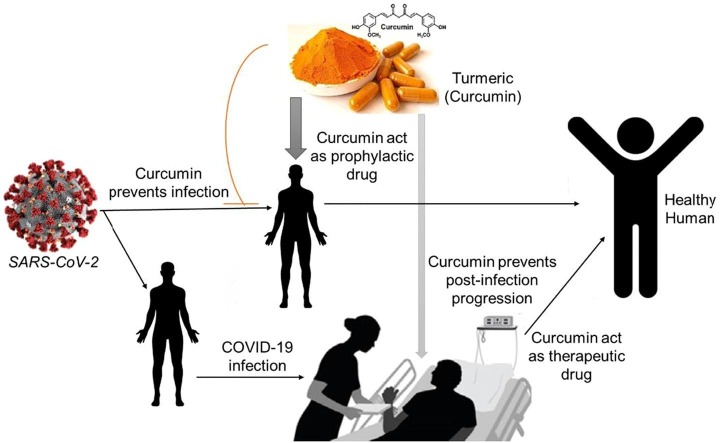
Graphical abstract
Keywords: COVID-19, Viral lifecycle, Curcumin, Immunomodulation, Prophylaxis, Therapeutic drug
Abstract
Curcumin has already acknowledged immense interest from both medical and scientific research because of its multifaceted activity. To date, the promising effects of curcumin were perceived against numerous inflammatory diseases. Besides, curcumin’s role as a medicine has been studied in many virus infections like influenza, HIV, etc. There is a need to analyze the cellular mechanisms of curcumin including host-pathogen interaction and immunomodulatory effects, to explore the role of curcumin against COVID-19. With this background, our study suggests that curcumin can prevent COVID-19 infections by inhibiting the pathogen entry, viral genome replication and steps in the endosomal pathway along with inhibition of T-cell signalling by impairing the autophagy-mediated antigen-presenting pathway. This review explicit the possible mechanisms behind curcumin-induced cellular immunity and a therapeutive dosage of curcumin suggesting a preventive strategy against COVID-19.
1. Background
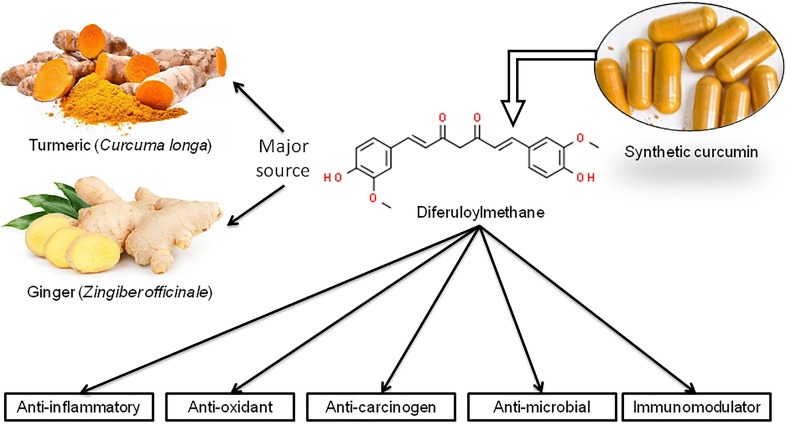
Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione), also known as Diferuloylmethane, is a polyphenol product derived from the rhizomes of plant turmeric and other Curcuma sp. (Fig. 1 ). Historically, curcumin, in the form of turmeric, is generally used for flavouring additives and food preservatives in Asiatic countries due to its anti-fungal and anti-oxidant property. The popular use of curcumin in research is mainly due to its pleiotropic properties including anti-inflammatory, anti-oxidant and anti-carcinogenic activities (Ahmad et al., 2020). Curcumin can inhibit the inflammatory mediators, oxidation processes, and oxidative stress thereby acts as an anti-inflammatory agent against many diseases (Wal et al., 2019). Praditya et al. (2019b) have shown curcumin as an anti-bacterial agent against several strains of Staphylococcus, Streptococcus, Helicobacter and Pseudomonas mainly by growth inhibition. They also reported the anti-fungal property of curcumin. Different studies identified the efficacy of curcumin against Human immunodeficiency virus (HIV), Herpes simplex virus (HSV), Hepatitis viruses etc. (Praditya et al., 2019a, Prasad and Tyagi, 2015, Vitali et al., 2020). Although there are some controversies, the majority of the studies support the potential role of curcumin in inhibiting viral replication and growth inhibition (Mathew & Hsu, 2018). Apart from these, long-term intake of curcumin can improve systolic blood pressure (Hadi et al., 2019), control obesity (Jarząb & Kukula-Koch, 2019), Type 2 Diabetes Mellitus (Pivari et al., 2019). Also, curcumin often acts as a cardio-protective, nephroprotective, anti-neoplastic, hepato-protective and anti-rheumatic compound (Pivari et al., 2019). In the present pandemic scenario, it would be of utmost importance to understand the effectiveness of this popular spice against SARS-CoV-2 (the causative microorganism of COVID-19) by extrapolating computational predictions and already published literature. The hallmark of the present review is to scrutinize the mechanistic insight of this traditional ethnomedicine curcumin concerning viral lifecycle within the host cell and immune-modulation within the cell.
Fig. 1.
Curcumin: Major sources, chemical structure and its medicinal properties.
2. Methodology
A systematic literature review was conducted considering published literature from 2010 to 2021. A number of search engines like Pubmed, Google scholars, Dimensions and their cross-references were thoroughly studied. In the ‘Dimensions’ platform, 1606 articles were available with the phrase ‘Curcumin and SARS-CoV-2′, followed by 1216, 811 and 569 articles were available with the phrase ‘Curcumin and COVID-19′, ‘Clinical trial of curcumin against COVID-19′ and ‘Immunomodulatory effect of curcumin against SARS-CoV-2′. Using VOSviewer software, a total of 4202 articles were exported and analysed the terms of co-occurrence (Supplementary Fig. 1). The result identified the novel areas to explore further. Considering the keywords “COVID-19”, “viral infection mechanism”, “viral lifecycle”, “immune pathogenicity”, “curcumin”, “antiviral mechanism”, etc., 205 published articles have been retrieved. Among them, the articles written in English, accessible full paper and relevance with the theme of the review, a total of 123 articles were finally selected for this comprehensive review.
3. COVID-19 – A briefing of pathophysiology
The preliminary symptoms like cough and cold along with fever in COVID-19, gradually develop into further complications like severe pneumonia, coagulopathy and acute respiratory distress syndrome (ARDS). Accumulating information suggest that SARS-CoV-2 principally targets the cells which overexpress angiotensin-converting enzyme 2 (ACE2) and such target cells are airway epithelial cells, alveolar epithelial cells, vascular endothelial cells and macrophages in the lung. In general, men are more susceptible to COVID 19 and it has been linked with higher ACE2 expression in them compared to women (Jin et al., 2020, Sharma et al., 2020, Tay et al., 2020). In addition to this, the risk of COVID 19 infections among children (Kelvin & Halperin, 2020) intrigued the need for host-pathogen interaction study in further detail.
The transmission of SARS-CoV-2 occurs via respiratory aerosols and binds to the nasal epithelial cells in the upper respiratory tract resulting in severe pneumonia-like symptoms. Subsequent migration towards the lower respiratory tract causes damage in alveolar cells leading to the development of ARDS (Parasher, 2020). Also, COVID-19-associated coagulopathy (CAC) presents many similarities with those of sepsis-induced coagulopathy (SIC)/disseminated intravascular coagulation (DIC), haemophagocytic syndrome (HPS)/haemophagocytic lymphohistiocytosis (HLH), antiphospholipid syndrome (APS), and thrombotic microangiopathy (TMA) (Iba et al., 2020). DIC like massive intravascular clot formation is frequently observed among severely ill COVID patients (Iba et al., 2020). Cytokine storm has been strongly associated with COVID-19 so far. Discharge of proinflammatory cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF- α, TGF β, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) from immune effector cells lead to such lethal uncontrolled systemic inflammatory response (Mehta et al., 2020). Although negligible changes in prothrombin time and platelet count were observed initially, unlike SIC/DIC, CAC displays increased D-dimer and fibrinogen levels. Venous thromboembolism and arterial thrombosis are more frequent in CAC (Iba et al., 2020, Iba et al., 2020). Interestingly, secondary HLH is associated with COVID-19 disease severity and is characterized by the profound release of several cytokines and chemokines, including interleukin IL-2, IL-7, granulocyte-colony stimulating factor, interferon-γ inducible protein 10, monocyte chemo-attractant protein (MCP) 1, macrophage inflammatory protein (MIP) 1-α, and tumour necrosis factor (TNF)-α. Nonetheless, while neutrophil, monocyte, high-sensitivity C-reactive protein (Hs-CRP), procalcitonin were increased, the amount of eosinophil, lymphocyte numbers, lymphocyte-immune subsets, IgM, and C3 are found to be decreased significantly in COVID-19 patients (Li et al., 2020, Li et al., 2020, Mehta et al., 2020).
4. Deciphering the mechanisms of COVID-19 infection
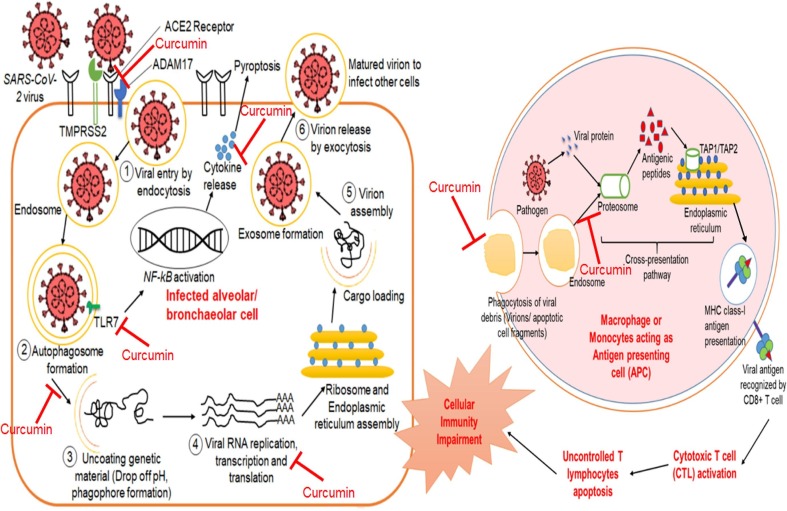
Several viral proteins encoded by the viral genome play a pivotal role in the viral lifecycle as well as immune evasion within the host cell (Astuti and Ysrafil, 2020, V’kovski et al.,, 2020). Viral invasion impairs the innate immune system and thus are more prone to viral infection. The mechanism of viral replication and disease progression has been schematically shown (Fig. 2 ) and highlighted the major viral proteins in Table 1 .
