MRI data was available thereafter
Anti-vector immunity after CNS administration of AAVrh.10hCLN2.
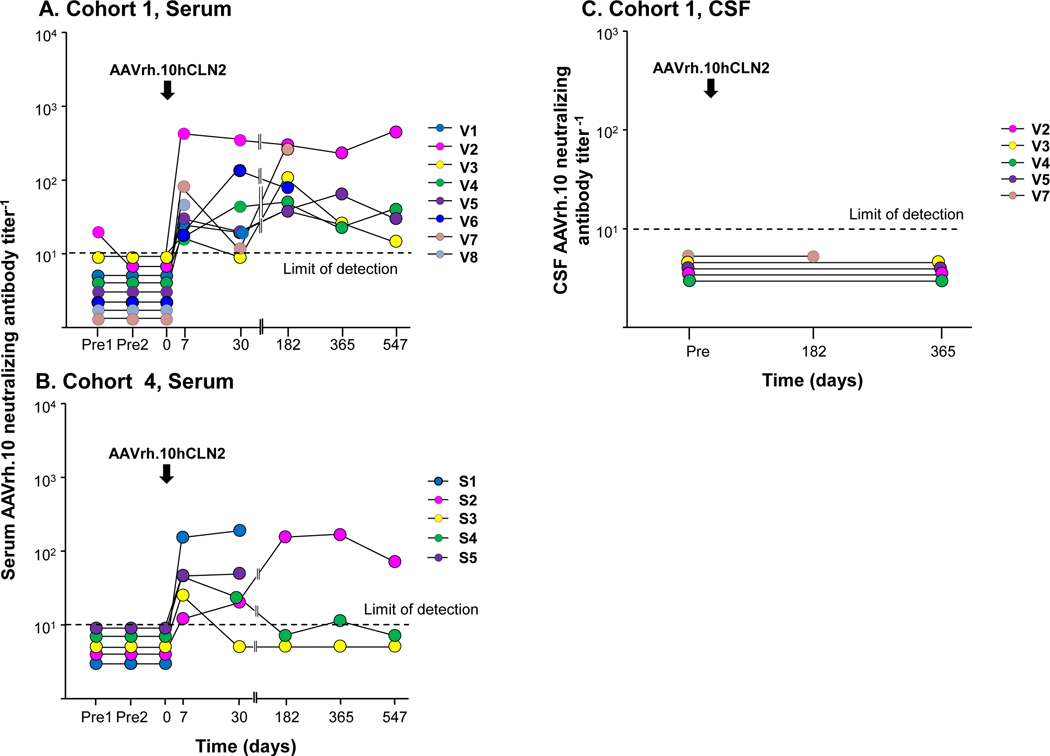
Before gene transfer, only one subject (V2) had mildly detectable serum anti-AAVrh.10 neutralizing antibodies, all others had undetectable neutralizing antibodies (fig. 2A). In cohort 1, 7 of 8 subjects developed mild increase in detectable anti-rh.10 capsid neutralizing antibodies; 1 subject (V2) developed higher neutralizing anti-capsid antibodies. At 18 months, the antibody responses persisted, but were mostly low. In cohort 4, CNS administration of the AAVrh.10hCLN2 vector also resulted in a mild, systemic anti-vector humoral immune response (fig. 2B). Four of 5 subjects developed a mild systemic humoral immune response while one subject (S1) developed a higher neutralizing antibody response to the AAVrh.10 capsid after CNS administration of the vector. At 18 months, the antibody response was slightly elevated for subject S2, but with a titer <100. Statistical comparisons of anti-vector neutralizing antibody responses to the dose were not possible due to small number of data points.
Figure 2. AAVrh.10 neutralizing antibody titers.
Titers are expressed as the reciprocal of dilution at which 50% inhibition of an AAVrh.10Luc reporter gene expression in 293 ORF6 cells in vitro. Samples were not available for some time points for some participants. Vector administration is indicated by an arrow. Dashed black line represents the limit of assay detection. A. Cohort 1 (V1-V8), serum. Participant V8 only had 1 time point (7 days) and then dropped out of the 18-month follow-up study. Participant V7 received the lower dose (2.85×1011 gc). B. Cohort 4 (S1-S5), serum. Participants S3-S5 received the lower dose (2.85×1011 gc). C. Cohort 1, cerebral spinal fluid (CSF). CSF samples were available from subjects V2-V5, V7.
For cohort 1, anti-AAVrh.10 neutralizing antibodies were also assessed in the CSF for whom pre- and post-treatment CSF samples were available (n=5). No detectable anti-rh.10 capsid neutralizing antibodies were observed (fig. 2C).
Blood T-cell responses to the AAVrh.10 capsid and the CLN2 transgene were assessed by IFN-γ ELISpot. Blood mononuclear cells obtained from subjects before therapy at screening and pre-vector administration and at days 7, 14 and months 1, 6, 12 and 18 after vector administration were stimulated with AAVrh.10 capsid and CLN2 transgene peptide library pools. There were sporadic, but not persistent, mild responses among the samples to the vector capsid or transgene, with no correlation to time after vector administration (fig. S4A, B). Subject S2 and S4 had mildly elevated ELISpots. This could be a function of the CLN2 genotype as these two subjects each have one allele that is unique among study participants. Qualitatively, there was no correlation of neutralizing antibodies titers and the minor, specific T cell responses, nor with vector dose.
Assessment of treatment efficacy
Follow-up data over the 18-month study period was available for analysis from 7 of the 8 treated children in cohort 1. The parameters used to assess efficacy included: (1) TPP1 amount in CSF; (2) assessment of MRI % grey matter volume; (3) vision parameters; and (4) neurologic clinical assessment of motor + language scale. All 3 CNS parameters suggested a positive treatment effect (fig. 3–5, fig. S5 and S6). The limited amount of vision-related data showed no treatment effect. The ITQoL-PF97 or CHQ-PF50 quality of life questionnaires and Mullen scales were used pre- and post-therapy to question the parents and assess the children, respectively. We found these scales to be highly variable, with no measurable differences between cohort 1 and the control cohort 2 (fig S7, S8; table S6).
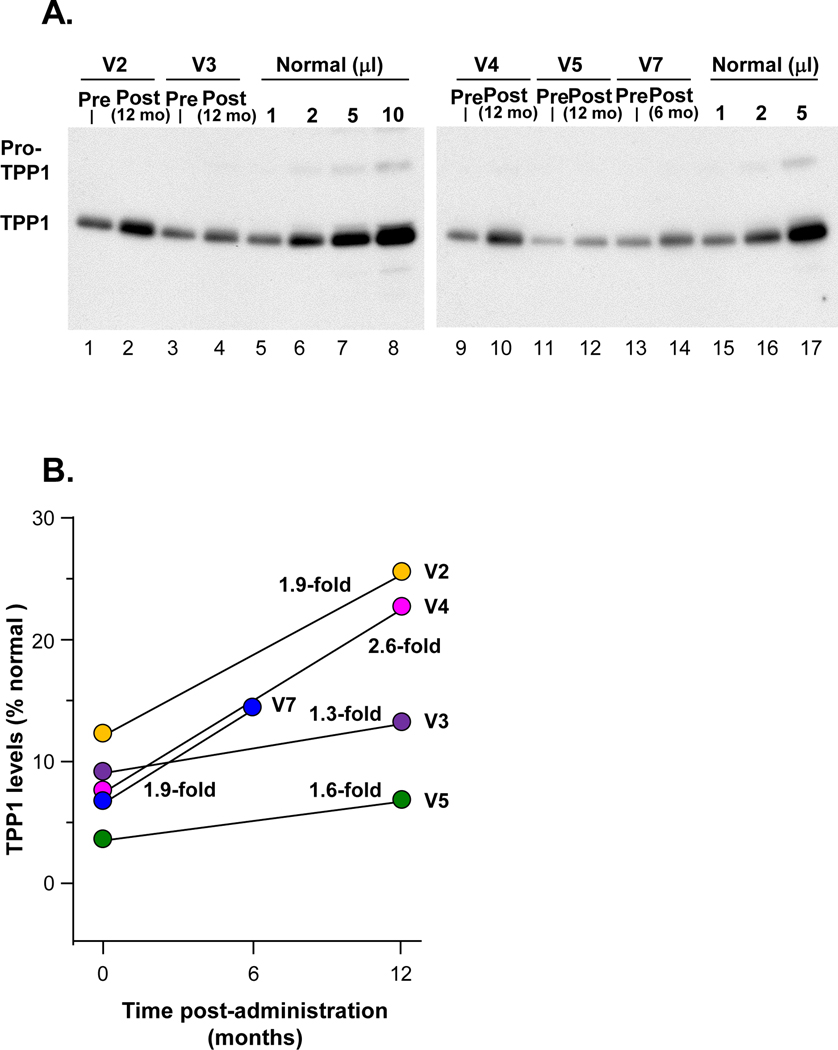
Figure 3. Human TPP1 in CSF following AAVrh.10hCLN2 administration to cohort 1.
