Abstract
Introduction
Diabetic foot infections (DFIs) are among the most severe complications of diabetes. The aim of this study was to determine the etiological pathogens of DFIs in different Wagner’s and IDSA/IWGDF grades, and to assess their antimicrobial susceptibility pattern together with molecular characterization of antibiotic resistance genes.
Methods
A prospective study was conducted on 120 DFI patients at Main Alexandria University Hospital, Egypt. The aerobic and anaerobic etiological pathogens were determined using semi-quantitative culture and PCR respectively. The antimicrobial susceptibility pattern was done according to Clinical Laboratory Standards Institute guidelines. Detection of carbapenemases and class-1 integron genes was carried out by polymerase chain reaction (PCR).
Results
A total of 178 (124 aerobic, 54 anaerobic) pathogens were identified from patients with DFI, with an average of 1.82 isolates/subject. Among aerobic pathogens, Gram-negative predominated (98/124; 79%), of which Pseudomonas spp. and Proteus spp. were the most common. MRSA constituted more than 50% of Gram-positive isolates. Polymicrobial infection was found in 42 (42.9%) subjects. The proportion of Gram-negative bacteria and anaerobes increased with increased DFI grades and severity. Multidrug and extensively drug resistant isolates were observed in 86 patients (87.7%). PCR identified carbapenemases genes in 14 (11.7%) and class 1 integron in 28 (23.3%) DFI cases. Vancomycin, teicoplanin, linezolid were the most effective antimicrobial agents against Gram-positive pathogens, while colistin, imipenem, meropenem, and piperacillin-tazobactam were effective against Gram-negative pathogens.
Conclusions
Multidrug and extensively drug resistant Gram-negative bacteria were the dominant pathogens among all DFI severity grades. However, the proportion of Gram-positive bacteria decreased with the severity of infection. The clinical role of our relatively high rate of anaerobes should be investigated. The results found in this study could be beneficial for designing future empiric antimicrobial protocols in relation to the severity of DFIs.
Keywords: Diabetic foot infection, diabetic foot ulcer, microbiology, multidrug-resistant, extensively drug-resistant, integron
Introduction
Diabetic foot ulcers (DFUs) and diabetic foot infections (DFIs) remain a major complication in patients with diabetes.1 Around 25% of patients with diabetes will develop a DFU in their lifetime, and more than 50% of these ulcers become infected.1
DFU results from repetitive trauma due to a combination of factors like loss of protective sensation, peripheral vascular disease, and impaired immunity. Several microorganisms would colonize and proliferate in the ulcer, accentuating tissue damage and resulting in infection.2
DFI is an infection in soft tissue or bone below the malleoli in a diabetic patient. DFIs range from superficial skin infections to chronic osteomyelitis ending by severe complications such as gangrene, and lower limb amputation in 15% of patients.3 DFI is usually monomicrobial and caused by Gram-positive cocci in the early acute stage, whereas it is mostly polymicrobial with a mixture of Gram-negative aerobes and anaerobes and very rarely fungi in the chronic stage.3
As the treatment of DFIs requires appropriate antimicrobial selection, continuous updates of the microorganisms responsible of infection and their resistance pattern remain a keystone in the management process, since infection with multidrug resistant strains is increasing and poses additional morbidity and mortality.3
The aim of the study was to determine the appropriate sampling technique from DFI ulcers, to identify the etiological pathogens of DFIs in different Wagner-Meggitt’s and IDSA/IWGDF (the Infectious Diseases Society of America/International Working Group on the Diabetic Foot) grades, and to assess their antimicrobial susceptibility pattern together with molecular characterization of antibiotic resistance genes, in an attempt to find out the best antimicrobial treatment options for those patients.
Methods
All patients [(120 patients; 110 with type 2 and ten with type 1 diabetes mellitus (DM)] with DFU showing clinical evidence of infection (redness, warmth, swelling, tenderness, pain, or purulent discharge) were recruited from the outpatient clinic and/or diabetic foot clinic of Main Alexandria University Hospital (MAUH) from the 1st of December 2019 until the end of May 2020 in a cross-sectional prospective observational study. Patients with no ulcers (Wagner’s grade 0) as well as patients with extensive gangrene involving the whole foot (Wagner’s grade 5) were excluded from the study.
The study was approved by the Ethics Committee of Alexandria Faculty of Medicine, and a written informed consent was obtained from every recruited patient.
All patients underwent the following:
Detailed history and physical examination including age, sex, previous hospitalization, type and duration of diabetes, body mass index (BMI), presence of diabetic complications such as retinopathy (assessed by an ophthalmologist), hypertension, nephropathy (micro or macro-albuminuria or creatinine >1.5 mg/dL).
Full clinical assessment of diabetic foot, including the presence of sensory neuropathy, peripheral vascular disease. DFUs were graded according to Wagner-Meggitt classification system.4 The DFUs were also classified according to IDSA/IWGD 5 classification. The presence of signs of inflammation/infection (erythema, hotness, swelling, tenderness, purulent discharge) was also evaluated.
Routine laboratory investigations including complete blood count, fasting blood glucose, HbA1c, inflammatory markers (C-reactive protein – CRP), serum creatinine, urinary albumin creatinine ratio were performed.
Microbiological investigations.
Sample collection
Swabs were collected from ulcers after cleansing using sterile saline and gauze and proper debridement. The swab was rotated over a 1 cm2 of the affected area for at least 10 seconds with sufficient pressure using Levine’s technique. Two tissue biopsies were also obtained from the base of the ulcer using a sterile punch biopsy needle (6 mm); one was used for microbiological culture and the second was used for PCR testing. Swabs and biopsies were placed into sterile phosphate buffered saline (PBS). All samples were transported immediately to the Microbiology Laboratory of MAUH for processing.
