Abstract
Adverse drug reactions (ADRs) are a major concern for patients, clinicians, and regulatory agencies. The discovery of serious ADRs leading to substantial morbidity and mortality has resulted in mandatory phase IV clinical trials, black box warnings, and withdrawal of drugs from the market. Real‐world data, data collected during routine clinical care, is being adopted by innovators, regulators, payors, and providers to inform decision making throughout the product life cycle. We outline several different approaches to modern pharmacovigilance, including spontaneous reporting databases, electronic health record monitoring and research frameworks, social media surveillance, and the use of digital devices. Some of these platforms are well‐established while others are still emerging or experimental. We highlight both the potential opportunity, as well as the existing challenges within these pharmacovigilance systems that have already begun to impact the drug development process, as well as the landscape of postmarket drug safety monitoring. Further research and investment into different and complementary pharmacovigilance systems is needed to ensure the continued safety of pharmacotherapy.
The safety of a drug continues to be monitored after approval and marketing in an ongoing process of pharmacovigilance. 1 This postmarket drug safety monitoring is especially important with regard to adverse drug reactions (ADRs) that are rare, only occurring in certain subgroups, and/or only develop after long‐term drug exposure. In some cases, serious ADRs are not recognized until long after a drug has been approved for market, as seen in the case of thalidomide where its use in pregnant women led to congenital malformations. Accordingly, the importance of postmarket monitoring is highlighted by the finding that one‐third of newly identified safety issues in the postmarketing period are added to the warnings and precautions section of the label, the second highest tier of severity, indicating the serious nature of newly identified ADRs. 2
The passage of the 21st Century Cures Act has modernized clinical trials and requires the evaluation of the potential use of real‐world data (RWD), data collected during routine clinical care in the form of electronic health records (EHRs), medical billing, and other data generating activities in the regulatory decision making and approval process. Real‐world evidence (RWE) is the evidence of the potential benefits of the medical product in a clinical setting derived from RWD. Results from various study designs and analyses, both prospective and retrospective, that use RWD are accepted as RWE. The US Food and Drug Administration (FDA) guidance on RWE describes several contexts in which it can be used during the product life cycle, such as proving an unmet medical need, substituting for a control group, as supporting evidence for a label expansion, and as a part of postmarketing studies. The multiple emergency use authorizations granted to drugs during the coronavirus disease 2019 (COVID‐19) pandemic highlights a situation where postmarket pharmacovigilance becomes pivotal to maintaining long‐term patient safety. Collectively, the legislative acts and regulatory practices have led to an increased reliance on postmarket pharmacovigilance to inform drug safety. Innovation in pharmacovigilance is needed to address these challenges and complement clinical trials by improving the sensitivity and specificity of ADR detection and streamlining the process of refining RWD into RWE that supports regulatory decision making.
ESTABLISHED PHARMACOVIGILANCE SYSTEMS
Published case reports have been circulated among physicians since the late 1960s and continue to serve an important role in pharmacovigilance. They are typically rich in information because physicians are trained in the rigorous evaluation of medical histories, drug exposures, and outcomes; additionally, peer review provides a form of quality control. However, case reports are fundamentally anecdotal data points, and as such cannot support conclusions in broader populations. The digitization of written media and advent of databases and search engines make it possible to collect, store, and rapidly retrieve relevant and comprehensive case series, but the data are unstructured text, which is not suitable for rigorous quantitative analysis. Despite these limitations, case reports published in journals are useful for generating hypotheses, and pharmacovigilance studies often start with a search of the relevant case literature.
Medwatch has been the principal means of collecting and analyzing information about ADRs since 1993 and is used by the FDA to collect information on both small molecule drugs and biologics. Data are collected using standardized individual case safety reports forms, which are submitted physically or electronically to the FDA Adverse Event Reporting System (FAERS). The aggregate data are then mined for safety signals, which generate hypotheses for further investigation. FAERS has successfully identified previously unreported ADRs, with FAERS data contributing to more than 50% of all postmarket safety‐related label changes. 3 Table 1 lists a selection of additional pharmacovigilance studies in which FAERS or other ADR databases have played a prominent role. In addition to FAERS, the FDA has event reporting systems for (1) foods, dietary supplements, and cosmetics, (2) medical devices, and (3) vaccines, via CAERS, MAUDE, and VAERS, respectively.
Table 1.
