Abstract
Cells in diverse organisms can store the information of previous environmental conditions for long periods of time. This form of cellular memory adjusts the cell’s responses to future challenges, providing fitness advantages in fluctuating environments. Many biological functions, including cellular memory, are mediated by specific recurring patterns of interactions among proteins and genes, known as “network motifs.” In this review, we focus on three well-characterized network motifs - negative feedback loops, positive feedback loops, and feedforward loops, which underlie different types of cellular memories. We describe the latest studies identifying these motifs in various molecular processes and discuss how the topologies and dynamics of these motifs can enable memory encoding and storage.
Living organisms can remember previous experience and adjust their behaviors to future challenges. The most well-known examples are neuronal memory and adaptive immunity in metazoans, both of which require interactions and cooperation among many different cells. Similarly, individual cells can also memorize prior environmental signals and modulate their responses to subsequent cues. This type of “cellular memory” is mediated by interactions and cooperation of different genes and molecules inside the cell [1]. How these biochemical interactions enable the storage of prior environmental information remains a challenging question. Several recurring patterns of regulatory interactions, defined as “network motifs” [2], have been found to confer memory behaviors. In this review, we focus on the latest progress in characterizing these network motifs in diverse cellular systems, as well as in understanding how their structures and dynamics contribute to the encoding and maintenance of cellular memories.
Negative feedback loops – desensitization
Odors and light become less noticeable after prolonged exposure. Similarly, continuous use of drugs and alcohol can cause refractoriness to their further administration. These familiar experiences that we have encountered in daily life stem from a general phenomenon - “desensitization,” in which a stimulus triggers a response inside the cell and, at the same time, it also induces a process that inhibits the response to a future exposure to the same stimulus [3]. From the network perspective, desensitization, in many cases, is mediated by a negative feedback loop after a delay (Fig. 1).
Figure 1. Negative feedback loops lead to desensitization.
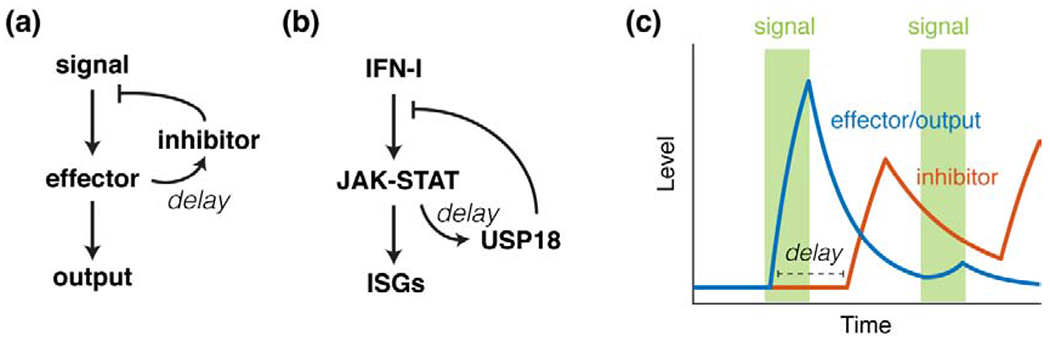
(a) Topology of a general negative feedback loop. (b) An example of negative feedback system that causes desensitization - the USP18-mediated negative feedback loop in the JAK-STAT pathway. (c) Time traces of a delayed negative feedback system in response to repetitive stimulations. Green – signal; blue – effector/output; red – inhibitor.
A classic example is the desensitization of G protein-coupled receptor (GPCR) signaling, which mediates the effects of many hormones and neurotransmitters [4]. Agonist binding to GPCRs increases the intracellular level of the second messenger cAMP, which in turn activates protein kinase A (PKA). PKA then phosphorylates downstream cellular substrates and regulates many aspects of cellular physiology. Once the external signal is transduced into the cell and elicits cellular responses, the desensitization process is initiated. In addition to the well-established feedback inhibition of GPCRs by β-arrestin [5,6], a recent study found a delayed negative feedback loop through chaperone-assisted ubiquitination and degradation of the activated PKA catalytic subunit, resulting in a refractory phase that uncouples further agonist stimulation from continuous signal propagation [7].
