Abstract
Isocitrate dehydrogenase (IDH) genes are mutated in a significant portion of gliomas, myeloid leukemias and chondroid neoplasms. In gliomas, IDH mutations are prognostic, as those tumors with the mutation are associated with a proneural subclass and have longer survival compared with those without the mutation. We developed a simple, PCR‐based SNaPshot® assay (Life Technologies, Carlsbad, CA, USA) to detect IDH1/2 mutations. This protocol combines a single, multiplexed PCR reaction using gene specific primers followed by a single, multiplexed SNaPshot reaction and detection by capillary electrophoresis. In a blinded study of 32 paraffin‐embedded glioma specimens previously screened for IDH mutations by a PCR/direct sequencing method, concordance of our IDH SNaPshot test with sequencing was 100%. We performed the assay on an additional 57 specimens submitted for diagnostic IDH mutation evaluation. Data analysis was much faster and easier to perform than analysis of the sequencing data, and results could be obtained in 1 day from DNA extraction to analysis. Furthermore, we could readily identify a mixture of 5% mutant allele vs. 95% wild‐type allele in our SNaPshot assay, in comparison to approximately 20% mutant allele in our PCR‐sequencing assay. Our assay represents a fast, sensitive, straightforward method of reliably detecting common mutations of IDH genes in glial neoplasms, or other tumors.
Keywords: diagnostic, IDH1, IDH2, mutation detection, SNaPshot®
INTRODUCTION
Abnormal carbohydrate metabolism in cancer was well described by Warburg (23) in the 1920s, although the mechanism and implications of altered cancer metabolism have been a focus of study since. Mutations of two genes encoding enzymes involved in the tricarboxylic acid (TCA) cycle have been previously identified in cancer—succinate dehydrogenase (5) and fumarate hydratase (1). Subsequently, mutations of two of the three isocitrate dehydrogenase (IDH1 and IDH2) genes were found—initially in carcinomas (21), then in a high proportion of gliomas (19), acute myelogenous leukemias (17) and, recently, chondroid tumors (2).
The isocitrate dehydrogenase enzymes are normally responsible for the metabolism of isocitrate to 2‐oxoglutarate (α‐ketoglutarate) in the TCA cycle. Because of the large free energy favoring this conversion from the reduction of NADPH, the reaction is usually nonreversible, although it does occur in cells, especially under hypoxic conditions, to drive α‐ketoglutarate to acetyl‐CoA and lipid biosynthetic pathways 18, 25. Mutant IDH, however, is better able to catalyze the reversible reaction of NADP+ to NADPH than normal IDH, hence can generate a novel metabolite R(‐)‐2‐hydroxyglutarate (2‐HG), although losing the ability to generate α‐ketoglutarate, reviewed in (9). Interestingly, mutations of IDH 1 and 2 are heterozygous, suggesting that a gain of function, rather than loss of function over wild type, may be driving the selection of mutant cells in oncogenesis (19). In vitro experiments have shown that the mutant IDH proteins can act as a dominant‐negative suppressor of wild‐type protein function (28). As might be expected, tumors with mutant IDH generate 2‐HG, which can be detected by liquid chromotography‐mass spectrometry (8). The oncometabolite 2‐HG is a competitive inhibitor of multiple α‐ketoglutarate‐dependent deoxygenases, including histone demethylases, leading to increased levels of histone methylation and decreased levels of 5‐methylcytosine in gliomas (26). Although the role of mutant IDH function in oncogenesis is still unclear, that IDH mutations are found across a spectrum of diverse cancer types suggests an important and common oncogenic mechanism.
Mutations of IDH genes are present in most World Health Organization (WHO) grades II and III gliomas, and in secondary glioblastomas, but are rare in other glioma types 13, 19, 27. IDH mutations in gliomas correlate with a better outcome, hence identification of IDH mutations is useful for treatment planning and patient counseling 16, 27.