Select examples of successful pharmacovigilance studies in which ADR and RWD database studies played a prominent role
| Drug(s) | Effect(s) | Source(s) | Citation |
|---|---|---|---|
| Acetaminophen | Liver injury | HER | 15 |
| Agomelatine | Liver injury | Lit review | 16 |
| Gabapentin, pregabalin | Liver injury, hematological disorders | ADR database | 17 |
| Apixaban | Liver injury | Case report, ADR database | 18 |
| Ketoconazole | Liver injury | Lit review, ADR database | 19 |
| Methadone | Arrhythmia | Lit review, ADR database | 20 |
| Ranolazine | Seizure | Sentinel | 21 |
| Levetiracetam, phenytoin | Angioedema | OHDSI | 22 |
| Citalopram | Arrhythmia | EHR | 23 |
| Hydroxyzine | Arrhythmia | Lit review, ADR database | 24 |
ADR, adverse drug reaction; EHR, electronic health record; OHDSI, Open Health Data Science Informatics; RWD, real‐world data.
However, FAERS case reports as a source of data are limited by incompleteness, bias, and inconsistency. Prescribing decisions are often influenced by factors that affect clinical outcomes, such as comorbidities, insurance, and access to primary care, information that is not available in the publicly available FAERS data. The Institute for Safe Medical Practices found that over half of the reports in FAERS were missing basic information, such as age, gender, exposure date, and outcome. Additionally, FAERS does not measure the total number of exposures in the population, so there is no “denominator” to estimate the frequency of adverse events. Although adverse events are generally under‐reported, stimulated reporting driven by news, social media, and advertising can increase reporting rates for certain drugs. Incorrect hypotheses generated from erroneous or incomplete adverse event report data can be costly, with false‐positives resulting in resources wasted on unnecessary studies and false‐negatives leading to harm to patients.
EMERGING PHARMACOVIGILANCE SYSTEMS
Another component of the data revolution within health care has been the adoption of information technology by the health insurance industry and the adoption of EHRs by healthcare systems as a result of the 2009 Health Information Technology for Economic and Clinical Health (HITECH) Act. Insurance claims capture prescription and medical diagnoses across healthcare providers, with the caveat that they do not directly measure outcomes. EHRs contain rich information, such as clinical notes, images, and laboratory test values; however, they are often locked within institutional silos on systems that are unique for each provider institution and suffer from bias related to their primary purpose, a clinical and legal record (Figure 1 ).
Figure 1.
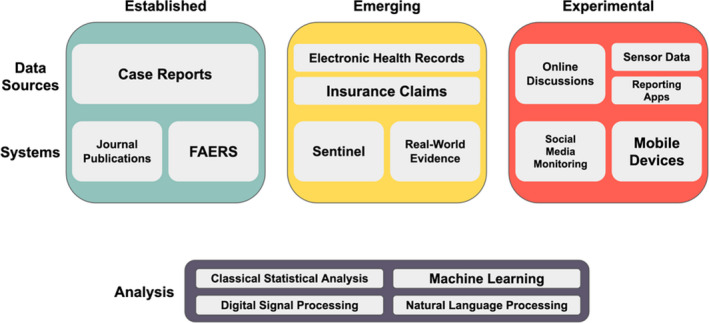
Overview of pharmacovigilance methods at varying stages of development. Established (green, left), emerging (yellow, middle), and experimental (red, right) pharmacovigilance data sources and systems are presented. Examples of methodological areas that are currently used and under active development for the analysis of these different data types are included in the bottom box. FAERS, FDA Adverse Event Reporting System.
The Sentinel initiative extends the pharmacovigilance capabilities of the FDA by leveraging EHR systems and insurance claims data in distributed data networks of partner institutions. 4 The Sentinel system is used to study specific drug‐event outcomes and, more recently, is being used to generate drug safety signals. Analyses can be submitted to the partner network and run independently at each site and results can then be combined to provide comprehensive safety profiles. The integration of these various data sources has allowed for a more comprehensive and synergistic pipeline and capabilities. A general workflow is presented in the top row of Figure 2 . Sentinel required the development and implementation of a common data model and data quality assurance standards to ensure interoperability of data and reliability of analytical findings. Current efforts have been primarily focused on billing and claims data. Several new data partnership networks and consortia have emerged, such as PedsNet and the Open Health Data Science Informatics (OHDSI) network, that are improving and extending the governance, interoperability, and data stewardship frameworks pioneered by Sentinel. For example, the OHDSI network has adopted the OMOP’s Common Data Model for standardizing identifiers for diseases, procedures, drugs, and other components of a patient’s health record and has created a network of hospitals standardized to this data model. This enables an analysis designed at one member institution to be quickly replicated in other healthcare systems within the OHDSI network with minimal need to readjust the analysis. For instance, an analysis designed at Stanford could be run at hospitals in Israel, South Korea, and Australia, quickly finding support for or discrepancies in the findings of a single institution. Patient Centered Outcome Research Institute (PCORI) is establishing data networks, as well as procedures for evaluating and ensuring the relevance and reliability of data. The FDA is piloting demonstration cases for the use of RWE in regulatory decision making.