Quantitative aerobic bacterial and fungal culture
All samples were processed as per our institution laboratory’s operating procedures. For culture, swabs in PBS were thoroughly vortexed then tenfold serially diluted in normal saline followed by inoculation of 10 µL of each dilution onto each of chocolate blood agar, blood agar, MacConkey’s agar (Thermo Fisher, UK) for isolation of aerobic bacteria, as well as onto Sabouraud dextrose agar (Thermo Fisher) for isolation of fungi. Tissues were weighed and properly homogenized then subjected to semi-quantitative culture after serial dilutions in the same manner as processing of swab samples. After 48 h incubation at 37°C, colonies of each type of bacteria were counted and identified according to standard microbiological methods.6
Antimicrobial susceptibility testing
It was carried out using the Kirby-Bauer disc diffusion / broth microdilution methods in accordance with the clinical and laboratory standards Institute (CLSI, 2020).7 Phenotypic detection of methicillin resistance in staphylococci, ESBL (extended spectrum β-lactamases) and carbapenemases in Gram-negative bacilli was carried out using cefoxitin 30 μg disc diffusion, ESBL combined disc diffusion test, and modified carbapenem inactivation method (mCIM) respectively.7 Multidrug resistant (MDR), and extensively drug resistant (XDR) isolates were defined according to standard international terminology presented by ECDC (European Centre for Disease Prevention and Control) and CDC (Centre for Disease Prevention and Control).8
Molecular analysis
DNA was extracted from tissue biopsies using the QIAamp DNA Mini kit (Qiagen GmbH, Germany) following manufacturer’s instructions. Three multiplex PCRs were performed. One multiplex PCR, optimized in the present study, was used to amplify genes of common anaerobes (Bacteroides spp., Clostridioides spp., Peptostreptococcus spp., Peptococcus spp., and Ruminococcus spp.).9 A second multiplex PCR, for amplification of genes of metallo-beta-lactamases,10 as well as of class-1 integron 11 of Gram-negative bacilli, was also optimized in the current study. The third PCR was done for amplification of carbapenemases genes of Enterobacterales as previously described.12 16SrDNA and beta globin genes amplification were included as internal controls in every PCR reaction.13 The amplification reactions were carried out in Applied Biosystems 2720, thermal cycler. PCR products were separated on 1.5% agarose gel (Bioline, UK) and detected using a gel documentation system (Bio-Rad Laboratories, USA). The DNA sequences of used primers and PCR reaction conditions are mentioned in Table 1.
Table 1.
Primers DNA sequences used in the study
| Primer sequence (5'- 3') | Target | PCR program | Size of amplicon (bp) |
|---|---|---|---|
| First multiplex PCR | |||
| GGGGTTCTGAGAGGAAG ACCCCCCATTGTACCAC |
Bacteroides spp.9 | 4 min at 94°C; 30 cycles (45 sec at 94°C, 45 sec at 60°C, and 1 min at 72°C); 7 min at 72°C (this study) | 950 |
| CTCAACTTGGGTGCTGCATTT ATTGTAGTACGTGTGTAGCCC |
Clostridioides spp.9 | 619 | |
| GGTGCCGCAGTAAACACAATAAGT AAGGCCCGGGAACGTATTCA |
Peptococcus spp.9 | 539 | |
| CCTCTGACCGCTCTTTAATCGGAGCTTTCCTTC CCAGTTATCGGTCCCACCTTCGGCAGCT |
Ruminococcus spp.9 | 482 | |
| AACTCCGGTGGTATCAGATG GGGGCTTCTGAGTCAGGTA |
Peptostreptococcus spp.9 | 270 | |
| Second multiplex PCR | |||
| GTTTGGTCGCATATCGCAAC AATGCGCAGCACCAGGATAG |
bla VIM variants10 | 4 min at 94°C; 30 cycles (45 sec at 94°C, 45 sec at 55°C, and 1 min at 72°C); 7 min at 72°C (this study) | 382 |
| GAAGGYGTTTATGTTCATAC GTAMGTTTCAAGAGTGATGC |
bla IMP variants10 | 587 | |
| GGGCAGTCGCTTCCAACGGT GTAGTGCTCAGTGTCGGCAT |
bla NDM-1 10 | 475 | |
| TCTCGGGTAACATCAAGG AGGAGATCCGAAGACCTC |
intI-1 11 | 234 | |
| Third multiplex PCR12 | |||
| GCAGCTTGTCGGCCATGCGGGC GGTCGCGAAGCTGAGCACCGCAT |
bla NDM-1 | 5 min at 95°C; 35 cycles (45 sec at 95°C, 45 sec at 60°C, and 1 min at 72°C); 8 min at 72°C | 782 |
| GCGTGGTTAAGGATGAACAC CATCAAGTTCAACCCAACCG |
bla OXA-48 | 438 | |
| TGTCACTGTATCGCCGTC CTCAGTGCTCTACAGAAAACC |
bla KPC | 900 | |
| GAAGGCGTTTATGTTCATAC GTACGTTTCAAGAGTGATGC |
bla IMP | 587 | |
| GTTTGGTCGCATATCGCAAC AATGCGCAGCACCAGGATAG |
bla VIM-1 | 389 | |
| Internal control genes | |||
| CAACTTCATCCACGTTCACC GAAGAGCCAAGGACAGGTAC |
Human β-globin13 | 268 | |
| AGTTTGATCCTGGCTCAG GGACTACCAGGGTATCTAAT |
Bacterial 16SrRNA13 | 798 | |
Statistical analysis
All statistical analyses were carried out using Statistical Package for Social Sciences (SPSS ver. 22, IBM Corp., USA). Data was processed as numerical or categorical as appropriate. Data was presented using mean, median, range and frequencies. Normally distributed variables were analysed using one-way ANOVA. Variables without normal distribution were compared using Kruskal-Wallis test. Qualitative variables were compared using Chi-square test. Statistically significant results were considered when p value was less than 0.05.
Results
A total of 120 DFI patients (64 males and 56 females) were studied; their average age was 56.1±9.9 years, with a male to female ratio of 1.1:1. Among all patients, 108 (90%) had type 2 DM, whereas only 12 (10%) patients had type 1 DM, and more than half of them (64; 53.3%) were smokers. The duration of diabetes ranged from 0.4-32 years with a mean of 14.6±6.4 years. The mean fasting blood glucose and HbA1c were 274.5±79.6 mg/dL and 10.3±2.1% respectively. More than half of the studied patients had diabetic retinopathy (68; 56.7%), while three quarters of them (90; 75.0%) had diabetic nephropathy. DFUs were graded according to Meggitt Wagner’s grade into grade 2 in 30 patients (25%), grade 3 in 60 patients (50%), and grade 4 in 30 patients (25%). DFUs were also classified into mild (30; 25%), moderate (48; 40%), and severe (42; 35%) according to IDSA/IWGDF classification. All patients received previous antibiotic therapy (ranging from 48 h to one month) before taking the samples (as fourth generation cephalosporins, carbapenems, quinolones, clindamycin, linezolid; monotherapy or in combination). All patients had at least one previous hospital admission.