Conventionally, mutational detection for IDH1 and IDH2 has been performed by PCR and direct Sanger sequencing. A limitation of conventional PCR sequencing, however, is sensitivity; at least 50% of the specimens must be composed of neoplastic cells to ensure reliable detection of mutant alleles. As these tumors are infiltrative and often admixed with normal cells, development of novel assays for IDH mutations that can provide more sensitivity than typical PCR sequencing would be an asset to the clinical laboratory. Recently, an IDH1 R132H mutation‐specific antibody was developed that shows high sensitivity and specificity 6, 7. Although the antibody detects the most common IDH1 mutation (IDH R132H), fulfilling some diagnostic needs, it will not detect the other described mutations in IDH1 or IDH2, which represent ∼15% of mutations in IDH1/2. In this brief report, we describe a simple, PCR‐based SNaPshot assay that can be used on formalin‐fixed paraffin‐embedded tissue to detect the more commonly encountered IDH1 and IDH2 mutations.
MATERIALS AND METHODS
Control DNA
Control DNA was generated by oligonucleotide‐based, site‐directed mutagenesis of wild‐type IDH1 and IDH2 cDNA generated from a lymph node tissue specimen and cloned into pBluescript® SK (Agilent Technologies, Santa Clara, CA, USA). Mutant IDH‐containing plasmids were generated for the common IDH‐mutant alleles: wild‐type IDH1, IDH1(R132H), IDH1(R132C), IDH1(R132L), IDH1(R132S), and IDH1(R132G), and wild‐type IDH2 and IDH2 (R172M) and mixed to generate a control pool of mutants.
Patient samples
Slides were reviewed by a neuropathologist for a diagnosis of glioma. DNA was extracted from formalin‐fixed paraffin‐embedded tissue slide scrapings using the QIAamp DNA micro kit (Qiagen GmbH, Hilden, Germany). For validation of our assay, we utilized DNA specimens from a set of 32 glioma samples, which had been previously screened for IDH mutations via a PCR/direct sequencing method (15). Testing was then performed on an extended set of 57 clinical samples referred to us for evaluation. Basic clinical and pathologic information for all 89 cases is summarized in Supporting Information Table S1. Our research using clinical specimens conformed to the policies of the Conjoint Health Ethics Review Board of the University of Calgary and Alberta Health Services, as granted through our protocol E‐23650.
PCR
A multiplex PCR was performed with 50 ng of sample DNA using a Qiagen Multiplex Master Mix kit and IDH1 and IDH2 specific primers (Table 1). Cycling conditions used were as follows: 95°C for 15 minutes; six touchdown cycles of 94°C for 30 s‐63°C for 90 s‐72°C for 45 s; 34 cycles of 94°C for 30 s‐57°C for 90 s‐72°C for 45 s; 72°C for 10 minutes; hold at 4°C. PCR products were checked on an agarose gel for IDH1(235 bp) and IDH2(293 bp) products, then 5.0 uL of PCR product was cleaned using 2.0 uL of Exo‐SAP‐IT™ (Affymetrix, Santa Clara, CA, USA) by incubating at 37°C for 15 minutes followed by 80°C for 15 minutes.
Table 1.
PCR primer and SNaPshot primer sequences. Abbreviation: IDH = isocitrate dehydrogenase
| Primer | Sequence |
|---|---|
| IDH1‐F2 | GGTTGAGGAGTTCAAGTTG |
| IDH1‐R2 | AGTTGGAAATTTCTGGGCCAT |
| IDH2‐F | GCTGCAGTGGGACCACTATT |
| IDH2‐R | TGTGGCCTTGTACTGCAGAG |
| IDH1sn1r | (A)12CTTACTTGATCCCCATAAGCATGA |
| IDH1sn2r | (A)19CTTACTTGATCCCCATAAGCATGAC |
| IDH2sn1 | (A)29GACCAAGCCCATCACCATTGGCA |
Summary of oligonucleotides used in this study. IDH F and R primers are the forward and reverse primers used for the PCR amplification of exon 4 from IDH1 or IDH2, respectively. IDH1 sn1r and IDH1 sn2r are SNaPshot primers evaluating R132X mutations, respectively. IDH2sn1 is the SNaPshot primer evaluating IDH2 R172X mutations.