Figure 2.
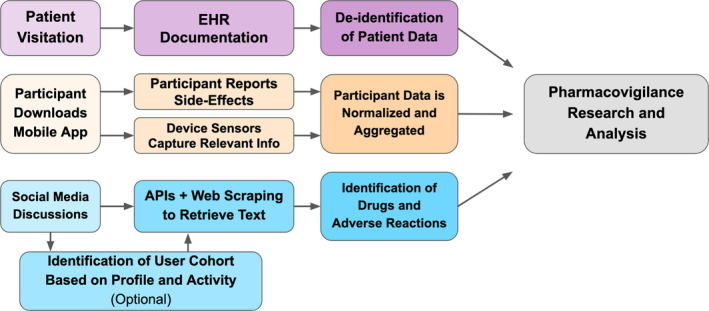
General pharmacovigilance workflows for emerging and experimental systems. EHR based pharmacovigilance workflow is shown in the purple top row. A mobile device‐based pharmacovigilance workflow is shown in the orange middle row. The social media‐based pharmacovigilance workflow is shown in the blue bottom row. These data can then be used separately or in combination to perform pharmacovigilance research and analysis. API, application programming interface; EHR, electronic health record.
An example of a new drug approval that relied on RWE, is Avelumab, a monoclonal antibody directed against programmed death ligand 1. Avelumab was approved based on a single arm, phase II trial where historical controls were identified from EHRs and were used to characterize the natural history of the disease. 5 Additionally, Aspirin Dosing: A Patient‐Centric Trial Assessing Benefits and Long‐Term Effectiveness (ADAPTABLE), a clinical trial evaluating the optimal dose of aspirin in patients with atherosclerotic cardiovascular disease, has utilized PCORnet EHRs and claims data at multiple stages of their study, from identifying patients who meet the inclusion/exclusion criteria to capturing primary and secondary study end points. 6 The ADAPTABLE trial represents the first randomized trial within PCORnet and, as such, has also developed new methodologies to take advantage of the data with the PCORnet data infrastructure.
The primary purpose of EHRs is to inform clinical decisions and/or support administrative functions (i.e., documentation to support billing). As a result, issues such as human/coding errors or bias may affect how information is captured prior to analysis. Additionally, the fractionalized nature of the US healthcare system makes it difficult to track patients across different healthcare systems resulting in incomplete data entries.
Clinical definitions, terminology, and note‐taking style vary between and within healthcare systems, making the extraction and transformation of clinical information to standardized elements, such as SNOMED codes, technically difficult. The challenging nature of clinical note processing has resulted in the majority of analyses to date primarily focusing on the billing related International Classification of Disease 10 codes. Last, unpredictability about patient compliance (i.e., even if a prescription is written does not mean the patient will pick it up) limits the use and extension of these data. These represent major obstacles to widespread pharmacovigilance using EHRs and future work will need to overcome these issues before the benefits of EHR data can be fully realized.
EXPERIMENTAL PHARMACOVIGILANCE SYSTEMS
Although Sentinel, PCORI, and OHDSI have greatly improved pharmacovigilance efforts, they rely on a constrained set of information within the healthcare system, that is, information in the EHR or in billing and claims data. 7 Outside the healthcare system, data from social media represent another key opportunity for pharmacovigilance. Social media data contains various data streams, potentially enabling us to identify patterns in behavior, environment, drug use, drug‐drug interactions, and ADRs. A general workflow for pharmacovigilance in social media data is presented in the bottom row of Figure 2 . The broad usage of social media by the public yields a massive dataset that is continuously growing and has huge potential for generating public health benefits. Individual experiences with a particular drug are often posted directly to social media. These testimonials can be found on both general platforms like Twitter and Reddit, as well as health‐oriented websites, such as AskaPatient.com, drugs.com, and iodine.com. Social media data often contain information critical to postmarket pharmacovigilance, such as individual experiences of ADRs, information about environmental factors, reports of pill diversions, and polypharmacy (both recreational and prescribed) that is often missed by other postmarketing surveillance systems.
