SUMMARY
The role of fatty acid synthesis in endothelial cells (ECs) remains incompletely characterized. We report that fatty acid synthase knockdown (FASNKD) in ECs impedes vessel sprouting by reducing proliferation. Endothelial loss of FASN impaired angiogenesis in vivo, while FASN blockade reduced pathological ocular neovascularization, at >10-fold lower doses than used for anti-cancer treatment. Impaired angiogenesis was not due to energy stress, redox imbalance, or palmitate depletion. Rather, FASNKD elevated malonyl-CoA levels, causing malonylation (a post-translational modification) of mTOR at lysine 1218 (K1218). mTOR K-1218 malonylation impaired mTOR complex 1 (mTORC1) kinase activity, thereby reducing phosphorylation of downstream targets (p70S6K/4EBP1). Silencing acetyl-CoA carboxylase 1 (an enzyme producing malonyl-CoA) normalized malonyl-CoA levels and reactivated mTOR in FASNKD ECs. Mutagenesis unveiled the importance of mTOR K1218 malonylation for angiogenesis. This study unveils a novel role of FASN in metabolite signaling that contributes to explaining the anti-angiogenic effect of FASN blockade.
INTRODUCTION
Fatty acid synthase (FASN) mediates de novo lipid synthesis by catalyzing the production of palmitate from acetyl-coenzyme A (CoA), malonyl-CoA, and NADPH. Palmitate is used for the synthesis of more complex fatty acids (FAs), plasma membrane structures, and post-translational protein palmitoylation (Rohrig and Schulze, 2016). In contrast to most non-transformed cells, which satisfy their demand from dietary lipids, cancer cells often synthesize up to 95% of FAs de novo despite sufficient dietary supply (Zaidi et al., 2013). Not surprisingly, lipogenesis has been implicated in cancer cell biology (Rohrig and Schulze, 2016).
Limited evidence has implicated FASN in angiogenesis. In vitro, the FASN blocker orlistat inhibits EC proliferation, associated with downregulation of vascular endothelial growth factor receptor 2 (Browne et al., 2006), but it is unknown whether the latter is cause or consequence of FASN inhibition. Constitutive FASN loss in ECs and hematopoietic cells decreases angiogenesis in ischemia (Wei et al., 2011). Nonetheless, the role of endothelial FASN loss in physiological development in vivo has not been studied, neither the therapeutic potential of FASN blockade in pathological ocular angiogenesis.
Malonyl-CoA participates in (patho-)physiological processes (Saggerson, 2008). While FASN inhibition impaired palmitoylation due to reduced palmitate synthesis (Wei et al., 2011), consequences of elevated levels of malonyl-CoA as a result of FASN inhibition in ECs were not considered. Malonyl-CoA can be non-enzymatically used for protein lysine malonylation, a recently identified post-translational modification (PTM) that can alter protein activity (Peng et al., 2011). Even though protein malonylation is evolutionarily conserved from bacteria to mammals, the functional consequences and stoichiometry of this PTM are poorly characterized (Du et al., 2015; Nishida et al., 2015), and have never been determined in a single study. Here, we explored whether FASN might regulate angiogenesis via malonylation of key targets.
RESULTS
Fatty Acid Synthesis in Endothelial Cells
We explored whether human umbilical venous ECs (referred to hereafter as ECs) synthesized palmitate de novo. To mimic in vivo physiological conditions, we used physiological concentrations of palmitate (100–120 μM), acetate (200–500 μM), and glucose (5.5 mM) and added trace amounts of 14C-labeled acetate (<0.1% of the concentration of the cold acetate). When quantifying the incorporation of labeled carbons into the FA fraction, we detected measurable levels of FA synthesis in ECs, despite available palmitate in the medium (Figures 1A and 1B).
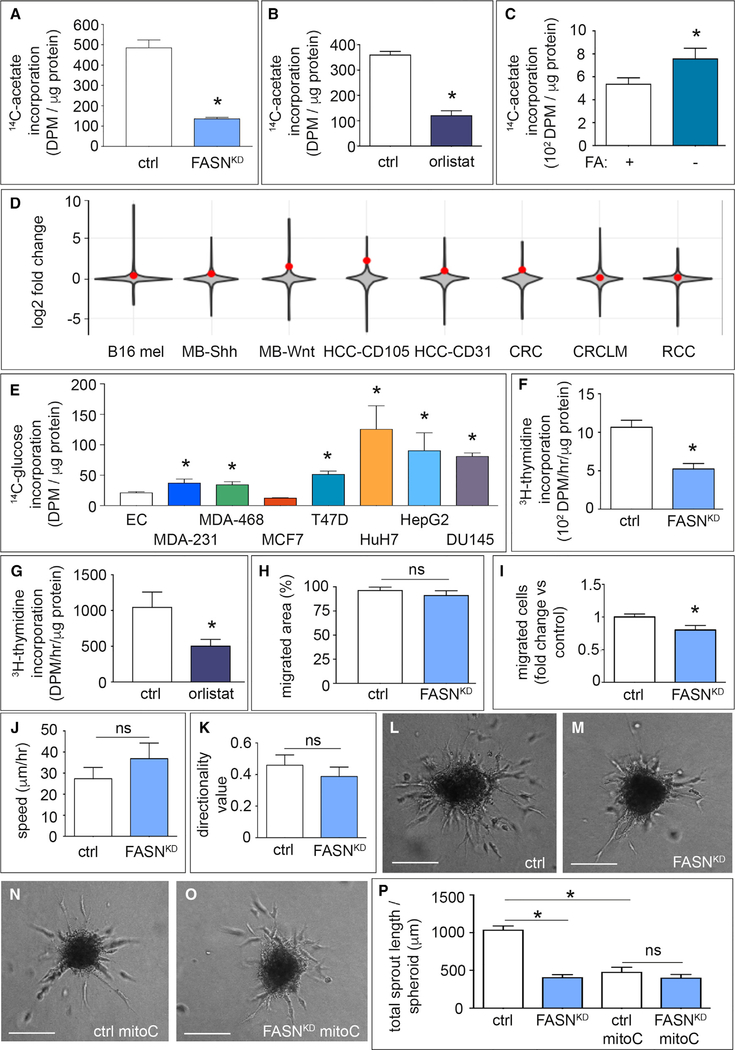
Figure 1. Role of Endothelial Fatty Acid Synthase in Vessel Sprouting.
(A and B) Incorporation of carbons from [U-14C]-acetate in the fatty acid (FA) fraction of control (ctrl) and FASNKD ECs (n = 3) (A) or control and orlistat-treated (10 μM) ECs (n = 4) (B). DPM, disintegrations per minute.
(C) Incorporation of carbons from [U-14C]-acetate in the FA fraction of ECs in the presence or absence of FAs in the medium (n = 3). DPM, disintegrations per minute.
(D) Violin plots visualizing the log2 fold-change distribution in gene expression (gray area) in murine and human tumor ECs versus their counterpart normal healthy ECs. The red dot indicates where FASN is located in the distribution. Data are based on a meta-analysis of publicly available transcriptome datasets of murine B16 melanoma (B16 mel); murine medulloblastoma (MB-Shh, MB-Wnt); human hepatocellular carcinoma (HCC-CD105, HCC-CD31); human colorectal cancer (CRC, CRCLM) and human renal cancer (RCC). p = 1.0 × 10−20 (Fisher’s method).
(E) Incorporation of carbons from [U-14C]-glucose in the FA fraction of ECs (EC), breast cancer cell lines MDA-MB-231 (MDA-231), MDA-MB-468 (MDA-468), T47D, and MCF7, hepatocellular carcinoma cells HuH7 and HepG2, and prostate cancer cells DU145 in their optimal growth medium (n = 3). DPM, disintegrations per minute.
(F and G) [3H]-Thymidine incorporation into DNA (proliferation assay) in control and FASNKD ECs (n = 4) (F), and in control or orlistat-treated ECs (n = 5) (G). DPM, disintegrations per minute.
(H) Scratch wound migration assay using mitotically inactivated (mitoC) control or FASNKD ECs (n = 3).
(I) Number of mitoC-treated control and FASNKD ECs that traversed the membrane in a Boyden chamber (expressed relative to control; n = 3).
(J and K) Analysis of random cell-motility tracks obtained by time-lapse imaging of GFP+ ECs (transduced with a lentiviral vector expressing GFP), for speed (J) or directional persistence of EC movement (K) in control and FASNKD ECs (n = 4).
(L–O) Representative phase-contrast images of EC spheroids of proliferating (L and M) and mitotically inactivated ECs (mitoC) (N and O) using control (L and N) or FASNKD ECs (M and O). Scale bars, 200 μm.
(P) Total sprout length of control and FASNKD EC spheroids with and without mitotic inactivation (mitoC) (n = 3).
Data are mean ± SEM. *p < 0.05 by Fisher’s combined probability test (D) and mixed models statistics for all other panels. ns, not significant. See also Figure S1.
To assess whether this signal was due to FASN activity, we silenced its expression in ECs using lentiviral vectors encoding two non-overlapping FASN-specific short hairpin RNAs (shRNAs) (shFASN) and non-overlapping RNAi oligomers, which lowered FASN levels (Figures S1A–S1F). FASN knockdown (FASNKD) or use of the FASN blocker orlistat reduced 14C label incorporation into FAs in ECs (Figures 1A and 1B), similarly as in other cell types (Kridel et al., 2004). Notably, orlistat inhibited FASN in ECs at concentrations 10- to 20-fold lower than those used to inhibit FASN in cancer cells (Yang et al., 2015). When using FA-depleted medium, ECs upregulated FASN protein (Figure S1G) and FA synthesis levels (Figure 1C). We reanalyzed in-house transcriptomics data of murine tumor ECs (TECS) versus normal ECs (NECs) (Cantelmo et al., 2016), as well as published transcriptomics studies of murine and human TECs versus NECs (Phoenix et al., 2016; Roudnicky et al., 2013; Wragg et al., 2016). This meta-analysis revealed that FASN was overexpressed in TECs (Figure 1D; p = 1.0 × 10−20). Compared with cancer cells, known to express FASN (Hou et al., 2008; Hunt et al., 2007), ECs had lower FASN protein levels (Figure S1H). Incorporation of 14C label from [U-14C]-glucose into FAs was lower in ECs than in cancer cells, except for MCF7 cells (Figure 1E).
FASN Promotes Vessel Sprouting In Vitro
FASNKD or a low concentration of orlistat (10 μM) decreased EC proliferation (Figures 1F, 1G, and S1I–S1L). As the outcome of EC migration assays is influenced by EC proliferation, we analyzed the migratory behavior of ECs upon mitotic inactivation by mitomycin C (mitoC) (De Bock et al., 2013). FASNKD did not impair migration of mitoC-treated ECs, as measured in the scratch wound assay (Figure 1H) and only minimally reduced EC motility in the Boyden chamber assay (Figure 1I). Analysis of the velocity and directionality of spontaneous migration confirmed that FASNKD did not reduce EC motility (Figures 1J and 1K). Using the EC spheroid model (Schoors et al., 2015), FASNKD or orlistat reduced EC sprouting (Figures 1L–1P and S1M–S1P). MitoC impaired vessel sprouting, but FASNKD was unable to further reduce sprouting upon mitoC treatment, showing that FASNKD impaired vessel sprouting primarily by reducing EC proliferation, not migration (Figures 1N–1P and S1M).
FASN Gene Deletion in ECs Impairs Physiological Angiogenesis In vivo
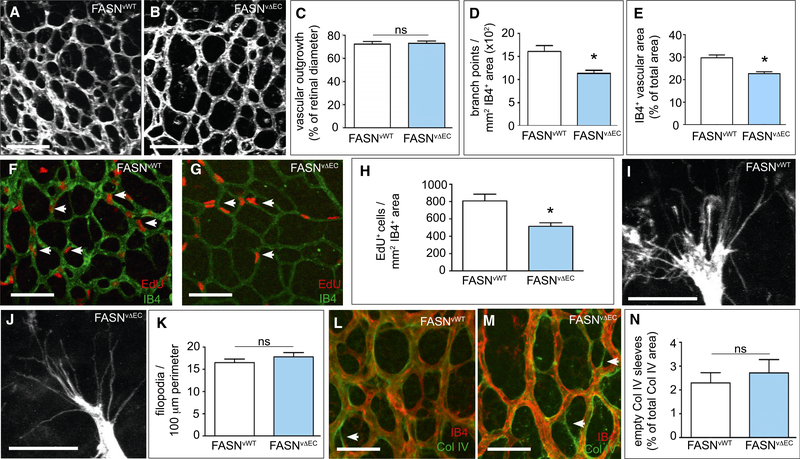
To study the in vivo relevance of endothelial FASN deficiency, we crossed FASNlox/lox mice (Wei et al., 2011) with VE-cadherin(PAC)-CreERT2 mice, a tamoxifen-inducible EC-specific Cre-driver line (Benedito et al., 2009). Treatment of double-transgenic pups with tamoxifen at postnatal day 1 (P1) to P3 yielded viable FASNΔvEC mice, in which FASN was deleted in ECs (Figures S2A and S2B). Staining of retinal vessels with isolectin-B4 (IB4) revealed that EC loss of FASN reduced the number of vascular branch points and total vascular area (Figures 2A–2E). The vascular defects in FASNΔvEC mice were due to reduced EC proliferation (Figures 2F–2H), and FASNΔvEC mice had a normal number of filopodia per 100-μm vascular front perimeter (Figures 2I–2K). The vascular defect in FASNΔvEC mice was not due to vessel regression, as staining for IB4 and collagen IV did not reveal a change in the fraction of empty sleeves (Figures 2L–2N). Vessel maturation was normal in FASNΔvEC mice (Figures S2C–S2E). Similar effects were seen in conditional FASNΔEC mice when a tamoxifen-inducible platelet-derived growth factor Cre-recombinase line was used as EC-specific driver line (FASNΔpEC) (Figures S2F–S2Q). Thus, impaired angiogenesis in FASNΔpEC mice was also due to EC proliferation defects.
Figure 2. Effect of FASN Gene Deletion in ECs on Physiological Angiogenesis In vivo.
(A and B) Representative confocal images of isolectin-B4 (IB4)-stained retinal vessels of neonatal mice at postnatal day 5 (P5), showing reduced vascular branching upon EC-specific (VE-Cadherin-Cre) deletion of FASN (FASNvΔEC) (B) as compared with wild-type (FASNvWT) (A) pups. Scale bars, 100 μm.
(C) Quantification of retinal vascular outgrowth (radial expansion in percent of retinal diameter) in FASNvWT (n = 17) and FASNvΔEC (n = 20) mice.
(D) Number of branch points in the retinal vasculature of FASNvWT (n = 17) and FASNvΔEC (n = 20) mice.
(E) IB4+ retinal vascular area (expressed as percent of total area) in FASNvWT (n = 7) and FASNvΔEC mice (n = 7).
(F and G) Representative confocal images of the vascular front in the retinal vasculature stained for 5-ethynyl-2′-deoxyuridine (EdU) (red) and isolectin-B4 (IB4) (green) in FASNvWT (F) and FASNvΔEC (G) mice. Arrows denote EdU+ ECs. Scale bars, 100 μm.
(H) Number of proliferating EdU+ ECs in the retinal vasculature of FASNvWT (n = 9) and FASNvΔEC (n = 7) mice.
(I and J) Representative high-magnification confocal images of ECs at the retinal vascular front (isolectin-B4 staining) in FASNvWT (I) and FASNvΔEC (J) pups. Scale bars. 25 μm.
(K) Number of filopodia per 100-μm perimeter of the retinal vascular front in FASNvWT (n = 14) and FASNvΔEC (n = 16) pups.
(L and M) Representative confocal images of the retinal vasculature stained for isolectin-B4 (red) and the basement membrane marker collagen IV (green) in FASNvWT (L) and FASNvΔEC (M) mice. Arrows denote isolectin-B4− collagen IV+ empty sleeves (green vascular profiles consisting only of green basement membrane without red ECs). Scale bars, 50 μm.
(N) Percentage of isolectin-B4− collagen IV+ empty sleeves compared with the total collagen IV+ vascular area in FASNvWT (n = 7) and FASNvΔEC (n = 9) mice.
Data are mean ± SEM. *p < 0.05 by standard two-tailed t test; ns, not significant. See also Figure S2.
Effects of FASN Silencing on EC Metabolism
FASNKD did not affect glycolysis, glucose, or glutamine oxidation (Figures 3A–3C), but reduced FA oxidation (FAO), though by no more than 30% (Figure 3D), consistent with the fact that malonyl-CoA inhibits carnitine palmitoyltransferase 1A (CPT1A, a rate-controlling enzyme of FAO) and malonyl-CoA levels were elevated in FASNKD cells (see below). FASNKD did not affect label incorporation from [U-13C]-glucose or [U-13C]-glutamine into tricarboxylic acid (TCA) cycle intermediates, but slightly reduced label incorporation from [U-13C]-palmitate, in line with the reduced FAO (Figures S3A–S3C).