Mutation detection
About 1.0 uL of Exo‐SAP‐IT cleaned PCR product served as template for the SNaPshot reactions. SNaPshot reactions used IDH1 and IDH2 unique primers with the following conditions: 25 cycles of 96°C for 10 s‐50°C for 5 s‐60°C for 30 s; then hold at 4°C. The SNaPshot primers were designed with varying length of poly A tails to allow for multiplex size discrimination during electrophoresis (Table 1). The SNaPshot multiplex consisted of two IDH1 primers to interrogate the two base positions of interest at the IDH1 R132 codon, and one IDH2 primer to assess nucleotide of interest at the R172 codon of IDH2. Post‐SBE reaction cleanup to remove unincorporated nucleotides was achieved using BioSpin™ columns (Bio‐Rad, Hercules, CA, USA). About 1.0 uL of a 1:10 dilution of cleaned SNaPshot product was separated on a 3500 Genetic Analyzer (Life Technologies) with GS120LIZ size ladder. The raw data from capillary electrophoresis were analyzed using Gene Mapper™ v4.1 software (Applied Biosystems) to generate interpretable electropherograms.
RESULTS
We established a SNaPshot assay for IDH1 and IDH2 mutations as described in Methods. This assay utilizes the SNaPshot Multiplex Kit (Life Technologies) to extend the SNaPshot primers by single base extension (SBE) technology. The reaction adds a fluorescently labeled dideoxynucleotide at the 3′ end of the unlabelled SNaPshot primer, so these primers are designed to end one nucleotide before the nucleotide of interest. To ensure detection of common IDH‐mutant alleles, we cloned mutant oligonucleotides into IDH cDNA plasmids using oligonucleotide‐based, site‐directed mutagenesis. A control panel was generated for the assay by mixing all wild‐type and mutant clones at a 1000‐fold dilution into a single tube (Figure 1). Figure 1 shows the capillary electropherograms of our control (panel D) which represents a pool of possible IDH1/2 mutations, and some example cases demonstrating wild‐type, IDH1 R132H or IDH1 R132S mutations (panels B and C). We found that our assay was able to detect all the mutant alleles in our control plasmids.
Figure 1.
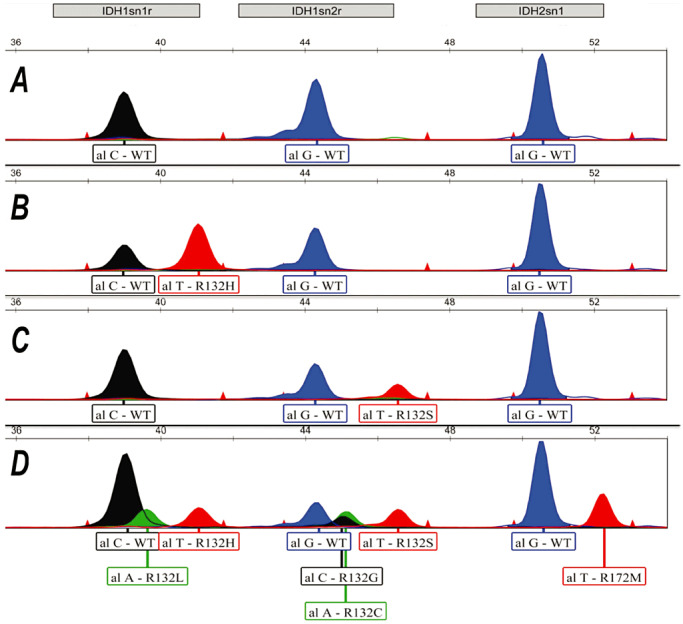
Representative electropherograms of IDH1 R132 mutant SNaPshot products. Panel A. Clinical sample with no isocitrate dehydrogenase (IDH) mutations. Panel B. Patient sample containing the IDH1 R132H mutation. Panel C. Patient sample showing the R132S mutation. Panel D. Control panel showing the common IDH1 and IDH2 mutant and wild‐type peaks.
