Abstract
The elaboration of novel pathogenic aspects of multiple sclerosis (MS) requires the analysis of well‐defined stages of lesion development. However, specimens of certain stages and lesion types are either present in small brain biopsies, insufficient in size for further molecular studies or available as formalin‐fixed and paraffin‐embedded (FFPE) material only. Therefore, application of current molecular biology techniques to FFPE tissue is warranted. We compared FFPE and frozen tissue by using quantitative polymerase chain reaction and report: (1) FFPE material is highly heterogeneous regarding the utility for transcript profiling of mRNAs; well‐preserved FFPE samples had about a 100‐fold reduced sensitivity compared with frozen tissue, but gave similar results for genes of sufficient abundance; (2) FFPE samples not suitable for mRNA analysis are still highly valuable for miRNA quantification; (3) the length of tissue fixation greatly affects utility for mRNA but not for miRNA analysis; (4) FFPE samples can be processed via a hot water bath for dissection of defined lesion areas; and (5) in situ hybridization for proteolipid protein (PLP) helps to identify samples not suitable for mRNA amplification. In summary, we present a detailed protocol how to use autoptic FFPE tissue for transcript profiling in dissected tissue areas.
Keywords: FFPE, microRNA, molecular analysis, multiple sclerosis, quantitative PCR, transcriptome
INTRODUCTION
Recent advances in molecular biological techniques offer attractive tools to investigate pathogenetic mechanisms of multiple sclerosis (MS) or any other human disease. However, this requires that the appropriate tissue and analytical techniques can be brought together 9, 17, 33, 35, 36. Particularly, the initial stages of MS are characterized by a focal pathology and pronounced temporal dynamic, and these are therefore of special interest. The majority of tissue specimens containing these lesion stages and types (eg, fulminant active MS lesions), are either available as small brain biopsies, where lesion size is often insufficient for further molecular studies, or available as formalin‐fixed and paraffin‐embedded (FFPE) material only. Whereas good preservation of tissue structures in FFPE material allows for precise histopathological and immunohistochemical evaluation, investigation of RNA expression levels with modern techniques is challenging because of impaired RNA quality. Also, FFPE specimens could be useful for investigation of rare diseases, of which samples are difficult to obtain for prospective brain banks. Therefore it is interesting to see to which extent FFPE material can be used for such an approach and to provide a practical protocol for routine screening for the utility of FFPE material, in which conditions of tissue conservation are unknown.
Most of the previous studies on RNA expression analysis in FFPE tissue have used shortly fixed biopsy material 1, 10, 18, 25, 31, 34, 39, 40, 42. In contrast, archival brain tissues are obtained at autopsy and conditions of tissue conservation often variable and undefined. In these tissues RNA degradation by terminal hypoxia and post‐mortem autolysis add to the inevitable RNA destruction and modification during tissue fixation and paraffin embedding 3, 6, 8, 20, 22, 26. However, studies using frozen specimens of brain tissue obtained by autopsy showed considerable stability of messenger RNA (mRNA) up to 48 h post‐mortem 7, 12, 29, 43.
In addition to mRNA, non‐coding RNA, for example, microRNAs (miRNAs), have increasingly been recognized as important players in physiology and pathology of many diseases including MS (14). Both microarray and quantitative polymerase chain reaction (qPCR) have been used to analyze miRNA expression in human tissue 34, 40, 42.
The purpose of this study was to elucidate to what extent FFPE MS tissue specimens of autoptic origin can be used for reliable transcript profiling using qPCR technology and to present a detailed protocol how to do this. We specifically addressed the following issues: (i) RNA yield in FFPE compared with frozen brain tissue; (ii) ability to amplify transcripts coding for proteins and miRNAs in randomly selected archival brain specimens; (iii) effect of duration of tissue fixation on the amplification of protein coding mRNA and miRNAs in brain tissue; (iv) application of a more complex experimental setup including dissection of defined tissue areas from FFPE brain tissue sections and flattening in a hot water bath; (v) linkage of signal intensity of in situ hybridization to subsequent utility for qPCR amplification; (vi) comparison of FFPE vs. frozen tissue performance in analysis of gene expression profiles by TaqMan low density arrays (LDAs). For this purpose 84 genes coding for extracellular matrix (ECM)‐related proteins were selected given their differential regulation in MS lesions and their wide spectrum of abundance in brain tissue (23).
MATERIALS AND METHODS
Analyzed tissue
In total, we analyzed 74 autoptic tissue samples (frozen n = 22, FFPE n = 52) of 19 MS patients, one patient with Alzheimer's disease, one patient with meningitis tuberculosis and 12 healthy control donors by qPCR and in situ hybridization. Frozen samples were snap frozen in liquid nitrogen and then stored at −80°C. FFPE specimens had been formalin fixed under variable conditions over different time periods before paraffin embedding. Tissue samples were kindly provided by the Center for Brain Research in Vienna, the Netherlands Brain Bank, the NeuroResource at UCL Institute of Neurology in London and the Neurobiobank Munich (Supporting Information Table S1).
Tissue conservation for experiments analyzing the effect of fixation time
To analyze the effect of fixation time on transcript amplification, 30 tissue specimens from six MS patients and three healthy donors were fixed in 4% paraformaldehyde (PFA) at 4°C for time periods ranging from 2 days to 3 years. After fixation, all samples were embedded in paraffin and stored at room temperature. One additional tissue specimen of each of these nine donors was snap frozen in liquid nitrogen and stored at −80°C. Three FFPE tissue specimens of three further MS patients were fixed in buffered formalin at room temperature for 1 month before paraffin embedding. From these three specimens also snap frozen mirror blocks were available.
Tissue processing and MS lesion dissection
FFPE tissue sections were obtained using a sliding microtome (SM 200R, Leica, Wetzlar, Germany). In one set of experiments the sections were directly collected in RNAse free reaction tubes. Subsequently, the tissue was deparaffinized by incubation in xylol for 2 × 10 minutes followed by centrifugation (5 minutes at 12 000 rounds per minute). Supernatant was discarded before rehydration in descending dilutions of ethanol (∼100% → 90% → 70%) for 7 minutes each followed by centrifugation (5 minutes at 12 000 rounds per minute) and discarding of the supernatant. To optimize RNA yield from FFPE tissue, we compared efficiency of RNA isolation in 4‐, 6‐ and 8‐µm sections and found 6‐µm sections to give the most efficient RNA yield. Therefore, 6‐µm
In another set of experiments, specific tissue areas were sections were used in subsequent experiments.dissected. For this purpose, 6‐µm FFPE tissue sections were flattened in a 50°C DEPC water bath, taken onto polyethylene (PEN) membrane covered object slides (P.A.L.M Microsystems, Bernried, Germany) and air dried at 37°C overnight. Every 10th section underwent Luxol Fast Blue (LFB) staining and was used as a model for proper lesion dissection. All other sections were deparaffinized and rehydrated as described earlier. Then the MS lesion or the healthy white matter was excised with a scalpel and tissue was collected in RNase free reaction tubes for further processing. The total tissue amount ranged from 180 to 300 µm × 0.5 cm × 0.5 cm.
Of the frozen specimens, 10‐ to 20‐µm‐thick sections were obtained and mounted onto PEN membrane‐coated object slides. Every 6th section was stained with LFB. Then MS lesions and healthy white matter were excised as described for FFPE tissue. The total tissue amount ranged from 200 to 300 µm × 0.5 cm × 0.5 cm.
This experimental setup was applied to 10 MS lesions (FFPE n = 3, frozen n = 7) and 8 white matter control samples (FFPE n = 2, frozen n = 6).