Fig. 2.
Potential therapeutic targets of curcumin against SARS-CoV-2 suggesting its role in COVID-19 management.
Table 1.
Description of the viral proteins encoded by SARS-CoV-2 genome highlighting their role in the viral lifecycle and immune pathogenicity.
| Viral proteins | Characteristics | Molecular functions | References | |
|---|---|---|---|---|
| Structural protein | Spike-like surface glycoprotein | MW ~ 150 kDa; 1273 amino acids; Rich in glutamine, asparagine, leucine, phenylalanine and serine amino acids | S1 subunit facilitates hACE2-mediated virus attachment; S2 subunit promotes membrane fusion process into the host cell; help in promoting adhesion of infected cells with adjacent non-infected cells | Astuti & Ysrafil, 2020; Bianchi et al., 2020; Chen et al., 2020; Satarker & Nampoothiri, 2020 |
| Envelope glycoprotein | MW ~ 8–12 kDa; 76–109 amino acids; Contains N Terminal Domain (NTD) and hydrophilic C Terminal Domain (CTD) | Forms viriporins which is required for virion assembly and release | ||
| Membrane glycoprotein | 220–260 amino acids; Contains hydrophilic C terminal and amphipathic N terminal | Determine the shape of the virus envelope; helps to stabilize nucleocapsids and promotes completion of viral assembly by stabilizing N protein-RNA complex, inside the internal virion | ||
| Nucleocapsid protein | Composed of a serine-rich linker region between NTD and CTD | NTD forms orthorhombic crystals and binds to the viral genome; linker region regulate self- functioning; CTD promotes nucleocapsid formation; promotes the activation of COX-2 leading to inflammation in the lungs; inhibits IFN-1 causing restrictions in immune responses | ||
| Non-structural proteins | NSP1 (Leader protein) | Inhibit host mRNA translation, antagonize IFN signalling and induce inflammatory cytokines and chemokines | Astuti & Ysrafil, 2020; Min et al., 2020; Samaddar et al., 2020; Yoshimoto, 2020, Zhang et al., 2020 | |
| NSP2 | Binding to prohibitin-1 (PHB1) and prohibitin-2 (PHB2) leads to disruption of the host cell environment | |||
| NSP3 | Encode papain-like protease (PLpro) that helps to cleave the site between NSP2 and NSP3 and release essential viral proteins for viral activity | |||
| NSP4 (Transmembrane domain 2) | Interacting with NSP3 causes rearrangement of the host cell membrane | |||
| NSP5 | Encode chymotrypsin-like protease (3CLpro/Mpro) that cleaves at 11 different sites to produce mature and intermediate viral polyproteins | |||
| NSP6 (Putative Transmembrane domain) | Restricting autophagolysosome development cause hindrance of autophagosomes from transporting viral components for degradation in lysosomes | |||
| NSP7 (Peptide cofactor) | Forms a complex NSP8 and NSP12 to yield RNA polymerase activity | |||
| NSP8 (Peptide cofactor) | Forms hexadecameric complex of RNA polymerase | |||
| NSP9 | Encode RNA-binding protein phosphatase that helps in viral genome replication and transcription | |||
| NSP10 | Interacting with NSP14 and NSP16 to stimulate SAM-dependent methyltransferase activity and 2′-O-Ribosemethyltransferase activity respectively | |||
| NSP11 | Unknown activity yet | |||
| NSP12 (RdRP) | Encodes RNA-dependent RNA polymerase that replicates viral RNA | |||
| NSP13 (Helicase) | Unwinds duplex RNA; elicits 5′-triphosphatase activity to introduce a 5′-terminal cap of mRNA | |||
| NSP14 (N7-methyltransferase) | Proofreading of viral genome by endonuclease and methyltransferase activity | |||
| NSP15 (Endoribonuclease) | Cleaves RNA at 3′-uridylates to form a 2′-3′ cyclic phosphodiester product that protects viral RNA from host recognition and inhibit innate response | |||
| NSP16 (2′-O-Ribosemethyltransferase) | Methylate 2′-hydroxyadenine using SAM as methyl pool thereby avoid MDA5 recognition of viral RNA and inhibit innate immunity regulation | |||
4.1. Virus lifecycle within the host cell
Like other viral infection, the lifecycle of SARS-CoV-2 involves four major steps i.e. viral entry by attachment and fusion into a host cell, endocytosis of the virus, genome expansion and virion release.
4.1.1. Viral attachment
Attachment is the initial event that brings viruses to the cell membrane surface of the target cell through a receptor. Several studies have already established the importance of host cell receptors in viral infections, for example, hemagglutinin (HA) of Influenza virus binds with sialic acid (Samji, 2009), Human Immune deficiency Virus (HIV) binds to CD4 receptor (Wilen et al., 2012). Globally, a variety of studies have already confirmed that the S protein of the β-Coronavirus is a significant determinant of virus entry into host cells (Ou et al., 2020). Previous studies have shown that envelope spike glycoprotein of SARS-CoV binds to CD209L (a C-type lectin) and angiotensin-converting enzyme 2 (ACE2) whereas MARS-CoV binds to Dipeptidyl peptidase 4 (DPP4, also known as CD26). Similar to SARS-CoV, SARS-CoV-2 also uses the angiotensin-converting enzyme 2 (ACE2) as a cellular receptor to penetrate the host system (Li et al., 2020).
For an enveloped virus, delivery of the viral genome requires membrane fusion reaction between the lipid bilayer of the virus and a host target cell membrane (Doms, 2016). In SARS-COV-2, this membrane fusion process is initiated by serine protease 2 (TMPRSS2) which activate the spike protein (S) through specific cleavage (Heurich et al., 2014, Hirano and Murakami, 2020, Hoffmann et al., 2020). The binding of this spike protein to the receptor triggers the ADAM17-mediated proteolytic “shedding” of ACE2 at the ectodomain site for the promotion of viral uptake into the cell (Ciaglia et al., 2020, Scialo et al., 2020). Therefore, TMPRSS2 and ADAM17 both augment the viral entry into host cells and follow the clathrin-dependent and –independent endosomal pathway (Li et al., 2020).
4.1.2. Endocytosis of the virus
Following attachment and receptor clustering, many viruses are endocytosed and transported to endosomes where the low pH induces alterations in structural proteins of the virus that facilitate membrane fusion by protonating acidic residues. Generally, endocytic pathways include clathrin-mediated endocytosis, caveolae, macropinocytosis, and novel clathrin- and caveolae-independent pathways (Wang et al., 2008). In the case of SARS-CoV-2, clathrin-mediated endocytosis occurs in nasal epithelial cells because of the abundance of GTPase, dynamin whereas clathrin-independent endocytosis occurs in pneumocytes. However, due to comparatively higher expression of C‐terminal binding protein (CtBP) 1 & 2 and P21‐activated kinase 1, macropinocytosis also occurs in pneumocytes (Glebov, 2020). The virus-containing vacuoles undergo acidification, maturation and fusion with late endosomes or lysosomes. In the case of Influenza A virus, neuraminidase (NA) binds to lysosome-associate membrane proteins (LAMPs) and ruptures the membrane of the autophagosomes by deglycosylation (Ju et al., 2015).
Likewise in SARS-CoV-2 infection, the ACE2-Spike protein complex is cleaved by cathepsin L protease within the autophagosomes leading to the fusion of viral and host cell membrane (Blaess et al., 2020). The acidic environment initiates conformational modifications in the viral capsid resulting in exposure of hydrophobic domains within the endosomal membrane and forming a protein pore through which the viral genome can exit and enter the cytoplasm (Doms, 2016, Yamauchi and Helenius, 2013). In this regards, nonstructural viral proteins like (NSP2, NSP3, NSP4 and NSP6) play a crucial role in virus escaping through pore formation in late endosome (discussed in Table 1).
4.1.3. Viral genome expansion
The literature review suggests that RNA-dependent RNA polymerase (RdRp) plays an important role in the replication process of RNA viruses like SARS-CoV-2 (Mathew & Hsu, 2018). After escaping from the autophagolysosome, genomic RNA undergoes an immediate translation. To avoid host detection, several non-structural proteins form a replication-transcription complex (RTC) as well as viral replication organelles like perinuclear double-membrane vesicles, convoluted membranes and small open double-membrane spherules (V’kovski et al., 2020). Two overlapping open reading frame (ORF1a and ORF1b) translated into the viral enzymes 3C-like protease (3CLpro) and papain-like protease (PLpro), help in viral replication via proteolytic cleavage. Another segment of RNA encodes structural proteins of the virus, such as the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins (Chen et al., 2020, Sala de Oyanguren et al., 2020, Shereen et al., 2020). Translated structural proteins translocate into endoplasmic reticulum (ER) membranes and transport through the ER-to-Golgi intermediate compartment (ERGIC). Freshly formed genomic RNA undergoes N-encapsidation, resulting in budding into the lumen of secretory vesicular compartments (V’kovski et al.,, 2020, Zhang et al., 2020, Zhang et al., 2020).
4.1.4. Virion release
Pathogen-associated molecular patterns (PAMPs) of the evolutionarily conserved microbes can be recognized by pattern recognition receptors (PRRs). Interestingly both SARS-CoV and MERS-CoV can induce the production of double-membrane vesicles that lack PRRs and then replicate in these vesicles, thereby avoiding the host detection of their dsRNA (Li et al., 2020). In this regards, viral single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) are recognized by extracellular as well as endosomal Toll-like receptors (TLRs) and stimulate downstream signalling cascades (Vabret et al., 2020). It has been found that TLR-7 plays a crucial role in SARS-CoV-2 genome recognition and clearance through exocytosis (Onofrio et al., 2020). Also, an in-silico experiment reported that extracellular TLR-4 is most likely to be involved in recognizing molecular patterns from SARS‐CoV‐2 (Choudhury & Mukherjee, 2020). However, the freshly formed envelope glycoproteins are inserted into the membrane of the endoplasmic reticulum or Golgi, and the Nucleocapsid is shaped by the combination of genomic RNA and Nucleocapsid protein. The virions germinate into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). Finally, the vesicles containing the virus particles fuse with the plasma membrane to release the virus through exocytosis and infect surrounding cells (Li et al., 2020; Shereen et al., 2020).