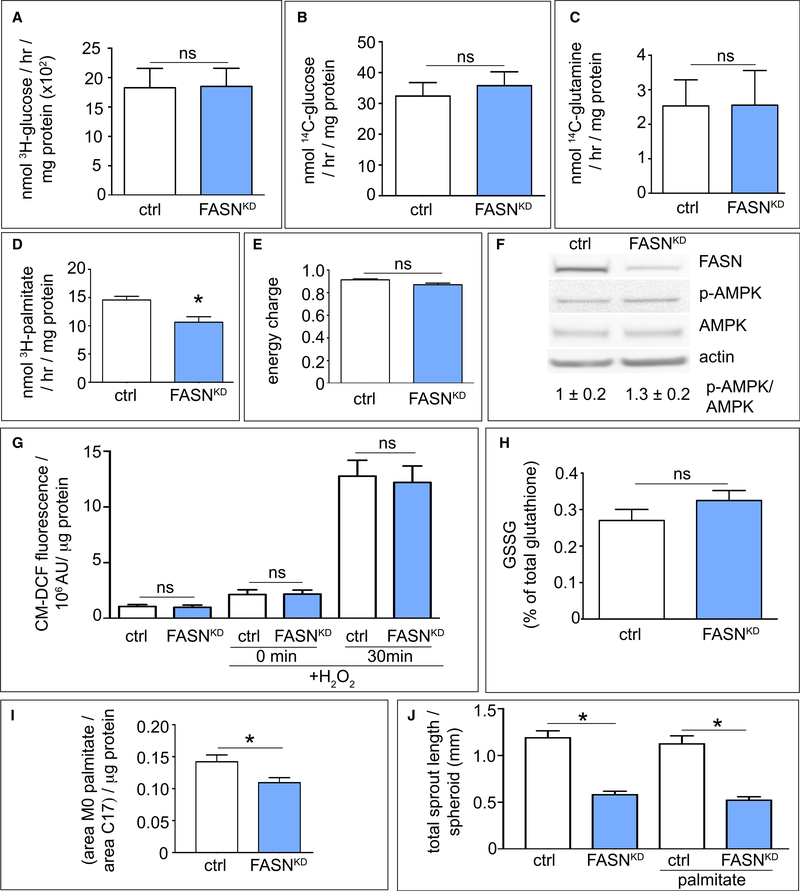
Figure 3. Effect of FASN Silencing on EC Metabolism.
(A–D) Effect of FASN silencing (FASNKD) in ECs on glycolysis (A), glucose oxidation (B), glutamine oxidation (C), or fatty acid oxidation (D) (A–C: n = 3, D: n = 4). (E and F) Effect of FASN silencing (FASNKD) in ECs on energy charge (([ATP] + 1/2 [ADP])/([ATP] + [ADP] + [AMP])) (E), and on AMPK phosphorylation as verified by immunoblotting (F) (n = 3). Actin was used as loading control in (F). Densitometric quantification of the p-AMPK/AMPK ratio, expressed relative to control, is shown beneath the blot.
(G) Intracellular ROS levels (CM-DCF fluorescence) in control and FASNKD ECs in baseline and upon supplementation with 50 μM H2O2 for 30 min (n = 3). AU, arbitrary units.
(H) Oxidized glutathione levels (percent of GSH + GSSG) in control and FASNKD ECs (n = 3).
(I) Quantification of intracellular palmitate levels (normalized to the standard C17 peak area) in control and FASNKD ECs (n = 3).
(J) Quantification of total sprout length of control (ctrl) and FASNKD spheroids grown in medium without or with supplementation with palmitate (50 μM) (n = 3). Data are mean ± SEM. *p < 0.05 by standard two-tailed t test with Welch’s correction (A–F and H–I) or ANOVA (G and J); ns, not significant. See also Figures S3 and S4.
FASN silencing or blockade did not cause energy distress, as determined by measuring the energy charge or p-AMPK levels (Figures 3E, 3F, and S3D). This is not surprising, since FAO contributes only minimally (<5%) to the total ATP production in ECs, which relies primarily on glycolysis (De Bock et al., 2013), and FAO was reduced only moderately. Measurements of cellular reactive oxygen species (ROS) levels in baseline and upon challenge with exogenous H2O2 (Figure 3G), in combination with quantification of oxidized glutathione (GSSG) levels (Figure 3H), revealed that FASNKD did not cause redox imbalance or the ability of ECs to cope with exogenous oxidative stress. Even though FAO supports EC proliferation by providing acetyl-CoA to sustain the TCA cycle for dNTP synthesis in conjunction with an anaplerotic carbon source (Schoors et al., 2015), the modest reduction of FAO in FASNKD cells did not likely explain their proliferation defect (Figures S3E–S3N).
FASN Silencing Moderately Affects Palmitate Levels in ECs
Impairment of proliferation and viability of cancer cells upon inhibition of FASN or acetyl-CoA carboxylase (ACC) has been attributed to depletion of cellular palmitate pools by 50%–97%, sufficient to cause ER stress and apoptosis (Ventura et al., 2015). FASNKD lowered palmitate levels only by ±20% in ECs (Figure 3I), likely because ECs can take up FAs from the medium or recycle membrane lipids (Rohrig and Schulze, 2016). Import of [U-14C]-palmitate was not compensatorily increased by FASNKD (Figure S4A) or orlistat, in contrast to sulfo-N-succinimidyl oleate (SSO), an inhibitor of FA translocase CD36 (Kuda et al., 2013) (Figure S4B), suggesting that the palmitate uptake capacity of ECs is sufficient to maintain cellular palmitate pools. Consistent with reduced de novo palmitate synthesis, the fraction of monounsaturated fatty acyl chains was moderately decreased in FASNKD ECs (Figures S4C and S4D).
The reduced pool of palmitate in FASNKD ECs was insufficient to cause ER stress and cell death (Figures S4E–S4I and S5A). Unlike cancer cells (Svensson et al., 2016), supplementation of palmitate, elevating cellular palmitate pools in FASNKD ECs even slightly above the levels in control cells (Figure S5B), did not rescue the FASNKD sprouting defect (Figure 3J). Thus, the sprouting defect of FASNKD ECs was not due to the depletion of cellular palmitate pools.
FASN Silencing Increases Malonyl-CoA Levels
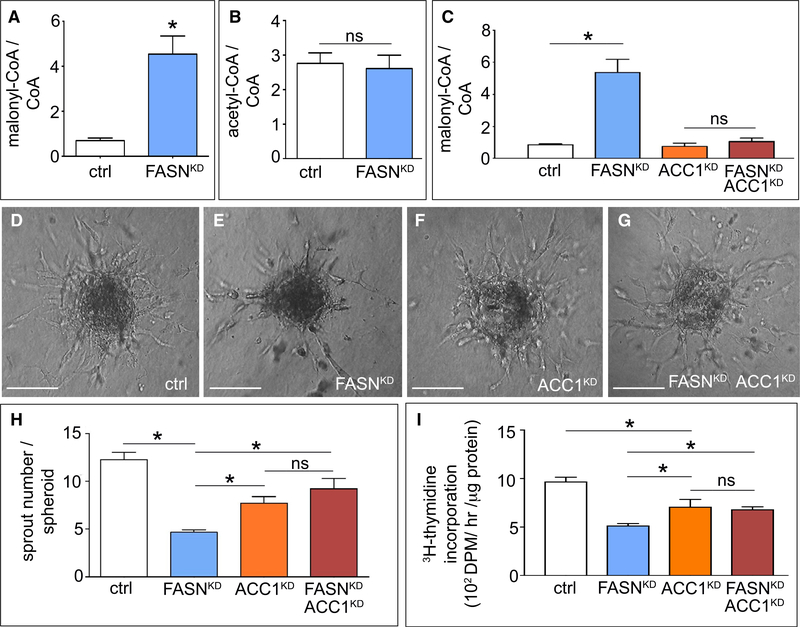
We then explored whether FASNKD enhanced the accumulation of its substrate malonyl-CoA. Lipogenesis involves conversion of acetyl-CoA to malonyl-CoA by ACC, and the use of malonyl-CoA and acetyl-CoA by FASN to synthesize palmitate. FASNKD or orlistat substantially elevated malonyl-CoA levels, without affecting acetyl-CoA levels (Figures 4A, 4B, and S5C). To explore whether the elevated malonyl-CoA levels contributed to the sprouting defects, we silenced the expression of acetyl-CoA carboxylase 1 (ACC1), which is expressed more abundantly than its isoenzyme ACC2 in ECs (Figure S5D). ACC1 knockdown (ACC1KD; lowering ACC1 mRNA levels by 83%) alone did not affect malonyl-CoA levels, but when both FASN and ACC1 were silenced, malonyl-CoA levels were no longer increased (Figure 4C). Silencing of malonyl-CoA decarboxylase (catalyzing the reverse reaction of ACC1) and of SIRT5 (a de-malonylase [Nishida et al., 2015]) did not induce changes in protein malonylation in ECs (not shown).
Figure 4. Effect of FASN Silencing on Malonyl-CoA Levels.
(A) Malonyl-CoA/CoA ratio in control and FASNKD ECs (n = 4).
(B) Acetyl-CoA/CoA ratio in control and FASNKD ECs (n = 4).
(C) Malonyl-CoA/CoA levels in ECs upon single or combined knockdown of ACC1 (ACC1KD) and FASN (FASNKD) (n = 3).
(D–G) Representative phase-contrast images of EC spheroids using control ECs (D) or ECs with single (E and F) or combined (G) silencing of ACC1 (ACC1KD) and FASN (FASNKD). Scale bars, 150 μm.
(H) Sprout number of EC spheroids as in (D) to (G) (n = 4).
(I) [3H]-Thymidine incorporation into DNA (proliferation assay) in control ECs and ECs with single or combined knockdown of ACC1 (ACC1KD) and FASN (FASNKD) (n = 4). DPM, disintegrations per minute.
Data are mean ± SEM. *p < 0.05 by ANOVA (C, H, and I) or standard two-tailed t test with Welch’s correction (A and B); ns, not significant. See also Figures S4 and S5.
ACC1KD alone reduced vessel sprouting and EC proliferation (Figures 4D, 4F, 4H, and 4I) (a more prominent anti-mitogenic effect was reported for cancer cells [Svensson et al., 2016]). More importantly, however, when both FASN and ACC1 were silenced together, EC proliferation and vessel sprouting were similar relative to ACC1KD cells, showing that the anti-proliferative effect of FASNKD was abrogated by ACC1KD (Figures 4D–4I and S5E). Thus, FASNKD impaired sprouting and EC proliferation partly by elevating malonyl-CoA levels.
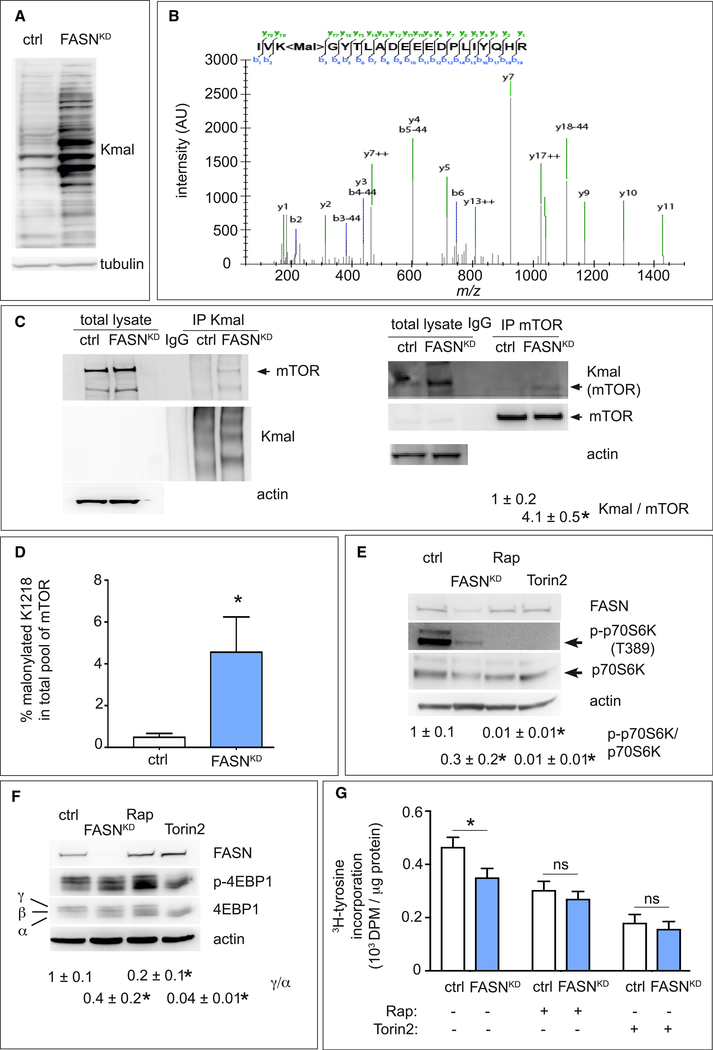
FASN Silencing Induces Malonylation of mTOR
We explored whether the increased malonyl-CoA levels influenced protein malonylation, a poorly studied non-enzymatic PTM driven by substrate levels (Peng et al., 2011). Immunoblotting of lysates for malonylated lysine residues (Kmal) revealed that FASNKD or orlistat elevated general Kmal levels in ECs (Figures 5A and S5F). To identify malonylated proteins, we performed a proteomics screen using an anti-Kmal antibody to enrich Kmal peptides and analyzed Kmal peptides as reported by Colak et al. (2015). We identified nearly 100 Kmal sites in 64 different proteins, of which 62 were reported in other malonylome screens, validating our approach (Table S1). Among the proteins with Kmal sites, we identified targets involved in pathways previously shown to be modified by lysine malonylation, including glycolysis, pentose phosphate pathway, and cytoskeleton remodeling (Table S1) (Nishida et al., 2015).
Figure 5. Effect of FASN Silencing on mTOR Malonylation and Activity.
(A) Representative immunoblot of lysine malonylated (Kmal) proteins in control and FASNKD ECs. Tubulin was used as loading control.
(B) MS/MS validation of the mTOR K1218 malonylation site. The fragmentation spectrum of the tryptic mTOR peptide H2N-1216IVK<mal>COOH (825.7395 m/z, 3+) carrying the modification is shown with annotated y-ions (green) and b-ions (blue). The b-ion series covers the malonylation site and displays the typical neutral loss of 44 Da (corresponding to the loss of CO2).
(C) Representative western blots of immunoprecipitated (IP) endogenous malonylated proteins (Kmal; left blot) or mTOR (right blot) and immunoblotted for the reciprocal proteins in control and FASNKD ECs. Actin was used as loading control. Arrowheads indicate malonylated (Kmal) or total mTOR band. Densitometric analysis of the Kmal mTOR to total mTOR ratio, expressed relative to control, is shown beneath the blots (n = 3).
(D) Stoichiometry of mTOR-K1218 malonylation determined by novel LC-MS/MS methods, in control and FASNKD ECs (n = 4).
(E) Representative immunoblot of total (p70S6K) and phosphorylated (p-p70S6K threonine 389 [T389]) p70S6K in control (ctrl) and FASNKD ECs. Actin was used as loading control. The mTORC1 inhibitor rapamycin (Rap) (20 nM) and mTORC1/2 inhibitor Torin2 (100 nM) were used as positive controls. Densitometric quantification of the p-p70S6K/p70S6K ratio, expressed relative to ctrl, is shown beneath the blots (n = 3).
(F) Representative immunoblot of total 4EBP1 and phosphorylated p-4EBP1 in control and FASNKD ECs. Actin was used as loading control (ctrl). The mTORC1 inhibitor rapamycin (Rap) (20 nM) and mTORC1/2 inhibitor Torin2 (100 nM) were used as positive controls. Densitometric analysis of the ratio between the high phosphorylated γ over low phosphorylated α form of p-4EBP1, expressed relative to ctrl, is shown beneath the blots (n = 3).
(G) [3H]-Tyrosine incorporation (protein synthesis) in control (ctrl) and FASNKD ECs, treated with control vehicle, or 20 nM rapamycin (Rap) or 100 nM Torin2 (n = 4). DPM, disintegrations per minute.
Data are mean ± SEM. *p < 0.05 by ANOVA (G) or standard two-tailed t test with Welch’s correction (C–F); ns, not significant. See also Figures S5 and S6; Tables S1 and S2.
Among the identified Kmal proteins, we focused on the mechanistic target of rapamycin (mTOR), a serine/threonine kinase activated by anabolic signals (Saxton and Sabatini, 2017), as it controls lipid synthesis, cell growth, metabolism, and other cell processes in response to nutrition (Saxton and Sabatini, 2017). A report that mTOR complex 1 (mTORC1) blockade by rapamycin inhibits ocular angiogenesis (Yagasaki et al., 2014b) primed our interest to focus on this target. The mTOR kinase nucleates two protein complexes named mTORC1 and mTORC2. The proteomic analysis identified only one malonylated lysine residue in mTOR (K1218), showing a characteristic tandem mass spectrometry (MS/MS) spectrum including C-terminal y-ions and amino-terminal b-ions (Table S1 and Figure 5B).
Immunoprecipitation of endogenous mTOR followed by immunoblotting for Kmal (also in the reverse order for FASNKD cells) confirmed that mTOR was malonylated, and that malonylated mTOR levels were increased in FASNKD or orlistat-treated ECs (Figures 5C and S5G). The weak immunoblot signals are attributable to the fact that only one lysine is malonylated in mTOR and, possibly, that only a fraction of the endogenous mTOR pool in ECs is malonylated. Indeed, when generating a tryptic peptide containing a stably malonylated K1218 of mTOR to measure the malonylation stoichiometry via MS, we found 4.5% of the mTOR pool to be malonylated in FASNKD ECs (Figure 5D).
Malonylation of mTOR Reduces mTORC1 Activity
To assess whether mTOR malonylation affected the activity of its complexes, we analyzed phosphorylation of the downstream targets of mTORC1 (eukaryotic translation initiation factor 4E-binding protein 1 [4EBP1]; p70 S6 kinase [p70S6K]) and mTORC2 (Akt), using the mTORC1 inhibitor rapamycin and the mTORC1/mTORC2 inhibitor torin2 as positive controls (Saxton and Sabatini, 2017). FASNKD (inducing mTOR malonylation) reduced mTORC1’s activity to phosphorylate p70S6K and 4EBP1 (Figures 5E and 5F). In line with reports that 4EBP and p70S6K regulate protein synthesis downstream of mTORC1 (Saxton and Sabatini, 2017), de novo protein synthesis was reduced upon FASNKD (Figure 5G). Treatment of control cells with rapamycin or torin2 reduced protein synthesis, which could not be further decreased by FASNKD (Figure 5G). In agreement with findings that p70S6K regulates cell size (Saxton and Sabatini, 2017), FASNKD reduced EC size (Figure S5H).