Tissue lysis and RNA isolation
For tissue lysis, 30 µL lysis buffer [50 mMTris, 25 mM ethylenediaminetetraacetic acid (EDTA), 500 mMNaCl, 0.1% Nonidet® P‐40, 1% sodium dodecyl sulfate (SDS)] and 300 µL Proteinase K (from Tritirachium album, c = 20 mg/mL, molecular grade, RNAse free, Sigma, Munich, Germany) were added per 1.5 mL reaction tube and incubated at 60°C for 14 h. To improve tissue lysis, reaction tubes were vortexed every 30 minutes at 1000 rpm for 5 minutes. RNA was isolated by: (i) adding 1 mL TRI®Reagent (Sigma) and 200 µL chloroform; (ii) collecting the aequous supernatant; and (iii) precipating RNA with isopropanol. This was done twice to obtain high RNA purity. Finally, RNA was washed with 75% ethanol. RNA amount and purity [optical density (OD) 260/280 ratio] were assessed with the Nanodrop® ND‐1000 Spectrophotometer. For some samples, RNA was extracted using the Roche high pure FFPE RNA micro kit (Roche Diagnostics GmbH, Mannheim, Germany). To determine the RNA quality, one µL of the test RNA (1–5 ng RNA/µL) was applied to an RNA Pico Chip (Agilent Technologies, Böblingen, Germany), which was then run on Agilent 2100 bioanalyzer.
Reverse Transcription, quantitative PCR, TaqMan® LDAs
For manually pipetted qPCR assays, cDNA from mRNA was synthesized using random hexamer primers (Roche Diagnostics GmbH) and Moloney Murine Leukemia Virus Reverse Transcriptase (Promega, Madison, WI, USA) or the ABI cDNA kit (Applied Biosystems, Darmstadt, Germany). MiRNAs were reversely transcribed with the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems) and miRNA specific RT primers (TaqMan® miRNA Assays, Applied Biosystems). Quantitative PCR for the housekeeping genes peptidyl‐prolyl‐isomerase A (PPIA) and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as well as miR‐181a and miR‐124 was performed using the qPCR Core Kit (RT‐QP 73‐05, Eurogentec, Seraing, Belgium) together with TaqMan® PCR Assays and TaqMan® miRNA Assays (both Applied Biosystems), respectively (Supporting Information Table S2) TaqMan®qPCR was performed on the 7900 HT Fast Real‐Time PCR System (Applied Biosystems). The amount of cDNA used per manually pipetted qPCR reaction was usually 25 ng RNA equivalents for coding genes, but 9 ng in one case (MS 38). In this case, the Ct value for PPIA used in the analysis comparing qPCR and in situ hybridization results was corrected arithmetically according to the exponential nature of the amplification: Ctcorr = Ctmeasured + log2(actually used ng/25 ng). For miRNA analysis, 5 ng RNA equivalents were employed. Technical duplicates were measured and the standard deviation was calculated as 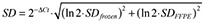 based on 16, 24.
based on 16, 24.
For theTaqMan® LDA analysis, cDNA was synthesized using the High‐Capacity cDNA Reverse Transkription Kit (Applied Biosystems). TaqMan® LDA reactions contained Gene Expression Mastermix (Applied Biosystems) and a RNA equivalent of 3–9.5 ng per reaction well. We conducted gene expression profiling in 18 tissue specimens (10 MS lesions (FFPE n = 3, frozen n = 7) and 8 healthy controls (FFPE n = 2, frozen n = 6) for 84 ECM‐related genes (50 ECM components and 34 ECM modifying enzymes) on custom made TaqMan® LDAs (Applied Biosystems). PPIA and GAPDH were included as housekeeping genes (Supporting Information Table S2).
Data were analyzed with SDS 2.3 and RQ Manager 1.2 Software (Applied Biosystems). After comparing the ability to amplify of GAPDH and PPIA in the five selected FFPE samples, PPIA was selected as the reference housekeeping gene because it was more abundant than GAPDH: mean Ct (PPIA, FFPE specimens) = 26.54; mean Ct (GAPDH, FFPE specimens) = 29.53. For normalization, relative gene expression with respect to the housekeeping gene PPIA was calculated and is shown as percent PPIA: % PPIA = 2−ΔCt · 100 with ΔCt = Ct (target gene) – Ct (PPIA).
In situ hybridization
In situ hybridization was performed essentially as described before (2): After deparaffinization and rehydration, RNA was fixed in 4% PFA for 20 minutes. Sections were rinsed with Tris buffered saline (TBS), followed by protein denaturation by incubation with 0.2 M HCl. Sections were again rinsed in TBS and then the tissue was partially digested with proteinase K (in TBS, supplemented with CaCl2) to expose the RNA. Afterwards the slides were washed in TBS and left at 4°C for 5 minutes. To avoid non‐specific binding, the slides were incubated with 0.5% acetic anhydride (in TBS, pH 8) for 10 minutes. Slices were dehydrated and put into a wet chamber (50°C, 30 minutes) to facilitate the dispersal of the hybridization mix containing the proteolipid protein 1 (PLP1) probe labeled with digoxigenin. The 1.4 kb RNA probe was constructed from a plasmid containing the DM20 variant of the mouse PLP1 sequence. Because of the high degree of sequence homology, this 1.4 kb RNA probe also binds to human PLP1/DM20. After application of the probe, a coverslip was put for protection and the slide was heated to 95°C for 4 minutes to linearize the RNA. Afterwards the hybridization was allowed to take place at 65°C over night. The coverslip was removed by incubation in 2× saline sodium citrate (SSC) followed by three highly stringent washing steps with 50% formamide in 1× SSC for 20 minutes each at 55°C. Sections were then washed thrice with 1× SSC (15 minutes per washing cycle), then rinsed with TBS and incubated with Boehringer Blocking Reagent containing 10% FCS for 15 minutes. Anti‐digoxigenin antibodies, which were coupled to alkaline phosphatase (Roche) and dissolved in blocking reagent, were applied to the slices for 1 h. After five final washing steps with TBS, the development was performed with NBT/BCIP at 4°C, up to 140 h. For assessment of RNA quality, the signal appearance was controlled microscopically. Signal strength and distribution was evaluated semiquantitatively after 26 or 70 h of incubation as described detailed in the legend of Figure 4.
Figure 4.
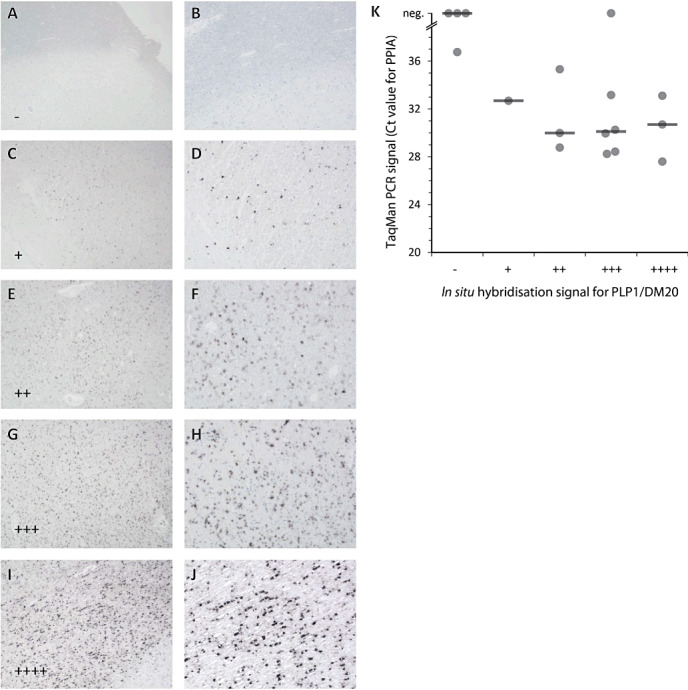
Comparison of in situ hybridization for PLP1/DM20 and quantitative polymerase chain reaction (qPCR) signal. Strength of the in situ hybridization signal of different tissue blocks was compared after 26 h of substrate reaction. Five different signal intensities were defined: (A,B) no signal (−); (C,D) weak signal in focal cells (+); (E,F) weak signal in most oligodendrocytes (++); (G,H) strong signal focally and moderate signal in most oligodendrocytes (+++); (I,J) strong signal in most/all oligodendrocytes (++++). Magnification: A, C, E, G, I: ×32; B, D, F, H, J: 80. K. qPCR Ct values for PPIA were plotted against the PLP1/DM20 in situ hybridization signal. A negative qPCR signal (“neg.”) was defined as no signal after 40 cycles. Horizontal lines indicate medians. The negative correlation between in situ hybridization signal and raw Ct value was statistically significant (P = 0.01, Spearman's σ = −0.6).
