Supplemental Digital Content is available in the text.
Keywords: atrial fibrillation, calibration, general surgery, likelihood functions, thoracic surgery
Abstract
Background:
Postoperative atrial fibrillation (PoAF) remains a significant risk factor for increased morbidity and mortality after cardiac surgery. The ability to accurately identify patients at risk through clinical risk factors is limited. There is growing evidence that polygenic risk contributes significantly to PoAF and incorporating measures of genetic risk could enhance prediction.
Methods:
A retrospective cohort study of 1047 patients of White European ancestry who underwent either coronary artery bypass grafting or valve surgery at a tertiary academic center and were free from a history or persistent preoperative atrial fibrillation. The primary outcome was defined as PoAF based on postoperative ECG reports, medical record documentation, and changes in medication. The exposure was a polygenic risk score (PRS) comprising 2746 single-nucleotide polymorphisms previously associated with atrial fibrillation risk. The prediction of PoAF risk was assessed using measures of model discrimination, calibration, and net reclassification improvement.
Results:
A total of 259 patients (24.7%) developed PoAF. The PRS was significantly associated with a higher risk for PoAF (odds ratio, 1.63 per SD increase in PRS [95% CI, 1.41–1.90]). Addition of PRS to patient- and procedure-related predictors of PoAF significantly increased the C statistic from 0.742 to 0.782 (change in C statistic, 0.040 [95% CI, 0.021–0.060]) while maintaining good calibration. The addition of the PRS to patient- and procedure-related predictors of PoAF improved model fit (likelihood ratio test, P=2.8×10−15) and significantly improved measures of reclassification (net reclassification improvement, 0.158 [95% CI, 0.066–0.274]).
Conclusions:
The PRS for PoAF was associated with improved discrimination, calibration, and risk reclassification compared with conventional clinical predictors suggesting that a PoAF PRS may enhance risk prediction of PoAF in patients undergoing coronary artery bypass grafting or valve surgery.
Postoperative atrial fibrillation (PoAF) is a common and significant complication following cardiac surgery that occurs in as many as 30% to 50% of patients.1–3 It is associated with an increased risk for postoperative neurological events, congestive heart failure, myocardial infarction, perioperative mortality, prolonged hospital length of stay, and increased hospital costs.4 Therefore, accurately identifying patients at high risk for PoAF would allow development of targeted treatment modalities and reduce the risk for subsequent complications and mortality.5,6
Older age, a history of atrial fibrillation (AF), chronic obstructive pulmonary disease, valve surgery, and discontinuation of β-blocker or angiotensin-converting enzyme inhibitor therapy are risk factors for PoAF after cardiac surgery.4,7 Several comprehensive clinical risk indices incorporating these risk factors have been developed to predict PoAF risk and identify potential preventative strategies.7–10 However, the performance and the generalizability of these risk indices for PoAF is modest.10 Previous candidate gene and genome-wide association studies have identified multiple genetic loci and common single-nucleotide polymorphism (SNP) variants that predispose to PoAF.11–13 However, the contributions of these individual common genetic variants beyond conventional clinical risk factors for PoAF has been minimal.11–13 Recently, there has been a growing interest in using polygenic risk scores (PRSs) that incorporate multiple common genetic variants associated with AF to identify individuals in the general population who are at increased risk for developing AF.14 To date, the relationship between polygenic variation to postcardiac surgery AF susceptibility has not been studied. Moreover, the additional predictive value of such a PRS in addition to conventional clinical risk factors for predicting the risk of PoAF has remained unexplored.
To address these knowledge gaps, using a large, real-world clinical data set of patients who underwent cardiac surgery at the Vanderbilt University Medical Center, we tested the hypothesis that a PRS for AF risk would enhance risk prediction of PoAF, as compared with a validated clinical predictive model.
Methods
The authors will make the data, methods used in the analysis, and materials used to conduct the research available to any qualified researcher trained in human subject confidentiality protocols for purposes of reproducing the results or replicating the procedure. This study was approved by the Institutional Review Board at the Vanderbilt University Medical Center. Given the retrospective design of the study and the use of deidentified data, the need for an informed patient consent was waived by the Vanderbilt University Medical Center Institutional Review Board Committee. Full methods are available in the Data Supplement.
