Abstract
Lung cancer has become the leading cause of cancer-associated mortality worldwide. However, the under- lying mechanisms of lung cancer remain poorly understood. DnaJ heat shock protein family (HSP40) member C12 (DNAJC12) is a type III member belonging to the HSP40/DNAJ family. The role of DNAJC12 in numerous types of cancer has been previously reported; however, the effect of DNAJC12 in lung cancer remains unknown. The results of the present study indicated that DNAJC12 may be involved in lung cancer proliferation and migration by regulating the β-catenin signaling pathway. Data generated in the present study and from The Cancer Genome Atlas revealed that the DNAJC12 expression levels were significantly upregulated in lung cancer tissues compared with non-cancer lung tissues. The expression of DNAJC12 was subsequently knocked down in A549 and NCI-H1975 lung cancer cells using lentiviral transfections and further experiments demonstrated that the knockdown of DNAJC12 inhibited the proliferation, colony formation, migration and invasion of lung cancer cells. The results of flow cytometric assays also revealed that the knockdown of DNAJC12 induced the apoptosis of lung cancer cells. In addition, the effects of DNAJC12 knockdown on the in vivo growth of lung cancer cells were observed. Signaling pathway analysis revealed that the knockdown of DNAJC12 expression suppressed the phosphorylation of p65 NF-κB, downregulated the expression levels and inhibited the subsequent activation of β-catenin, and downregulated the expression levels of vimentin. Rescue experiments demonstrated that the over- expression of β-catenin, but not that of NF-κB or vimentin, reversed the effects of DNAJC12 knockdown on the proliferation and invasion of lung cancer cells. On the whole, the findings of the present study suggest that DNAJC12 may play a crucial role in lung cancer tumorigenesis by regulating the expression and activation of β-catenin. Therefore, DNAJC12 may represent a novel target for the treatment of lung cancer.
Keywords: DnaJ heat shock protein family (Hsp40) member C12, lung cancer, proliferation, migration, apoptosis, β-catenin
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide (1-3), and the morbidity and mortality rates for the disease remain high. Over the past two decades, significant advancements have been achieved in the diagnosis and treatment of lung cancer, which has relied on the improved understanding of the disease biology and the underlying mechanisms of lung cancer progression and metastasis, as well as the advancements in early detection methods and multimodal care (1). However, despite this progress, the prognosis of lung cancer remains unsatisfactory and the underlying mechanisms of lung cancer remain poorly understood. Therefore, further studies investigating the underlying mechanisms of lung cancer and identifying novel targets to devise novel drugs, as well as combination therapies, are required to improve the outcomes of patients with lung cancer.
DnaJ heat shock protein family (HSP40) member C12 (DNAJC12) is a J domain-containing protein that belongs to the DnaJ homology C (DNAJC) family, which is a subclass of heat shock proteins (4,5). To the best of our knowledge, the current physiological and pathological functions of DNAJC12 remain unclear. During endoplasmic reticulum (ER) stress, the binding of the cochaperone DNAJC12 and heat shock protein family A (HSP70) member 8 is enhanced (6). Several genetic studies have revealed that DNAJC12 participates in the development of numerous types of human disease. For example, biallelic mutations in the DNAJC12 gene in humans have been shown to lead to dystonia and intellectual disability (7). In addition, DNAJC12 mutations identified by whole-exome sequencing have been observed in patients with mild hyperphenylalaninemia (8,9). In the Chinese Han population, DNAJC12 mutations have also been found to be associated with Parkinson's disease (10).
Over the past few years, the roles of DNAJC12 in cancer biology have also been reported. For instance, in patients with rectal cancer, upregulated expression levels of DNAJC12 have been shown to predict a poor response to neoadjuvant concurrent chemoradiotherapy (11). The aggressive phenotype of gastric cancer has also been found to be associated with the upregulation of DNAJC12 expression levels (12). In addition, in breast cancer, DNAJC12 expression has been found to be associated with the estrogen receptor status (13). However, the association between DNAJC12 and lung cancer remains unknown. Moreover, although the association between DNAJC12 and several types of cancer has been reported, to the best of our knowledge, the biological functions of DNAJC12 in cancer development and metastasis also remain unknown.
β-catenin is a 90 kDa multifunctional protein that participates in cell development under normal physiological conditions (14,15). β-catenin is a pivotal transcriptional factor for Wnt signaling and plays an important role in stem cell self-renewal and organ regeneration, in addition to cancer development and drug resistance (16). The roles of β-catenin in lung cancer have been well established. For example, β-catenin has been discovered to promote tumorigenesis, cancer stem cell self-renewal, drug resistance and metastasis in lung cancer cells by promoting the activation of Wnt signaling and the expression of downstream target genes (17,18). In addition, β-catenin expression has been reported to be induced by epidermal growth factor receptor (EGFR) mutations and to contribute to lung cancer development (19). However, the mechanisms through which β-catenin responds to upstream regulators remain only partially understood and upstream regulators of β-catenin also remain to be identified.
The present study thus aimed to investigate the roles of DNAJC12 in lung cancer. The findings presented herein demonstrate that DNAJC12 promotes lung cancer cell tumorigenesis by regulating the expression and activation of β-catenin.
Materials and methods
Patient samples
A total of 15 pairs of lung cancer and adjacent non-cancer tissues were obtained from patients with lung cancer (age range, 45-76 years; sex, 9 males and 6 females) between January, 2019 and February, 2020 from the Shandong Provincial Hospital (Jinan, China). The samples were stored at −80°C until required for subsequent experimentation. Written informed consent was obtained from each patient prior to participation and the clinical study was approved by the Ethics Committee of Clinical Research of Shandong Provincial Hospital.