Statistical analysis
We tested the hypothesis that in situ hybridization signals and qPCR Ct values were negatively correlated (higher in situ hybridization signal ∼ lower Ct value) using one‐sided Spearman's rank correlation in R version 2.12.1 (R Development Core Team (2010). R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria. ISBN 3‐900051‐07‐0, URL http://www.R‐project.org/) (28). cor.test Function was used with option exact = F as ties were present, resulting in P = 0.008 and σ = −0.57. To further address the extent of uncertainty introduced by ties, we applied a random tie breaking approach. In brief, ties were resolved by adding a random jitter that was smaller than the smallest non‐zero difference between data points, allowing us to apply cor.test function with option exact = TRUE in an iteration loop (i = 100 000). The median P (0.012) and σ (−0.55) values were only slightly different from the original ones, and 98.7% of all iterative P‐values were <0.05. Therefore we concluded that the correlation between in situ hybridization signal and qPCR Ct value has to be regarded as significant with rounded P = 0.01 and rounded σ = −0.6 (Figure 4).
RESULTS
RNA yield from FFPE tissue is comparable with frozen tissue
In order to optimize the RNA yield from FFPE tissue blocks, we compared variations of section thickness and number of sections per reaction tube. Thereby we found that the RNA yield was optimal with 6‐µm sections at a maximum of 7 sections per 1.5 mL reaction tube. For average sized tissue blocks, a total tissue amount of 126 µm × 1–1.5 cm × 1–1.5 cm, which was processed at a maximum of 7 × 6‐µm sections per reaction tube was most efficient with respect to RNA yield and technical convenience. This was applied to 38 FFPE samples (nine MS and seven healthy donors) and the mean RNA yield was 5.8 µg (range 1.5–15.5 µg). This is comparable with the average RNA yield of 4.7 µg (range 2.4–7.0 µg) in 10 frozen samples (six MS and four healthy donors) of which similar total tissue amounts were used. RNA purity was acceptable for FFPE samples with a mean OD260/280 ratio of 1.82 (range 1.57–1.97) and for frozen specimens with a mean OD260/280 ratio of 1.91 (range 1.84–1.99).
We have evaluated RNA quality of three FFPE samples with the Agilent Bioanalyzer. Neither the 28S (4718 nucleotides) nor the 18S (1874 nucleotides) bands could be detected (data not shown), which is in harmony with previous observations 4, 5. The calculated RNA integrity numbers were 2.3–2.4. Nevertheless we were able to detect amplicons with a length <150 bp and all the more miRNAs in these samples. The Ct value of the housekeeping gene GAPDH was 30.3–30.7 in these samples.
Heterogenous (about 105‐fold different) amplifiability of coding RNA in various FFPE specimens
We evaluated utility of randomly selected FFPE samples for expression analysis by qPCR. To this end, we performed qPCR for the housekeeping gene PPIA in 27 FFPE specimens from 11 MS donors and compared results for each sample to mean expression level in frozen tissue (mean expression in six frozen samples from six MS donors). For this, an expression ratio FFPE/frozen was calculated as 2−ΔCt. ΔCt was determined as difference between the Ct value for PPIA in each FFPE tissue specimen and the mean Ct for PPIA in six frozen samples [ΔCt = Ct (FFPE) − mean Ct (frozen)] (Figure 1).
Figure 1.
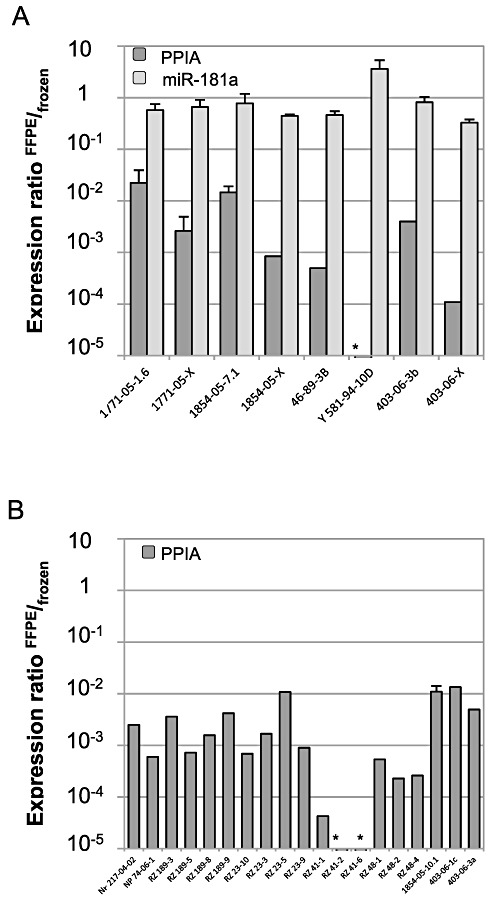
Amplification of peptidyl‐prolyl‐isomerase A (PPIA) and miR‐181a in randomly selected archival multiple sclerosis (MS) tissue samples. A. Amplification of PPIA (dark gray columns) and miR‐181a (light gray columns) was analyzed in eight formalin‐fixed and paraffin‐embedded (FFPE) specimens of five MS patients. Amplification in each FFPE specimen was compared with mean amplification in six frozen specimens (standard error of the mean < 2%). The expression ratio FFPE/frozen was calculated as 2−ΔCt with ΔCt = Ct (individual FFPE specimen) − mean Ct (n = 6 frozen). An expression ratio of 1 indicates comparable amplification in FFPE compared with frozen tissue. B. Similarly, PPIA was analyzed in 19 additional FFPE specimens of eight MS patients (dark gray columns). PPIA was not detected in samples marked with *. Error bars represent standard deviation for technical duplicates.
In all FFPE samples, amplification of the housekeeping gene PPIA was considerably reduced compared with frozen tissue. However, the extent of reduced amplification was highly different between the randomly selected FFPE samples and ranged from 45‐fold to more than 200 000‐fold (Figure 1A).
The analysis of archival tissue specimens from healthy control donors yielded similar results: we analyzed eight FFPE specimens from seven donors and compared it with three frozen samples from three donors of whom also FFPE samples were obtained. The ability to amplify PPIA was reduced by a mean of 132‐fold (data not shown).
We further had the chance to analyze three frozen MS tissue samples along with mirrored FFPE specimens reflecting the same tissue components. The ability to amplify the housekeeping gene PPIA was reduced by a mean of 277‐fold (range 81‐ to 620‐fold) (left part of Figure 2). This is in contrast to the largely unaltered amplification of miR‐181a (right part of Figure 2).