4.2. Cellular immunity against infection
A large group of scientists have tried to understand the immunological perspective of COVID-19. Usually soon after the virus enters into the host cell, the cellular immune response is initiated by PPRs recognition of PAMPs. This ligand binding activates several signalling pathways like NF-kB, IRF3 and AP-1 which synergistically stimulate IFN-1 production thereby limit virus entry as well as restrict viral replication (Taefehshokr et al., 2020). In a healthy individual, a well-coordinated immune response characterizes the first line of defence against viral infection, while on the contrary, extreme inflammatory innate response and dysregulated adaptive immune system ensue detrimental tissue damage at the systemic level (Catanzaro et al., 2020). Although coronaviruses have a preferential tropism for lung cells, it mostly impairs the innate immune machinery by directly infecting T cells (Wang et al., 2020). SARS-CoV-2 targets multiple aspects of innate immunity like cytokine storm, impairing interferon signalling and suppression of antigen presentation on both MHC class I and II (Taefehshokr et al., 2020). At the same time, an accumulation of mononuclear cells in the lungs coupled with a low level of hyperactive T-cells in the peripheral blood indicates that T-cells are attracted away from blood towards the infected sites to control the viral infection (Li et al., 2020; Tay et al., 2020). Both B-cells and T-cells responses against SARS-CoV-2, are diagnosed in the blood around 7 days after the onset of symptoms.
In the lung/alveolar epithelial cells, SARS-CoV-2 mainly deploy cellular immune responses and interfere with the IFN-signalling pathway with the membrane protein (M), Nucleocapsid protein (N) and non-structural proteins like NSP1, NSP3, PLpro acting as IFN antagonist (Taefehshokr et al., 2020, Tay et al., 2020). During infection, non-structural proteins of this virus protect themselves from recognition by PRRs and inhibit the transcription of interferons leading to modulate IFN response in plasmacytoid dendritic cells (pDC) and other immune cells. This alters the cytokine secretion profile of infected cells to enhance the recruitment of myeloid immune cells over NK cells, which in turn produce more cytokines, creating a cycle of inflammation that damages the lung (Taefehshokr et al., 2020). Apart from this interferon signalling, antigen presentation is an important antiviral mechanism that is being affected in the case of COVID-19. In this regards, Macrophages in the lung and upper respiratory tract act as sentinel cells and are among the first immune cells to encounter incoming virion whereas DCs are key players in antigen presentation, cytokine production, instructing specific T cell responses (Kumar et al., 2020, Li et al., 2020, Li et al., 2020, Taefehshokr et al., 2020). Nonetheless, coronaviruses trigger different complement pathways producing C3a and C5a components which then combine with dysregulated neutrophilia, endothelial injury and hyper-coagulopathy seem to be entangled to drive the severity of COVID-19 (Java et al., 2020, Liu and Ying, 2020).
After an encounter with the infected cells, the viral antigen, through the endosomal cross-presentation pathway, will be presented to the antigen presentation cells (APC) and its antigenic peptides are accessible by major histocompatibility complex (MHC-I) or human leukocyte antigen (HLA) in humans thereby recognized by virus-specific cytotoxic T (CD8+ T) lymphocytes. Antigen presentation subsequently stimulates the body’s humoral and cellular immunity, which are mediated by virus-specific B and T cells (Li et al., 2020). In the case of a healthy immune system, T cells become activated when CD8+ T cells recognize viral antigens by MHC-I molecules present on the infected cells. Then activated T cells undergo clonal expansion to proliferate into a large number of progeny T cells with identical receptors. This ultimately lyses the infected cells by inducing apoptosis and secrete pro-inflammatory cytokines (Gutierrez et al., 2020). As a result, uncontrolled apoptosis of T lymphocytes occurs with the release of pro-inflammatory cytokines leads to local tissue damage and impairment of the immune system.
5. Chemistry of curcumin
5.1. Sources & chemical composition
Curcumin is the main natural polyphenol found in turmeric (Curcuma longa) and other species of the Zingiberaceae family (Amalraj et al., 2017, Hewlings and Kalman, 2017). The powdered extracts of turmeric-dried roots may contain essential oils, proteins, fat, minerals, carbohydrates, moisture, and curcuminoids (Ahmad et al., 2020, Prasad et al., 2014, Prasad et al., 2014). The aroma of this spice is principally derived from sesquiterpenes which is a mixture of α- and β-turmerones with aromatic turmerone (Amalraj et al., 2017). The polyphenols (curcuminoids) are a mixture of 71.5% curcumin, chemically a diferuloylmethane [1,7-bis(4-hydroxy-3-methoxy-phenyl)-hepta-1,6-diene-3,5-dione] mixed with its two derivatives, 19.4% demethoxycurcumin [4-hydroxycinnamoyl-(4-hydroxy-3-methoxycinnamoyl) methane] and 9.1% bisdemethoxycurcumin [bis-(4-hydroxy cinnamoyl) methane], defining the chemical formulae as C21H20O6, C20H18O5 and C19H16O4 respectively (Amalraj et al., 2017, Gupta et al., 2013, Gupta et al., 2013, Hewlings and Kalman, 2017, Prasad et al., 2014, Prasad et al., 2014).
5.2. Pharmacokinetic property
Curcumin can be delivered by oral intake, subcutaneous, intraperitoneal, intravenous injection, topical and nasal treatment (Prasad et al., 2014, Prasad et al., 2014). However, pieces of the literature suggest that curcumin has poor absorption, biodistribution, metabolism, and bioavailability despite having vast pharmacological significance (Dei Cas and Ghidoni, 2019, Prasad et al., 2014, Prasad et al., 2014). After oral intake, curcumin gets metabolized within the liver through extensive phase I and II biotransformation with the help of gut microbiota (Dei Cas & Ghidoni, 2019). In phase I reaction, double bonds of curcumin is reduced by alcohol dehydrogenase into dihydro-curcumin, tetrahydro-curcumin, hexahydro-curcumin, and octahydro-curcumin (Dei Cas and Ghidoni, 2019, Nelson et al., 2017). Later in phase II reaction, curcumin and its phase I metabolites are rapidly conjugated with glucuronic acid by UGTs (Uridine 5′-diphospho-glucuronosyltransferases) and sulfate by SULTs (Sulfotransferases) at the phenol position (Dei Cas & Ghidoni, 2019).
6. Major targets of curcumin against COVID-19
Based on the above-explained mechanisms of infection pathogenesis, two major targets can be considered for the therapeutic approach - one is the lifecycle of the virus within the host cell and another part is the ability of the innate immune system to defend against infection.
6.1. Targeting virus lifecycle within the host cell
Combating viral infection, viral entry and its replication within the host cell are the most important events and have always been a challenge. While non-enveloped viruses (e.g. Adenovirus, Poliovirus, Rotavirus, Hepatitis A virus, Coxsackievirus) can directly or indirectly penetrate the plasma membrane, an enveloped virus (e.g. Hepatitis C virus, Rubella virus, HIV, Ebola virus, Influenza virus, Coronavirus, etc.) requires the fusion of the viral envelope with a cellular membrane (Thorley et al., 2010).
6.1.1. Inhibition of viral attachment with the host cell
Evidences suggest the mechanism behind curcumin-induced inhibition of enveloped virus entry into a host cell. Several in vitro experiments have been conducted to explore the role of curcumin and its analogues on viral entry inhibition (Anggakusuma et al., 2014, Mathew and Hsu, 2018). For instance, membrane fluidity experiments indicate that curcumin impairs both viral binding and fusion in the case of HCV and thus inhibit cell-to-cell transmission independent of the viral genotype (Anggakusuma et al., 2014). Also, curcumin has a direct effect on viral particle infectivity redirected by the inhibition of haemagglutination in H1N1 as well as in H6N1 subtype and treatment with 30 μM curcumin reduced more than 90% virus yield (Praditya et al., 2019a). Consistently, both the zika and chikungunya virus was responded to 5 µM curcumin without effecting the cellular viability. In a dose- and time-dependent manner, curcumin can reduce the infectivity of the enveloped viruses without disturbing the integrity of the viral RNA (Mounce et al., 2017).
Recently literature revealed that curcumin holds the better binding capability to the ACE2 receptors and may hinder the entry of the COVID‐19 virus (Zahedipour et al., 2020). In-vivo studies revealed that curcumin derivatives at a comparatively low dose can decrease the level of Ang II thereby upregulate ACE2 protein (Pang et al., 2015, Xu et al., 2018). Also, scientists have tried to explore the viral entry inhibition efficiency of curcumin using an insilico simulation study. It has been found that curcumin can bind to the human ACE2 and receptor-binding domain of the viral S-protein with a binding energy of −7.8 Kcal/mol and −7.9 Kcal/mol respectively (Jena, 2019). Later, it has been found that keto and enol form of curcumin interacts (binding energy −20.753 Kcal/mol and −16.08067 Kcal/mol respectively) with the key residues (Q493, N501, Y505, Y489 and Q498) of the receptor-binding motif (RBM) in the spike glycoprotein (Shanmugarajan et al., 2020). Also, curcumin may play important role in ADAM17 inhibition (Borah et al., 2016). Recently curcumin possesses a strong binding affinity (19.86 kJ/mol) with TMPRSS2 (Motohashi et al., 2020).
6.1.2. Inhibition of endocytic pathway within the host cell
To date, no shreds of evidence were reported on the direct effect of curcumin on the viral genome released into the host cell. But there is plenty of literature that supported the impact of curcumin on lysosomal function (Rainey et al., 2020, Zhang et al., 2016). Studies found that curcumin induces endoplasmic reticulum stress which cause an unfolded protein response and Ca2+ release thereby enhances apoptosis through destabilizing the mitochondrial compartment in the cancerous cell (Moustapha et al., 2015, Sala de Oyanguren et al., 2020). Besides this, lysosomal membrane permeabilization is involved in curcumin-induced programmed cell death in the lung cancer cell, where curcumin pretreatment causes lysosome rupture and subsequent release of several lysosomal proteins and this cytosolic relocation of lysosomal proteins leads to cellular apoptosis (Chen et al., 2012). But apoptosis seems to be a “devil” in the case of cytopathic virus like SARS-CoV-2 because this phenomenon causes extreme inflammation leading to vascular leakage (Tay et al., 2020). In the non-cancerous cell, curcumin can stabilize the pathogen containing vacuolar membrane and prevents the escape of cytosolic pathogens, thereby inhibit the fusion with lysosomes (Marathe et al., 2012). Emerging evidence indicates the function of curcumin may vary depending on the cellular environment. In the case of COVID-19, curcumin may block autophagosomes-lysosome fusion by stabilizing pathogen containing vacuole and restrict genome replication.