To evaluate the performance of assay on formalin‐fixed paraffin‐embedded (FFPE) material, we tested 32 sequential cases previously screened for IDH mutations by PCR/direct sequencing method with the SNaPshot assay in a blinded study. In our study, concordance with sequencing data for the 32 random FFPE specimens was 100%, as we perform the assay with >50% cancer cells in the specimen. Throughput for the IDH SNaPshot assay was similar to that of PCR/direct sequencing with results being available within 1 day. In our first test group of patients, we detected 11 samples with the common R132H mutation of IDH1 (Figure 1, panel B), a single specimen with the R132S IDH1 (Figure 1, panel C) mutation as well as two samples with the R172K IDH2 mutation (Figure 2, panels B and C).
Figure 2.
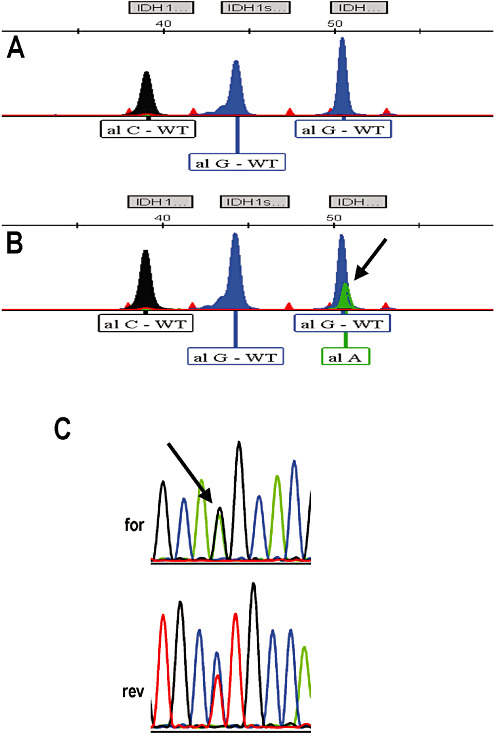
Representative electropherograms of IDH2 R172 mutant SNaPshot products. Panel A. Clinical sample with no isocitrate dehydrogenase (IDH) mutations. Panel B. Patient sample containing the IDH2 R172K mutation. Panel C. Sequencing data (forward and reverse) showing heterozygosity of the IDH2 R172K mutation in the same patient specimen as above.
Next, we performed the assay on additional 57 specimens, usually primary glioblastoma cases, that were submitted to the laboratory for IDH mutation analysis. Of these, we identified a further nine cases of the R132H and two cases of the R132S IDH mutations. No examples of IDH2 mutation could be identified in the additional 57 glioma specimens; however, the IDH2 control reaction shows the ability to identify the R172M peak (Figure 1, panel D), so both IDH R172K and M mutations would have been detected if they were present. An aggregate data set of the clinical characteristics and results of our tests are presented in Table 2.
Table 2.
Summary of histologic subtype and isocitrate dehydrogenase (IDH) mutation status. Abbreviations: GBM = glioblastoma; PMA = pilomyxoid astrocytoma; DNT = dysembryoplastic neuroepithelial tumor
| Diagnosis | IDH mutant | IDH wild type | % Mutated |
|---|---|---|---|
| Primary GBM | 3 | 47 | 6 |
| Secondary GBM | 2 | 0 | 100 |
| Astrocytoma, grades II and III | 4 | 1 | 80 |
| Oligodendroglioma, grades II and III | 11 | 1† | 92 |
| Oligoastrocytoma | 5 | 0 | 100 |
| Pilocytic astrocytoma/PMA | 0 | 8 | 0 |
| Ganglioglioma | 0 | 4 | 0 |
| Miscellaneous (meningioma, DNT) | 0 | 3 | 0 |
| Totals | 25 | 64 | 28 |
Tumors are grouped by WHO 2007 histological classification. IDH mutations include both IDH1 and IDH2 mutated genes.
Denotes a case of IDH1 wild‐type pediatric oligodendroglioma.
