Abstract
Background:
The development of severe coronavirus disease 2019 (COVID-19) is associated with systemic hyperinflammation, which drives multi-organ failure and death. Disease deterioration tends to occur when the virus is receding; however, whether other factors besides viral products are involved in the inflammatory cascade remains unclear.
Methods:
Twenty-eight COVID-19 patients with laboratory-confirmed SARS-CoV-2 infection hospitalized at the Fifth Medical Center of Chinese PLA General Hospital from January 23 to February 20, 2020 and nine healthy donors during the same period were recruited in the study. COVID-19 patients were grouped as mild, moderate, severe based on disease severity. Plasma damage-associated molecular patterns (DAMPs), including high mobility group box 1 (HMGB1), calprotectin (S100A8/A9), surfactant protein A (SP-A), cold-inducible RNA-binding protein (CIRBP), and Histone H4 were detected by ELISA assay, and analyzed in combination with clinical data. Plasma cytokines, chemokines and lymphocytes were determined by flow cytometry.
Results:
Plasma levels of HMGB1 (38292.3 ± 4564.4 vs. 32686.3 ± 3678.1, P = 0.002), S100A8/A9 (1490.8 ± 819.3 vs. 742.2 ± 300.8, P = 0.015), and SP-A (6713.6 ± 1708.7 vs. 5296.3 ± 1240.4, P = 0.048) were increased in COVID-19 patients compared to healthy donors, while CIRBP (57.4 ± 30.7 vs. 111.9 ± 55.2, P = 0.004) levels decreased. Five DAMPs did not vary among mild, moderate, and severe patients. Moreover, SP-A levels correlated positively with inflammatory cytokines and negatively with time elapsed after symptom onset, whereas CIRBP showed an opposite pattern.
Conclusions:
These findings suggest SP-A may involve in the inflammation of COVID-19, while CIRBP likely plays a protective role. Therefore, DAMPs represent a potential target in the prevention or treatment of COVID-19.
Keywords: COVID-19, DAMP, Immune response, Inflammation, SARS-CoV-2
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread globally, already infecting tens of millions of people worldwide.[1] COVID-19 patients present with a variety of clinical manifestations, ranging from asymptomatic or mild respiratory illness to fulminant severe acute respiratory distress syndrome (ARDS) with extra-pulmonary complications.[2–4] The invasion and pathogenesis of SARS-CoV-2 are associated with the host immune response. Although the anti-viral immune response is crucial for eliminating SARS-CoV-2, a robust and persistent anti-viral immune response can also cause a massive production of inflammatory cytokines and damage to host tissues.[5–8] Indeed, the overproduction of cytokines caused by aberrant immune activation (termed a cytokine storm) is hypothesized to be a major cause of disease progression and eventual death in patients with COVID-19.[9]
There is an urgent need to better understand the host-pathogen interaction in COVID-19 for improved treatment and management of the disease. Danger-associated molecular patterns (DAMPs) are host-derived molecules that are released from damaged or dying cells and can initiate and perpetuate a non-infectious inflammatory response by interacting with pattern recognition receptors (PRRs).[10] DAMPs originate from different sources, including the nucleus (ie, histones and high mobility group box 1 [HMGB1]), cytosol (ie, cold-inducible RNA-binding protein [CIRBP] and calprotectin [S100A8/A9]), extracellular matrix (ie, short fragment hyaluronan and heparan sulfate), mitochondria (eg, formyl peptide and mitochondrial DNA [mtDNA]), granules (ie, defensins and granulysin), and the plasma membrane (ie, glypicans).[10] Some of these molecules play a critical role in modulating the lung injury response.[11] For example, influenza-infected patients who develop severe pneumonia express elevated circulating HMGB1.[12] Additionally, treatment with anti-HMGB1 mAb provided partial protection against both pneumonia and encephalopathy in murine models of influenza infection without affecting virus propagation in the lungs.[13] Nonetheless, the controlled production of DAMPs is needed to achieve full homeostatic restoration and repair from tissue injury. Moreover, in COVID-19, the roles of DAMPs are not yet fully understood.
Therefore, to better understand the potential roles of DAMPs in COVID-19, we measured plasma DAMPs (including HMGB1, S100A8/A9, SP-A, CIRBP, and Histone H4) in patients with mild, moderate and severe COVID-19, and determined the relationships to the severity of the illness. Our data imply the potential role of endogenous danger signals in modulating the lung injury response in COVID-19 patients.
Methods
Ethical approval
This study was approved by the ethics committee of the Fifth Medical Center of Chinese PLA General Hospital (S2020-044-02). Informed consent forms were obtained from all enrolled participants or their legal guardians.
Study participants
From January 23 to February 20, 2020, 28 patients with laboratory-confirmed SARS-CoV-2 infection hospitalized at the Fifth Medical Center of Chinese PLA General Hospital in Beijing, China, were enrolled. Nine healthy donors were recruited during the same period. Patients were classified into three clinical groups (mild, moderate, or severe) based on the guidelines for the diagnosis and treatment of Coronavirus Disease 2019 issued by the National Health Commission of China (7th edition) (http://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_11815/202003/t20200305_214142.html). Patients in the mild disease group had mild clinical symptoms and no pneumonia. Those in the moderate disease group had obvious clinical symptoms and pneumonia, and were admitted to general wards but did not require intensive care. Those in the severe disease group required critical care and met one or more of these criteria: dyspnea and respiratory rate ≥30/minute, blood oxygen saturation ≤93%, PaO2/FiO2 ratio <300 mmHg, 1 mmHg = 0.133 KPa, and lung infiltrates on CT scan >50% within 24 to 48 hours, or who exhibited respiratory failure, septic shock, and/or multiple organ dysfunction/failure.