6.1.3. Inhibition of viral genome expansion into host cell
Previous literature suggested that curcumin plays important role in hindering viral genome expansion through several mechanisms either within virus particle or host cells. For instance, curcumin was found to have an inhibitory effect against the Hepatitis B virus (HBV). It has been observed that PGC1-α, co-activator of HBV transcription, was downregulated by curcumin treatment (Mouler Rechtman et al., 2010). On the other hand, curcumin downregulates HCV gene expression via suppression of the Akt-SREBP-1 activation (Kim et al., 2010). However, curcumin can potentially inhibit HBV gene replication via down-regulation of covalently closed circular DNA-bound histone acetylation (Wei et al., 2017). A comprehensive analysis suggested that curcumin inhibits HIV-1 and HIV-2 proteases at concentration IC50 100 µM and 250 µM respectively by substrate binding activity while HIV-1 integrase was inhibited at concentration IC50 40 µM. In addition to this, curcumin also hinders transcription elongation by inhibiting the Tat protein of HIV-1 (Ali and Banerjea, 2016, Kumari et al., 2015, Prasad and Tyagi, 2015). Recently an insilico analysis revealed that curcumin has a potential binding affinity with Ebola viral protein through which viral entry and replication might be inhibited (Noor et al., 2018).
Recently some molecular docking studies reported that curcumin can possess an inhibitory effect on viral protein translation by binding with the active sites of 3CLPro/chymotrypsin-like protease (NSP3) and PLPro/papain-like protease (NSP5) (Das et al., 2020, Linda Laksmiani et al., 2020).
6.2. Role of curcumin in immunomodulation
Apart from targeting the viral lifecycle, the immune system plays an important role against infection. The role of curcumin as an immune-modulator against a plethora of viral and bacterial infection has been extensively studied (Lai et al., 2020, Mathew and Hsu, 2018, Sordillo and Helson, 2015). Several mechanisms of cellular immunity against pathogen have been observed.
6.2.1. Autophagy and apoptosis
Autophagy and apoptosis are two major interconnected host cell responses to viral infection. Autophagy is a catabolic process that degrades and recycles cytosolic materials while apoptosis is a highly regulated form of cell death in which the cell contains the necessary information to die on its own (Mehrbod et al., 2019). Earlier kinds of the literature confirmed that curcumin can induce G2/M cell cycle arrest, autophagy, stimulate apoptosis, and interrupt molecular signalling (Rainey et al., 2020). Several in-vitro studies also confirmed that curcumin causes apoptosis by blocking the PI3K-Akt pathway (Kuttikrishnan et al., 2019), by inhibiting the NF-kB activation in tumour cells and releasing cytochrome c (Araveti & Srivastava, 2019).
Interestingly, these two processes play a negative role in viral infection. For example, autophagy stimulates the replication of Influenza A virus (Wang et al., 2018), HIV-1 and HIV disease progression (Killian, 2012). Also, autophagy is involved in antigen preparation and presentation (Crotzer & Blum, 2009) which produce viral antigen on the cell surface and eventually triggers T-cell mediated signalling pathway. Therefore, inhibition of normal autophagy and apoptosis can be the target to combat viral infection. In this favour, curcumin potentially inhibits cellular apoptosis and autophagy by inhibiting caspase-3 activation (Li et al., 2017), inducing the expression of Bcl‑2 and inhibiting the expression levels of Bax, beclin‑1, BNIP3 and SIRT1 thereby show a protective effect against hypoxia/reoxygenation (HUANG et al., 2015), uncontrolled tissue damage (Zhao et al., 2017).
6.2.2. TLRs activation
Activation of Toll‐like receptors (TLRs) play a pivotal role in the activation of innate immune response and inflammation. It has been found that curcumin can inhibit extracellular TLR2, TLR4 and intracellular TLR9 (Boozari et al., 2019). Curcumin attenuates activation of TLR4 acting directly on the receptor or by its downstream pathway and thus suppress the release of cytokines and chemokines (Panaro et al., 2020). In the case of Influenza A virus, curcumin can inhibit virus-induced activation of TLR2 and TLR4 thereby block the NF-kB signalling pathway (Dai et al., 2018). A mouse model experiment revealed that post-infection intravenous treatment of 200 mg/kg of body weight can significantly reduce pro-inflammatory cytokines level and augment anti-inflammatory cytokines by inhibiting TLR2, TLR4 and TLR9 (Abo-Zaid et al., 2020). However, another study suggested that curcumin may play an inhibitory role against the activation of TLR7 (Lai et al., 2017) thereby reducing the production of INF-α and TNF-α. In this regards, curcumin can inhibit ROS generation by increasing glutathione (GSH) activity as well as modulating several signalling pathways like PPARγ, JNK, NF-kB and Nrf2 (Kim et al., 2020, Liu and Ying, 2020).
6.2.3. T-cell Regulation
Regulation of T-cell is one of the most important mechanisms in cellular immunity, which can be modulated by curcumin and its derivatives (Bhattacharyya et al., 2010, Chai et al., 2020, Kliem et al., 2012). Curcumin thwarts T cell-activation-induced Ca2+ mobilization with IC50 of 12.5 µM and thereby impedes Nuclear Factor of Activated T Cells (NFAT) activation and NFAT-regulated cytokine expression (Kliem et al., 2012). Besides this, curcumin abolishes CD2/CD3/CD28-initiated T-helper cell activation by constraining cell proliferation, differentiation and cytokine production. Also, a spontaneous decline of CD69 expression and upregulation of CCR7, L-selectin, and TGF-β1 was observed (Kim et al., 2013). This may lead to inhibit the suppressive activity of T-regulatory cells (Bhattacharyya et al., 2010). A more recent study revealed that curcumin can ease the grade of severity of ALI/ARDS and unrestrained inflammation by promoting the delineation of immature CD4+ T cells to CD4+ CD25+ FOXP3+ Treg (T-regulatory) cells (Chai et al., 2020).
6.2.4. Cytokine regulation
Viral infection is mostly resulted in a discrepancy in pro-and anti-inflammatory cytokines as a part of innate immunity activation to eliminate viruses. A large number of evidence suggest that curcumin potentially regulate cytokine release (Bereswill et al., 2010, Mathew and Hsu, 2018, Sordillo and Helson, 2015). Curcumin inhibited the production of IL-8, MIP-1α, MCP-1, IL-1β, and TNF-α by PMA- or LPS-stimulated monocytes and alveolar macrophages in a concentration- and a time-dependent manner (Mathew & Hsu, 2018) while an integrated analysis revealed that five asymmetric mono-carbonyl analogues of curcumin can hinder the release of TNF-α and IL-6 in a dose-dependent pattern (Liang et al., 2014). It has been shown that curcumin significantly increases the level of anti-inflammatory cytokine IL-10 and decrease pro-inflammatory cytokine expression in the mouse intestine (Bereswill et al., 2010). Another study found that IL‐10 concentrations were significantly increased in curcumin-treated (average concentration of 200 mg/kg) hepatic cirrhosis mouse when compared to the non-treated group (Abo-Zaid et al., 2020). Decreased serum level of TNF-α and IL-6 was also found in rat with acute renal injury after treatment of curcumin at a concentration of 100 µg/g body weight (Zhu et al., 2017). A structure–activity relationship coupled with a molecular docking study showed that most of the curcumin derivatives effectively inhibit H1N1 neuraminidase activity. Along with this, significant attenuation of pro-inflammatory cytokines (TNF-α, IL-6 and IFN-γ) levels was observed in mice after curcumin treatment (Lai et al., 2020). Even so, a recent study on the mouse with an acute lung infection (ALI/ARDS) suggested that IL-17A, MPO-producing neutrophils decreased after pretreatment with 50 µL of curcumin at a concentration of 20 mg/mL (Chai et al., 2020). Also, curcumin-stabilized silver nanoparticles (Cur-AgNP) significantly reduced HIV replication within the host cell by inhibition of pro-inflammatory cytokines level (Sharma et al., 2017). However, a mouse model study revealed that curcumin potentially decreases the systematic as well as local myocardial expression of pro-inflammatory cytokines (TNF- α, IL-6, IL-1β) resulting in inhibition of NF-kB activation in Coxsackievirus infection (Song et al., 2013). In contrast to this, a reduction of only local tissue inflammation was found in case of HSV-2 infection resulting decrease the risk of HIV acquisition in the female genital tract of women (Vitali et al., 2020). Furthermore, Sordillo & Helson (2015) also showed a number of evidences regarding the action of curcumin against cytokine storm. They found that curcumin can effectively block the release of pro-inflammatory cytokines and thereby can be suggested as useful for treatment of Ebola patients.
6.2.5. Complement system
Curcumin significantly inhibit the classical pathway and zymosan-induced activation of the alternate pathway of the complement system in a dose-dependent manner (Kulkarni, 2005). In support of this, curcumin is also shown to decrease C3 activity with increase IgG and IgM, thereby trigger humoral immunity (Çiftçi, 2011).
7. Potential role of curcumin in COVID-19 management
Since mid-December of 2019, the pandemic COVID-19 has been spreading exponentially and shook the global health scenario. The symptoms of SARS-CoV-2 infection develop ARDS by manifesting Acute Lung Infection (ALI) which was also found in previous infections like SARS-CoV, MERS-CoV, H5N1, H7N9. However, ‘cytokine storm’ is the common phenomenon that plays a crucial role in the development and progression of fatal pneumonia (Liu and Ying, 2020, Mehta et al., 2020). Hence, apart from the inhibition of viral entry into the host cell, targeting the regulation of the cytokine is one of the most important mechanisms of combating COVID-19 infection. Many studies have highlighted the usefulness of herbal medicines like Phytoextracts, fruit extracts, traditional spices, etc. in the improvement of severe illness. Curcumin is one of the extensively studied compounds that has already claimed for its benefits in human health and disease prevention. Supplementary Table 1 shows that curcumin can potentially block pathogen entry into host cell irrespective of the presence of envelope, along with inhibiting the viral genome expansion while it can also inhibit virion release mechanisms in some cases. Comparing with the available literature, it has been suggested that CoVs infection can be prevented by inhibition of viral entry, viral genome release and replication within the host cell. In addition to this, curcumin induced immunomodulation plays an important role in viral infection. The present review portraits the possible underlying molecular aspects of the disease and how curcumin can play role in the prophylaxis of COVID-19 (Fig. 2).
