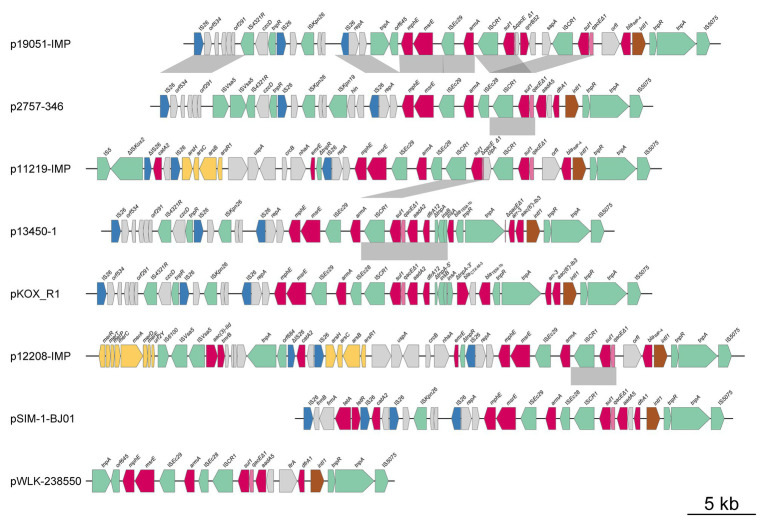Abstract
Aminoglycosides are important options for treating life-threatening infections. However, high levels of aminoglycoside resistance (HLAR) among Klebsiella pneumoniae isolates have been observed to be increasing frequently. In this study, a total of 292 isolates of the K. pneumoniae complex from a teaching hospital in China were analyzed. Among these isolates, the percentage of HLAR strains was 13.7% (40/292), and 15 aminoglycoside resistance genes were identified among the HLAR strains, with rmtB being the most dominant resistance gene (70%, 28/40). We also described an armA-carrying Klebsiella variicola strain KP2757 that exhibited a high-level resistance to all aminoglycosides tested. Whole-genome sequencing of KP2757 demonstrated that the strain contained one chromosome and three plasmids, with all the aminoglycoside resistance genes (including two copies of armA and six AME genes) being located on a conjugative plasmid, p2757-346, belonging to type IncHI5. Comparative genomic analysis of eight IncHI5 plasmids showed that six of them carried two copies of the intact armA gene in the complete or truncated Tn1548 transposon. To the best of our knowledge, for the first time, we observed that two copies of armA together with six AME genes coexisted on the same plasmid in a strain of K. variicola with HLAR. Comparative genomic analysis of eight armA-carrying IncHI5 plasmids isolated from humans and sediment was performed, suggesting the potential for dissemination of these plasmids among bacteria from different sources. These results demonstrated the necessity of monitoring the prevalence of IncHI5 plasmids to restrict their worldwide dissemination.
Keywords: plasmid, IncHI5, armA, high-level aminoglycoside resistance, Klebsiella pneumoniae
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the most common pathogens responsible for human infections. The emergence of highly antibiotic-resistant K. pneumoniae has become a major challenge facing clinical management and global public health (Kathryn et al., 2015).
Aminoglycosides are important options for treating life-threatening infections and are generally administered in combination with β-lactam agents (Becker and Cooper, 2013). However, increasing rates of aminoglycoside resistance in K. pneumoniae have been reported in recent years (Nasiri et al., 2018). Moreover, an increase in the aminoglycoside resistance rate in Escherichia coli isolates has been reported in France, leading to global concerns (Caméléna et al., 2020). Studies of aminoglycoside resistance in different species or populations have subsequently been conducted. A high level of aminoglycoside resistance in Acinetobacter baumannii and Pseudomonas aeruginosa isolated from a hospital in Brazil was observed (Ballaben et al., 2018), and an alarmingly high resistance to aminoglycosides has also been reported in diseased animals and clinical isolates of Enterobacterales from China (Fang-you et al., 2010; Yu-Ting et al., 2013).
The most common mechanism of resistance to aminoglycosides involves aminoglycoside-modifying enzymes (AMEs). These enzymes include acetyltransferases (AACs), nucleotidyltransferases (ANTs), and phosphotransferases (APHs), which differ in their ability to modify aminoglycosides (Miró et al., 2013). Other mechanisms contributing to aminoglycoside resistance involve the upregulation of efflux pumps (Poole, 2004) and reduced intake of antibiotics into the bacterial cell (Nakamatsu et al., 2007); however, the production of 16S rRNA methyltransferase (16S-RMTase) is another important mechanism mediating resistance to nearly all clinically available aminoglycosides. Different types of 16S-RMTase genes, including rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, armA, and npmA, have been identified (Doi et al., 2016). Of these genes, rmtB and armA represent the most widespread 16S rRNA methylase genes (Caméléna et al., 2020). Genes encoding AMEs or methylases can be carried in integrons or transposons located in a variety of plasmids, which may facilitate the rapid spread of 16S-RMTase genes and AME genes among bacterial species (Nagasawa et al., 2014).
The H incompatibility (IncH) group is composed of IncHI and IncHII. IncHI plasmids can be further divided into five subgroups, IncHI1 to IncHI5, based on the different replication genes they contain (Liang et al., 2018). Currently, only 10 armA-carrying IncHI5 plasmids have been sequenced (last accessed on June 1st, 2020): pWLK-238,550 (CP038277), p13450-1 (CP026014), an unnamed plasmid (CP022441), pRo24724 (CP021328), pSIM-1-BJ01 (MH681289), p19051-IMP (MF344565), p13450-IMP (MF344564), p12208-IMP (MF344562), p11219-IMP (MF344561; Liang et al., 2018), and pKOX_R1 (CP003684; Huang et al., 2013). In this work, based on antimicrobial susceptibility testing, molecular biological methods, and whole-genome sequencing, the molecular resistance mechanisms of the clinical K. pneumonia complex, especially a high-level aminoglycoside-resistant strain that carries two copies of armA and an additional six AME genes, were analyzed.