Similarly, the Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling that mediates innate immunity is also subject to desensitization. In mammals, type I interferon (IFN-I) is secreted upon pathogen infection and binds to the IFN-I receptor on the cell membrane, leading to activation of JAK and tyrosine kinase 2 (TYK2) that in turn phosphorylate transcription factors STAT1 and STAT2 and trigger the formation of IFN-stimulated gene factor 3 (ISGF3) complex. ISGF3 translocates to the nucleus and induces the expression of over 300 IFN-stimulated genes (ISGs), exerting anti-proliferative and anti-pathogenic effects [8,9]. Previous studies showed that one of the ISGs encodes the ubiquitin-specific peptidase 18 (USP18), which is a major inhibitor of JAK-STAT signaling that acts at the receptor level [10,11]. IFN pretreatment induces a delayed upregulation of USP18, resulting in long-lasting refractoriness to further stimulations [12,13] (Fig. 1b).
In both examples, desensitization is mediated by negative feedback loops with a delay (Fig. 1c). This delay is functionally relevant as it allows efficient signal propagation and response activation upon the initial stimulus and, importantly, it can enable signal processing of the initial stimulus for memory encoding. For instance, the delay enables a “persistence detector” that can filter out transient inputs: a brief pulse of stimulus that is shorter than the delay time does not induce pathway inhibition, whereas only persistent signals can lead to strong desensitization. This function is particularly crucial for cells in a fluctuating environment to avoid inappropriately entering a refractory phase triggered by spurious signals. Many mechanisms can give rise to a delay in feedback inhibition, including extensive nucleosome occupancy at the promoter regions [14–16], multi-layer activation cascades [17,18], or specific gene network topologies (e.g. feedforward loops) [19,20]. We recently found that cell cycle regulation and DNA methylation can also contribute to the delay in feedback inhibition (e.g. in the case of USP18 upregulation), which leads to stimulus duration-dependent refractoriness [12].
Positive feedback loops – hysteresis and priming
As opposed to desensitization, in many biological systems, previous exposure to a stimulus can accelerate or boost the responses to subsequent stimulations. This process is often mediated by positive feedback loops, which are well-known to give rise to bistability that enables long-lasting or permanent memories of past environmental conditions [21–23]. Increasing evidence has shown that, in the contexts of metabolic shifts or animal development, an identical environmental signal can lead to distinct outcomes in genetically identical cells. For example, a stimulus that causes some cells to differentiate can induce proliferation in other cells. The outcome upon a stimulus is generally determined by the metabolic or developmental state of the cell, which depends on the environmental history of the cell. This type of history-dependent behavior, defined as “hysteresis,” often arises from the bistability mediated by positive feedback loops, in which a previous environmental condition can permanently switch the cell to an induced state, resulting in different response dynamics and steady state, from those of uninduced cells, upon future stimuli [24,25] (Fig. 2).