Out of 120 DFI patients, 98 cases (81.7%) were culture-positive and 22 cases (18.3%) were culture-negative using the biopsy culture technique. In comparison to the biopsy culture, the swab culture technique was positive in 94 cases (true positive) and negative in 26 cases (22 true negative and 4 false negative). This results in 95.9% sensitivity, 100% specificity, 100% positive predictive value (PPV), 84.6% negative predictive value (NPV) and 96.7% accuracy of swab culture relative to the biopsy culture technique. The measure of agreement [kappa; (po – pe) / (1– pe)] between the results of swab culture and biopsy culture was 0.89 resulting in perfect agreement (96.7%).
A total of 124 aerobic pathogens were isolated from the 98 DFI patients; of these 98 (79%) were Gram-negative and 26 (21%) were Gram-positive. No fungal organisms were recovered from any case. Additionally, 54 anaerobic bacteria (54/178; 30.3%) were identified by PCR. Therefore, a total of 178 bacterial isolates (aerobic and anaerobic) were isolated from the 98 patients, accounting for an average of 1.82 isolates per subject. The contribution of anaerobic bacteria was about 0.5 per case (Table 2).
Table 2.
The distribution of identified microorganisms in patients with DFI
| Organisms | No | % |
|---|---|---|
| Gram-positive isolates | ||
| MRSA | 14 | 53.8 |
| Enterococcus spp. | 6 | 23.1 |
| viridans streptococci | 4 | 15.4 |
| MS CoNS | 2 | 7.7 |
| Total | 26 | 100.0 |
| Gram-negative isolates | ||
| Non-fermenters | ||
| Pseudomonas spp. | 20 | 20.4 |
| Acinetobacter spp. | 12 | 12.2 |
| Enterobacterales | ||
| Proteus spp. | 20 | 20.4 |
| Klebsiella spp. | 18 | 18.4 |
| Morganella spp. | 10 | 10.2 |
| E. coli | 8 | 8.2 |
| Enterobacter spp. | 8 | 8.2 |
| Citrobacter spp. | 2 | 2 |
| Total | 98 | 100.0 |
| Anaerobes | ||
| Ruminococcus spp. | 20 | 37.1 |
| Bacteroides spp. | 18 | 33.3 |
| Peptococcus spp. | 8 | 14.8 |
| Clostridioides spp. | 4 | 7.4 |
| Peptostreptococcus spp. | 4 | 7.4 |
| Total | 54 | 100 |
Out of the 98 culture-positive DFI patients, 56 (57.1%) had monomicrobial infection and 42 (42.9%) had polymicrobial infection. The majority of monomicrobial infections were caused by Gram-negative pathogens (34/56; 60.7%). Eighty-six patients (87.7%) had an MDR and/or XDR pathogen (58 with MDR, and 28 patients with XDR).
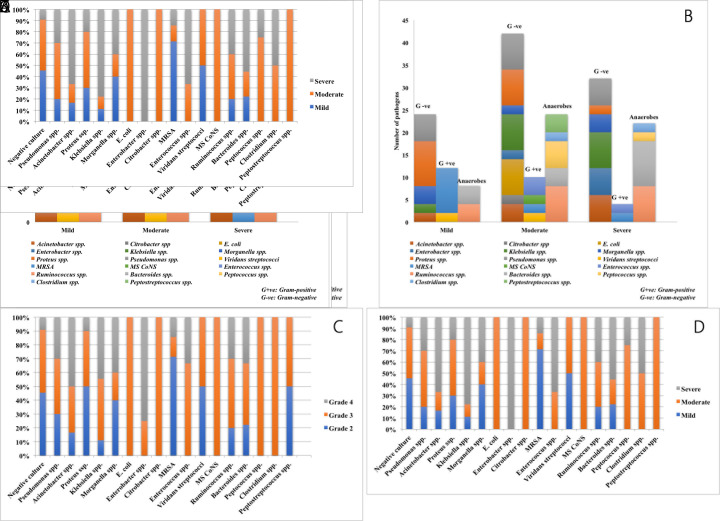
The distribution of various bacterial isolates in different Meggitt Wagner’s grades and IDSA/IWGDF classes of DFU are shown in Figure 1. Gram-negative pathogens constituted the major pathogens in all grades. The proportion of Gram-positive pathogens decreased with the severity of infection. On the contrary, anaerobes increased in frequency with increased severity of DFIs. The distribution pattern of bacteria was nearly similar in both Meggitt Wagner’s and IDSA/IWGDF classification.
Figure 1. The distribution of the number of pathogens isolated from patients with DFI in different grades.
(A) The number of pathogens in different Meggitt Wagner’s grades. G+ve – Gram positive; G-ve – Gram negative
(B) The number of pathogens in different IDSA/IWGDF infection severity. G+ve – Gram positive; G-ve – Gram negative
(C) The distribution of each pathogen in different Meggitt Wagner’s grade.
(D) The distribution of each pathogen in different IDSA/IWGDF infection severity.
The mean age of patients with moderate and severe infection (IDSA/IWGDF) was significantly higher than that of patients with mild infection (p=0.037). With increasing Wagner’s grades and severity of IDSA/IWGDF, the C-reactive protein (CRP) level and the absolute neutrophil count (ANC) were increasing (p<0.001). The presence of callosities on examination of the diabetic foot was more common in lower grades and mild infection than in higher grades and severe infection (p=0.017, p=0.007 respectively). No significant differences were observed in other clinical characteristics. When comparing microbiological data in different severity grades, Gram-negative bacteria were statistically significantly predominant in all grades compared to Gram-positive ones (p=0.01). This finding emphasizes the importance of covering Gram-negative bacteria in empiric treatment protocols in our hospital. There was no statistically significant difference regarding absence or presence of monomicrobial/polymicrobial and anaerobic infection between different grades (Table 3). Additionally, there was no statistically significant association between the presence of monomicrobial/polymicrobial infection and certain age (p=0.388).
Table 3.