Materials and Methods
Bacterial Strains
A total of 292 nonrepetitive isolates of the K. pneumonia complex were isolated from sputum, urine, blood, secretion, and pus samples for routine examination from 2015 to 2017 in an affiliated hospital of Wenzhou Medical University in Zhejiang Province, China. These strains were identified using a VITEK 2 automated microbiology analyzer (BioMerieux Corporate, Craponne, France). Given that the strains could not be distinguished into exact species on the basis of their biological phenotypes and multilocus sequence typing (MLST; Long et al., 2017; Yatao et al., 2018), we referred to them collectively as the K. pneumonia complex.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) were determined by the agar dilution method and interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines. As no breakpoint criteria for streptomycin were established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) or CLSI, the corresponding results were interpreted according to the criteria of the US Food and Drug Administration (Ya et al., 2017). High-level resistance of the bacterium to aminoglycosides was defined according to the MICs of certain antibiotics. For streptomycin (Pericàs et al., 2017) and kanamycin, MIC was ≥1,024 mg/L. For netilmicin, the MIC was ≥512 mg/L, while for gentamicin, amikacin, and tobramycin, the MIC was ≥256 mg/L (Caméléna et al., 2020). E. coli ATCC 25922 was used as a quality control strain.
Genome Sequencing, Assembly, Annotation, and Bioinformatic Analysis
In order to identify aminoglycoside resistance genes that were carried by all strains, we performed pooled genomic DNA sequencing for 292 isolates. Each strain was cultured in 5 ml of lysogeny broth (LB) at 37°C for 16 h, and the strains were subsequently pooled together. Whole-cell DNA was extracted using an AxyPrep Bacterial Genomic DNA Miniprep Kit (Axygen Scientific, Union City, CA, United States) and pooled DNA sequencing was performed at Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China). Megahit (Li et al., 2015) was used to assemble the pooled sequencing data. In addition, three strains (KP2757, KP2723, and KP1878) showing high-level resistance to aminoglycosides were selected for whole-genome sequencing (WGS). Genomic DNA was extracted using the AxyPrep Bacterial Genomic DNA Miniprep kit and sequenced with Illumina NovaSeq 6,000 with 2 × 150-bp paired-end libraries (Illumina, Inc., San Diego, CA, United States) and PacBio RS II (Pacific Biosciences) systems with a 20-kb library at Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China). The Illumina reads were subjected to adaptor trimming and quality filtering (Phred quality score >20). Reads from PacBio RS II sequencing were assembled using the Canu v1.8 pipeline (Koren et al., 2017), and the quality of the genomic sequence was further corrected by reads from Illumina NovaSeq 6,000 sequencing.
Open reading frame (ORF) prediction of the pooled sequence data was performed using Prodigal, with the default parameters. Antibiotic resistance genes were predicted using both the Comprehensive Antibiotic Resistance Database (CARD; Moura et al., 2009) and the ResFinder (Jia et al., 2017) database. The relative abundance (sequencing depth) of a certain gene was calculated using BBMap.1 Potential ORFs of the whole genome of an isolate were predicted using the RAST pipeline (Overbeek et al., 2014) and annotated against the UniProt/SwissProt and nonredundant protein databases using the BLASTX program, with an e-value threshold of 1e-5. Annotation of mobile genetic elements and resistance genes was performed using ISfinder (Siguier et al., 2006), INTEGRALL, CARD, and ResFinder, with the default parameters being employed accordingly.
Basic genomic features were constructed using BLAST Ring Image Generator (BRIG; Nabil-Fareed et al., 2011). The comparison of nucleotide sequences was visualized using the genoPlotR package.2 A SNP tree was reconstructed with the pipeline CSI phylogeny accessible from the Center for Genomic Epidemiology.3
Detection of Aminoglycoside Resistance Genes by PCR
The distribution of 16 aminoglycoside resistance genes was screened via PCR. The primers used for the resistance genes and their expected amplicon sizes and annealing temperatures are shown in Supplementary Table S1. The PCR products were sequenced, and the sequences were compared with those in the NCBI nucleotide database using BLAST.4
Conjugation Experiments
Conjugation experiments were performed with sodium azide-resistant E. coli J53 as the recipient and the armA-positive isolate KP2757 as the donor. Briefly, a mixture of the donor and recipient strains was placed onto a sterile nitrocellulose filter, which was subsequently placed on a brain heart infusion (BHI) agar plate and incubated at 24°C for 24 h. The transconjugant was selected on a Muller-Hinton (MH) agar plate containing 256 mg/L sodium azide plus 64 mg/L amikacin. The existence of armA and additional AMEs in the transconjugant was verified by PCR and Sanger sequencing. Antimicrobial susceptibility testing was further performed for the transconjugant.
Nucleotide Sequence Accession Numbers
The complete nucleotide sequences of chromosome KP2757 and plasmids p2757-138, p2757-40, and p2757-346 were submitted to GenBank under accession numbers CP060807–CP060810.