Figure 2. Positive feedback loops enable hysteresis and priming effects.
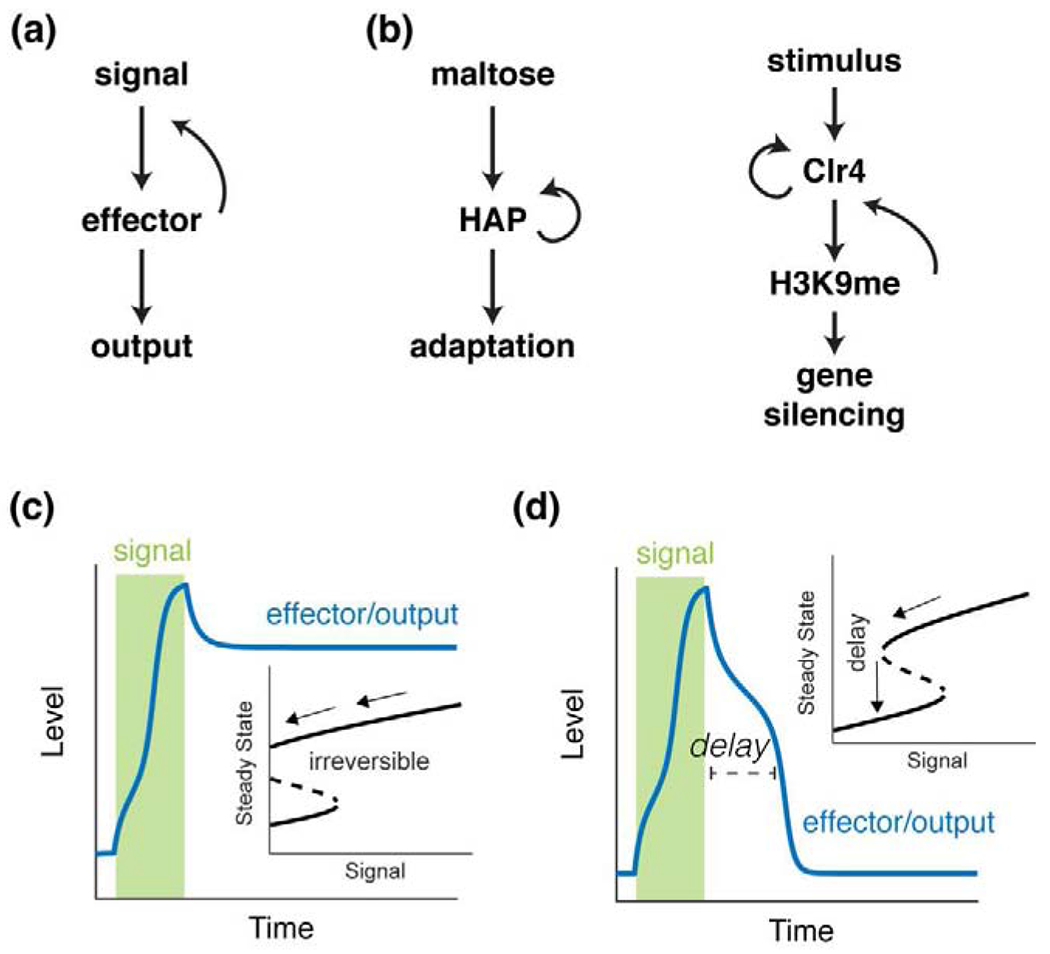
(a) Topology of a general positive feedback loop. (b) Examples of positive feedback loops that generate cellular memories. Left: a diagram of the regulatory network underlying the adaptation to carbon source switch from glucose to maltose. Right: a diagram of the epigenetic regulatory network underlying H3K9me3-mediated gene silencing. (c) The time trace of a positive feedback system that induces an irreversible state transition in response to a transient stimulus. Inset: a bifurcation diagram that shows the steady state values as a function of the signal level, illustrating the origin of irreversibility. (d) The time trace of a positive feedback system that induces a reversible response upon a transient stimulus. Inset: a bifurcation diagram that shows the steady state values as a function of the signal level, illustrating the origin of the delay in recovery time.
Metabolic regulatory networks underlying the adaptation of microorganisms to carbon source changes, e.g. the lac operon in E. coli, are classic examples of such systems [26,27]. A recent study in yeast [28] showed that previous exposure to maltose can switch the cell to a stable respiratory state through positive feedback loops, such as those mediated by the heme-activated protein complex [29], which can shorten the lag time needed for adapting to a recurring carbon source switch from glucose to maltose, even in daughter cells that have not experienced the first change (Fig. 2b, left). Along the same line, researchers built a semi-synthetic regulon in yeast that controls the utilization of xylose, a nutrient that is non-native for yeast, and showed that positive feedback loops are sufficient to generate bistability and hysteresis, enabling history-dependent behaviors [30].
Epigenetic modification, a major mechanism underlying persistent cellular memory and cell differentiation, is also driven by positive feedback loops that enable stable and heritable states of DNA regions [31,32]. Such positive feedback loops involve the recruitment of specific enzymes to a modified histone, which catalyze the same modifications on neighboring histones [33]. For instance, a recent study showed that this type of modification-based positive feedback of histone H3 lysine 9 methylation (H3K9me), coupled with the autoregulation of the H3K9 methylthansferase Clr4, can reinforce the maintenance of gene silencing across many cell cycles [34] (Fig. 2b, right). Inspired by the self-propagating mode of regulation observed in nature, researchers also developed a synthetic epigenetic system in mammalian cells that can read and write N6-methyladenine (m6A), a DNA modification not commonly found in metazoans [35]. The incorporation of a positive feedback loop in the system enables a persistent epigenetic memory of transcriptional states.