CSF was analyzed from TPP1 by Western analysis. The IRB approved protocol allowed for CSF sampling before therapy and only 1 time after therapy. A. Western analysis. Lanes 1–2, participant V2; lanes 3–4, participant V3; lanes 5–8, 1, 2, 5 and 10 μl, respectively, of combined CSFs of three healthy children (1:1:1 volume mix) as a positive control. Lanes 9–10, participant V4; lanes 11–12, participant V5; lanes 13–14, participant V7; lanes 15–17, 1, 2 and 5 ml, respectively, of combined CSFs of three healthy subjects (1:1:1 volume mix) as a positive control. B. Quantitation of TPP1 in CSF before and after therapy expressed as percent normal TPP1 in CSF following AAVrh.10hCLN2 administration compared to pre-administration (pre vs post % normal, p<0.03, paired two-tailed t-test). V7 received the lower dose (2.85×1011 gc).
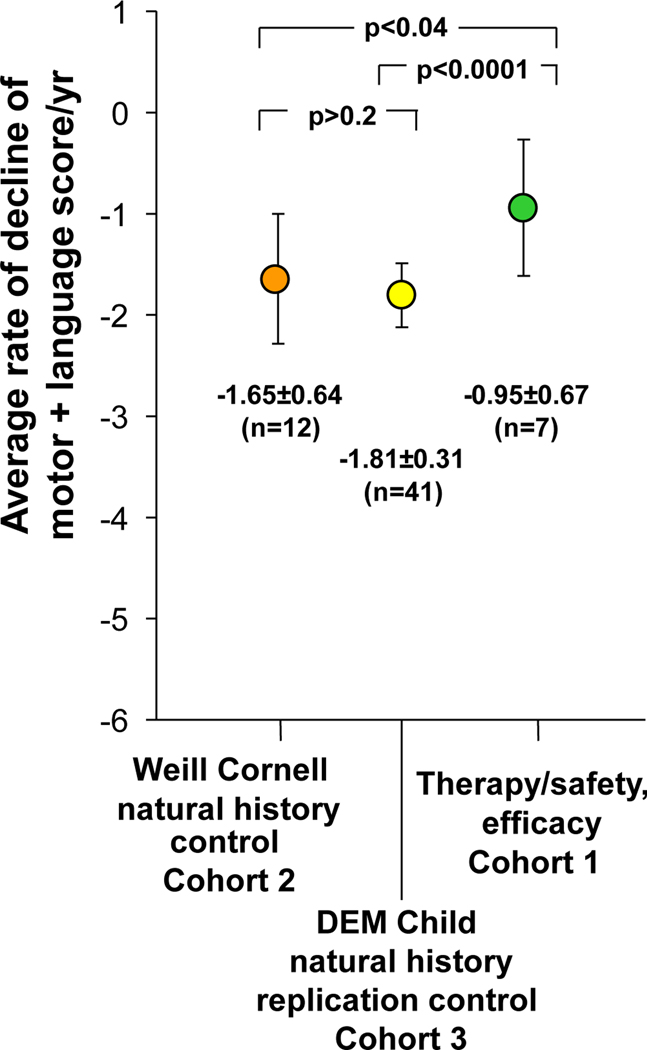
Figure 5. Quantitation of the rate of decline of motor and language assessment in the therapy cohort (cohort 1) compared to Weill Cornell natural history control cohort (cohort 2) and the DEM Child natural history replication control (cohort 3).
For cohorts 1 and 2, linear regression was taken for each participant’s motor and language assessment over time to calculate the individual rate of decline. The individual rates of decline for all children within a cohort were then averaged to calculate the rate of decline/year for each individual cohort. The rates of decline/year for each cohort are plotted as a mean rate of decline with the error bars representing ± one SD from the mean. The raw data for cohorts 1 and 3 are in Table 2 and table S7. For cohort 3, the sample size, mean, and standard deviation were derived from Nickel et al (5); p values were determined using a two-tailed unpaired Student t-test.
All AAVrh.10hCLN2-treated patients for whom pre- and post-treatment CSF samples were available (n=5) had increased TPP1 in the CSF compared to the pre-treatment values (fig. 3A). Quantification of CSF TPP1 6 to 12 months after therapy demonstrated a 1.3- to 2.6-fold increase over pre-therapy values. When compared to TPP1 in normals, the pre-treatment values ranged from 5–13% of normal and the post-treatment values ranged from 8–26% (p<0.03, fig. 3B).
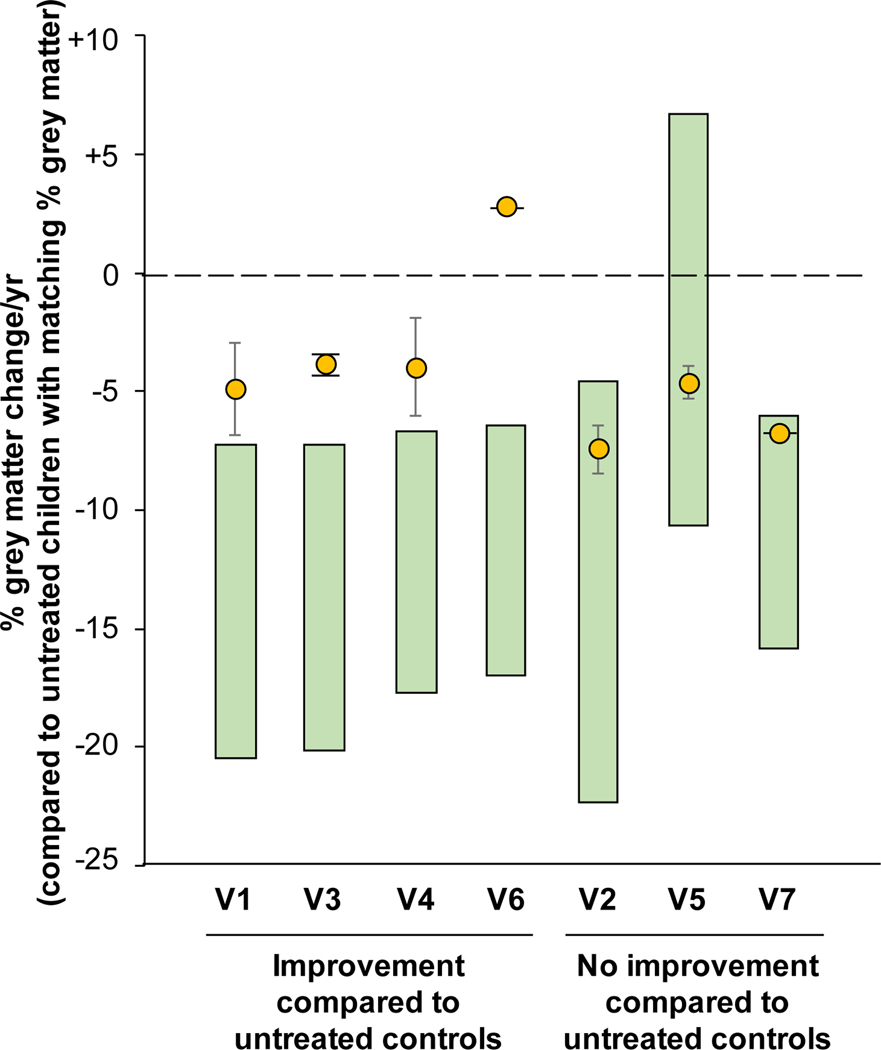
Untreated children with CLN2 disease have a decrease in % grey matter volume from ages 2 to 6 (36, 37). The % grey matter volume over time of the treated children in cohort 1 was compared to the untreated children with CLN2 disease (fig. 4, fig S4, S5). Two of the 7 treated children had only a single scan following treatment, insufficient to calculate the standard error of the rate of decline of % grey matter. For the 5 children with 2 or more post-therapy scans, 3 had rates of decline in % grey matter volume less than the untreated CLN2 children. Of the 2 children with only 1 post therapy scan, 1 of 2 had % grey matter decline slower than that of the untreated children. Two children who were the youngest in the cohort may not yet have been of age for rapid change in grey matter to occur were likely to have indistinguishable effects.
