Abstract
Gliosarcoma is a rare variant of glioblastoma characterized by a biphasic pattern of glial and mesenchymal differentiation. It is unclear whether mesenchymal differentiation in gliosarcomas is because of extensive genomic instability and/or to a mechanism similar to epithelial–mesenchymal transition (EMT). In the present study, we assessed 40 gliosarcomas for immunoreactivity of Slug, Twist, matrix metalloproteinase‐2 (MMP‐2) and matrix metalloproteinase‐9 (MMP‐9), which are involved in EMT in epithelial tumors. Nuclear Slug expression was observed in >50% of neoplastic cells in mesenchymal tumor areas of 33 (83%) gliosarcomas, but not in glial areas (P < 0.0001). Nuclear Twist expression was observed in >50% of neoplastic cells in mesenchymal tumor areas of 35 (88%) gliosarcomas, but glial tumor areas were largely negative except in four cases (P < 0.0001). Expression of MMP‐2 and MMP‐9 was also significantly more extensive in mesenchymal than in glial tumor areas. None of 20 ordinary glioblastomas showed Slug or Twist expression in >10% neoplastic cells. Thus, expression of Slug, Twist, MMP‐2 and MMP‐9 was characteristic of mesenchymal tumor areas of gliosarcomas, suggesting that mechanisms involved in the EMT in epithelial neoplasms may play roles in mesenchymal differentiation in gliosarcomas.
Keywords: epithelial to mesenchymal transition, gliosarcoma, MMP, Slug, Twist
INTRODUCTION
The epithelial–mesenchymal transition (EMT) is the conversion of cells with an epithelial phenotype into cells with a highly motile fibroblastoid or mesenchymal phenotype 23, 33. EMT is a critical mechanism in embryonic development, chronic inflammation and fibrosis 10, 33, 36, and is considered to play a key role in tumor progression 6, 25, 41 and metastasis 38, 42. Several transcription factors, including Slug, Snai1, Twist, matrix metalloproteinases (MMPs), have been reported to be involved in EMT 11, 26, 27, 33.
Gliosarcoma is a variant of glioblastoma that is characterized by biphasic neoplastic tissue composed of glial and mesenchymal components (14). The mesenchymal component shows long bundles of spindle cells with malignant features, and abundant deposition of collagen. Despite the presence of these two distinct histological components, gliosarcomas are considered to be monoclonal tumors, as genetic analyses have shown that glial and mesenchymal tumor areas are usually genetically identical in terms of TP53 mutations, PTEN mutations, p16INK4a deletion, CDK4 amplification and MDM2 amplification 2, 28.
It is unclear whether mesenchymal differentiation in gliosarcoma simply reflects the extensive genomic instability of glioblastomas and/or whether mesenchymal differentiation is caused by a mechanism similar to that involved in EMT in epithelial neoplasms 16, 33. Yang et al (37) showed that Slug expression facilitates cellular invasion and angiogenesis as well as increasing spindle morphology in a mouse intracranial human glioma. Mikheeva et al (19) revealed that Twist1 promotes invasion through mesenchymal changes in glioblastoma.
In the present study, we carried out immunohistochemistry for Slug, Twist, matrix metalloproteinase‐2 (MMP‐2) and matrix metalloproteinase‐9 (MMP‐9) in 40 gliosarcomas, to assess whether there is evidence of expression of genes associated with EMT, suggesting that mechanisms similar to EMT may be operative in gliosarcomas.
MATERIALS AND METHODS
Tumor samples
Formalin‐fixed tissue samples of 40 gliosarcomas and 20 primary (de novo) glioblastomas were obtained from the Department of Neuropathology, University Hospital Zürich, Switzerland; the Institute of Neuropathology, University Hospital Munster, Germany; the Institute of Neuroscience, Bordeaux, France; the Department of Neuropathology, University Hospital Rome, Italy; and the Department of Pathology, Gunma University, Japan. Gliosarcomas from patients with recurrent disease were not included in this study. The median age at histological diagnosis was 59 ± 11 years (range, 32–82 years) and the male/female ratio was 1.22. Survival data were available for 16 patients. Mean follow‐up time was 10 months, and 2 out of 16 (13%) patients were still alive at the time of the study. Mean overall survival among the 14 patients who died was 8.5 ± 7.5 months (range, 3–29.7 months).