There has been progress in developing new methods for postmarketing surveillance in social media data through the use of statistical models, machine learning, and deep neural network architectures. The annual Social Media Mining for Health Applications (SMM4H) workshop has resulted in algorithms capable of identifying drug mentions with high precision and recall, even in situations where these mentions are informal slang terms or misspelled drug names. However, high performance of ADRs continues to present a challenge as text descriptions of a particular ADR might vary greatly in written language, for instance “stomach” may be expressed as “stomach ache,” “stomach pain,” “abdominal pain,” “tummy ache,” etc. Additionally, classifying a particular tweet for first‐person vs. secondary reports of medication ingestion presents another challenge and has also been featured as challenges for the community with varying levels of success. Ideally, these efforts will culminate in systems capable of actively monitoring social media data and generating real‐time statistics relevant to pharmacovigilance efforts.
Although social media can provide a large volume of easily accessible data, the nature of social media presents several challenges for the extraction of signals related to pharmacovigilance. The first set of these challenges are that: (1) very few social media posts are relevant to pharmacovigilance, ~ 0.2% of tweets mention a medication 8 ; (2) information is represented in unstructured text; (3) drugs and medical conditions are often misspelled, abbreviated, or discussed using slang 9 ; (4) mentions of medical events may not be firsthand accounts; and (5) social media reports will contain false‐positives, but often provide less information than clinical case reports and so the reliable identification of true drug side effects from these data will be difficult. Recent work, as mentioned above, indicates that many of these problems may be overcome in the near future. Once these systems can produce robust ADR event statistics, further work may extend their functionality through analysis of the individual testimonies found within social media data. Social media data often contains lifestyle information like exercise patterns, eating habits, socio‐economic issues, and/or drug abuse behavior that will be missing from the EHR for the foreseeable future. For example, systems may find indications of relative quality of life improvements given a particular medication, patient preferences, or capture additional demographic information that could be key to protecting at risk populations, such as pregnant women and children.
In a demonstration of the value of general social media, recent efforts using Twitter have focused on vulnerable populations, such as pregnant women, that are often excluded from clinical trials, and, as a result, drug safety is not typically established in these groups in the premarket space. Although there are methods to gather this information postapproval, such as pregnancy registries, these databases are often constrained by issues, such as attrition, cost, and patient compliance. A recent study using data from Twitter accounts of pregnant women observed a higher medication intake in women who reported birth defects. 10 Similarly, another study developed a natural language processing method to identify tweets by users whose child had a birth defect. 11 These preliminary studies demonstrate how social media, such as Twitter, might help supplement existing resources, especially in vulnerable populations. Thus, it represents an exciting source of potentially complementary information for postmarket pharmacovigilance efforts.
A recent effort questioned the overall value proposition of social media data, citing the low prevalence of posts relevant to pharmacovigilance and low coverage for many drugs. 12 The analysis compared ADR signals from social media to Vigibase report statistics, focusing on the FDA drug labeling changes or “validated” safety signals, where there is evidence the drug has a causal relationship with the ADR. However, Vigibase report statistics may not be an appropriate evaluation baseline because the FDA labeling changes and/or the “validated” safety signal may have resulted from signals within the spontaneous reporting systems, likely inflating the baseline performance. Additionally, this evaluation effort did not adequately address the noisy nature of social media drug reports, failing to include drug misspellings or slang terms in their search queries, potentially missing a substantial number of reports. 9 It is likely that more advanced report identification methods would increase the value of social media data. The overall lack of social media discussions surrounding some drugs will continue to pose a challenge. Although the authors did not recommend the use of general social media data for pharmacovigilance, they indicated that social media generated in the context of a drug or health‐oriented platform (e.g., drugs.com) vs. a general platform (e.g., Twitter) may still hold value.