Figure 4. Quantitative MRI assessment of grey matter decline in treated vs untreated CLN2 children.
The reduction in the % grey matter, assessed by MRI, is shown for each treated participant (yellow circle) above the range of grey matter decline (green bar) for untreated CLN2 children matched by % grey matter. The range of the grey matter decline is derived from the data in figures S5 and S6. Treated children with grey matter decline above the range for the untreated cohort indicates a decline that is slower and outside the 95% confidence interval for untreated children. Participants V2 and V5 were the youngest trial participants and had slow rates of decline at the time of treatment such that the effect of therapy was not yet apparent. Participant V6 had only one post-treatment scan and therefore error bars could not be calculated. Participant V7 received the lower dose (2.85×1011 gc).
The ocular and CNS disorders associated with CLN2 disease are distinct and develop independently. The ocular findings are typified by a gradually progressive retinal degeneration, commencing at the outer retina (specifically in the retinal pigmented epithelium and photoreceptors in a bull’s eye pattern), and progressing from the central macula to the peripheral retina, symmetrically between the two eyes (38, 39). This retinal degeneration ultimately results in widespread retinal atrophy encompassing the entire fundus. None of the eyes of the CLN2 subjects exhibited any anterior segment abnormalities, regardless of the severity of the retinal degeneration, the advancing age of the subjects or the extent of neurological deterioration.
One subject (V3) evaluated prior to CNS directed gene therapy at age 53 months was found to have a central macular thickness (CMT) in the right eye of 295 μm and in the left eye of 282 μm. Seven months after CNS directed CLN2 gene therapy, at age 60 months, the CMT of the right eye was 266 μm and CMT of the left eye was 267 μm: despite the CNS gene therapy, the ophthalmic degeneration in both eyes followed the same accelerated decline as was seen with the natural history of untreated patients. The progressive retinal changes noted on exam and on dilated fundus photography in all eyes demonstrated continued degeneration despite the CNS-directed gene therapy.
The primary efficacy parameter was the neurologic rating scale of assessment of motor + language. The reproducibility of the measurement of the motor and language parameters demonstrated an average coefficient of variation among the assessors of 0.07±0.14 for the motor domain and 0.16±0.18 for the language domain (table S7). To determine the reproducibility of the motor + language assessment, based on the knowledge that our primary rating scale should not change in a short period, we assessed data from n=6 children that had repeat assessments <1.5 months apart. The data demonstrated excellent reproducibility in the repeat assessments of the subjects for motor (p>0.9), language (p>0.6) and for the combined score of motor and language (p>0.7; table S8).
The consensus motor + language neurologic parameter of the therapy/safety and efficacy group (cohort 1, table 2) was compared with the Weill Cornell natural history control cohort (cohort 2, table S9) and to the European DEM-Child natural history replication (cohort 3) data sets (fig. 5). The difference in the annual rate of decline between the 2 control groups was not significant (p>0.2). In contrast, the decline per year in the treated subjects was slower in comparison to the decline for both control groups. The annual rate of decline for the treated cohort (cohort 1) was −0.95±0.67 (mean ± SD, n=7). In comparison, the annual rate of decline for the control cohort (cohort 2) was −1.65±0.64 (mean ± SD, n=12). The treatment yielded a 42.4% slowing in the rate of decline in motor + language assessment of treated to untreated children, which was statistically significant (p<0.04). For the replication untreated European cohort (cohort 3), the annualized rate of decline was −1.81±0.31 (mean ± SD, n=41). Comparison of the treated group (cohort 1) to cohort 3 demonstrated 47.5% slowing in the rate of decline (p<0.0001).
Due to the small study population, it was not possible to reach definitive conclusions regarding the response to therapy of the different genotypes or age of treatment. With this caveat, as this information may be useful for the design of a larger study, we are presenting the data. There was no correlation between the annual rate of decline and the genotype of the treated subjects (fig. S9A) or between the age at vector administration and the rate of decline post treatment (fig. S9B). Similarly, assessment of impact of the motor + language score at the time of vector administration on the rate of decline did not demonstrate a correlation (fig. S9C). Finally, we evaluated the impact of the peak neutralizing antibody response post-vector administration and its impact on the rate of decline; there was no correlation among these parameters (fig. S9D). Interestingly, the subject who had the highest systemic anti-AAVrh.10 neutralizing capsid antibody response (V2), had the best clinical response, with the resulting rate of motor + language decline of −0.3 units/year.
In a study carried out by BioMarin Pharmaceuticals, with biweekly intraventricular infusions of cerliponase alfa™ (recombinant TPP1) in 23 subjects, the mean (±SD) rate of decline using the same motor + language score per 48-week period was −0.27±0.35 in treated patients (31), extrapolated to an annual rate of decline of −0.29±0.38. Compared to the natural history control groups, this represents an improvement of 82.4 – 84.0% in slowing the rate of decline, compared to our gene therapy improvement of 42.4 – 47.5%.
Discussion
In the present study, we used direct CNS administration of a serotype AAVrh.10 gene transfer vector to deliver the normal coding sequence of human CLN2 to the CNS of children with CLN2 disease. The administration of the vector and subsequent follow-up over 18 months demonstrated that the therapy was safe, with minimal serious adverse events, and presented preliminary measures of efficacy. The challenges of testing the gene therapy for CLN2 disease include the highly variable clinical phenotype in a relatively small target population which makes statistically relevant conclusions difficult. Further, the complex array of clinical sequelae which include seizures, motor neuron dysfunction and cognitive impairment, which differ among individuals, create a barrier to evaluating safety as well.
