Abstract
Deep learning (DL) based auto-segmentation has the potential for accurate organ delineation in radiotherapy applications but requires large amounts of clean labeled data to train a robust model. However, annotating medical images is extremely time-consuming and requires clinical expertise, especially for segmentation that demands voxel-wise labels. On the other hand, medical images without annotations are abundant and highly accessible. To alleviate the influence of the limited number of clean labels, we propose a weakly-supervised DL training approach using deformable image registration (DIR)-based annotations, leveraging the abundance of unlabeled data. We generate pseudo-contours by utilizing DIR to propagate atlas contours onto abundant unlabeled images and train a robust DL-based segmentation model. With 10 labeled TCIA dataset and 50 unlabeled CT scans from our institution, our model achieved Dice similarity coefficient of 87.9%, 73.4%, 73.4%, 63.2% and 61.0% on mandible, left & right parotid glands and left & right submandibular glands of TCIA test set and competitive performance on our institutional clinical dataset and a third party (PDDCA) dataset. Experimental results demonstrated the proposed method outperformed traditional multi-atlas DIR methods and fully-supervised limited data training and is promising for DL-based medical image segmentation application with limited annotated data.
1. Introduction
Deep neural networks (DNNs), which have gained great interest in recent years, have the ability of learning and summarizing the knowledge automatically from the training datasets and the ability of rapid and efficient inference (LeCun et al., 2015). Lots of deep learning (DL) studies accomplished promising performance in many specific tasks, such as image classification (He et al., 2016; Huang et al., 2017), object detection (Ren et al., 2015), instance segmentation (He et al., 2017), natural language processing (Radford et al., 2019), etc. The applications of DL in medical fields have gradually attracted researchers’ attention and made significant progress (Litjens et al., 2017; Hosny et al., 2018).
In radiation oncology, accurate image segmentation is crucial for treatment planning and treatment plan adaptation (Gu et al., 2010). Recently, DL-based segmentations have achieved great success in medical fields. U-Net (Ronneberger et al., 2015), which used convolution layers to take the full context of images into account in an end-to-end manner, made a breakthrough in this domain. The down-sampling and up-sampling layers as encoder-decoder extract the high-level and low-level features while skip connections concatenate these extracted features. Furthermore, numerous network structures similar to U-Net were developed and used for medical image segmentation (Çiçek et al., 2016; Milletari et al., 2016). Chen et al. (2019) combined clinical prior knowledge to localize small-volume organs with the coordinates of surrounding organs and segmented head and neck (H&N) organs in a coarse-to-fine manner. Xu and Niethammer (2019) jointly optimized their convolution networks for registration and segmentation and achieved improvements for both tasks compared with separate training. Zhou et al. (2019) proposed a semi-supervised method to fine-tune their pre-trained lesion segmentation network by semi-supervised learning from large quantities of lesion grading data with an attention mechanism. FocusNet (Kaul et al., 2019) solved the imbalance of the large and small H&N organs by concentrating on patches of small organs with a subnetwork.
To a certain extent, the great achievements of these DL methods in medical fields can be attributed to their large scale of training data (Sun et al., 2017). ChestXNet detected 14 pathologies on ChestXray-14, which contains over 100,000 frontal view X-ray images, and outperformed the practicing radiologists on F1 score (Rajpurkar et al., 2017). However, it is usually very time-consuming and costly to annotate such an enormous medical image dataset. For most daily life applications, where it is easy to obtain annotations like objects’ category or their bounding boxes, crowdsourcing would be a wise choice. However, annotating medical images requires clinical expertise from practicing doctors or radiologists to ensure the validity and reliability of the annotations, which restricts the size of labeled medical datasets for deep learning. Image-level or volume-wise labels for disease diagnosis may be extracted from medical reports using natural language processing (Rajpurkar et al., 2017), while voxel-wise labels for medical segmentation is very scarce. Therefore, it is challenging to obtain a large dataset to train robust segmentation models. In medical image segmentation fields, the problem of training deep neural networks with limited labeled data still needs to be addressed.
In this paper, we proposed a weakly-supervised DL algorithm for medical image segmentation with very limited labels. We innovatively used our in-house developed Demons based free-form deformable image registration (DIR) algorithm to transfer the contours of organs from labeled moving images to unlabeled fixed images to generate a large database and then trained the recursive ensemble organ segmentation (REOS) model (Lu et al., 2004; Lu et al., 2006; Gu et al., 2010; Chen et al., 2019). Though the contours generated with a DIR approach are not as accurate as manual delineated ground truth (Thor et al., 2011; Guy et al., 2019), DL models can learn and summarize the potential ground-truth from multiple noisy contours. We collected The Cancer Imaging Archive (TCIA) dataset, our clinical dataset from the University of Texas Southwestern Medical Center (UTSW), and the Public Domain Database for Computational Anatomy (PDDCA) dataset for training and evaluation. Ablation studies were conducted to explore how the number of training data and quality of the generated contours affect the segmentation performance. Finally, we compared the qualitative and quantitative performance between the proposed method and baselines that employ conventional multi-atlas DIR method or fully-supervised DNN.
2. Methods and materials
2.1. Weakly-supervised Framework
Due to lack of adequate clean annotations in organ segmentation, it is difficult to train a robust DL model for clinical applications. Therefore, we propose a weakly-supervised DL algorithm to alleviate the dependency of the segmentation model on such enormous clean labels. Consider a small labeled dataset with images Il and segmentation annotations Yl, and a large unlabeled dataset with only images Iu. It is common in medical applications that rare clean medical annotations are available (i.e. N ≪ M).
Our weakly-supervised method is illustrated in figure 1. The entire approach consists of two steps: 1) DIR propagates contours from the atlases to unlabeled images to generate pseudo-contours. 2) A 3D segmentation network is trained with generated pseudo-contours supervision.
Figure 1.
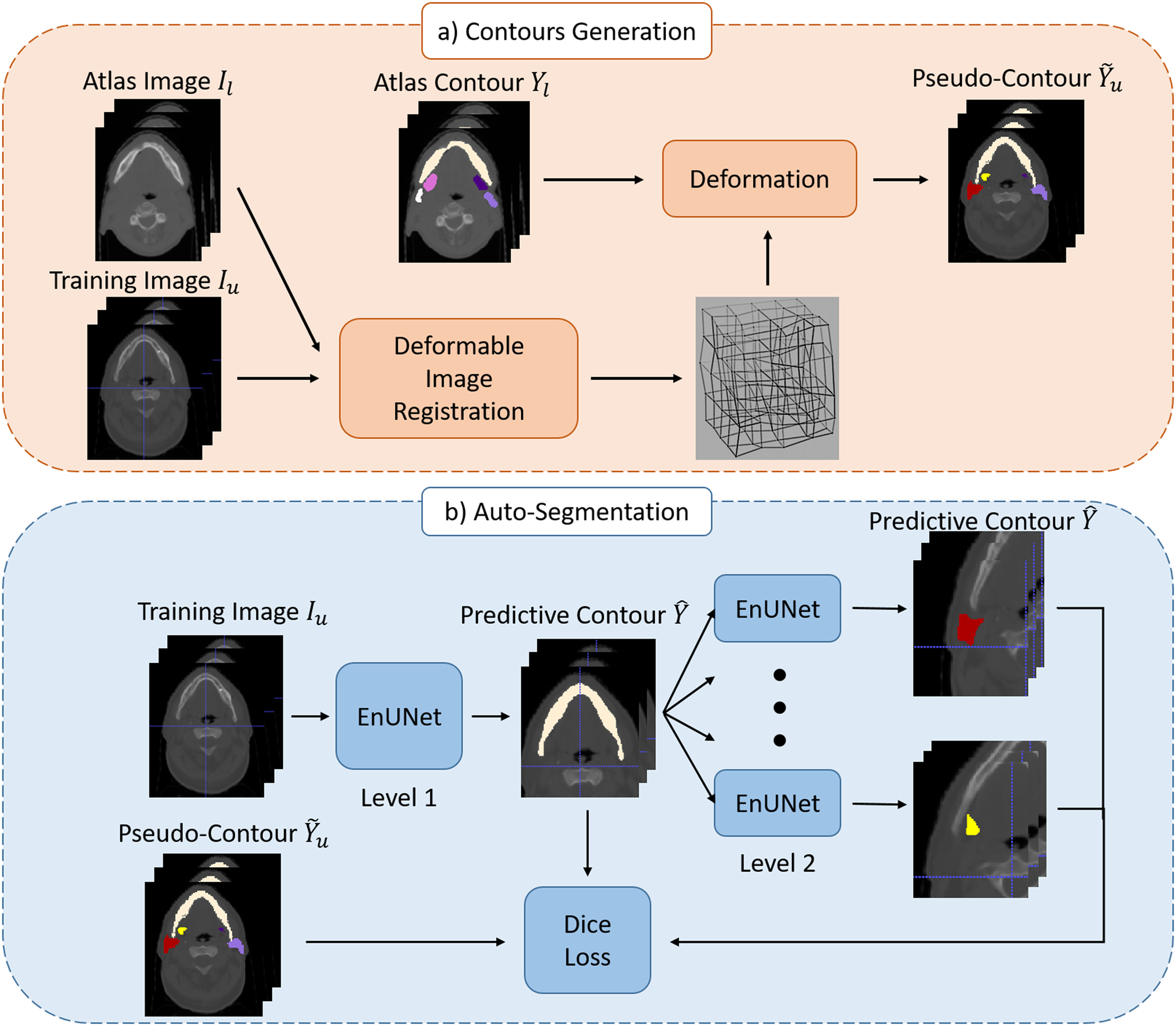
Illustration of the proposed method: a) pseudo-contour generation; b) auto-segmentation model trained with pseudo-contours.