We note that the irreversible state transition (Fig. 2c) is not required for memory generation by positive feedback. In reversible systems, positive feedback loops can still function to prolong the duration of priming effects from prior treatments (Fig. 2d). For example, in the JAK-STAT pathway, an IFN-I pretreatment induces the accumulation of STAT1, STAT2, and IRF9, the components of ISGF3 mediating IFN-driven gene expression [12,13,36], which primes the cell for accelerated responses to future stimulations. In this case, the positive feedback loop produces a time delay in the decay of cellular memory, extending the period that priming effects can last after the removal of pretreatment (Fig. 2d).
Feedforward loops – phase separation and information storage
A growing number of recent studies have revealed that, in addition to feedback loops, feedforward loops also play important roles in encoding memories of environmental signals (Fig. 3a). Intriguingly, many of these feedforward loops involve signal-induced formation of phase-separated assemblies or granules, resulting in a period of desensitization or priming phase depending on the nature of the aggregates formed. For example, Caudron et al showed that, whereas pheromone stimulation leads to cell cycle arrest in yeast, prior signal exposures from an unsuccessful mating partner encounter can trigger aggregation and thereby inactivation of Whi3, an RNA-binding protein required for the G1/S arrest, thereby causing a stable pheromone-refractory state that lasts for many cell cycles [37].
Figure 3. Feedforward loops allow the spreading of signal effects in time.

(a) Topology of a general feedforward loop. (b) An example of a feedforward loop that generates cellular memory through signal-induced protein aggregation. (c) The time trace of a feedforward system that enables a long-lasting response to a transient signal. Green – signal; blue – effector; red – effector 2 (slow); yellow - output. (d) An example of a variant of feedforward loops that generates cellular memory through signal-induced mRNA storage in PBs/SGs. (e) The time trace of the feedback forward system in (d) that enables long-lasting plateau of gene expression response and acquired stress resistance upon a transient stimulus. Green – signal; red – mRNAs stored in PBs/SGs; blue – stress-induced functional mRNAs/acquired stress resistance.
It is generally believed that protein aggregates are detrimental to the cell. However, emerging evidence reveals that they can also function as beneficial storage devices for cellular memories (Fig. 3, b and c). For instance, in bacteria, proteotoxic stresses induce protein aggregates that can be asymmetrically inherited for many generations, conferring a significantly increased resistance to future stress [38]. Similarly, the cytoskeleton-associated protein Lsb2 forms a metastable prion in yeast under heat shock, which further promotes the conversion of other proteins into prions. These prions can be passed across several generations after the initial stress and can help enhance cell survival against future challenges [39]. Another study showed that stress can induce phase separation and sequestering of the yeast translation termination factor Sup35 through its prion-like domain, providing a heritable fitness advantage after the stress [40]. The [SMAUG+] prion has been identified in yeast to encode memories of recurring environmental fluctuations from the ancestors and to regulate gene expression, growth and stress resistance, enabling an anticipation of future environmental changes [41,42].
Processing bodies (PBs) and stress granules (SGs) are stress-induced cytoplasmic messenger ribonucleoprotein (mRNP) granules, conserved from yeast to mammals. In response to stress, the mRNAs of some stress-responsive genes can be localized in PBs and SGs, which regulate the translation, degradation and storage of these mRNAs [43,44]. In our recent work [45], we found that the storage of newly synthesized mRNAs in PBs and SGs upon an initial stress, constituting a variant of feedforward loops, enables a long-lasting plateau of acquired stress resistance that accelerates the adaptation to future stresses (Fig. 3, d and e). The initiation of the storage process is elicited by protein kinase A (PKA) signals, whereas the duration of the memory depends on the amount of mRNA being stored in PBs/SGs and hence depends on both the amplitude and duration of the initial stress. This regulatory scheme allows the cell to determine how long the memory can last, based on the severity and period of the initial stress.