The current US- and Europe-approved treatment option for the CNS manifestations of CLN2 disease is recombinant TPP1 administered every other week via a subcutaneous reservoir with a catheter into a CNS ventricle (30–32). While AAVrh.10hCLN2 therapy slowed progression of the CNS disease, using the same control group (the European DEM Child natural history replication control cohort) as the comparator, recombinant TPP1 therapy was more efficacious, with a greater reduction in the rate of decline of the same neurologic parameters compared to gene therapy. Recombinant TPP1 therapy provided an 84.0% decrease in the rate of neurological decline, compared to 47.5% for this gene therapy. If the gene therapy could be improved, the theoretical advantage is that it could potentially be efficacious with a single administration, while the recombinant TPP1 therapy requires administration every other week (31, 40, 41). Gene therapy would also substantially reduce the costs over a lifetime and assure 100% compliance.
There are several approaches to improve the gene therapy. First would be to combine the direct parenchymal administration route with additional routes such as intracerebroventricular and/or intracisternal delivery which have also been shown to lead to widespread distribution of transgene products (42–47). TPP1 is a secreted protein and is capable of cross-correcting neighboring cells mediated via the mannose-6-receptor pathway (27–29). It is not necessary to transfer the normal CLN2 cDNA to all of the cells in the CNS; corrected cells will secrete TPP1 which is then endocytosed via the mannose-6-phosphate receptor pathway by neighboring cells for therapeutic correction (12, 28). AAV-based gene therapy, which would be unlikely to transduce every cell with the normal therapeutic gene, is the source of corrective enzymes for even the non-infected neighboring cells, and thus additional routes of administrations may provide a greater potential for success of the therapy (12, 28, 46). To inform future clinical development of AAVrh.10hCLN2, we are planning nonhuman primate studies of direct comparison of the distribution of vector expression by each of the different routes of administration or in combination.
Second, an alternative approach would be to combine the intraparenchymal gene therapy strategy together with recombinant TPP1, likely leading to greater efficacy and possibly reducing the frequency of administration and/or dose of recombinant TTP1 to achieve maximal efficacy.
Third, we chose the AAVrh.10 vector based on effective experimental animal studies in rodents and nonhuman primates (33–35). While the AAVrh.10hCLN2 vector was efficacious in slowing the rate of progression of the disease, it is possible that improvements in vector and/or expression cassette design could provide better distribution and higher concentrations of TPP1 throughout the brain.
Fourth, despite the fact that no immunosuppression was used in the current study, there was little evidence of systemic anti-capsid neutralizing antibodies or anti-capsid/anti-transgene cellular immunity generated by the CNS gene therapy. No subjects had evidence of inflammation in CSF. Most had CNS MRI lesions localized to the region at the tip of the catheter. While CNS anti-vector immunity could lead to lack of efficacy, all of the subjects assessed had increased expression of TPP1 in the CSF 6 month to 1 year after administration, and there was no capsid anti-neutralizing antibody in the CSF. The lack of anti-capsid neutralizing antibodies in the CSF suggests that readministration of the gene therapy vector could be used to boost the response.
Fifth, the dose could be increased. While this is unlikely to be safe with intraparenchymal administration, it might be done by the intracerebroventricular or intracisternal routes.
Finally, in the children screened, the average time from 1st symptom to diagnosis of CLN2 disease was 19 months. Anecdotally, the youngest treated child had the best reduction in slowing of the rate of decline on the neurologic rating scale, and studies in murine knockout model of CLN2 disease demonstrated that earlier treatment is more effective (35, 48). In this context, it is likely that there would be improvement in efficacy with earlier diagnosis and treatment.
In conclusion, direct intraparenchymal AAVrh.10hCLN2 vector administration is safe and there is a therapeutic benefit over 18 months, slowing the rate of neurological decline from CLN2 disease. However, while direct comparisons have not been made, compared to the same control group, the gene therapy is not as effective as recombinant TPP1 therapy administered biweekly. This provides the rationale for developing future clinical trials of AAV gene transfer in children with CLN2 disease using more effective CNS delivery strategies, consideration of immunotherapy and possibility of higher doses. To achieve maximal effectiveness, gene therapy for CLN2 disease should begin early, ideally before symptom onset. This will require earlier diagnosis and emphasizes the importance of adding CLN2 disease to universal newborn screening.
Methods
Study design
The goal of this study was to assess the safety and preliminary efficacy of intraparenchymal delivery of AAVrh.10hCLN2, in a non-randomized trial consisting of 2 cohorts assessed over 18 months: cohort 1 (mild to moderate neurologic disease, treated with AAVrh.10hCLN2) and cohort 2 (mild to moderate disease, no therapy; FDA BB IND13591; ClinicalTrials.gov identifiers: NCT01035424, NCT01161576; Table 1). As per the inclusion/exclusion criteria (table S10), the genotype of each subject had to include at least 1 of the 5 following CLN2 mutant genotypes: C3670T (nonsense Arg208 to stop), G3556C (intron 7 splice), G5271C (Gln422His), T4396G (aberrant splicing, intron 8) and G4655A (Cys365Tyr) (1, 2, 5, 11). If either parental allele was R447H (Arg447His, a known “slow progression” genotype), the subject was not included (11, 49, 50). Pre-therapy, all children had confirmation of their CLN2 mutations, routine blood and urine studies, comprehensive neurological assessments (CNS Magnetic Resonance Imaging ((MRI) and neurologic rating scale), assessment of anti-AAVrh.10 neutralizing antibodies, and assessment of relative quantity (pre/post) of TPP1 in the CSF. Cohort 1 included n=8 children (V1-V8) and cohort 2 included n=12 children (C1-C12).
Cohort 1 received 2.85–9.0 ×1011 genome copies (gc) of AAVrh.10hCLN2 (6 received 9.0×1011 gc, 2 received 2.85×1011 gc) delivered directly via catheter into the CNS via 6 burr holes (3 bilaterally), with equal doses to 2 sites/burr hole (51–53). No immunosuppression was used. One child (V-8) did not return for any follow up visit in the 18-month study and was excluded from the analysis of efficacy parameters.
The primary efficacy parameter was a CLN2 neurologic rating scale assessing motor and language parameters (table S1). As a replication control cohort for the CLN2 neurologic rating scale, comparison was also made to cohort 3 (Table 1), a control cohort from the published European CLN2 neurologic rating scale DEM Child Natural History study of n=41 untreated children with CLN2 disease followed longitudinally an average of 28 assessments over the life of the child (5). Details regarding cohort 3 are in Nickel et al (5). Secondary efficacy parameters included assessment of TPP1 in CSF and MRI assessment of % grey matter volume. For comparison of the MRI % grey matter volume of the treated children, the control data included data from n=62 MRI obtained from 47 children with CLN2 disease, including the untreated controls (cohort 2), the pre-therapy time points for the children in cohort 1 and 4, and children in the screening study that did not participate in the therapy vs no therapy study. Additional secondary efficacy parameters included parental assessment using the Child Health Questionnaire (CHQ) or Infant Toddler Quality of Life (ITQoL) quality of life questionnaire (depending on age) and the Mullen scale. See below for details regarding the efficacy parameters, table S10 for inclusion/exclusion criteria for cohorts 1 and 2 and table S11 for the timeline for cohorts 1 and 2.
The safety data also included cohort 4, a therapy/safety only cohort of n=5 children who were not eligible for the “therapy vs no therapy” study based on disease severity and/or genotype (see table S12 for inclusion/exclusion criteria for cohort 4). Cohort 4 was treated with AAVrh.10hCLN2 in the identical fashion as cohort 1, with n=2 at 9.0×1011 gc total dose divided int0 12 sites and n=3 at 2.85×1011 gc total dose. The timeline for cohort 4 was similar to that of cohort 1 (table S11). Because the children did not fit the eligibility criteria for the “therapy vs no therapy” study, the data was only used for safety comparisons. No immunosuppression was used.