For the analysis using public data, 57 pairs of lung cancer and adjacent non-cancer tissues were analyzed using a dataset from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases. In addition, the expression levels of DNAJC12 were also analyzed in 54 control and 498 cancer unpaired tissues.
Cell lines and culture
Lung cancer cell lines (A549, NCI-H1299, NCI-H1975 and 95D) and 293T cells were purchased from the American Type Culture Collection (ATCC). Mycoplasma testing was performed for all cell lines and all cell lines were authenticated using STR profiling. Cells were cultured in DMEM (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Thermo Fisher Scientific, Inc.). All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cancer tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1 μg) was reverse transcribed into cDNA using a cDNA synthesis kit (cat. no. 6130, Takara Bio, Inc.). qPCR was subsequently performed using an SYBR-Green II reagent kit (cat. no. RR820A; Takara Bio, Inc.) as previously described (20). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec and 70°C for 10 sec. The following primer pairs were used for the qPCR: DNAJC12 forward, 5′-AATGGTTGGCACCTTCGTTTC-3′ and reverse, 5′-GTTGGCAGCATAGGGGACAG-3′; CTNNB1 forward, 5′-AGCTTCCAGACACGCTATCAT-3′ and reverse, 5′-CGGTACAACGAGCTGTTTCTAC-3′; VIM forward, 5′-AGTCCACTGAGTACCGGAGAC-3′ and reverse, 5′-CATTTCACGCATCTGGCGTTC-3′; RELA forward, 5′-GTGGGGACTACGACCTGAATG-3′ and reverse, 5′-GGGGCACGATTGTCAAAGATG-3′; and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. The mRNA expression levels were quantified using the 2−ΔΔCq method as previously described (21).
Lentiviral transfection
Short hairpin RNA (shRNA/sh) targeting DNAJC12 (shDNAJC12; 5′-GGATGTGATGAACTATCTT-3′) and control shRNA (shCtrl; 5′-TTCTCCGAACGTGTCACGT-3′) were cloned into GV115 plasmids (Shanghai GeneChem Co., Ltd.). For overexpression, the coding sequences of CTNNB1 (NM_001904.4), p65 (NM_021975.4), VIM (NM_003380.5) were cloned into GV610 vectors (Shanghai GeneChem Co., Ltd.). The GV115 or GV610 plasmids (20 μg) were subsequently co-transfected into 293T cells alongside pHelper1.0 (15 μg) and pHelper2.0 (10 μg) packaging vectors using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) as previously described (22). Following transfection with the lentiviruses for 48 h (MOI=10), the transduced cells were selected with 5 μg/ml puromycin (Sigma-Aldrich; Merck KGaA).
High-content screening (HCS) assay
A HCS assay was performed to determine the cell number by counting the GFP-expressing cells, as previously described (22), following the culture of 2×103 A549 cells in 96-well plates. ArrayScan™ HCS software (Cellomics Inc.) was used to analyze the cell proliferation of each well every 24 h.
MTT proliferation assay
The proliferation of the A549 and NCI-H1975 cells seeded into 96-well plates at a density of 2×103 cells/well was analyzed using an MTT assay kit (Gen-view Scientific, Inc.). MTT solution was added to each well and incubated for 4 h at 37°C. Subsequently, the solution was replaced with 100 μl/well DMSO, and the plates were agitated for 5 min. The optical density of each well was read at 490 nm using a microplate spectrophotometer (Tecan infinite, M2009PR; Tecan Group, Ltd.).
Colony formation assay
A total of 1×103 A549 and NIC-H1975 cells with or without DNAJC12 knockdown were plated into 6-well plates and cultured for 2 weeks. Colonies were subsequently stained with crystal violet (Sangon Biotech Co., Ltd.) and stained colonies were visualized, followed by evaluation of colony number and size.
Wound healing assay
A549 and NCI-H1975 cell migration was determined in 6-well plates (105 cells/well) using a wound healing assay, as previously described (23). Briefly, the cells were used in the wound healing assay when the confluency of the cells was closed to 100%. After the cells were scratched using 10 μl pipette tips, the medium was replaced with FBS-free medium. The images were captured at 0, 24 and 48 h using an inverted fluorescence microscope (Olympus Corporation; magnification, ×100).
Cell migration and invasion assay
A Transwell migration and Matrigel invasion assay was used to determine the migration and invasive ability of the A549 and NCI-H1975 cells. For migration, 100 μl FBS free medium were added onto the upper surface of the migration chambers (cat. no. 3422; Corning, Inc.) and incubated at 37°C for 30 min. Subsequently, the FBS-free medium was discarded and a total of 1×105 cells in 100 μl FBS-free medium was added onto the upper surface of the migration chambers. The lower chambers were filled with 30% FBS medium. For invasion assay, a total of 500 μl of the mixture was added onto the upper surface of the invasion chambers (cat. no. 354480; Corning, Inc.), while the lower chambers were filled with 30% FBS medium. Following incubation for 24 or 48 h at 37°C, the culture medium and the cells attached on the upper surface were removed. Cells attached on the lower surface were fixed with 4% paraformaldehyde for 30 min at room temperature and stained with Giemsa stain (cat. no. 32884; Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. The images of migratory and invasive cells were collected using a light microscope (Olympus Corporation; magnification, ×100).