Characteristics of patients with DFI in different grades of Meggitt Wagner’s and IDSA/IWGDF
| Parameters | Meggitt Wagner’s grade | p value | IDSA/IWGDF infection severity | p value | ||||
|---|---|---|---|---|---|---|---|---|
| Grade 2 | Grade 3 | Grade 4 | Mild | Moderate | Severe | |||
| No of patients (n=120) | 30 (25%) | 60 (50%) | 30 (25%) | 30 (25%) | 48 (40%) | 42 (35%) | ||
| Clinical and laboratory characteristics | ||||||||
| Age (years) | 50.1±9.9 | 58.2±8.35 | 57.73±10.8 | 0.051 | 51.8±11.2 | 59.7± 8.3 | 54.9±9.4 | 0.037 |
| BMI | 29.1±0.35 | 29.7±5.7 | 28.63±5.7 | 0.908 | 30±5.9 | 28.3±5.9 | 29.7±5.8 | 0.619 |
| SBP | 131.3±18 | 138.3±16.6 | 134±15.4 | 0.391 | 131.3±17.3 | 137.5±15.4 | 136.2±18.0 | 0.568 |
| DBP | 84.6±10.6 | 86±9.6 | 84.6±9.9 | 0.923 | 84±9.8 | 87.1±9.5 | 84.3±10.3 | 0.643 |
| Duration of DM (years) | 12.9±4.18 | 13.9±6.9 | 17.4±6.6 | 0.127 | 14±5.5 | 13.5±5.4 | 16.2±7.9 | 0.346 |
| HbA1c (%) | 10.2±2.02 | 9.8±2.05 | 11.44±2.1 | 0.053 | 10.3±1.9 | 10±2.3 | 10.8±2.1 | 0.494 |
| Fasting blood glucose (mg/dL) | 254.8±80.7 | 273.1±81.1 | 296.8±74.8 | 0.356 | 273.7±82.1 | 273.7±82.1 | 279.9±77.9 | 0.968 |
| Serum creatinine (mg/dL) | 1.4±1.3 | 0.9±0.3 | 0.9±0.3 | 0.062 | 1.4±1.3 | 0.9±0.3 | 0.9±0.3 | 0.015 |
| eGFR | 49.2±30.0 | 55.9±19.8 | 50.4±23.1 | 0.237 | 45.7±29.3 | 52.9±20.0 | 57.9±21.9 | 0.053 |
| UACR | 41.9±23.3 | 42.3±18.4 | 41.6±20.1 | 0.853 | 42.9±24.2 | 40.8±16.3 | 43±21.3 | 0.956 |
| CRP (mg/dL) | 25.1±21.5 | 84.9±61.5 | 123.5±80.9 | <0.001 | 30±25.1 | 68.9±38.7 | 127.4±87.03 | <0.001 |
| TLC (g/L) | 98.4±4.6 | 97.2±4.1 | 98.7±5.02 | 0.312 | 98±4.4 | 98.3±4.2 | 97.5±5.0 | 0.956 |
| ANC (g/L) | 68.6±8.3 | 76.9±9.4 | 81±6.19 | 0.001 | 70.2±10.1 | 76.9±8.9 | 78.7±8.4 | 0.021 |
| Diabetic foot characteristics | ||||||||
| Reflexes (intact) | 28 (93.3%) | 36 (60%) | 20 (66.7%) | 0.070 | 24 (80%) | 36 (75%) | 24 (57.1%) | 0.265 |
| Presence of callosities | 26 (86.7%) | 26 (43.3%) | 14 (46.7%) | 0.017 | 26 (86.7%) | 26 (54.2%) | 14 (33.3%) | 0.007 |
| Presence of inter-digital infection | 14 (46.7%) | 36 (60%) | 18 (60%) | 0.665 | 14 (46.7%) | 28 (58.3%) | 26 (61.9%) | 0.646 |
| Microbiological data | ||||||||
| Type of infection (n=98) | ||||||||
| Monomicrobial (n=56) | 12 (40%) | 24 (40%) | 20 (66.7%) | 0.420 | 10 (33.3%) | 22 (45.8%) | 24 (57.1%) | 0.756 |
| Polymicrobial (n=42) | 12 (40%) | 20 (33.3%) | 10 (33.3%) | 10 (33.3%) | 16 (33.3%) | 16 (38.1%) | ||
| Aerobic bacteria (n=124) | ||||||||
| Gram-positive (n=26) | 12 | 10 | 4 | 0.063 | 12 | 8 | 6 | 0.010 |
| Gram-negative (n=98) | 24 | 42 | 32 | 18 | 36 | 44 | ||
| Anaerobic bacteria (n=120) | ||||||||
| Present (n=26) | 6 | 14 | 6 | 0.906 | 4 | 12 | 10 | 0.437 |
| Absent (n=94) | 24 | 46 | 24 | 26 | 36 | 32 | ||
Data is presented as mean±SD or n (%).
BMI – body mass index; SBP – systolic blood pressure; DBP – diastolic blood pressure; DM – diabetes mellitus; HbA1c – glycosylated hemoglobin; eGFR – estimated glomerular filtration rate; UACR – urinary albumin creatinine ratio; CRP – C-reactive protein; TLC – total leucocyte count; ANC – absolute neutrophil count.
The results of antimicrobial susceptibility testing are summarized in Table 4 and 5. Resistance of Gram-negative bacteria to most antibiotic classes exceeded 60%, in addition, 55.1% of the isolates were MDR and 34% were XDR. PCR identified class 1 integron in 28 (23.3%) DFI cases, blaNDM genes in six (5%) cases, blaVIM and blaOXA in four (3.3%) cases each.
Table 4.