While discussing the role of curcumin on COVID-19 treatment, insilico studies play important role in understanding the possible mechanism. Recently multi-omics based study has identified curcumin as a candidate prophylactic agent (Barh et al., 2020). Curcumin possesses the binding efficiency to the RBD of the viral spike protein as well as human ACE2 causing the blockage of ACE2 receptor resulting in inhibition of the viral attachment with the host cell (Jena, 2019, Shanmugarajan et al., 2020). Also, curcumin potentially binds to TMPRSS2 and ADAM17 leading to mispriming of the S-protein of the virus and hACE2 (Borah et al., 2016, Motohashi et al., 2020). This possibly inhibits the membrane fusion process of viral entry into the lung/alveolar epithelial cells. In addition to this, curcumin can potentially increase soluble ACE2 protein which may competitively bind with SARS-CoV-2 not only to neutralize the virus but also rescue cellular ACE2 activity which negatively regulates the renin-angiotensin system (RAS) to protect the lung from injury (Zhang et al., 2020). Not only entry inhibition, but curcumin can also impair the endosome-lysosomal fusion process by endosomal membrane stabilization (Marathe et al., 2012) and lysosomal membrane permeabilization (Chen et al., 2012) that eventually blocks the viral genome released into the host cell and prevent replication. Insilico simulation suggests the binding efficiency of curcumin with the NSP3 and NSP5 may affect the RdRP activity of viral genome replication and DMV formation (Astuti and Ysrafil, 2020, Das et al., 2020, Linda Laksmiani et al., 2020, Soni et al., 2020). Also, impairment of the PLpro activity eventually protects the host cell environment by blocking NSP2 function and prevents the endosomal escape of essential non-structural proteins (Astuti and Ysrafil, 2020, Yoshimoto, 2020). Hence, it can be proposed that curcumin may inhibit several stages of the viral lifecycle within the host cell.
Earlier pieces of literature have already established the fact that curcumin acts as an immunomodulatory agent against viral infections and other inflammatory diseases. It was found that curcumin binds to a series of enzymes (carbonyl reductase, glutathione-S-transferase, glyoxalase, etc.) involved in reactive oxygen species (ROS) metabolic pathway and upregulate their expression (Larasati et al., 2018). In oxidative stress condition, curcumin decreased malondialdehyde and nitric oxide levels by increasing thiol, superoxide dismutase, and catalase levels (Memarzia et al., 2021). In the case of COVID-19, curcumin-induced inhibition of the autophagolysosome fusion may disturb MHC class-II antigen preparation and processing in the infected alveolar/lung epithelial cells. Thus, CD4+ T cells cannot be recognized by the APC (dendritic cell) thereby become unsuccessful to differentiate into T-helper cell. This inactivation of T cells is one of the primary mechanism behind the inhibition of pro-inflammatory cytokines release. Apart from this, curcumin interferes with TLRs leading to the NF-kB signalling pathway (Kim et al., 2020, Liu and Ying, 2020). This also activates IFN-1 signalling cascades that restricts viral genome replication and viral entry into the T cells. Overall, curcumin pro‐inflammatory cytokine interleukin 4 (IL)‐4, transforming growth factor‐beta, IL‐17, interferon‐gamma levels, and type 1/type 2 helper cells (Th1)/(Th2) ratio in conditions with disturbance in the immune system (Memarzia et al., 2021). Recently an integrated insilico simulation study revealed that hydrazinocurcumin act as a warrior against COVID-19 by altering the cytokine regulation (H. Noor et al., 2021). The immunomodulatory role of curcumin against this infection understood so far, supports its potential prophylactic use, which either directly inactivates T-cell or regulate cytokine production.
The present review attempted to understand the molecular mechanism of COVID-19 pathogenesis. A comparative analysis has been performed (Table 2 ) considering several viral diseases and highlighted the dose-dependent response of curcumin in multiple test system (in vitro, in vivo and human population-based studies). It has been suggested that pre-treatment of curcumin may regulate lysosomal functions in a dose-dependent manner through ER/calcium/mitochondrial pathway (Rainey et al., 2020). While most of the studies showed that curcumin can inhibit viral transcription when treated after exposure, some studies indicate that it can regulate cytokine release in case of pre-exposure treatment. However, a major criticism of curcumin has been raised regarding its poor bioavailability. Though dose-escalating studies have indicated the safety of curcumin at doses as high as 12 g/day over 3 months (Gupta et al., 2013, Gupta et al., 2013), curcumin possesses very poor bioavailability and often found in undetectable concentrations in blood and extra intestinal tissue due to its chemical instability, rapid metabolism, poor absorption and rapid systemic elimination (Dei Cas and Ghidoni, 2019, Lopresti, 2018, Prasad et al., 2014, Prasad et al., 2014). Since most of the studies revealed the oral delivery of curcumin, it has been suggested that subcutaneous treatment of formulated curcumin provide effective and sustained tissue concentration (Prasad et al., 2014). In addition to this, it has been found that different novel delivery systems such as solid lipid particle, micellar system, or hydrophilic nanoparticles could increase curcumin concentration up to 15–20 fold (Dei Cas & Ghidoni, 2019).
Table 2.
The dose-dependent response of curcumin in multiple test system (in vitro, in vivo and population based studies).
| Study system | Dose | Time of administration | Route of administration | Mechanism of action | References | |
|---|---|---|---|---|---|---|
| In vitro study | ||||||
| Human Huh-7 cells | 10–20 µM | Pre- transfection | NA | Activates lysosomal destabilization, induces apoptosis | Oyanguren et al., 2020 | |
| HeLa, BHK-21, and Vero-E6 cells | 5 µM | Pre-transfection | NA | Reduce the infectivity of the Chikungunya and Zika viruses without disturbing the integrity of the viral RNA | Mounce et al., 2017 | |
| HepG2, HepG2.2.15 and HEK293 cells | 50–150 μM | Pre-transfection | NA | Inhibits HBV transcription by downregulation of PGC1-α | Mouler Rechtman et al., 2010 | |
| Human Huh-7 cells | 5–15 mM | Pre-transfection | NA | Inhibits HCV gene expression by suppressing Akt-SREBP-1 activation | Kim et al., 2010 | |
| PBMC cell line | IC50 100 µM | Post-transfection | NA | Attenuates viral replication by inhibiting HIV-1 protease | Kumari et al., 2015, Prasad and Tyagi, 2015 | |
| IC50 250 µM | Post-transfection | NA | Attenuates viral replication by inhibiting HIV-2 protease | |||
| IC50 40 µM | Post-transfection | NA | Attenuates viral replication by inhibiting HIV-1 integrase | |||
| HEK-293 T cells | 80 µM | Post-transfection | NA | Inhibits HIV-1 virus transcription by promoting Tat protein degradation | Ali & Banerjea, 2016 | |
| HepG2.2.15 cells | 20 μmol/L | Post-transfection | NA | Inhibits HBV gene replication via down-regulation of cccDNA-bound histone acetylation | Wei et al., 2017 | |
| PBMC cells | IC50 12.5 µM | Post-transfection | NA | Inhibit T cell activation by blocking Ca2+ mobilization and NFAT activation | Kliem et al., 2012 | |
| PBMC cells | 20 µg/ml | Post- transfection | NA | Decline of CD69 expression and upregulation of CCR7, L-selectin, TGF-β1; attenuates T cell activation | Kim et al., 2013 | |
| In vivo study | ||||||
| ALI/ARDS Mouse | 50 µL (20 mg/ml) | Pre-infection | Intra-peritonial | IL-17A ↓, MPO-producing neutrophils ↓, NF-κB p65↓ in lungs, IL-10 ↑ | Chai et al., 2020 | |
| Mice with liver injury | 200 mg/kg body weight | Post- infection | Intra-venous | Reduce pro-inflammatory cytokines level and augment anti-inflammatory cytokines by inhibiting TLR2, TLR4 and TLR9 | Abo-Zaid et al., 2020 | |
| H1N1 infected mouse | 25 mg/kg body weight /day | Post-infection | Intra-peritonial | TNFα↓, IL-6↓, INF-γ↓ | Lai et al., 2020 | |
| Rat with acute kidney injury | 100 mg/kg body weight | Post-infection | Intra-peritonial | TNFα↓, IL-6↓ | Zhu et al., 2017 | |
| Human population study | ||||||
| Healthy male | 500 mg/3days | Pre-exercise | Oral intake | IL-6↓ | Sciberras et al., 2015 | |
| Osteoarthritis patients | 80 mg daily | Intervention | Oral intake | Decrease the level of C-reactive protein (CRP), CD4 + and CD8 + T cells, Th17 cells and B cells frequency | Atabaki et al., 2020 | |
| COVID-19 patient | 160 mg curcumin daily for 2 weeks | Intervention | Oral intake (nano-micelles) | IL-6↓, IL-1β↓ | Hassaniazad et al., 2020, Saber-Moghaddam et al., 2021, Valizadeh et al., 2020 | |
| 1 g curcumin daily for 2 weeks | Intervention | Oral intake (Curcumin-piperine capsule) | Trial continue | Miryan et al., 2020 | ||
Recent clinical trial showed that daily four times oral intake of 40 mg nanocurcumin in micelles formation for 2 weeks can significantly decrease IL-6 and IL-1β expression and secretion in COVID-19 patients (Hassaniazad et al., 2020, Saber-Moghaddam et al., 2021, Valizadeh et al., 2020). However, another trial of curcumin-piperine co-supplementation against COVID-19 is still being continued (Miryan et al., 2020). Thimmulappa et al., (2021) suggested the nasal delivery of liposomal curcumin may inhibit SARS-CoV-2 infectivity by deposition of curcumin in the lower airways at a higher concentration and consequently mitigate pulmonary inflammation and the progression to ARDS. In summary, accumulated evidences suggested that curcumin can be effective against SARS-CoV-2 as supportive therapy. Table 2 provides a systemic representation of effective curcumin dose and hence can be utilized as a reference frame for validation in future.
8. Conclusion
While the entire world is putting efforts into the discovery of a vaccine against coronavirus infection, the antiviral potential of curcumin against SARS-CoV-2 shows a promising role in COVID-19 management. Since ancient time, curcumin possesses a multifaceted role in several disease management. As an antiviral agent, existing literature suggested that targeting viral lifecycle and cellular responses are important strategies to combat viral infection. The present review emphasized the molecular mechanism of host-pathogen interaction and subsequent immune response in the host. In this background, curcumin can potentially inhibit the SARS-CoV-2 entry within human mainly by blocking the ACE2 receptor, hinder viral genome replication by altering viral non-structural protein activity. Not only viral entry, curcumin indeed can prohibit ‘cytokine storm’-induced multi-organ failure by constraining the inflammatory response and other cellular immune response. However, having such potency, it can be suggested that curcumin must be administered as solid lipid particle, micellar system, or hydrophilic nanoparticles due to its poor bioavailability. Hence, further studies warrant detailed dose–response analysis for validation in the large-scale human population.