Sections were routinely stained with hematoxylin and eosin (HE), glial fibrillary acidic protein (GFAP) and reticulin. The glial areas with astrocytic features showed GFAP expression without reticulin staining. The mesenchymal areas showed typical reticulin expression in bundles of neoplastic spindle cells.
Immunohistochemistry
Immunohistochemistry was carried out using the Ultravision Quanto detection system (Thermo Scientific, Fremont, CA, USA) or Vectorstain ABC kit (Vector Laboratories, Burlingame, CA, USA). Briefly, paraffin sections were deparaffinized in xylene and rehydrated through a graded series of ethanol dilutions. Endogenous peroxidase activity was blocked by incubation in 0.3% hydrogen peroxidase for 30 minutes. After pretreatments performed according to manufacturer's instructions, sections were incubated at room temperature for 1 h with primary antibody, and visualized with diaminobenzidine. Antibodies were as follows: rabbit monoclonal to Slug (9585, Cell Signalling, Boston, MA, USA; dilution, 1:100), mouse monoclonal to Twist (ab50887, Abcam, Cambridge, UK; dilution, 1:250), mouse monoclonal to MMP‐2 (MS804, ThermoFisher Scientific, Cheshire, UK; dilution, 1:100) and rabbit polyclonal to MMP‐9 (RB9234, ThermoFisher Scientific; dilution, 1:500).
Immunoreactivity to each antibody was evaluated in glial and mesenchymal tumor areas separately. Staining was defined as nuclear (Slug, Twist) or cytoplasmic (MMP‐2, MMP‐9) immunoreactivity in neoplastic cells, and was scored as: −, <10% of positive cells; +, 10%–50%; ++, 51%–90%; and +++, >90%. The number of neoplastic cells with immunoreactivity for Slug, Twist, MMP‐2 or MMP‐9 was counted in 15–20 fields per tumor area by light microscopy at ×400 magnification. For Twist, vascular epithelial cells on the same histological slide were used as internal positive controls. Neoplastic cells with expression of Twist that was similar or higher than that in vascular epithelial cells were considered to be positive for Twist.
To assess co‐localization of Slug and GFAP, or Twist and GFAP, in tumor cells, double immunohistochemical staining was performed with antibodies to GFAP and Slug, or GFAP and Twist, in four gliosarcomas. Sections were first immunostained for Slug or Twist. The epitopes were then visualized with diaminobenzidine (brown staining). After completion of the first staining, sections were treated in Linblock (AbCys, Paris, France) for a total of 4 minutes at room temperature to remove the Slug or Twist antibody complex. Sections were then incubated with antibodies to GFAP overnight at 4°C and visualized with Vector VIP (purple staining; Vector Laboratories). After washing sections with phosphate‐buffered saline, a slight green counterstaining of the nuclei was performed for 5 minutes in a 0.5% solution of methyl green (Gurr, Bucks, UK).
Statistical analyses
Chi‐squared test or Fisher's exact test were used to assess the group differences analysis of qualitative features, and a Kaplan–Meier curve with log‐rank test was used for survival analysis. For comparative analysis of immunohistochemical expression in all protein groups (Slug, Twist, MMP2 and MMP9), data were analyzed using post hoc Bonferroni–Dunn tests to compare individual protein groups. We declared statistical significance at the traditional threshold of P = 0.05. However, when multiple comparisons were made, we also included Bonferroni‐corrected P‐values.
RESULTS
Immunohistochemistry to Slug, TwisT, MMP‐2 and MMP‐9
Nuclear Slug expression was observed in >50% neoplastic cells in mesenchymal tumor areas of 33 (83%) gliosarcomas (Table 1; Figure 1). In contrast, neoplastic cells in glial tumor areas were largely negative (P < 0.0001).
Table 1.