The antimicrobial resistance pattern of Gram-negative bacterial isolates from patients with DFI
| Antibiotics | Gram-negative non-fermenters | Enterobacterales | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pseudomonas spp. (n=20) | Acinetobacter spp. (n=12) | Proteus spp. (n=20) | Klebsiella spp. (n= 18) | Morganella spp. (n=10) | E. coli (n=8) | Enterobacter spp. (n=8) | Citrobacter spp. (n=2) | Total (n=98) | ||
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||
| Penicillins | Ampicillin | 20 (100) | 12 (100) | 20 (100) | 18 (100) | 10 (100) | 8 (100) | 8 (100) | 2 (100) | 98 (100) |
| Piperacillin | 16 (80) | 12 (100) | 8 (40) | 16 (88.9) | 4 (40) | 8 (100) | 6 (75) | 2 (100) | 72 (73.5) | |
| Penicillins/beta lactamase inhibitors | Amoxicillin/clavulanic acid | 20 (100) | 12 (100) | 8 (40) | 18 (100) | 10 (100) | 6 (75%) | 8 (100) | 2 (100) | 84 (85.7) |
| Ampicillin/sulbactam | 20 (100) | 12 (100) | 6 (30) | 18 (100) | 6 (60) | 6 (75%) | 8 (100) | 2 (100) | 78 (79.6) | |
| 1st generation cephalosporins | Cefazolin | 20 (100) | 12 (100) | 20 (100) | 18 (100) | 10 (100) | 8 (100) | 8 (100) | 2 (100) | 98 (100) |
| Cephalexin | 20 (100) | 12 (100) | 20 (100) | 18 (100) | 10 (100) | 8 (100) | 8 (100) | 2 (100) | 98 (100) | |
| Cephamycins | Cefoxitin | 20 (100) | 12 (100) | 18 (90) | 18 (100) | 8 (80) | 6 (75) | 8 (100) | 2 (100) | 92 (93.9) |
| 2nd generation cephalosporins | Cefotetan | 20 (100) | 12 (100) | 18 (90) | 18 (100) | 8 (80) | 6 (75) | 8 (100) | 2 (100) | 94 (95.9) |
| Cefuroxime | 20 (100) | 12 (100) | 20 (100) | 18 (100) | 10 (100) | 6 (75) | 8 (100) | 2 (100) | 96 (98) | |
| 3rd generation cephalosporins | Cefoperazone | 20 (100) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 56 (57.1) |
| Ceftriaxone | 20 (100) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 56 (57.1) | |
| Cefotaxime | 20 (100) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 56 (57.1) | |
| Ceftazidime | 12 (60) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 48 (49) | |
| 4th generation cephalosporins | Cefepime | 10 (100) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 46 (46.9) |
| Cephalosporins/beta lactamase inhibitors | Piperacillin/tazobactam | 8 (40) | 10 (83.3) | 0 | 14 (77.8) | 0 | 6 (75) | 0 | 0 | 38 (38.8) |
| Cefoperazone/sulbactam | 14 (70) | 10 (83.3) | 0 | 14 (77.8) | 0 | 4 (50) | 2 (25) | 0 | 44 (44.9) | |
| Monobactams | Aztreonam | 10 (50) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 4 (50) | 2 (100) | 48 (49) |
| Carbapenems | Imipenem | 10 (50) | 10 (83.3) | 0 | 12 (66.7) | 0 | 4 (50) | 0 | 0 | 36 (36.7) |
| Meropenem | 8 (40) | 10 (83.3) | 0 | 14 (77.8) | 0 | 4 (50) | 0 | 0 | 36 (36.7) | |
| Ertapenem | 20 (100) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 56 (57.1) | |
| Aminoglycosides | Gentamicin | 16 (80) | 12 (100) | 12 (60) | 14 (77.8) | 8 (80) | 6 (75) | 2 (25) | 2 (100) | 72 (73.5) |
| Amikacin | 14 (70) | 12 (100) | 8 (40) | 14 (77.8) | 6 (60) | 6 (75) | 0 | 2 (100) | 62 (63.3) | |
| Tobramycin | 14 (70) | 12 (100) | 8 (40) | 14 (77.8) | 6 (60) | 6 (75) | 2 (25) | 2 (100) | 64 (65.3) | |
| Quinolones | Ciprofloxacin | 14 (70) | 10 (83.3) | 10 (50) | 14 (77.8) | 4 (40) | 8 (100) | 4 (50) | 0 | 64 (65.3) |
| Levofloxacin | 14 (70) | 10 (83.3) | 10 (50) | 12 (66.7) | 4 (40) | 8 (100) | 4 (50) | 0 | 62 (63.3) | |
| Ofloxacin | 16 (80) | 12 (100) | 12 (60) | 16 (89) | 4 (40) | 8 (100) | 4 (50) | 2 (100) | 74 (75.5) | |
| Tetracyclines | Tetracycline | 20 (100)* | 12 (100) | 20 (100) | 16 (89) | 10 (100) | 8 (100) | 4 (50) | 2 (100) | 92 (93.9) |
| Doxycycline | 20 (100)* | 8 (66.7) | 20 (100) | 10 (55.6) | 10 (100) | 8 (100) | 4 (50) | 2 (100) | 82 (83.7) | |
| Minocycline | 20 (100)* | 8 (66.7) | 20 (100) | 12 (66.7) | 10 (100) | 8 (100) | 4 (50) | 2 (100) | 84 (85.7) | |
| Tigecycline | 20 (100)* | 6 (50) | 20 (100) | 4 (44.4) | 10 (100) | 4 (50) | 0 | 2 (100) | 66 (67.3) | |
| SMX/TMP | SMX/TMP | 20 (100)* | 12 (100) | 14 (70) | 16 (88.9) | 8 (80) | 8 (100) | 6 (75) | 2 (100) | 86 (87.8) |
| Polymyxins | Colistin | 0 | 0 | 20 (100)* | 0 | 10 (100)* | 0 | 0 | 0 | 30 (30.6) |
| Polymyxin B | 0 | 0 | 20 (100)* | 0 | 10 (100)* | 0 | 0 | 0 | 30 (30.6) | |
| MDR | 12 (60) | 2 (16.7) | 12 (60) | 6 (33.3) | 8 (80) | 4 (50) | 8 (100) | 2 (100) | 54 (55.1) | |
| XDR | 8 (40) | 10 (83.3) | 0 | 12 (66.7) | 0 | 4 (50) | 0 | 0 | 34 (34.7) | |
| ESBL | 12 (60) | 12 (100) | 0 | 14 (77.8) | 0 | 6 (75) | 2 (25) | 2 (100) | 48 (49) | |
| CPO | 10 (50) | 10 (83.3) | 0 | 14 (77.8) | 0 | 6 (75) | 0 | 0 | 40 (40.8) | |
*The 100% resistance rate is related to intrinsic resistance of Pseudomonas spp. to tetracyclines, sulfamethoxazole-trimethoprim, and Proteus spp., Morganella spp. to colistin.
SMX/TMP – sulfamethoxazole/trimethoprim; MDR – multidrug resistant; XDR – extensively drug resistant; ESBL – extended spectrum beta lactamase; CPO – carbapenemase-producing organism.
Table 5.