2.1.1. Pseudo-contour Generation
In general, image registration is to find a transformation ϕ that maximizes the similarity between the moving image Im and fixed images If, which is defined as follows:
where is the similarity measure function and Im ∘ϕ is the warped moving image by the transformation ϕ. Besides translation and rotation in rigid registration, DIR warps moving images with deformation vector fields (DVF) to maximize the similarity between the warped moving image and the fixed image.
Since the voxel in the image and its corresponding segmentation mask represent homologous biological locations, they will remain homologous after applying the same DVF. If DIR can perfectly deform the moving image into the fixed image, the warped segmentation mask of the moving image should match the manual segmentation annotation of the fixed image. However, at present, DIR algorithms often cause certain errors between the warped image and fixed image, consequently inducing noise in the warped masks. In our approach, these warped segmentation masks from registration are regarded as pseudo-labels that deviate from ground-truth labels.
Our method exploits the Demons algorithm (Gu et al., 2010) to accomplish free-form DIR for generating pseudo-labels as shown in figure 2. Basically, the Demons algorithm associates each voxel in the fixed image with a Demon’s local force to deform the moving image. It iterates between the estimation of the local force and the transformation of the moving image with the force. We use labeled images Il from as moving images and unlabeled images Iu from as fixed images. In each iteration, the demon force is calculated as follows:
where is the warped moving image in the nth iteration and is the original moving image.
Figure 2.
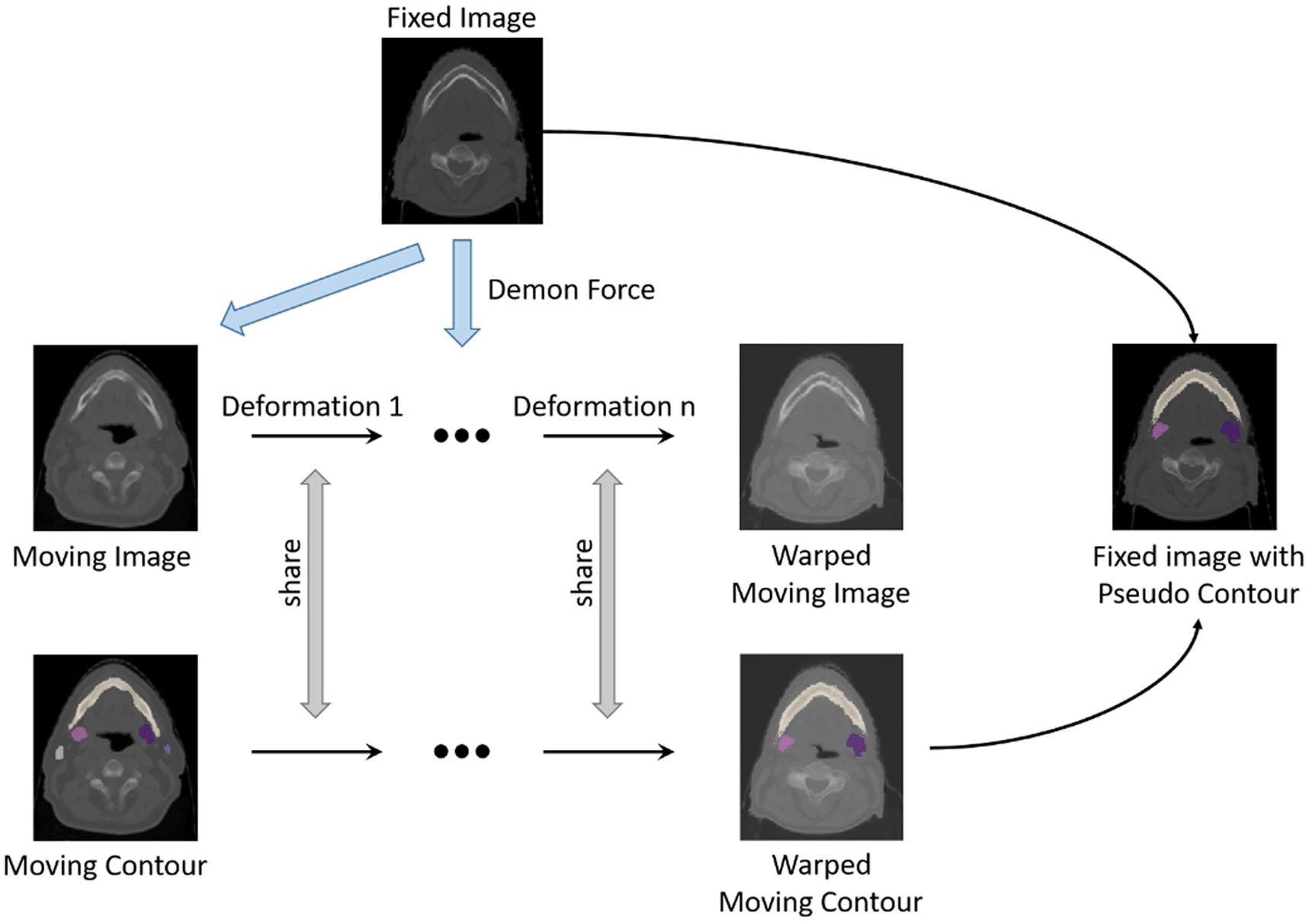
The procedure of generating pseudo contours with Demons DIR algorithm.
For each unlabeled image Iu, we performed DIR using each of the N labeled image Il as the moving image and generated N pseudo contours from the ground truth atlas, one for each of the N labeled images. There were M*N pseudo contours generated in total and used to train the segmentation neural network. Each DIR segmentation result transformed from different ground truth contours can be regarded as the actual contour plus random noise. With the N pseudo-labels for each unlabeled image, the DL model becomes robust to random noise and learns the ground truth through their common grounds (Vandat, 2017; Rolnick et al., 2017; Yu et al., 2020).
2.1.2. Segmentation model
We devised a deep neural network based on the REOS framework (Chen et al., 2019). Briefly, the REOS framework assigns different organs to different levels with their own regions of interest because neighbor organs can share location information for better delineation. Organs at low levels will facilitate localizations of the organs at high levels via their predicted segmentation mask. In this study, we focused on studying the segmentation of organs, including the mandible, left & right parotid glands, and left & right submandibular glands. According to their volume size and contrast, we assigned the mandible to level 1 and others to level 2, as shown in table 1. The intact volume of training data passes through the DNN of large organs in level 1. And the input of small organs in level 2 is cropped based on the location of organs segmented in level 1, discarding redundant information for better feature extraction.
Table 1.