Apoptosis assay
The apoptosis of the lung cancer cells was analyzed using an Annexin V-allophycocyanin (APC) apoptosis detection kit (eBioscience; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, the A549 and NCI-H1975 cells were infected with lentiviruses expressing shCtrl or shDNAJC12 for 24 h. The cells were then harvested (1,000 × g, 5 min, room temperature) and washed with PBS. The cells were subsequently resuspended in a staining buffer to a final density of 1×106 cells/ml. Finally, 100 μl cell suspension was incubated with 5 μl Annexin V-APC at room temperature for 15 min. The stained cells were subjected to flow cytometry using a FACSCalibur flow cytometer (BD Biosciences) and data were analyzed with FlowJo software (version 7.6.1; FlowJo LLC).
In vivo tumor growth experiment
Female BALB/c athymic nude mice (age, 4 weeks; weight, 15-17 g) were obtained from Charles River Laboratories, Inc. and were randomly divided into the following two groups (10 mice/group): i) ShCtrl; and ii) shDNAJC12. Mice were healthy and housed under SPF-grade conditions. A total of 1×107 shCtrl- or shDNAJC12-transfected A549 cells were subcutaneously implanted into the right flanks of the nude mice, according to the protocol as described previously (24-26). The tumor volume was measured every 3 days starting at 30 days following transplantation and the mice were sacrificed by cervical dislocation at day 42. The animal experiments were performed according to the indicated protocols and were approved by the Animal Research Ethics Committee of Shandong Provincial Hospital.
Western blot analysis
Total protein was extracted from the lung cancer tissues and A549 and NCI-H1975 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitor cocktail. The protein concentration was determined using a BCA assay kit (Thermo Fisher Scientific, Inc.) and 40 μg protein/lane was separated via 12% SDS-PAGE. The separated proteins were subsequently transferred onto PDVF membranes and blocked with 5% fat-free milk in TBS-Tween-20 (TBST) for 1 h at room temperature. The membranes were then washed with TBST and incubated with the following primary antibodies at 4°C overnight: Anti-DNAJC12 (Abcam; cat. no. ab167425), anti-GAPDH (Santa Cruz Biotechnology, Inc.; cat. no. sc-32233), anti-p65 (Cell Signaling Technology, Inc.; cat. no. 8242), anti-phosphorylated (p)-p65 (Cell Signaling Technology, Inc.; cat. no. 3033), anti-β-catenin (Cell Signaling Technology, Inc.; cat. no. 8480), anti-p-β-catenin (Cell Signaling Technology, Inc.; cat. no. 2009) and anti-vimentin (Cell Signaling Technology, Inc.; cat. no. 3932). Following the primary antibody incubation, the membranes were washed with TBST and incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (Cell Signaling Technology, Inc.; cat. nos. 7076 and 7074) at room temperature for 2 h. The membranes were subsequently washed with TBST and protein bands were visualized using Pierce™ ECL Western Blotting substrate (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were performed at least in triplicate and data are presented as the mean ± SEM. Statistical differences between two groups were determined using a Student's t-test, while statistical differences between ≥3 groups were determined using a one-way ANOVA followed by a Tukey's post hoc test. Categorical data were analyzed using a χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
DNAJC12 expression levels are upregulated in lung cancer tissues
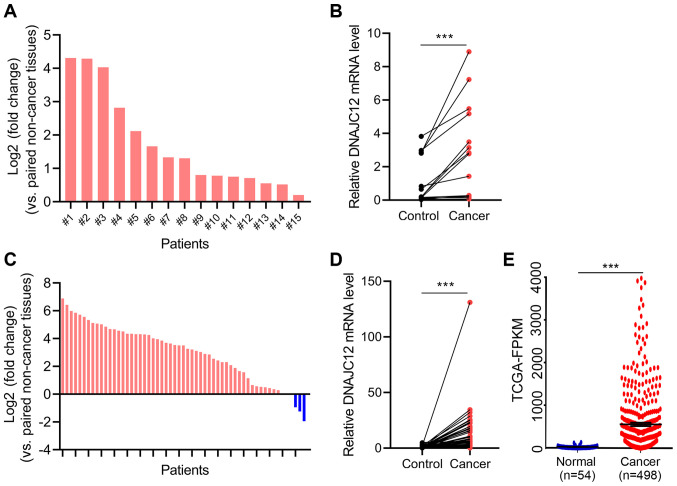
To determine the potential roles of DNAJC12 in lung cancer, the expression levels of DNAJC12 in lung cancer and adjacent non-cancer tissues were analyzed by RT-qPCR. The expression levels of DNAJC12 were significantly upregulated in the lung cancer tissues compared with the adjacent non-cancer tissues (Fig. 1A and B). In addition, the expression levels of DNAJC12 in 57 pairs of lung cancer and control tissues from TCGA database were analyzed. The results also revealed that the expression levels of DNAJC12 were upregulated in lung cancer tissues compared with non-cancer control lung tissues (Fig. 1C and D). Furthermore, analysis of a large unpaired TCGA dataset (54 control and 498 cancer tissues) also demonstrated that the expression levels of DNAJC12 were upregulated in lung cancer tissues (Fig. 1E). In addition, the association between patient clinicopathological features and DNAJC12 expression in 498 lung cancer tissues from TCGA database is presented in Table I. The expression of DNAJC12 was not associated with age, sex, T stage, N metastasis and M metastasis. Therefore, these findings suggested that DNAJC12 may be involved in the development of lung cancer.
Figure 1.
DNAJC12 expression levels are upregulated in human lung cancer tissues. (A) Fold change in DNAJC12 expression levels in 15 lung cancer tissues compared with paired adjacent non-cancer lung tissues were analyzed using reverse transcription-quantitative PCR. The fold change in the expression levels of each patient are presented. (B) Analysis of data from part (A) was performed using a paired Student's t-test. (C) Fold change in DNAJC12 expression levels in lung cancer tissues compared with paired non-cancer lung tissues obtained from TCGA database; n=57. (D) Analysis of data from part (C) was performed using a paired Student's t-test. ***P<0.001. (E) Expression levels of DNAJC12 in lung cancer tissues compared with unpaired non-cancer lung tissues obtained from TCGA database. Control tissues, n=43; lung cancer tissues, n=498. ***P<0.001. Data were analyzed using an unpaired Student's t-test. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; TCGA, The Cancer Genome Atlas.