Fraction of immunoreactive cells to moleculaes associated with EMT in glial and mesenchymal tumor areas of gliosarcomas
| Slug | Twist | MMP‐2 | MMP‐9 | |||||
|---|---|---|---|---|---|---|---|---|
| Glial | Mesenchymal | Glial | Mesenchymal | Glial | Mesenchymal | Glial | Mesenchymal | |
| Gliosarcomas | ||||||||
| GS1 | − | + | − | + | − | − | − | − |
| GS2 | − | ++ | − | ++ | − | + | − | + |
| GS3 | − | +++ | − | + | − | ++ | − | − |
| GS4 | − | + | − | ++ | − | + | − | − |
| GS5 | − | + | − | ++ | + | + | − | + |
| GS6 | − | +++ | + | +++ | + | ++ | − | +++ |
| GS7 | − | ++ | − | +++ | − | + | + | +++ |
| GS8 | + | +++ | − | +++ | + | + | + | + |
| GS9 | − | ++ | − | +++ | − | + | − | + |
| GS10 | − | − | − | +++ | − | ++ | + | ++ |
| GS11 | − | +++ | ++ | +++ | − | +++ | +++ | +++ |
| GS12 | − | + | − | +++ | − | ++ | ++ | ++ |
| GS13 | − | +++ | ++ | +++ | − | + | + | +++ |
| GS14 | − | +++ | − | +++ | + | +++ | − | +++ |
| GS15 | − | ++ | − | +++ | + | + | − | ++ |
| GS16 | − | +++ | − | +++ | − | ++ | − | +++ |
| GS17 | − | +++ | − | +++ | − | ++ | − | +++ |
| GS18 | − | +++ | − | + | + | +++ | + | ++ |
| GS19 | − | ++ | − | +++ | + | + | + | ++ |
| GS20 | − | +++ | − | +++ | − | +++ | − | +++ |
| GS21 | − | +++ | − | +++ | + | ++ | + | ++ |
| GS22 | − | +++ | + | +++ | − | ++ | − | ++ |
| GS23 | − | ++ | − | + | − | ++ | + | ++ |
| GS24 | − | ++ | − | +++ | + | ++ | ++ | ++ |
| GS25 | − | ++ | − | +++ | − | ++ | − | ++ |
| GS26 | − | +++ | − | +++ | + | + | + | +++ |
| GS27 | − | +++ | − | +++ | + | + | + | + |
| GS28 | − | +++ | − | +++ | − | ++ | − | +++ |
| GS29 | − | + | − | +++ | + | + | ++ | +++ |
| GS30 | − | ++ | − | +++ | − | ++ | + | ++ |
| GS31 | − | +++ | − | +++ | + | + | + | ++ |
| GS32 | − | +++ | − | +++ | + | +++ | − | + |
| GS33 | − | +++ | − | +++ | − | ++ | ++ | ++ |
| GS34 | − | +++ | − | ++ | − | ++ | ++ | +++ |
| GS35 | − | +++ | − | +++ | + | ++ | + | ++ |
| GS36 | − | ++ | − | +++ | − | ++ | + | ++ |
| GS37 | − | ++ | − | + | + | ++ | + | + |
| GS38 | − | + | − | +++ | − | ++ | − | +++ |
| GS39 | − | +++ | − | +++ | − | ++ | − | ++ |
| GS40 | − | ++ | − | +++ | − | ++ | − | + |
| Glioblastomas | ||||||||
| G1 | − | − | − | − | ||||
| G2 | − | − | − | − | ||||
| G3 | − | − | + | + | ||||
| G4 | − | − | + | + | ||||
| G5 | − | − | − | − | ||||
| G6 | − | − | + | + | ||||
| G7 | − | − | − | − | ||||
| G8 | − | − | − | − | ||||
| G9 | − | − | − | − | ||||
| G10 | − | − | + | − | ||||
| G11 | − | − | + | − | ||||
| G12 | − | − | − | − | ||||
| G13 | − | − | + | + | ||||
| G14 | − | − | + | − | ||||
| G15 | − | − | + | − | ||||
| G16 | − | − | ++ | + | ||||
| G17 | − | − | − | − | ||||
| G18 | − | − | − | − | ||||
| G19 | − | − | − | − | ||||
| G20 | − | − | + | − | ||||
The number of Slug‐, Twist‐, MMP‐2‐ and MMP‐9‐positive cells in immunohistochemistry was estimated using a four‐tiered scale; −, <10%; +, 10%–50%; ++, 51%–90%; +++, >90%.
Figure 1.