Among the patients included in the study, two patients died and the rest were discharged from hospital. The discharge criteria were the same as those described in the guidelines (7th version). Briefly, the resolution of respiratory symptoms, substantial improvement of chest CT images, afebrile for more than three days, and two negative RT-PCR results on sequential respiratory tract swab samples taken at least 24 hours apart. All patients had routine laboratory investigations (full blood count, liver and renal function tests, and coagulation tests).
Lymphocyte counts and subsets
Absolute lymphocyte counts and subsets were determined using the lymphocyte detection kit (Beijing Tongshengshidai Biotechnology Co., LTD, Beijing, China) following the manufacturer's instructions.
Sample collection
Blood samples from COVID-19 patients and healthy donors were collected within 24 hours of admission or before discharge, and centrifuged at 400 g for 5 min at room temperature. Plasma samples were collected and stored at -80°C until use. After plasma collection, peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation.
Enzyme-linked immunosorbent assay (ELISA)
The plasma DAMPs were assessed by commercially available ELISA kits, according to the manufacturer's instructions, as follows: the S100A8/A9 ELISA kit (DS8900) from R&D Systems (Minneapolis, MN, USA), the HMGB1 ELISA kit (NBP2-62766) and SP-A ELSA kit (NBP2-76692) from Novus Biologicals (Centennial, CO, USA), the CIRBP ELISA kit (CY-8103) from MBL Life Sciences (Nagoya, Japan), and the Histone H4 ELISA kit (Ab156909) from Abcam (Cambridge, UK).
Cytokine and chemokine measurement
Ten different cytokines and chemokines (interleukin-1 receptor antagonist [IL-1RA], monocyte chemoattractant protein [MCP]-1, macrophage inflammatory protein [MIP]-1α, interferon gamma-induced protein 10 [IP-10], interleukin [IL]-1β, IL-2, IL-6, IL-12P70, tumor necrosis factor [TNF]-α, interferon [IFN]-γ) in the plasma of 28 patients infected with SARS-CoV-2 and nine healthy donors were determined by flow cytometry using an Aimplex kit (Beijing Quantobio, China), as per the manufacturer's instructions.
Flow cytometry
PBMC samples were stained with the following antibodies for T cell activation analysis: CD3-BV510 (clone OKT3), CD4-BV421 (clone OKT4), CD8-PE-Cy7 (clone SK1), HLA-DR-FITC (clone L243; Biolegend, San Diego, CA, USA); CD38-APC (clone HIT2; BD Biosciences, San Diego, CA, USA). The BD Canto II instrument (BD Biosciences, San Diego, CA, USA) was used for data collection, and the data was analyzed using FlowJo software V10 (Tree star Inc., Ashland, OR, USA).
Statistical analysis
The GraphPad Prism statistical software version 8.0 (GraphPad Software, San Diego, CA) and SPSS version 25 (SPSS Inc., Chicago, IL, USA) were used for statistical analysis. Pearson correlation analysis was performed by R software using the ggcor package. Categorical variables were reported as the counts and percentages, and significance was detected by the χ2 test or the Fisher's exact test. Continuous variables were compared using the Mann-Whitney U test and the Kruskal-Wallis H test, and displayed as median and interquartile range (IQR). A two-tailed P-value of < 0.05 was considered statistically significant.
Results
Patients characteristics
Twenty-eight laboratory-confirmed acute COVID-19 patients and nine healthy donors (HDs group) were enrolled for analysis. Patients were classified as mild (n = 9), moderate (n = 10), and severe (n = 9) according to the guidelines for diagnosis and treatment of Coronavirus Disease 2019 issued by the National Health Commission of China (7th edition). The demographics and baseline characteristics of these patients are shown in Table 1.
Table 1.
Demographics and baseline characteristics of study participants.
| Variable | HDs (n = 9) | Total (n = 28) | Mild (n = 9) | Moderate (n = 10) | Severe (n = 9) | P value |
|---|---|---|---|---|---|---|
| Characteristics | ||||||
| Age (years) | 38.0 (35.0–42.0) | 45.0 (34.8–59.0) | 39.8 (27.5–54.5) | 41.9 (32.0–54.8) | 61.1 (43.5–78.5) | 0.020∗ |
| Sex | 0.265 | |||||
| Male | 6 | 17 | 4 | 8 | 5 | |
| Female | 3 | 11 | 5 | 2 | 4 | |
| Days from illness | NA | 4.0 (3.0–6.8) | 4.0 (2.5–4.5) | 4.0 (3.8–5.3) | 7.0 (3.5–10.0) | 0.153 |
| Exposure to Wuhan | NA | 16 | 7 | 5 | 4 | 0.306 |
| Any comorbidity | 0 | 6 | 0 | 2 | 4 | 0.071 |
| Hypertension | 0 | 5 | 0 | 1 | 4 | 0.035∗ |
| Diabetes | 0 | 3 | 1 | 1 | 1 | 0.996 |
| Malignancy | 0 | 1 | 0 | 0 | 1 | 0.335 |
| Chronic liver disease | 0 | 1 | 0 | 1 | 0 | 0.393 |
| Signs and symptoms | ||||||
| Fever (°C) | 0.152 | |||||
| <37.3 | NA | 1 | 1 | 0 | 0 | |
| 37.3–38.0 | NA | 10 | 5 | 4 | 1 | |
| 38.1–39.0 | NA | 15 | 2 | 5 | 8 | |
| >39.0 | NA | 2 | 1 | 1 | 0 | |
| Cough | 0 | 16 | 4 | 6 | 6 | 0.619 |
| Expectoration | 0 | 9 | 1 | 3 | 5 | 0.128 |
| Myalgia or fatigue | 0 | 13 | 3 | 2 | 8 | 0.007∗ |
| Sore throat | 0 | 2 | 0 | 1 | 1 | 0.598 |
| Shortness of breath | 0 | 6 | 0 | 1 | 5 | 0.009∗ |
| Chest pain | 0 | 4 | 0 | 0 | 4 | 0.007∗ |
| Diarrhea | 0 | 1 | 1 | 0 | 0 | 0.335 |
Categorical variables are presented as number. Continuous variables are shown as median (interquartile range). P values indicate the comparison between the mild, moderate, and severe groups, and were calculated by the Fisher's exact test or ANOVA, as appropriate.