Table I.
Association between patient clinicopathological features and DNAJC12 expression in 498 lung cancer tissues from TCGA database.
| Characteristics | DNAJC 12
|
Total | P-value | |
|---|---|---|---|---|
| Low | High | |||
| Sex | 0.151 | |||
| Male | 107 | 123 | 230 | |
| Female | 142 | 126 | 268 | |
| Total | 249 | 249 | 498 | |
| Age, years | 0.744 | |||
| ≤65 | 119 | 113 | 232 | |
| >65 | 123 | 124 | 247 | |
| Total | 242 | 237 | 479 | |
| T stage | 0.852 | |||
| T1/2 | 214 | 218 | 432 | |
| T3/4 | 32 | 31 | 63 | |
| Total | 246 | 249 | 495 | |
| N metastasis | 0.876 | |||
| N0 | 160 | 161 | 321 | |
| N1/2/3 | 82 | 85 | 167 | |
| Total | 242 | 246 | 488 | |
| M metastasis | 0.555 | |||
| M0 | 166 | 167 | 333 | |
| M1 | 10 | 13 | 23 | |
| Total | 176 | 180 | 356 | |
Knockdown of DNAJC12 suppresses lung cancer cell proliferation in vitro
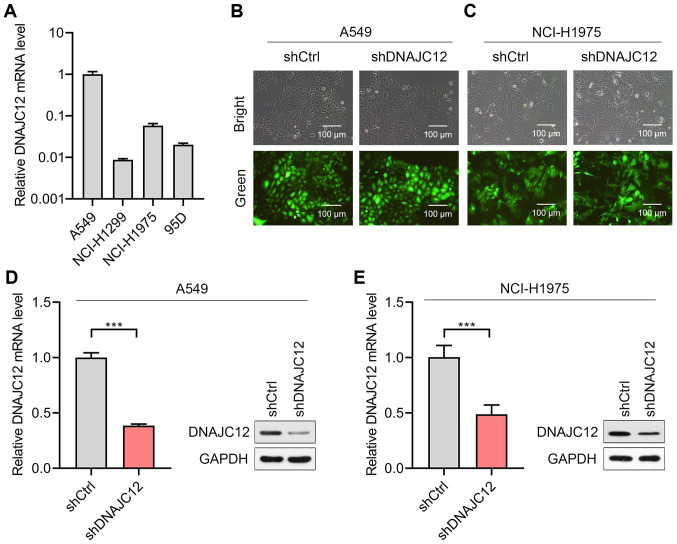
To determine the potential roles of DNAJC12 in lung cancer cells, the expression levels of DNAJC12 were first analyzed in four lung cancer cell lines. The results revealed that the A549 and NCI-H1975 cells expressed higher expression levels of DNAJC12 compared with the NCI-H1299 and 95D cells (Fig. 2A). Therefore, the A549 and NCI-H1975 cells were selected for use in further experiments. Lentiviruses expressing shRNA targeting DNAJC12 (shDNAJC12) and control shRNA (shCtrl) were subsequently successfully infected into A549 and NCI-H1975 cells (Fig. 2B and C). The results of RT-qPCR and western blot analysis demonstrated that the lentivirus-mediated knockdown of DNAJC12 significantly downregulated the expression levels of DNAJC12 in the A549 and NCI-H1975 lung cancer cells (Fig. 2D and E). Thus, A549 and NCI-H1975 cells with/without stable DNAJC12 knockdown were successfully generated for use in further experiments.
Figure 2.
Knockdown of DNAJC12 in lung cancer cells. (A) Relative mRNA expression levels of DNAJC12 were analyzed in the four lung cancer cell lines, A549, NCI-H1299, NCI-1975 and 95D by RT-qPCR. Fluorescence images demonstrating the infection efficiency of lentivirus expressing shDNAJC12 or shCtrl following 24 h of infection in (B) A549 and (C) NCI-H1975 cells. RT-qPCR and western blot analysis were used to determine DNAJC12 knockdown efficiency in (D) A549 and (E) NCI-H1975 cells transfected as described in parts (B and C), respectively. ***P<0.001. Data were analyzed using a Student's t-test. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin RNA; Ctrl, control.
The present study first determined the effects of the knockdown of DNAJC12 on the proliferation rate of A549 and NCI-H1975 lung cancer cells using HCS and MTT assays. The results determined that the knockdown of DNAJC12 significantly reduced the proliferation rate of the A549 and NCI-H1975 cells (Fig. 3). Colony formation is a key feature of cancer cells (27); therefore, the present study also analyzed the effects of DNAJC12 on the colony formation of lung cancer cells. The results revealed that DNAJC12 knockdown reduced the number and size of colonies formed from the A549 and NCI-H1975 lung cancer cells (Fig. 4). Taken together, these findings suggest that the knockdown of DNAJC12 may suppress the proliferation of lung cancer cells.
Figure 3.
Knockdown of DNAJC12 inhibits the proliferation of lung cancer cells. HCS and MTT assays were used to determine the effects of DNAJC12 on A549 and NCI-H1975 cell proliferation. (A and B) Representative images of the HCS assay using A549 cells with/without DNAJC12 knockdown (A) and the statistical result (B). The relative cell number was analyzed using a one-way ANOVA. ***P<0.001. (C and D) MTT assay was used to demonstrate that DNAJC12 knockdown inhibited the proliferation of (C) A549 and (D) NCI-H1975 cells with/without DNAJC12 knockdown. The relative cell number was analyzed using a one-way ANOVA. ***P<0.001. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; HCS, high-content screening.