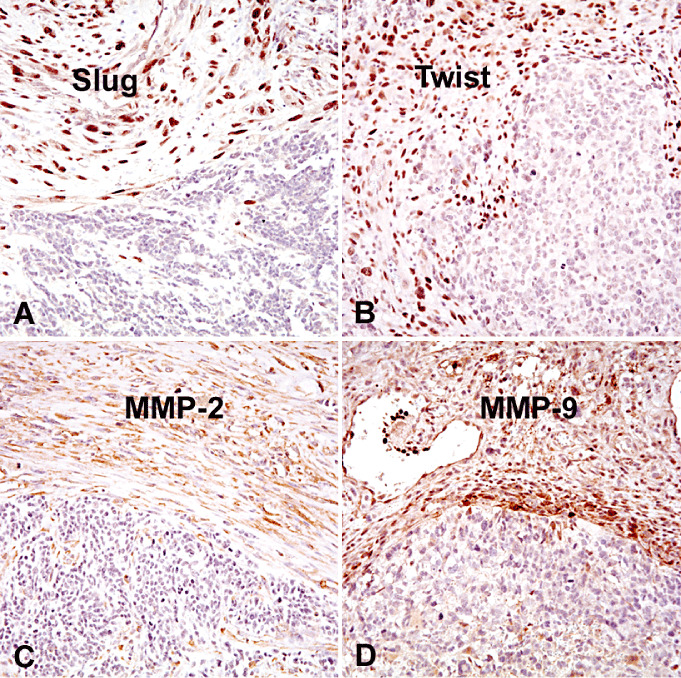
A border between mesenchymal and glial tumor areas of gliosarcomas. Note that Slug (A), Twist (B), MMP‐2 (C) and MMP‐9 (D) are selectively expressed in mesenchymal tumor areas of gliosarcomas.
Nuclear Twist expression was observed in >50% of neoplastic cells in mesenchymal tumor areas of 35 (88%) gliosarcomas (Table 1). In contrast, neoplastic cells in glial tumor areas were largely negative, except for four cases (P < 0.0001).
Cytoplasmic MMP‐2 expression was detected in >50% neoplastic cells in mesenchymal tumor areas of 26 (65%) gliosarcomas, but not in glial areas of any gliosarcoma (Table 1) (P < 0.0001). Cytoplasmic MMP‐9 expression was seen in >50% of neoplastic cells in mesenchymal tumor areas of 29 (73%) gliosarcomas, but in glial tumor areas of only six cases (15%, P < 0.0001) (Table 1).
In gliosarcomas, Bonferroni's correction revealed significant correlations between expression of Twist and MMP‐9 (P = 0.0002), Slug and MMP‐2 (P = 0.0005), and MMP‐2 and Twist expression (P < 0.0001), but there was no significant correlation between MMP2 and MMP9 (P = 0.16), MMP9 and Slug (P = 0.035), or Slug and Twist (P = 0.09).
We correlated survival of patients with gliosarcoma and the fraction of mesenchymal cells expressing Slug, Twist, MMP‐2 or MMP‐9 (>90%, 10%–90% or <10%). There was no significant difference in survival among gliosarcoma patients whose tumors showed different fractions of Slug, Twist, MMP‐2 and MMP‐9 immunoreactivity (data not shown).
None of the 20 ordinary glioblastomas showed Slug or Twist expression in >10% neoplastic cells (Table 1). The fraction of cells expressing MMP‐2/MMP‐9 in ordinary glioblastomas was similar to those in glial tumor areas of gliosarcomas (Table 1).
Double staining of GFAP and Slug, or GFAP and TWIST
In the majority (36 cases; 90%) of gliosarcomas, the mesenchymal components typically lacked GFAP expression. In four cases, focal mesenchymal tumor areas with co‐expression of GFAP and reticulin were detected (Figure 2); double staining demonstrated co‐expression of GFAP and Slug in 2 of these cases and co‐expression of GFAP and Twist in three cases (Figure 2).
Figure 2.