In the examples described above, cellular memories arise from feedforward loops through signal-induced phase-separation, featuring a reversible process with slow kinetics. In particular, after stimulus removal, the slow release of functional monomers from aggregates is crucial for the persistence of memories. This slow releasing kinetics allows the spreading of signal effects in time, maintaining sustained acquired functions after the initial stimulus is gone (Fig. 3, c and e). A recent study, combining mathematical modeling and optogenetic stimulus experiments, showed that the inherent physics of protein droplets (e.g. Ostwald ripening) can also enable the cell to memorize the spatial patterns of clustered regulatory factors induced by transient localized stimuli [46].
Feedforward loops that are unrelated to phase separation can also generate memory. For example, in Drosophila, odor or electric shock signals activate PKA to phosphorylate the transcription factor CREBB to enable long-term neuronal memory. PKA also activates a conserved kinase, Meng-Po, which inhibits the degradation of CREBB, contributing to maintenance of the CREBB level in the mushroom bodies of the brain without the continuous presence of the initial signal [47].
Conclusions and perspectives
Recent progress in quantitative cell biology has dramatically pushed forward our understanding of the mechanisms underlying cellular memory. These studies integrate new measurement technologies, such as microfluidics and advanced time-lapse imaging, with computational modeling, which provides a powerful suite of new tools to elucidate how genes and molecules interact dynamically to generate sustained cellular responses to transient stimuli, enabling long-lasting memories. As described above, multiple network motifs have been identified, each with different dynamic behaviors and contributing to different types of memory effects. Emerging questions along this direction include the combined effects of multiple network motifs operating in a single pathway and the role of heterogeneity in network dynamics and memory encoding.
In many biological systems, a single regulatory pathway often contains several network motifs that are coupled to one another. For example, the JAK-STAT pathway is comprised of multiple positive and negative feedback loops, which act collectively to modulate the cellular responses to varying interferon signals [12,13]. Future studies will be needed to investigate how the functions of these network motifs are dynamically coordinated and what are the functional benefits of the coupling and cooperation among these motifs, e.g. plasticity [48], robustness [49], multistability [50–52], synergy [53,54], or redundancy over different time scales [55,56]. Unravelling such complexity will require systematic genetic perturbation analyses in combination with dynamic measurements and computational modeling.
Recent single-cell analyses demonstrated the existence of substantial clonal heterogeneity in cellular responses to signals [57]. Interesting questions that deserve further investigation are whether individual cells differ in their abilities to memorize the same environmental signals and, if that is the case, what the mechanisms are which underlie the cell-to-cell variabilities, how different network motifs influence the stochasticity in responses, and how these variabilities contribute to biological functions in the physiological contexts. Advances in single-cell technologies will enable us to track the dynamic responses of individual cells and analyze the source, the control and the consequence of the cell-to-cell heterogeneity in memorizing environmental signals.
Acknowledgements
We thank Dr. Lorraine Pillus for carefully reading the manuscript and providing insightful comments. This work was supported by NIH R01 GM111458.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References:
* of special interest
- 1.Burrill DR, Silver PA: Making cellular memories. Cell 2010, 140:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon U: Network motifs: theory and experimental approaches. Nat Rev Genet 2007, 8:450–461. [DOI] [PubMed] [Google Scholar]
- 3.Dohlman HG: Diminishing returns. Nature 2002, 418:591. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowitz RJ: A brief history of G-protein coupled receptors (Nobel Lecture). Angew Chem Int Ed Engl 2013, 52:6366–6378. [DOI] [PubMed] [Google Scholar]
- 5.Gurevich VV, Gurevich EV: GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front Pharmacol 2019, 10:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luttrell LM, Lefkowitz RJ: The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci 2002, 115:455–465. [DOI] [PubMed] [Google Scholar]
- 7.Rinaldi L, Delle Donne R, Catalanotti B, Torres-Quesada O, Enzler F, Moraca F, Nistico R, Chiuso F, Piccinin S, Bachmann V, et al. : Feedback inhibition of cAMP effector signaling by a chaperone-assisted ubiquitin system. Nat Commun 2019, 10:2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivashkiv LB, Donlin LT: Regulation of type I interferon responses. Nat Rev Immunol 2014, 14:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider WM, Chevillotte MD, Rice CM: Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014, 32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arimoto KI, Lochte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, Wilmes S, Fan JB, Heinisch JJ, Li Z, et al. : STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol 2017, 24:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE: UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 2006, 25:2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12.Mudla A, Jiang Y, Arimoto KI, Xu B, Rajesh A, Ryan AP, Wang W, Daugherty MD, Zhang DE, Hao N: Cell-cycle-gated feedback control mediates desensitization to interferon stimulation. eLife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used a combination of single-cell imaging analyses and mathematical modeling and found the opposing effects of IFN-I, priming versus desensitization, depending on the duration of its pretreatment. Experimental and modeling analyses revealed that a regulatory network with a fast-acting postive feedback loop and a delayed negative feedback loop mediated by upregulation of a negative regulator USP18 can account for the opposite effects from different input durations. Further, the delay in USP18 upregulation can be attributed to a cell cycle gating mechanism at the single-cell level. This study discovered a cell cycle-dependent temporal compartmentalization of feedback processes with different functions, which is fundamentally important for understanding the complex IFN signaling.