The antimicrobial resistance pattern of Gram-positive bacterial isolates from patients with DFI
| Antibiotics | Gram-positive isolates | Total (n=26) | ||||
|---|---|---|---|---|---|---|
| MRSA (n=14) | Enterococcus spp. (n=6) | viridans streptococci (n=4) | MS CoNS (n=2) | |||
| N (%) | N (%) | N (%) | N (%) | N (%) | ||
| Penicillins | Penicillin G | 14 (100) | 2 (33.3) | 2 (50) | 2 (100) | 20 (76.9) |
| Ampicillin | NA | 2 (33.3) | NA | NA | 2/6 (33.3) | |
| Cephamycins | Cefoxitin | 14 (100) | NA | NA | 0 | 14/16 (87.5) |
| 3rd generation cephalosporins | Ceftriaxone | 14 (100) | NA | 2 (50) | 0 | 16/20 (80) |
| Cefotaxime | 14 (100) | NA | 2 (50) | 0 | 16/20 (80) | |
| 4th generation cephalosporins | Cefepime | 14 (100) | NA | 2 (50) | 0 | 16/20 (80) |
| Oxazolidinones | Linezolid | 0 | 0 | 0 | 0 | 0 |
| Glycopeptides | Vancomycin | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 0 | 0 | |
| Quinolones | Ciprofloxacin | 2 (14.3) | 0 | 0 | 0 | 2 (7.7) |
| Levofloxacin | 2 (14.3) | 0 | 0 | 0 | 2 (7.7) | |
| Ofloxacin | 2 (14.3) | 0 | 0 | 0 | 2 (7.7) | |
| Tetracyclines | Tetracycline | 6 (42.9) | 0 | 2 (50) | 0 | 8 (30.8) |
| Doxycycline | 6 (42.9) | 0 | 2 (50) | 0 | 8 (30.8) | |
| Minocycline | 6 (42.9) | 0 | 2 (50) | 0 | 8 (30.8) | |
| Tigecycline | 2 (14.3) | 0 | 0 | 0 | 2 (7.7) | |
| Lincomycin | Clindamycin | 6 (42.9) | NA | 0 | 0 | 6/20 (30) |
| Macrolides | Erythromycin | 8 (57.1) | 2 (33.3) | 2 (50) | 0 | 12 (46.1) |
| Azithromycin | 8 (57.1) | 2 (33.3) | 2 (50) | 0 | 12 (46.1) | |
| Clarithromycin | 8 (57.1) | 2 (33.3) | 2 (50) | 0 | 12 (46.1) | |
| Aminoglycosides | Gentamicin | 10 (71.4) | NA | NA | 0 | 10/16 (62.5) |
| Amikacin | 8 (57.1) | NA | NA | 0 | 8/16 (50) | |
| Gentamicin (HLA) | NA | 0 | NA | NA | 0 | |
| SMX/TMP | SMX/TMP | 4 (28.6) | NA | 0 | 0 | 4/16 (25) |
| Rifamycins | Rifampicin | 2 (14.3) | 0 | 0 | 0 | 2 (7.7) |
| MDR | 12 (85.6) | 0 | 0 | 0 | 12 (46.1) | |
| XDR | 2 (100) | 0 | 0 | 0 | 2 (7.7) | |
MRSA – methicillin resistant Staphylococcus aureus; MS CoNS – methicillin sensitive coagulase-negative staphylococci; SMX/TMP – sulfamethoxazole/trimethoprim; HLA – high level aminoglycosides; MDR – multidrug resistant; XDR – extensively drug resistant; NA – not applicable (the organism is not tested against this antibiotic).
When comparing the clinical characteristics of patients having (MDR and/or XDR) pathogens (86 patients) or not (12 patients), no statistically significant difference was found in any parameter. Additionally, no association was found between the presence of MDR/XDR pathogen and the grade or the severity of DFIs. The low number of non-MDR/XDR group may have had an impact on analysis of factors that could be associated with the presence of MDR/XDR infection (data not shown).
Discussion
DFIs continue to be a major health problem worldwide. In the present study, the clinical and laboratory results of DFI patients (age, male predominance, elevated inflammatory markers, inadequate glucose level control) are similar to those previously reported.13-17
DFI appears approximately after 17-20 years of the initial diagnosis of DM.15 The mean duration of diabetes (14.6±6.4 years) in our study was shorter than reported in a previous study.15 This is an alarming finding in our population that should be clinically investigated, as early diagnosis of DM can reduce the rate of complications of this disease and increase the quality of life.
According to Wagner’s grading and IDSA/IWGDF classification, the majority of the patients (75%) in the current study, had higher grades of DFU (grade 2-4). This was in accordance with the results of studies conducted in other countries.14,16 The presentation with higher grade ulcers could be explained by the poor educational level of the population, self-medication, and delay in seeking medical advice.
In the present study, we reported a perfect agreement (96.7%) between the results of biopsy and swab cultures. Similarly, several studies found no significant difference in the isolates between swabbing and deep tissue biopsy culture technique.18,19 On the other hand, swab cultures were found highly sensitive but less specific both in patients with neuropathic and neuroischemic DFUs in the study of Demetriou et al.20 They attributed the low specificity to the presence of bacterial flora contaminating swab cultures. The perfect agreement reported in the current study might be explained by the meticulous cleansing and debridement step preceding sample collection reducing superficial contamination.
Although a lower frequency (42; 42.9%) of polymicrobial infection was reported in the present study and some other reports including reports from Egypt,21,22 DFI is usually considered a polymicrobial infection. Several studies showed predominance of polymicrobial infections in DFI patients.16,23-25 The lower prevalence of polymicrobial infections in our study may be again attributed to the proper sampling.