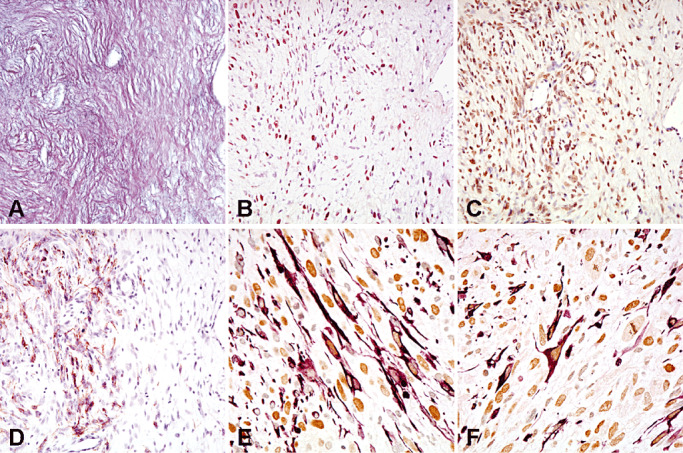
Consecutive sections of the mesenchymal tumor area of a gliosarcoma co‐expressing reticulin (A), Slug (B) and Twist (C). Note GFAP expression (D) in a focal area. Double stainings show co‐expression of GFAP (purple, cytoplasms) and Slug (brown nuclei) (E), or GFAP (purple, cytoplasms) and Twist (brown nuclei) (F) at the single‐cell level.
Analyses of TCGA data
We assessed mRNA expression of Slug, Twist, MMP‐2 and MMP‐9 in glioblastomas in the TCGA database (http://cancergenome.nih.gov). Z scores of > +2 were considered as overexpression. A small fraction of glioblastomas (4%) showed Slug (Snai2) overexpression, and this was significantly associated with poorer outcome of glioblastoma patients (mean survival, 11.6 vs. 14.3 months; P = 0.032). Twist1 overexpression was found in 9% of glioblastomas; this was not prognostic for patient survival. Overexpression of MMP‐2 and MMP‐9 was detected in 3.6% and 1.4% of glioblastomas, respectively.
We correlated mRNA expression of Slug, Twist, MMP‐2 and MMP‐9 in glioblastomas with their transcriptional expression profiles, that is, proneural, neural, classical, mesenchymal subgroups, reported by Verhaak et al (35). The mesenchymal subgroup was characterized by gene expression typical of mesenchymal tumors, but molecules involved in EMT, such as Slug, Twist, MMP‐2 or MMP‐9, were not used to define the mesenchymal subgroup (35). Slug (Snai2) and Twist1 were significantly overexpressed in mesenchymal subgroups compared with the other subgroups (proneural, neural, classical) (5/56, 9%, P = 0.007 for Slug; and 13/56, 23%, P = 0.0001 for Twist). Overexpression of MMP‐9 was found in 2/138 cases, and these two positive cases showed a mesenchymal expression profile. Overexpression of MMP‐2 was found in 3 of 56 cases in the mesenchymal subgroup, and 2 of 54 in the classical subgroup, but in no tumors in the neural (29 cases) or proneural (56 cases) subgroups.
DISCUSSION
We show that expression of Slug, Twist, MMP‐2, MMP‐9 is characteristic of mesenchymal tumor areas of gliosarcomas, suggesting that similar mechanisms involved in the EMT in epithelial neoplasms play roles in mesenchymal differentiation in gliosarcoma.
In gliosarcoma, GFAP expression in glial cells and reticulin staining in mesenchymal‐shaped cells are typically mutually exclusive (14). However, we may encounter foci of GFAP‐expressing fibroblastic‐like neoplastic cells in mesenchymal tumor areas in gliosarcomas. In the present study, four gliosarcomas with such foci of GFAP‐expressing cells in mesenchymal tumor areas showed co‐expression of GFAP/Slug and GFAP/Twist (Figure 2). These foci may represent the initial step in glial–mesenchymal transition in gliosarcoma.
Slug was first identified in the neural crest and in mesodermal cells emigrating from the primitive streak in chick embryos and is expressed by cells undergoing EMT during embryonic development (20). Overexpression of Slug has been reported in a variety of neoplasms, including gastric cancer (29), ovarian cancer 4, 39, breast cancer 4, 17, bladder cancer (40) and lung cancer (8). Yang et al (37) subcutaneously injected U251 glioblastoma cells overexpressing Slug using Slug lentivirus, into nude mice, and found that glioblastomas overexpressing Slug displayed spindle‐like morphology, suggesting mesenchymal differentiation (37).