Figure 2.
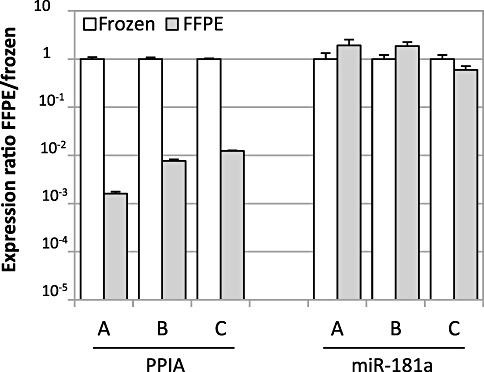
Comparative amplification of peptidyl‐prolyl‐isomerase A (PPIA) and miR‐181a in formalin‐fixed and paraffin‐embedded (FFPE)‐frozen mirror blocks. Each of three multiple sclerosis (MS) lesions (A = S06/126‐11.3, B = S04/247‐3 and C = S03/222‐9.2) was represented by a FFPE (gray columns) and a frozen (white columns) tissue part. Transcript amplification of PPIA and miR‐181a in the FFPE part was compared with the frozen part and is shown as expression ratio FFPE/frozen, which was calculated as 2−ΔCt with ΔCt = Ct (individual FFPE specimen) − Ct (corresponding frozen sample). An expression ratio of 1 indicates comparable transcript amplification in FFPE compared with frozen tissue. Error bars represent standard deviation for technical duplicates.
We considered a FFPE specimen as suitable for further qPCR analysis, if the Ct value for PPIA was <30, because this allows detection of genes with an expression level of >3% PPIA if the detection limit is set as Ct = 35. Applying this, 31% (12/38) of the analyzed archival brain specimens were classified suitable.
Amplification of miRNAs in different FFPE specimens is remarkably stable
Analogous to PPIA, we also analyzed the ability to amplify miR‐181a and miR‐124 in a total of 36 samples (FFPE n = 27, frozen n = 9) from 14 MS and 7 healthy donors. We selected these two miRNAs because they are relatively abundant in the brain (13).
Amplification of miR‐181a was analyzed in eight FFPE samples of five MS donors and compared with mean expression in six frozen samples of three overlapping MS donors. Again, the expression ratio FFPE/frozen was calculated as 2−ΔCt for each analyzed sample (Figure 1). In contrast to PPIA, the ability to amplify of miR‐181a in FFPE tissue compared with frozen tissue was found to be largely unaltered with a mean reduction by 1.7‐fold only (range 0.3‐ to 3.1‐fold, Figure 1A). Importantly, even in FFPE samples that are barely suitable for quantification of genes coding for proteins, the miRNA can be quantified without evident loss of sensitivity (eg, samples Y581‐94‐10D and 403‐06‐X in Figure 1).
This is further supported by an only slightly reduced amplification of miR‐181a in our mirrored FFPE‐frozen MS lesions. In these samples, the ability to amplify miR‐181a was reduced by a mean of 0.9‐fold (range 0.5‐ to 1.7‐fold) (Figure 2, right part). Similar results were obtained in healthy brain tissue (three FFPEs compared with three frozen samples of the same donors), where the ability to amplify of miR‐181a was reduced by a mean of 1.6‐fold (range 1‐ to 2.4‐fold) (data not shown). Similarly, miR‐124 was analyzed in 16 randomly selected archival brain samples of eight MS donors. Expression of miR‐124 was considerably robust in all 16 samples with a mean Ct of 23.3 (SEM 0.32). This is in accordance with the stable ability to amplify of miR‐181a (n = 8 FFPE samples, mean Ct 21.7, SEM 0.26).
Transcript amplification in FFPE tissue depends on duration of tissue fixation
To investigate the role of fixation time on amplification of gene transcripts we fixed tissue specimens for different periods of time before paraffin embedding and subsequent RNA analysis. For this purpose, we used 30 autoptic brain specimens (from three healthy donors and six MS patients). Of each donor one tissue sample was snap frozen in liquid nitrogen and 1–4 tissue samples were kept in 4% PFA over different time periods before embedding in paraffin. FFPE samples were grouped according to duration of formalin fixation: “days” (n = 9, fixation for 2–21 days), “months” (n = 9, fixation for 1–4 months) and “years” (n = 3, fixation for 2.5–3 years). Results in FFPE samples were compared with corresponding frozen specimens (n = 9). Expression of PPIA and miR‐181a was measured in all 30 specimens. At first, the expression ratio FFPE/frozen = 2−ΔCt was calculated for each FFPE‐frozen pair. Then, the mean expression within each group was determined. Thereby we noted that the length of fixation has a strong impact on the ability to amplify of mRNAs, but had little effect on miRNAs. Fixation for days reduced amplifiable PPIA transcripts by a mean of 15‐fold, fixation for months by 44‐fold and fixation for years by about 600‐fold. In contrast, amplification of miR‐181a was largely unaltered by duration of fixation in 4% PFA (maximal 1.7‐fold difference with year‐long fixation (Figure 3). Although the fixation period did affect the ability to amplify RNA, it did not affect the RNA yield.
Figure 3.
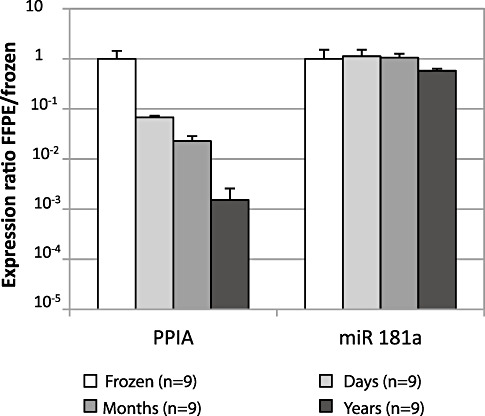
Effect of formalin fixation on amplification of PPIA and miR‐181a. Thirty FFPE samples were grouped according to three time periods of fixation in 4% PFA: “days”: (n = 9, formalin fixed for 2–21 days), “months” (n = 9, formalin fixed for 2–4 months), “years” (n = 3, formalin fixed for 2–3 years) and transcript amplification was compared with “frozen” specimens of the same donors (n = 9). ΔCt was calculated for each FFPE and the corresponding frozen sample. Mean amplification in each of the three FFPE subgroups (columns in different shades of gray) is shown in relation to the mean amplification in the group “frozen” (white columns, SEM < 2%). The expression ratio FFPE/frozen was calculated as 2−ΔCt with ΔCt = mean Ct of the FFPE subgroup − mean Ct of the frozen samples. An expression ratio of 1 indicates comparable transcript amplification in FFPE compared with frozen tissue. Error bars represent standard error of the mean (SEM).
Unaltered RNA yield and transcript amplification after dissection from membrane‐coated slides
We were looking for a suitable approach to dissect defined areas in FFPE tissue without impairment of subsequent RNA analysis. In this regard, we considered tissue flattening in a hot water bath and tissue dissection from membrane‐coated slides as critical steps. In pilot experiments with human tonsils RNA yield and the ability to amplify GAPDH was remarkably reduced if the FFPE tissue had been exposed to tap water. In contrast, if exposed to DEPC treated water, RNA yield was comparable with tissue that had been processed without exposure to water at all. In following experiments, GAPDH amplification was unaltered in archival white matter specimens, of which sections were flattened in a 50°C water bath and then dissected from PEN membrane‐coated object slides as compared with directly processed white matter (data not shown).