Ethical statement
This material is the authors' own original work, which has not been previously published elsewhere.
The paper is not currently being considered for publication elsewhere.
The paper reflects the authors' own research and analysis in a truthful and complete manner.
The paper properly credits the meaningful contributions of co-authors and co-researchers.
The results are appropriately placed in the context of prior and existing research.
All sources used are properly disclosed (correct citation). Literally copying of text must be indicated as such by using quotation marks and giving proper reference.
All authors have been personally and actively involved in substantial work leading to the paper, and will take public responsibility for its content.
Credit Author Statement
SD: Literature search, Manuscript writing, Figure and table preparing; PB: Supervision, Manuscript writing and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors are thankful to Arijit Reeves, MSc student, Department of Environmental Science, University of Calcutta for helping in the VOSviewer analysis process and Ankita Das, Research scholar, Department of Environmental Science, University of Calcutta for putting necessary inputs to enrich the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jff.2021.104503.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary Fig. 1.
VOSviewer analysis of novelty screening based on published literatures.
The role of curcumin against viral disease management.
References
- Abo-Zaid M.A., Shaheen E.S., Ismail A.H. Immunomodulatory effect of curcumin on hepatic cirrhosis in experimental rats. Journal of Food Biochemistry. 2020:e13219. doi: 10.1111/jfbc.13219. [DOI] [PubMed] [Google Scholar]
- Ahmad R.S., Hussain M.B., Sultan M.T., Arshad M.S., Waheed M., Shariati M.A.…Hashempur M.H. Biochemistry, Safety, Pharmacological Activities, and Clinical Applications of Turmeric: A Mechanistic Review. Evidence-Based Complementary and Alternative Medicine. 2020;2020:1–14. doi: 10.1155/2020/7656919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A., Banerjea A.C. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Scientific Reports. 2016;6:1–9. doi: 10.1038/srep27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalraj A., Pius A., Gopi S., Gopi S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives – A review. Journal of Traditional and Complementary Medicine. 2017;7(2):205–233. doi: 10.1016/j.jtcme.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggakusuma, Colpitts C.C., Schang L.M., Rachmawati H., Frentzen A., Pfaender S.…Steinmann E. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2014;63(7):1137–1149. doi: 10.1136/gutjnl-2012-304299. [DOI] [PubMed] [Google Scholar]
- Atabaki M., Shariati-Sarabi Z., Tavakkol-Afshari J., Mohammadi M. Significant immunomodulatory properties of curcumin in patients with osteoarthritis; a successful clinical trial in Iran. International Immunopharmacology. 2020;85 doi: 10.1016/j.intimp.2020.106607. [DOI] [PubMed] [Google Scholar]
- Araveti P.B., Srivastava A. Curcumin induced oxidative stress causes autophagy and apoptosis in bovine leucocytes transformed by Theileria annulata. Cell Death Discovery. 2019;5(1):100. doi: 10.1038/s41420-019-0180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barh D., Tiwari S., Weener M.E., Azevedo V., Góes-Neto A., Gromiha M.M., Ghosh P. Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19. Computers in Biology and Medicine. 2020;126:104051. doi: 10.1016/j.compbiomed.2020.104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereswill S., Muñoz M., Fischer A., Plickert R., Haag L.-M., Otto B.…Heimesaat M.M. Anti-Inflammatory Effects of Resveratrol, Curcumin and Simvastatin in Acute Small Intestinal Inflammation. PLoS ONE. 2010;5(12):e15099. doi: 10.1371/journal.pone.0015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Md Sakib Hossain D., Mohanty S., Sankar Sen G., Chattopadhyay S., Banerjee S.…Sa G. Curcumin reverses T cell-mediated adaptive immune dysfunctions in tumor-bearing hosts. Cellular and Molecular Immunology. 2010;7(4):306–315. doi: 10.1038/cmi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 envelope and membrane proteins: Structural differences linked to virus characteristics? BioMed Research International. 2020;1–6 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess M., Kaiser L., Sauer M., Csuk R., Deigner H.-P. COVID-19/SARS-CoV-2 Infection: Lysosomes and Lysosomotropism Implicate New Treatment Strategies and Personal Risks. International Journal of Molecular Sciences. 2020;21(14):4953. doi: 10.3390/ijms21144953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozari M., Butler A.E., Sahebkar A. Impact of curcumin on toll-like receptors. Journal of Cellular Physiology. 2019;234(8):12471–12482. doi: 10.1002/jcp.28103. [DOI] [PubMed] [Google Scholar]
- Borah P.K., Chakraborty S., Jha A.N., Rajkhowa S., Duary R.K. In silico approaches and proportional odds model towards identifying selective ADAM17 inhibitors from anti-inflammatory natural molecules. Journal of Molecular Graphics and Modelling. 2016;70:129–139. doi: 10.1016/j.jmgm.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduction and Targeted Therapy. 2020;5(1):84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Chen Y., Lin S., Xie K., Wang C., Yang Y., Xu F. Curcumin regulates the differentiation of naïve CD4+T cells and activates IL-10 immune modulation against acute lung injury in mice. Biomedicine & Pharmacotherapy. 2020;125:109946. doi: 10.1016/j.biopha.2020.109946. [DOI] [PubMed] [Google Scholar]
- Chen Q.-Y., Shi J.-G., Yao Q.-H., Jiao D.-M., Wang Y.-Y., Hu H.-Z.…Wu L.-J. Lysosomal membrane permeabilization is involved in curcumin-induced apoptosis of A549 lung carcinoma cells. Molecular and Cellular Biochemistry. 2012;359(1–2):389–398. doi: 10.1007/s11010-011-1033-9. [DOI] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., Qian C., Yang Y., You R.…Ye L. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cellular and Molecular Immunology. 2020 doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. Journal of Medical Virology. 2020;92(10):2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaglia E., Vecchione C., Puca A.A. COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children. Frontiers in Pediatrics. 2020;8 doi: 10.3389/fped.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiftçi O. Curcumin prevents toxic effects of 2,3,7,8-tetrachlorodibenzo- p -dioxin (TCDD) on humoral immunity in rats. Food and Agricultural Immunology. 2011;22(1):31–38. doi: 10.1080/09540105.2010.517308. [DOI] [Google Scholar]
- Crotzer V.L., Blum J.S. Autophagy and Its Role in MHC-Mediated Antigen Presentation. The Journal of Immunology. 2009;182(6):3335–3341. doi: 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Gu L., Su Y., Wang Q., Zhao Y., Chen X.…Li K. Inhibition of curcumin on influenza A virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-κB pathways. International Immunopharmacology. 2018;54:177–187. doi: 10.1016/j.intimp.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics. 2020;1–11 doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dei Cas M., Ghidoni R. Dietary Curcumin: Correlation between Bioavailability and Health Potential. Nutrients. 2019;11(9):2147. doi: 10.3390/nu11092147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms, R. W. (2016). Basic Concepts. In Viral Pathogenesis (pp. 29–40). https://doi.org/10.1016/B978-0-12-800964-2.00003-3.
- Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. The FEBS Journal. 2020;287(17):3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.C., Patchva S., Aggarwal B.B. Therapeutic Roles of Curcumin: Lessons Learned from Clinical Trials. The AAPS Journal. 2013;15(1):195–218. doi: 10.1208/s12248-012-9432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.C., Sung B., Kim J.H., Prasad S., Li S., Aggarwal B.B. Multitargeting by turmeric, the golden spice: From kitchen to clinic. Molecular Nutrition & Food Research. 2013;57(9):1510–1528. doi: 10.1002/mnfr.201100741. [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Beckford J., Alachkar H. Deciphering the TCR Repertoire to Solve the COVID-19 Mystery. Trends in Pharmacological Sciences. 2020;41(8):518–530. doi: 10.1016/j.tips.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi A., Pourmasoumi M., Ghaedi E., Sahebkar A. The effect of Curcumin/Turmeric on blood pressure modulation: A systematic review and meta-analysis. Pharmacological Research. 2019;150:104505. doi: 10.1016/j.phrs.2019.104505. [DOI] [PubMed] [Google Scholar]
- Hassaniazad M., Inchehsablagh B.R., Kamali H., Tousi A., Eftekhar E., Jaafari M.R.…Nikpoor A.R. The clinical effect of Nano micelles containing curcumin as a therapeutic supplement in patients with COVID-19 and the immune responses balance changes following treatment: A structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):876. doi: 10.1186/s13063-020-04824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. Journal of Virology. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlings S., Kalman D. Curcumin: A Review of Its Effects on Human Health. Foods. 2017;6(10):92. doi: 10.3390/foods6100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Murakami M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity. 2020 doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S.…Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Ye B., Dai Z., Wu X., Lu Z., Shan P., Huang W. Curcumin inhibits autophagy and apoptosis in hypoxia/reoxygenation-induced myocytes. Molecular Medicine Reports. 2015;11(6):4678–4684. doi: 10.3892/mmr.2015.3322. [DOI] [PubMed] [Google Scholar]
- Iba T., Levy J.H., Connors J.M., Warkentin T.E., Thachil J., Levi M. The unique characteristics of COVID-19 coagulopathy. Critical Care. 2020;24(1):360. doi: 10.1186/s13054-020-03077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba T., Levy J.H., Levi M., Thachil J. Coagulopathy in COVID-19. Journal of Thrombosis and Haemostasis. 2020;18(9):2103–2109. doi: 10.1111/jth.14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarząb A., Kukula-Koch W. Recent Advances in Obesity: The Role of Turmeric Tuber and Its Metabolites in the Prophylaxis and Therapeutical Strategies. Current Medicinal Chemistry. 2019;25(37):4837–4853. doi: 10.2174/0929867324666161118095443. [DOI] [PubMed] [Google Scholar]
- Java A., Apicelli A.J., Liszewski M.K., Coler-Reilly A., Atkinson J.P., Kim A.H.J., Kulkarni H.S. The complement system in COVID-19: Friend and foe? JCI. Insight. 2020;5(15) doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena A.B. Catechin and Curcumin interact with corona (2019- nCoV / SARS-CoV2) viral S protein and ACE2 of human cell membrane : Insights from Computational study and implication for intervention CURRENT STATUS : UNDER REVIEW. Nature Research. 2019:1–19. doi: 10.21203/rs.3.rs-22057/v1. [DOI] [Google Scholar]
- Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M.…Yang J.-K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Frontiers in Public Health. 2020;8 doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X., Yan Y., Liu Q., Li N., Sheng M., Zhang L.…Jiang C. Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture. Journal of Virology. 2015;89(20):10347–10358. doi: 10.1128/JVI.01411-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelvin A.A., Halperin S. COVID-19 in children: The link in the transmission chain. The Lancet Infectious Diseases. 2020;2(20):2019–2020. doi: 10.1016/S1473-3099(20)30236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian M. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Research and Therapy. 2012;9(1):16. doi: 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G., Jang M.S., Son Y.M., Seo M.J., Ji S.Y., Han S.H.…Yun C.H. Curcumin Inhibits CD4+ T Cell Activation, but Augments CD69 Expression and TGF-β1-Mediated Generation of Regulatory T Cells at Late Phase. PLoS One. 2013;8(4):1–12. doi: 10.1371/journal.pone.0062300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Oh J.-M., Choi H., Kim S.W., Kim S.W., Kim B.G.…Lee D.C. Activation of the Nrf2/HO-1 pathway by curcumin inhibits oxidative stress in human nasal fibroblasts exposed to urban particulate matter. BMC Complementary Medicine and Therapies. 2020;20(1):101. doi: 10.1186/s12906-020-02886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim K.H., Kim H.Y., Cho H.K., Sakamoto N., Cheong J. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Letters. 2010;584(4):707–712. doi: 10.1016/j.febslet.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Kliem C., Merling A., Giaisi M., Köhler R., Krammer P.H., Li-Weber M. Curcumin suppresses T cell activation by blocking Ca 2+ mobilization and nuclear factor of activated T cells (NFAT) activation. Journal of Biological Chemistry. 2012;287(13):10200–10209. doi: 10.1074/jbc.M111.318733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A.P. Curcumin Inhibits the Classical and the Alternate Pathways of Complement Activation. Annals of the New York Academy of Sciences. 2005;1056(1):100–112. doi: 10.1196/annals.1352.007. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Nyodu, R., Maurya, V. K., & Saxena, S. K. (2020). Host Immune Response and Immunobiology of Human SARS-CoV-2 Infection. https://doi.org/10.1007/978-981-15-4814-7_5.