- *13.Kok F, Rosenblatt M, Teusel M, Nizharadze T, Goncalves Magalhaes V, Dachert C, Maiwald T, Vlasov A, Wasch M, Tyufekchieva S, et al. : Disentangling molecular mechanisms regulating sensitization of interferon alpha signal transduction. Mol Syst Biol 2020, 16:e8955. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study established a computational model of the JAK-STAT pathway based on quantitative time-resolved data from a hepatoma cell line. Through experimental and modeling analyses, the authors showed that a low dose of IFN-I prestimulation hypersensitizes the pathway through STAT2 and IRF9, whereas a high dose of prestimulation desensitizes the pathway through negative feedback loops by USP18 and SOCS1. Further use of the model revealed that the basal levels of USP18 and STAT2 in primary hepatocytes from patients can predict the responsiveness to IFN treatments, suggesting potential diagnostic markers for personalized medicine. This study, together with Mudla et al [12], provides a comprehensive picutre about how interconnected feedback loops govern the dynamics of JAK-STAT responses to repetitive IFN simulations.
- 14.Sen S, Cheng Z, Sheu KM, Chen YH, Hoffmann A: Gene Regulatory Strategies that Decode the Duration of NFkappaB Dynamics Contribute to LPS- versus TNF-Specific Gene Expression. Cell Syst 2020, 10:169–182 e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao N, O’Shea EK: Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol 2012, 19:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen AS, O’Shea EK: cis Determinants of Promoter Threshold and Activation Timescale. Cell Rep 2015, 12:1226–1233. [DOI] [PubMed] [Google Scholar]
- 17.Korsbo N, Jonsson H: It’s about time: Analysing simplifying assumptions for modelling multi-step pathways in systems biology. PLoS Comput Biol 2020, 16:e1007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beguerisse-Diaz M, Desikan R, Barahona M: Linear models of activation cascades: analytical solutions and coarse-graining of delayed signal transduction. J R Soc Interface 2016, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangan S, Zaslaver A, Alon U: The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol 2003, 334:197–204. [DOI] [PubMed] [Google Scholar]
- 20.Thurley K, Wu LF, Altschuler SJ: Modeling Cell-to-Cell Communication Networks Using Response-Time Distributions. Cell Syst 2018, 6:355–367 e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitrophanov AY, Groisman EA: Positive feedback in cellular control systems. Bioessays 2008, 30:542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balazsi G, van Oudenaarden A, Collins JJ: Cellular decision making and biological noise: from microbes to mammals. Cell 2011, 144:910–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golding I: Decision making in living cells: lessons from a simple system. Annu Rev Biophys 2011, 40:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Walker BL, Iannaccone S, Bhatt D, Kennedy PJ, Tse WT: Bistable switches control memory and plasticity in cellular differentiation. Proc Natl Acad Sci U S A 2009, 106:6638–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong W, Ferrell JE Jr.: A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 2003, 426:460–465. [DOI] [PubMed] [Google Scholar]
- 26.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A: Multistability in the lactose utilization network of Escherichia coli. Nature 2004, 427:737–740. [DOI] [PubMed] [Google Scholar]
- 27.Acar M, Becskei A, van Oudenaarden A: Enhancement of cellular memory by reducing stochastic transitions. Nature 2005, 435:228–232. [DOI] [PubMed] [Google Scholar]
- *28.Cerulus B, Jariani A, Perez-Samper G, Vermeersch L, Pietsch JM, Crane MM, New AM, Gallone B, Roncoroni M, Dzialo MC, et al. : Transition between fermentation and respiration determines history-dependent behavior in fluctuating carbon sources. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors investigated molecular mechanisms underlying history-dependent behaviors in yeast during carbon source changes and showed that the accelerated adaptation was not due to the persistance of chromatin modifications or metabolic proteins induced by previous changes. Instead, the history-dependent adaptation can be attributed to slow transitions between fermentation and respiration states, mediated by the HAP-regulated network with positive feedback loops. This study advances the mechanistic understanding about transgenerational cellular memory of environmental changes.