In contrast, FASNKD lowered phosphorylation of mTORC2’s target Akt (Figure S6A), but less prominently than of mTORC1 targets, and not consistently across different assays (see below). Possibly, the effect by FASNKD on p-Akt relates to the reduced confluence of these cells (cell-cell contacts activate Akt [Taddei et al., 2008]). Though not extensively documented, certain PTMs of mTOR correlate with changes in the activity of mTORC1, not of mTORC2 (Rosner et al., 2010). While remaining unexplained, the more selective regulation of mTORC1, rather than mTORC2, pathway activity by mTOR malonylation may thus not be surprising. Additional ACC1KD in FASNKD ECs normalized the malonylated mTOR levels to those in control cells and restored mTORC1’s activity (Figures S6B–S6D); FASN overexpression yielded similar results (Figures S6E and S6F).
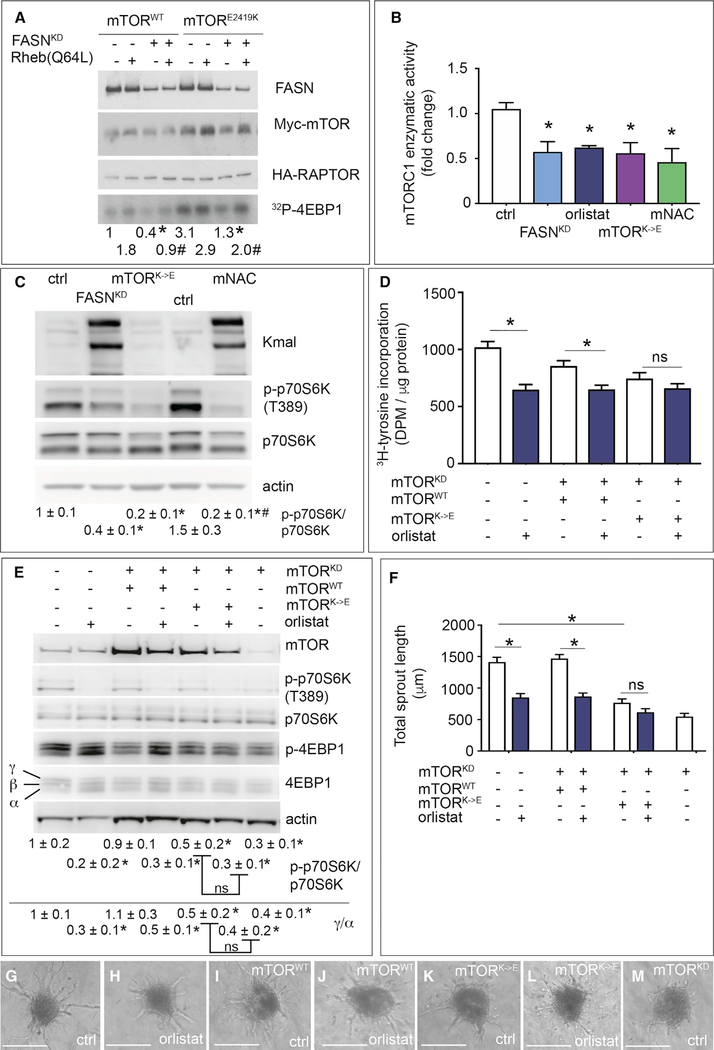
Malonylation of mTOR upon FASNKD reduced the enzymatic activity of mTORC1, in two complementary assays (radioactive phosphor donor [Dunlop et al., 2009]; commercial ELISA-based kit) (Figures 6A and 6B), without, however, affecting its stability or intracellular localization (Figures S6G, S6H, and S7A). FASN inhibition by orlistat yielded largely similar results (Figure 6B), although phosphorylation of Akt was unchanged (see below).
Figure 6. Role of Lysine 1218 in the Regulation of mTORC1 Activity upon FASN Blockade.
(A) Enzymatic mTORC1 kinase assay measuring phosphorylation of recombinant 4EBP1 ([32P]-4EBP1) in HEK293T cells overexpressing myc-mTOR and HA-RAPTOR. Top: immunoblotting for FASN of total lysates of control or FASN-silenced (FASNKD) HEK293T cells overexpressing either myc-tagged wild-type mTOR (mTORWT) (first 4 columns) or a myc-tagged highly active E2419K mutant of mTOR (last 4 columns), with or without addition in the kinase reaction mixture of recombinant Rheb Q64L, a highly active mutant of the mTORC1 activator Rheb. Middle (rows 2 and 3): immunoblotting for myc-mTOR (row 2) or HA-RAPTOR (row 3) of lysates of the above listed cells after immunoprecipitation of the mTORC1 complex using anti-myc antibodies. Immunoblotting for myc-mTOR is shown to confirm mTOR overexpression; immunoblotting for HA-raptor is shown as loading control for the kinase assay. Bottom: representative autoradiograms of [32P]-4EBP1. Densitometric quantification of the [32P]-4EBP1 bands, expressed as fold change relative to control (column 1) is shown beneath the autoradiogram. The more active E2419K mutant of mTOR was overexpressed to enhance the AR signal (last 4 lanes).
(B) mTORC1 enzymatic activity (K-LISA mTOR activity kit [Millipore]) in control (ctrl), FASN-silenced (FASNKD), orlistat-treated ECs, ECs expressing mTOR harboring a K1218E mutation (mTORK→E, malonylation mimicking), and ECs treated with malonyl-NAC (mNAC, a cell-permeable malonyl-CoA mimic) (n = 3). Values were normalized to protein input and are expressed as fold change relative to control.
(C) Representative immunoblot of Kmal, total and phosphorylated p70S6K, and actin as loading control, in control (ctrl) and FASN-silenced (FASNKD) ECs, ECs expressing mTOR harboring a K1218E mutation (mTORK→E, malonylation mimicking), and ECs treated with malonyl-NAC (mNAC, a cell-permeable malonyl-CoA mimic). Densitometric quantification of the p-p70S6K/p70S6K ratio, expressed relative to control, is shown beneath the blot (n = 3).
(D) [3H]-Tyrosine incorporation in control and orlistat (10 μM)-treated ECs, silenced for endogenous mTOR (mTORKD) as indicated and expressing a wild-type mTOR (mTORWT) or mTOR harboring a K1218E (mTORK→E) mutation (both resistant to the mTOR shRNA) (n = 4). DPM, disintegrations per minute.
(E) Representative immunoblot of total and phosphorylated levels of the mTORC1 targets p70S6K and 4EBP1 in control ECs and ECs silenced for endogenous mTOR (mTORKD) with or without overexpression of wild-type mTOR (mTORWT) or mTOR harboring a K1218E (mTORK→E) mutation (both resistant to the mTOR shRNA) and with or without FASN inhibition (orlistat). Densitometric quantification of the p-p70S6K/p70S6K ratio or of the ratio between the high phosphorylated γ over low phosphorylated α form of 4EBP1 (expressed relative to ctrl ECs) is shown beneath the blots (n = 3).
(F) Quantification of vessel sprouting from spheroids shown in (G) to (M) (n = 3).
(G–M) Representative phase-contrast images of control (ctrl) and orlistat-treated EC spheroids using wild-type ECs (G and H), or ECs silenced for endogenous mTOR (mTORKD) and expressing mTOR WT (mTORWT) (I and J) or mTOR K1218 mutant (mTORK→E) (both resistant to the mTOR shRNA) (K and L). (M) shows a representative spheroid using mTORKD ECs. Scale bars, 200 μm.
Data are mean ± SEM. In (A), *p < 0.05, #p < 0.05 for FASNKD versus its corresponding FASN wild-type in each condition. In (B), (D), and (F), *p < 0.05 by ANOVA. In (E), *p < 0.05 for orlistat versus its corresponding control, or for mTORKD versus control (column 1), by standard two-tailed t test with Welch’s correction. In (C), *p < 0.05 versus its respective control and #p < 0.05 for mNAC versus FASNKD, determined by ANOVA followed by t test. ns, not significant. See also Figure S7.
We also assessed whether mTOR malonylation decreased mTORC1’s activity independently of FASN inhibition by employing malonyl-NAC (N-acetylcysteamine), a cell-permeable agent capable of directly inducing cellular protein malonylation (Kulkarni et al., 2017). Similar to FASNKD cells, malonyl-NAC treatment of control ECs reduced mTORC1’s activity (Figure 6B) and target phosphorylation (Figure 6C), illustrating that direct malonylation of mTOR, independently of FASN manipulation, sufficed to reduce mTORC1’s activity.
mTOR Mutagenesis Studies
To test whether the increased mTOR malonylation contributed to the vascular defects upon FASN inhibition, we mutated K1218 of mTOR into glutamic acid (E) in order to mimic constitutive malonylation (mTORK→E) (Nishida et al., 2015). To avoid any confounding interpretation resulting from endogenous wild-type (WT) mTOR expression, we silenced mTOR (lowering mRNA levels by 60%–70%; Figure S7B) and expressed wild-type mTOR (mTORWT) or mTORK→E, both resistant to the mTOR-specific shRNA (Figure S7B). To avoid additional stress for ECs resulting from multiple viral transductions, we used orlistat to inhibit FASN. Compared with mTORWT, mTORK→E expression reduced protein synthesis in control cells to similar levels as in orlistat-treated mTORWT cells, while protein synthesis was not further reduced by orlistat in mTORK→E cells (Figure 6D). Similar results were obtained when analyzing phosphorylation of p70S6K and 4EBP1 (Figure 6E) and vessel sprouting (Figures 6F–6M and S7C). Also, mTORK→E reduced mTORC1’s enzymatic activity to levels similar to those of FASNKD or orlistat (Figure 6B).
To confirm specificity, we mutated mTOR K1218 into arginine (mTORK→R) in order to prevent malonylation, an approach widely used in the acetylation field (Baeza et al., 2016), though never performed in the malonylation field to date (in higher eukaryotes). Overexpression of mTORWT in control cells (in which endogenous mTOR expression was not silenced) elevated mTOR malonylation upon FASNKD (Figure S7D). In contrast, overexpression of mTORK→R in control cells elevated mTOR malonylation much less upon FASNKD (Figure S7D), the residual increase resulting from endogenous WT mTOR. Using a similar strategy as employed for mTORK→E, we observed that mTORK→R abrogated the decrease of p-p70S6K/p70S6K levels upon FASNKD (Figure S7E). When analyzing vessel sprouting, overexpression of mTORK→R in control cells induced a small baseline effect (consistent with reports of mTORK→R in other assays [Gorsky et al., 2016]), but abrogated the decrease upon FASNKD as observed in mTORWT-expressing cells (Figure S7F). As mentioned, orlistat did not reduce p-Akt/Akt levels in control ECs or ECs re-expressing mTORWT, mTORK→E, or mTORK→R (Figure S7G).
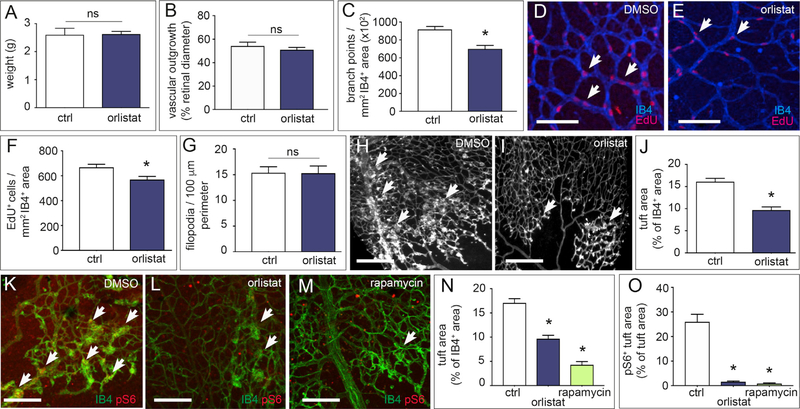
Pharmacological FASN Blockade Reduces Angiogenesis and Vascular Defects
Pharmacological compounds blocking FASN (orlistat) are being used to treat obesity (Point et al., 2016). However, in preclinical models, orlistat induces weight loss when used at a very high dose (240 mg/kg/day) (Kridel et al., 2004). We therefore used orlistat at a low dose (10 mg/kg/day) that did not cause body weight loss (Figure 7A). Treatment of pups with orlistat from P1 to P4 caused qualitatively similar vascular changes as observed in FASNΔEC mice, including reduced vessel branching and EC proliferation, without affecting EC migration (Figures 7B–7G).
Figure 7. Effect of FASN Blockade on Physiological and Pathological Angiogenesis In Vivo.
(A) Body weights of DMSO-treated (ctrl; n = 5) and orlistat-treated (10 mg/kg/day; n = 8) pups at P5.
(B) Quantification of retinal vascular outgrowth (radial expansion in percent of retinal diameter) in pups treated with vehicle DMSO (ctrl; n = 5) or orlistat (n = 8).
(C) Number of branch points in the retinal vasculature of pups treated with vehicle DMSO (ctrl; n = 5) or orlistat (n = 7).
(D and E) Representative confocal images of isolectin-B4 (IB4, blue) and EdU (red) stained retinal vessels at P5 in DMSO-treated (D) and orlistat-treated (E) pups to visualize branching and IB4+ EdU+ ECs (arrows). Scale bars, 100 μm.
(F) Number of IB4+ EdU+ ECs per mm2 of the retinal vasculature of pups treated with vehicle DMSO (n = 5) or orlistat (n = 8).
(G) Number of filopodia per 100 μm perimeter retinal vascular front in vehicle DMSO-treated (ctrl; n = 5) and orlistat-treated (n = 8) pups.
(H and I) Representative images of retinal flat mounts of retinopathy of prematurity (ROP) mice treated with vehicle DMSO (H) or orlistat (I). Arrows denote vascular tufts. Scale bars, 250 μm.
(J) Quantification of vascular tufts in ROP mice treated with vehicle DMSO (ctrl; n = 5) or orlistat (n = 6).
(K–M) Representative confocal images of pS6 and isolectin-B4 (IB4) staining of retinas from ROP mice treated with vehicle (DMSO), orlistat (10 mg/kg/day), or rapamycin (10 mg/kg/day) as positive control. Arrows denote hyperproliferative tufts. Scale bars, 200 μm (n = 4).
(N) Quantification of vascular tufts in ROP mice treated with vehicle (DMSO; ctrl), orlistat, or rapamycin (n = 4).
(O) pS6+ area in vascular tufts in ROP mice treated with vehicle (DMSO; ctrl), orlistat, or rapamycin (n = 4).
Data are mean ± SEM. *p < 0.05 by standard two-tailed t test with Welch’s correction; ns, not significant.
To test the therapeutic potential of FASN blockade in pathological angiogenesis, we used the model of retinopathy of prematurity (ROP), since the vascular tufts induced by ischemia primarily consist of proliferating ECs (Schoors et al., 2015). Treatment of pups with a low dose of orlistat during the vascular proliferation phase reduced vascular tuft formation (Figures 7H–7J). Immunostaining revealed reduced phosphorylated S6 immunoreactive levels in vascular tufts in orlistat-treated mice similar to rapamycin-treated mice (Figures 7K–7O). Additional staining experiments confirmed reduced levels of phosphorylated p70S6K and unchanged levels of phosphorylated Akt in ECs freshly isolated from FASNvΔEC mice, and in pulmonary ECs on fixed lung from FASNvΔEC mice (Figures S7H–S7L). Thus, FASN blockade with a low dose of orlistat can inhibit pathological ocular neovascularization by reducing mTOR activity.
DISCUSSION
In addition to mediating de novo lipid synthesis, FASN in ECs indirectly controls PTM of target proteins via lysine malonylation, in particular mTOR, a prime regulator of metabolism. mTOR malonylation upon FASN inhibition reduced mTORC1’s enzymatic activity, contributing to vascular defects.
Identification of mTOR K1218 Malonylation
Using an unbiased proteomic approach in FASNKD ECs, along with complementary biochemical validation, we established that mTOR is malonylated at K1218, embedded in the ICRIVKGYTLA amino acid sequence, containing amino acids (K, R, G, A, V, Y, I) that are often enriched in a lysine malonylation consensus site (Nishida et al., 2015). K1218 in mTOR is acetylated in cancer cells (Choudhary et al., 2009). Since acetylation and malonylation modify lysine residues (Nishida et al., 2015), this finding raises the question of whether mTOR K1218 may be a key node for mTORC1 activity regulation by metabolite second messengers such as malonyl-CoA and acetyl-CoA (Pietrocola et al., 2015). Notably, K1218 in mTOR is conserved in Homo sapiens, Mus musculus, and multiple other species (Table S2). Nonetheless, we acknowledge that increased malonylation of other targets upon FASN inhibition may contribute to the impaired neovascularization.
Stoichiometry of mTOR K1218 Malonylation
Quantification of the absolute levels of stoichiometry of protein malonylation has never been successfully achieved, due to spontaneous decarboxylation of the malonyl moiety on standard peptides. To date, only one study reported an indirect estimate of malonylation stoichiometry using SILAC ratios (Colak et al., 2015). We developed a targeted liquid chromatography-MS assay for quantification of absolute levels of malonylated versus total mTOR, employing a malonylated stable isotope labeled reference peptide (synthesized with Fmoc-Lys(mono-tert-butylmalonate)-OH as stable precursor for incorporation of the malonylated lysine) and five additional non-modified mTOR peptides as internal standards. Using this assay (useful for future malonylation stoichiometry studies), we reliably quantified the absolute malonylation stoichiometry. This analysis showed mTOR malonylation levels of 4.5% upon FASNKD.