The organ-at risk (OARs) to be studied and their mean volume (mm3) in the TCIA dataset. In our segmentation model, the mandible is segmented in level 1 and others in level 2.
| OARs | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|
| Volume (mm3) | 61093.46 | 30289.71 | 29345.19 | 9059.90 | 8846.28 |
Further, segmentation in each level has three modules: 1) A localization module segments target organs roughly to localize their positions and extract global features. 2) A refinement module focuses on local details to refine the segmentation mask. In this step, we take the image and its corresponding feature map from the localization module as the input of the refinement module. 3) An ensemble module concatenates the feature maps from the above modules to yield final precise contours.
Both the localization and contour modules use a 3D U-Net structure with two symmetric paths. The contracting path contains four convolutional blocks, each followed by a max-pooling layer as a down-sampling operation. In the expanding path, we up-sample feature maps to the same size as the contracting path. Skip connections are applied to concatenate the feature maps of the same depth in the two paths before each convolutional block to integrate high-level and low-level information. After the expanding path, we add an additional convolutional block to predict the contours. Moreover, the ensemble module is comprised of three consecutive convolutional layers. Details of the REOS algorithm can be found in the reference (Chen et al., 2019).
Since organs usually occupy a small region compared with the whole image, we choose the Dice loss for training to avoid the network bias towards the background. The Dice loss is defined as follows:
where YP and YG are the predictive and manual segmentation with K voxels, respectively.
2.2. Experimental Data
We demonstrate the proposed method for head and neck CT scans from public labeled datasets TCIA (Nikolov et al., 2018), PDDCA (Raudaschl et al., 2017), and an in-house UTSW dataset.
The TCIA dataset contains 31 annotated H&N CT scans from The Cancer Imaging Archive (Clark et al., 2013). Twenty-one organs at risk (OARs, normal organs) were delineated by a radiographer and arbitrated by another radiographer and a radiation oncologist, according to the consensus guideline Brouwer atlas (Brouwer et al., 2015). The in-plane pixel resolution of CT scans varies between 0.94 to 1.27 mm, and their slice thickness is 2.5 mm. The TCIA dataset is randomly split into three subsets: training (10), validation (10), and test set (11).
The PDDCA dataset contains 48 H&N CT images, 15 of which were used only for testing to exhibit the generalization ability of our model in this study. Nine OARs were manually delineated for each scan, including mandible, parotid glands, submandibular glands, brainstem, optic chiasm and optic nerves. Because PDDCA and TCIA datasets both contain CT images from the Radiation Therapy Oncology Group (RTOG) 0522 study, three common cases were excluded from both PDDCA test set and TCIA training/validation set.
The UTSW dataset contains 230 planning CT scans from different H&N cancer patients. The in-plane voxel resolution of CT scans varies between 1.17 to 1.37 mm and their slice thickness is 3 mm. The majority of UTSW images are unlabeled. Thus, they are used for generating pseudo contours from DIR and consequently for model training. Only six scans from the UTSW dataset were used as test images. For these 6 scans, a radiographer examined the original clinically-used contours and manually corrected five OAR contours, which were then defined as the ground truth labels for evaluation. One and two cases were excluded for right parotid gland and right submandibular gland, respectively, since these cases underwent surgical resection.
The total numbers of each organ for testing are shown in table 2. The size of training set in each experiment is determined by the number of the moving images from TCIA dataset and the fixed images from UTSW dataset. For a fair comparison, the test sets were only accessed during the final evaluation of segmentation model performance, and the validation set was used for model selection.
Table 2.
The numbers of CT images on TCIA, PDDCA and UTSW test sets.
| Dataset | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|
| TCIA | 11 | 11 | 11 | 11 | 11 |
| PDDCA | 12 | 12 | 12 | 12 | 12 |
| UTSW | 6 | 5 | 6 | 4 | 6 |
2.3. Experiment Design and Evaluation Metric
For all experiments, we sampled moving images from the TCIA training set and fixed images from the UTSW dataset to generate the pseudo labels via DIR. The UTSW images with pseudo labels were then used to train our segmentation model for H&N organs at risk. We experimented on five OARs: mandible, left & right parotid glands (Parotid-L and Parotid-R), and left & right submandibular glands (Submand-L and Submand-R). The Dice similarity coefficient (DSC), 95 percentile Hausdorff distance (95% HD) and average symmetric surface distance (ASD) were applied to evaluate the segmentation performances of our method and the baselines.
Our DL model was compared to the performance of multi-atlas DIR (Rohlfing et al., 2004; Iglesias and Sabuncu, 2015). Since multiple contours were generated for each test image via registration with several moving images, both DIR-average and DIR-majority-vote were taken into consideration. DIR-average calculates the mean DSC of the N generated contours, while DIR-majority-vote evaluates the mask that is the aggregate of voxels from N pseudo contours based on majority votes. Specifically, the mask of DIR-majority-vote can be described as follows:
where is the mask of voxel K of pseudo contours and [∙] is the rounding operation.
Since our method depends on the labels generated by registration, we conducted the experiment about the influence of the number of moving/fixed images and registration algorithms on the segmentation model. We also compared the proposed method and fully-supervised method.
To explore the effect of the number of moving images, 1/2/5/10 TCIA scans with contours were used for registration while keeping the number of UTSW scans M = 100 and other settings fixed.
To explore the effect of the number of fixed images, 10/50 unlabeled UTSW scans were used for registration while keeping the number of labeled TCIA scans N = 10 and other settings fixed.
Deep neural networks are susceptible to the quality of training data. The pseudo labels generated by registration are primarily affected by the registration algorithm. A rigid-body registration can be described with translations and rotations while DIR exploits complicated deformation vector fields to match two images after rigid registration. Therefore, DIR is more accurate than rigid body registration. Here, we explore the effect of quality of training data on segmentation performance by comparing models trained with contours transformed from rigid registration vs DIR warped contours. In this experiment, 10 TCIA scans and 50 UTSW scans were used as moving images and fixed images for registration, respectively.
To further demonstrate the performance of our method, we trained our segmentation model with only 10 labeled TCIA scans (fully-supervised model) for comparison. Our weakly-supervised segmentation method used 10 TCIA scans and 50 unlabeled UTSW scans for training.
2.4. Implementation
The CT images and their segmentation masks were pre-processed as follows. To make the spatial resolution consistent among all training data, both images and corresponding masks were resampled to the CT voxel size of 1.17mm × 1.17mm × 3mm. Before conducting deformable registration for generating pseudo contours, all the images and their masks were aligned to a standard CT atlas and cropped to the volume size of 256 × 256 × 128. To avoid gradient explosion during DNN training, the Hounsfield Units V of the CT images were normalized via .
Experiments were performed on the platform with an NVIDIA GeForce 2080 Ti GPU with 12 GB memory, and our segmentation model was implemented with the deep learning library Keras (version 2.2.4) using the backend of Tensorflow (version 1.13.1) in Python (version 3.7.3). We trained our neural networks with the Adam optimizer and learning rate of 10−4 until convergence. The best models on validation were saved for the final evaluation on the test sets. Due to the limited GPU memory, the input size of the model was 128 × 128 × 64, and the batch size was 2.
3. Results
3.1. Quantitative Results
We conducted experiments to explore the influence of different factors on the proposed weakly-supervised segmentation model.
The comparison of DIR-average and DIR-majority-vote using different numbers of moving images for registration is shown in table 3. When using only one moving image, DIR-majority-vote is the same as DIR-average, and their results are omitted in the table. When using two moving images, DIR-majority-vote predicts a voxel belonging to the OAR using one of the two warped labels, while DIR-average used the average DSC of the two warped labels. Thus, in the case of two moving images, the performance of DIR-majority-vote is slightly worse than DIR-average. However, as a result of its ability to exclude outliers, DIR-majority-vote outperforms DIR-average by 4.4% and 5.7% on average while using 5 and 10 TCIA scans as moving images, respectively. Therefore, for brevity, we will primarily compare our proposed method with the DIR-majority-vote in the following experiments.
Table 3.