HDs: Healthy donors; NA: Not applicable.
P < 0.05.
The median age of the COVID-19 patients was 45.0 (IQR, 34.8–59.0) years, and 17 (61%) patients were male. The median age of the HDs group was 38.0 (IQR, 35.0–42.0) years, and six of them (66.7%) were male. No significant differences were found between the HDs and COVID-19 patients in terms of age and gender. Fever (100%) and cough (57%) were the most frequently reported symptoms. The median number of days from disease onset to admission was 4.0 (IQR, 3.0–6.8); two patients died, and all other patients were discharged. The median number of days from admission to discharge was 26.0 (IQR, 17.5–32.0).
Patients with severe COVID-19 were older than those with mild or moderate disease (P = 0.020). Compared with mild and moderate cases, severe cases reported a higher proportion of symptoms, including myalgia or fatigue, shortness of breath, chest pain [shown in Table 1]. Severe patients also showed more severe clinical symptoms, such as more severe lymphopenia and higher D-dimer and serum ferritin levels (P = 0.004, 0.014, 0.038, respectively). Other laboratory parameters, including routine blood tests, coagulation function, and biochemistry tests, are shown in Table 2.
Table 2.
Laboratory findings of hospitalized patients infected with SARS-CoV-2.
| Variablea | Total (n = 28) | Mild (n = 9) | Moderate (n = 10) | Severe (n = 9) | P value |
|---|---|---|---|---|---|
| Blood routine | |||||
| Leukocyte (×109/L; 3.97–9.15) | 4.59 (3.88–6.73) | 4.27 (3.85–5.99) | 4.59 (3.90–6.65) | 5.51 (3.90–9.57) | 0.757 |
| Normal or decreased | 26 | 9 | 9 | 7 | 0.312 |
| Neutrophil (×109/L; 2.0–7.0) | 2.9 (2.2–4.8) | 2.3 (2.0–3.8) | 2.9 (2.4–4.5) | 4.8 (2.8–8.5) | 0.104 |
| Increased | 3 | 0 | 1 | 2 | 0.312 |
| Lymphocyte (×109/L; 0.8–4.0) | 1.4 (0.9–1.6) | 1.5 (1.3–1.6) | 1.6 (1.2–2.1) | 0.6 (0.5–1.3) | 0.004∗ |
| Decreased | 6 | 0 | 0 | 6 | <0.001∗ |
| Monocytes (×109/L; 0.12–1.00) | 0.40 ± 0.17 | 0.43 ± 0.19 | 0.42 ± 0.12 | 0.34 ± 0.19 | 0.446 |
| Eosinophil (×109/L; 0.02–0.50) | 0.01 (0.00–0.03) | 0.01 (0.01–0.10) | 0.01 (0–0.08) | 0.00 (0.00–0.01) | 0.032∗ |
| Decreased | 19 | 5 | 6 | 8 | 0.032∗ |
| Basophil count (×109/L; 0.00–1.00) | 0.01 (0.01–0.02) | 0.01 (0.01–0.02) | 0.01 (0.01–0.03) | 0.01 (0.05–0.02) | 0.443 |
| Platelet (×109/L; 85–303) | 192 ± 80 | 168 ± 36 | 168 ± 70 | 250 ± 103 | 0.046∗ |
| T cell count (690–2540/μL)b | 731 (382–988) | 931 (613–1219) | 875 (666–1324) | 321 (258–393) | 0.005∗ |
| CD4+ T cell count (410–1590/μL)b | 411 (231–656) | 520 (342–834) | 431 (353–662) | 132 (123–283) | 0.016∗ |
| CD8+ T cell count (190–1140/μL)b | 329 ± 197 | 339 ± 108 | 446 ± 213 | 225 ± 45 | 0.005∗ |
| B cell count (90–660/μL)b | 148 ± 75 | 168 ± 65 | 172 ± 75 | 99 ± 59 | 0.111 |
| Natural Killer cell count (90–590/μL)b | 162 (122–229) | 232 (134–404) | 182 (151–306) | 119 (52–181) | 0.113 |
| CD4/CD8 ratio (0.68–2.47)b | 1.11 (0.89–1.66) | 1.50 (1.26–2.12) | 1.07 (0.93–1.35) | 0.79 (0.67–2.28) | 0.195 |
| Hemoglobin (131–172 g/L) | 138 ± 14 | 141 ± 9 | 146 ± 10 | 126 ± 15 | 0.003∗ |
| Coagulation function | |||||
| Activated partial thromboplastin time (23–42 s) | 31 (26–33) | 32 (27–35) | 30 (25–34) | 31 (26–33) | 0.768 |
| Prothrombin time (10.2–14.3 s) | 11.9 (11.1–12.3) | 12.1 (11.5–12.4) | 12.0 (11.0–12.7) | 11.7 (11.2–12.1) | 0.612 |
| D-dimer (<0.55 m/L) | 0.28 (0.21–0.53) | 0.23 (0.19–0.30) | 0.28 (0.16–0.50) | 0.73 (0.30–3.10) | 0.014∗ |
| Blood biochemistry | |||||
| Albumin (35–55 g/L) | 38 ± 5 | 40 ± 3 | 39 ± 5 | 34 ± 4 | 0.003∗ |
| Alanine aminotransferase (5.0–40.0 U/L) | 24.5 (15.8–52.5) | 27.0 (18.0–42.5) | 21.5 (15.0–57.5) | 25.0 (14.5–72.5) | 0.839 |