Figure 4.
Knockdown of DNAJC12 reduces the colony formation of lung cancer cells. (A) A549 and (B) NCI-H1975 cells with/without DNAJC12 knockdown were subjected to a colony formation assay and the number of colonies was analyzed 2 weeks later. DNAJC12 knockdown reduced the number of colonies formed in both lung cancer cell lines. ***P<0.001. Data were analyzed using a Student's t-test. DNAJC12 knockdown reduced the size of the colonies from (C) A549 and (D) NCI-H1975 lung cancer cells. Representative colony size of A549 cells is shown in (A) and the representative colony size of NCI-H1975 cells is shown in (B). DNAJC12, DnaJ heat shock protein family (Hsp40) member C12.
Knockdown of DNAJC12 suppresses lung cancer cell growth in vivo
To determine whether DNAJC12 regulates lung cancer cell growth in vivo, xenograft experiments were performed. A549 cells expressing shCtrl and shDNAJC12 lentiviruses were subcutaneously transplanted into the right flanks of 4-week-old female nude mice. Tumor growth was monitored for 42 days following implantation. The results revealed that the knockdown of DNAJC12 reduced the growth rate, size and weight of the lung cancer tumors (Fig. 5). Thus, these results indicate that DNAJC12 may contribute to lung cancer growth in vivo.
Figure 5.
Knockdown DNAJC12 represses lung cancer cell growth in vivo. (A) Microscopy images of xenograft tumors grown in nude mice that were transplanted with 1×107 shCtrl- and shDNAJC12-transfected A549 cells. (B) Tumor volume was monitored for 42 days after transplantation. ***P<0.001. Data were analyzed using a one-way ANOVA. The maximum tumor diameter obtained was 14.85 mm. The maximum tumor volume obtained was 1,247.66 mm3. (C) Tumors were weighed immediately after sacrificing the mice. ***P<0.001. Data were analyzed using a Student's t-test. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; sh, short hairpin RNA; Ctrl, control.
Knockdown of DNAJC12 suppresses lung cancer migration and invasion
Metastasis is a feature of human lung cancer cells (27). To determine whether DNAJC12 regulates the invasion of lung cancer cells, the Transwell assay and Transwell Matrigel assay was performed using the A549 and NCI-H1975 cells. The results revealed that A549 and NCI-H1975 cell invasion was significantly suppressed following the knockdown of DNAJC12 (Fig. 6A and B). Matrigel invasion assay also demonstrated that DNAJC12 knockdown suppressed the invasion of A549 and NCI-H1975 cells (Fig. 6C and D). The present study subsequently analyzed the effects of DNAJC12 on the migration of lung cancer cells by performing wound healing assays. The results revealed that the migration of the A549 and NCI-H1975 cells, which was observed within 48 h, was inhibited by DNAJC12 knockdown (Fig. 6E and F). Taken together, these findings suggest that DNAJC12 regulates the invasion and migration of lung cancer cells.
Figure 6.
Knockdown of DNAJC12 inhibits the migration and invasion of lung cancer cells. The knockdown of DNAJC12 inhibited the migration of (A) A549 and (B) NCI-H1975 lung cancer cells. Number of migratory cells per field were analyzed. ***P<0.001. Data were analyzed using a Student's t-test. Invasion of A549 and NCI-H1975 cells with/without DNAJC12 knockdown was analyzed using a Matrigel invasion assay. DNAJC12 knockdown inhibited the invasion of (C) A549 and (D) NCI-H1975 lung cancer cells. Number of invasive cells per field were analyzed. ***P<0.001. Data were analyzed using a Student's t-test. Migration of A549 or NCI-H1975 cells with/without DNAJC12 knockdown was analyzed using a wound-healing assay. The knockdown of DNAJC12 inhibited the migration of (E) A549 and (F) NCI-H1975 lung cancer cells. The migratory rate was analyzed at the end of the experiment. ***P<0.001. Data were analyzed using a Student's t-test. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12.
Knockdown of DNAJC12 induces the apoptosis of lung cancer cells
The suppression of cancer growth and metastasis may result from cancer cell apoptosis induced by clinical drug treatment (27). Thus, the present study aimed to determine whether the DNAJC12-induced effects on the proliferation, migration and invasion of lung cancer cells were associated with the apoptosis of lung cancer cells. Cell apoptosis was analyzed by flow cytometry in the A549 and NCI-H1975 cells. The results revealed that the knockdown of DNAJC12 induced the apoptosis of A549 and NCI-H1975 cells (Fig. 7), which may be associated with the effects of DNAJC12 on the proliferation, migration and invasion of lung cancer cells.
Figure 7.
Knockdown of DNAJC12 induces the apoptosis of lung cancer cells. (A) A549 and (B) NCI-H1975 cells were infected with lentivirus expressing shDNAJC12 or shCtrl for 24 h, then the cells were harvested and subjected to apoptosis analysis with flow cytometry. Knockdown of DNAJC12 was discovered to induce the apoptosis of A549 and NCI-H1975 cells, respectively. ***P<0.001. Data were analyzed using a Student's t-test. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; short hairpin RNA; Ctrl, control.