The SNaPshot assay should theoretically provide a better signal to noise detection of a mutant allele over wild‐type (as the detection of a mutant peak is a de novo peak above background whereas a sequencing peak is a ratio between wild‐type and mutant allele peaks) compared to conventional PCR and direct sequencing. To assess the sensitivity of our IDH SNaPshot assay, we titrated the PCR products generated from mutant or wild‐type specimens and performed a direct comparison. Figure 3 demonstrates such a titration, and shows that the detection of a mutant allele in the SNaPshot assay is still easily discerned (eg, panels C and D) while the mutant allele of the sequencing assay disappears into background. Further evaluation (Figure 4) shows that between 2.5 and 5% mutant allele can be easily observed with the SNaPshot assay.
Figure 3.
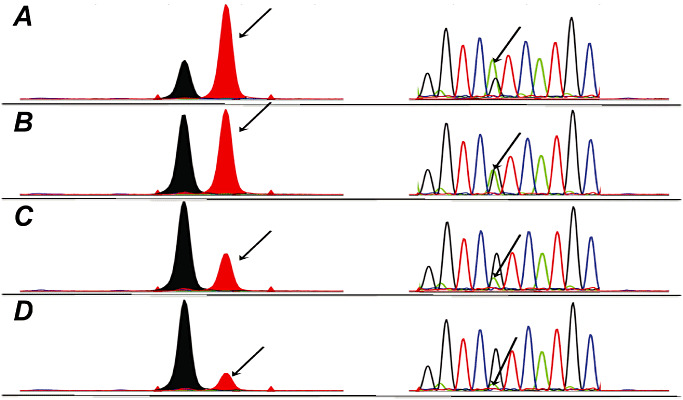
Comparison of sequencing and SNaPshot assay analytical sensitivity. Panel A. 60% R132H mutant(Mut):40% wild type(WT); Panel B. 40% Mut:60% WT; Panel C. 20% Mut:80% WT; Panel D. 10% Mut:90% WT. Arrow indicates position of the R132H mutant peak.
Figure 4.
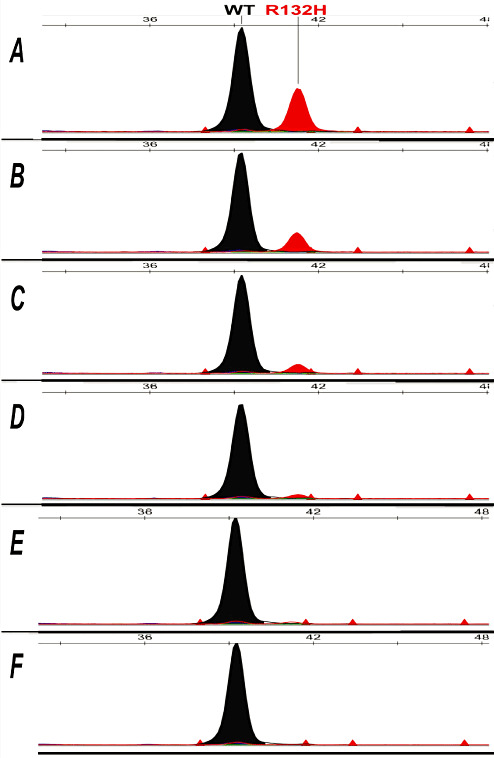
Titration of SNaPshot assay to assess assay maximum analytical sensitivity. Panel A. 20% R132H mutant, 80% wild‐type PCR product. Panel B. 10% mutant, 90% wild type. Panel C. 5% mutant, 95% wild type. Panel D. 2.5% mutant, 97.5% wild type. Panel E. 1% mutant, 99% wild type. Panel F. 100% wild type.
DISCUSSION
Alterations of IDH1 in glial tumors were first identified in a genome‐wide mutation screen of glioblastoma specimens (19). Of these, approximately 12% exhibited a mutation of IDH1. Several large series of gliomas characterized the frequency of mutations among many glial tumors and found striking differences in frequency corresponding to histological subtypes across the entire spectrum of glioma (27), (24) and, additionally, corresponding to pediatric vs. adult diffuse gliomas 3, 4, 20. For example, adult grade II astrocytomas exhibit an ∼90% frequency of IDH mutations whereas pilocytic astrocytomas, ependymomas and pediatric diffuse astrocytomas (of all grades) virtually never carry IDH mutations. Interestingly, adult primary glioblastomas had a frequency of less than 5% vs. secondary glioblastomas, which showed a frequency of ∼85%, and oligodendrogliomas, which had a very high frequency of mutations—>80% regardless of grade. More importantly, IDH mutations in glioma patients have been shown to be associated with a better outcome for the patients 11, 24. As well, IDH mutations have been recently reported to be associated with leukemic transformation in myelofibrosis patients, and a worse outcome (22), suggesting clinical utility for IDH genotyping in other neoplasms.