Results
Susceptibility Testing
MICs of six aminoglycosides against 292 K. pneumonia complex isolates showed that the resistance rates for gentamicin, kanamycin, tobramycin, netilmicin, amikacin, and streptomycin were 14.73% (43/292), 14.38% (42/292), 12.33% (36/292), 11.3% (33/292), 9.93% (29/292), and 9.59% (28/292), respectively. Although the resistance percentages for these six antibiotics did not exhibit notable differences (ranging from 9.59 to 14.73%), MICs, especially MIC90 values, differed considerably (>64-fold difference) among the antibiotics. MIC90 values for amikacin (16 mg/L) and streptomycin (32 mg/L) were considerably lower than those of gentamicin (>256 mg/L) and kanamycin (>1,024 mg/L), respectively (Table 1). Of the 292 KPC isolates, 62 were resistant to at least one of the aminoglycosides, and the percentage of high levels of aminoglycoside resistance (HLAR) was 13.7% (40/292). All 40 high-level aminoglycoside-resistant strains showed resistance to kanamycin, with 97.2% (39/40) showing MICs of >1,024 mg/L and the remaining strain showing a MIC of 128 mg/L. In addition, 70% (28/40) showed high MICs for five aminoglycosides (MICs of ≥1,024 mg/L for kanamycin; ≥256 mg/L for gentamicin, amikacin, and tobramycin; and ≥512 mg/L for netilmicin). Of note, KP2757 was the only strain showing high MICs for all six antibiotics (including MICs ≥1,024 mg/L for streptomycin). Ten strains showed a high MIC (>1,024 mg/L) for one antibiotic (kanamycin), while one strain (KP2723) showed a high MIC (256 mg/L) for gentamicin (Supplementary Table S2). The MICs of other classes of antimicrobials are shown in Supplementary Table S3.
Table 1.
In vitro susceptibility to six aminoglycoside antibiotics for 292 isolates of the Klebsiella pneumoniae complex (mg/L).
| Antibiotics | Breakpoints (CLSI) | Range | MIC50 | MIC90 | Resistance (%) |
|---|---|---|---|---|---|
| Amikacin | ≥64 | 0.125–>1,024 | 1 | 16 | 9.93 |
| Gentamicin | ≥16 | 0.125–>256 | 0.25 | >256 | 14.73 |
| Kanamycin | ≥64 | 0.25–>1,024 | 1 | >1,024 | 14.38 |
| Netilmicin | ≥32 | 0.125–>512 | 0.25 | 128 | 11.3 |
| Tobramycin | ≥16 | 0.125–>256 | 0.25 | >256 | 12.33 |
| Streptomycin | ≥64a | 0.125–>1,024 | 2 | 32 | 9.59 |
MIC50/90, MIC that inhibits 50/90% of the isolates, respectively.
The clinical breakpoint is defined by the US Food and Drug Administration.
Detection of Aminoglycoside Resistance Genes
The pooled DNA sequencing of the 292 isolates predicted 16 aminoglycoside resistance genes, of which 15 were identified among the 40 HLAR strains [the most dominant resistance gene was rmtB (70%, 28/40)]. In addition, 55% (22/40) of the isolates carried two or more resistance genes. The most prevalent combination of the resistance genes was rmtB and aadA2, and 15% (6/40) of the strains had this combination of genes. Of the 29 16S-RMTase gene-positive strains, strain KP2757 carried the most aminoglycoside resistance genes, namely, aac(3)-IId, aac(6´)-Ib3, aph(3´´)-Ib, aph(6)-Id, aadA2, aadA5, and armA (free of rmtB), while the other 28 isolates were rmtB positive. Ten of 16S-RMTase gene-negative strains showed a high MIC (>1,024 mg/L) for kanamycin, which carried at least one aac(3)-IId or aph(3´)-Ia gene. The aminoglycoside resistance genes of the 40 high-level aminoglycoside-resistant strains are shown in Supplementary Table S2.
Overview of the Whole-Genome Sequence of KP2757
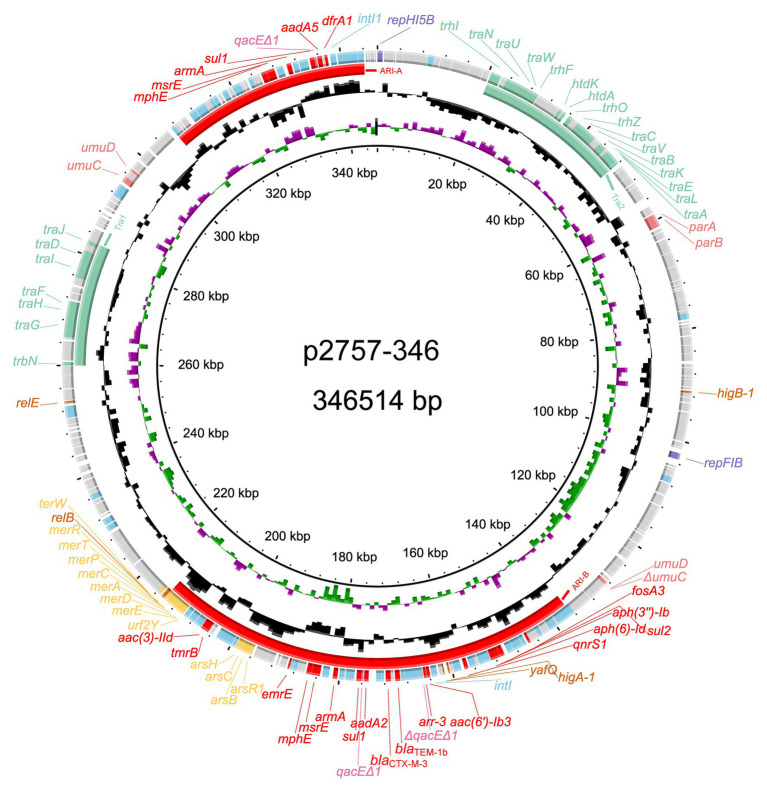