DNAJC12 regulates NF-κB, β-catenin and vimentin signaling
The biological pathways regulated by DNAJC12 were subsequently investigated. The growth and metastasis of lung cancer cells are regulated by pivotal intracellular transcriptional regulators, such as NF-κB and β-catenin, as well as key extracellular matrix regulators, such as matrix metalloproteinase (MMP) and vimentin (14,28,29). The present study analyzed the effects of DNAJC12 on the expression levels of these proteins by western blotting. The knockdown of DNAJC12 reduced the phosphorylation of NF-κB p65 and downregulated the expression levels and activation of β-catenin in A549 cells (Fig. 8A and B). The knockdown of DNAJC12 also downregulated the expression levels of vimentin (Fig. 8C), which is a factor of the extracellular matrix. Therefore, these findings suggest that DNAJC12 may target key regulators of lung cancer growth and metastasis.
Figure 8.
DNAJC12 regulates NF-κB, β-catenin and vimentin signaling pathways. Total protein extracted from A549 cells with/without DNAJC12 knockdown was analyzed using western blotting. (A) DNAJC12 knockdown reduced the phosphorylation of NF-κB p65 in lung cancer cells. (B) DNAJC12 knockdown reduced the phosphorylation and downregulated the expression levels of β-catenin in lung cancer cells. (C) DNAJC12 knockdown downregulated vimentin expression levels in lung cancer cells. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12.
Overexpression of β-catenin blocks the effects of DNAJC12 knockdown
To determine whether p65, β-catenin and vimentin are involved in the effects of DNAJC12 on lung cancer cells, p65, β-catenin and vimentin were overexpressed in A549 cells in which DNAJC12 expression was knocked down. RT-qPCR analysis indicated that DNAJC12 was successfully knockdown, while p65, β-catenin and vimentin were effectively overexpressed in the A549 cells (Fig. 9A). An HCS assay was subsequently performed and the results revealed that the overexpression of β-catenin, but not that of p65 or vimentin, reversed the suppressive effects of DNAJC12 knockdown on the proliferation of A549 cells (Fig. 9B). The involvement of β-catenin in the regulatory role of DNAJC12 over A549 cell proliferation was also validated using an MTT assay (Fig. 9C). Furthermore, the reduced invasive capacity of the A549 cells induced by DNAJC12 knockdown was also reversed by the overexpression of β-catenin (Fig. 9D). Taken together, these findings suggest that DNAJC12 may regulate the proliferation and invasion of lung cancer cells in a β-catenin-dependent manner.
Figure 9.
β-catenin is involved in the effect of DNAJC12 on lung cancer cells. (A) RT-qPCR analysis was used to determine DNAJC12 knockdown efficiency and overexpression efficiency of CTNNB1, VIM and RELA in A549 cells. ***P<0.001. Data were analyzed using one-way ANOVA followed by a Tukey's post hoc test. (B) A549 cells with knocked down DNAJC12 expression were infected with lentivirus overexpressing p65, β-catenin, or vimentin. Cells were analyzed using a HCS assay. The overexpression of β-catenin reversed the DNAJC12-induced inhibition of proliferation. (C) Cells were infected with lentivirus overexpressing β-catenin and proliferation was analyzed using an MTT assay. The overexpression of β-catenin reversed the DNAJC12-induced inhibition of proliferation. (D) Cells were infected with lentivirus overexpressing β-catenin and invasion was analyzed using a Transwell invasion assay. The overexpression of β-catenin reversed the migration of A549 lung cancer cells. **P<0.01. Data were analyzed using a one-way ANOVA. DNAJC12, DnaJ heat shock protein family (Hsp40) member C12; HCS, high-content screening.
Discussion
The present study provides evidence to suggest that DNAJC12 may promote lung cancer proliferation, invasion, migration and survival, which may be partially mediated by β-catenin. Using data generated in the present study and public data, the DNAJC12 expression levels were found to be upregulated in lung cancer tissues compared with non-cancerours tissues. The lentivirus-mediated knockdown of DNAJC12 demonstrated that DNAJC12 was important for lung cancer cell proliferation and growth in vitro and in vivo, respectively. In addition, the knockdown of DNAJC12 reduced the migration and invasion, and induced the apoptosis in lung cancer cells. Signaling pathway analysis revealed that DNAJC12 targeted NF-κB, β-catenin and vimentin signaling. Finally, rescue experiments revealed that β-catenin was essential for the function of DNAJC12 in regulating the proliferation, migration and invasion of lung cancer cells.
DNAJC12 belongs to the DNAJC protein family of heat shock proteins. The potential association between DNAJC12 and cancer biology has been previously reported (11,12). However, to the best of our knowledge, the role of DNAJC12 in lung cancer remains unknown. Therefore, the present study investigated the potential roles of DNAJC12 in lung cancer. Using patient lung cancer samples and public data from the TCGA database, the present results demonstrated that DNAJC12 mRNA expression levels were significantly upregulated in lung cancer tissues compared with adjacent non-cancer lung tissues. Furthermore, DNAJC12 expression was significantly elevated in lung cancer patients with higher stage.
Although previous studies have reported the potential association between DNAJC12 and numerous types of cancer (11-13), the role of DNAJC12 in cancer biology remains unknown. The present study performed HCS and MTT assays, which revealed that the knockdown of DNAJC12 reduced the proliferative rate of lung cancer cells. Colony formation is a key feature of cancer cells (27). The present study also demonstrated that DNAJC12 not only regulated the number of colonies formed, but it also altered the size of the colonies formed from A549 and NCI-H1975 cells. Notably, the data presented herein also demonstrated that DNAJC12 knockdown reduced lung cancer cell growth in vivo. Taken together, these findings suggested that DNAJC12 may be crucial for lung cancer cell proliferation and growth in vitro and in vivo, respectively.