| Aspartate aminotransferase (8.0–40.0 U/L) | 27.0 (18.0–38.0) | 27.0 (25.5–37.0) | 25.5 (16.0–37.5) | 23 .0 (15.5–80.5) | 0.570 |
| Total bilirubin (3.40–20.50 μmol/L) | 11.58 ± 5.44 | 9.24 ± 3.53 | 12.99 ± 5.27 | 12.34 ± 6.86 | 0.296 |
| Serum creatinine (62.0–115.0 μmol/L) | 78.5 ± 10.9 | 80.0 ± 12.4 | 81.4 ± 11.9 | 73.8 ± 6.9 | 0.284 |
| Lactate dehydrogenase (109.0–245.0 U/L) | 210 .0 (162.0–259.0) | 211.0 (203.0–237.5) | 195.0 (148.8–250.0) | 209.0 (159.0–367.5) | 0.800 |
| Interleukin-6 (0.0–7.0 pg/mL) | 11.0 (5.7–26.5) | 11.0 (6.7–15.0) | 8.6 (6.6–18.8) | 26.4 (4.8–50.4) | 0.601 |
| C-reactive protein (0.068–8.20 mg/L) | 7.55 (4.23–15.23) | 7.40 (2.60–9.00) | 7.46 (4.29–27.13) | 12.60 (4.83–28.23) | 0.273 |
| Procalcitonin (0.00–0.50 ng/mL) | 0.05 (0.04–0.07) | 0.04 (0.03–0.06) | 0.06 (0.05–0.07) | 0.04 (0.04–0.13) | 0.361 |
| ESR (0.0–15.0 mm/h) | 13.0 (8.0–43.3) | 8.0 (6.0–20.5) | 12.0 (7.8–34.5) | 53.0 (16.0–86.0) | 0.011∗ |
| Serum ferritin (30.0–400.0 ng/mL) | 445.0 (317.7–800.0) | 311.2 (64.0–457.2) | 492.3 (414.1–1024.3) | 836.4 (425.0–1280.0) | 0.038∗ |
| Glucose (3.9–6.1 mmol/L) | 5.3 (5.0–7.3) | 5.1 (4.6–5.5) | 5.2 (4.9–7.9) | 7.3 (5.3–11.7) | 0.069 |
| Lactate (0.6–2.2 mmol/L) | 2.3 ± 1.0 | 2.0 ± 0.5 | 2.4 ± 1.2 | 2.5 ± 1.1 | 0.584 |
Categorical variables are presented as number. Continuous variables are shown as mean ± standard deviation (SD) or median (interquartile range) depending on whether they are normally distributed. P-values indicate the comparison between the mild, moderate, and severe groups, and were calculated by the Fisher's exact test or ANOVA, as appropriate.
Value in bracket indicates reference range.
Data available for 22 patients.
P < 0.05.
ESR: Erythrocyte sedimentation rate.
Plasma levels of DAMPs in COVID-19 patients and HDs
ELISA assays were used to determine the concentration of plasma DAMPs. Five DAMP proteins were chosen in this study considering the availability of commercial kits at the time of the COVID-19 outbreak. Three of these proteins (S100A8/A9 heterodimer, HMGB1, and histone H4) are classical DAMPs that have been well studied in other inflammatory diseases.[10] Additionally, CIRBP was reported to play a pathogenic role in a mouse model of sepsis by inducing TNF-α and HMGB1.[14] Finally, SP-A is preferentially expressed in lung epithelial cells and is released into the blood during lung injury.[15]
We found plasma levels of the S100A8/A9 heterodimer (1490.8 ± 819.3 vs. 742.2 ± 300.8, P = 0.015) and HMGB1 (38292.3 ± 4564.4 vs. 32686.3 ± 3678.1, P = 0.002) were higher in COVID-19 patients compared to HDs [Figure 1A, B]. Moreover, plasma S100A8/A9 heterodimer levels did not return to normal after discharge (ie, during the convalescent phase) [Figure 1A], whereas HMGB1 levels decreased significantly compared to admission levels (P = 0.049) [Figure 1B]. As expected, plasma SP-A levels in COVID-19 patients were higher than that in HDs (6713.6 ± 1708.7 vs. 5296.3 ± 1240.4, P = 0.048) and decreased during the recovery stage [Figure 1C]. Surprisingly, plasma CIRBP levels in COVID-19 patients were dramatically lower than that in HDs (57.4 ± 30.7 vs. 111.9 ± 55.2, P = 0.004) and did not return to their normal levels during the convalescent phase [Figure 1D]. No significant difference in plasma Histone H4 levels was observed between COVID-19 patients and HDs (Supplemental Digital Content, Figure 2A, B). Furthermore, the plasma levels of these five DAMPs did not vary among mild, moderate, and severe patients, indicating these proteins are not affected by disease severity (Supplemental Digital Content, Figure 1).
Figure 1.