These stoichiometry levels may appear low at first sight, but they are nonetheless consistent with reports for other PTMs, in particular acetylation. Indeed, most acetylation sites have very low stoichiometry, not higher than 0.5% (Weinert et al., 2015). An acetylation stoichiometry of 1% is already considered “high,” with only five out of thousands of sites having a stoichiometry >5%, and only two sites with a stoichiometry of >10% (Weinert et al., 2015). Studies in other organisms confirmed these data (Meyer et al., 2016). Malonylation cannot be compared with phosphorylation as PTM, as the latter can occur at a much higher stoichiometry, especially during the cell cycle (80%–90% [Olsen et al., 2010]), although the stoichiometry of phosphorylation in response to other stimuli is at most 25% (Sharma et al., 2014).
Though not fully appreciated, subpools of a single protein can participate in different biological processes. To induce a biologically relevant effect, PTMs thus may only need to affect the active fraction, not the total protein pool (Baeza et al., 2016). This active fraction might be much smaller than the total pool. Future work will be required to resolve these outstanding questions.
Functional Validation of mTOR K1218 Malonylation, Independent of FASN Inhibition
Protein lysine malonylation was only recently discovered (2011); by transferring a bulky, negatively charged acyl group to a lysine residue, malonylation has been proposed to alter the structure and function of the target protein (Nishida et al., 2015). Indeed, we observed a reduction in mTORC1 pathway activation and, in particular, of mTORC1’s enzymatic activity, as determined by two independent cell-free assays.
Site-specific mutagenesis of K1218 showed that malonylation of this residue regulates mTORC1 pathway activation and mTORC1’s enzymatic activity, as well as vascular responses. In addition, use of malonyl-NAC revealed that direct malonylation of mTOR, independently of FASN inhibition, was sufficient to induce similar results. All this is consistent with a model whereby inhibition of mTORC1’s activity upon FASN silencing or blockade is not an indirect epiphenomenon of FASN inhibition, but a direct effect of mTOR malonylation. The findings also suggest that a partial stoichiometry of mTOR malonylation may suffice and a high stoichiometry of mTOR malonylation is not required to induce a biological effect (reduced mTORC1 activity). Regardless, to the best of our knowledge, no previous report in the malonylation field determined both the stoichiometry and the functional importance of a malonylation site through mutagenesis and functional validation.
Possible Translational Implications
Treatment with orlistat, the most widely used drug for obesity treatment (Point et al., 2016), reduced ocular neovascularization. The more prominent angiogenesis inhibition in pathological than physiological settings may relate to the fact that ECs more actively proliferate in disease. Increased mTOR and S6 activity levels have been documented in retinal ECs in the ROP model, while mTORC1 blockade reduces ocular angiogenesis (Yagasaki et al., 2014a). In agreement with our model, we observed a decrease of phosphorylated S6 levels in orlistat-treated ROP lesions. A more specific FASN blocker confirmed the orlistat results in the experiments tested (not shown).
Confirming our earlier reports (De Bock et al., 2013; Huang et al., 2017; Schoors et al., 2015), emerging evidence shows that EC metabolism differs from that of cancer and immune cells. First, orlistat induces anti-angiogenic effects at >10-fold lower doses than needed to impair tumor growth, offering opportunities to reconsider the use of specific FASN inhibitors as anti-angiogenic agent in cancer patients. Second, in cancer cells, orlistat caused cell death by inhibiting mTORC1 (Yang et al., 2015), although in most studies the underlying molecular mechanism was not revealed. Our data identified a mechanism of FASN-dependent regulation of mTOR signaling. Third, ECs differ from cancer cells, in which FASN inhibition depletes the cellular palmitate content (Ventura et al., 2015). These EC-specific properties warrant consideration of FASN as an attractive target to inhibit pathological angiogenesis.
Limitations of the Study
While our study highlights that a low mTOR malonylation stoichiometry suffices to affect mTORC1’s enzymatic activity, the precise molecular details of this phenomenon remain to be unveiled. Our data do not exclude that increased malonylation of other proteins upon FASN inhibition may be involved in the impaired angiogenic response. Nevertheless, the data obtained using ECs expressing mTORK→E and mTORK→R mutants, in which malonylation of other proteins was not affected, support a role of mTOR signaling in the angiogenesis phenotype. Furthermore, it would also be insightful to evaluate more specific inhibitors of FASN, once available, for preclinical testing. In conclusion, we discovered a role and therapeutic potential of targeting FASN for pathological neovascularization.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Carmeliet (peter.carmeliet@kuleuven.vib.be).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animal housing and all experimental animal procedures were approved by the Ethical Committee Animal Experimentation of the KU Leuven (Belgium).
Generation of EC Specific FASN Conditional Knockout Mice
We crossed FASNlox/lox mice (Wei et al., 2011) with the tamoxifen-inducible EC-specific Cre-driver line VE-cadherin(PAC)-CreERT2 mice (Benedito et al., 2009) to obtain endothelial cell (EC) specific FASN deletion upon tamoxifen treatment, referred to as FASNΔvEC. A second tamoxifen-inducible EC-specific FASN knock out line was generated by intercrossing the FASNlox/lox mice with PDGFb(PAC)-CreERT2 mice (Claxton et al., 2008) referred to as FASNΔpEC mice. Correct Cre-mediated excision of the floxed FASN segment in tamoxifen-treated Cre+ mice (named FASNΔEC mice) was confirmed via PCR analysis of genomic DNA using primers spanning the floxed region, by the appearance of a 300-bp band (Figure S1L). FASNΔEC mice were always compared with tamoxifen-treated Cre− FASNlox/lox littermates (denoted as WT). C57BL/6 wild type mice were obtained from the KU Leuven Animal Facility.
Cell Lines
Primary Human Endothelial Cells
Human umbilical vein endothelial cells (HUVECs) were freshly isolated from umbilical cords obtained from multiple donors (with approval from the Ethics Committee Research UZ/KU Leuven and informed consent obtained from all subjects) as previously described (Jaffe et al., 1973), regularly tested for mycoplasma. HUVECs were maintained in endothelial cell basal medium (EGM2; containing 2% fetal bovine serum (FBS)) supplemented with endothelial cell growth medium supplement pack (ECGM-2; Promocell) or 50:50 volumetric dilution with M199 (Gibco) supplemented with 10% FBS (final concentration) (Biochrom BmgH), one vial of EC growth factor mix and heparin (Promocell), 100 IU/mL penicillin and 100 μg/mL streptomycin. In all experiments, HUVECs were always used as single-donor cultures and were used between passage (p) 1 and 4. For treatment with acetate, the spheroids were incubated for 24 hr with 10 mM acetate (Sigma-Aldrich). Supplementation of spheroids with the nucleotide mix (dNTPs 100 mM; Life Technologies) was done for 24 hr with 500 μM of each dNTP. Palmitate supplementation of the media was carried out by first conjugating (ethanol dissolved) palmitate to 10% fatty acid free BSA (Sigma-Aldrich) for 1 hr at 55°C in a 50:50 volumetric ratio. Conjugated palmitate was diluted in EGM2 media to 50 μM and incubated on cells and spheroids for 24 hr. Sulfosuccinimidyl oleate (SSO; Cayman Chemical) was used at 100 μM for 24 hr. Orlistat (Sigma-Aldrich or Cayman Chemical) was used at 10 μM for 24 hr. Rapamycin and torin2 (Sigma-Aldrich) were used at 20 nM and at 100 nM, respectively, for 24 hr.
Mouse Endothelial Cells
Mouse endothelial cells were isolated from perfused healthy livers of FASNΔvEC mice and their wild-type littermates as described previously (Cantelmo et al., 2016). Briefly, mice were anesthetized using Nembutal (60 mg/kg) and perfused with PBS followed by perfusion with 5 mL of a water based perfusion buffer containing 1.7 M NaCl, 84 mM KCl, 120 mM HEPES and 1 mM NaOH followed by perfusion with 5 mL of a PBS based digestion buffer containing 0.1% collagenase II (Life Technologies, Ghent, Belgium), collagenase I (Life Technologies, Ghent, Belgium), 2 mM CaCl2, 1% antibiotic-antimycotic (Life Technologies, Ghent, Belgium) at a perfusion rate of 2 mL/min. The organs were dissected and placed into a 50 mL conical tube containing 5 mL of the digestion buffer, and incubated in a water bath at 37°C for approximately 30 min. Next, tissue was homogenously dissociated and the reaction was stopped by adding 10 mL PBS/0.1% BSA. Subsequently, the cell suspension was filtered through a 100 μm cell strainer (Corning) and ECs were isolated by magnetic bead sorting using Dynabeads (CELLection Biotin Binder Kit, Life Technologies) coated with anti-mouse CD31 (Anti-mouse CD31 Clone 390, eBioscience), according to the manufacturer’s procedure. CD31+ ECs were collected by placing the tubes on a DynaMagTM-50 Magnet (Life Technologies, Ghent, Belgium) and ECs were resuspended in 50% EGM2:50% M199 and plated at the appropriate density on gelatin-precoated (0.1% in PBS) cell culture plates in 50:50 ratio of M199: EGM2 medium for subsequent RNA extraction the next day.
Cancer Cell Lines
Breast cancer cell lines (MCF-7 (Levenson and Jordan, 1997), MDA-MB-231, MDA-MB-468 (Cailleau et al., 1978), T47D (Keydar et al., 1973), hepatocellular carcinoma (HepG2 (Aden et al., 1979), HuH7 (Nakabayashi et al., 1982)) and prostate cancer (DU145) (Stone et al., 1978) were a kind gift from Prof. Sarah-Maria Fendt (VIB, Leuven, Belgium). All cancer cell lines were cultured in DMEM (4.5 mg/mL D-glucose) from Gibco (Invitrogen, Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Biochrom BmgH), 2 mM L-glutamine, 100 IU/mL penicillin and 100 μg/mL streptomycin.
METHOD DETAILS
Mouse Models of Physiological and Pathological Angiogenesis
Analysis of Postnatal Retinal Angiogenesis
Genetic endothelial cell specific inactivation of FASN in neonatal mice was achieved by intraperitoneal injection of 100 mg/kg tamoxifen solution (Sigma-Aldrich T5648; dissolved in 1:10 EtOH:corn oil) once daily from postnatal day (P) 1 to P3 (FASNΔvEC) or only at P2 (FASNΔpEC) in Cre+ FASNlox/lox mice (yielding FASNΔEC mice) and Cre− FASNlox/lox mice (WT controls). C57BL/6 wild type mice were injected with orlistat (Sigma-Aldrich) (10 mg/kg/day) from P1-P3 and eyes were collected at P5 for subsequent immunohistochemistry and analysis. For detection of cell proliferation, 5-ethynyl-2′-deoxyuridine (EdU) (Invitrogen) was injected 2 hr before dissection. At P5, pups were euthanized and eyes were enucleated, fixed with 4% PFA for 2 hr at 4°C and prepared for vascular analysis. As all animal treatments were done in baseline conditions, no randomization was required. Retinal whole mounts were prepared for vascular analysis as described (De Bock et al., 2013). Only litters for which the WT littermates (for FASNΔEC mice) reached normal outgrowth and body weight at P5 were included for analysis. Radial vascular outgrowth (radial vascular length in % of retinal diameter), branching points, EdU incorporation and number of filopodia were analyzed on isolectin-B4 stained retinas (see below) using the NIH ImageJ software package and Leica LASAF-MMAF morphometric analysis software (MetaMorph) (Leica Microsystems, Mannheim, Germany) with in-house developed macros and normalized to the vascular area. EdU incorporation was analyzed at the vascular front of the retinal plexus. Vessel maturation was evaluated by quantification of NG2+ pericyte coverage; vessel regression by quantification of empty sleeves (collagen IV-stained basement membrane without isolectin-B4+ ECs; see below).
Oxygen-Induced Retinopathy Model
Oxygen induced retinopathy (ROP) was performed by exposing C57BL/6 pups and their mother to 70% oxygen from P7-P12. Subsequent exposure to normoxia induces artificial hypoxia resulting in vascular tuft formation in the retina. In this period pups were treated to 10 mg/kg/day orlistat or DMSO vehicle from P12-P17. On P17, pups were euthanized, eyes enucleated, and retina dissected prior to isolectin B4 staining (see below). Tuft area was analyzed using the NIH ImageJ software package and normalized to vascular area. No statistical method was used to predetermine the sample size. For all mouse experiments, data analysis was done by experimenters blinded to the group allocation.
In vivo Analysis of MTORC1 Target Phosphorylation
Immunohistochemistry on lung sections was performed as previously reported (Wenes et al., 2016). In brief, for serial sections of lungs from FASN cut at 7 μm thickness, tissue samples were fixed in 2% PFA overnight at 4°C, dehydrated and embedded in paraffin. Paraffin slides were first rehydrated to further proceed with antigen retrieval in citrate solution (DAKO). Sections were then fixed in 100% methanol. If necessary, 0.3% hydrogen peroxide was added to methanol, to block endogenous peroxidases. The sections were blocked with the appropriate serum (DAKO) and incubated overnight with the following antibodies: rabbit anti-FASN (Abcam ab 99359), rat anti-CD34 (BD Pharmingen), rabbit anti-phoshpo-p70S6K (T389) (Cell Signaling, 9205), rabbit anti-phospho-4EBP1 (Cell Signaling 2855. Appropriate biotin-labeled secondary antibodies (Jackson Immunoresearch) 1:300 were used, along with streptavidin-bound peroxidase. When necessary, TCA fluoresceine-tyramine or TSA Plus Cyanine 3 system amplification (Perkin Elmer, Life Sciences) were performed according to the manufacturer’s instructions. The sections were subsequently stained with Hoechst. ProLong Gold mounting medium without DAPI (Invitrogen) was used. Microscopic analysis was done with an Olympus BX41 microscope and CellSense imaging software.
Meta-Analysis of FASN Expression
We aimed to determine FASN expression in previously published murine and human tumor ECs (TEC) vs normal ECs (NEC) transcriptomics datasets (Cantelmo et al., 2016). To this end we screened PubMed, ArrayExpress and the GEO database for relevant studies and identified 5 studies comprising 8 distinct TEC vs NEC datasets. For RNA-sequencing data (E-MTAB-4842), the raw sequenced reads were mapped to the mouse reference transcriptome and genome (GRCm38/mm10) using the Bowtie TopHat pipeline (Langmead and Salzberg, 2012). Mapped reads were assigned to ensemble gene IDs by HTSeq resulting in on average 14,814,487 ± 5,948,836 counts per sample. Genes expressed at a level of at least 1 count per million reads in at least three of eight samples were filtered out with the EdgeR package (Robinson et al., 2010). Data from Affymetrix genechips (GSE5401, GSE73752) were preprocessed and Robust Multichip Average (RMA) normalized using the R-packages affy and oligo. Data from Agilent chips (GSE77199) was preprocessed and normalized using a custom script provided with the data. We then performed pair-wise, TEC vs NEC, differential expression analysis for each dataset independently as described previously (Cantelmo et al., 2016). Differentially expressed genes and their false discovery rate (FDR) corrected p values were identified by the Limma package (Ritchie et al., 2015). The log2 fold change distribution was visualized using violin plots (using the R-package ggplot2), the location of FASN in this distribution is indicated by a red dot. Finally, we used Fisher’s combined probability test (metap R-package) to combine the unidirectional p values of each individual dataset. We performed the same analysis for adjusted p values.
Mutagenesis, Knockdown and Overexpression Strategies
Site-Directed and Knockdown Resistant Mutagenesis of mTOR
For the mTOR mutation pcDNA3-FLAG mTOR WT (Addgene #26603) was used for mutagenesis of lysine 1218. An EcoR1 fragment comprising nucleotide (nt) 3118 to nt 4004 of the pcDNA3-FLAG mTOR WT (Addgene #26603) was cloned in the pGEM-T Easy vector (Promega). Lysine to glutamic acid (K->E) or arginine (K->R) mutation were done with Quickchange site-directed mutagenesis kit (Stratagene). An NsiI fragment comprising the mTOR cDNA with the correct mutation was replaced in the vector pRRLsinPPT.CMV.MCS MM WPRE vector (Michieli et al., 2004) to obtain pRRL-mTORWT, pRRL-mTORK->E, or pRRL-mTORK->R. To achieve the shRNA mTOR resistance, a mutation was introduced on the pgem 9Zf plasmid in which an XbaI fragment containing nt 4240 to nt 7650 of the mTOR cDNA was cloned. Thereafter, a BstE2-BspE1 fragment containing the shRNA resistance mutation was replaced in pRRL-mTORWT, pRRL-mTORK->E, and pRRL-mTORK->R
Knockdown-Resistant Mutagenesis of FASN
We cloned full-length human FASN cDNA from the plasmid pCMV6-XL4 into the vector pRRLsinPPT.CMV.MCS MM WPRE vector (Michieli et al., 2004) to obtain pRRL-FASN. To achieve the shRNA FASN resistance, a 3.3 kb MluI fragment of FASN was cloned into a bluescript vector. Thereafter, Quickchange site-directed mutagenesis kit (Stratagene) was used to introduce the shRNA resistant mutation. Subsequently, a BspE1 fragment of the bluescript-MluI-FASN vector containing the shRNA resistance mutation was replaced in pRRL-FASN.