The underlying mechanisms of lung cancer metastasis remain poorly understood (30). To determine the potential role of DNAJC12 in the metastasis of lung cancer cells, the present study performed in vitro Transwell and wound healing assays to assess cell invasion and migration, respectively. The results revealed that the knockdown of DNAJC12 significantly reduced the invasive and migratory capacities of A549 and NCI-H1975 cells. These findings indicated that the upregulated expression of DNAJC12 in lung cancer tissues might be one of the underlying mechanisms of metastasis in lung cancer cells.
Following clinical drug treatment, the suppression of cancer growth and metastasis occurs as a result of cancer cell apoptosis (27). Therefore, the present study also aimed to determine whether the effects of DNAJC12 on lung cancer proliferation, invasion and migration were associated with cell apoptosis in A549 and NCI-H1975 cells using flow cytometry. The present data demonstrated that DNAJC12 knockdown significantly induced the apoptosis of A549 and NCI-H1975 cells. These results suggested that DNAJC12 may regulate the apoptosis of lung cancer cells to modulate cell proliferation, migration and invasion.
NF-κB plays a crucial role in lung cancer by regulating growth and apoptosis (29). The findings of the present study demonstrated that the knockdown of DNAJC12 reduced the phosphorylation levels of the NF-κB p65 subunit, suggesting that DNAJC12 may promote the activation of NF-κB. β-catenin has been reported to regulate several functions of lung cancer, including growth, stemness, metastasis and drug resistance (resistance to apoptosis) (17). It was also found that DNAJC12 knockdown downregulated the expression levels and phosphorylation of β-catenin. Vimentin is a mesenchymal marker that participates in the remodeling of the extracellular matrix and contributes to the metastasis of numerous types of cancer, including lung cancer (28). Thus, the present study also aimed to determine the effects of DNAJC12 on the expression levels of vimentin. The knockdown of DNAJC12 also downregulated the expression levels of vimentin. These data suggested that DNAJC12 may regulate several key modulators of proliferation, migration and invasion in lung cancer cells. To investigate which of these factors was a key downstream target gene of DNAJC12 in lung cancer, rescue experiments were performed by overexpressing these factors in lung cancer cells following DNAJC12 knockdown. Further rescue experiments indicated that β-catenin, but not NF-κB or vimentin, may be involved in DNAJC12 function in lung cancer cell proliferation, invasion and migration. However, the biological mechanisms through which DNAJC12 regulated β-catenin expression and activation remain to be determined in further studies. A previous study reported that DNAJC12 participated in ER stress (6). ER stress is an important regulator involved in the activation of β-catenin (31), thus DNAJC12 may regulate β-catenin via activating ER stress. Further study is required to determine this hypothesis.
In conclusion, the findings of the present study suggested that DNAJC12 may promote lung cancer development and metastatic features, at least in part, by activating β-catenin. Thus, DNAJC12 may serve as a potential prognostic factor and therapeutic target in human lung cancer. However, several limitations exist for the present study. First, the mechanism through which DNAJC12 was regulated in lung cancer remains unknown. Second, the potential effects of DNAJC12 on the prognosis of patients with lung cancer were not determined. Finally, the underlying mechanisms of DNAJC12 regulation over β-catenin remain unknown. Further studies addressing these points would improve the current understanding of the role and mechanism of DNAJC12 in lung cancer.
Acknowledgments
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL designed the study, and drafted and revised the manuscript. ML and FJ performed the majority of the experiments. JL and MC analyzed and interpreted the data. JY performed the animal experiments. All authors have read and approved the final manuscript. All authors confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The clinical study was approved by the Ethics Committee of Clinical Research of Shandong Provincial Hospital and written informed consent was obtained from each patient prior to participation. The animal experiments were performed according to the indicated protocols and were approved by the Animal Research Ethics Committee of Shandong Provincial Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Long K, Suresh K. Pulmonary toxicity of systemic lung cancer therapy. Respirology. 2020;25(Suppl 2):72–79. doi: 10.1111/resp.13915. [DOI] [PubMed] [Google Scholar]
- 2.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/S1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.Tang X, Mo C, Wang Y, Wei D, Xiao H. Anti-tumour strategies aiming to target tumour-associated macrophages. Immunology. 2013;138:93–104. doi: 10.1111/imm.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roosen DA, Blauwendraat C, Cookson MR, Lewis PA. DNAJC proteins and pathways to parkinsonism. Febs J. 2019;286:3080–3094. doi: 10.1111/febs.14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zylicz M, Yamamoto T, McKittrick N, Sell S, Georgopoulos C. Purification and properties of the dnaJ replication protein of Escherichia coli. J Biol Chem. 1985;260:7591–7598. doi: 10.1016/S0021-9258(17)39649-7. [DOI] [PubMed] [Google Scholar]