Mean DSC (%) and standard deviation (in parentheses) of DIR-average and DIR-majority-vote with different numbers of moving images on the TCIA test set.
| Method | Moving images | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L | |
|---|---|---|---|---|---|---|---|
| DIR-average | 2 | 81.1 (±7.4) | 65.0 (±9.2) | 64.4 (±8.1) | 50.5 (±14.0) | 51.8 (±11.3) | |
| 5 | 80.4 (±6.9) | 62.7 (±8.6) | 62.6 (±8.2) | 49.4 (±12.7) | 50.6 (±11.8) | ||
| 10 | 78.7 (±8.0) | 61.9 (±9.3) | 63.6 (±8.6) | 46.5 (±14.1) | 48.7 (±11.9) | ||
| DIR-majority-vote | 2 | 79.9 (±8.0) | 61.8 (±8.9) | 60.8 (±6.3) | 45.4 (±14.4) | 50.0 (±10.4) | |
| 5 | 86.7 (±3.7) | 66.0 (±8.4) | 66.1 (±7.7) | 54.2 (±10.3) | 54.7 (±11.4) | ||
| 10 | 87.2 (±2.7) | 67.8 (±7.3) | 69.1 (±7.1) | 49.9 (±12.2) | 54.0 (±9.4) |
Figure 3 and figure 4 compare the mean DSC of our segmentation algorithm and those of DIR-majority-vote on the TCIA test set and UTSW test set, respectively, for different numbers of moving images. When using 10 moving images, the Dice scores of our model on Mandible, Parotid-R, Parotid-L, Submand-R and Submand-L of TCIA test set were 85.9%, 73.0%, 71.7%, 58.9%, 62.6%, respectively. And the Dice scores were 87.2%, 67.8%, 69.1%, 49.9%, 54.0%, respectively, for DIR-majority-vote. Overall, our segmentation model achieved better performance than DIR-majority-vote on 5 target OARs for both test sets, except the case on mandible when using 10 moving images. Our method improved segmentation capability with the DSC above 80%, 65%, and 50% on mandible, parotid glands and submandibular glands, respectively, throughout experiments with different numbers of moving images on the TCIA test set. In addition, more labeled moving images further facilitated the segmentation of our model, especially for small-volume OARs like submandibular glands. Similar conclusions can be observed on the UTSW test set.
Figure 3.
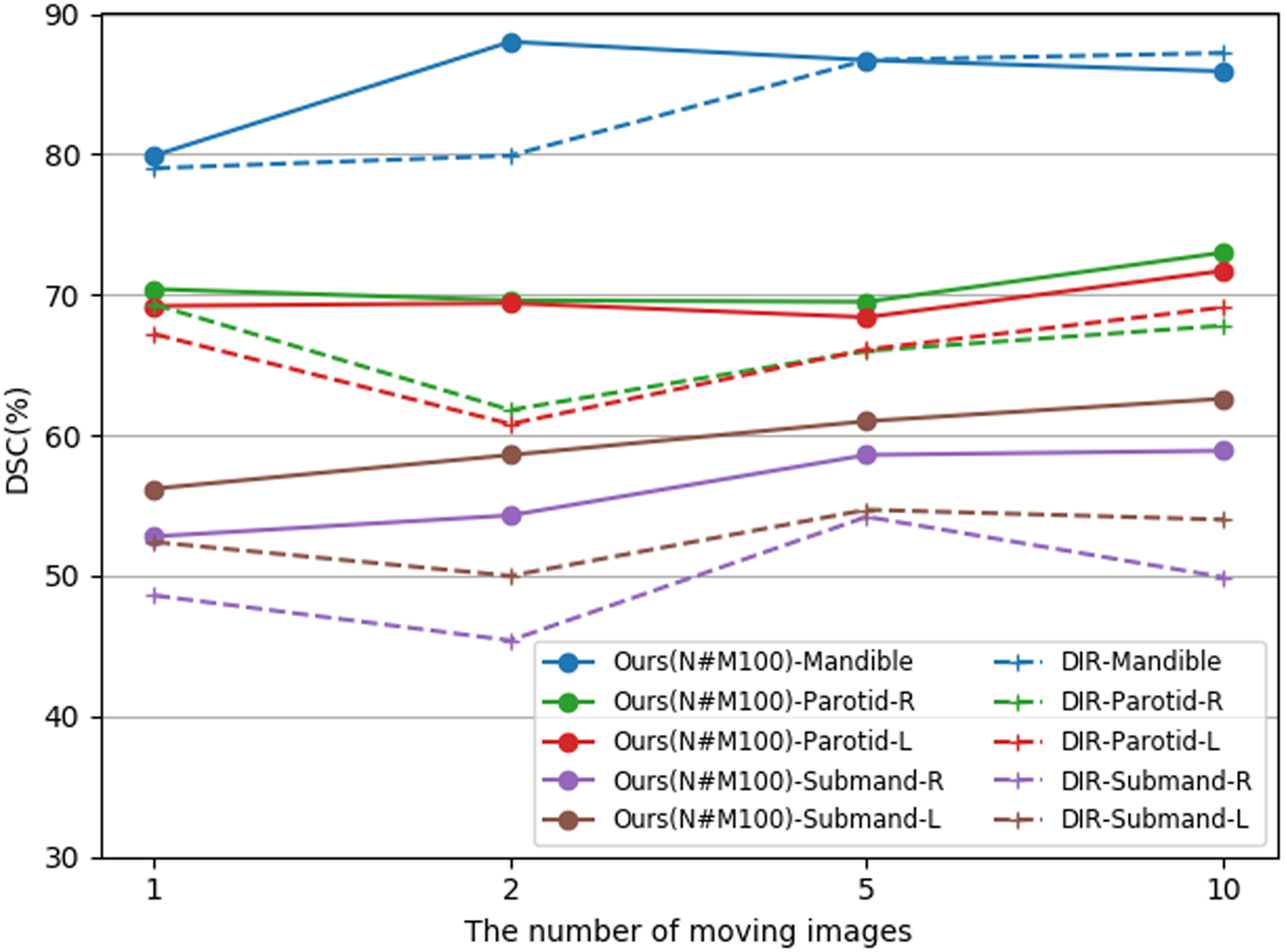
Mean DSC (%) of the proposed method (Ours(N#M100)-, solid lines) and DIR-Majority-vote (DIR-, dashed lines) on the TCIA test set while using different numbers of moving images and 100 fixed images for generating pseudo-labels. Here, the notation ‘N#M100’ refers to the number of labeled (moving) images N and the number of unlabeled (fixed) images M. Contours of mandible, left & right parotid glands (Parotid-L and Parotid-R), and left & right submandibular glands (Submand-L, Submand-R) are measured in this experiment.
Figure 4.
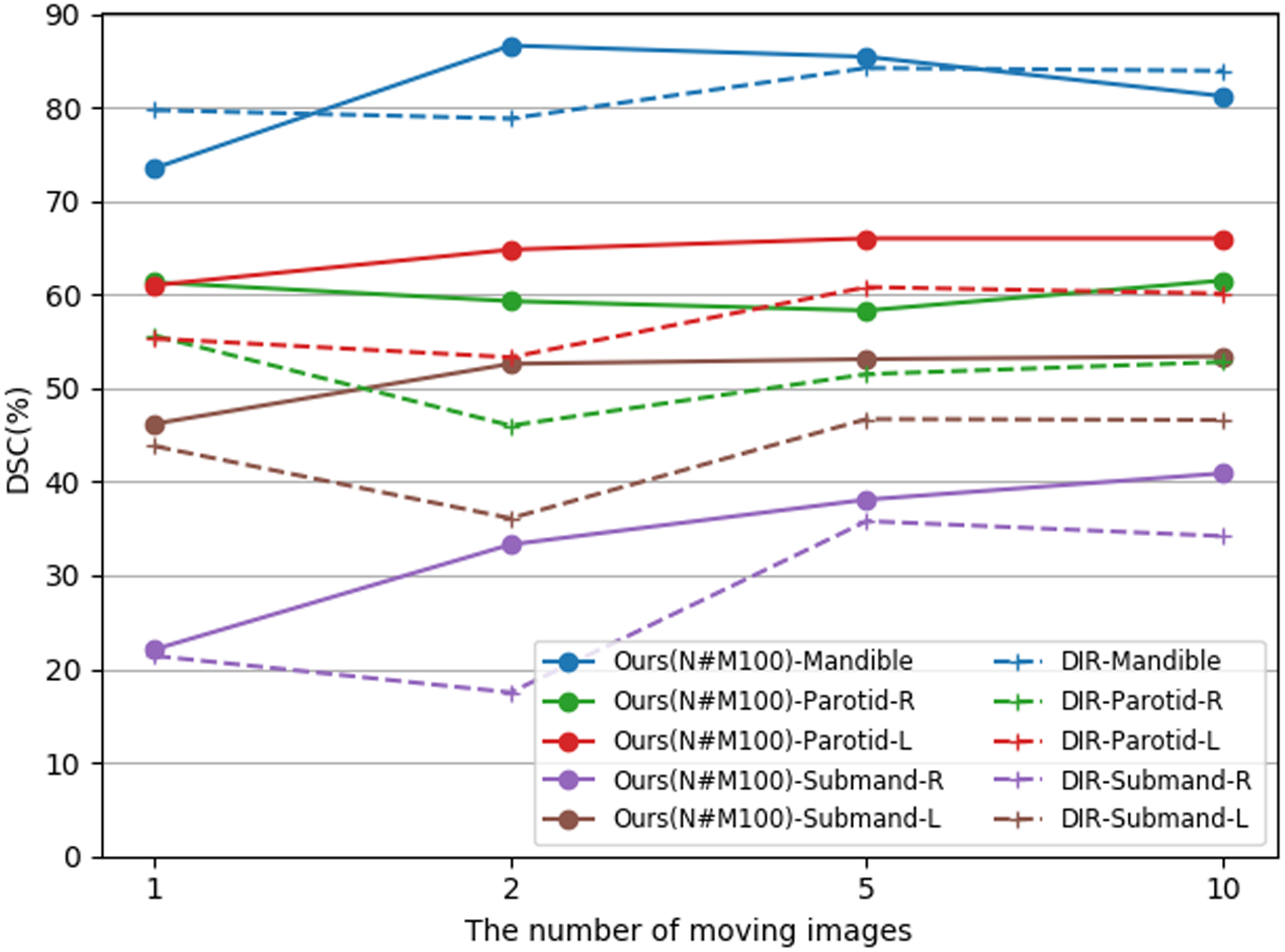
Mean DSC (%) of the proposed method (Ours(N#M100)-, solid lines) and DIR-Majority-vote (DIR-, dashed lines) on the UTSW test set while using different numbers of moving images and 100 fixed images for generating pseudo-labels.