The complete genome sequences of three strains were obtained, and based on average nucleotide identity (ANI) analysis, KP2757 was identified as Klebsiella variicola, while the remaining strains were K. pneumonia strains. Annotation results of the genome sequences revealed the unique structure of the armA-related sequence in a plasmid of KP2757, so we focused on the analysis of KP2757 in this work. KP2757 contained a 5.21-Mb chromosome and three plasmids of different replicon types. Plasmid p2757-138 was an IncFII(K)/IncR plasmid, while p2757-40 was a nontypable plasmid according to PlasmidFinder analysis.5 Plasmid p2757-346 was 346,514 bp in size and was assigned to the IncHI5 group (Liang et al., 2018). This plasmid had an average G + C content of 47.25% and harbored 387 predicted ORFs. Sequence analysis demonstrated that p2757-346 contained a typical IncHI5 backbone (Liang et al., 2018), including genes involved in plasmid replication (repHI5B and repFIB-like), partitioning (parA, parB), and conjugative transfer (transfer regions Tra1 and Tra2; Figure 1). Notably, in addition to genes providing resistance to β-lactams (bla CTX-M-3 and bla TEM-1b), fosfomycin (fosA3), macrolides (mphE and msrE), rifampicin (arr-3), quinolones (qnrS1), sulfonamides (sul1 and sul2), and trimethoprim (dfrA1), p2757-346 carried eight aminoglycoside resistance genes [two copies of armA, aac(3)-IId, aac(6´)-Ib3, aph(3´´)-Ib, aph(6)-Id, aadA2, and aadA5]. Heavy metal resistance genes were also identified on plasmid p2757-346. Sequence analysis showed that plasmid p2757-346 carried different types of mobile genetic elements (MGEs), including a number of IS elements (e.g., IS5075, IS26, IS903, ISEc33, ISEc28, ISEc29, ISCR1, ISVsa5, IS6100, ISKpn28, ISKpn26, ISKpn21, ISKpn19, IS4321R, and ISBcen27) and class 1 integrons (Figure 1).
Figure 1.
Schematic diagram of the p2757-346 plasmid. The various colors indicate different gene functions, including replication (in purple), transfer (in green), maintenance (in pink), and resistance (in red), and mobile elements (in blue). The circles show (from outside to inside) predicted coding sequences, the GC content, the GC skew, and the scale (in kilobases).
Comparative Genomic Analyses of the Backbone Sequences of armA-Carrying IncHI5 Plasmids
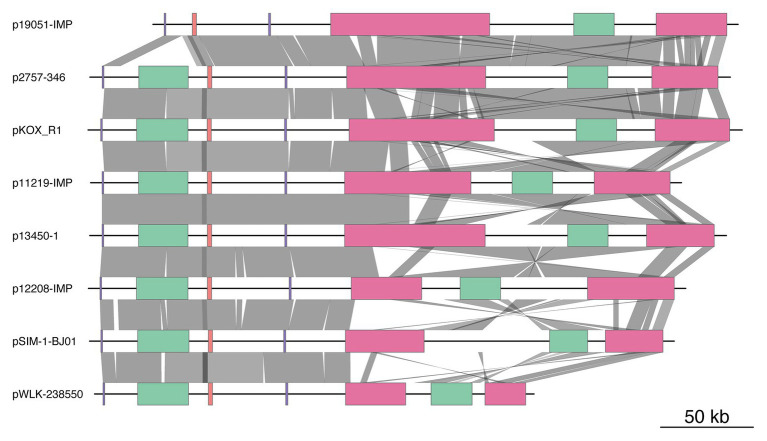
By searching the NCBI nucleotide database and using sequences of the combination of replicon genes of IncHI5 plasmids and the armA gene as queries, we obtained the sequences of 10 armA-carrying IncHI5 plasmids. These plasmids were detected in K. pneumoniae [p11219-IMP, p12208-IMP, p13450-IMP, pSIM-1-BJ01, p19051-IMP, and unnamed plasmid (CP022441)], K. variicola (p13450-1), Klebsiella michiganensis (pKOX-R1), and Raoultella ornithinolytica (pWLK-238,550 and pRo24724). Sequences of all plasmids were complete, except for pRo24724. Sequence alignment revealed that two pairs of plasmids [pSIM-1-BJ01/unnamed plasmid (CP022441) and p13450-IMP/p13450-1] shared identical sequences. Therefore, three plasmids [pRo24724, unnamed plasmid (CP022441), and p13450-IMP] were omitted, and eight plasmid sequences were used for the comparative genomic analysis (Figure 2 and Supplementary Table S4).
Figure 2.
Comparison of the genome structure of eight plasmids. The color-coded boxes indicate different gene functions, including replication (in purple), plasmid transfer (in green), plasmid partition (in pink), and antibiotic resistance island (ARI; in red). Orthologous regions are connected and color coded. The plasmids and their accession numbers are p2757-346 (CP060810), pWLK-238,550 (CP038277), p13450-1 (CP026014), pSIM-1-BJ01 (MH681289), p19051-IMP (MF344565), p12208-IMP (MF344562), p11219-IMP (MF344561), and pKOX_R1 (CP003684). The shading indicates 90% nucleotide sequence identity.