In situ hybridization identifies tissue specimens not suitable for further PCR analysis
To determine whether in situ hybridization is a potential tool to identify archival specimens with good RNA quality we compared performance of 17 MS brain samples using an in situ hybridization for PLP and qPCR for PPIA. We found that specimens with low raw Ct values for PPIA (ie, better RNA quality) did also have stronger signals in the in situ hybridization for PLP (Figure 4). This inverse correlation between the signal strength of in situ hybridization and raw Ct values was statistically significant (P = 0.01, Spearman's σ = −0.6). Statistical analysis was performed as described in materials and methods.
About 200 archival blocks from MS patients and control were examines by in situ hybridization in Vienna and classified for further molecular analysis based on the intensity of the in situ signal with the PLP probe after 70 h. From controls 30/47 blocks (22/27 cases) and from MS cases 45/152 blocks (27/57 cases) gave a high in situ signal. This means 50–60% of the MS blocks in a large archival collection have to be excluded from further molecular analysis. The ratios of controls and MS blocks are slightly different, because all available MS cases were analyzed, whereas in controls mainly samples with a known short fixation period were included. In 33 cases, different blocks from the same donor could be analyzed. Typically, the RNA quality was similar in blocks from the same donor, but we also noted exceptions: The ratio of concordant/discordant was 8/2 in controls and 25/10 in MS cases.
Transcript profiling in FFPE compared with frozen tissue: similar pattern despite sensitivity loss
To define sensitivity and reliability of qPCR studies in FFPE compared with frozen tissue we quantified the expression of 84 ECM‐related genes in five FFPE samples (three demyelinated MS lesions and two healthy white matter controls) which were selected based on good amplification of PPIA seen in previous experiments (Table 1). The ECM‐related genes are particularly suitable in this regard, because the ECM is altered in MS lesions 23, 38 and they comprise a spectrum of genes ranging from high to low abundance. Results of FFPE samples were compared with values obtained with 13 frozen specimens (seven demyelinated MS lesions and six healthy white matter controls) (23).
Table 1.
Expression ratios MS/healthy of 84 extracellular matrix (ECM)‐related genes in formalin‐fixed and paraffin‐embedded (FFPE) tissue compared with frozen tissue. Abbreviations: ↑ = upregulated; ↓ = downregulated; ↔ = not regulated; nd = not detected; * = fold expression change not determinable; › = not detected in MS group; ‹ = not detected in healthy group; ‹› = not detected in MS and healthy group
| Genes | FFPE | Frozen | ||||
|---|---|---|---|---|---|---|
| % PPIA | % PPIA | Expression ratio | Expression ratio | |||
| Healthy | MS | |||||
| MS/healthy | MS/healthy | |||||
| Fibrillar collagens | ||||||
| COL1A1 | 0.10 | 10.62 | 104.69 | ↑ | 7.01 | ↑ |
| COL1A2 | 0.09 | 3.10 | 34.27 | ↑ | 4.47 | ↑ |
| COL3A1 | 0.16 | 1.33 | 8.43 | ↑ | 10.85 | ↑ |
| COL5A1 | 0.04 | 1.55 | 34.72 | ↑ | 16.83 | ↑ |
| COL5A2 | nd | nd | * | 6.51 | ↑ | |
| COL5A3 | 1.36 | 4.30 | 3.17 | ↑ | 6.64 | ↑ |
| Basement membrane collagens | ||||||
| COL4A1 | 0.33 | 14.89 | 45.32 | ↑ | 14.55 | ↑ |
| COL4A2 | nd | nd | * | 5.49 | ↑ | |
| COL4A3 | 0.07 | nd | * | 1.21 | ↔ | |
| COL4A4 | nd | nd | * | * | › | |
| COL4A5 | 0.94 | 2.67 | 2.84 | ↑ | 2.67 | ↑ |
| COL4A6 | 0.14 | nd | * | 2.22 | ↑ | |
| Anchoring collagen | ||||||
| COL7A1 | 1.77 | 5.45 | 3.08 | ↑ | 3.63 | ↑ |
| Nidogens | ||||||
| NID1 | 0.64 | 4.68 | 7.28 | ↑ | 3.08 | ↑ |
| NID2 | 0.38 | 6.89 | 18.04 | ↑ | 2.42 | ↑ |
| Laminins | ||||||
| LAMA1 | 0.66 | 1.37 | 2.09 | ↑ | 4.55 | ↑ |
| LAMA2 | 0.28 | 2.13 | 7.51 | ↑ | 1.63 | ↔ |
| LAMA3 | 0.29 | nd | * | 2.50 | ↑ | |
| LAMA4 | 1.48 | 4.19 | 2.84 | ↑ | 6.25 | ↑ |
| LAMA5 | 0.04 | nd | * | 23.77 | ↑ | |
| LAMB1 | 0.06 | nd | * | 8.67 | ↑ | |
| LAMB2 | 3.71 | 40.61 | 10.93 | ↑ | 9.04 | ↑ |
| LAMB3 | 0.52 | nd | * | 2.38 | ↑ | |
| LAMC1 | 0.32 | 2.68 | 8.35 | ↑ | 9.69 | ↑ |
| LAMC2 | nd | nd | * | * | ‹› | |
| Lecticans | ||||||
| AGC1 | 0.06 | nd | * | 2.25 | ↑ | |
| BCAN | 34.34 | 47.22 | 1.37 | ↔ | 2.43 | ↑ |
| CSPG2 | nd | nd | * | * | › | |
| CSPG3 | 3.63 | nd | * | 0.87 | ↔ | |
| Small leucine rich proteoglycans (SLRPs) | ||||||
| BGN | 4.21 | 13.05 | 3.10 | ↑ | 9.41 | ↑ |
| DCN | 2.52 | 31.12 | 12.34 | ↑ | 3.56 | ↑ |
| FMOD | nd | nd | * | 10.37 | ↑ | |
| LUM | nd | nd | * | 7.42 | ↑ | |
| Hyaluronan and proteoglycan link proteins (HAPLNs) | ||||||
| HAPLN1 | 0.12 | nd | * | * | › | |
| HAPLN2 | 22.28 | 16.96 | 0.76 | ↔ | 0.15 | ↓ |
| HAPLN3 | 0.28 | 9.46 | 33.66 | ↑ | 9.87 | ↑ |
| HAPLN4 | nd | nd | * | 6.71 | ↑ | |
| Heparan sulfate proteoglycan (HSPG) | ||||||
| HSPG2 | 0.71 | 11.81 | 16.69 | ↑ | 5.49 | ↑ |
| Tenascins | ||||||
| TNC | 3.98 | 5.49 | 1.38 | ↔ | 2.14 | ↑ |
| TNR | 4.55 | 3.79 | 0.83 | ↔ | 0.93 | ↔ |
| Thrombospondins | ||||||
| THBS1 | nd | 22.28 | * | 11.90 | ↑ | |
| THBS2 | 1.29 | 4.19 | 3.25 | ↑ | 2.84 | ↑ |
| THBS3 | nd | nd | * | 3.34 | ↑ | |
| THBS4 | 0.60 | nd | * | 2.71 | ↑ | |
| Fibrillins | ||||||
| FBN1 | 4.92 | 4.52 | 0.92 | ↔ | 0.92 | ↔ |
| FBN2 | 0.06 | nd | * | 1.20 | ↔ | |
| FBN3 | 0.25 | nd | * | 1.33 | ↔ | |
| Others | ||||||
| FN1 | 7.38 | 68.23 | 9.25 | ↑ | 3.36 | ↑ |
| RELN | 0.28 | nd | * | 4.16 | ↑ | |
| VTN | nd | nd | * | 2.39 | ↑ | |
| A disintegrin and metalloproteinase domains (ADAMs) | ||||||
| ADAM8 | 0.24 | nd | * | 1.82 | ↔ | |
| ADAM10 | 7.27 | 13.07 | 1.80 | ↔ | 1.52 | ↔ |
| ADAM12 | 0.26 | nd | * | 2.12 | ↑ | |
| ADAM17 | 0.22 | nd | * | 1.76 | ↔ | |
| A disintegrin‐like and metalloproteinase with thrombospondin type 1 motifs (ADAMTs) | ||||||
| ADAMTS1 | 2.29 | 21.03 | 9.19 | ↑ | 1.73 | ↔ |
| ADAMTS4 | 14.82 | 28.36 | 1.91 | ↔ | 0.22 | ↓ |