Out of the 120 patients, 22 (18.3%) had negative results for both aerobic and anaerobic organisms despite the clinical confirmation of DFIs. On the contrary, Hefni et al.22 reported a lower percentage of negative cultures (8%). This high rate of negative cultures in the present study could be attributed to the injudicious use of antibiotics, that all patients received antibiotic therapy before taking the sample. The average number of bacterial isolates per patient was 1.82 isolates, which was in agreement with the results of previous studies,16,25 but higher than that reported in others.22,24
The bacterial aerobic profile of the 98 culture-positive DFIs in the current study confirmed a predominance of Gram-negative bacteria (79%), and absence of fungal organisms. These results opposed that of previous studies, which reported the predominance of Gram-positive bacteria, mainly S. aureus.14,21 However, several reports, especially from developing countries, were in agreement with our results regarding the Gram-negative bacterial predominance.16,24,26 Additionally, in a study conducted in Egypt, Dwedar et al. reported that the Gram-negative bacteria constituted (56.08%) of isolated pathogens.23 Another Egyptian study revealed that out of 98 isolates, Gram-negative bacteria (67.3%) were more commonly isolated.22
Pseudomonas spp., Proteus spp., Klebsiella spp. and MRSA were the most commonly isolated germs in our study. Similarly, Dwedar et al.23 found that Proteus mirabilis was the most common. Choucair et al.15 and Abd-El Mohsen 17 also found that P. aeruginosa was the most commonly isolated pathogen in their studies. However, among the aerobic pathogens isolated in an Indian study, the most predominant were S. aureus, followed by P. aeruginosa and E. coli.21
A high frequency of anaerobic bacteria (30.3%) was detected by PCR in the present study, and mostly recovered in moderately and severely infected ulcers combined with aerobic bacteria. A much lower rate was reported previously in Egypt.23 This may be due to differences in the sensitivity of detection methods (PCR and culture). The role of anaerobes in DFIs is not totally clear, assuming antibiotics given for aerobic bacteria are also effective against anaerobes, and given the fact that previous studies did not determine their clinical role in a more severe outcome.
The majority of studies did not associate the severity or grade of the ulcer with the type of microorganism. The differences in microbiology were assessed in two studies from Egypt 27 and China,26 where predominance of Gram-positive bacteria was observed in mild cases, while Gram-negatives predominated in moderate and severe cases. These studies also reported that with increasing severity, infection tends to be polymicrobial. The same observation was confirmed in the present study, in which Gram-negatives and enterococci were associated with moderate-severe cases, while streptococci, MRSA were mostly associated with mild cases. Although, polymicrobial infection was distributed mainly in moderate-severe infection, this was not statistically significant.
Concerning the antimicrobial susceptibility pattern of the isolated pathogens, out of all Gram-negative isolates, 55.1% and 34.7% were MDR and XDR respectively, where 49% were ESBL and 40.8% were carbapenemase producers. Similar results of high rates of MDR and ESBL were obtained from previous studies in Egypt and elsewhere.16,23,24 However, the rates of XDR as well as of carbapenemases were much higher than the rates reported previously.23,26 The rate of resistance in Gram-negative bacteria in this work reached an alarming rate making the choice of appropriate empiric therapy problematic. Currently, there is paucity of data in literature concerning the prevalence of carbapenemases genes and class 1 integron in DFIs, making comparison to previous work difficult. Class I integron was found in relatively lower frequency (23.3%) in the present study.
Moreover, the fact that all S. aureus in our study were methicillin resistant is truly alarming because this may denote the absence of strict antibiotic prescription polices, in addition to poor infection control procedures. The methicillin resistance rate in S. aureus in the present study is almost 3-4 times higher than that reported previously.21 On the other hand, all enterococci and streptococci in the present study were found to be susceptible to most antibiotics.
The high rate of antibiotic resistance in this study could be attributed to prolonged use of broad-spectrum antibiotics, previous hospitalization, that predispose patients to colonization and infection with antibiotic-resistant organisms. Additionally, increased antibiotic resistance could be attributed to horizontal transfer of antibiotic resistant genes.
In the current study, Gram-negative bacteria were mostly susceptible to colistin, imipenem, meropenem, piperacillin-tazobactam (69.4%, 63.3%, 63.3%, 61.2%), while Gram-positive bacteria were mostly susceptible to vancomycin (100%), teicoplanin (100%), linezolid (100%). It is obvious from our results that all penicillins, cephalosporins, aminoglycosides, quinolones, sulfamethoxazole-trimethoprim and tetracyclines cannot be recommended for empiric antibiotic therapy especially in moderate to severe cases where the Gram-negative pathogens predominate. However, it should be mentioned that some microorganisms showed a high rate of resistance to certain antibiotics just because of their intrinsic resistance to this particular antibiotic (e.g., Pseudomonas spp. and tetracyclines, sulfamethoxazole-trimethoprim, Proteus spp. and Morganella spp. and colistin). MRSA represented 11.3% of total aerobic isolates in this study. IDSA recommended empiric MRSA coverage only if the local prevalence exceeds 50% for mild infection and 30% of moderate infection. Thus, the low prevalence may eliminate the need for empiric MRSA coverage in our hospital, except for patients with predisposing factors. The continuous monitoring and awareness of the antimicrobial susceptibility pattern of the DFI isolates is crucial for tailoring appropriate empiric antimicrobial therapy.
According to the resistance pattern of the isolates in the current study, and taking into consideration the predominant Gram-negative (Pseudomonas spp., Proteus spp., Klebsiella spp. and Acinetobacter spp.) and Gram-positive isolates (MRSA), the suggested empiric treatment regimen should cover XDR resistant Gram-negative pathogens and MRSA (only in patients with predisposing factors). We suggest a resistance rate of 40% or less to define an agent as potential empiric antibiotic. Colistin, imipenem, meropenem, piperacillin-tazobactam and vancomycin, teicoplanin, linezolid, quinolones, sulfamethoxazole-trimethoprim, clindamycin, tetracyclines were the most active against Gram-negative and Gram-positive isolates, respectively.
Nevertheless, the current study has some limitations: all patients received antibiotic therapy prior to sample collection, hence the microbiology and the antimicrobial profile might not be accurately representative. Moreover, the culture of anaerobic bacteria was not performed, thus our PCR results might have overestimated their clinical importance.
Conclusions
DFIs in our patients were commonly associated with MDR and XDR Gram-negative bacterial etiology. The antimicrobial susceptibility pattern suggests that colistin, imipenem, meropenem, or piperacillin-tazobactam may be appropriate agents for empirical coverage. Vancomycin should be added only if there are any predisposing factors. Continuous surveillance of resistant bacteria is crucial to provide the guidance for empirical therapy. Additionally, collection of sample for culture could be performed by swabbing technique after proper debridement without the need of invasive biopsy.
Further multicenter studies are indicated to determine the national microbial and antimicrobial profile of DFI in Egypt, to help accurate design of antimicrobial treatment guidelines for DFI to prevent further complications and to reduce the abuse of last-line antimicrobials.