Beyond the technical challenges of working with social media data, its pseudonymous, open, and ephemeral nature creates new challenges in ethics, law, and reproducibility that must be navigated. Many platforms limit the sharing of data collected from their users and require that content be deleted upon user request. Social media posts experience high deletion rates with more than 40% of posts from one study being deleted from the platform after the study was published. 13 Researchers must preserve their own copies of data used for a particular study to ensure reproducibility. The publishing of the contents of social media posts in scientific journals may disclose potentially sensitive information about users, such as illicit drug use or mental health issues. Researchers must balance between making research reproducible and the ethical concerns of the risk of making research datasets freely available, which might increase the risk of abuse.
Mobile devices are a recent innovation in capturing information about ADRs, again providing another avenue of data collection in an uncontrolled setting. A general workflow for pharmacovigilance using mobile devices is presented in the middle row of Figure 2 . MyHeart Counts is used to do a 6‐minute walk test, which can be done daily in an in‐home setting. MedWatcher was a mobile application version of the FDA 3500 form for medical devices and is currently undergoing implementation in the European Union. Hugo platform for postmarket surveillance is under development at the Yale‐Mayo Center of Excellence in Regulatory Science and Innovation, Yale‐Mayo, which can collect electronic patient‐reported outcomes outside of the hospital. 14 Next steps include interfacing with connected devices to measure end points; however, the strides made in this more recent area of pharmacovigilance are very promising.
These are two modalities among many that researchers are investigating as potential new means of pharmacovigilance. Through the FDA funded Centers of Excellence in Regulatory Science and Innovation (CERSI), other databases and methodologies are being studied as potential pharmacovigilance systems, for examples see https://pharm.ucsf.edu/cersi/research.
CONCLUSION
Clearly, the development of these massive sources of data for future pharmacovigilance efforts creates an opportunity for capitalizing on recent advances in deep learning and anomaly detection. A continuously learning artificial intelligence system could not only learn to integrate these heterogeneous data sources for real‐time ADR detection, but could help identify potential cases and interface with members of the pharmacotherapy community to gather more information when needed. The field of pharmacovigilance is rapidly evolving, however, the resources we have highlighted are only part of the solution; the FDA and National Institutes of Health (NIH) will need to continue their funding of research that focuses on how to effectively analyze these data streams. Ideally, funding mechanisms will ensure interdisciplinary teams of experts from epidemiology, sociology, statistics, and computer science among others. Collaborative interdisciplinary efforts will ensure both institutional buy‐in as well as methodological rigor. Ultimately, the combination of various data sources and expertise will result in safer and more effective pharmacotherapy for everyone.
Funding
A.L. is supported by the National Science Foundation Graduate Research Fellowship, DGE – 1656518. B.V. is supported by an Oak Ridge Institute for Science and Education (ORISE) Fellowship, and is the recipient of an Achievement Rewards for College Scientists (ARCS) Scholarship. R.B.A. is supported by NIH GM102365, HG010615, and the Chan‐Zuckerberg Biohub. This work was partially supported by Grant Number U01FD004979/U01FD005978 from the FDA, which supports the UCSF‐Stanford Center of Excellence in Regulatory Sciences and Innovation.
Conflict of Interest
The authors declare no conflict of interest.
Disclaimer
As Deputy Editor‐in‐Chief of Clinical Pharmacology & Therapeutics, Kathleen M. Giacomini was not involved in the review or decision process for this paper.
ACKNOWLEDGMENTS
The authors would like to thank the reviewers for their excellent feedback during the review process and the editorial board for their understanding during the COVID‐19 pandemic. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the Health and Human Services (HHS) or the FDA.
Contributor Information
Russ Altman, Email: Russ.Altman@stanford.edu.
Stefano Rensi, Email: srensi@stanford.edu.
References
- 1. Giacomini, K.M. et al. When good drugs go bad. Nature 446, 975–977 (2007). [DOI] [PubMed] [Google Scholar]
- 2. Pinnow, E. et al. Postmarket safety outcomes for new molecular entity (NME) drugs approved by the Food and Drug Administration between 2002 and 2014. Clin. Pharmacol. Ther. 104, 390–400 (2018). [DOI] [PubMed] [Google Scholar]
- 3. Lester, J. et al. Evaluation of FDA safety‐related drug label changes in 2010. Pharmacoepidemiol. Drug Saf. 22, 302–305 (2013). [DOI] [PubMed] [Google Scholar]
- 4. Platt, R. et al. The FDA Sentinel Initiative—an evolving national resource. N. Engl. J. Med. 379, 2091–2093 (2018). [DOI] [PubMed] [Google Scholar]
- 5. Center for Drug Evaluation Research FDA D.I.S.C.O. Avelumab in Merkel Cell Carcinoma Transcript <https://www.fda.gov/drugs/resources‐information‐approved‐drugs/fda‐disco‐avelumab‐merkel‐cell‐carcinoma‐transcript> (2020).