- 6.Choi J, Djebbar S, Fournier A, Labrie C. The co-chaperone DNAJC12 binds to Hsc70 and is upregulated by endoplasmic reticulum stress. Cell Stress Chaperones. 2014;19:439–446. doi: 10.1007/s12192-013-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anikster Y, Haack TB, Vilboux T, Pode-Shakked B, Thöny B, Shen N, Guarani V, Meissner T, Mayatepek E, Trefz FK, et al. Biallelic mutations in DNAJC12 cause hyperphenylalaninemia, dystonia, and intellectual disability. Am J Hum Genet. 2017;100:257–266. doi: 10.1016/j.ajhg.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Yang Q, Yi S, Qin Z, Luo J, Fan X. Two novel mutations in DNAJC12 identified by whole-exome sequencing in a patient with mild hyperphenylalaninemia. Mol Genet Genomic Med. 2020;8:e1303. doi: 10.1002/mgg3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego D, Leal F, Gámez A, Castro M, Navarrete R, Sanchez- Lijarcio O, Vitoria I, Bueno-Delgado M, Belanger-Quintana A, Morais A, et al. Pathogenic variants of DNAJC12 and evaluation of the encoded cochaperone as a genetic modifier of hyperphenylalaninemia. Human Mutation. 2020;41:1329–1338. doi: 10.1002/humu.24026. [DOI] [PubMed] [Google Scholar]
- 10.Fan Y, Yang Zh, Li F, Hu XC, Yue YW, Yang J, Liu YT, Liu H, Wang YL, Shi CH, Xu YM. DNAJC12 mutation is rare in Chinese Han population with Parkinson's disease. Neurobiol Aging. 2018;68:159.e1–159.e2. doi: 10.1016/j.neurobiolaging.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 11.He HL, Lee YE, Chen HP, Hsing CH, Chang IW, Shiue YL, Lee SW, Hsu CT, Lin LC, Wu TF, Li CF. Overexpression of DNAJC12 predicts poor response to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Exp Mol Pathol. 2015;98:338–345. doi: 10.1016/j.yexmp.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Uno Y, Kanda M, Miwa T, Umeda S, Tanaka H, Tanaka C, Kobayashi D, Suenaga M, Hattori N, Hayashi M, et al. Increased expression of DNAJC12 is associated with aggressive phenotype of gastric cancer. Ann Surg Oncol. 2019;26:836–844. doi: 10.1245/s10434-018-07149-y. [DOI] [PubMed] [Google Scholar]
- 13.De Bessa SA, Salaorni S, Patrão DF, Neto MM, Brentani MM, Nagai MA. JDP1 (DNAJC12/Hsp40) expression in breast cancer and its association with estrogen receptor status. Int J Mol Med. 2006;17:363–367. [PubMed] [Google Scholar]
- 14.Martin-Orozco E, Sanchez-Fernandez A, Ortiz-Parra I, Ayala-San Nicolas M. WNT Signaling in tumors: The way to evade drugs and immunity. Front Immunol. 2019;10:2854. doi: 10.3389/fimmu.2019.02854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peifer M, McCrea PD, Green KJ, Wieschaus E, Gumbiner BM. The vertebrate adhesive junction proteins beta-catenin and plakoglobin and the Drosophila segment polarity gene armadillo form a multigene family with similar properties. J Cell Biol. 1992;118:681–691. doi: 10.1083/jcb.118.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haseeb M, Pirzada RH, Ain QU, Choi S. Wnt signaling in the regulation of immune cell and cancer therapeutics. Cells. 2019;8:1380. doi: 10.3390/cells8111380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 18.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayama S, Sng N, Carretero J, Welner R, Hayashi Y, Yamamoto M, Tan AJ, Yamaguchi N, Yasuda H, Li D, et al. β-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 2014;74:5891–5902. doi: 10.1158/0008-5472.CAN-14-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X, Ma H, Han L, Zheng W, Lu YB, Chen XF, Liang ST, Wei GH, Zhang ZQ, Chen HZ, Liu DP. SIRT1 deacetylates the cardiac transcription factor Nkx2.5 and inhibits its transcriptional activity. Sci Rep. 2016;6:36576. doi: 10.1038/srep36576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expres- sion data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Cui X, Song L, Bai Y, Wang Y, Wang B, Wang W. Stromal interaction molecule 1 regulates growth, cell cycle, and apoptosis of human tongue squamous carcinoma cells. Biosci Rep. 2017;37:BSR20160519. doi: 10.1042/BSR20160519. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Wu R, Zhao B, Ren X, Wu S, Liu M, Wang Z, Liu W. MiR-27a-3p targeting GSK3β promotes triple-negative breast cancer proliferation and migration through Wnt/β-catenin pathway. Cancer Manag Res. 2020;12:6241–6249. doi: 10.2147/CMAR.S255419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JY, Kim HJ, Jung CW, Lee TS, Kim EH, Park MJ. CXCR4 uses STAT3-mediated slug expression to maintain radioresistance of non-small cell lung cancer cells: Emerges as a potential prognostic biomarker for lung cancer. Cell Death Dis. 2021;12:48. doi: 10.1038/s41419-020-03280-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sivaraman A, Kim D, Bhattarai D, Kim M, Lee HY, Lim S, Kong J, Goo JI, Shim S, Lee S, et al. Synthesis and structure-activity relationships of arylsulfonamides as AIMP2-DX2 inhibitors for the development of a novel anticancer therapy. J Med Chem. 2020;63:5139–5158. doi: 10.1021/acs.jmedchem.9b01961. [DOI] [PubMed] [Google Scholar]
- 26.Liang G, Meng W, Huang X, Zhu W, Yin C, Wang C, Fassan M, Yu Y, Kudo M, Xiao S, et al. miR-196b-5p-mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc Natl Acad Sci USA. 2020;117:4347–4357. doi: 10.1073/pnas.1917531117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci. 2019;20:4947. doi: 10.3390/ijms20194947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong KK, Jacks T, Dranoff G. NF-kappaB fans the flames of lung carcinogenesis. Cancer Prev Res (Phila) 2010;3:403–405. doi: 10.1158/1940-6207.CAPR-10-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016;35:75–91. doi: 10.1007/s10555-016-9618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia Z, Wu S, Wei X, Liao Y, Yi P, Liu Y, Liu J, Liu J. Hypoxic ER stress suppresses β-catenin expression and promotes cooperation between the transcription factors XBP1 and HIF1α for cell survival. J Biol Chem. 2019;294:13811–13821. doi: 10.1074/jbc.RA119.008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.