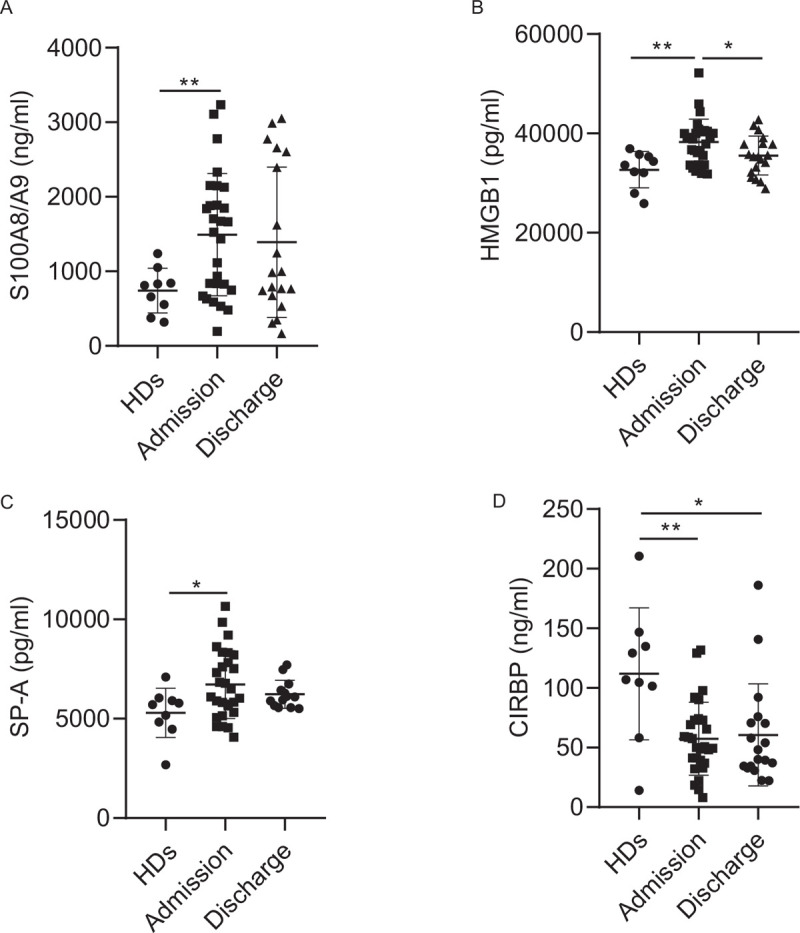
Plasma DAMPs were differently regulated in COVID-19 patients. Plasma concentration of S1008/A9 (A), HMGB1 (B), SP-A (C), and CIRBP (D) in healthy donors (HDs, n = 9) or COVID-19 patients at admission (n = 28) and discharge (n = 19; sample at discharge stage for SPA-A analysis: n = 13) as analyzed by ELISA. Data are expressed as mean ± SD. Two-tailed Mann-Whitney U test, ∗P < 0.05; ∗∗P < 0.01. HMGB1: histones and high mobility group box 1; SP-A: sufactant protein A; CIRBP: cold-inducible RNA-binding protein; HD: Health Donors.
Receiver operating characteristic analysis of plasma DAMPs from COVID-19 patients and HDs
Next, we analyzed whether DAMPs could be used as predictors of SARS-CoV-2 infection. The receiver operating characteristic (ROC) curve for each DAMP molecule was calculated using the plasma levels at admission. As shown in Figure 2, the ROC curves of the S100A8/A9 heterodimer, HMGB1, and CIRBP were able to distinguish COVID-19 patients from HDs. The area under the curve (AUC) with 95% confidence interval were 0.8429 for HMGB1 (P = 0.0015), 0.7893 for S100A8/A9 heterodimer (P = 0.0073), 0.6571 for SP-A (P = 0.15), and 0.8161 for CIRBP (P = 0.0033), respectively [Figure 2A–D]. The AUC for Histone H4 was also analyzed; however, it could not discriminate COVID-19 patients from HDs (Supplemental Digital Content, Figure 2C).
Figure 2.
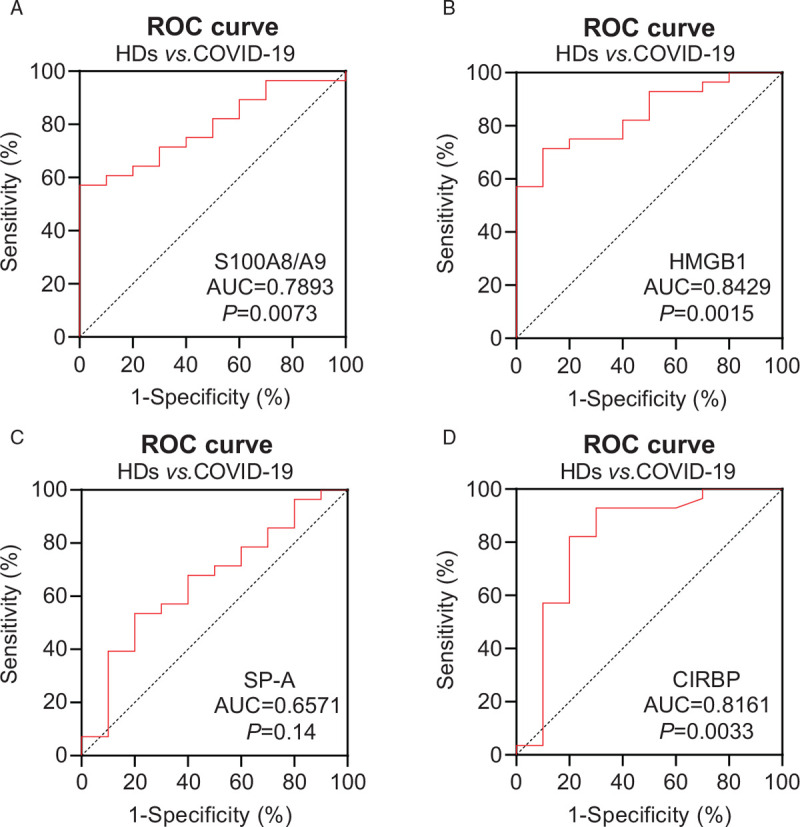
Receiver operating characteristic (ROC) analysis of plasma DAMPs in COVID-19 patients and healthy donors (HDs). The sensitivity and specificity for both COVID-19 patients and HDs were calculated. The ROC curves of S100A8/A9 (A), HMGB1(B), SP-A (C), CIRBP(D) were plotted and the area under the curve (AUC) was calculated with the 95% confidence intervals (CIs) based on DeLong's method.