Twist is a helix‐loop‐helix transcription factor and plays a central role in determining cell fate in mesoderm 12, 43. Overexpression of Twist has been observed in a variety of tumors, including gastric cancer (29), lung cancer (8), breast cancer (17), bladder cancer (40), ovarian cancer (39) and choroid plexus papillomas (7). In SNB19 glioblastoma cells, overexpression of Twist1 produced mesenchymal cellular changes (19). Furthermore, Twist1 significantly increased invasion in orthotopic xenotransplants and increased expression of genes that are involved in mesenchymal phenotypes in SNB19 and T98G glioblastoma cells (19).
MMPs are proteolytic enzymes that play key roles in tumor invasion, metastasis and angiogenesis (3). MMP‐2 5, 26, 34 and MMP‐9 26, 32, 34 are often up‐regulated during EMT. MMP‐2 and MMP‐9 up‐regulation has been associated with poor prognosis in various tumors 13, 18.
In the present study, nuclear Slug expression was observed in >50% of neoplastic cells in mesenchymal tumor areas of 33 (83%) gliosarcomas, but not in glial areas. Nuclear Twist expression was observed in >50% of neoplastic cells in mesenchymal tumor areas of 35 (88%) gliosarcomas, but glial tumor areas were largely negative except in four cases. None of the 20 ordinary glioblastomas showed Slug or Twist expression in >10% neoplastic cells. Similarly, the fractions of neoplastic cells expressing MMP‐2 and MMP‐9 in ordinary glioblastomas were similar to those in the glial tumor areas of gliosarcomas, that is, much less extensive than in mesenchymal tumor areas of gliosarcomas. Consistent with this finding, TCGA data show that overexpression of Slug, Twist, MMP‐2 and MMP‐9 in glioblastomas is rare (1.4%–9% of all cases).
The other widely used markers for EMT in epithelial tumors appear to be loss of E‐cadherin and up‐regulation of N‐cadherin (23). We carried out E‐cadherin immunohistochemistry to assess whether there are differences in E‐cadherin expression between glial and mesenchymal tumor areas of gliosarcomas (data not shown). However, strong E‐cadherin expression was not detectable in any of the glioblastomas or gliosarcomas analyzed, and therefore it was not possible to reliably assess whether there was down‐regulation of E‐cadherin in mesenchymal tumor areas of gliosarcomas. It has been also reported that, distinct from EMT in epithelial neoplasms, TWIST1 did not generate an E‐ to N‐cadherin “switch” in glioblastoma cells (19).
Several studies have shown that glioblastomas may be classified according to their expression profiles 24, 35. Verhaak et al (35) classified 204 glioblastomas from the TCGA project into four subgroups, that is, mesenchymal, proneural, neural and classical; the mesenchymal subgroup, for example, was characterized by expression of genes typical to mesenchymal tumors. However, Slug, Twist, MMP‐2 or MMP‐9 were not included among the genes defining mesenchymal subgroup of glioblastomas (35). Our analyses using the dataset from Verhaak et al (35) showed that Slug and Twist were significantly overexpressed in mesenchymal subgroups in glioblastoma compared with the other subgroups (proneural, neural, classical), and that MMP‐2 and MMP‐9 were also selectively expressed in glioblastomas with a mesenchymal phenotype.
There is evidence that EMT is associated with tumor metastasis in human cancer 9, 38, 42. Extracranial metastasis of glioblastomas is extremely rare (31), although it may occur in cases of secondary glioblastoma with longer survival (15). Beaumont et al reported extracranial metastasis in 18 of 162 (11%) gliosarcomas (1), which was more frequent than in ordinary glioblastomas (0.4%–0.5%) 22, 31. Sites of metastasis of gliosarcoma were the lung, liver and lymph nodes (1). Interestingly, metastatic foci from gliosarcomas were typically composed of tumor cells showing mesenchymal differentiation, but not of glial cells 1, 21, 30.
In summary, we show that mesenchymal differentiation in gliosarcoma is associated with up‐regulation of several transcriptional factors known to be involved in EMT. Our results suggest that mesenchymal differentiation in gliosarcomas is at least partly attributable to mechanisms similar to EMT, and gliosarcoma may be a useful model to study glial–mesenchymal transition in glioblastoma.
ACKNOWLEDGMENTS
The authors thank Mrs. Christine Carreira for her technical assistance.