| ADAMTS5 | 0.09 | nd | * | 4.04 | ↑ | |
| Matrix metalloproteinases (MMPs) | ||||||
| MMP1 | nd | nd | * | * | ‹› | |
| MMP2 | 0.73 | 8.37 | 11.51 | ↑ | 6.41 | ↑ |
| MMP3 | nd | nd | * | * | › | |
| MMP7 | nd | nd | * | * | ‹› | |
| MMP8 | nd | nd | * | * | ‹› | |
| MMP9 | 0.06 | nd | * | 12.46 | ↑ | |
| MMP10 | nd | nd | * | * | ‹› | |
| MMP11 | nd | nd | * | 6.43 | ↑ | |
| MMP12 | nd | nd | * | * | ‹› | |
| MMP13 | nd | nd | * | * | ‹› | |
| MMP14 | 7.31 | 22.15 | 3.03 | ↑ | 11.59 | ↑ |
| MMP15 | 0.87 | 1.57 | 1.80 | ↔ | 1.93 | ↔ |
| MMP16 | 1.43 | nd | * | 2.14 | ↑ | |
| MMP17 | 0.64 | nd | * | 11.64 | ↑ | |
| MMP19 | nd | nd | * | 10.71 | ↑ | |
| MMP20 | nd | nd | * | * | ‹› | |
| MMP21 | nd | nd | * | * | › | |
| MMP23B | nd | nd | * | 2.37 | ↑ | |
| MMP24 | 0.45 | nd | * | 4.51 | ↑ | |
| MMP25 | nd | nd | * | * | › | |
| MMP26 | nd | nd | * | * | ‹› | |
| MMP27 | nd | nd | * | * | ‹› | |
| MMP28 | nd | nd | * | 3.76 | ↑ | |
| Tissue inhibitor of metalloproteinases (TIMPs) | ||||||
| TIMP1 | 1.13 | 6.00 | 5.32 | ↑ | 11.94 | ↑ |
| TIMP2 | 34.80 | 35.65 | 1.02 | ↔ | 2.30 | ↑ |
| TIMP3 | 9.48 | 191.91 | 20.24 | ↑ | 4.83 | ↑ |
| TIMP4 | 2.65 | 4.37 | 1.65 | ↔ | 3.71 | ↑ |
Data calculation: A Ct value < 35 was set as detection limit and the gene expression ratio % PPIA (MS)/%PPIA (healthy) was determined. A total of five FFPE tissue specimens (n = 3 MS, n = 2 healthy) and 13 frozen tissue specimens (n = 7 MS, n = 6 healthy) were analyzed. Expression of the ECM‐related genes in frozen tissue using glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as a housekeeping gene were published in a different context (23).
We considered a gene reliably detected if the measured Ct value was <35. Using this as a detection limit, we noted a reduced sensitivity in FFPE material. Of all 84 ECM‐related genes we were able to detect and quantify the expression of 68 of 84 genes (81%) in frozen tissue of both MS and controls (74/84 genes in the control and 68/84 genes in the MS group) and 36 genes (43%) in FFPE tissue (57/84 genes in the control group and 37/84 in the MS group). All the 36 genes detected in FFPE tissue were also detected in frozen tissue.
When evaluating the expression data of Table 1 in detail, we have to consider that in addition to the FFPE‐frozen comparison, we have a biological variability of different samples. We compared the direction of the ratios MS/healthy and this showed that the analysis of FFPE tissue can give reliable results. To compare gene regulation results (in terms of up‐, down‐ or no regulation) for the 36 genes detected in FFPE tissue, we calculated the gene expression ratio as % PPIA (MS)/%PPIA (healthy). A gene was considered as up‐ or downregulated if the gene expression ratio was >2 or <0.5, respectively. This was done separately within FFPE and the frozen tissue before results were compared. The direction of the detected gene regulation (MS/healthy) in the frozen and the FFPE group were largely similar (Table 1): in the frozen MS lesions, 28 genes were up‐ and two were downregulated. Twenty‐four of these 28 genes seen upregulated in frozen MS lesions were also classified as upregulated in FFPE MS lesions; the other four genes were also induced in the FFPE MS lesions but less than twofold. The two downregulated genes in frozen MS lesions (HAPLN2, ADAMTS4) were not regulated in FFPE MS lesions; of the six genes that were classified as not regulated in frozen tissue, four genes were not regulated in FFPE tissue and the other two genes (LAMA2, ADAMTS1) were considered as upregulated. In summary, 28 of these 36 genes were classified similarly in both, FFPE and frozen tissue (Table 1).
DISCUSSION
Here we present a detailed protocol describing how to use autoptic FFPE samples for mRNA and miRNA quantification and a quantitative comparison of transcript amplification in FFPE and frozen tissue (Supporting Information Table S3). We found that the utility of FFPE material for the analysis of protein coding genes is highly heterogeneous and that the length of tissue fixation before embedding in paraffin is a key factor in this regard. Remarkably, specimens that were poor candidates for mRNA analysis were still highly valuable for miRNA quantification. In an expression analysis of a larger set of ECM‐related genes, FFPE tissue processing including a hot water bath and subsequent dissection of MS lesion areas did not impair results. Compared with data obtained in frozen tissue, a similar pattern of gene expression was found in FFPE tissue—provided that the target genes were of sufficient abundance.
In our screening of archival MS brain samples for possible utility in coding gene expression analysis only about a third of all analyzed samples showed acceptable quality [defined as Ct (PPIA) < 30]. This is in concordance with a study in which utility of 157 archival tumor specimens for microarray analysis with respect to tumor classification was analyzed and in which only 25% of the performed arrays were informative (27).
In our autoptic FFPE brain specimens, the ability to amplify of PPIA was very heterogeneously distributed and ranged from very poor [ΔCt (CtFFPE‐Ctfrozen) > 15] in some old archival tissue specimens to adequate (ΔCt = 5) in other specimens (eg, samples 1771‐05‐1.6, 1854‐05‐7.1, RZ 23‐5, 1854‐05‐10.1 and mirror samples A, B, C in Figure 2). An average increase of Ct values by five cycles (which means a reduced sensitivity by a factor of 25 = 32) has also been observed by different studies using bioptic FFPE specimens 10, 19. The decline of retrievable RNA copies in archival tissue has been explained in depth by: (i) chemical RNA destruction/modification by formalin fixation 8, 22, 26; (ii) variable conditions during tissue fixation such as pH (15), temperature (32) and length 3, 6, 22; (iii) level of tissue penetration by the fixative (31) and period of storage in paraffin 6, 20, 34.