Correlation analysis of DAMPs and clinical parameters
We performed a correlation analysis of DAMPs with plasma cytokines, immune cell counts, T cell activation status, and other clinical parameters. Plasma levels of S100A8/A9 heterodimer showed a significant negative correlation with eosinophil cell count, T cell count, and CD4+ T cell count, and a positive correlation with lactic dehydrogenase (LDH) concentration [Figure 3]. Meanwhile, HMGB1 levels positively correlated with plasma IP-10 and IFN-γ, two well-known predictors of COVID-19 progression.[16] Interestingly, HMGB1 levels positively correlated with T cell count, CD4+ T cell, and CD8+ T cell count. Moreover, HMGB1 levels positively correlated with the number of active CD4+ T cells (ie, CD4+HLA-DR+ T cells). HMGB1 levels also negatively correlated with blood glucose and lactate concentrations.
Figure 3.
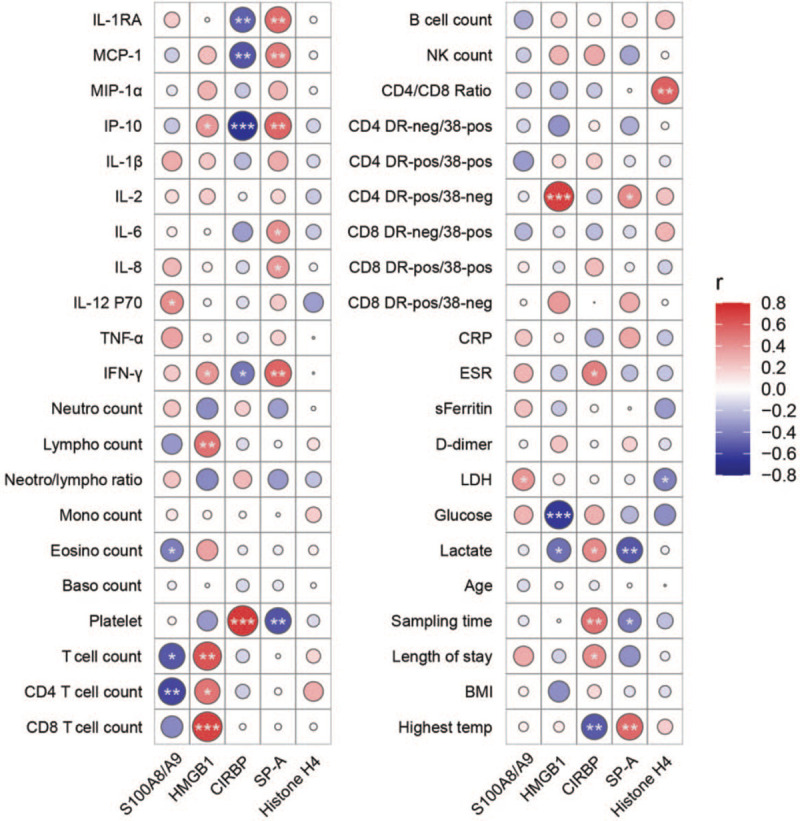
Correlation analysis of plasma concentrations of danger-associated molecular patterns and clinical parameters of COVID-19 patients. Pearson's correlation analysis of DAMPs and clinical parameters performed by the ggcor package of the R software. ∗P < 0.05; ∗∗P < 0.01, ∗∗∗P < 0.001. IL-1RA: Interleukin1 receptor antagonist; MCP-1: Macrophage chemoattractant protein-1; MIP-1α: Macrophage inflammatory protein-1 alpha; IP-10: Interferon gamma-induced protein 10; TNF-α: Tumor necrosis factor alpha; IFN-γ: Interferon γ; Neutro: Neutrophil; Lympho: Lymphocyte; Mono: Monocyte; Eosino: Eosinophil; Baso: Basophil; NK: Natural killer cell; DR: HLA-DR; neg: Negative; 38: CD38; pos: Positive; CRP: C-reactive protein; ESR: Erythrocyte sedimentation rate; sFerritin: Serum ferritin; LDH: Lactate dehydrogenase; Sampling time: Time after symptom onset; BMI: Body-mass index.
CIRBP levels negatively correlated with levels of IL-1RA, MCP-1, IP-10, IFN-γ, and the highest body temperature. Meanwhile, CIRBP levels positively correlated with platelet levels, erythrocyte sedimentation rate (ESR), and lactate concentration. CIRBP levels also positively correlated with time after symptom onset and length of hospital stay. We also found a strong negative correlation between CIRBP and SP-A levels. SP-A levels were positively related to IL-1RA, MCP-1, IP-10, and IFN-γ levels. Moreover, SP-A levels showed strong positive correlation with IL-6, IL-8, C-reactive protein (CRP) levels, and the highest body temperature. Finally, SP-A levels negatively correlated with time elapsed after symptom onset and the length of hospital stay.