Although we have a much smaller series of patient specimens, as the intent of our study was to develop and validate a new method, our data reflect similar results. As expected, no pilocytic astrocytomas or gangliogliomas were found to be mutant for IDH. Also, of specimens that were classified as primary glioblastoma, only 6% exhibited a mutation. In contrast, 100% of secondary glioblastomas, 80% of WHO grades II and III diffuse astrocytomas, 92% of WHO grades II and III oligodendrogliomas, and 100% of oligoastrocytomas exhibited IDH mutations (Table 2). Of note, the one case of oligodendroglioma that we found to be IDH wild type occurred in a child—if this case were excluded, the mutation rate for oligodendrogliomas in our study would increase to 100%. The rationale for suggesting this is that it has been reported that pediatric gliomas may differ biologically from adult gliomas (10). Of course, the frequency of types of specimens that we tested reflects the practice patterns of our referring neuropathologists and is not representative of the overall population‐based frequency of central nervous system neoplasms diagnosed in our center. Overall, however, our results are in accordance with the results of other groups (27).
In this manuscript, we describe the development of a simple and sensitive test for mutations of IDH1 and IDH2 genes which will be useful for treatment decisions in cases of glioma, and likely other tumors such as myeloid leukemias and some sarcomas. Other assays for IDH mutations have also been described. The most simple of these, a specific antibody to IDH1 proteins which bear the common R132H mutation has been shown to be highly sensitive and specific in detecting the mutant protein by Western blotting or immunohistology (7). This antibody has been shown to be very useful in identifying even single tumor cells bearing the mutation, and is a very useful diagnostic tool for identifying very low levels of tumor cell infiltration in tumor types which have a high frequency of this mutation such as oligodendroglial neoplasms (6). Nonetheless, other mutations of IDH1 as well as mutations of IDH2 will not be detected.
Pyrosequencing can theoretically identify all, including nonconventional, IDH mutations at higher sensitivity than conventional sequencing (12). Fluorescent melting curve analysis also showed high fidelity to sequencing and a significant increase in analytical sensitivity (14). Like the others, the SNaPshot assay, identifying the most common mutations of IDH1 and IDH2, can be performed from paraffin‐embedded specimens. Results can be obtained in 1 day from DNA extraction to analysis. Data analysis was much faster and easier to perform than analysis of the sequencing data. The analytical sensitivity of the new test was considerably better than that using a conventional “gold standard” genomic PCR and sequencing method. As well, the analytical sensitivity of the SNaPshot assay of 2.5%–5% may be slightly better than that of fluorescence melting curve analysis (reported as 10%) (14) and also than that reported for pyrosequencing (reported as ∼7%) (12), allowing potentially better detection of mutant alleles when tumor cell frequency is low. Furthermore, the SNaPshot assay allows multiplexing of the assays for the common mutations of IDH1 and IDH2 into a single reaction. Primers to detect additional mutants could be added relatively easily to our multiplex assays. Our assay also allows for rapid and automated reaction setup and rapid data analysis, especially compared with sequencing methods.
Supporting information
Table S1. Clinicopathologic characteristics of patients.
Supporting info item
ACKNOWLEDGMENTS
All authors receive salary support from Calgary Laboratory Services (CLS). JAC acknowledges salary support from an Alberta Innovates‐Health Solutions Clinical Investigator award. CLS is also acknowledged for the operating support of this project. Robert Hay is acknowledged for initially establishing the PCR‐sequencing assay for IDH1 exon 4 and IDH2 exon 4.