Figure 5 illustrates the comparison between the proposed method with 10 or 50 fixed images, DIR-majority-vote and DIR-average on the TCIA test set. Compared with DIR-majority-vote and DIR-average, our weakly-supervised segmentation model improved the performance significantly for most cases, except for the DSC of mandible when using 10 fixed images. Moreover, the model of 50 fixed images ameliorated the segmentation performance on mandible, Parotid-L and Submand-R, remaining on a par with the model of 10 fixed images for Parotid-R and Submand-L. Exploiting more fixed images can promote the DL-assisted delineation of OARs without additional labeled moving images. And the model with 50 fixed images also decreased the standard deviation of DSC, especially for L&R submandibular glands.
Figure 5.
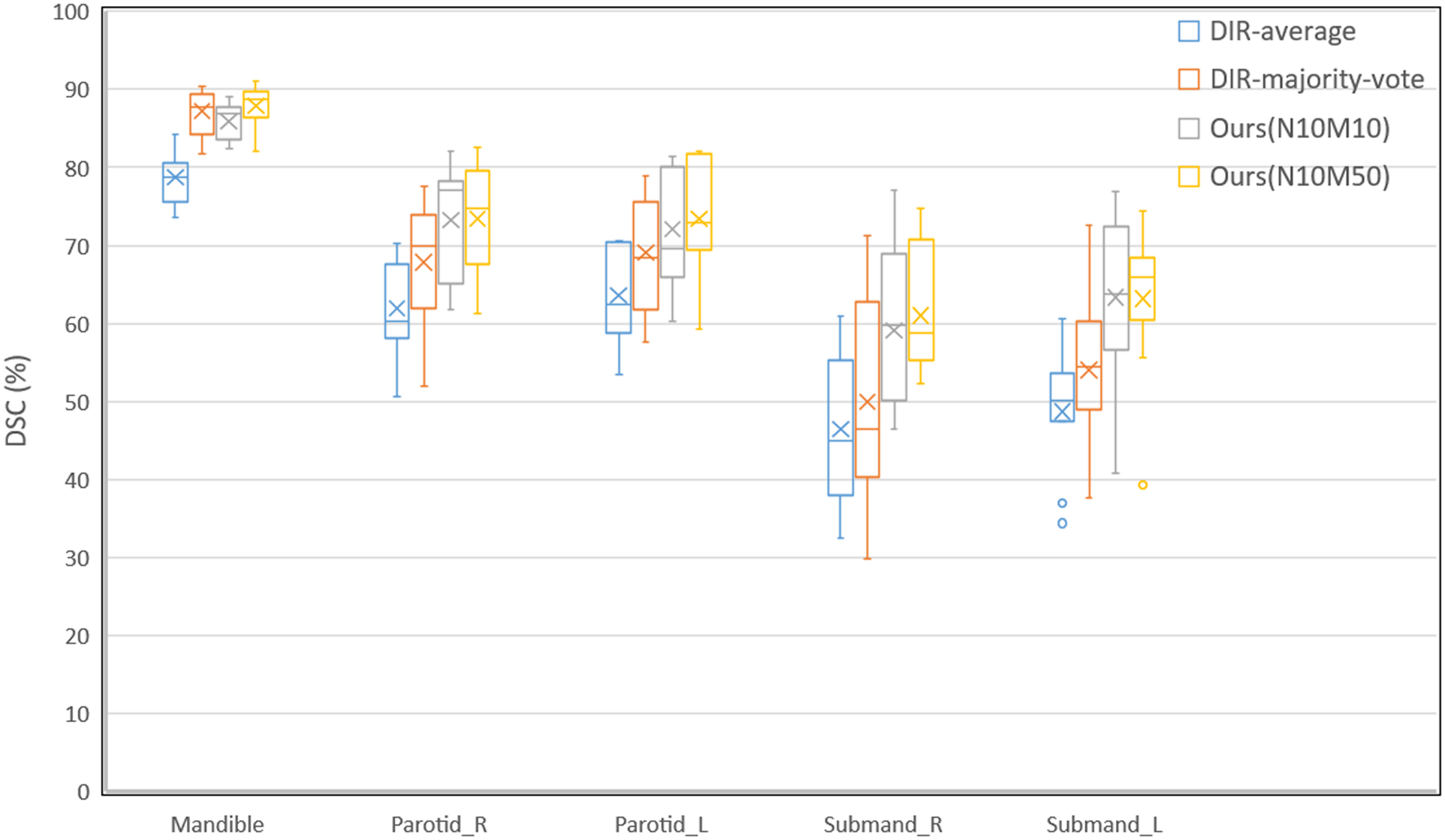
Comparison among the proposed method with 10 (Ours(N10M10)) and 50 fixed images (Ours(N10M50)), DIR-average, and DIR-majority-vote on the TCIA test set in box plots. The box shows the interquartile range (IQR) and the whiskers are 1.5× IQR. The horizontal line represents the median, the cross the mean Dice score, and the circle the outlier. All the experiments exploited 10 TCIA scans as moving images.
We further explored the effect of registration algorithms on our weakly-supervised method. Table 4 indicates the mean DSC of our method with rigid registration and DIR on TCIA and UTSW test sets. Compared with rigid registration, DIR notably increased the DSC of mandible, Parotid-R, Parotid-L, Submand-R, Submand-L by 36.4%, 14.3%, 15.1%, 6.2% and 10.4%, respectively, on the TCIA test set. More significant improvements were accomplished on the UTSW test set. Since the rigid registration with limited parameters could not match two images precisely and generate plausible labels for training, it will inevitably degenerate the generalization of deep neural networks.
Table 4.