Discussion
While much investigation has focused on external triggers of hyperinflammation, our study focused on the potential role of endogenous danger signals in modulating the lung injury response during COVID-19. After analyzing the circulating levels of five DAMPs (HMGB1, S100A8/A9, SP-A, CIRBP, and Histone H4) in 28 COVID-19 patients (nine mild, ten moderate, and nine severe cases), we found that plasma levels of the S100A8/A9 heterodimer, HMGB1, and CIRBP were able to distinguish COVID-19 patients from HDs. Moreover, S100A8/A9 heterodimer, HMGB1, and CIRBP plasma levels correlated with lymphopenia, inflammatory cytokines, and T cell activation status in COVID-19 patients. Furthermore, our findings suggest SP-A may involve in the inflammation of COVID-19, while CIRBP likely plays a protective role.
HMGB1 was one of the first members of the DAMP family to be identified. HMGB1 triggers inflammatory signals in multiple ways, by directly activating the Toll-like receptor (TLR) 4 or forming complexes with extracellular DNA, RNA, and other DAMPs or pathogen-associated molecular patterns (PAMPs).[17] Thus, HMGB1 plays a crucial role as a sterile inflammatory mediator. Notably, a recent study found that CD24Fc treatment, which is able to quell HMGB1 signaling,[18] protected against viral pneumonia in simian immunodeficiency virus-infected Chinese rhesus monkeys.[19] Hence CD24Fc is being tested as an immunomodulator drug in COVID-19 patients in a phase 3 clinical trial (NCT04317040). Whether HMGB1 contributes to the harmful hyper-inflammation loop induced by PAMPs and DAMPs in COVID-19 patients would be worth investigating.[20]
S100A8 (MRP8) and S100A9 (MRP14) are calcium-binding proteins that belong to the S100 family, and mainly exist as a heterodimer known as S100A8/A9 or calprotectin. Calprotectin is found in abundance in neutrophils and is released extracellularly upon neutrophil activation or death, where it serves as a pro-inflammatory ligand for TLR4. High levels of calprotectin have been found in many types of infectious and inflammatory diseases, including sepsis, myocardial infarction, and inflammatory bowel disease.[21] We found that the plasma S100A8/A9 level was elevated in COVID-19 patients, and negatively correlated with CD4+ T cell count and positively correlated with LDH concentration. Similar to our findings, Chen et al. reported that serum levels of S100A8/A9 at admission positively correlated with peak CT score and oxygen demand in patients with COVID-19, which is indicative of the severity of lung injury.[22] In another study, Shi et al. found that COVID-19 patients have markedly elevated peripheral levels of S100A8/A9, which was suggested to be a predictive marker of severe disease.[21] In addition, Silvin et al. showed that a burst of plasma S100A8/A9 precedes cytokine release syndrome and could be used to discriminate patients who develop the severe form of COVID-19.[23]
SP-A is a pulmonary surfactant that is synthesized and secreted into the alveoli by type II epithelial cells. SP-A mainly regulates surfactant metabolism and exerts immune-modulatory functions.[24] Indeed, circulating SP-A has been used as a prognostic biomarker for interstitial lung diseases[25] and ARDS.[26,27] We observed that plasma SP-A concentrations were significantly higher in COVID-19 patients, and correlated positively with inflammatory cytokines and negatively with time after symptom onset. Therefore, SP-A may be a useful biomarker for SARS-CoV-2 infection and may play a role in amplifying inflammation.
CIRBP is an RNA-binding protein located in the nucleus that is constitutively expressed at a low level in the steady state, but is up-regulated during mild hypothermia, exposure to UV irradiation, and hypoxia.[14] Under cold stress, CIRBP translocates from the nucleus to the cytoplasm to regulate its target mRNAs and protects the cell from apoptosis.[28] However, if secreted outside the cell, CIRBP can modulate the inflammatory response through the TLR4/myeloid differentiation protein-2 (MD2) signaling pathway.[29] We found that plasma CIRBP levels were significantly decreased in COVID-19 patients compared with HDs. This finding is reminiscent of a recent study that linked chronic hypoxia to CIRBP depression, thereby explaining why patients with chronic hypoxia have worse myocardial reperfusion injury than normoxic patients after similar periods of ischemic cardioplegic arrest.[30] Indeed, hypoxia is one of the most frequent symptoms in COVID-19 patients, and mortality rates tend to be higher in individuals with cardiac comorbidities.[2,31] We found CIRBP levels positively correlated with time elapsed after symptom onset and length of hospital stay, and negatively correlated with inflammatory cytokines. These observations suggest CIRBP might be protective in COVID-19. Therefore, strategies aimed at compensating for the loss of CIRBP would likely be helpful in the treatment of COVID-19.
There are several limitations to this study. First, only five DAMPs were included in the current study, and it is not clear whether other DAMPs are also involved. Second, further research is needed to clarify the upstream and downstream mechanism(s) of DAMPs in COVID-19. Third, the sample size is small, and we failed to differentiate between different symptoms. Therefore, a larger sample size with longitudinal data collection is warranted in further studies.
Acknowledgements
We thank Jin-Hong Yuan for flow cytometry analysis.
Funding
This work was supported by the Innovation Groups of the National Natural Science Foundation of China (No. 81721002) and the National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2017ZX10202102-004-002).