Detailed methods regarding AAVrh.10hCLN2 administration, post-administration assessments including safety and efficacy parameters are provided in supplemental methods.
AAVrh.10hCLN2
AAVrh.10hCLN2 is composed of the capsid of AAVrh.10, a clade E AAV derived from rhesus macaque (33, 54, 55) and a genome composed of 5’ and 3’ AAV2 inverted terminal repeats surrounding an expression cassette including: the enhancer from human cytomegalovirus; promoter, splice donor and left hand intron sequence from chicken β-actin (CAG); the splice acceptor from rabbit β-globin; the normal human CLN2 coding sequence cDNA; and the rabbit β-globin polyA sequence (34, 56) (fig. S1). AAVrh.10hCLN2 was produced by a two-plasmid co-transfection under Good Manufacturing Practice (GMP) conditions at the Belfer Gene Therapy Core Facility, Weill Cornell Medical College. This co-transfection of an expression cassette plasmid (pAAV2-CAG-hCLN2) and an adenovirus/AAVrh.10 helper plasmid (pPAK-MArh.10) was carried out in a certified 293T cell line using PolyFect reagent (Qiagen Sciences). The helper plasmid included the AAVrh.10 cap gene and AAV2 rep gene necessary for viral reproduction and capsid production. The vector was released from cells following three freeze-thaw cycles 72 hr post-transfection, and a crude viral lysate (CVL) was generated. Benzonase (Sigma-Aldrich) was used to remove any contaminant genomic DNA. The remaining CVL was centrifuged and the supernatant applied to a discontinuous iodixanol gradient, and then purified by Q-HP ion exchange chromatography and centrifugally concentrated into phosphate buffered saline, pH 7.4 (PBS). Vector concentration in genome copies was determined by TaqMan real time PCR. To confirm functionality, HEK293-ORF6 cells were infected with AAVrh.10hCLN2, and TPP1 enzymatic activity was verified in the cell supernatant 72 hr post-infection (33, 57). Full characterization of the final product included U.S. FDA-approved lot release assays to ensure identity, purity and function.
Statistical analysis
Association of Post-operative T2 flair, ADC or DWI, with dose was performed by a two-tailed Fisher Exact test. For the Mullen Scale and Child Health Questionnaires, data was using a two-tailed unpaired Student t-test. In the assessment of the primary efficacy parameter, the CLN2 neurologic rating scale, the p value was calculated using a standard t-test assuming equal variance. For cohort 3, the sample size, mean, and standard deviation were derived from Nickel et al. (5); a two-tailed unpaired Student t-test was used to compare cohort 1 and cohort 2 individually against cohort 3.
Supplementary Material
Figure S1. AAVrh.10hCLN2 vector.
Figure S2. Trajectory planning for gene therapy infusions.
Figure S3. Axial T2 FLAIR (T2 FLAIR) assessment of participants post-therapy.
Figure S4. T cell responses to AAVrh.10 capsid and CLN2 transgene.
Figure S5. MRI assessment of the treated CLN2 children vs untreated CLN2 children.
Figure S6. Quantitative MRI assessment of the treated CLN2 children (cohort 1) vs untreated CLN2 children.
Figure S7. Mullen scale quantitation of the rate of decline for cohorts 1 (red, treated) and 2 (blue, control).
Figure S8. Impact of treatment on quality of life as assessed by age-dependent quality of life questionnaires.
Figure S9. Correlation of various parameters to the rate of decline of motor + language of cohort 1.
Table S1. CLN2 Disease Severity Clinical Rating Scales
Table S2. Vector Infusion Time and Operating Room Surgery and Anesthesia Duration in Cohort 1 Participants
Table S3. Vector Infusion Time and Operating Room Surgery and Anesthesia Duration in Cohort 4 Participants
Table S4. Cerebral Spinal Fluid Nucleated Cells
Table S5. Percent Volume of the Brain with MRI T2 Hyperintensity
Table S6. Quality of Life Questionnaires
Table S7. Coefficient of Variation Among Observers in the CLN2 Disease Motor + Language Neurologic Rating Scale
Table S8. Reproducibility of Motor and Language Assessment
Table S9. Assessments of Motor and Language Parameters for Cohort 2
Table S10. Inclusion/Exclusion Criteria for Cohorts 1 and 2
Table S11. Timeline of the Clinical Study
Table S12. Inclusion/Exclusion Criteria for Cohort 4
Acknowledgments.
We thank A. Mushlin (Healthcare Policy and Research, Weill Cornell Medicine) for discussions relating to the trial design, J. Greenfield (Department of Neurological Surgery, Weill Cornell Medicine) for surgical assistance, R. Calcedo and J.Wilson, Gene Therapy Program, University of Pennsylvania for assistance with cellular immunity assays, and N. Mohamed for help in preparing this manuscript.
Funding: This study was supported, in part, by NIH 1R01NS061848 (RGC), NIH U54NS065768 (RGC), and UL1 TR000457 (JI-M); and Nathan’s Battle, Cures Within Reach (RGC), Noah’s Hope (RGC), and Hope4Bridget foundations (RGC).
Footnotes
Competing interests: RGC is a consultant to BioMarin, a biotechnology company that markets Brineura®, a recombinant TPP1 enzyme used to treat CLN2 disease. BioMarin was not involved in this study. After study completion, AAVrh.10hCLN2, the gene therapy vector used in this study, was licensed by Cornell University to LEXEO Therapeutics, a gene therapy biotechnology company founded by Dr. Crystal. LEXEO had no role, financial or otherwise, in the design, conduct, analysis, or write-up of this study. Drs. Crystal, Sondhi and Kaminsky hold equity in, and are advisors to LEXEO. All other authors declare no conflict of interest.
Data and materials availability: All data associated with this study are available in the main text or the supplementary materials. The vector construct AAVrh.10hCLN2 can be provided to academic researchers via an MTA upon request to Cornell’s Center for Technology Licensing at Weill Cornell Medical College.