REFERENCES
- 1. Alam NA, Rowan AJ, Wortham NC, Pollard PJ, Mitchell M, Tyrer JP et al (2003) Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum Mol Genet 12:1241–1252. [DOI] [PubMed] [Google Scholar]
- 2. Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F et al (2011) IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224:334–343. [DOI] [PubMed] [Google Scholar]
- 3. Antonelli M, Buttarelli FR, Arcella A, Nobusawa S, Donofrio V, Oghaki H, Giangaspero F (2010) Prognostic significance of histological grading, p53 status, YKL‐40 expression, and IDH1 mutations in pediatric high‐grade gliomas. J Neurooncol 99:209–215. [DOI] [PubMed] [Google Scholar]
- 4. Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A (2008) Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602. [DOI] [PubMed] [Google Scholar]
- 5. Baysal BE, Ferrell RE, Willett‐Brozick JE, Lawrence EC, Myssiorek D, Bosch A et al (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287:848–851. [DOI] [PubMed] [Google Scholar]
- 6. Capper D, Sahm F, Hartmann C, Meyermann R, von Deimling A, Schittenhelm J (2010) Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy‐induced changes. Am J Surg Pathol 34:1199–1204. [DOI] [PubMed] [Google Scholar]
- 7. Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D et al (2010) Characterization of R132H mutation‐specific IDH1 antibody binding in brain tumors. Brain Pathol 20:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM et al (2009) Cancer‐associated IDH1 mutations produce 2‐hydroxyglutarate. Nature 462:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dang L, Jin S, Su SM (2010) IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med 16:387–397. [DOI] [PubMed] [Google Scholar]
- 10. De Carli E, Wang X, Puget S (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:2248; author reply 9. [DOI] [PubMed] [Google Scholar]
- 11. Dubbink HJ, Taal W, van Marion R, Kros JM, van Heuvel I, Bromberg JE et al (2009) IDH1 mutations in low‐grade astrocytomas predict survival but not response to temozolomide. Neurology 73:1792–1795. [DOI] [PubMed] [Google Scholar]
- 12. Felsberg J, Wolter M, Seul H, Friedensdorf B, Goppert M, Sabel MC, Reifenberger G (2010) Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol 119:501–507. [DOI] [PubMed] [Google Scholar]
- 13. Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A et al (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1010 diffuse gliomas. Acta Neuropathol 118:469–474. [DOI] [PubMed] [Google Scholar]
- 14. Horbinski C, Kelly L, Nikiforov YE, Durso MB, Nikiforova MN (2010) Detection of IDH1 and IDH2 mutations by fluorescence melting curve analysis as a diagnostic tool for brain biopsies. J Mol Diagn 12:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horbinski C, Kofler J, Kelly LM, Murdoch GH, Nikiforova MN (2009) Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin‐fixed, paraffin‐embedded glioma tissues. J Neuropathol Exp Neurol 68:1319–1325. [DOI] [PubMed] [Google Scholar]
- 16. Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J et al (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low‐grade gliomas. Neurology 75:1560–1566. [DOI] [PubMed] [Google Scholar]
- 17. Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K et al (2011) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paugh BS, Qu C, Jones C, Liu Z, Adamowicz‐Brice M, Zhang J et al (2010) Integrated molecular genetic profiling of pediatric high‐grade gliomas reveals key differences with the adult disease. J Clin Oncol 28:3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD et al (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274. [DOI] [PubMed] [Google Scholar]
- 22. Tefferi A, Jimma T, Sulai NH, Lasho TL, Finke CM, Knudson RA et al (2012) IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia 26:475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warburg O, Wind F, Negelein E (1927) The metabolism of tumors in the body. J Gen Physiol 8:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J et al (2009) Molecular predictors of progression‐free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol 27:5743–5750. [DOI] [PubMed] [Google Scholar]
- 25. Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM et al (2011) Hypoxia promotes isocitrate dehydrogenase‐dependent carboxylation of alpha‐ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A 108:19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH et al (2011) Oncometabolite 2‐hydroxyglutarate is a competitive inhibitor of alpha‐ketoglutarate‐dependent dioxygenases. Cancer Cell 19:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P et al (2009) Glioma‐derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF‐1alpha. Science 324:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinicopathologic characteristics of patients.
Supporting info item