- 29.Zhang T, Bu P, Zeng J, Vancura A: Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J Biol Chem 2017, 292:16942–16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Endalur Gopinarayanan V, Nair NU: A semi-synthetic regulon enables rapid growth of yeast on xylose. Nat Commun 2018, 9:1233. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors investigated the functional benefits from nutrient-responsive gene regulatory netowrks, as opposed to simple constitutive gene expression. They built semi-synthetic networks in yeast that sense and metabolize a non-native nutrient for yeast, xylose, and showed that the postive feedback loops, synthetically implemented from the native galacotose-responsive pathway, are sufficient to generate bistability and hysteresis, supporting high growth rate and cell density, independent of galactose metabolism. This study highlights the functional relevance of network motifs in regulating cell physiology and provides a paradigm for engineering nutrient sensing netowrks in microorganisms.
- 31.Dodd IB, Micheelsen MA, Sneppen K, Thon G: Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 2007, 129:813–822. [DOI] [PubMed] [Google Scholar]
- 32.Moazed D: Mechanisms for the inheritance of chromatin states. Cell 2011, 146:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang T, Cooper S, Brockdorff N: The interplay of histone modifications - writers that read. EMBO Rep 2015, 16:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, Moazed D: Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature 2018, 560:504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors investigated the biochemical regulation of Clr4, the methyltransferase that catalyzes histone 3 lysine 9 methylation (H3K9me) in the fission yeast. They found that automethylation of Clr4 mediates a conformational switch between inhibited and activated states. The intrinsic inhibition and auto-activation are important in preventing aberrant gene silencing, and function together with the positive feedback mediated by histone post-translational modifications to reinforce the maintenance of epigenetic memory across generations.
- *35.Park M, Patel N, Keung AJ, Khalil AS: Engineering Epigenetic Regulation Using Synthetic Read-Write Modules. Cell 2019, 176:227–238 e220. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors developed a synthetic epigenetic system in mammalian cells to decipher the principles underlying epigenetic regulation in natural systems. The system contains synthetic protein factors that can read and write N6-methyladenine (m6A), a DNA modification not commonly found in metazoans, which are coupled with synthetic transcriptional regulators to control reporter gene expression. Guided by modeling, they further constructed minimal regulatory circuits, e.g. positive feedback loops, to enable spatial propagation and epigenetic memory, the behaviors observed in natural epigenetic systems. This study establishes an orthogonal synthetic platform to systematically examine the core circuit architectures and regulatory principles underlying epigenetic regulation in eukaryotes.
- 36.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR: IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J 2013, 32:2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caudron F, Barral Y: A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell 2013, 155:1244–1257. [DOI] [PubMed] [Google Scholar]
- *38.Govers SK, Mortier J, Adam A, Aertsen A: Protein aggregates encode epigenetic memory of stressful encounters in individual Escherichia coli cells. PLoS Biol 2018, 16:e2003853. [DOI] [PMC free article] [PubMed] [Google Scholar]; By tracking the dynamics of protein aggregates (PAs) and cell viability under heat shock, the authors found that the segregation of stress-induced PAs is asymmetric between two sister cells during cell division and the PA-bearing cell has a higher survival rate than the PA-free cell. This study revealed that the asymmetric inheritance of PAs and their associated protein quality control components enable a multi-generational memory of the stress responses.