References
- 1.Sleat DE, Donnelly RJ, Lackland H, Liu CG, Sohar I, Pullarkat RK, Lobel P, Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science 277, 1802–1805 (1997); published online EpubSep 19 ( 10.1126/science.277.5333.1802). [DOI] [PubMed] [Google Scholar]
- 2.Williams RE, Gottlob I, Lake BD, Goebel HH, Wheeler W, Wheeler RB, in The Neuronal Ceroid Lipofuscinoses (Batten Disease), Goebel HH, Mole SE, Lake BD, Eds. (IOS Press, Amsterdam, 1999), pp. 37–54. [Google Scholar]
- 3.Haltia M, The neuronal ceroid-lipofuscinoses. J Neuropathol Exp Neurol 62, 1–13 (2003); published online EpubJan ( 10.1093/jnen/62.1.1). [DOI] [PubMed] [Google Scholar]
- 4.Worgall S, Kekatpure MV, Heier L, Ballon D, Dyke JP, Shungu D, Mao X, Kosofsky B, Kaplitt MG, Souweidane MM, Sondhi D, Hackett NR, Hollmann C, Crystal RG, Neurological deterioration in late infantile neuronal ceroid lipofuscinosis. Neurology 69, 521–535 (2007); published online EpubAug 7 ( 10.1212/01.wnl.0000267885.47092.40). [DOI] [PubMed] [Google Scholar]
- 5.Nickel M, Simonati A, Jacoby D, Lezius S, Kilian D, Van de Graaf B, Pagovich OE, Kosofsky B, Yohay K, Downs M, Slasor P, Ajayi T, Crystal RG, Kohlschutter A, Sondhi D, Schulz A, Disease characteristics and progression in patients with late-infantile neuronal ceroid lipofuscinosis type 2 (CLN2) disease: an observational cohort study. Lancet Child Adolesc Health 2, 582–590 (2018); published online EpubAug ( 10.1016/S2352-4642(18)30179-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakheja D, Bennett MJ, Neuronal ceroid-lipofuscinoses. Transl Sci Rare Dis 3, 83–95 (2018). [Google Scholar]
- 7.Fietz M, AlSayed M, Burke D, Cohen-Pfeffer J, Cooper JD, Dvorakova L, Giugliani R, Izzo E, Jahnova H, Lukacs Z, Mole SE, Noher de Halac I, Pearce DA, Poupetova H, Schulz A, Specchio N, Xin W, Miller N, Diagnosis of neuronal ceroid lipofuscinosis type 2 (CLN2 disease): Expert recommendations for early detection and laboratory diagnosis. Mol Genet Metab 119, 160–167 (2016); published online EpubSep ( 10.1016/j.ymgme.2016.07.011). [DOI] [PubMed] [Google Scholar]
- 8.Williams RE, Adams HR, Blohm M, Cohen-Pfeffer JL, de Los Reyes E, Denecke J, Drago K, Fairhurst C, Frazier M, Guelbert N, Kiss S, Kofler A, Lawson JA, Lehwald L, Leung MA, Mikhaylova S, Mink JW, Nickel M, Shediac R, Sims K, Specchio N, Topcu M, von Lobbecke I, West A, Zernikow B, Schulz A, Management Strategies for CLN2 Disease. Pediatr Neurol 69, 102–112 (2017); published online EpubApr ( 10.1016/j.pediatrneurol.2017.01.034). [DOI] [PubMed] [Google Scholar]
- 9.Jalanko A, Braulke T, Neuronal ceroid lipofuscinoses. Biochim Biophys Acta 1793, 697–709 (2009); published online EpubApr ( 10.1016/j.bbamcr.2008.11.004). [DOI] [PubMed] [Google Scholar]
- 10.Mole SE, Haltia M, in Rosenberg’s molecular and genetic basis of neurological and psychiatric disease, Rosenberg RN, Pascual JM, Eds. (Academic Press, Cambridge, MA, 2015), chap. 70, pp. 16. [Google Scholar]
- 11.Gardner E, Bailey M, Schulz A, Aristorena M, Miller N, Mole SE, Mutation update: Review of TPP1 gene variants associated with neuronal ceroid lipofuscinosis CLN2 disease. Hum Mutat 40, 1924–1938 (2019); published online EpubNov ( 10.1002/humu.23860). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sondhi D, Hackett NR, Apblett RL, Kaminsky SM, Pergolizzi RG, Crystal RG, Feasibility of gene therapy for late neuronal ceroid lipofuscinosis. Arch Neurol 58, 1793–1798 (2001); published online EpubNov ( 10.1001/archneur.58.11.1793). [DOI] [PubMed] [Google Scholar]
- 13.Sondhi D, Rosenberg JB, Van de Graaf B, Kaminksy SM, Crystal RG, Advances in the treatment of neuronal ceroid lipofuscinosis. Expert Opin Orphan Drugs 1, 951–975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarczyluk MA, Cooper JD, Investigative and emerging treatments for Batten disease. Expert Opinion on Orphan Drugs 3, 1031–1045 (2015). [Google Scholar]
- 15.Donsante A, Boulis NM, Progress in gene and cell therapies for the neuronal ceroid lipofuscinoses. Expert Opin Biol Ther 18, 755–764 (2018); published online EpubJul ( 10.1080/14712598.2018.1492544). [DOI] [PubMed] [Google Scholar]
- 16.Kohlschutter A, Schulz A, Bartsch U, Storch S, Current and Emerging Treatment Strategies for Neuronal Ceroid Lipofuscinoses. CNS Drugs 33, 315–325 (2019); published online EpubApr ( 10.1007/s40263-019-00620-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson TB, Cain JT, White KA, Ramirez-Montealegre D, Pearce DA, Weimer JM, Therapeutic landscape for Batten disease: current treatments and future prospects. Nat Rev Neurol 15, 161–178 (2019); published online EpubMar ( 10.1038/s41582-019-0138-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mole SE, Anderson G, Band HA, Berkovic SF, Cooper JD, Kleine Holthaus SM, McKay TR, Medina DL, Rahim AA, Schulz A, Smith AJ, Clinical challenges and future therapeutic approaches for neuronal ceroid lipofuscinosis. Lancet Neurol 18, 107–116 (2019); published online EpubJan ( 10.1016/S1474-4422(18)30368-5). [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg JB, Chen A, Kaminksy SM, Crystal RG, Sondhi D, Advances in the treatment of neuronal ceroid lipofuscinosis. Expert Opin Orphan Drugs, 473–500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray SJ, Gene therapy and neurodevelopmental disorders. Neuropharmacology 68, 136–142 (2013); published online EpubMay ( 10.1016/j.neuropharm.2012.06.024). [DOI] [PubMed] [Google Scholar]
- 21.Weinberg MS, Samulski RJ, McCown TJ, Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology 69, 82–88 (2013); published online EpubJun ( 10.1016/j.neuropharm.2012.03.004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hocquemiller M, Giersch L, Audrain M, Parker S, Cartier N, Adeno-Associated Virus-Based Gene Therapy for CNS Diseases. Hum Gene Ther 27, 478–496 (2016); published online EpubJul ( 10.1089/hum.2016.087). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murlidharan G, Samulski RJ, Asokan A, in Translational Neuroscience fundamental approaches for neurological disorders, Tuszynski MH, Ed. (Springer, Boston, MA, 2016), chap. 2, pp. 24. [Google Scholar]
- 24.Deverman BE, Ravina BM, Bankiewicz KS, Paul SM, Sah DWY, Gene therapy for neurological disorders: progress and prospects. Nat Rev Drug Discov 17, 641–659 (2018); published online EpubSep ( 10.1038/nrd.2018.110). [DOI] [PubMed] [Google Scholar]
- 25.Hudry E, Vandenberghe LH, Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron 101, 839–862 (2019); published online EpubMar 6 ( 10.1016/j.neuron.2019.02.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sleat DE, Gin RM, Sohar I, Wisniewski K, Sklower-Brooks S, Pullarkat RK, Palmer DN, Lerner TJ, Boustany RM, Uldall P, Siakotos AN, Donnelly RJ, Lobel P, Mutational analysis of the defective protease in classic late-infantile neuronal ceroid lipofuscinosis, a neurodegenerative lysosomal storage disorder. Am J Hum Genet 64, 1511–1523 (1999); published online EpubJun ( 10.1086/302427). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neufeld EF, Fratantoni JC, Inborn errors of mucopolysaccharide metabolism. Science 169, 141–146 (1970); published online EpubJul 10 ( 10.1126/science.169.3941.141). [DOI] [PubMed] [Google Scholar]