Footnotes
Authors’ contributions statement: MAM, AAI, and HSK contributed to the study conception and design. MAM performed all microbiological and molecular laboratory work and wrote the manuscript. All authors (AAI, MAM, TAAE, NEAA and HSK) performed material preparation, data collection, and analysis. All authors revised and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: None to declare.
References
- 1.Armstrong DG, Boulton AJM, Bus SA. Diabetic foot ulcers and their recurrence. N Engl J Med. 2017;376:2367–75. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 2.Noor S, Zubair M, Ahmad J. Diabetic foot ulcer – a review on pathophysiology, classification and microbial etiology. Diabetes Metab Syndr. 2015;9:192–9. doi: 10.1016/j.dsx.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Pitocco D, Spanu T, Di Leo M, et al. Diabetic foot infections: a comprehensive overview. Eur Rev Med Pharmacol Sci. 2019;23;2(Suppl):26–37. doi: 10.26355/eurrev_201904_17471. [DOI] [PubMed] [Google Scholar]
- 4.Wagner FW., Jr The dysvascular foot: a system for diagnosis and treatment. Foot Ankle. 1981;2:64–122. doi: 10.1177/107110078100200202. [DOI] [PubMed] [Google Scholar]
- 5.Monteiro-Soares M, Russell D, Boyko EJ, et al. Guidelines on the classification of diabetic foot ulcers (IWGDF 2019) Diabetes Metab Res Rev. 2020;36(Suppl 1):e3273. doi: 10.1002/dmrr.3273. [DOI] [PubMed] [Google Scholar]
- 6.Tille P. Bailey and Scott's Diagnostic Microbiology. 14th Edition. St Louis, Missouri: Mosby Elsevier; 2017. [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 30th ed. CLSI supplement M100. Wayne, Pennsylvania, USA: CLSI; 2020. [Google Scholar]
- 8.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 9.Rekha R, Rizvi MA, Jaishree P. Designing and validation of genus-specific primers for human gut flora study. Electron J Biotechnol. 2006;9:505–11. doi: 10.2225/vol9-issue5-fulltext-2. [DOI] [Google Scholar]
- 10.Jovcic B, Lepsanovic Z, Suljagic V, et al. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011;55:3929–31. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao AN, Barlow M, Clark LA, Boring JR, 3rd, Tenover FC, McGowan JE, Jr, et al. Class 1 integrons in resistant Escherichia coli and Klebsiella spp., US hospitals. Emerg Infect Dis. 2006;12:1011–4. doi: 10.3201/eid1206.051596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50:3877–80. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane TD, Alexander JW, Johannigman JA. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cervantes-García E, Salazar-Schettino PM. Clinical and surgical characteristics of infected diabetic foot ulcers in a tertiary hospital of Mexico. Diabet Foot Ankle. 2017;8:1367210. doi: 10.1080/2000625X.2017.1367210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choucair J, Saliba G, Chehata N, Nasnas R, Saad NR. Epidemiology of the diabetic foot infection in a tertiary care hospital in the Lebanon: a retrospective study between 2000 and 2011. J Infect Dis Ther. 2018;6:381. doi: 10.4172/2332-0877.1000381. [DOI] [Google Scholar]
- 16.Kathirvel M, Prabakaran V, Jayarajan J, Sivakumar A, Govinda V. Risk factors for the diabetic foot infection with multidrug-resistant microorganisms in South India. Int Surg J. 2018;5:675–82. doi: 10.18203/2349-2902.isj20180374. [DOI] [Google Scholar]
- 17.Abd-El Mohsen SA. Diabetic foot ulcer infection rate, bacterial etiology and antibiotic susceptibility: a cross sectional study. Biomed Pharmacol J. 2020;13(1) doi: 10.13005/bpj/1854. [DOI] [Google Scholar]
- 18.Huang Y, Cao Y, Zou M, et al. A Comparison of tissue versus swab culturing of infected diabetic foot wounds. Int J Endocrinol. 2016;2016:8198714. doi: 10.1155/2016/8198714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gjødsbøl K, Skindersoe ME, Christensen JJ, et al. No need for biopsies: comparison of three sample techniques for wound microbiota determination. Int Wound J. 2012;9:295–302. doi: 10.1111/j.1742-481X.2011.00883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demetriou M, Papanas N, Panopoulou M, Papatheodorou K, Bounovas A, Maltezos E. Tissue and swab culture in diabetic foot infections: neuropathic versus neuroischemic ulcers. Int J Low Extrem Wounds. 2013;12:87–93. doi: 10.1177/1534734613481975. [DOI] [PubMed] [Google Scholar]
- 21.Otta S, Debata NK, Swain B. Bacteriological profile of diabetic foot ulcers. CHRISMED J Health Res. 2019;6:7–11. doi: 10.4103/cjhr.cjhr_117_17. [DOI] [Google Scholar]
- 22.Hefni A, Ibrahim AR, Attia KM, et al. Bacteriological study of diabetic foot infection in Egypt. J Arab Soc Med Res. 2013;8:26–32. [Google Scholar]
- 23.Dwedar R, Ismail DK, Abdulbaky A. Diabetic foot infection: microbiological causes with special reference to their antibiotic resistance pattern. Egypt J Med Microbiol. 2015;24:95–102. doi: 10.12816/0024935. [DOI] [Google Scholar]
- 24.Miyan Z, Fawwad A, Sabir R, Basit A. Microbiological pattern of diabetic foot infections at a tertiary care center in a developing country. J Pak Med Assoc. 2017;67:665–9. [PubMed] [Google Scholar]
- 25.Ogba OM, Nsan E, Eyam ES. Aerobic bacteria associated with diabetic foot ulcers and their susceptibility pattern. Biomed Dermatol. 2019;3:1. doi: 10.1186/s41702-019-0039-x. [DOI] [Google Scholar]
- 26.Xie X, Bao Y, Ni L, et al. Bacterial profile and antibiotic resistance in patients with diabetic foot ulcer in Guangzhou, Southern China: focus on the differences among different Wagner’s grades, IDSA/IWGDF grades, and ulcer types. Int J Endocrinol. 2017;2017:8694903. doi: 10.1155/2017/8694903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed MB, Said AMI, Naira EA, Fatani AJ, Emad EM, Soma AE. Infection in diabetic foot. Menoufia Med J. 2013;26:49–53. [Google Scholar]