The major IncHI5 backbone genes for replication, partition, and conjugal transfer were conserved among these eight plasmids. Two conjugal transfer regions, Tra1 and Tra2, were found in seven IncHI5 plasmids, but p19051-IMP had only a Tra1 region. Plasmid p2757-346 contained more than 20 conjugal transfer genes, including traA, traB, traC, traD, traE, traF, trhF, htdK, and htdA. Comparative genomic analysis indicated that the transfer regions of the four plasmids, pKOX-R1, p13450-1, p12208-IMP, and p11219-IMP, showed high similarity to that of p2757-346 (>95% coverage and >99% identity).
Phylogenetic analysis of these eight IncHI5 plasmids indicated that p2757-346 was closely related to plasmid pKOX-R1, the first sequenced IncHI5 plasmid, isolated from K. michiganensis (Huang et al., 2013) and plasmid p19051-IMP from K. pneumoniae (Liang et al., 2018; Figure 3).
Figure 3.
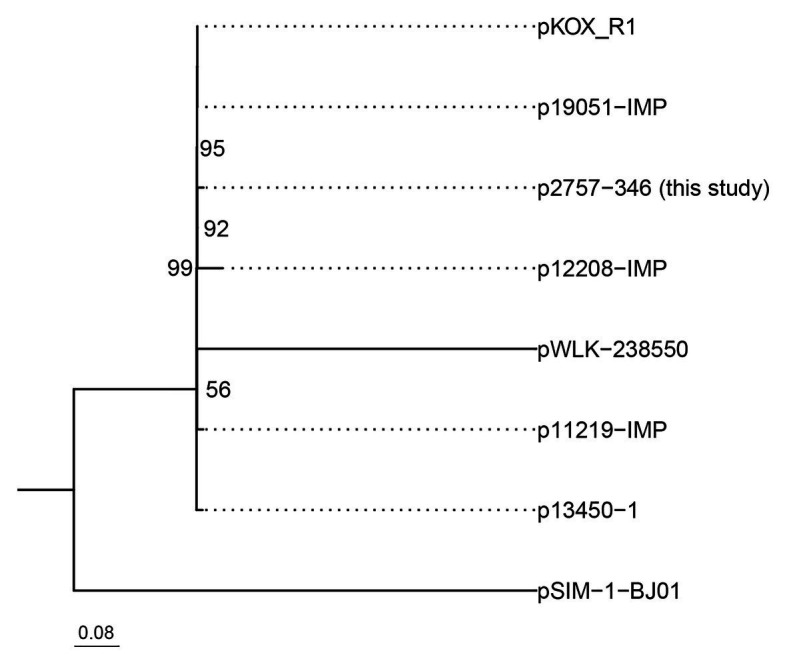
Phylogenetic tree of eight IncH5 plasmids. The bootstrap values are given at the nodes of the tree; values <50% are not shown. The bar corresponds to the scale of sequence divergence.
Comparative Genomic Analysis of Antibiotic Resistance Island
Plasmid p2757-346 contained two antibiotic resistance islands (ARIs), designated ARI-A and ARI-B (Figures 1, 4, 5). ARI-A measured 36,950 bp in length and was bracketed by IS26 and IS5075 elements from positions 307,130 to 344,080, in which six resistance genes (dfrA1, aadA5, sul1, armA, msrE, and mphE) were encoded. ARI-B was 77,593 bp in size and started from the truncated transposase gene ΔtnpA of ISEC63 at nucleotide sequence position 136,625 and ended in merR at 214,218. ARI-B encoded resistance genes providing resistance against various classes of antibiotics (aminoglycosides, β-lactams, fosfomycin, macrolides, rifampicin, quinolones, and sulfonamides), mer (merRTPCADEY) genes, and ars (arsR1BCH) resistance genes (Figure 1). Pairwise sequence comparisons via BLASTN showed that significant differences between the eight plasmids were observed in the ARIs. In addition, ARI-B in p19051-IMP had high similarity with that in p2757-346 (approximately 89% coverage and 99% identity).
Figure 4.
Comparative genomic analysis of antibiotic resistance island A (ARI-A). Mobile elements are shown by green, brown, and blue arrows. The red arrows indicate antibiotic resistance genes, the pink arrows denote biocide resistance genes, and the gray arrows represent genes of other functions. The shading indicates 100% nucleotide sequence identity.
Figure 5.
Comparative genomic analysis of antibiotic resistance island B (ARI-B). Mobile elements are shown by green, brown, and blue arrows, respectively. The red arrows indicate antibiotic resistance genes, the pink arrows denote biocide resistance genes, and the gray arrows represent genes of other functions. The shading indicates 100% nucleotide sequence identity.
All eight plasmids carried two copies of the intact armA gene (one encoded in ARI-A and the other in ARI-B), with the exceptions of pSIM-1-BJ01, which did not have an armA gene in the ARI-B region, and p12208-IMP, which had a truncated armA in the ARI-B region. Among these plasmids, the armA gene was located in the Tn1548 or truncated Tn1548 transposon (Figures 4, 5).