- 28.Sondhi D, Crystal RG, Kaminksy SM, in Translational Neuroscience fundamental approaches for neurological disorders, Tuszynski MH, Ed. (Springer, Boston, MA, 2016), chap. 7, pp. 19. [Google Scholar]
- 29.Platt FM, d’Azzo A, Davidson BL, Neufeld EF, Tifft CJ, Lysosomal storage diseases. Nat Rev Dis Primers 4, 27 (2018); published online EpubOct 1 ( 10.1038/s41572-018-0025-4). [DOI] [PubMed] [Google Scholar]
- 30.Markham A, Cerliponase Alfa: First Global Approval. Drugs 77, 1247–1249 (2017); published online EpubJul ( 10.1007/s40265-017-0771-8). [DOI] [PubMed] [Google Scholar]
- 31.Schulz A, Ajayi T, Specchio N, de Los Reyes E, Gissen P, Ballon D, Dyke JP, Cahan H, Slasor P, Jacoby D, Kohlschutter A, Group CLNS, Study of Intraventricular Cerliponase Alfa for CLN2 Disease. N Engl J Med 378, 1898–1907 (2018); published online EpubMay 17 ( 10.1056/NEJMoa1712649). [DOI] [PubMed] [Google Scholar]
- 32.Schulz A, Specchio N, Gissen P, De Los Reyes E, Cahan H, Slasor P, Jacoby D, Ajayi T, Persistent treatment effect of cerliponase alfa in children with CLN2 disease: A 3 year update from an ongoing multicenter extension study. Neuropediatrics 50, S1–S55 (2019). [Google Scholar]
- 33.Sondhi D, Hackett NR, Peterson DA, Stratton J, Baad M, Travis KM, Wilson JM, Crystal RG, Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector. Mol Ther 15, 481–491 (2007); published online EpubMar ( 10.1038/sj.mt.6300049). [DOI] [PubMed] [Google Scholar]
- 34.Sondhi D, Johnson L, Purpura K, Monette S, Souweidane MM, Kaplitt MG, Kosofsky B, Yohay K, Ballon D, Dyke J, Kaminksy SM, Hackett NR, Crystal RG, Long-term expression and safety of administration of AAVrh.10hCLN2 to the brain of rats and nonhuman primates for the treatment of late infantile neuronal ceroid lipofuscinosis. Hum Gene Ther Methods 23, 324–335 (2012); published online EpubOct ( 10.1089/hgtb.2012.120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondhi D, Peterson DA, Edelstein AM, del Fierro K, Hackett NR, Crystal RG, Survival advantage of neonatal CNS gene transfer for late infantile neuronal ceroid lipofuscinosis. Exp Neurol 213, 18–27 (2008); published online EpubSep ( 10.1016/j.expneurol.2008.04.022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dyke JP, Sondhi D, Voss HU, Shungu DC, Mao X, Yohay K, Worgall S, Hackett NR, Hollmann C, Yeotsas ME, Jeong AL, Van de Graaf B, Cao I, Kaminsky SM, Heier LA, Rudser KD, Souweidane MM, Kaplitt MG, Kosofsky B, Crystal RG, Ballon D, Assessment of disease severity in late infantile neuronal ceroid lipofuscinosis using multiparametric MR imaging. AJNR Am J Neuroradiol 34, 884–889 (2013); published online EpubApr ( 10.3174/ajnr.A3297). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyke JP, Sondhi D, Voss HU, Yohay K, Hollmann C, Mancenido D, Kaminsky SM, Heier LA, Rudser KD, Kosofsky B, Casey BJ, Crystal RG, Ballon D, Brain Region-Specific Degeneration with Disease Progression in Late Infantile Neuronal Ceroid Lipofuscinosis (CLN2 Disease). AJNR Am J Neuroradiol 37, 1160–1169 (2016); published online EpubJun ( 10.3174/ajnr.A4669). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovacs KD, Patel S, Orlin A, Kim K, Van Everen S, Conner T, Sondhi D, Kaminsky SM, D’Amico DJ, Crystal RG, Kiss S, Symmetric Age Association of Retinal Degeneration in Patients with CLN2-Associated Batten Disease. Ophthalmol Retina 4, 728–736 (2020); published online EpubJul ( 10.1016/j.oret.2020.01.011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlin A, Sondhi D, Witmer MT, Wessel MM, Mezey JG, Kaminsky SM, Hackett NR, Yohay K, Kosofsky B, Souweidane MM, Kaplitt MG, D’Amico DJ, Crystal RG, Kiss S, Spectrum of ocular manifestations in CLN2-associated batten (Jansky-Bielschowsky) disease correlate with advancing age and deteriorating neurological function. PLoS One 8, e73128 (2013) 10.1371/journal.pone.0073128). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zairi F, Le Rhun E, Bertrand N, Boulanger T, Taillibert S, Aboukais R, Assaker R, Chamberlain MC, Complications related to the use of an intraventricular access device for the treatment of leptomeningeal metastases from solid tumor: a single centre experience in 112 patients. J Neurooncol 124, 317–323 (2015); published online EpubSep ( 10.1007/s11060-015-1842-x). [DOI] [PubMed] [Google Scholar]
- 41.Slavc I, Cohen-Pfeffer JL, Gururangan S, Krauser J, Lim DA, Maldaun M, Schwering C, Shaywitz AJ, Westphal M, Best practices for the use of intracerebroventricular drug delivery devices. Mol Genet Metab 124, 184–188 (2018); published online EpubJul ( 10.1016/j.ymgme.2018.05.003). [DOI] [PubMed] [Google Scholar]
- 42.Gray SJ, Nagabhushan Kalburgi S, McCown TJ, Jude Samulski R, Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther 20, 450–459 (2013); published online EpubApr ( 10.1038/gt.2012.101). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samaranch L, Salegio EA, San Sebastian W, Kells AP, Foust KD, Bringas JR, Lamarre C, Forsayeth J, Kaspar BK, Bankiewicz KS, Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther 23, 382–389 (2012); published online EpubApr ( 10.1089/hum.2011.200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzaki Y, Konno A, Mukai R, Honda F, Hirato M, Yoshimoto Y, Hirai H, Transduction Profile of the Marmoset Central Nervous System Using Adeno-Associated Virus Serotype 9 Vectors. Mol Neurobiol 54, 1745–1758 (2017); published online EpubApr ( 10.1007/s12035-016-9777-6). [DOI] [PubMed] [Google Scholar]
- 45.Naidoo J, Stanek LM, Ohno K, Trewman S, Samaranch L, Hadaczek P, O’Riordan C, Sullivan J, San Sebastian W, Bringas JR, Snieckus C, Mahmoodi A, Mahmoodi A, Forsayeth J, Bankiewicz KS, Shihabuddin LS, Extensive Transduction and Enhanced Spread of a Modified AAV2 Capsid in the Non-human Primate CNS. Mol Ther 26, 2418–2430 (2018); published online EpubOct 3 ( 10.1016/j.ymthe.2018.07.008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg JB, Kaplitt MG, De BP, Chen A, Flagiello T, Salami C, Pey E, Zhao L, Ricart Arbona RJ, Monette S, Dyke JP, Ballon DJ, Kaminsky SM, Sondhi D, Petsko GA, Paul SM, Crystal RG, AAVrh.10-mediated APOE2 central nervous system gene therapy for APOE4-associated Alzheimer’s disease. Hum Gene Ther Clin Dev 29, 24–47 (2018); published online EpubMar ( 10.1089/humc.2017.231). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katz ML, Tecedor L, Chen Y, Williamson BG, Lysenko E, Wininger FA, Young WM, Johnson GC, Whiting RE, Coates JR, Davidson BL, AAV gene transfer delays disease onset in a TPP1-deficient canine model of the late infantile form of Batten disease. Sci Transl Med 7, 313ra180 (2015); published online EpubNov 11 ( 10.1126/scitranslmed.aac6191). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabrera-Salazar MA, Roskelley EM, Bu J, Hodges BL, Yew N, Dodge JC, Shihabuddin LS, Sohar I, Sleat DE, Scheule RK, Davidson BL, Cheng SH, Lobel P, Passini MA, Timing of therapeutic intervention determines functional and survival outcomes in a mouse model of late infantile batten disease. Mol Ther 15, 1782–1788 (2007); published online EpubOct ( 10.1038/sj.mt.6300249). [DOI] [PubMed] [Google Scholar]