- Kumari N., Kulkarni A.A., Lin X., McLean C., Ammosova T., Ivanov A.…Nwulia E. Inhibition of HIV-1 by curcumin A, a novel curcumin analog. Drug Design, Development and Therapy. 2015;9:5051–5060. doi: 10.2147/DDDT.S86558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttikrishnan S., Siveen K.S., Prabhu K.S., Khan A.Q., Ahmed E.I., Akhtar S.…Uddin S. Curcumin Induces Apoptotic Cell Death via Inhibition of PI3-Kinase/AKT Pathway in B-Precursor Acute Lymphoblastic Leukemia. Frontiers in Oncology. 2019;9 doi: 10.3389/fonc.2019.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-Y., Su Y.-W., Lin K.-I., Hsu L.-C., Chuang T.-H. Natural Modulators of Endosomal Toll-Like Receptor-Mediated Psoriatic Skin Inflammation. Journal of Immunology Research. 2017;2017:1–15. doi: 10.1155/2017/7807313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yan Y., Liao S., Li Y., Ye Y., Liu N.…Xu P. 3D-quantitative structure–activity relationship and antiviral effects of curcumin derivatives as potent inhibitors of influenza H1N1 neuraminidase. Archives of Pharmacal Research. 2020:0123456789. doi: 10.1007/s12272-020-01230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larasati Y.A., Yoneda-Kato N., Nakamae I., Yokoyama T., Meiyanto E., Kato J. Curcumin targets multiple enzymes involved in the ROS metabolic pathway to suppress tumor cell growth. Scientific Reports. 2018;8(1):2039. doi: 10.1038/s41598-018-20179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Chen Y., Liu H., Jia Y., Li F., Wang W.…Zeng R. Immune dysfunction leads to mortality and organ injury in patients with COVID-19 in China: Insights from ERS-COVID-19 study. Signal Transduction and Targeted Therapy. 2020;5(1):62. doi: 10.1038/s41392-020-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Feng K., Li J., Yu D., Fan Q., Tang T.…Wang X. Curcumin Inhibits Apoptosis of Chondrocytes through Activation ERK1/2 Signaling Pathways Induced Autophagy. Nutrients. 2017;9(4):414. doi: 10.3390/nu9040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Liu Z., Wang Z., Zhang Y., Xiao B., Fang Q.…Yang S. Discovery and evaluation of asymmetrical monocarbonyl analogs of curcumin as anti-inflammatory agents. Drug Design, Development and Therapy. 2014;373 doi: 10.2147/DDDT.S58168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linda Laksmiani, N. P., Febryana Larasanty, L. P., Gde Jaya Santika, A. A., Andika Prayoga, P. A., Intan Kharisma Dewi, A. A., & Ayu Kristiara Dewi, N. P. (2020). Active Compounds Activity from the Medicinal Plants Against SARS-CoV-2 using in Silico Assay. Biomedical and Pharmacology Journal, 13(02), 873–881. https://doi.org/10.13005/bpj/1953.
- Liu Z., Ying Y. The Inhibitory Effect of Curcumin on Virus-Induced Cytokine Storm and Its Potential Use in the Associated Severe Pneumonia. Frontiers in Cell and Developmental Biology. 2020;8 doi: 10.3389/fcell.2020.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopresti A.L. The Problem of Curcumin and Its Bioavailability: Could Its Gastrointestinal Influence Contribute to Its Overall Health-Enhancing Effects? Advances in Nutrition. 2018;9(1):41–50. doi: 10.1093/advances/nmx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe S.A., Sen M., Dasgupta I., Chakravortty D. Differential Modulation of Intracellular Survival of Cytosolic and Vacuolar Pathogens by Curcumin. Antimicrobial Agents and Chemotherapy. 2012;56(11):5555–5567. doi: 10.1128/AAC.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, D., & Hsu, W. L. (2018). Antiviral potential of curcumin. Journal of Functional Foods, 40(September 2017), 692–699. https://doi.org/10.1016/j.jff.2017.12.017.
- Mehrbod P., Ande S.R., Alizadeh J., Rahimizadeh S., Shariati A., Malek H.…Ghavami S. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence. 2019;10(1):376–413. doi: 10.1080/21505594.2019.1605803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarzia, A., Khazdair, M. R., Behrouz, S., Gholamnezhad, Z., Jafarnezhad, M., Saadat, S., & Boskabady, M. H. (2021). Experimental and clinical reports on anti‐inflammatory, antioxidant, and immunomodulatory effects of <scp> Curcuma longa </scp> and curcumin, an updated and comprehensive review. BioFactors, biof.1716. https://doi.org/10.1002/biof.1716. [DOI] [PubMed]
- Miryan M., Bagherniya M., Sahebkar A., Soleimani D., Rouhani M.H., Iraj B., Askari G. Effects of curcumin-piperine co-supplementation on clinical signs, duration, severity, and inflammatory factors in patients with COVID-19: A structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):1027. doi: 10.1186/s13063-020-04924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y.-Q., Mo Q., Wang J., Deng F., Wang H., Ning Y.-J. SARS-CoV-2 nsp1: Bioinformatics, potential structural and functional features, and implications for drug/vaccine designs. Frontiers in Microbiology. 2020;11 doi: 10.3389/fmicb.2020.587317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Vanam A., Gollapudi R. In Silico Study of Curcumin and Folic Acid as Potent Inhibitors of Human Transmembrane Protease Serine 2 in the Treatment of COVID-19. INNOSC Theranostics and Pharmacological Sciences. 2020:3–9. doi: 10.36922/itps.v3i2.935. [DOI] [Google Scholar]
- Mouler Rechtman M., Har-Noy O., Bar-Yishay I., Fishman S., Adamovich Y., Shaul Y.…Shlomai A. Curcumin inhibits hepatitis B virus via down-regulation of the metabolic coactivator PGC-1α. FEBS Letters. 2010;584(11):2485–2490. doi: 10.1016/j.febslet.2010.04.067. [DOI] [PubMed] [Google Scholar]
- Mounce B.C., Cesaro T., Carrau L., Vallet T., Vignuzzi M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral Research. 2017;142:148–157. doi: 10.1016/j.antiviral.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Moustapha A., Pérétout P., Rainey N., Sureau F., Geze M., Petit J.-M.…Petit P. Curcumin induces crosstalk between autophagy and apoptosis mediated by calcium release from the endoplasmic reticulum, lysosomal destabilization and mitochondrial events. Cell Death Discovery. 2015;1(1):15017. doi: 10.1038/cddiscovery.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K.M., Dahlin J.L., Bisson J., Graham J., Pauli G.F., Walters M.A. The Essential Medicinal Chemistry of Curcumin. Journal of Medicinal Chemistry. 2017;60(5):1620–1637. doi: 10.1021/acs.jmedchem.6b00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A., Gunasekaran S., Vijayalakshmi M.A. Article in Pharmacognosy Research · October 2017. Pharmacognosy Research. 2018;10(October):24–30. doi: 10.4103/pr.pr. [DOI] [Google Scholar]
- Noor H., Ikram A., Rathinavel T., Kumarasamy S., Nasir Iqbal M., Bashir Z. Immunomodulatory and anti-cytokine therapeutic potential of curcumin and its derivatives for treating COVID-19 – a computational modeling. Journal of Biomolecular Structure and Dynamics. 2021;1–16 doi: 10.1080/07391102.2021.1873190. [DOI] [PubMed] [Google Scholar]
- Onofrio L., Caraglia M., Facchini G., Margherita V., De Placido S., Buonerba C. Toll-like receptors and COVID-19: A two-faced story with an exciting ending. Future Science OA. 2020;6(8):FSO605. doi: 10.2144/fsoa-2020-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L.…Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaro M.A., Corrado A., Benameur T., Paolo C.F., Cici D., Porro C. The Emerging Role of Curcumin in the Modulation of TLR-4 Signaling Pathway: Focus on Neuroprotective and Anti-Rheumatic Properties. International Journal of Molecular Sciences. 2020;21(7):2299. doi: 10.3390/ijms21072299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X.F., Zhang L.H., Bai F., Wang N.P., Garner R.E., McKallip R.J., Zhao Z.Q. Attenuation of myocardial fibrosis with curcumin is mediated by modulating expression of angiotensin II AT1/AT2 receptors and ACE2 in rats. Drug Design, Development and Therapy. 2015;9:6043–6054. doi: 10.2147/DDDT.S95333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasher, A. (2020). COVID-19: Current understanding of its pathophysiology, clinical presentation and treatment. Postgraduate Medical Journal, postgradmedj-2020-138577. https://doi.org/10.1136/postgradmedj-2020-138577. [DOI] [PMC free article] [PubMed]
- Pivari F., Mingione A., Brasacchio C., Soldati L. Curcumin and Type 2 Diabetes Mellitus: Prevention and Treatment. Nutrients. 2019;11(8) doi: 10.3390/nu11081837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praditya D., Kirchhoff L., Brüning J., Rachmawati H., Steinmann J., Steinmann E. Anti-infective properties of the golden spice curcumin. Frontiers in Microbiology. 2019;10(MAY):1–16. doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praditya D., Kirchhoff L., Brüning J., Rachmawati H., Steinmann J., Steinmann E. Anti-infective Properties of the Golden Spice Curcumin. Frontiers in Microbiology. 2019;10 doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S., Gupta S.C., Tyagi A.K., Aggarwal B.B. Curcumin, a component of golden spice: From bedside to bench and back. Biotechnology Advances. 2014;32(6):1053–1064. doi: 10.1016/j.biotechadv.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Prasad S., Tyagi A.K. Curcumin and its analogues: A potential natural compound against HIV infection and AIDS. Food and Function. 2015;6(11):3412–3419. doi: 10.1039/c5fo00485c. [DOI] [PubMed] [Google Scholar]