Both the ARI-A and ARI-B regions contained class 1 integrons, which carried genes that provided resistance to aminoglycosides [aadA2, aadA5, and aac(6)-Ib3], rifampicin (arr-3), trimethoprim (dfr1), and sulfonamides (sul1). The class 1 integron of ARI-B carried bla TEM-1b and bla CTX-M-3. In addition, sul2, aph(3´´)-Ib, and aph(6)-Id were flanked by tnpR and IS5075, and this fragment was found in p19051-IMP, p11219-IMP, p13450-1, and pKOR-R1. The aac(3)-IId gene may have been mobilized by ISVsa5 in p2757-346, p19051-IMP, p13450-1, p12208-IMP, and pKOR-R1. The fosA3 gene was associated with IS26, which was also found in p19051-IMP, p11219-IMP, p13450-1, pKOR-R1, and p12208-IMP. The distributions of aminoglycoside resistance genes in all plasmids are shown in Supplementary Table S4.
Dissemination of armA-Carrying IncHI5 Plasmids
p2757-346 is a conjugative plasmid. The antibiotic susceptibilities of the transconjugant are shown in Table 2. Moreover, bacteria carrying the armA-encoding IncHI5 plasmids were isolated from humans and sediment (no data about bacteria of animal origin available), suggesting the potential of this plasmid in mediating the dissemination of the 16S-RMTase-encoding genes among humans and throughout the environment (Supplementary Table S4).
Table 2.
Antibiotic susceptibilities of KP2757, the transconjugant, the recipient control Escherichia coli J53, and the quality control strain E. coli ATCC 25922 (mg/L).
| Antibiotic | KP2757 | p2757-346/E. coli J53 | E. coli J53 | E. coli ATCC 25922 |
|---|---|---|---|---|
| Amikacin | >1,024 | >1,024 | 0.5 | 2 |
| Gentamicin | >256 | >256 | 0.25 | 0.5 |
| Kanamycin | >1,024 | >1,024 | 1 | 4 |
| Netilmicin | >512 | >512 | 0.25 | 0.5 |
| Tobramycin | >256 | >256 | 0.25 | 0.5 |
| Streptomycin | 1,024 | 256 | 2 | 4 |
Discussion
In this work, 13.7% (40/292) of clinical K. pneumoniae complex isolates collected in the period of 2015–2017 were high-level aminoglycoside-resistant strains. Most of them showed high MICs for aminoglycosides, especially kanamycin, with 97.2% (39/40) showing an MIC of >1,024 mg/L, and 9.9% (29/292) of strains showed high MICs for gentamicin, amikacin, and tobramycin. A previous study conducted in the same district (Wenzhou, China) found that only 6.2% (21/337) of the K. pneumoniae isolates collected from 2006 to 2007 showed high MICs to gentamicin, amikacin, and tobramycin (MIC ≥ 256 mg/L; Fangyou et al., 2009). The number of K. pneumoniae complex isolates resistant to all three antibiotics increased by approximately 3.7% (a relative increase of 59.7%) during the past 10 years. High-level aminoglycoside-resistant K. pneumoniae isolates have been reported in different districts and countries (Sabtcheva et al., 2008; Qiong et al., 2009). However, to the best of our knowledge, no other publication has reported the percentage of K. pneumoniae with HLAR, although high levels of aminoglycoside resistance in other species have been reported. The percentages of HLAR (MICs for both gentamicin and amikacin were higher than 512 mg/L) of the clinical A. baumannii isolates from Beijing, China, in 2006, 2007, 2008, and 2009 were 52.63, 65.22, 51.11, and 70.83%, respectively (Lu et al., 2014). Thirty of the 137 aminoglycoside-resistant E. coli collected from nine hospitals in France showed high-level resistance (MIC ≥ 256 mg/L) to gentamicin, amikacin, and tobramycin (Caméléna et al., 2020). The prevalence of high-level aminoglycoside-resistant bacteria is worrisome.
16S-RMTase genes (rmtB and armA) were identified among 72.5% (29/40) of high-level aminoglycoside-resistant strains in this study, of which rmtB (70%, 28/40) was much more prevalent than armA (2.5%, 1/40). These findings are in accordance with the observations of other studies in China, which were conducted in Wenzhou and Shanghai by other researchers (Fangyou et al., 2009; Qiong et al., 2009). Ten 16S-RMTase gene-negative strains showing a high MIC (>1,024 mg/L) for kanamycin carried at least one aac(3)-IId or aph(3´)-Ia gene, indicating that aac(3)-IId and aph(3´)-Ia may provide high-level resistance against kanamycin. Similarly, the production of 16S-RMTase or a combination of multiple AMEs was shown to be the major mechanism underlying high-level aminoglycoside-resistant gram-negative bacilli in Chengdu, China, and Brazil (Wang et al., 2016; Ballaben et al., 2018).
In this work, two copies of armA and multiple AME genes were found in a transferable IncHI5 plasmid. armA genes have previously been identified in IncL/M, IncA/C, IncX, IncHI1, and IncF plasmids (Dolejska et al., 2013; Caméléna et al., 2020). Compared with armA-positive IncHI5 plasmids, to the best of our knowledge, IncL/M, IncA/C, IncX, IncHI1, and IncF plasmids have never been reported to carry two copies of armA.
Comparative genomic analysis revealed that the genetic context of the armA gene was similar in IncHI5 plasmids from K. pneumoniae, K. variicola, and K. michiganensis. The module of armA-related transposable elements was highly similar to those of armA harboring IncFII plasmids (Xiang-Dang et al., 2012) and IncA/C plasmids (Patrick et al., 2012), suggesting that the armA module could disseminate among plasmids of different incompatibility groups.