Knockdown and Overexpression Strategies
To generate shRNA vectors against FASN, ACC1 or mTOR oligonucleotides were cloned into the pLKO- shRNA2 vector (No. PT4052–5; Clontech). A nonsense scrambled shRNA sequence was used as a negative control. Oligonucleotides are listed in the Key Resources Table. Production of lentiviruses by transfection into 293T cells was performed as described (De Bock et al., 2013). For transductions, a multiplicity of infection (MOI) of 10 was used in all FASNKD, ACC1KD and mTORKD experiments. For transductions with mTOR overexpression, a MOI of 20 was used. Cells were transduced overnight and re-fed with fresh medium the next day. Transduced cells were used in functional assays at least 3 to 4 days post-transduction. To silence FASN expression by siRNA, cells were transfected with a validated pool of siRNA duplexes directed against human FASN (SMARTpool, Dharmacon). A scrambled siRNA pool was used as control. Cells were transfected with the indicated siRNA (50 nM) using Lipofectamine RNAi Max (Invitrogen).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-4EBP1 | Cell Signaling | Cat# 9452; RRID: AB_10693791 |
| Rabbit anti-p-4EBP1 | Cell Signaling | Cat# 2855; RRID:AB_560835 |
| Rabbit anti-ACC1 | Cell Signaling | Cat# 3662; RRID:AB_2219400 |
| Rabbit anti-Akt | Cell Signaling | Cat# 9272S; RRID:AB_329827 |
| Rabbit anti-p-Akt(S473) | Cell Signaling | Cat# 4058; RRID:AB_331169 |
| Alexa 405-conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat# A31553; RRID:AB_221604 |
| Alexa 405-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A31556; RRID:AB_221605 |
| Alexa 488-conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat# A11034; RRID:AB_2576217 |
| Alexa 488-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A32723; RRID:AB_2633275 |
| Alexa 568-conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat# A11004; RRID:AB_2534072 |
| Alexa 568-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat# A11036; RRID:AB_10563566 |
| Mouse anti-α-Tubulin | Sigma-Aldrich | Cat# T6199; RRID:AB_477583 |
| Rabbit anti-α-Tubulin | Cell Signalling | Cat# 2144S; RRID:AB_2210548 |
| Rabbit anti-AMPKα | Cell signaling | Cat# 2532S; RRID:AB_330331 |
| Rabbit anti-p-AMPKα(Thr172) | Cell signaling | Cat# 2531S; RRID:AB_330330 |
| Rabbit anti-β-Actin | Cell Signaling | Cat# 4970; RRID:AB_2223172 |
| Rat anti-mouse CD31 Clone 390 | eBioscience | Cat# 14-0311-85; RRID:AB_467202 |
| Rat anti-mouse CD34 | BD Biosciences | Cat# 553731; RRID:AB_395015 |
| Rabbit anti-Cleaved Caspase-3 | Cell Signalling | Cat# 9664; RRID:AB_2070042 |
| Donkey anti-rabbit IgG, HRP linked | Jackson Immunoresearch | Cat# 711-036-152; RRID:AB_2340590 |
| Donkey anti-rat IgG, HRP linked | Jackson Immunoresearch | Cat# 712-036-180; RRID: N/A |
| Anti-collagen IV | AbD Serotec | Cat# 2150-1470; RRID:AB_2082660 |
| Mouse anti-FASN | Santa Cruz | Cat# Sc-55580; RRID:AB_2231427 |
| Rabbit anti-FASN | Abcam | Cat# ab 99359; RRID:AB_10697253 |
| Goat anti-Mouse IgG, HRP-linked | Cell Signaling | Cat# 7076; RRID:AB_330924 |
| Goat anti-Rabbit IgG, HRP-linked | Cell Signaling | Cat# 7074; RRID:AB_2099233 |
| Mouse anti-LAMP1 | Abcam | Cat# ab25630; RRID:AB_470708 |
| Rabbit anti-malonylated lysine | PTM Biolabs | Cat# PTM-901; RRID:AB_2687947 |
| Rabbit anti-malonylated lysine | Cell Signaling | Cat# 14942; RRID:AB_2687627 |
| Rabbit anti-mLST8 | Cell Signaling | Cat# 3274; RRID:AB_823685 |
| Mouse anti-mTOR | Life Technologies | Cat# AH01232; RRID: N/A |
| Rabbit anti-mTOR | Cell Signaling | Cat# 2972; RRID:AB_330978 |
| Rabbit anti-Na+/K+ ATPase | Cell Signaling | Cat# 3010S; RRID:AB_2060983 |
| Anti-NG2 Chondroitin Sulfate Proteoglycan | Merck Millipore | Cat# AB5320; RRID:AB_11213678 |
| Rabbit anti-p-p70S6K | Cell Signaling | Cat# 9205; RRID:AB_330944 |
| Rabbit anti-p70S6K | Cell Signaling | Cat# 9202; RRID:AB_331676 |
| Rabbit anti-RAPTOR | Cell Signaling | Cat# 2280; RRID:AB_561245 |
| Rabbit anti-RICTOR | Cell Signaling | Cat# 9476; RRID:AB_10612959 |
| Rabbit anti-S6 ribosomal protein | Cell Signaling | Cat# 4858; RRID:AB_916156 |
| Bacterial and Virus Strains | ||
| XL10-gold ultracompetent bacteria | Stratagene | 200314 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| [U-14C] acetate | Perkin Elmer | NEC553050UC |
| Alexa 405-conjugated streptavidin | Thermo Fisher Scientific | S32351 |
| Alexa 488-conjugated isolectin GS-IB4 | Thermo Fisher Scientific | I21411 |
| Alexa 488-conjugated phalloidin | Thermo Fisher Scientific | A12379 |
| Alexa 568-conjugated phalloidin | Thermo Fisher Scientific | A12380 |
| [γ-32P]-ATP | Perkin Elmer | BLU002Z250UC |
| Biotinylated Griffonia Simplicifolia Lectin I isolectin B4 | Vector Laboratories | B-1205 |
| 2,6-di-tert-butyl-4-methylphenol (BHT) | Sigma-Aldrich | 34750 |
| L-Carnitine | Sigma-Aldrich | C0283 |
| CHAPS | Sigma-Aldrich | C-3023 |
| CM-H2DCFDA | Thermo Fisher Scientific | C6827 |
| Collagen type 1 (rat tail) | Millipore | 08-115 |
| Dimethyl Sulfoxide | Sigma-Aldrich | 673439 |
| dNTP mix 100 mM | Life Technologies | 10297018 |
| Gelatin from bovine skin | Sigma-Aldrich | G9391 |
| [U-13C]-D-Glucose | Cambridge Isotope Laboratories | CLM-139-1 |
| [5-3H]-D-glucose | Perkin Elmer | NET531001MC |
| [6-14C]-D-glucose | Perkin Elmer | NEC045X050UC |
| [U-14C]-glucose | Perkin Elmer | NEC042V250UC |
| L-Glutamine | Life Technologies | 25030024 |
| [U-13C]-L-Glutamine | Cambridge Isotope Laboratories | CLM-1166 |
| [U-14C]-glutamine | Perkin Elmer | NEC451050UC |
| Glutathione sepharose beads 4B | Sigma-Aldrich | GE17-0756-01 SIGMA |
| Heparin | Sigma-Aldrich | H3393 |
| Hoechst 33342 | Molecular Probes | H3570 |
| Hydroxide of Hyamine 10x | Perkin Elmer | 50-905-1737 |
| Malonyl-NAC | Kulkarni et al., 2017 | N/A |
| Methyl cellulose (4000cP) | Sigma-Aldrich | M0512-100MG |
| Mitomycin C | Sigma-Aldrich | Y0000378 |
| Nicotinamide | Sigma-Aldrich | 72340 |
| N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide | Sigma-Aldrich | 19915 |
| Orlistat | Sigma-Aldrich | 1478800 |
| Orlistat | Cayman Chemical | 10005426 |
| [9,10-3H]-palmitate | Perkin Elmer | NET043001MC |
| [U-14C]-palmitate | Perkin Elmer | NEC534050UC |
| [U-13C]-potassium palmitate | Cambridge isotope laboratories | CLM-1889-1 |
| PC25:0 standard | Avanti Polar Lipids | 12:0-13:0 PC | LM-1000 |
| PC43:6 standard | Avanti Polar Lipids | 21:0-22:6 PC | LM-1003 |
| PE25:0 standard | Avanti Polar Lipids | 12:0-13:0 PE | LM-1100 |
| PE43:6 standard | Avanti Polar Lipids | 21:0-22:6 PE | LM-1103 |
| Phosphatase inhibitors (PhosSTOP) | Roche | 04906837001 |
| PI25:0 standard | Avanti Polar Lipids | 12:0-13:0 PI | LM-1500 |
| PI31:1 standard | Avanti Polar Lipids | 17:0-14:1 PI | LM-1504 |
| PI43:6 standard | Avanti Polar Lipids | 21:0-22:6 PI | LM-1503 |
| Protease inhibitors (cOmplete, EDTA-free Protease Inhibitor Cocktail) | Roche | 11873580001 |
| PS25:0 standard | Avanti Polar Lipids | 12:0-13:0 PS | LM-1300 |
| PS31:1 standard | Avanti Polar Lipids | 17:0-14:1 PS | LM-1304 |
| PS37:4 standard | Avanti Polar Lipids | 17:0-20:4 PS | LM-1302 |
| Protein G sepharose | Sigma-Aldrich | P3296 |
| Rapamycin | InVivoGen | tlrl-rap |
| Recombinant dephosphorylated 4EBP1 | Dunlop et al., 2009 | N/A |
| S43:6 standard | Avanti Polar Lipids | 21:0-22:6 PS | LM-1303 |
| SM30:1 standard | Avanti Polar Lipids | (d18:1/12:0) | LM-2312 |
| Sodium acetate | Sigma-Aldrich | S2889 |
| Sodium palmitate | Sigma-Aldrich | P9767 |
| Sulfo-N-succinimydil oleate (SSO) | Cayman Chemical | 11211 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| TCA (trichloroacetic acid) | Sigma-Aldrich | T6399 |
| Tert-butyldimethylchlorosilane | Sigma-Aldrich | 8.18642 |
| Tetrabutylammonium chloride | Sigma-Aldrich | 86852-10G-F |
| Thapsigargin | Sigma-Aldrich | T9033 |
| [3H]-thymidine | Perkin Elmer | NET355L005MC |
| Torin 2 | Sigma-Aldrich | SML-1224 |
| Triethylammoniumbicarbonate buffer (TEAB) | Sigma-Aldrich | T7408 |
| [ring-3,5-3H]-Tyrosine | Perkin Elmer | NET127250UC |
| Ultima-gold scintillation | Perkin-Elmer | 6013329 |
| Critical Commercial Assays | ||
| Agencourt AMPure XP | Beckman Coulter | A63880 |
| Click-iT EdU Alexa Fluor 555 Imaging Kit | Thermo Fisher Scientific | C10338 |
| Cytotoxicity Detection KitPLUS (LDH) | Roche | 04744934001 |
| Dynabeads protein A | Thermo Fisher Scientific | 10002D |
| Dynabeads protein G | Thermo Fisher Scientific | 10004D |
| In Situ Cell Death Detection Kit, TMR red | Roche | 12156792910 |
| iScript cDNA synthesis kit | Bio-Rad | 170-8891 |
| K-LISA mTOR activity kit | Merck Milipore | CBA055 |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 |
| Lipofectamine RNAi Max transfection | Thermo Fisher Scientific | 13778030 |
| Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | 32106 |
| Protein membrane extraction kit | 101 Biosciences | P503L |
| PureLink RNA Mini Kit | Thermo Fisher Scientific | 12183018A |
| Quikchange site-directed mutagenesis kit | Agilent | 200524 |
| SuperSignal West Femto Maximum Sensitivity substrate | Thermo Fisher Scientific | 34095 |
| TSA fluoresceine-tyramine system | Perkin Elmer | NEL701A001KT |
| TSA Plus Cyanine 3 system | Perkin Elmer | NEL744001KT |
| TaqMan Fast Universal PCR Master Mix (2x) | Thermo Fisher Scientific | 4352042 |
| Deposited Data | ||
| Malonylation screen raw and analyzed data | This paper | PRIDE: PXD006503 |
| RNA-sequencing data from TEC and NEC | Cantelmo et al., 2016 | ArrayExpress: E-MTAB-4842 |
| Experimental Models: Cell Lines | ||
| 293T cells | De Bock et al., 2013 | N/A |
| DU145 cell line | Stone et al., 1978 | N/A |
| HepG2 cell line | Aden et al., 1979 | N/A |
| HuH7 cell line | Nakabayashi et al., 1982 | N/A |
| Human umbilical vein endothelial cells | Jaffe et al., 1973 | N/A |
| MCF7 cell line | Levenson and Jordan, 1997 | N/A |
| MDA-MB231 cell line | Cailleau et al., 1978 | N/A |
| MDA-MB468 cell line | Cailleau et al., 1978 | N/A |
| Murine primary endothelial cells | This paper | N/A |
| T47D cell line | Keydar et al., 1973 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: FASNlox/lox C57Bl/6 | Wei et al., 2011 | N/A |
| Mouse: Cre-driver line PDGFb(PAC)-CreERT2 mice | Claxton et al., 2008 | N/A |
| Mouse: Cre-driver line VE-cadherin(PAC)-CreERT2 C57Bl/6 | Benedito et al., 2009 | N/A |
| Oligonucleotides | ||
| On-TARGET plus SMARTpool siRNA J-003954-11, FASN: UGACAUCGUCCAUUCGUUU | Dharmacon | L-003954-00-0005 |
| On-TARGET plus SMARTpool siRNA J-003954-12, FASN: GAAGCACAUUGGCAAAGUC | Dharmacon | L-003954-00-0005 |
| On-TARGET plus SMARTpool siRNA J-003954-13, FASN: GGUAUGCGACGGGAAAGUA | Dharmacon | L-003954-00-0005 |
| On-TARGET plus SMARTpool siRNA J-003954-14, FASN: CUUCCGAGAUUCCAUCCUA | Dharmacon | L-003954-00-0005 |
| Short-hairpin ACC1 RNA sequence: CCGGCTGCTTCTGTTGGCTCAGATACTCGAGTATCTGAGCCAACAGAAGCAGTTTTT | Broad Institute - Genetic perturbation platform | TRCN0000004769 |
| Short-hairpin FASN RNA sequence 1: CCGGCATGGAGCGTATCTGTGAGAACTCGAGTTCTCACAGATACGCTCCATGTTTTT | Broad Institute - Genetic perturbation platform | TRCN0000003127 |
| Short-hairpin FASN RNA sequence 2: CCGGGCTACGACTACGGCCCTCATTCTCGAGAATGAGGGCCGTAGTCGTAGCTTTTT | Broad Institute - Genetic perturbation platform | TRCN0000003126 |
| Short-hairpin mTOR RNA sequence: CCGGGCTGTGCTACACTACAAACATCTCGAGATGTTTGTAGTGTAGCACAGCTTTTTG | Broad Institute - Genetic perturbation platform | TRCN0000039785 |
| Nonsense scrambled short-hairpin FASN RNA sequence: AACAAGATGAAGAGCACCAA | Sigma-Aldrich | SHC002 |
| TaqMan primer-probe-Mix human ACC1: NM_198836 | IDT | Hs.PT.56a.1301053.g |
| TaqMan primer-probe-Mix human ACC2: NM_001093 | IDT | Hs.PT.56a.2559541 |
| TaqMan primer-probe-Mix human FASN: NM_004104 | IDT | Hs.PT.58.1757439 |
| TaqMan primer-probe-Mix murine FASN: NM_007988 | IDT | Mm.PT.58.11264416 |
| TaqMan primer-probe-Mix human HPRT1: NM_000194 | IDT | Hs.PT.58.2145446 |
| TaqMan primer-probe-Mix murine HPRT1: NM_013556 | IDT | Mm.PT.42.12662529 |
| TaqMan primer-probe-Mix human mTOR: NM_004958 | IDT | Hs.PT.58.1509098 |
| Recombinant DNA | ||
| mTOR K1218E mutant cDNA | This paper | N/A |
| mTOR K1218R mutant cDNA | This paper | N/A |
| pBluescript II vector | Agilent Technologies | 212205 |
| pcDNA3-FLAG mTOR wt | Addgene | Plasmid # 26603 |
| pCMV6-XL4-FASN | Vazquez-Martin et al., 2008 | N/A |
| pGEM-9Zf vector | Promega | P2391 |
| pGEM-T easy vector | Promega | A1360 |
| pLKO-shRNA2 vector | Clontech | No. PT4052-5 |
| pRRLsinPPT.CMV.MCS MM WPRE vector | Michieli et al., 2004 | N/A |
| Software and Algorithms | ||
| Adobe Illustrator CC (Adobe Systems Incorporated) | https://www.adobe.com/be_en/products/illustrator.html | N/A |
| Adobe InDesign CC (Adobe Systems Incorporated) | https://www.adobe.com/be_en/products/indesign.html | N/A |
| Agilent MassHunter Quantitative Analysis software | Agilent Technologies | N/A |
| Bowtie TopHat pipeline | Langmead and Salzberg, 2012 | N/A |
| Distiller version 2.5.1.0 (Matrix Science) | www.matrixscience.com/Distiller | N/A |
| EdgeR package | Robinson et al., 2010 | N/A |
| Fiji (ImageJ) | https://fiji.sc | N/A |
| FlowJo (LLC) | https://www.flowjo.com/solutions/flowjo | N/A |
| GraphPad Prism 6 software | http://www.graphpad.com/ | N/A |
| Imaris | http://www.bitplane.com/imaris-for-cell-biologists | N/A |
| LASAF-MMAF morphometric analysis software (MetaMorph) | Leica Microsystems | N/A |
| limma package | Ritchie et al., 2015 | N/A |
| Mascot search engine | www.matrixscience.com | N/A |
| RStudio | https://www.rstudio.com | N/A |
| Skyline v3.6 | https://skyline.ms/wiki/home/software/Skyline/page.view?name=default | N/A |
| Xcalibur Software | Thermo Fisher Scientific | N/A |
| Other | ||
| Antibiotic-antimycotic | Life Technologies | 15240062 |
| Cell lysis buffer (10x) | Cell Signaling | 9803 |
| Collagenase I | Life Technologies | 17100017 |
| Dispase | Life Technologies | 17105-041 |
| DNase1 from bovine pancreas | Sigma-Aldrich | D4527-10KU |
| Dulbecco’s Modified Eagle Medium (DMEM) | Thermo Fisher Scientific | 11965092 |
| DynaMagTM-50 Magnet | Life Technologies | 12302D |
| EGM2 (Endothelial growth medium) | Promocell | C22011 |
| FBS (Fetal Bovine Serum), charcoal stripped | Sigma-Aldrich | F6765 |
| FBS (Fetal Bovine Serum) | Biochrom BmgH | S0115 |
| Heparin/ECGS (Endothelial cell growth factor supplements) | Promocell | C30140 |
| Medium 199, HEPES | Thermo Fisher Scientific | 22340020 |
| Penicillin/streptomycin | Life Technologies | 15140122 |
| ProLong gold antifade mountant | Thermo Fisher Scientific | P36934 |
| RIPA lysis buffer | Thermo Fisher Scientific | 89901 |
| Sep-Pak C18 cartridges | Waters | WAT051190 |
| Transwell permeable supports | Costar | 3422 |
| Trypsin-EDTA (0.25%) | Life Technologies | 25200056 |
| Trypsin-sequencing grade, modified | Promega | V5117 |
Metabolism Assays and Metabolomics
Fatty Acid Synthesis
ECs and cancer cells were incubated in full growth medium supplemented with [U-14C]-Acetate or [U-14C]-D-glucose for 24 hr followed by snap freezing and methanol-water-chloroform extraction. Phase separation was achieved by centrifugation at 4°C and the methanol-water phase containing polar metabolites was used as negative control. Radioactivity in the chloroform phase containing fatty acids was quantified by liquid scintillation counting and values were normalized to protein concentration determined in the dried protein interphase.