Author Contributions
Fu-Sheng Wang and Chao Zhang conceived and designed the study; Xing Fan, Jin-Wen Song, Wen-Jing Cao, Xiu-Wen Wang, Ming-Ju Zhou, Tao Yang, Chun-Bao Zhou and Chao Zhang performed the experiments; Xing Fan, Chao Zhang and Fu-Sheng Wang wrote the manuscript; Si-Yu Wang and Fan-Ping Meng took care of patients and provided the clinical information; Jun Hou provided the safety guide for handling COVID-19 patients’ samples; Ji-Yuan Zhang and Ming Shi provided comments and feedback. All authors read and approved the final manuscript.
Conflicts of interest
None.
Supplementary Material
References
- [1].Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- [5].Tay MZ, Poh CM, Renia L, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Song JW, Zhang C, Fan X, et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun 2020;11(1):3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang JY, Wang XM, Xing X, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- [8].Kahn R, Schmidt T, Golestani K, et al. Mismatch between circulating cytokines and spontaneous cytokine production by leukocytes in hyperinflammatory COVID-19. J Leukoc Biol 2021;109:115–120. doi: 10.1002/jlb.5covbcr0720-310rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID-19. Immunity 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol 2020;15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- [11].Tolle LB, Standiford TJ. Danger-associated molecular patterns (DAMPs) in acute lung injury. J Pathol 2013;229(2):145–156. doi: 10.1002/path.4124. [DOI] [PubMed] [Google Scholar]
- [12].Hou X, Qin J, Zheng X, et al. Potential role of high-mobility group box 1 protein in the pathogenesis of influenza H5N1 virus infection. Acta Virol 2014;58(1):69–75. doi: 10.4149/av_2014_01_69. [DOI] [PubMed] [Google Scholar]
- [13].Nosaka N, Yashiro M, Yamada M, et al. Anti-high mobility group box-1 monoclonal antibody treatment provides protection against influenza A virus (H1N1)-induced pneumonia in mice. Crit Care 2015;19(1):249. doi: 10.1186/s13054-015-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qiang X, Yang WL, Wu R, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med 2013;19(11):1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kuroki Y, Tsutahara S, Shijubo N, et al. Elevated levels of lung surfactant protein A in sera from patients with idiopathic pulmonary fibrosis and pulmonary alveolar proteinosis. Am Rev Respir Dis 1993;147(3):723–729. doi: 10.1164/ajrccm/147.3.723. [DOI] [PubMed] [Google Scholar]
- [16].Chi Y, Ge Y, Wu B, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis 2020;222(5):746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang H, Wang H, Ju Z, et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med 2015;212(1):5–14. doi: 10.1084/jem.20141318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen GY, Tang J, Zheng P, et al. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 2009;323(5922):1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tian RR, Zhang MX, Liu M, et al. CD24Fc protects against viral pneumonia in simian immunodeficiency virus-infected Chinese rhesus monkeys. Cell Mol Immunol 2020;17(8):887–888. doi: 10.1038/s41423-020-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Andersson U, Ottestad W, Tracey KJ. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol Med 2020;26(1):42. doi: 10.1186/s10020-020-00172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shi H, Zuo Y, Yalavarthi S, et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol 2021;109:67–72. doi: 10.1002/jlb.3covcra0720-359r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen L, Long X, Xu Q, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol Immunol 2020;17(9):992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Silvin A, Chapuis N, Dunsmore G, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Papaioannou AI, Konstantelou E, Papaporfyriou A, et al. Serum surfactant protein levels in patients admitted to the hospital with acute COPD exacerbation. Lung 2018;196(2):201–205. doi: 10.1007/s00408-018-0099-5. [DOI] [PubMed] [Google Scholar]
- [25].Takahashi H, Kuroki Y, Tanaka H, et al. Serum levels of surfactant proteins A and D are useful biomarkers for interstitial lung disease in patients with progressive systemic sclerosis. Am J Respir Crit Care Med 2000;162(1):258–263. doi: 10.1164/ajrccm.162.1.9903014. [DOI] [PubMed] [Google Scholar]
- [26].Eisner MD, Parsons P, Matthay MA, et al. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax 2003;58(1):983–988. doi: 10.1136/thorax.58.11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Greene KE, Wright JR, Steinberg KP, et al. Serial changes in surfactant-associated proteins in lung and serum before and after onset of ARDS. Am J Respir Crit Care Med 1999;160(6):1843–1850. doi: 10.1164/ajrccm.160.6.9901117. [DOI] [PubMed] [Google Scholar]
- [28].Zhang HT, Xue JH, Zhang ZW, et al. Cold-inducible RNA-binding protein inhibits neuron apoptosis through the suppression of mitochondrial apoptosis. Brain Res 2015;1622:474–483. doi: 10.1016/j.brainres.2015.07.004. [DOI] [PubMed] [Google Scholar]
- [29].Aziz M, Brenner M, Wang P. Extracellular CIRP (eCIRP) and inflammation. J Leukoc Biol 2019;106(1):133–146. doi: 10.1002/JLB.3MIR1118-443R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu Y, Xing J, Li Y, et al. Chronic hypoxia-induced Cirbp hypermethylation attenuates hypothermic cardioprotection via down-regulation of ubiquinone biosynthesis. Sci Ttransl Med 2019;11(489):eaat8406. doi: 10.1126/scitranslmed.aat8406. [DOI] [PubMed] [Google Scholar]
- [31].Zhang C, Wang FS, Silvestre JS, et al. Is aberrant CD8+ T cell activation by hypertension associated with cardiac injury in severe cases of COVID-19? Cell Mol Immunol 2020;17(6):675–676. doi: 10.1038/s41423-020-0454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