Mean DSC (%) and standard deviation (in parentheses) of the proposed method with different registration algorithms for generating pseudo-labels. All the experiments employ 10 TCIA scans as moving images and 50 UTSW scans as fixed images.
| Test set | Registration algorithm | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L | |
|---|---|---|---|---|---|---|---|
| TCIA | Rigid registration | 51.5 (±11.7) | 59.1 (±11.3) | 58.3 (±12.5) | 54.8 (±10.9) | 52.8 (±8.9) | |
| DIR | 87.9 (±2.5) | 73.4 (±6.9) | 73.4 (±6.9) | 61.0 (±7.6) | 63.2 (±8.8) | ||
| UTSW | Rigid registration | 41.5 (±13.2) | 43.7 (±12.2) | 50.9 (±12.0) | 29.4 (±19.4) | 31.5 (±16.5) | |
| DIR | 84.5 (±3.9) | 62.1 (±11.4) | 68.1 (±10.9) | 42.0 (±29.1) | 54.4 (±19.1) |
To demonstrate the efficacy of the proposed method, a fully-supervised segmentation model was trained with manually labeled contours while our method only used the warped contours from registration. The comparisons of the proposed method, DIR-majority-vote and fully-supervised model in DSC, 95% HD and ASD are shown in table 5, 6 and 7, respectively. With 10 labeled and 50 unlabeled CT scans, our model achieved DSC of 87.9%, 73.4%, 73.4%, 63.2% and 61.0% for mandible, left & right parotid glands and left & right submandibular glands, respectively, on the TCIA test set. The mean DSC of our model on the TCIA test set was improved by 4.1%, 2.8%, 1.9% on Parotid-R, Parotid-L, Submand-R, while comparable on mandible and Submand-L, compared to the fully-supervised model. On the other two test sets, our model outperformed or was comparable to the fully-supervised model for all OARs except Submand-R of PDDCA dataset.
Table 5.
Mean DSC (%) and standard deviation (in parentheses) of the proposed method and the fully-supervised method (FS-model) trained with ground truth labels. Our segmentation model was trained with pseudo-labels registered from 10 TCIA scans to 50 UTSW scans, while the fully-supervised method was trained with 10 labeled TCIA scans.
| Test set | Method | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|---|
| TCIA | DIR-majority-vote | 87.2 (±2.7) | 67.8 (±7.3) | 69.1 (±7.1) | 49.9 (±12.2) | 54.0 (±9.4) |
| FS-model | 88.2 (±1.6) | 69.3 (±11.7) | 70.6 (±9.0) | 59.1 (±9.6) | 65.4 (±6.8) | |
| Ours(N10M50) | 87.9 (±2.5) | 73.4 (±6.9) | 73.4 (±6.9) | 61.0 (±7.6) | 63.2 (±8.8) | |
| UTSW | DIR-majority-vote | 83.9 (±2.5) | 52.8 (±10.2) | 60.1 (±8.0) | 34.2 (±23.3) | 46.6 (±14.5) |
| FS-model | 84.3 (±3.9) | 61.8 (±9.7) | 68.3 (±7.9) | 41.4 (±27.7) | 49.6 (±19.1) | |
| Ours(N10M50) | 84.5 (±3.9) | 62.1 (±11.4) | 68.1 (±10.9) | 42.0 (±29.1) | 54.4 (±19.1) | |
| PDDCA | DIR-majority-vote | 87.6 (±3.3) | 73.9 (±8.5) | 73.5 (±5.6) | 54.7 (±14.5) | 59.4 (±11.1) |
| FS-model | 88.1 (±1.6) | 76.2 (±8.7) | 75.8 (±4.7) | 61.8 (±9.3) | 63.4 (±8.6) | |
| Ours(N10M50) | 88.1 (±2.2) | 77.2 (±8.1) | 76.6 (±4.4) | 61.0 (±10.5) | 63.9 (±10.1) |
Table 6.
Ninety-five percentile Hausdorff distance (mm) and standard deviation (in parentheses) of the proposed method and the fully-supervised method (FS-model) trained with ground truth labels.
| Test set | Method | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|---|
| TCIA | DIR-majority-vote | 2.8 (±0.8) | 9.2 (±3.9) | 9.3 (±2.8) | 8.2 (±1.8) | 9.2 (±4.0) |
| FS-model | 2.6 (±0.7) | 10.3 (±5.5) | 10.5 (±5.3) | 7.7 (±3.5) | 5.8 (±1.7) | |
| Ours(N10M50) | 2.7 (±1.3) | 8.2 (±3.4) | 8.2 (±2.4) | 7.1 (±1.7) | 7.4 (±3.6) | |
| UTSW | DIR-majority-vote | 4.2 (±2.3) | 12.1 (±4.2) | 21.8 (±25.8) | 16.33 (±13.5) | 10.3 (±2.4) |
| FS-model | 3.5 (±1.5) | 11.9 (±5.0) | 8.2 (±1.7) | 16.09 (±15.6) | 8.4 (±3.4) | |
| Ours(N10M50) | 3.6 (±2.2) | 10.5 (±4.3) | 9.4 (±3.0) | 14.65 (±12.9) | 7.5 (±2.7) | |
| PDDCA | DIR-majority-vote | 2.6 (±1.0) | 7.8 (±2.9) | 8.3 (±3.2) | 7.2 (±2.9) | 7.3 (±2.0) |
| FS-model | 2.5 (±0.7) | 7.6 (±3.0) | 8.1 (±2.1) | 6.5 (±1.7) | 6.6 (±2.1) | |
| Ours(N10M50) | 2.2 (±0.7) | 7.3 (±2.7) | 7.8 (±2.6) | 6.8 (±2.5) | 7.1 (±2.5) |
Table 7.
Average symmetric surface distance (mm) and standard deviation (in parentheses) of the proposed method and the fully-supervised method (FS-model) trained with ground truth labels.
| Test set | Method | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|---|
| TCIA | DIR-majority-vote | 0.43 (±0.15) | 2.45 (±0.90) | 2.34 (±0.82) | 2.85 (±0.89) | 2.71 (±1.13) |
| FS-model | 0.40 (±0.08) | 2.56 (±1.33) | 2.49 (±1.21) | 2.39 (±0.78) | 1.86 (±0.61) | |
| Ours(N10M50) | 0.40 (±0.14) | 2.11 (±0.77) | 2.15 (±0.82) | 2.30 (±0.59) | 2.20 (±1.04) | |
| UTSW | DIR-majority-vote | 0.83 (±0.23) | 3.33 (±0.86) | 3.48 (±1.35) | 7.71 (±8.49) | 3.10 (±0.93) |
| FS-model | 0.77 (±0.32) | 2.79 (±0.97) | 2.25 (±0.44) | 7.85 (±9.52) | 2.91 (±1.25) | |
| Ours(N10M50) | 0.74 (±0.26) | 2.77 (±0.95) | 2.47 (±0.75) | 6.76 (±7.57) | 2.63 (±1.12) | |
| PDDCA | DIR-majority-vote | 0.44 (±0.15) | 2.03 (±0.97) | 2.05 (±0.60) | 2.38 (±1.06) | 2.14 (±0.74) |
| FS-model | 0.45 (±0.10) | 1.99 (±1.01) | 1.95 (±0.53) | 2.01 (±0.58) | 1.98 (±0.56) | |
| Ours(N10M50) | 0.44 (±0.13) | 1.87 (±0.85) | 1.92 (±0.46) | 2.14 (±0.74) | 2.03 (±0.72) |
Our model shows promising performances of 95% HD and ASD, which aligns with the results given by the DSC. Our DL model obtained average 95% HD of 6.72 mm, 9.13 mm and 6.24 mm on TCIA, UTSW and PDDCA test set, respectively, while the fully-supervised model obtained 7.38 mm, 9.62 mm and 6.26 mm, respectively. The proposed method had 10 lowest average surface distances out of 15 cases (5 OARs for each of three test sets). Moreover, the proposed method shows better performance of DSC, 95% HD and ASD than DIR-majority-vote on all three test sets.
Compared with conventional DIR and the fully-supervised model on three test sets, our approach manifests the superiority on the segmentation of medical images with limited annotations. The performances on the PDDCA test set show the generalization of our model to external datasets without overfitting the training data. It indicates the feasibility of our weakly-supervised segmentation method in the case where only a handful of images with manual annotations are available for DL.
3.2. Qualitative Results
Figure 6 shows the visualization comparison among the proposed method (trained with 10 moving images and 50 fixed images), DIR-majority-vote and fully-supervised model on the TCIA test set. In most cases, our model can accurately delineate the contour of mandible. However, the fully-supervised model failed to predict marginal parts of mandible and parotids as pointed by the red arrows. In addition, most of the pseudo contours mistakenly classified the teeth into mandible, resulting in the false delineation with DIR-majority-vote. For small-volume OARs, our segmentation model generated rough contours around the ground truth and performed better than the others.
Figure 6.