Fatty Acid Uptake
Cells were seeded in a 12-well format (100,000 cells) and incubated for 3 hr until attachment. The cells were subsequently incubated in full growth medium supplemented with [U-14C]-palmitate for 15 min followed by 5 washing steps with PBS. Cells were lysed using 200 μL 0.2 N NaOH. Radioactivity was quantified by liquid scintillation counting.
Fatty Acid Oxidation and Glycolysis
ECs were incubated in fully supplemented EGM2 medium with 100 μM unlabeled palmitate and 50 μM carnitine. Cells were incubated for 2 hr in growth medium containing 2 μCi/mL [9,10-3H]-palmitate Thereafter, the supernatant was transferred into glass vials sealed with rubber stoppers. 3H2O was captured in hanging wells containing a piece of Whatman paper soaked with H2O over a period of 48 hr at 37°C to reach saturation (Schoors et al., 2015). Radioactivity was determined by liquid scintillation counting. Glycolysis was measured analogously to fatty acid oxidation (cf supra) using 80 mCi/mmol [5-3H]-D-glucose (Perkin Elmer) (De Bock et al., 2013).
14C-glucose and 14C-glutamine Oxidation
Cells were incubated for 6 hr in growth medium containing 100 μCi/mmol [6-14C]-D-glucose. Thereafter, 250 μL of 2 M perchloric acid was added to each well to stop cellular metabolism and the wells were immediately covered with a 1× hyamine hydroxide-saturated piece of Whatman paper. Overnight absorption into the paper of 14CO2 released during oxidation of glucose was performed at room temperature, and radioactivity in the paper was determined by liquid scintillation counting. Glutamine oxidation was performed similarly as glucose oxidation, using 0.5 μCi/mL [U-14C]-glutamine as tracer.
14C-palmitate Carbon Incorporation into RNA/DNA
De novo DNA synthesis was measured by the incorporation of 14C into DNA or RNA using 100 μCi/mmol [U-14C]-palmitate and was corrected for the total amount of DNA or RNA per sample. Total RNA and DNA were isolated using TRIzol reagent (Thermo Fisher Scientific).
Protein Synthesis Assay
Protein synthesis was measured via the tyrosine incorporation assay. In brief, 1 × 106 cells were incubated in full growth media supplemented with 80 mCi/mmol [3H]-tyrosine (Perkin Elmer) for 6 hr. Cells were washed 3 times with PBS followed by precipitation with 10% TCA overnight. Protein lysates were scraped, transferred into an Eppendorf tube and centrifuged at 14,000 rpm for 10 min. Protein pellets were lysed in 500 μL 0.5 M NaOH, 0.1% TritonX at 50°C for 1 hr. 400 μL of lysate were neutralized with 200 μL 1 N acetic acid and radioactivity was determined by liquid scintillation counting. 100 μL of the remaining solution were used to determine protein concentrations for normalization.
Energy Charge Assessment
200,000 cells were extracted in 300 μL of a 50:30:20 (methanol:acetonitrile:10 mM Tris, pH 9.3) extraction buffer. Extraction samples were then centrifuged for 5 min at 20,000 × g and the supernatant was transferred to LC-MS vials. Measurement of ATP, ADP and AMP was performed using a Dionex UltiMate 3000 LC System (Thermo Fisher Scientific) coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) operated in negative ionization mode. Practically, 20 μL of sample was injected on a SeQuant ZIC/pHILIC Polymeric column (Merck Millipore). The gradient started with 10% of solvent B (10 mM NH4-acetate in mqH2O, pH 9.3) and 90% solvent A (acetonitrile, LC-MS grade) and remained at 10% B until 2 min post injection. Next, a linear gradient to 80% B was carried out until 29 min. At 38 min, the gradient returned to 40% B followed by a decrease to 10% B at 42 min. The chromatography was stopped at 58 min. The flow was maintained at 100 μL/min and the column was kept at 25°C throughout the analysis. The MS (Q Exactive, Thermo Fisher Scientific) operated both in full scan mode using a spray voltage of 3.2 kV, capillary temperature of 320°C, sheath gas at 20.0, auxiliary gas at 5.0. For Full scan - SIM mode, AGC target was set at 1e6 using a resolution of 140,000, with a maximum IT of 512 ms. Data collection was performed using Xcalibur software (Thermo Fisher Scientific), values of ATP, ADP, and AMP represent the area under the curve of the measured ions (m/z ATP 505.98793; ADP 426.02160; AMP 346.05526) with a maximum allowed relative mass deviation of 5 ppm. Energy charge was calculated as ([ATP] + 1/2 [ADP])/([ATP] + [ADP] + [AMP]).
Glutathione Species Measurement
Samples were collected in 300 μL 5% trichloro-acetic acid (Sigma-Aldrich). 50 μL was loaded onto an Ultimate 3000 UPLC (Thermo Fisher Scientific, Bremen, Germany) equipped with a Acquity UPLC HSS T3 column (cat # 186003976; 2.1 × 5 mm; 1.8 μm particles; Waters) in line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). A linear gradient was carried out using solvent A (0.05% formic acid) and solvent B (60% methanol, 0.05% formic acid). Practically, samples were loaded at 99% solvent A and from 10 to 12 min a ramp to 100% solvent B was carried out. From 15 to 16 min the column returned to 99% solvent A and the run was stopped at 21 min. Elution of GSH and GSSG occurred at 3 and 5.5 min respectively (isocratic separation). Flow rate was constant at 250 μL/min and the column temperature was kept constant at 37°C. The mass spectrometer operated in targeted SIM mode following the ions m/z 311.11456 and 308.59499 (GSH and GSSG respectively) using the ion 445.12003 as lock mass. The mass spectrometer ran in positive polarity, the source voltage was 3.0 kV, and the capillary temperature was set at 350°C. Additional sheath gas flow was put at 35 and auxiliary gas flow rate at 10. Auxiliary gas heater temperature was put at 60°C. AGC target was put at 1e5 ions with a maximum ion injection time of 200 ms) acquired at a resolution of 70,000. For the data analysis we manually integrated the peaks representing GSH and GSSG using the Thermo XCalibur Qual Browser software (Thermo Fisher Scientific) and data are represented as area of the respective GSH and GSSG peaks.
Detection of dNTPs
ECs were grown on a 6 well plate (200 000 cells) and were extracted in 300 μL of a 50:30:20 (methanol: acetonitrile: 20 mM Tris, pH 9.3) extraction buffer. Extraction samples were then centrifuged for 5 min at 15,000 × g and the supernatant was transferred to LC-MS vials. Targeted measurements of dATP, dTTP and dCTP were performed using a Dionex UltiMate 3000 LC System (Thermo Fisher Scientific) coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) operated in negative mode. Due to co-elution of dGTP with ATP the levels of dGTP cannot be purely distinguished from ATP by mass. Thus, dGTP levels were not included in the analysis. Practically, 35 μL of sample was injected on a SeQuant ZIC/pHILIC Polymeric column (Merck Millipore). The gradient started with 20% of solvent B (10 mM NH4-acetate in MQH2O, pH 9.3) and 80% solvent A (LC-MS grade acetonitrile) and remained at 20% B until 2 min post injection. Next, a linear gradient to 80% B was carried out until 29 min. At 38 min the gradient returned to 40% B followed by a decrease to 20% B at 42 min. The chromatography was stopped at 58 min. The flow was kept constant at 100 μL/min and the column was placed at 25°C throughout the analysis. The MS was operated both in targeted MS2 mode using a spray voltage of 3.5 kV, capillary temperature of 320°C, sheath gas at 10.0, auxiliary gas at 5.0. For the targeted MS2 mode, AGC was set at 2e5, maximum IT at 100 ms, a resolution of 17,500 and an isolation window of 1.2 m/z. Data collection was performed using Xcalibur software (Thermo Fisher Scientific).
Acetyl-CoA, Malonyl-CoA Levels (LC-MS Analysis)
Cells from a 6-well (200,000 cells) were extracted in 110 μL 5% PCA (perchloric acid, Sigma-Aldrich). Following extraction, samples were centrifuged for 10 min at 20×103 × g (cooled at 4°C). The supernatant was transferred to a vial. 90 μL of the extract was loaded onto an Ultimate 3000 UPLC (Thermo Fisher Scientific, Bremen, Germany) equipped with an Atlantis dC18 column (5 um particle, 2.1×150 mm, cat # 186001301, Waters) in line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). The analysis was performed according to a previously described protocol (Basu and Blair, 2011). Briefly, a linear gradient was carried out using solvent A (5 mM ammonium acetate) and solvent B (95% acetonitrile, 5 mM ammonium acetate). Practically, samples were loaded at 98% solvent A and from 1.5 to 5 min a ramp to 25% solvent B was carried out. From 5 min to 5.5 min a linear gradient to 100% solvent B was used, followed by an isocratic flow of 100% B until 14 min. From 14 to 16 min the solvent switched to 100% C (80% acetonitrile, 0.1% formic acid) and maintained at 100% C until 20 min. The gradient returned to 98% solvent A at 26 min and the run was stopped at 32 min. The flow rate was kept constant at 200 μL/min and the column temperature was set at 40°C throughout the run. The mass spectrometer operated in targeted MS2 mode following the masses with m/z 768.12248 (CoA), 809.13361 (acetyl-CoA), 854.12288 (malonyl-CoA) and fragmenting it at a collision energy (NCE) of 35. The mass spectrometer ran in positive polarity, the source voltage was 5.0 kV, and the capillary temperature was set at 450°C. Additional sheath gas flow was put at 60 and auxiliary gas flow rate at 20. Auxiliary gas heater temperature was put at 350°C. AGC target was put at 2e5 ions with a maximum ion injection time of 256 ms acquired at a resolution of 70,000.
For the data analysis, we integrated the peak areas using the Thermo XCalibur Quan Browser software (Thermo Fisher Scientific), the following transitions were used to quantify the aforementioned metabolites: CoA: 768.12248 —>261.1270, acetyl-CoA: 809.13361 —> 303.1385 and malonyl-CoA: 854.12288 —> 347.1280. Abundances are expressed as area under the curve.
Palmitate Measurement on GC-QQQ
Extracted fatty acids (chloroform fraction) were dried and re-dissolved in 75 μL of N-Methyl-N-trimethylsilylfluoroacetamide (Sigma-Aldrich, Bornem, Belgium). The mixture was heated at 60°C for 60 min. Reaction mixtures were centrifuged for 15 min at 20,000 × g at 4°C in order to remove insolubilities, the supernatant was transferred to a glass vial with conical insert (Agilent). GC-MS analyses were performed using an Agilent 7890A GC equipped with a HP-5 ms 5% Phenyl Methyl Silox (30 m - 0.25 mm i.d. - 0.25 μm; Agilent Technologies, Santa Clara, California, USA) capillary column, interfaced with a triple quadruple tandem mass spectrometer (Agilent 7000B, Agilent Technologies) operating under ionization by electron impact at 70 eV. The injection port, interface, and ion source temperatures were kept at 230°C. Temperature of the quadrupoles was maintained at 150°C. The injection volume was 1 μL, and samples were injected at 1:10 split ratio. Helium flow was kept constant at 1 mL/min. GC oven temperature was held at 60°C for 1 min, increased to 300°C at 10°C/min, and kept for 5 min. Next, the temperature was increased to 325°C and kept for 5 min. The mass spectrometer operated in SIM mode focusing on the 12C palmitic acid (m/z 313.3, retention time (RT) 17.82) and heptadecanoic acid standard (C17) (m/z 327.3, RT 18.75). For the data analysis we integrated the peak areas using the Agilent MassHunter Quantitative Analysis software, and normalized the palmitate peak area by the C17 standard peak area and the protein content per sample.
Determination of 13C-palmitate, Glucose and Glutamine Incorporation in TCA Intermediates
For 13C-carbon incorporation from palmitate, glucose, and glutamine in metabolites, cells were incubated for 48 hr with labeled substrates. For ECs, 13C-palmitate labeling was done in “100% labeling” conditions, whereby all 12C-palmitate in EGM2 culture medium (120 μM) was replaced by 120 μM [U-13C]-palmitate using EGM2 medium, containing charcoal stripped serum (which does not contain any fatty acids). Similar labeling methods were used for glucose (5.5 mM) and glutamine (2 mM). Metabolites for the subsequent mass spectrometry analysis were prepared by snap-freezing the cells in liquid nitrogen followed by a cold two phase methanol-water-chloroform extraction (Fendt et al., 2013). Phase separation was achieved by centrifugation at 4°C and the methanol-water phase containing polar metabolites was separated and dried using a vacuum concentrator (Fendt et al., 2013). The dried metabolite samples were stored at −80°C (Fendt et al., 2013). Polar metabolites were derivatized for 90 min at 37°C with 7.5 μL of 20 mg/mL methoxyamine in pyridine and subsequently for 60 min at 60°C with 15 μL of N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide, with 1 % tert-butyldimethylchlorosilane (Fendt et al., 2013). Isotopomer distributions and metabolite levels were measured with a 7890A GC system (Agilent Technologies) combined with a 5975C Inert MS system (Agilent Technologies). One microliter of sample was injected onto a DB35MS column in splitless mode using an inlet temperature of 270°C (Fendt et al., 2013). The carrier gas was helium with a flow rate of 1 mL/min. Upon injection, the GC oven was held at 100°C for 3 min and then ramped to 300°C with a gradient of 2.5°C/min. The MS system was operated under electron impact ionization at 70 eV and a mass range of 100–650 amu was scanned. Isotopomer distributions were extracted from the raw ion chromatograms using a custom Matlab M-file, which applies consistent integration bounds and baseline correction to each ion (Antoniewicz et al., 2007). In addition, we corrected for naturally occurring isotopes using the method of Fernandez et al (Fernandez et al., 1996). For relative metabolite levels, the total ion count was normalized to the internal standards norvaline and glutarate and to the protein content (Fendt et al., 2013). To correct for enrichment dilution, we used previously reported methods (Fendt et al., 2013). i.e. we divided the fractional contribution of a labeled metabolite of interest by the fractional contribution of its precursor (calculated by the formula below).
The total contribution of carbon was calculated using the following equation (Fendt et al., 2013):
Herewith, “n” is the number of C atoms in the metabolite, “i” represents the different mass isotopomers and “m” refers to the abundance of a certain mass. Glycolytic carbon contribution was calculated based on 13C-glucose labeling and label dilution in pyruvate (Fendt et al., 2013). For total metabolite levels, arbitrary units of the metabolite of interest were normalized to the protein content.
Lipidomics Analysis
Lipid composition and characterization has been performed as previously described (Rysman et al., 2010). In brief, cell pellets were diluted in PBS. 700 μl of the sample was mixed with 800 μl 1 N HCl:CH3OH 1:8 (v/v), 900 μl CHCl3 and 200 μg/ml of the antioxidant 2,6-di-tert-butyl-4-methylphenol (BHT; Sigma-Aldrich). The organic fraction was evaporated using a Savant Speedvac spd111v (Thermo Fisher Scientific) at room temperature and the remaining lipid pellet was stored at −20°C under argon.
Mass spectrometry
Just prior to mass spectrometry analysis, lipid pellets were reconstituted in running solution (CH3OH:CHCl3:NH4OH; 90:10:1.25; v:v:v). Phospholipid species were analyzed by electrospray ionization tandem mass spectrometry (ESI-MS/MS) on a hybrid triple quadrupole/linear ion trap mass spectrometer (4000 QTRAP system; Applied Biosystems SCIEX) equipped with a TriVersa NanoMate (Advion Biosciences) robotic nanosource for automated sample injection and spraying. Phospholipid profiling was executed by (positive or negative) precursor ion or neutral loss scanning at a collision energy of 50 eV/45 eV, 35 eV, −35 eV, and −60 eV for precursor 184 (sphingomyelin(SM)/phosphatidylcholine (PC)), neutral loss 141 (phosphatidylethanolamine (PE)), neutral loss 87 (phosphatidylserine (PS)) and precursor 241 (phosphatidylinositol (PI)), respectively. Phospholipid quantification was performed by multiple reactions monitoring (MRM), the transitions being based on the neutral losses or the typical product ions as described above. Lipid standards PC25:0, PC43:6, SM30:1, PE25:0, PE43:6, PI25:0, PI31:1, PI43:6, PS25:0, PS31:1 and PS37:4 (Avanti Polar Lipids) were added based on the amount of DNA in the original sample. Typically, a 3-minute period of signal averaging was used for each spectrum. The data were corrected for carbon isotope affects and chain length and analyzed using in house-developed software (RALP).