REFERENCES
- 1. Beaumont TL, Kupsky WJ, Barger GR, Sloan AE (2007) Gliosarcoma with multiple extracranial metastases: case report and review of the literature. J Neurooncol 83:39–46. [DOI] [PubMed] [Google Scholar]
- 2. Biernat W, Aguzzi A, Sure U, Grant JW, Kleihues P, Hegi ME (1995) Identical mutations of the p53 tumor suppressor gene in the gliomatous and the sarcomatous components of gliosarcomas suggest a common origin from glial cells. J Neuropathol Exp Neurol 54:651–656. [DOI] [PubMed] [Google Scholar]
- 3. Egeblad M, Werb Z (2002) New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2:161–174. [DOI] [PubMed] [Google Scholar]
- 4. Elloul S, Elstrand MB, Nesland JM, Trope CG, Kvalheim G, Goldberg I et al (2005) Snail, Slug, and Smad‐interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer 103:1631–1643. [DOI] [PubMed] [Google Scholar]
- 5. Gordon KJ, Kirkbride KC, How T, Blobe GC (2009) Bone morphogenetic proteins induce pancreatic cancer cell invasiveness through a Smad1‐dependent mechanism that involves matrix metalloproteinase‐2. Carcinogenesis 30:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grunert S, Jechlinger M, Beug H (2003) Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol 4:657–665. [DOI] [PubMed] [Google Scholar]
- 7. Hasselblatt M, Mertsch S, Koos B, Riesmeier B, Stegemann H, Jeibmann A et al (2009) TWIST‐1 is overexpressed in neoplastic choroid plexus epithelial cells and promotes proliferation and invasion. Cancer Res 69:2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS, Wu KJ (2009) Prognostic significance of hypoxia‐inducible factor‐1alpha, TWIST1 and Snail expression in resectable non‐small cell lung cancer. Thorax 64:1082–1089. [DOI] [PubMed] [Google Scholar]
- 9. Ishii K, Shimoda M, Sugiura T, Seki K, Takahashi M, Abe M et al (2011) Involvement of epithelial‐mesenchymal transition in adenoid cystic carcinoma metastasis. Int J Oncol 38:921–931. [DOI] [PubMed] [Google Scholar]
- 10. Kalluri R, Neilson EG (2003) Epithelial‐mesenchymal transition and its implications for fibrosis. J Clin Invest 112:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang Y, Massague J (2004) Epithelial‐mesenchymal transitions: twist in development and metastasis. Cell 118:277–279. [DOI] [PubMed] [Google Scholar]
- 12. Li L, Cserjesi P, Olson EN (1995) Dermo‐1: a novel twist‐related bHLH protein expressed in the developing dermis. Dev Biol 172:280–292. [DOI] [PubMed] [Google Scholar]
- 13. Lin CY, Tsai PH, Kandaswami CC, Lee PP, Huang CJ, Hwang JJ, Lee MT (2011) Matrix metalloproteinase‐9 cooperates with transcription factor Snail to induce epithelial‐mesenchymal transition. Cancer Sci 102:815–827. [DOI] [PubMed] [Google Scholar]
- 14. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (eds) (2007) WHO Classification of Tumours of the Central Nervous System. IARC: Lyon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lun M, Lok E, Gautam S, Wu E, Wong ET (2011) The natural history of extracranial metastasis from glioblastoma multiforme. J Neurooncol 105:261–273. [DOI] [PubMed] [Google Scholar]
- 16. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al (2008) The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin TA, Goyal A, Watkins G, Jiang WG (2005) Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol 12:488–496. [DOI] [PubMed] [Google Scholar]
- 18. Mikami S, Katsube KI, Oya M, Ishida M, Kosaka T, Mizuno R et al (2011) Expression of Snail and Slug in renal cell carcinoma: E‐cadherin repressor Snail is associated with cancer invasion and prognosis. Lab Invest 91:1443–1458. [DOI] [PubMed] [Google Scholar]
- 19. Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L et al (2010) TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer 9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nieto MA, Sargent MG, Wilkinson DG, Cooke J (1994) Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science 264:835–839. [DOI] [PubMed] [Google Scholar]
- 21. Ojeda VJ, Sterrett GF (1984) Cerebral gliosarcoma, pulmonary adenoid‐cystic carcinoma, and pulmonary metastatic gliosarcoma: report of an untreated case. Pathology 16:217–221. [DOI] [PubMed] [Google Scholar]