- Prasad S., Tyagi A.K., Aggarwal B.B. Recent Developments in Delivery, Bioavailability, Absorption and Metabolism of Curcumin: The Golden Pigment from Golden Spice. Cancer Research and Treatment. 2014;46(1):2–18. doi: 10.4143/crt.2014.46.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey N.E., Moustapha A., Petit P.X. Curcumin, a Multifaceted Hormetic Agent, Mediates an Intricate Crosstalk between Mitochondrial Turnover, Autophagy, and Apoptosis. Oxidative Medicine and Cellular Longevity. 2020;2020:1–23. doi: 10.1155/2020/3656419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber-Moghaddam N., Salari S., Hejazi S., Amini M., Taherzadeh Z., Eslami S.…Elyasi S. Oral nano-curcumin formulation efficacy in management of mild to moderate hospitalized <scp>coronavirus disease</scp> -19 patients: An open label nonrandomized clinical trial. Phytotherapy Research. 2021;ptr.7004 doi: 10.1002/ptr.7004. [DOI] [PubMed] [Google Scholar]
- Sala de Oyanguren F.J., Rainey N.E., Moustapha A., Saric A., Sureau F., O’Connor J.-E., Petit P.X. Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization. Cells. 2020;9(2):361. doi: 10.3390/cells9020361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaddar A., Gadepalli R., Nag V.L., Misra S. The enigma of low COVID-19 fatality rate in India. Frontiers in Genetics. 2020;11 doi: 10.3389/fgene.2020.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samji T. Influenza A: Understanding the viral life cycle. The Yale Journal of Biology and Medicine. 2009;82(4):153–159. http://www.ncbi.nlm.nih.gov/pubmed/20027280 Retrieved from. [PMC free article] [PubMed] [Google Scholar]
- Satarker S., Nampoothiri M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Archives of Medical Research. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialo F., Daniele A., Amato F., Pastore L., Matera M.G., Cazzola M.…Bianco A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung. 2020;198(6):867–877. doi: 10.1007/s00408-020-00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciberras J.N., Galloway S.D.R., Fenech A., Grech G., Farrugia C., Duca D., Mifsud J. The effect of turmeric (Curcumin) supplementation on cytokine and inflammatory marker responses following 2 hours of endurance cycling. Journal of the International Society of Sports Nutrition. 2015;12(1) doi: 10.1186/s12970-014-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugarajan, D., P., P., Kumar, B. R. P., & Suresh, B. (2020). Curcumin to inhibit binding of spike glycoprotein to ACE2 receptors: computational modelling, simulations, and ADMET studies to explore curcuminoids against novel SARS-CoV-2 targets. RSC Advances, 10(52), 31385–31399. https://doi.org/10.1039/D0RA03167D. [DOI] [PMC free article] [PubMed]
- Sharma, G., Volgman, A. S., & Michos, E. D. (2020). Sex Differences in Mortality from COVID-19 Pandemic: Are Men Vulnerable and Women Protected? JACC: Case Reports. https://doi.org/10.1016/j.jaccas.2020.04.027. [DOI] [PMC free article] [PubMed]
- Sharma R.K., Cwiklinski K., Aalinkeel R., Reynolds J.L., Sykes D.E., Quaye E.…Schwartz S.A. Immunomodulatory activities of curcumin-stabilized silver nanoparticles: Efficacy as an antiretroviral therapeutic. Immunological Investigations. 2017;46(8):833–846. doi: 10.1080/08820139.2017.1371908. [DOI] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Ge W., Cai H., Zhang H. Curcumin Protects Mice From Coxsackievirus B3-Induced Myocarditis by Inhibiting the Phosphatidylinositol 3 kinase/Akt/Nuclear Factor-κB Pathway. Journal of Cardiovascular Pharmacology and Therapeutics. 2013;18(6):560–569. doi: 10.1177/1074248413503044. [DOI] [PubMed] [Google Scholar]
- Soni V.K., Mehta A., Ratre Y.K., Tiwari A.K., Amit A., Singh R.P.…Vishvakarma N.K. Curcumin, a traditional spice component, can hold the promise against COVID-19? European Journal of Pharmacology. 2020;886:173551. doi: 10.1016/j.ejphar.2020.173551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordillo P.P., Helson L. Curcumin suppression of cytokine release and cytokine storm. A potential therapy for patients with Ebola and other severe viral infections. Vivo. 2015;29(1):1–4. [PubMed] [Google Scholar]
- Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: Perspectives on Innate Immune Evasion. Frontiers in Immunology. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nature Reviews Immunology. 2020 doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa R.K., Mudnakudu-Nagaraju K.K., Shivamallu C., Subramaniam K.J.T., Radhakrishnan A., Bhojraj S., Kuppusamy G. Antiviral and immunomodulatory activity of curcumin: A case for prophylactic therapy for COVID-19. Heliyon. 2021;7(2):e06350. doi: 10.1016/j.heliyon.2021.e06350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley J.A., McKeating J.A., Rappoport J.Z. Mechanisms of viral entry: Sneaking in the front door. Protoplasma. 2010;244(1–4):15–24. doi: 10.1007/s00709-010-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., & Thiel, V. (2020). Coronavirus biology and replication: implications for SARS-CoV-2. Nature Reviews Microbiology. https://doi.org/10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M.…Laserson U. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valizadeh H., Abdolmohammadi-vahid S., Danshina S., Ziya Gencer M., Ammari A., Sadeghi A.…Ahmadi M. Nano-curcumin therapy, a promising method in modulating inflammatory cytokines in COVID-19 patients. International Immunopharmacology. 2020;89:107088. doi: 10.1016/j.intimp.2020.107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali D., Bagri P., Wessels J.M., Arora M., Ganugula R., Parikh A.…Kaushic C. Curcumin can decrease tissue inflammation and the severity of HSV-2 infection in the female reproductive mucosa. International Journal of Molecular Sciences. 2020;21(1) doi: 10.3390/ijms21010337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wal P., Saraswat N., Pal R.S., Wal A., Chaubey M. A Detailed Insight of the Anti-inflammatory Effects of Curcumin with the Assessment of Parameters, Sources of ROS and Associated Mechanisms. Open Medicine Journal. 2019;6(1):64–76. doi: 10.2174/1874220301906010064. [DOI] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Research. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhu Y., Zhao J., Ren C., Li P., Chen H.…Zhou H. Autophagy Promotes Replication of Influenza A Virus In Vitro. Journal of Virology. 2018;93(4) doi: 10.1128/JVI.01984-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu W., Hu G., Xia S., Sun Z., Liu Z.…Lu L. RETRACTED ARTICLE: SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cellular & Molecular Immunology. 2020 doi: 10.1038/s41423-020-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wei Z.-Q., Zhang Y.-H., Ke C.-Z., Chen H.-X., Ren P., He Y.-L.…Meng Z.-J. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World Journal of Gastroenterology. 2017;23(34):6252. doi: 10.3748/wjg.v23.i34.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen C.B., Tilton J.C., Doms R.W. HIV: Cell Binding and Entry. Cold Spring Harbor Perspectives in Medicine. 2012;2(8):a006866. doi: 10.1101/cshperspect.a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Cai Y., Yu Y. Effects of a novel curcumin derivative on the functions of kidney in streptozotocin-induced type 2 diabetic rats. Inflammopharmacology. 2018;26(5):1257–1264. doi: 10.1007/s10787-018-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Helenius A. Virus entry at a glance. Journal of Cell Science. 2013;126(6):1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- Yoshimoto F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein Journal. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedipour F., Hosseini S.A., Sathyapalan T., Majeed M., Jamialahmadi T., Al-Rasadi K.…Sahebkar A. Potential effects of curcumin in the treatment of <scp>COVID</scp> -19 infection. Phytotherapy Research. 2020;ptr.6738 doi: 10.1002/ptr.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Medicine. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang J., Xu J., Lu Y., Jiang J., Wang L.…Xia D. Curcumin targets the TFEB-lysosome pathway for induction of autophagy. Oncotarget. 2016;7(46):75659–75671. doi: 10.18632/oncotarget.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lan Y., Sanyal S. Membrane heist: Coronavirus host membrane remodeling during replication. Biochimie. 2020;179:229–236. doi: 10.1016/j.biochi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Gu Q., Xiang L., Dong X., Li H., Ni J.…Chen G. Curcumin inhibits apoptosis by modulating Bax/Bcl-2 expression and alleviates oxidative stress in testes of streptozotocin-induced diabetic rats. Therapeutics and Clinical Risk Management. 2017;13:1099–1105. doi: 10.2147/TCRM.S141738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Zhang C., Weng Q., Ye B. Curcumin protects against acute renal injury by suppressing JAK2/STAT3 pathway in severe acute pancreatitis in rats. Experimental and Therapeutic Medicine. 2017;14(2):1669–1674. doi: 10.3892/etm.2017.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The role of curcumin against viral disease management.