RNA Isolation and Quantitative RT-PCR
RNA was collected and purified using the PureLink RNA Mini Kit (Invitrogen, Life Technologies) and converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad). RNA expression analysis was performed by Taqman quantitative RT-PCR as described (Carmeliet et al., 2001) using premade primer sets (IDT). Premade primer set ID numbers are listed in the Key Resources Table. For comparison of gene expression between conditions, expression (normalized to HPRT as endogenous control) is expressed relative to control condition.
Immunoblot and Co-immunoprecipitation Analysis
Immunoblotting
Protein extraction and immunoblot analysis were performed using a modified Laemmli sample buffer (125 mM Tris-HCl, pH 6.8 buffer containing 2% SDS and 10% glycerol), cell lysis buffer (Cell Signaling Technology) or RIPA lysis and extraction buffer (Thermo Fisher Scientific) in the presence of protease and phosphatase inhibitors (Roche, Vilvoorde, Belgium). Lysates were separated by SDS-PAGE under reducing conditions, transferred to a nitrocellulose membrane, and analyzed by immunoblotting. Primary antibodies used were rabbit anti-FASN (No. Santa Cruz Biotechnology, the Netherlands), rabbit tubulin (Sigma-Aldrich, #T6199), ACC1 (Cell Signaling Technology, #3662), RAPTOR (Cell Signaling Technology, #2280), RICTOR (Cell Signaling Technology, #9476), mLST8 (GβL)(Cell Signaling Technology, #3274), phospho-p70S6K (Cell Signaling Technology, #9205), p70S6K (Cell Signaling Technology, #9202), phospho4EBP1 (Cell Signaling Technology, #2855), 4EBP1 (Cell Signaling Technology, #9452), phospho-Akt(S473) (Cell Signaling Technology, #4058), Akt (Cell Signaling Technology, #9272) phospho-AMPKα (Cell Signalling Techonology, #2531), AMPKα (Cell Signaling Technology, #2532). Antibodies used for immunoprecipitation were rabbit anti-malonylated lysine (PTM Biolabs, No. PTM-901, Illinois, USA), mouse anti-mTOR (Life technologies, #AH01232), and rabbit anti-mTOR (Cell Signaling Technology, #2972). Equal loading was verified by beta-actin immunoblotting (rabbit anti-β Actin, Cell Signaling Technology, #4970) and Ponceau Red staining. All antibodies were used in a 1:1000 dilution in 5% bovine serum albumin (BSA). Appropriate secondary antibodies were from Cell Signaling Technology (Anti-Rabbit IgG HRP linked #7074; Anti-Mouse IgG HRP linked, #7076) in a 1:2000 dilution in 5% bovine serum albumin (BSA). Signal was detected using the ECL and/or Femto systems (Amersham Biosciences, GE Healthcare, Diegem, Belgium) according to the manufacturer’s instructions. Densitometric quantifications of bands were done with Fiji software (https://fiji.sc).
Detection of mTOR Malonylation
For detection of endogenous malonylated mTOR in HUVEC, cells were lysed in Cell signaling lysis buffer (Cell Signaling Technology) supplemented with 10 mM nicotinamide (NAM) and protease and phosphatase inhibitors. After sonication and pre-clearing (1 hr incubation of protein lysate with Dynabead protein A or G slurry (Life Technologies) on a rotating platform), a lysate volume containing 800–1000 μg of protein per sample was submitted to immunoprecipitation using Dynabead protein A or G slurry (Life Technologies) and 10 μL of either anti-mTOR (Life Technologies) or anti-malonylated lysine antibody (PTM Biolabs). The immune complexes were analyzed by western blotting using the reciprocal antibody.
Co-immunoprecipitation
HUVECs were lysed using a modified coIP buffer (40mM HEPES pH 7.5, 120 mM NaCl, 0.3% CHAPS, 2 mM EDTA, and phosphatase and protease inhibitors). The lysate was incubated with gentle shaking for 3 hr at 4°C. No shearing or sonication was performed, not to compromise the integrity of mTORC1 and mTORC2 complexes. After pre-clearing (1 hr incubation of protein lysate with Dynabead protein A or G slurry (Life Technologies) on a rotating platform), a volume of lysate containing 1500 μg of protein was submitted to immunoprecipitation using Dynabead protein A slurry (Life Technologies) and 10 μL of anti-mTOR antibody. The immune complexes were analyzed by western blotting using anti-RAPTOR, anti-RICTOR and anti-mLST8.
Protein Membrane Extraction
Protein membrane extraction was performed on HUVEC lysates using the Plasma membrane protein extraction kit (#P503L; 101 Biosciences) according to the manufacturer’s protocol. The fractions for cytoplasmic, plasma membrane, and organelle membrane proteins were analysed by western blotting. To assess the quality of the fractionation, immunoblotting for α-tubulin (Cell Signaling Technology, 2144S) and Na+/K+ ATP-ase (Cell Signaling Technology, 3010S) was performed, respectively markers enriched in the cytoplasm and plasma membrane fraction.
Determination of MTORC1 Kinase Activity
Recombinant Assay
The was performed as previously described (Dunlop et al., 2009). Briefly, HEK293 cells transfected with Myc-mTOR, HA-Raptor and FASN shRNA (by lipofectamine 2000, Thermo Fisher Scientific) were lysed in mTORC1 Lysis Buffer (40 mM HEPES pH 7.4, 2 mM EDTA, 10 mM β-glycerophosphate, 0.3% (w/v) CHAPS and protease inhibitors). Insoluble material was removed by centrifugation at 13,000 rpm for 8 min. mTORC1 complex was purified using anti-Myc antibodies coupled to Protein-G Sepharose beads by immunoprecipitation for 4 hr at 4°C. Beads were washed once in low salt buffer (40 mM HEPES pH 7.4, 2 mM EDTA,10 mM β-glycerophosphate,150 mM NaCl, 0.3% (w/v) CHAPS and protease inhibitors), then twice in high salt buffer (40 mM HEPES pH 7.4, 2 mM EDTA, 10 mM β-glycerophosphate, 400 mM NaCl, 0.3% (w/v) CHAPS and protease inhibitors), and twice in HEPES/KCl buffer (25 mM HEPES pH 7.4, 20 mM KCl) and then subjected to in vitro mTORC1 assay with and without purified GTP-loaded GST-Rheb(Q64L).
GST-Rheb(Q64L) (GTP-loaded) was purified from transfected HEK293 cells (lipofectamine 2000, Thermo Fisher Scientific) after lysis in Rheb lysis buffer (40 mM HEPES pH 7.4,10 mM, β-glycerophosphate, 10 mM pyrophosphate, 5 mM MgCl2, 0.3% (w/v) CHAPS and protease inhibitors (but no DTT)). GST-Rheb(Q64L) was captured on a GST spin trap module (GE Healthcare, UK), washed three times in Rheb lysis buffer, then Rheb storage buffer (20 mM HEPES pH 8, 200 mM NaCl, 5 mM MgCl2) and then eluted in Rheb storage buffer with 10 mM reduced glutathione.
mTORC1 in vitro kinase assay was performed in 25 mM HEPES pH 7.4, 10 mM MgCl2, 140 mM KCl, 0.5 mM ATP, and 0.3 μCi of [γ−32P]ATP (Perkin Elmer) using 150 ng dephosphorylated recombinant 4E-BP1 as substrate per assay. Reaction was performed at 30°C for 30 min with constant shaking and stopped by the addition of 4× sample buffer (0.5 M Tris pH 6.8, 2.86 M β-mercaptoethanol, 30% (v/v) glycerol, 0.4 mM bromophenol blue). Samples were resolved by SDS-PAGE, and the gel was then fixed in 10 % (v/v) acetic acid and 45 % (v/v) methanol, dried under vacuum and the level of [32P]-radiolabel incorporation into 4E-BP1 was determined by autoradiography.
K-LISA mTOR Activity Kit
The assay was carried out according to the manufacturer’s protocol. Briefly, this ELISA-based activity assay utilizes the phosphorylation of recombinant p70S6K-GST fusion protein as readout for mTORC1 activity. mTORC1 was immunoprecipitated from endothelial cell lysate. Endothelial cells were lysed in coIP buffer (40mM HEPES pH 7.5, 120 mM NaCl, 0.3% CHAPS, 2mM EDTA, and phosphatase and protease inhibitors) in order to retain mTORC1 integrity. The lysates were clarified by centrifugation (16000g for 10 min) and total protein was adjusted to 2–3mg. Immunoprecipitation was performed by overnight incubation with anti-mTOR antibody (Life technologies) at 4°C, followed by 1 hr of incubation with 20 μL of Dynabeads protein G antibody at room temperature. The immunoprecipitates were washed twice with lysis buffer and once with kinase buffer.
After washes, the mTORC1-bound beads were incubated with recombinant de-phosphorylated p70S6K along with ATP. After 30 min at 30°C, the supernatants were collected and added into wells of a glutathione-coated 96-well plate. The phosphorylated substrate was detected with Anti-p70S6K-T389 secondary antibody, followed by detection with HRP-antibody conjugate and TMB substrate. The absorbance of each well at a wavelength of 450 nm was measured against a reference wavelength of 595 nm.
Detection of Malonylated Proteins (KMAL Peptide Screen)
Cells were lysed in 50 mM Triethylammonium bicarbonate (TEAB, Sigma-Aldrich) pH 8, protease inhibitors, and 10mM Nicotinamide, followed by sonication and centrifugation. Reduction was done with 5 mM DTT at 50°C for 30 min, and afterwards alkylation was done using 15 mM of iodoacetamide at room temperature for 30 min in darkness. Digestion to peptides was performed using trypsin (Promega), keeping an enzyme-to-substrate ratio of 1/50 (w/w). The digestion was performed at 37°C with gentle shaking overnight and stopped by boiling at 95°C for 10 min. For desalting the tryptic peptides, SPE C18 cartridges (Waters) were used. Prior to loading the samples, the cartridges were activated by passing 3 mL of Conditioning /Elution Solution (70% acetonitrile/water with 0.1% TFA (v/v/v)) through the packing bed. The cartridges were then equilibrated by passing 6 mL of Equilibration /Loading solution (0.1% TFA in water (v/v)) through the packing bed. The pH of each tryptic peptide sample was adjusted to 3 with formic acid, and subsequently loaded in the SPE cartridges (up to 1 mL) by passing slowly (no faster than 1 drop/second) into the cartridge packing. Loading was completed by slowly passing 1 mL of Equilibration/Load solution. Each sample was then washed by passing 6 mL desalting solution (2% acetonitrile/water with 0.1% TFA (v/v/v)) through the packing bed. Finally, each sample was eluted by slowly (less than 1 drop/second) passing 1 mL of Conditioning /Elution Solution through the cartridge bed. The eluates were dried in a Speed-Vac overnight, and resuspended in NETN buffer (50 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5% NP40, pH 8). The resuspended peptides were immunoprecipitated using Protein A dynabeads conjugated to anti-Pan Malonyl Lysine (Kmal) (PTM Biolabs) antibody. The peptides were incubated with the dynabeads for 4 hours at 4°C, washed twice in NETN buffer, and once in NETN buffer without NP40. Finally, the peptides were eluted in 3 fractions in 0,1% TFA, and dried overnight using a vacuum concentrator.
The dried peptide mixtures were redissolved in 20 μL 0.1% TFA in water/ acetonitrile, 2/98 (v/v). 2.5 μL was loaded on an Ultimate 3000 RSLC nano LC (Thermo Fisher Scientific, Bremen, Germany) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific) to perform an LC-MS/MS analysis. The peptides were first loaded onto a trapping column (made in-house, 100 μm internal diameter (I.D.) × 20 mm, 5 μm beads C18 Reprosil-HD, Dr. Maisch, Ammerbuch-Entringen, Germany) and after flushing from the trapping column, peptides were loaded on an analytical column (made in-house, 75 μm I.D. × 150 mm, 3 μm beads C18 Reprosil-HD, Dr. Maisch) packed in the needle (PicoFrit SELF/P PicoTip emitter, PF360–75-15-N-5, New Objective, Woburn, MA, USA). Peptides were loaded with loading solvent and separated with a linear gradient from 98% solvent A’ (0.1% formic acid in water) to 55% solvent B′ (0.1% formic acid in water/acetonitrile, 20/80 (v/v)) in 30 min at a flow rate of 300 nL/min, followed by a 9 min wash reaching 99% solvent B’ ending with an equilibration of 9 min with 98% solvent A’. The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and MS/MS acquisition for the 10 most abundant peaks in a given MS spectrum. The source voltage was 3.5 kV, and the capillary temperature was 275°C. One MS1 scan (m/z 400−2,000, AGC target 3 × 106 ions, maximum ion injection time 80 ms), acquired at a resolution of 70,000 (at 200 m/z), was followed by up to 10 tandem MS scans (resolution 17,500 at 200 m/z) of the most intense ions fulfilling predefined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 60 ms, isolation window 2 m/z, fixed first mass 140 m/z, spectrum data type: centroid, underfill ratio 2%, intensity threshold 1.7×E4, exclusion of unassigned, 1, 5–8, >8 positively charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time 50 s). The HCD collision energy was set to 25% Normalized Collision Energy and the polydimethylcyclosiloxane background ion at 445.120025 Da was used as internal calibration (lock mass).
Mascot Generic Files were created using Distiller software (version 2.5.1.0 Matrix Science, www.matrixscience.com/Distiller). Next, these peak lists were searched with the Mascot search engine using the Mascot Daemon interface (version 2.5.1, Matrix Science) against the SwissProt database restricted to human proteins (release-2015_04, 20273 entries). Variable modifications were set to pyro-glutamate formation of amino terminal glutamine, oxidation of methionine and malonylation of lysine embedding a neutral loss scoring of CO2. Carbamidomethylation of cysteine was set as a fixed modification. Mass tolerance on MS was set to 10 ppm (with Mascot’s C13 option set to 1), MS/MS tolerance at 20 mmu. The peptide charge was set to 1+,2+,3+ and instrument setting was put on ESI-QUAD. Enzyme was set to trypsin/P, allowing 2 missed cleavages. Only peptides that were ranked one and scored above the threshold score, set at 99% confidence, were withheld.
Flow Cytometry
Cell Cycle Analysis
Cells in G1 were identified as a 2N DNA population including 5-ethynyl-2 ′- deoxyuridine (EdU, Invitrogen, Life Technologies) incorporation. Briefly, cells were labeled with 10 μM EdU over 24 hr, collected by trypsinization and fixed (4% PFA). The incorporated EdU was detected by a ‘click-It reaction’ with Alexa Fluor 647 according to the manufacturer’s instructions (Invitrogen, Life Technologies). Alexa Fluor 647 was excited with a 604 nm red laser and emission was recorded at 660 nm. Data were recorded by the flow cytometer, and resultant data were analyzed with the FlowJo 8.8.6 software (https://www.flowjo.com).
Determination of MTOR Malonylation Stoichiometry
For quantification of mTOR malonylation by targeted LC-MS, Fmoc-Lys(mono-tert-butylmalonate)-OH was synthesized according to (Peng et al., 2011). The stable isotope-labelled (SIL) tryptic peptides IVK(mal)GYTLADEEEDPLIYQHR, AALETVDR, DASAVSLSESK, ETSFNQAYGR, ILLNIEHR, TLDQSPELR of mTOR were synthesized in-house using a Syro I synthesis unit (MultiSynTech, Witten, Germany) and Fmoc chemistry. A “heavy”-labelled arginine or lysine was incorporated at the C terminus resulting in a mass shift of +10.0083 or +8.0142 Da, respectively. Peptides were purified by preparative RP-LC coupled via a 1:20 split to an MS-Q (Thermo Fisher Scientific, Bremen, Germany) mass spectrometer for targeted fractionation. After lyophilization, peptides were solubilized in 0.1 % TFA and concentration was determined using amino acid analysis according to Cohen et al. (Cohen and Michaud, 1993). Quantification was conducted against a five-point calibration curve of derivatized amino acids in the range from (5 −25 pmol/μL).
mTOR was immunoprecipitated from EC lysate as described above, starting with 3mg of protein. The samples were eluted in 3 % SDS, reduced with 10 mM DTT (30 min at 56°C) and alkylated using 20 mM IAA (30 min in the dark). For precipitation, 9 volumes of ice-cold ethanol were added followed by incubation at −40°C for 1 hr. After centrifugation at 20,000 × g for 45 min, the protein pellet was washed with ice-cold acetone followed by another incubation and centrifugation. The protein pellet was resolubilized in 3 μL 2 M GuHCL and digested with 500 ng trypsin (sequencing grade – modified, Promega, Mannheim, Germany) in 17 μL 50 mM ABC and 1 mM CaCl2 at 37°C for 15 hr. The digestion was stopped by adding TFA to a final concentration of 1 %. 0.5 fmol of the KMal SIL peptide and 10 fmol of non-modified SIL peptides were spiked into 33% of digest before LC-MS analysis.
Targeted nanoLC-MS/MS was done in parallel reaction monitoring (PRM) mode to absolutely quantify the levels of malonylated and non-modified mTOR peptides against the known concentrations of the SIL peptides. PRM was performed using a Q Exactive HF mass spectrometer online coupled to a U3000 RSLC nanoHPLC equipped with an Acclaim PepMap trap-column (100 μm × 2 cm, 5 μm particles, 100 Å pores) and an Acclaim PepMap main column (75 μm × 50 cm, 3 μm particles, 100 Å pores, all from Thermo Fisher Scientific, Bremen Germany). Peptides were injected onto the trap column in 0.1 % TFA using a flow rate of 10 μL/min. After 5 min the trap column was switched in-line and peptides were separated using a 52 min gradient ranging from 2.5 to 35 % ACN in 0.1 % FA at a flow rate of 250 nL/min (at 60°C).