- 39.Chernova TA, Kiktev DA, Romanyuk AV, Shanks JR, Laur O, Ali M, Ghosh A, Kim D, Yang Z, Mang M, et al. : Yeast Short-Lived Actin-Associated Protein Forms a Metastable Prion in Response to Thermal Stress. Cell Rep 2017, 18:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nuske E, Richter D, Baumeister W, Grill SW, Pappu RV, et al. : Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359. [DOI] [PubMed] [Google Scholar]; This study showed that stress induces the phase separation of a yeast translation termination factor, Sup35, through its prion-like domain. This phase separation is fully reversible, distinct from the formation of pathological amyloid-like aggregates, and serves a protective and storage role for functional Sup35, promoting cell adaptation and fitness after stress. These findings suggest that, prion-like domains, given their prevalence in the proteome, may function as environmental stress sensors with beneficial roles for cell survivial in flutuating environments.
- *41.Itakura AK, Chakravarty AK, Jakobson CM, Jarosz DF: Widespread Prion-Based Control of Growth and Differentiation Strategies in Saccharomyces cerevisiae. Mol Cell 2020, 77:266–278 e266. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study, together with Chakravarty et al [42], revealed that the [SMAUG+] prion in yeast encodes a transgenerational memory of recurring environmental fluctuations in natural habitats and regulates a gene expression program underlying a fate decision between growth and meiosis/stress resistance upon starvation. This finding elucidates a mechanism that underlies how cells can predict future environemntal changes to obtain a selective advantage.
- 42.Chakravarty AK, Smejkal T, Itakura AK, Garcia DM, Jarosz DF: A Non-amyloid Prion Particle that Activates a Heritable Gene Expression Program. Mol Cell 2020, 77:251–265 e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decker CJ, Parker R: P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 2012, 4:a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, Spang A: Context-dependent deposition and regulation of mRNAs in P-bodies. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Jiang Y, AkhavanAghdam Z, Li Y, Zid BM, Hao N: A protein kinase A-regulated network encodes short- and long-lived cellular memories. Sci Signal 2020, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study combined microfluidics and computational modeling to examine the cellular memory of prior stress in yeast. Two phases of memory, both regulated by protein kinase A (PKA), were identified: short-lived memory mediated by trehalose metabolism and long-lived memory mediated by stress-induced transcripts sequestered in P-bodies/stress granules. This work revealed a PKA-mediated regulatory network that enables memory encoding based on the dynamics of intial stresses and reported a new physiological role of messenger ribonucleoprotein granules in mediating long-lasting memory of previous stresses.
- 46.Dine E, Gil AA, Uribe G, Brangwynne CP, Toettcher JE: Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Syst 2018, 6:655–663 e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee PT, Lin G, Lin WW, Diao F, White BH, Bellen HJ: A kinase-dependent feedforward loop affects CREBB stability and long term memory formation. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettensohn CA, Kitazawa C, Cheers MS, Leonard JD, Sharma T: Gene regulatory networks and developmental plasticity in the early sea urchin embryo: alternative deployment of the skeletogenic gene regulatory network. Development 2007, 134:3077–3087. [DOI] [PubMed] [Google Scholar]
- 49.Macneil LT, Walhout AJ: Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res 2011, 21:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Jiang Y, Paxman J, O’Laughlin R, Klepin S, Zhu Y, Pillus L, Tsimring LS, Hasty J, Hao N: A programmable fate decision landscape underlies single-cell aging in yeast. Science 2020, 369:325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guantes R, Poyatos JF: Multistable decision switches for flexible control of epigenetic differentiation. PLoS Comput Biol 2008, 4:e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Su RQ, Lai YC, Wang X: Engineering of a synthetic quadrastable gene network to approach Waddington landscape and cell fate determination. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anastassiou D: Computational analysis of the synergy among multiple interacting genes. Mol Syst Biol 2007, 3:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu R, Wang X, Moazed D: Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 2018, 558:615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kafri R, Levy M, Pilpel Y: The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A 2006, 103:11653–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.AkhavanAghdam Z, Sinha J, Tabbaa OP, Hao N: Dynamic control of gene regulatory logic by seemingly redundant transcription factors. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeknic S, Kudo T, Covert MW: Techniques for Studying Decoding of Single Cell Dynamics. Front Immunol 2019, 10:755. [DOI] [PMC free article] [PubMed] [Google Scholar]