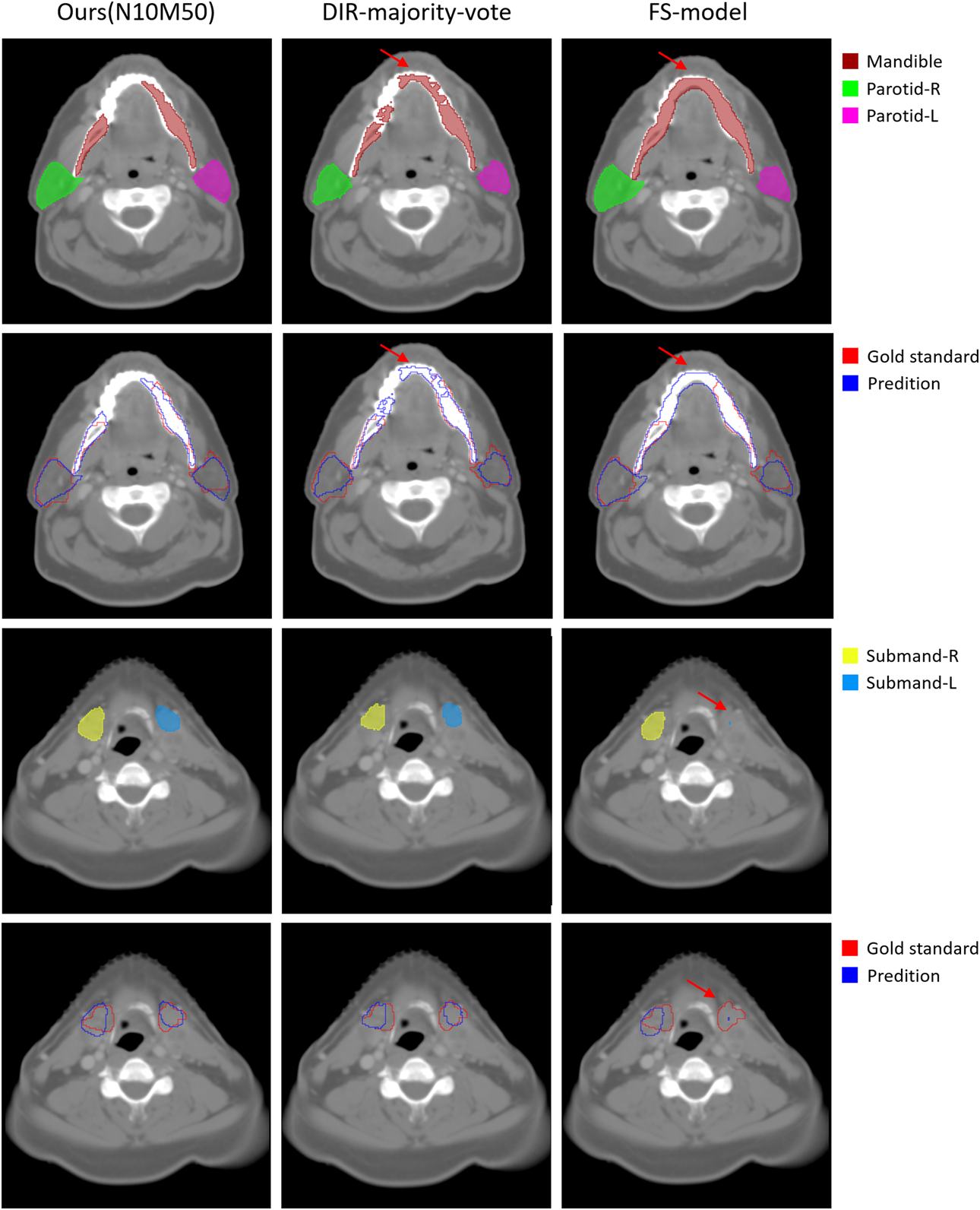
Qualitative comparison among the proposed method, DIR-majority-vote and fully-supervised model (FS-model) on a patient of the TCIA test set. The first and third rows are the predictive contours of each method while the second and fourth rows are the comparison between predictive contours and gold standards.
4. Discussion
In this paper, we proposed a DL-based algorithm weakly-supervised with DIR to automatically delineate OARs for radiotherapy treatment planning.
Because the pseudo-labels for training are generated from image registration, the number of moving images and fixed images will definitely affect our neural network. As seen in figure 3 and figure 4, our model achieved higher DSC of five organs when more labeled TCIA scans were exploited. Nevertheless, the improvement from 5 to 10 moving images was smaller than that from 1 to 2 moving images, except for left & right parotid glands, suggesting the slowing-down in the rate of improvement. After arriving at the bottleneck, the model does not substantially benefit from more moving images. Due to the limited labeled TCIA scans, we have not explored the optimal number of moving images. Generating pseudo-contours with more fixed images also increases the segmentation capacity of our DL model according to the experimental results. Nevertheless, the mean DSC with 100 fixed images is approximately similar to that with only 50 fixed images, which means that adding endless fixed images is not conducive to further improvement.
Table 8 shows the results on the TCIA and UTSW test sets when the training had the same number of pseudo contours but different combinations of the numbers of moving/fixed images. The model with 10/50 moving/fixed images increased the DSC of 5 OARs by 2.9% on average on the TCIA test set, which demonstrates the importance of moving images over fixed images. It is mainly because the pseudo contours propagated from the same moving image remain in a certain relation to the original contour of the moving image and thus lack diversity for training. In spite of this, the model with only one moving image performed delineation better than DIR-average and DIR-majority-vote (Figure 3 and 4).
Table 8.
Mean DSC (%) and standard deviation (in parentheses) of the proposed method trained with 500 pseudo contours, which are generated by 5/10 moving images and 100/50 fixed images.
| Test set | Method | Mandible | Parotid-R | Parotid-L | Submand-R | Submand-L |
|---|---|---|---|---|---|---|
| TCIA | Ours(N5M100) | 86.7 (±2.8) | 69.5 (±8.5) | 68.4 (±8.6) | 58.6 (±7.7) | 61.0 (±7.5) |
| Ours(N10M50) | 87.9 (±2.5) | 73.4 (±6.9) | 73.4 (±6.9) | 61.0 (±7.6) | 63.2 (±8.8) | |
| UTSW | Ours(N5M100) | 85.4 (±2.9) | 58.3 (±10.4) | 66.0 (±11.5) | 38.1 (±26.1) | 53.1 (±20.1) |
| Ours(N10M50) | 84.5 (±3.9) | 62.1 (±11.4) | 68.1 (±10.9) | 42.0 (±29.1) | 54.4 (±19.1) |
Image registration was applied to align the interpatient CT images in our experiments, and its precision affected the quality of the contours for training. Even if DNNs can converge and overfit the noisy training data, its capacity will inevitably decline due to the noise (Zhang et al., 2016; Yu et al., 2020). Since the organ shapes of different patients are of great discrepancy, rigid registration could not handle these cases well with the operation of translation and rotation. On the other hand, DIR generated more plausible labels to teach the segmentation model and resulted in substantial improvement even over 40% on the mandible. In addition, our method can further benefit from state-of-the-art registration algorithms.
Because of the ability to learn from the feedback iteratively, our weakly-supervised method for OARs segmentation manifests its superior performance over DIR-average, DIR-majority-vote and the fully-supervised model. The majority vote seeks intra-patient consensus among different observers, while the fully supervised model finds the interpatient relations. Our weakly-supervised method, in a sense, explores interpatient and intra-patient information simultaneously, leveraging the abundance of unlabeled data. Instead of using a fixed threshold (50%) in DIR-majority-vote, the deep neural networks can automatically trade off the accuracy and reliability of noisy contours and learn the best segmentation standard according to the training set. And our model is less prone to overfitting the training data due to multiple coarse contours for the same image, while the fully-supervised model might overfit these 10 training samples and degenerate its generalizability. Moreover, the fully-supervised model with limited ground truth dataset was inferior to the proposed model trained with the dataset composed of the synthetic data, which indicates that the model enhancement is due to the addition of synthetic data.
The proposed method can be applied for automatic contouring in adaptive radiotherapy (ART) application. For example, in a CBCT-driven ART, each study usually comprises CT scans with manual delineation (ground truth labels) for treatment planning and cone-beam CT (CBCT) scans acquired before each treatment for image guidance. The labeled CT images and unlabeled CBCT images can be used as moving images and fixed images of DIR, respectively, to generate pseudo labels for CBCT images. Once training the DL model with CBCT images and corresponding pseudo labels is finished, the model can predict the contours efficiently for the new coming CBCT images, which is essential for CBCT based adaptive planning.