- 6. Marquis‐Gravel, G. et al. Rationale and design of the Aspirin Dosing‐A Patient‐Centric Trial Assessing Benefits and Long‐term Effectiveness (ADAPTABLE) trial. JAMA Cardiol. 5, 598–607 (2020). [DOI] [PubMed] [Google Scholar]
- 7. Freifeld, C.C. Digital pharmacovigilance: the Medwatcher system for monitoring adverse events through automated processing of internet social media and crowdsourcing <http://search.proquest.com/openview/3b48f9adb40db928bb56ec6501f3a4fa/1?pq‐origsite=gscholar&cbl=18750&diss=y&casa_token=v2llyFuk8wgAAAAA:kgT9vdU4v1l9jzSRhqiNX2AOnXdlSf3BqshqFfAFvYkFW_4h69ZPbjOeVVSxMEzuJe5eMfBe7A> (2014).
- 8. Weissenbacher, D. et al. Deep neural networks ensemble for detecting medication mentions in tweets. J. Am. Med. Inform. Assoc. 26, 1618–1626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lavertu, A. & Altman, R.B. RedMed: extending drug lexicons for social media applications. J. Biomed. Inform. 99, 103307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Golder, S. et al. Pharmacoepidemiologic evaluation of birth defects from health‐related postings in social media during pregnancy. Drug Saf. 42, 389–400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klein, A.Z. , Sarker, A. , Weissenbacher, D. & Gonzalez‐Hernandez, G. Towards scaling Twitter for digital epidemiology of birth defects. NPJ Digit. Med. 2, 96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Stekelenborg, J. et al. Recommendations for the use of social media in pharmacovigilance: lessons from IMI WEB‐RADR. Drug Saf. 42, 1393–1407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magge, A. , Sarker, A. , Nikfarjam, A. & Gonzalez‐Hernandez, G. Comment on: ‘deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts’. J. Am. Med. Inform. Assoc. 26, 577–579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhruva, S.S. et al. Aggregating multiple real‐world data sources using a patient‐centered health‐data‐sharing platform. NPJ Digit. Med. 3, 60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tong, H.Y. et al. Hepatotoxicity induced by acute and chronic paracetamol overdose in adults. Where do we stand? Regul. Toxicol. Pharmacol. 72, 370–378 (2015). [DOI] [PubMed] [Google Scholar]
- 16. Freiesleben, S.D. & Furczyk, K. A systematic review of agomelatine‐induced liver injury. J. Mol. Psychiatry 3, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuzier, R. et al. Adverse drug reactions to gabapentin and pregabalin. Drug Saf. 36, 55–62 (2013). [DOI] [PubMed] [Google Scholar]
- 18. Clarke, S.‐A. , Alsaad, A.A. , Mack, A. & Phillips, M.B. Apixaban‐induced liver injury. BMJ Case Rep. 2016, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raschi, E. , Poluzzi, E. , Koci, A. , Caraceni, P. & Ponti, F.D. Assessing liver injury associated with antimycotics: concise literature review and clues from data mining of the FAERS database. World J. Hepatol. 6, 601–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kao, D.P. , Haigney, M.C.P. , Mehler, P.S. & Krantz, M.J. Arrhythmia associated with buprenorphine and methadone reported to the food and drug administration. Addiction 110, 1468–1475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eworuke, E. , Welch, E.C. , Tobenkin, A. & Maro, J.C. Use of FDA’s sentinel system to quantify seizure risk immediately following new ranolazine exposure. Drug Saf. 42, 897–906 (2019). [DOI] [PubMed] [Google Scholar]
- 22. Duke, J.D. et al. Risk of angioedema associated with levetiracetam compared with phenytoin: findings of the observational health data sciences and informatics research network. Epilepsia 58, e101–e106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castro, V.M. et al. QT interval and antidepressant use: a cross sectional study of electronic health records. BMJ 346, f288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schlit, A.‐F. et al. Risk of QT prolongation and torsade de pointes associated with exposure to hydroxyzine: re‐evaluation of an established drug. Pharmacol. Res. Perspect. 5, e00309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