- 49.Lin L, Lobel P, Expression and analysis of CLN2 variants in CHO cells: Q100R represents a polymorphism, and G389E and R447H represent loss-of-function mutations. Hum Mutat 18, 165 (2001); published online EpubAug ( 10.1002/humu.1170). [DOI] [PubMed] [Google Scholar]
- 50.Kousi M, Lehesjoki AE, Mole SE, Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum Mutat 33, 42–63 (2012); published online EpubJan ( 10.1002/humu.21624). [DOI] [PubMed] [Google Scholar]
- 51.Crystal RG, Sondhi D, Hackett NR, Kaminsky SM, Worgall S, Stieg P, Souweidane M, Hosain S, Heier L, Ballon D, Dinner M, Wisniewski K, Kaplitt M, Greenwald BM, Howell JD, Strybing K, Dyke J, Voss H, Clinical protocol. Administration of a replication-deficient adeno-associated virus gene transfer vector expressing the human CLN2 cDNA to the brain of children with late infantile neuronal ceroid lipofuscinosis. Hum Gene Ther 15, 1131–1154 (2004); published online EpubNov ( 10.1089/hum.2004.15.1131). [DOI] [PubMed] [Google Scholar]
- 52.Worgall S, Sondhi D, Hackett NR, Kosofsky B, Kekatpure MV, Neyzi N, Dyke JP, Ballon D, Heier L, Greenwald BM, Christos P, Mazumdar M, Souweidane MM, Kaplitt MG, Crystal RG, Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum Gene Ther 19, 463–474 (2008); published online EpubMay ( 10.1089/hum.2008.022). [DOI] [PubMed] [Google Scholar]
- 53.Souweidane MM, Fraser JF, Arkin LM, Sondhi D, Hackett NR, Kaminsky SM, Heier L, Kosofsky BE, Worgall S, Crystal RG, Kaplitt MG, Gene therapy for late infantile neuronal ceroid lipofuscinosis: neurosurgical considerations. J Neurosurg Pediatr 6, 115–122 (2010); published online EpubAug ( 10.3171/2010.4.PEDS09507). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM, Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A 99, 11854–11859 (2002); published online EpubSep 3 ( 10.1073/pnas.182412299). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cearley CN, Wolfe JH, Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther 13, 528–537 (2006); published online EpubMar ( 10.1016/j.ymthe.2005.11.015). [DOI] [PubMed] [Google Scholar]
- 56.Daly TM, Okuyama T, Vogler C, Haskins ME, Muzyczka N, Sands MS, Neonatal intramuscular injection with recombinant adeno-associated virus results in prolonged beta-glucuronidase expression in situ and correction of liver pathology in mucopolysaccharidosis type VII mice. Hum Gene Ther 10, 85–94 (1999); published online EpubJan 1 ( 10.1089/10430349950019219). [DOI] [PubMed] [Google Scholar]
- 57.Lin L, Lobel P, Production and characterization of recombinant human CLN2 protein for enzyme-replacement therapy in late infantile neuronal ceroid lipofuscinosis. Biochem J 357, 49–55 (2001); published online EpubJul 1 ( 10.1042/0264-6021:3570049). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De BP, Heguy A, Hackett NR, Ferris B, Leopold PL, Lee J, Pierre L, Gao G, Wilson JM, Crystal RG, High levels of persistent expression of alpha1-antitrypsin mediated by the nonhuman primate serotype rh.10 adeno-associated virus despite preexisting immunity to common human adeno-associated viruses. Mol Ther 13, 67–76 (2006); published online EpubJan ( 10.1016/j.ymthe.2005.09.003). [DOI] [PubMed] [Google Scholar]
- 59.Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, Conlon TJ, Boye SL, Flotte TR, Byrne BJ, Jacobson SG, Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther 19, 979–990 (2008); published online EpubOct ( 10.1089/hum.2008.107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Brady M, Smith S, Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20, 45–57 (2001); published online EpubJan ( 10.1109/42.906424). [DOI] [PubMed] [Google Scholar]
- 61.Smith SM, Fast robust automated brain extraction. Hum Brain Mapp 17, 143–155 (2002); published online EpubNov ( 10.1002/hbm.10062). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voevodskaya O, Simmons A, Nordenskjold R, Kullberg J, Ahlstrom H, Lind L, Wahlund LO, Larsson EM, Westman E, Alzheimer’s Disease Neuroimaging I, The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front Aging Neurosci 6, 264 (2014) 10.3389/fnagi.2014.00264). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinfeld R, Heim P, von Gregory H, Meyer K, Ullrich K, Goebel HH, Kohlschutter A, Late infantile neuronal ceroid lipofuscinosis: quantitative description of the clinical course in patients with CLN2 mutations. Am J Med Genet 112, 347–354 (2002); published online EpubNov 1 ( 10.1002/ajmg.10660). [DOI] [PubMed] [Google Scholar]
- 64.Wyrwich KW, Schulz A, Nickel M, Slasor P, Ajayi T, Jacoby DR, Kohlschutter A, An adapted clinical measurement tool for the key symptoms of CLN2 disease. JIEMS 6, 1–7 (2018). [Google Scholar]
- 65.Mullen EM, Mullen scales of early learning: AGS edition. (American Guidance Service, Circle Pines, MN, 1995). [Google Scholar]
- 66.Akshoomoff N, Use of the Mullen Scales of Early Learning for the assessment of young children with Autism Spectrum Disorders. Child Neuropsychol 12, 269–277 (2006); published online EpubAug ( 10.1080/09297040500473714). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raat H, Landgraf JM, Oostenbrink R, Moll HA, Essink-Bot ML, Reliability and validity of the Infant and Toddler Quality of Life Questionnaire (ITQOL) in a general population and respiratory disease sample. Qual Life Res 16, 445–460 (2007); published online EpubApr ( 10.1007/s11136-006-9134-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCullough N, Parkes J, White-Koning M, Beckung E, Colver A, Reliability and validity of the Child Health QuestionnairePF-50 for European children with cerebral palsy. J Pediatr Psychol 34, 41–50 (2009); published online EpubJan-Feb ( 10.1093/jpepsy/jsn048). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. AAVrh.10hCLN2 vector.
Figure S2. Trajectory planning for gene therapy infusions.
Figure S3. Axial T2 FLAIR (T2 FLAIR) assessment of participants post-therapy.
Figure S4. T cell responses to AAVrh.10 capsid and CLN2 transgene.
Figure S5. MRI assessment of the treated CLN2 children vs untreated CLN2 children.
Figure S6. Quantitative MRI assessment of the treated CLN2 children (cohort 1) vs untreated CLN2 children.
Figure S7. Mullen scale quantitation of the rate of decline for cohorts 1 (red, treated) and 2 (blue, control).
Figure S8. Impact of treatment on quality of life as assessed by age-dependent quality of life questionnaires.
Figure S9. Correlation of various parameters to the rate of decline of motor + language of cohort 1.
Table S1. CLN2 Disease Severity Clinical Rating Scales
Table S2. Vector Infusion Time and Operating Room Surgery and Anesthesia Duration in Cohort 1 Participants
Table S3. Vector Infusion Time and Operating Room Surgery and Anesthesia Duration in Cohort 4 Participants
Table S4. Cerebral Spinal Fluid Nucleated Cells
Table S5. Percent Volume of the Brain with MRI T2 Hyperintensity
Table S6. Quality of Life Questionnaires
Table S7. Coefficient of Variation Among Observers in the CLN2 Disease Motor + Language Neurologic Rating Scale
Table S8. Reproducibility of Motor and Language Assessment
Table S9. Assessments of Motor and Language Parameters for Cohort 2
Table S10. Inclusion/Exclusion Criteria for Cohorts 1 and 2
Table S11. Timeline of the Clinical Study
Table S12. Inclusion/Exclusion Criteria for Cohort 4