Except for armA and multiple AME genes, extended-spectrum β-lactamase (ESBL) genes were colocalized with genes conferring resistance to fosfomycin, macrolides, rifampicin, quinolones, and sulfonamides on the IncHI5 plasmid p2757-346. Acquired 16S-RMTase genes were mainly identified in transferable plasmids and were always associated with ESBL and carbapenemase genes or other resistance genes (Laurent et al., 2014). Plasmids carrying resistance genes may cause the global spread of resistance determinants, constituting an emerging global threat. In recent years, in addition to the phenomenon of resistance genes of various antibiotic classes being identified in a single bacterium, bacteria with multiple copies of identical resistance genes or gene arrays have been detected more often (Naas et al., 2001; Ding-Qiang et al., 2016; Teng et al., 2018). This phenomenon makes the treatment of infectious diseases more difficult and threatens human health.
Conclusion
To the best of our knowledge, we first described that two copies of armA together with six AME genes coexisted on one plasmid in a HLAR K. variicola isolate. Comparative genomic analysis showed that most armA-positive IncHI5 plasmids harbored two copies of the armA gene, which had not been previously reported in IncL/M, IncA/C, IncX, IncHI1, and IncF plasmids. Eight armA-encoding IncHI5 plasmids were isolated from humans and sediment, suggesting the potential for dissemination of this armA-carrying plasmid among bacteria of different sources. This result demonstrates the necessity of monitoring the prevalence of IncHI5 plasmids to restrict their worldwide dissemination.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HZ, CL, and QB designed the study. XZ, QL, HaL, LL, HoL, JL, and XL acquired the data. WZ, CQ, and ZS performed the results analysis and interpreted the data, and XZ wrote the first draft of the paper. KL, TX, and QB revised the draft critically for important intellectual content. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the colleagues and the reviewers who helped with this work.
Funding. This study was supported by the National Natural Science Foundation of China (81973382, 81960381, and 81700011); the Natural Science Foundation of Zhejiang Province, China (LQ17H010003, LY19C060002, and LQ17H190001); the Science and Technology Project of Inner Mongolia Autonomous Region, China (201802125); and the Science and Technology Project of Wenzhou City, China (Y20170205, Y20170625).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.636396/full#supplementary-material
References
- Ballaben A. S., Andrade L. N., Galetti R., Ferreira J. C., McElheny C. L., Mettus R. T., et al. (2018). Diversity of high-level aminoglycoside resistance mechanisms among gram-negative nosocomial pathogens in Brazil. Antimicrob. Agents Chemother. 62, e01550–e01618. 10.1128/AAC.01550-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B., Cooper M. A. (2013). Aminoglycoside antibiotics in the 21st century. ACS Chem. Biol. 8, 105–115. 10.1021/cb3005116, PMID: [DOI] [PubMed] [Google Scholar]
- Caméléna F., Morel F., Merimèche M., Decousser J. W., Jacquier H., Clermont O., et al. (2020). Genomic characterization of 16S rRNA methyltransferase-producing Escherichia coli isolates from the Parisian area, France. J. Antimicrob. Chemother. 75, 1726–1735. 10.1093/jac/dkaa105, PMID: [DOI] [PubMed] [Google Scholar]
- Ding-Qiang C., Ai-Wu W., Ling Y., Dan-Hong S., Yong-Ping L., Yan-Wei H., et al. (2016). Emergence and plasmid analysis of Klebsiella pneumoniae KP01 carrying blaGES-5 from Guangzhou, China. Antimicrob. Agents Chemother. 60, 6362–6364. 10.1128/AAC.00764-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y., Wachino J. I., Arakawa Y. (2016). Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 30, 523–537. 10.1016/j.idc.2016.02.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Poirel L., Nordmann P., Carattoli A. (2013). Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68, 34–39. 10.1093/jac/dks357, PMID: [DOI] [PubMed] [Google Scholar]
- Fang-you Y., Dan Y., Jing-ye P., Chong C., Zhi-qiang Q., Chris P., et al. (2010). High prevalence of plasmid-mediated 16S rRNA methylase gene rmtB among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect. Dis. 10:184. 10.1186/1471-2334-10-184, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangyou Y., Liangxing W., Jingye P., Dan Y., Chan C., Tao Z., et al. (2009). Prevalence of 16S rRNA methylase genes in Klebsiella pneumoniae isolates from a Chinese teaching hospital: coexistence of rmtB and armA genes in the same isolate. Diagn. Microbiol. Infect. Dis. 64, 57–63. 10.1016/j.diagmicrobio.2009.01.020, PMID: [DOI] [PubMed] [Google Scholar]
- Huang T. W., Wang J. T., Lauderdale T. L., Liao T. L., Lai J. F., Tan M. C., et al. (2013). Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob. Agents Chemother. 57, 4072–4076. 10.1128/AAC.02266-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. 10.1093/nar/gkw1004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathryn E. H., Heiman W., Ruth N. Z., Stephen B., Chris A. W., David D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U. S. A. 112, E3574–E3581. 10.1073/pnas.1501049112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., Phillippy A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. 10.1101/gr.215087.116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent P., Jaime L., Helia B., Maria Luisa R., Sandrine B., Patrice N. (2014). Emergence of the 16S rRNA methylase RmtG in an extended-spectrum-β-lactamase-producing and colistin-resistant Klebsiella pneumoniae isolate in Chile. Antimicrob. Agents Chemother. 58, 618–619. 10.1128/AAC.02059-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C. M., Luo R., Sadakane K., Lam T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. 10.1093/bioinformatics/btv033, PMID: [DOI] [PubMed] [Google Scholar]