However, there are some limitations in this study. 1) The results of the proposed method, DIR-average and DIR-majority-vote on the UTSW test set were worse than on the TCIA test set, except for mandible. This is because of the different procedures and standards for delineating the ground truth contours of these two test sets. In general, it is a challenging obstacle that most deep neural networks that perform well on the benchmarked datasets may fail on realistic images outside the training set (Yuille and Liu, 2018). 2) Due to the limited labeled CT images, our method is not competitive with state-of-the-art methods. Nikolov et al. (2018) trained a 3D U-Net network with 663 manually annotated CT scans from 389 patients and achieved mean DSC of 93.8%, 84.0%, 83.2%, 76.0% and 80.3% for mandible, Parotid-R, Parotid-L, Submand-R and Submand-L, respectively, on their test set. However, it is unfair to directly compare our results with these state-of-the-art DNN methods. These methods are fully-supervised and trained on large amounts of labeled data, and the performances of DNNs can easily benefit from large amounts of training data. We will dedicate to resolve the above limitations in the future.
5. Conclusion
In this study, we purpose a novel weakly-supervised training method for image segmentation to address the label-starving issue in the medical image fields. Experimental results demonstrated its superior performance for fast segmentation of head and neck anatomy. This training method can be easily generalized to other OARs and modalities like MRI.
Acknowledgments
This work was partially supported by NIH R01 CA218402 and R01 CA235723. The authors acknowledge Dr. Haibin Chen for assisting on the REOS algorithm/code, and Dr. Mingkui Tan for general comments on the manuscript.
References
- Brouwer CL, Steenbakkers RJHM, Bourhis J, Budach W, Grau C, Gregoire V, van Herk M, Lee A, Maingon P, Nutting C, O’Sullivan B, Porceddu SV, Rosenthal DI, Sijtsema NM and Langendijk JA 2015. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines Radiother Oncol 117 83–90 [DOI] [PubMed] [Google Scholar]
- Chen HB, Lu WG, Chen ML, Zhou LH, Timmerman R, Tu D, Nedzi L, Wardak Z, Jiang S, Zhen X and Gu XJ 2019. A recursive ensemble organ segmentation (REOS) framework: application in brain radiotherapy Phys Med Biol 64 025015. [DOI] [PubMed] [Google Scholar]
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T and Ronneberger O 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 424–32 [Google Scholar]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L and Prior F 2013. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository J Digit Imaging 26 1045–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XJ, Pan H, Liang Y, Castillo R, Yang DS, Choi DJ, Castillo E, Majumdar A, Guerrero T and Jiang SB 2010. Implementation and evaluation of various demons deformable image registration algorithms on a GPU Phys Med Biol 55 207–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Weiss E, Che SM, Jan N, Zhao S and Rosu-Bubulac M 2019. Evaluation of Image Registration Accuracy for Tumor and Organs at Risk in the Thorax for Compliance With TG 132 Recommendations Adv Radiat Oncol 4 177–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He KM, Gkioxari G, Dollar P and Girshick R 2017. Mask R-CNN IEEE International Conference on Computer Vision (ICCV) 2980–8 [Google Scholar]
- He KM, Zhang XY, Ren SQ and Sun J 2016. Deep Residual Learning for Image Recognition IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–8 [Google Scholar]
- Hosny A, Parmar C, Quackenbush J, Schwartz LH and Aerts HJWL 2018. Artificial intelligence in radiology Nat Rev Cancer 18 500–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, van der Maaten L and Weinberger KQ 2017. Densely Connected Convolutional Networks IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2261–9 [Google Scholar]
- Iglesias JE and Sabuncu MR 2015. Multi-atlas segmentation of biomedical images: A survey Med Image Anal 24 205–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul C, Manandhar S and Pears N 2019. FocusNet: an attention-based fully convolutional network for medical image segmentation IEEE 16th International Symposium on Biomedical Imaging (ISBI) 455–8 [Google Scholar]
- LeCun Y, Bengio Y and Hinton G 2015. Deep learning Nature 521 436–44 [DOI] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B and Sanchez CI 2017. A survey on deep learning in medical image analysis Med Image Anal 42 60–88 [DOI] [PubMed] [Google Scholar]
- Lu WG, Chen ML, Olivera GH, Ruchala KJ and Mackie TR 2004. Fast free-form deformable registration via calculus of variations Phys Med Biol 49 3067–87 [DOI] [PubMed] [Google Scholar]
- Lu WG, Olivera GH, Chen Q, Chen ML and Ruchala KJ 2006. Automatic re-contouring in 4D radiotherapy Phys Med Biol 51 1077–99 [DOI] [PubMed] [Google Scholar]
- Milletari F, Navab N and Ahmadi SA 2016. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation Int Conf 3d Vision 565–71 [Google Scholar]
- Nikolov S, Blackwell S, Mendes R, De Fauw J, Meyer C, Hughes C, Askham H, Romera-Paredes B, Karthikesalingam A and Chu C 2018. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy arXiv preprint arXiv:1809.04430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A, Wu J, Child R, Luan D, Amodei D and Sutskever I 2019. Language models are unsupervised multitask learners OpenAI Blog 1 9 [Google Scholar]
- Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C and Shpanskaya K 2017. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning arXiv preprint arXiv:1711.05225 [Google Scholar]
- Raudaschl PF, Zaffino P, Sharp GC, Spadea MF, Chen AT, Dawant BM, Albrecht T, Gass T, Langguth C, Luthi M, Jung F, Knapp O, Wesarg S, Mannion-Haworth R, Bowes M, Ashman A, Guillard G, Brett A, Vincent G, Orbes-Arteaga M, Cardenas-Pena D, Castellanos-Dominguez G, Aghdasi N, Li YM, Berens A, Moe K, Hannaford B, Schubert R and Fritscher KD 2017. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015 Med Phys 44 2020–36 [DOI] [PubMed] [Google Scholar]
- Ren SQ, He KM, Girshick R and Sun J 2015. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks Advances in Neural Information Processing Systems (NeurIPS) 28 91–9 [Google Scholar]
- Rohlfing T, Brandt R, Menzel R and Maurer CR 2004. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocol microscopy images of bee brains Neuroimage 21 1428–42 [DOI] [PubMed] [Google Scholar]
- Rolnick D, Veit A, Belongie S and Shavit N 2017. Deep learning is robust to massive label noise arXiv preprint arXiv:1705.10694 [Google Scholar]
- Ronneberger O, Fischer P and Brox T 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 9351 234–41 [Google Scholar]
- Sun C, Shrivastava A, Singh S and Gupta A 2017. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era IEEE International Conference on Computer Vision (ICCV) 843–52 [Google Scholar]
- Thor M, Petersen JBB, Bentzen L, Hoyer M and Muren LP 2011. Deformable image registration for contour propagation from CT to cone-beam CT scans in radiotherapy of prostate cancer Acta Oncol 50 918–25 [DOI] [PubMed] [Google Scholar]
- Vandat A 2017. Toward Robustness against Label Noise in Training Deep Discriminative Neural Networks Advances in Neural Information Processing Systems (NeurIPS) 30 5596–605 [Google Scholar]
- Xu Z and Niethammer M 2019. Deepatlas: Joint semi-supervised learning of image registration and segmentation International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 420–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhang E, Wu J, Yu H, Yang Z, Ma L, Chen M, Gu X and Lu W 2020. Robustness study of noisy annotation in deep learning based medical image segmentation arXiv preprint arXiv:2003.06240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuille AL and Liu C 2018. Deep Nets: What have they ever done for Vision? arXiv preprint arXiv:1805.04025 [Google Scholar]
- Zhang C, Bengio S, Hardt M, Recht B and Vinyals O 2016. Understanding deep learning requires rethinking generalization Proceedings of the International Conference on Learning Representations (ICLR) [Google Scholar]
- Zhou Y, He X, Huang L, Liu L, Zhu F, Cui S and Shao L 2019. Collaborative learning of semi-supervised segmentation and classification for medical images Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2079–88 [Google Scholar]