- 22. Pasquier B, Pasquier D, Golet AN, Panh MH, Couderc P (1980) Extraneural metastases of astrocytomas and glioblastomas: clinicopathological study of two cases and review of the literature. Cancer 45:112–125. [DOI] [PubMed] [Google Scholar]
- 23. Peinado H, Portillo F, Cano A (2004) Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol 48:365–375. [DOI] [PubMed] [Google Scholar]
- 24. Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al (2006) Molecular subclasses of high‐grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173. [DOI] [PubMed] [Google Scholar]
- 25. Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9:265–273. [DOI] [PubMed] [Google Scholar]
- 26. Qiao B, Johnson NW, Gao J (2010) Epithelial‐mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor‐beta1 is Snail family‐dependent and correlates with matrix metalloproteinase‐2 and ‐9 expressions. Int J Oncol 37:663–668. [DOI] [PubMed] [Google Scholar]
- 27. Radisky ES, Radisky DC (2010) Matrix metalloproteinase‐induced epithelial‐mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia 15:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reis RM, Konu‐Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H (2000) Genetic profile of gliosarcomas. Am J Pathol 156:425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R et al (2002) Differential expression of the epithelial‐mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol 161:1881–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith DR, Hardman JM, Earle KM (1969) Contiguous glioblastoma multiforme and fibrosarcoma with extracranial metastasis. Cancer 24:270–276. [DOI] [PubMed] [Google Scholar]
- 31. Smith DR, Hardman JM, Earle KM (1969) Metastasizing neuroectodermal tumors of the central nervous system. J Neurosurg 31:50–58. [DOI] [PubMed] [Google Scholar]
- 32. Tan TK, Zheng G, Hsu TT, Wang Y, Lee VW, Tian X et al (2010) Macrophage matrix metalloproteinase‐9 mediates epithelial‐mesenchymal transition in vitro in murine renal tubular cells. Am J Pathol 176:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thiery JP (2002) Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer 2:442–454. [DOI] [PubMed] [Google Scholar]
- 34. Ullmann U, Gilles C, De RM, Van V, Sermon K, Liebaers I (2008) GSK‐3‐specific inhibitor‐supplemented hESC medium prevents the epithelial‐mesenchymal transition process and the up‐regulation of matrix metalloproteinases in hESCs cultured in feeder‐free conditions. Mol Hum Reprod 14:169–179. [DOI] [PubMed] [Google Scholar]
- 35. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallerand H, Cai Y, Wainberg ZA, Garraway I, Lascombe I, Nicolle G et al (2010) Phospho‐Akt pathway activation and inhibition depends on N‐cadherin or phospho‐EGFR expression in invasive human bladder cancer cell lines. Urol Oncol 28:180–188. [DOI] [PubMed] [Google Scholar]
- 37. Yang HW, Menon LG, Black PM, Carroll RS, Johnson MD (2010) SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer 10:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939. [DOI] [PubMed] [Google Scholar]
- 39. Yoshida J, Horiuchi A, Kikuchi N, Hayashi A, Osada R, Ohira S et al (2009) Changes in the expression of E‐cadherin repressors, Snail, Slug, SIP1, and Twist, in the development and progression of ovarian carcinoma: the important role of Snail in ovarian tumorigenesis and progression. Med Mol Morphol 42:82–91. [DOI] [PubMed] [Google Scholar]
- 40. Yu Q, Zhang K, Wang X, Liu X, Zhang Z (2010) Expression of transcription factors snail, slug, and twist in human bladder carcinoma. J Exp Clin Cancer Res 29:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeisberg M, Neilson EG (2009) Biomarkers for epithelial‐mesenchymal transitions. J Clin Invest 119:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S, Wang X, Osunkoya AO, Iqbal S, Wang Y, Chen Z et al (2011) EPLIN downregulation promotes epithelial‐mesenchymal transition in prostate cancer cells and correlates with clinical lymph node metastasis. Oncogene 30:4941–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuniga A, Quillet R, Perrin‐Schmitt F, Zeller R (2002) Mouse Twist is required for fibroblast growth factor‐mediated epithelial‐mesenchymal signalling and cell survival during limb morphogenesis. Mech Dev 114:51–59. [DOI] [PubMed] [Google Scholar]