Among these effects, we have analyzed the effect of fixation time in more detail. Under otherwise “standardized conditions” (PFA, 4°C) we found a progressive decline in the ability to amplify coding genes in correlation with progressive time of fixation leading to around 600‐fold loss of amplifiable transcripts with tissue fixation over years (Figure 3). However, this loss in amplification is relatively moderate compared with the about 105‐fold loss of amplification in some other archival brain samples of which fixation conditions were unknown.
We further noticed that 15 of 18 of the specimens which had been fixed under these “standardized conditions” were suitable for subsequent gene expression analysis [Ct value (PPIA) < 30]. All of this suggests that utility of archival MS brain specimens depends on the overall quality of tissue preservation including length of tissue fixation.
Despite the heterogeneous utility of the FFPE samples for the analysis of mRNA expression levels, all of our archival brain specimens were found to be useful for subsequent miRNA quantification. In the present study we observed a mean Ct increase by 0.5 for FFPE samples analyzed for miR‐181a compared with frozen tissue. We have already applied this observation to establish miRNA profiles of white matter lesions of MS patients (13). Our observation, that miRNAs can readily be quantified in FFPE tissue which is considered not useful for analysis of protein coding mRNA, does extend previous work on miRNA analysis in FFPE tissue 34, 40, 42. The small size (42) and protective miRNA–protein interactions 11, 21 might explain the enormous stability of miRNAs. All of this makes archival MS samples a valuable tissue resource for further investigation of the role of miRNAs in MS, for example, their impact on cortical demyelination (35) or the potential presence of viral agents 30, 41.
Five suitable FFPE specimens were used for gene expression analysis for a larger set of ECM related genes. This revealed an expression pattern similar to 13 frozen specimens, where fibrillar collagens, decorin and biglycan were found to be associated with infiltrating immune cells (23). These genes were also found to be strongly induced in FFPE tissue along with basement membrane collegens and laminins, the induction of which was previously described using immunostaining 23, 37. However, we were not able to detect especially low abundant genes in FFPE tissue, for example, fibromodulin (FMOD), lumican (LUM), reelin (RELN) and most of the metalloproteinases (MMP9, MMP11, MMP17, MMP19, MMP24, MMP28) (Table 1).
Archival FFPE specimes are of great value for research on the pathogenesis of MS because certain lesion types are rarely encountered in biopsy specimens or in frozen material from human brain banks. The molecular analysis of some stages of MS lesion development therefore depends on FFPE tissue samples. In such precious FFPE samples, post‐mortem time, pre‐mortem conditions and duration of formalin fixation are often unknown and molecular research on such archival material is very challenging. Therefore, a simple and reliable method to pre‐screen such material for its suitability for molecular studies is urgently required. Our present data show, that such a pre‐screening can be performed by screening the ability to amplify a housekeeping gene by qPCR or in situ hybridization for an abundantly expressed mRNA, for example, PLP. In particular, a lack of signal in the in situ hybridization (“‐” in Figure 4) identifies samples that are unlikely to perform well in qPCR studies of mRNA.
In summary, the present study shows that gene expression analysis for both mRNA and miRNA can be performed with FFPE specimens of good quality, even if a bioanalyzer indicates degraded RNA. Especially miRNA expression can be quantified in virtually all FFPE specimens with sensitivity that is comparable with frozen tissue, whim amplification of mRNA is only possible in a subset of FFPE specimens. The mRNA analysis of FFPE material by qPCR is less sensitive than of frozen tissue but gives similar results for genes of sufficient abundance. The utility of archival MS tissue specimens for mRNA analysis can be evaluted by in situ hybridization with an abundant probe and by qPCR for a housekeeping gene. Furthermore, defined tissue areas can be dissected in FFPE specimens without impairment of subseqent qPCR analysis of mRNAs or miRNAs using the protocol described here.
Supporting information
Table S1. Analyzed tissue specimens.
Table S2. Extracellular matrix and extracellular matrix related genes and corresponding TaqMan® PCR assays used for TaqMan® Low Density Array (LDA) analysis (Applied Biosystems, Darmstadt, Germany).
Table S3. Protocol for using archival FFPE specimen.
Supporting info item
ACKNOWLEDGMENTS
We are grateful to Heike Rübsamen, Angelika Henn and Ulrike Köck for technical advice and support. We are especially thankful to the Center for Brain Research in Vienna, the Netherlands Brain Bank, the NeuroResource at UCL Institute of Neurology in London, and the Neurobiobank Munich for providing valuable tissue specimens. We thank Drs. S. Rauskolb and G. Krishnamoorthy for helpful comments on the article. We are grateful to Prof. H. Wekerle for continuous support.
Grants and Support: The Deutsche Forschungsgemeinschaft (SFB 571), Herrmann and Lilly Schilling Foundation, Verein zur Therapieforschung für Multiple‐Sklerose‐Kranke, the Bundesministerium für Bildung und Forschung (“Krankheitsbezogenes Kompetenznetz Multiple Sklerose”), the Excellency Initiative of the Ludwig Maximilians University Munich and the scholarship program “Förderung für Forschung und Lehre (FöFoLe)” of the Ludwig Maximilians University Munich, Germany.
REFERENCES
- 1. Abrahamsen HN, Steiniche T, Nexo E, Hamilton‐Dutoit SJ, Sorensen BS (2003) Towards quantitative mRNA analysis in paraffin‐embedded tissues using real‐time reverse transcriptase‐polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J Mol Diagn 5:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breitschopf H, Suchanek G, Gould RM, Colman DR, Lassmann H (1992) In situ hybridization with digoxigenin‐labeled probes: sensitive and reliable detection method applied to myelinating rat brain. Acta Neuropathol (Berl) 84:581–587. [DOI] [PubMed] [Google Scholar]
- 3. Bresters D, Schipper ME, Reesink HW, Boeser‐Nunnink BD, Cuypers HT (1994) The duration of fixation influences the yield of HCV cDNA‐PCR products from formalin‐fixed, paraffin‐embedded liver tissue. J Virol Methods 48:267–272. [DOI] [PubMed] [Google Scholar]
- 4. Chung JY, Hewitt SM (2010) An optimized RNA extraction method from archival formalin‐fixed paraffin‐embedded tissue. Methods Mol Biol 611:19–27. [DOI] [PubMed] [Google Scholar]
- 5. Chung JY, Braunschweig T, Hewitt SM (2006) Optimization of recovery of RNA from formalin‐fixed, paraffin‐embedded tissue. Diagn Mol Pathol 15:229–236. [DOI] [PubMed] [Google Scholar]
- 6. Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC et al (2004) Measurement of gene expression in archival paraffin‐embedded tissues: development and performance of a 92‐gene reverse transcriptase‐polymerase chain reaction assay. Am J Pathol 164:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummings TJ, Strum JC, Yoon LW, Szymanski MH, Hulette CM (2001) Recovery and expression of messenger RNA from postmortem human brain tissue. Mod Pathol 14:1157–1161. [DOI] [PubMed] [Google Scholar]
- 8. Finke J, Fritzen R, Ternes P, Lange W, Dolken G (1993) An improved strategy and a useful housekeeping gene for RNA analysis from formalin‐fixed, paraffin‐embedded tissues by PCR. Biotechniques 14:448–453. [PubMed] [Google Scholar]