- Liang Q., Jiang X., Hu L., Yin Z., Gao B., Zhao Y., et al. (2018). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9:3318. 10.3389/fmicb.2018.03318, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. W., Sarah E. L., Matthew Ojeda S., Concepcion C., James J. D., Thomas B., et al. (2017). Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2, e00290–e00317. 10.1128/mSphereDirect.00290-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Yuemeng L., Min Y., Xinxin H., Tongying N., Xinyi Y., et al. (2014). Genetic basis of high level aminoglycoside resistance in Acinetobacter baumannii from Beijing, China. Acta Pharm. Sin. B 4, 295–300. 10.1016/j.apsb.2014.06.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miró E., Grünbaum F., Gómez L., Rivera A., Mirelis B., Coll P., et al. (2013). Characterization of aminoglycoside-modifying enzymes in enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microb. Drug Resist. 19, 94–99. 10.1089/mdr.2012.0125, PMID: [DOI] [PubMed] [Google Scholar]
- Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. 10.1093/bioinformatics/btp105, PMID: [DOI] [PubMed] [Google Scholar]
- Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183, 235–249. 10.1128/JB.183.1.235-249.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabil-Fareed A., Nicola K. P., Nouri L. B. Z., Scott A. B. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M., Kaku M., Kamachi K., Shibayama K., Arakawa Y., Yamaguchi K., et al. (2014). Loop-mediated isothermal amplification assay for 16S rRNA methylase genes in gram-negative bacteria. J. Infect. Chemother. 20, 635–638. 10.1016/j.jiac.2014.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Nakamatsu E. H., Fujihira E., Ferreira R. C., Balan A., Costa S. O., Ferreira L. C. (2007). Oligopeptide uptake and aminoglycoside resistance in Escherichia coli K12. FEMS Microbiol. Lett. 269, 229–233. 10.1111/j.1574-6968.2007.00634.x, PMID: [DOI] [PubMed] [Google Scholar]
- Nasiri G., Peymani A., Farivar T. N., Hosseini P. (2018). Molecular epidemiology of aminoglycoside resistance in clinical isolates of Klebsiella pneumoniae collected from Qazvin and Tehran provinces, Iran. Infect. Genet. Evol. 64, 219–224. 10.1016/j.meegid.2018.06.030, PMID: [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. 10.1093/nar/gkt1226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick M., Jun H., Robert J. C., Yu Y., Yoon I. K., Robert A. K., et al. (2012). Complete sequence of a novel 178-kilobase plasmid carrying Bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56, 1673–1679. 10.1128/AAC.05604-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericàs J. M., García-de-la-Mària C., Brunet M., Armero Y., García-González J., Casals G., et al. (2017). Early in vitro development of daptomycin non-susceptibility in high-level aminoglycoside-resistant Enterococcus faecalis predicts the efficacy of the combination of high-dose daptomycin plus ampicillin in an in vivo model of experimental endocarditis. J. Antimicrob. Chemother. 72, 1714–1722. 10.1093/jac/dkx016, PMID: [DOI] [PubMed] [Google Scholar]
- Poole K. (2004). Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10, 12–26. 10.1111/j.1469-0691.2004.00763.x, PMID: [DOI] [PubMed] [Google Scholar]
- Qiong W., Yibo Z., Lizhong H., Jingyong S., Yuxing N. (2009). Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob. Agents Chemother. 53, 271–272. 10.1128/AAC.00748-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabtcheva S., Saga T., Kantardjiev T., Ivanova M., Ishii Y., Kaku M. (2008). Nosocomial spread of armA-mediated high-level aminoglycoside resistance in Enterobacteriaceae isolates producing CTX-M-3 beta-lactamase in a cancer hospital in Bulgaria. J. Chemother. 20, 593–599. 10.1179/joc.2008.20.5.593, PMID: [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. 10.1093/nar/gkj014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X., Jian W., Jianchao Y., Tingyuan Z., Yabo L., Lei X., et al. (2018). Characterisation of a class 1 integron associated with the formation of quadruple blaGES-5 cassettes from an IncP-1β group plasmid in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 52, 485–491. 10.1016/j.ijantimicag.2018.07.002, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Shen M., Yang J., Dai M., Chang Y., Zhang C., et al. (2016). Prevalence of carbapenemases among high-level aminoglycoside-resistant Acinetobacter baumannii isolates in a university hospital in China. Exp. Ther. Med. 12, 3642–3652. 10.3892/etm.2016.3828, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang-Dang D., De-Xi L., Gong-Zheng H., Yang W., Yan-Hong S., Cong-Ming W., et al. (2012). Tn1548-associated armA is co-located with qnrB2, aac(6′)-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. J. Antimicrob. Chemother. 67, 246–248. 10.1093/jac/dkr407, PMID: [DOI] [PubMed] [Google Scholar]
- Ya H., Lu L., Xiaoxia Z., Yu F., Zhiyong Z. (2017). In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant Enterobacteriaceae clinical strains. Front. Microbiol. 8:2275. 10.3389/fmicb.2017.02275, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatao G., Yao Z., Zhao Z., Daixi L., Zhanwei W., Jingquan L., et al. (2018). Complete genomic analysis of a kingdom-crossing Klebsiella variicola isolate. Front. Microbiol. 9:2428. 10.3389/fmicb.2018.02428, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Ting D., Zhen-Ling Z., Wei T., Tong Y., Jian-Hua L. (2013). Prevalence and characteristics of rmtB and qepA in Escherichia coli isolated from diseased animals in China. Front. Microbiol. 4:198. 10.3389/fmicb.2013.00198, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.