For quantification PRM was conducted using the m/z of the +3 ion of malonylated and the +2 ions of non-modified peptides (SIL and endogenous). Fragmentation was done using 0.8 m/z isolation width and normalized collision energy of 27. Fragment ion spectra of the malonylated peptide were acquired at a resolution R = 240,000 with a maximum ion injection time (maxIT) of 1500 ms and non-modified peptides were acquired at R = 60,000 with 250 ms maxIT. Automatic gain control (AGC) target value was 106 for both.
Raw-files were imported into Skyline v3.6 and the 1–3 best transitions (y7+, y5+, y4+) of the malonylated and the 4–8 best transitions of the non-modified peptides AALETVDR, ETSFNQAYGR and ILLNIEHR were used for quantification. Peptide ratio results were exported to Microsoft Excel and the percentage of mTOR malonylation (site occupancy) was determined via:
In Vitro Functional Assays
Proliferation
EC Proliferation was quantified by incubating cells for 2 hr with 1 μCi/mL [3H]-thymidine. Thereafter, cells were fixed with 100% ethanol for 15 min at 4°C, precipitated with 10% TCA and lysed with 0.1 N NaOH. The amount of [3H]-thymidine incorporated into DNA was measured by scintillation counting.
Doubling Time
50,000 cells were seeded in a 24 well plate format and counted after 24, 48, and 72 hr. Doubling time was calculated using the following equation: (72–24)*(log(2)/(log(cell number 72 hr/cell number 24 hr)).
Intracellular ROS Analysis
Intracellular ROS levels were measured using 5-(and-6)-chloromethyl- 2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) according to the manufacturer’s instructions (Invitrogen, Life Technologies). CM-H2DCFDA is metabolized by intracellular esterases to a non-fluorescent molecule, which is oxidized by H2O2 to the fluorescence product CM-DCF. The intracellular ROS levels were determined by pre-incubation of the ECs for 30 min with 10 μM CM-H2DCFDA and H2O2 scavenging capacity was determined after a subsequent incubation for 2 hr with 50 μM H2O2 (Merck, Milipore) in serum free M199. The fluorescent intensity was measured using a spectrofluorometer (VICTORTM X2 Multilabel Plate Reader, Perkin Elmer).
Scratch Wound Migration Assay
A scratch wound was applied on confluent EC monolayers (pretreated with 1 μg/mL MitoC for 24 hr where indicated) using a 200 μL tip, 24 hr after seeding (100,000 cells per well in 24-well plates). After scratch wounding and photography (T0), the cultures were further incubated in fully supplemented EGM2 medium for 6 hr (until near closure was reached in the control condition), and photographed again after 6 hr (T6). Migration distance (gap area at T0 minus gap area at T6) was measured with NIH ImageJ software package and used to calculate the % migrated area.
Modified Boyden Chamber Migration
Assays were performed using MitoC-treated (24 hr treatment with 1 μg/mL MitoC) ECs. Therefore, 50,000 cells were seeded in fully supplemented EGM2 medium on 0.1% gelatin coated transwells (8-μ/mL MitoC) ECs. Upon adherence, the transwells were washed and re-fed with medium containing only 0.1% FBS and transferred to bottom wells containing medium with 0.1% FBS (baseline) or 5% FBS (migration stimulus). Transwells were incubated for 16 hr and processed and analysed for migrated cells as described (Schoors et al., 2015).
Spontaneous Motility Assay
Spontaneous motility was assessed using time-lapse video imaging of sparsely seeded cultures grown on glass bottom 24-well plates (PAA, Pashing, Austria). Cell movement was tracked using Imaris software by tracking nucleus position over time and by superimposing migration origin at the zero-cross point (De Bock et al., 2013). Velocity is expressed as μm/hr. Directionality of migration was assessed by analysis of the total length of the migration track of a cell. The directionality value was then calculated by dividing the distance between the start and end location of the cell by the total track length; a higher directionality value denotes higher migration directionality.
Spheroid Capillary Sprouting Assay
ECs were incubated overnight in hanging drops in EGM2 medium containing methylcellulose (20 volume% of a 1.2% solution of methylcellulose 4000 cP) (Sigma-Aldrich, Bornem, Belgium) to form spheroids. For mitotic inactivation, MitoC (1 μg/mL) was added to this medium. Spheroids were then embedded in collagen gel as described (Schoors et al., 2015) and cultured for 20 hr to induce sprouting. Cultures were fixed with 4% PFA at room temperature and imaged under bright field using a Motic AE 31 microscope (Motic Electric Group, Xiamen, China); or Leica DMI6000 microscope (Leica Microsystems, Mannheim, Germany). Analysis of the number of sprouts per spheroid and the total sprout length (cumulative length of primary sprouts and branches per spheroid) was done on phase contrast images using the NIH ImageJ software package.
Immunohistochemistry and Morphometry
Cell Size
Cells were fixed with 4% PFA and stained for F-actin using phalloidin-Alexa 488 (Molecular probes). Cell size was determined using the NIH ImageJ software package.
Apoptosis
Analysis of apoptosis was done by fluorescent staining for cleaved caspase 3 (Cell signaling Technology) and TUNEL (In situ cell death kit, Roche) in 4% PFA fixed monolayers combined with Hoechst 33342 (Molecular Probes, H3570) as nuclear staining and imaged with the Leica DMI6000 microscope (Leica Microsystems, Mannheim, Germany).
Analysis of mTOR and Lysosome Co-localization
Analysis of lysosomal localization of mTOR was done by fluorescent staining for mTOR (Cell signaling Technology, #2972) and LAMP1 (Abcam, ab25630) in 4% PFA fixed monolayers combined with Hoechst 33342 (Molecular Probes, H3570) as nuclear staining and imaged with the Leica DMI6000 microscope (Leica Microsystems, Mannheim, Germany).
Immunohistochemistry of In vivo Angiogenesis Models
All methods for histology and immunostainings have been described (Cantelmo et al., 2016; De Bock et al., 2013). Whole mount retinas were immediately fixed after dissection in 4% PFA overnight at 4°C. Retinas were subjected to immunofluorescence staining using the following isolectin conjugates or primary antibodies: isolectin Griffonia simplicifolia (GS)-IB4-Alexa 488 (Molecular Probes, Thermo Fischer Scientific), anti-NG2 (Merck Millipore), anti-ColIagen IV (Serotec). Retinas were then incubated with the appropriate fluorescently conjugated secondary antibodies (Alexa 488 or 546, Molecular Probes, Invitrogen, Life Technologies), followed by amplification with the proper tyramide signal amplification systems when needed (Perkin Elmer). For mice injected with EdU prior to euthanization, incorporated EdU was detected using the The Click-iT 5-ethynyl-2′-deoxyuridine (EdU) Alexa Fluor 555 Imaging Kit (Invitrogen). Morphometric analysis is described in the section Mouse models of physiological and pathological angiogenesis (see above).
QUANTIFICATION AND STATISTICAL ANALYSIS
The data in the figures and text represent mean ± SEM of at least three independent experiments with for the in vitro experiments each at least three technical replicates. In case of multiple comparisons statistical significance was calculated by ANOVA or for single comparison standard two-tailed t test with Welch’s correction was used (Prism v6.0f). When inter-experimental variability was large between experiments with isolated ECs from different umbilical cord donors, mixed model statistics (R version 3.2.4 using Kenward-Roger approximation) was used with experiment (i.e. donor) as random factor to correct for variation between individual experiments. For all western blot densitometry data, one sample t test was used. For the data in Figure 1D (violin plots of RNA-sequencing data), Fisher’s method combined probability test was used. For all other experiments, standard two-tailed t test with Welch’s correction was used (Prism v6.0f). P < 0.05 was considered statistically significant.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Data
The malonylated peptide screen proteomics data have been deposited in the PRIDE database under accession number PRIDE: PXD006503.
Raw RNA-sequencing data from NEC and TECs are available in the ArrayExpress database under accession number ArrayExpress: E-MTAB-4842.
Software
All software is freely or commercially available and is listed in the STAR Methods description and Key Resources Table.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help with the lipidomics of Etienne Waelkens. We thank Ralph Adams for providing the VE-cadherin(PAC)-CreERT2 mice. We thank John Blenis for constructive discussions. U.B. was an FWO fellow and Marie Curie-IEF fellow. K.C.S.Q. was funded by CAPES foundation, Brazil. F.M.-R., J.K., J.G., C.D., A.R.C., and B.G. are supported by FWO. The work of P.C. is supported by a Federal Government Belgium grant (IUAP7/03); long-term structural Methusalem funding by the Flemish Government; grants from the FWO and Foundation against Cancer; and a European Research Council Advanced Research grant (EU-ERC269073). X.L. is supported by the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center at the Sun Yat-Sen University and by the National Natural Science Foundation of China (81330021, 81670855).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.cmet.2018.07.019.
REFERENCES
- Aden DP, Fogel A, Plotkin S, Damjanov I, and Knowles BB (1979). Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 282, 615–616. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, and Stephanopoulos G (2007). Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, and Denu JM (2016). Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 41, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SS, and Blair IA (2011). SILEC: a protocol for generating and using isotopically labeled coenzyme A mass spectrometry standards. Nat. Protoc. 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, and Adams RH (2009). The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 137, 1124–1135. [DOI] [PubMed] [Google Scholar]
- Browne CD, Hindmarsh EJ, and Smith JW (2006). Inhibition of endothelial cell proliferation and angiogenesis by orlistat, a fatty acid synthase inhibitor. FASEB J. 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Cailleau R, Olive M, and Cruciger QV (1978). Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro 14, 911–915. [DOI] [PubMed] [Google Scholar]
- Cantelmo AR, Conradi LC, Brajic A, Goveia J, Kalucka J, Pircher A, Chaturvedi P, Hol J, Thienpont B, Teuwen LA, et al. (2016). Inhibition of the glycolytic activator PFKFB3 in endothelium induces tumor vessel normalization, impairs metastasis, and improves chemotherapy. Cancer Cell 30, 968–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Moons L, Luttun A, Vincenti V, Compernolle V, De Mol M, Wu Y, Bono F, Devy L, Beck H, et al. (2001). Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat. Med. 7, 575–583. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, and Mann M (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. [DOI] [PubMed] [Google Scholar]
- Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K, and Fruttiger M (2008). Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis 46, 74–80. [DOI] [PubMed] [Google Scholar]
- Cohen SA, and Michaud DP (1993). Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 211, 279–287. [DOI] [PubMed] [Google Scholar]
- Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H, Cheng Z, Park J, Wan X, Liu X, et al. (2015). Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteomics 14, 3056–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G, et al. (2013). Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 154, 651–663. [DOI] [PubMed] [Google Scholar]
- Du Y, Cai T, Li T, Xue P, Zhou B, He X, Wei P, Liu P, Yang F, and Wei T (2015). Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteomics 14, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Dodd KM, Seymour LA, and Tee AR (2009). Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell. Signal. 21, 1073–1084. [DOI] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, and Stephanopoulos G (2013). Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, and Brunengraber H (1996). Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262. [DOI] [PubMed] [Google Scholar]
- Gorsky MK, Burnouf S, Dols J, Mandelkow E, and Partridge L (2016). Acetylation mimic of lysine 280 exacerbates human Tau neurotoxicity in vivo. Sci. Rep. 6, 22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, et al. (2008). SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 283, 20015–20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Vandekeere S, Kalucka J, Bierhansl L, Zecchin A, Bruning U, Visnagri A, Yuldasheva N, Goveia J, Cruys B, et al. (2017). Role of glutamine and interlinked asparagine metabolism in vessel formation. EMBO J. 36, 2334–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DA, Lane HM, Zygmont ME, Dervan PA, and Hennigar RA (2007). MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines. Anticancer Res. 27, 27–34. [PubMed] [Google Scholar]
- Jaffe EA, Nachman RL, Becker CG, and Minick CR (1973). Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 52, 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keydar J, Gilead Z, Hartman JR, and Ben-Shaul Y (1973). In vitro production of mouse mammary tumor virus in a mouse mammary tumor ascites line. Proc. Natl. Acad. Sci. USA 70, 2983–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel SJ, Axelrod F, Rozenkrantz N, and Smith JW (2004). Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 64, 2070–2075. [DOI] [PubMed] [Google Scholar]
- Kuda O, Pietka TA, Demianova Z, Kudova E, Cvacka J, Kopecky J, and Abumrad NA (2013). Sulfo-N-succinimidyl oleate (SSO) inhibits fatty acid uptake and signaling for intracellular calcium via binding CD36 lysine 164: SSO also inhibits oxidized low density lipoprotein uptake by macrophages. J. Biol. Chem. 288, 15547–15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, et al. (2017). Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 24, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson AS, and Jordan VC (1997). MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 57, 3071–3078. [PubMed] [Google Scholar]
- Meyer JG, D’Souza AK, Sorensen DJ, Rardin MJ, Wolfe AJ, Gibson BW, and Schilling B (2016). Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J. Am. Soc. Mass Spectrom. 27, 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michieli P, Mazzone M, Basilico C, Cavassa S, Sottile A, Naldini L, and Comoglio PM (2004). Targeting the tumor and its microenvironment by a dual-function decoy Met receptor. Cancer Cell 6, 61–73. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Miyano K, Yamane T, and Sato J (1982). Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42, 3858–3863. [PubMed] [Google Scholar]
- Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, et al. (2015). SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. (2010). Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. (2011). The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix TN, Patmore DM, Boop S, Boulos N, Jacus MO, Patel YT, Roussel MF, Finkelstein D, Goumnerova L, Perreault S, et al. (2016). Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell 29, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, and Kroemer G (2015). Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821. [DOI] [PubMed] [Google Scholar]
- Point V, Benarouche A, Zarrillo J, Guy A, Magnez R, Fonseca L, Raux B, Leclaire J, Buono G, Fotiadu F, et al. (2016). Slowing down fat digestion and absorption by an oxadiazolone inhibitor targeting selectively gastric lipolysis. Eur. J. Med. Chem. 123, 834–848. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig F, and Schulze A (2016). The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749. [DOI] [PubMed] [Google Scholar]
- Rosner M, Siegel N, Valli A, Fuchs C, and Hengstschlager M (2010). mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids 38, 223–228. [DOI] [PubMed] [Google Scholar]
- Roudnicky F, Poyet C, Wild P, Krampitz S, Negrini F, Huggenberger R, Rogler A, Stohr R, Hartmann A, Provenzano M, et al. (2013). Endocan is upregulated on tumor vessels in invasive bladder cancer where it mediates VEGF-A-induced angiogenesis. Cancer Res. 73, 1097–1106. [DOI] [PubMed] [Google Scholar]
- Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniels VW, Machiels J, et al. (2010). De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 70, 8117–8126. [DOI] [PubMed] [Google Scholar]
- Saggerson D (2008). Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. 28, 253–272. [DOI] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, et al. (2015). Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 520, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, and Mann M (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594. [DOI] [PubMed] [Google Scholar]
- Stone KR, Mickey DD, Wunderli H, Mickey GH, and Paulson DF (1978). Isolation of a human prostate carcinoma cell line (DU 145). Int. J. Cancer 21, 274–281. [DOI] [PubMed] [Google Scholar]
- Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, et al. (2016). Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Giampietro C, Conti A, Orsenigo F, Breviario F, Pirazzoli V, Potente M, Daly C, Dimmeler S, and Dejana E (2008). Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nat. Cell Biol. 10, 923–934. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A, Colomer R, Brunet J, Lupu R, and Menendez JA (2008). Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell Prolif. 41, 59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura R, Mordec K, Waszczuk J, Wang Z, Lai J, Fridlib M, Buckley D, Kemble G, and Heuer TS (2015). Inhibition of de novo palmitate synthesis by fatty acid synthase induces apoptosis in tumor cells by remodeling cell membranes, inhibiting signaling pathways, and reprogramming gene expression. EBioMedicine 2, 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Schneider JG, Shenouda SM, Lee A, Towler DA, Chakravarthy MV, Vita JA, and Semenkovich CF (2011). De novo lipogenesis maintains vascular homeostasis through endothelial nitric-oxide synthase (eNOS) palmitoylation. J. Biol. Chem. 286, 2933–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, and Choudhary C (2015). Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 34, 2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenes M, Shang M, Di Matteo M, Goveia J, Martin-Perez R, Serneels J, Prenen H, Ghesquiere B, Carmeliet P, and Mazzone M (2016). Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 24, 701–715. [DOI] [PubMed] [Google Scholar]
- Wragg JW, Finnity JP, Anderson JA, Ferguson HJ, Porfiri E, Bhatt RI, Murray PG, Heath VL, and Bicknell R (2016). MCAM and LAMA4 are highly enriched in tumor blood vessels of renal cell carcinoma and predict patient outcome. Cancer Res. 76, 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagasaki R, Nakahara T, Mori A, Sakamoto K, and Ishii K (2014a). Effects of mTOR inhibition on normal retinal vascular development in the mouse. Exp. Eye Res. 129, 127–134. [DOI] [PubMed] [Google Scholar]
- Yagasaki R, Nakahara T, Ushikubo H, Mori A, Sakamoto K, and Ishii K (2014b). Anti-angiogenic effects of mammalian target of rapamycin inhibitors in a mouse model of oxygen-induced retinopathy. Biol. Pharm. Bull. 37, 1838–1842. [DOI] [PubMed] [Google Scholar]
- Yang CS, Matsuura K, Huang NJ, Robeson AC, Huang B, Zhang L, and Kornbluth S (2015). Fatty acid synthase inhibition engages a novel caspase-2 regulatory mechanism to induce ovarian cancer cell death. Oncogene 34, 3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, and Smans K (2013). Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 52, 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
Data
The malonylated peptide screen proteomics data have been deposited in the PRIDE database under accession number PRIDE: PXD006503.
Raw RNA-sequencing data from NEC and TECs are available in the ArrayExpress database under accession number ArrayExpress: E-MTAB-4842.
Software
All software is freely or commercially available and is listed in the STAR Methods description and Key Resources Table.