- 9. Fugger L, Friese MA, Bell JI (2009) From genes to function: the next challenge to understanding multiple sclerosis. Nat Rev Immunol 9:408–417. [DOI] [PubMed] [Google Scholar]
- 10. Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH (2000) Quantitative mRNA expression analysis from formalin‐fixed, paraffin‐embedded tissues using 5′ nuclease quantitative reverse transcription‐polymerase chain reaction. J Mol Diagn 2:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T et al (2009) Robust global micro‐RNA profiling with formalin‐fixed paraffin‐embedded breast cancer tissues. Lab Invest 89:597–606. [DOI] [PubMed] [Google Scholar]
- 12. Johnson SA, Morgan DG, Finch CE (1986) Extensive postmortem stability of RNA from rat and human brain. J Neurosci Res 16:267–280. [DOI] [PubMed] [Google Scholar]
- 13. Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R et al (2009) MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132(Pt 12):3342–3352. [DOI] [PubMed] [Google Scholar]
- 14. Junker A, Hohlfeld R, Meinl E (2011) Emerging role of miRNAs in multiple sclerosis. Nat Rev Neurol 7:56–59. [DOI] [PubMed] [Google Scholar]
- 15. Kingsbury AE, Foster OJ, Nisbet AP, Cairns N, Bray L, Eve DJ et al (1995) Tissue pH as an indicator of mRNA preservation in human post‐mortem brain. Brain Res Mol Brain Res 28:311–318. [DOI] [PubMed] [Google Scholar]
- 16. Krumbholz M, Theil D, Cepok S, Hemmer B, Kivisakk P, Ransohoff RM et al (2006) Chemokines in multiple sclerosis: CXCL12 and CXCL13 up‐regulation is differentially linked to CNS immune cell recruitment. Brain 129(Pt 1):200–211. [DOI] [PubMed] [Google Scholar]
- 17. Lassmann H (2011) The architecture of inflammatory demyelinating lesions: implications for studies on pathogenesis. Neuropathol Appl Neurobiol 37:698–710. [DOI] [PubMed] [Google Scholar]
- 18. Lehmann U, Glockner S, Kleeberger W, von Wasielewski HF, Kreipe H (2000) Detection of gene amplification in archival breast cancer specimens by laser‐assisted microdissection and quantitative real‐time polymerase chain reaction. Am J Pathol 156:1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Smyth P, Cahill S, Denning K, Flavin R, Aherne S et al (2008) Improved RNA quality and TaqMan Pre‐amplification method (PreAmp) to enhance expression analysis from formalin fixed paraffin embedded (FFPE) materials. BMC Biotechnol 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lisowski AR, English ML, Opsahl AC, Bunch RT, Blomme EA (2001) Effect of the storage period of paraffin sections on the detection of mRNAs by in situ hybridization. J Histochem Cytochem 49:927–928. [DOI] [PubMed] [Google Scholar]
- 21. Lu J, Getz G, Miska EA, Alvarez‐Saavedra E, Lamb J, Peck D et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838. [DOI] [PubMed] [Google Scholar]
- 22. Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K (1999) Analysis of chemical modification of RNA from formalin‐fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27:4436–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mohan H, Krumbholz M, Sharma R, Eisele S, Junker A, Sixt M et al (2010) Extracellular matrix in multiple sclerosis lesions: fibrillar collagens, biglycan and decorin are upregulated and associated with infiltrating immune cells. Brain Pathol 20:966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real‐time RT‐PCR. Biotechniques 32:1372–1379. Erratum in: Biotechniques 2002, 33:514. [PubMed] [Google Scholar]
- 25. Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z (2004) Microarray‐based, high‐throughput gene expression profiling of microRNAs. Nat Methods 1:155–161. [DOI] [PubMed] [Google Scholar]
- 26. Park YN, Abe K, Li H, Hsuih T, Thung SN, Zhang DY (1996) Detection of hepatitis C virus RNA using ligation‐dependent polymerase chain reaction in formalin‐fixed, paraffin‐embedded liver tissues. Am J Pathol 149:1485–1491. [PMC free article] [PubMed] [Google Scholar]
- 27. Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA et al (2007) RNA expression analysis of formalin‐fixed paraffin‐embedded tumors. Lab Invest 87:383–391. [DOI] [PubMed] [Google Scholar]
- 28. R Development Core Team (2010) R Foundation for Statistical Computing,Vienna, Austria. ISBN 3‐900051‐07‐0. Available at: http://www.R‐project.org/ (accessed 6 February 2012).
- 29. Schramm M, Falkai P, Tepest R, Schneider‐Axmann T, Przkora R, Waha A et al (1999) Stability of RNA transcripts in post‐mortem psychiatric brains. J Neural Transm 106:329–335. [DOI] [PubMed] [Google Scholar]
- 30. Serafini B, Rosicarelli B, Franciotta D, Magliozzi R, Reynolds R, Cinque P et al (2007) Dysregulated Epstein‐Barr virus infection in the multiple sclerosis brain. J Exp Med 204:2899–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H (2001) Quantitative gene expression analysis in microdissected archival formalin‐fixed and paraffin‐embedded tumor tissue. Am J Pathol 158:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srinivasan M, Sedmak D, Jewell S (2002) Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 161:1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steinman L, Zamvil S (2003) Transcriptional analysis of targets in multiple sclerosis. Nat Rev Immunol 3:483–492. [DOI] [PubMed] [Google Scholar]
- 34. Szafranska AE, Davison TS, Shingara J, Doleshal M, Riggenbach JA, Morrison CD et al (2008) Accurate molecular characterization of formalin‐fixed, paraffin‐embedded tissues by microRNA expression profiling. J Mol Diagn 10:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torkildsen O, Stansberg C, Angelskar SM, Kooi EJ, Geurts JJ, Myhr KM et al (2010) Upregulation of immunoglobulin‐related genes in cortical sections from multiple sclerosis patients. Brain Pathol 20:720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapp BD, Nave KA (2008) Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci 31:247–269. [DOI] [PubMed] [Google Scholar]
- 37. van Horssen J, Bo L, Vos CM, Virtanen I, de Vries HE (2005) Basement membrane proteins in multiple sclerosis‐associated inflammatory cuffs: potential role in influx and transport of leukocytes. J Neuropathol Exp Neurol 64:722–729. [DOI] [PubMed] [Google Scholar]
- 38. van Horssen J, Dijkstra CD, de Vries HE (2007) The extracellular matrix in multiple sclerosis pathology. J Neurochem 103:1293–1301. [DOI] [PubMed] [Google Scholar]
- 39. Waddell N, Cocciardi S, Johnson J, Healey S, Marsh A, Riley J et al (2010) Gene expression profiling of formalin‐fixed, paraffin‐embedded familial breast tumours using the whole genome‐DASL assay. J Pathol 221:452–461. [DOI] [PubMed] [Google Scholar]
- 40. Weng L, Wu X, Gao H, Mu B, Li X, Wang JH et al (2010) MicroRNA profiling of clear cell renal cell carcinoma by whole‐genome small RNA deep sequencing of paired frozen and formalin‐fixed, paraffin‐embedded tissue specimens. J Pathol 222:41–51. [DOI] [PubMed] [Google Scholar]
- 41. Willis SN, Stadelmann C, Rodig SJ, Caron T, Gattenloehner S, Mallozzi SS et al (2009) Epstein‐Barr virus infection is not a characteristic feature of multiple sclerosis brain. Brain 132:3318–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J (2007) Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin‐fixed paraffin‐embedded samples. RNA 13:1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yasojima K, McGeer EG, McGeer PL (2001) High stability of mRNAs postmortem and protocols for their assessment by RT‐PCR. Brain Res Brain Res Protoc 8:212–218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Analyzed tissue specimens.
Table S2. Extracellular matrix and extracellular matrix related genes and corresponding TaqMan® PCR assays used for TaqMan® Low Density Array (LDA) analysis (Applied Biosystems, Darmstadt, Germany).
Table S3. Protocol for using archival FFPE specimen.
Supporting info item