Results
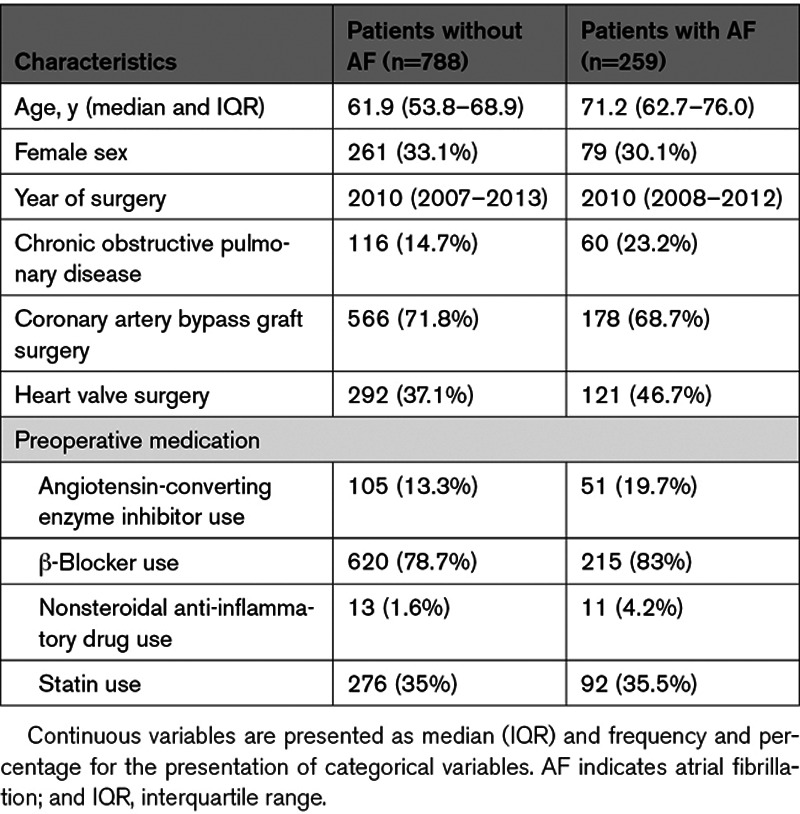
The final study population comprised 1047 subjects (Figure 1). The median age was 63.9 years (interquartile range, 55.6 – 71.6), 340 (39%) were women, and 744 (71%) underwent coronary artery bypass grafting (CABG) surgery. PoAF developed in 259 (24.7%) individuals. As compared with controls, patients with PoAF were older, had a history of chronic obstructive pulmonary disease, underwent heart valve surgery, and had a history of preoperative angiotensin-converting enzyme inhibitor use and nonsteroidal anti-inflammatory drug use compared with patients without PoAF (Table 1).
Figure 1.
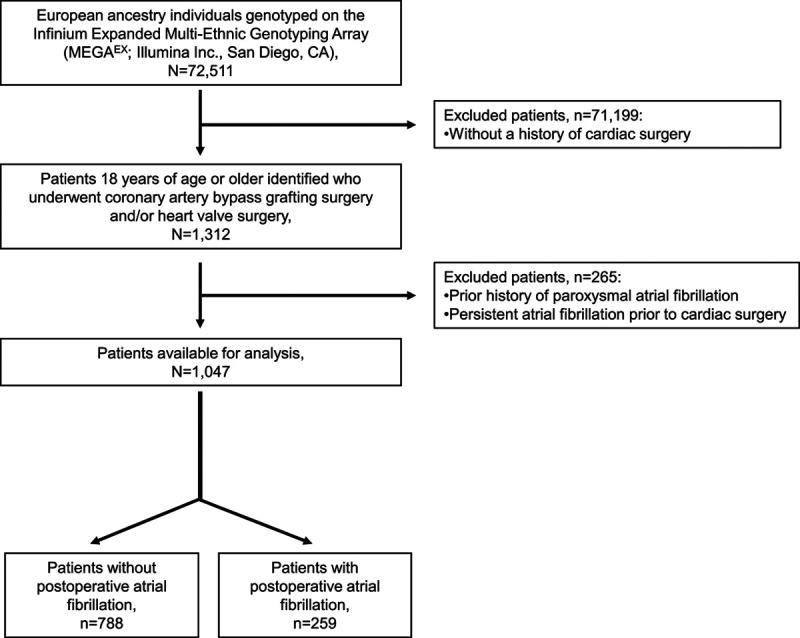
Flowchart of the study population with inclusion and exclusion criteria.
Table 1.
Characteristics of the Study Population
Models for Predicting PoAF
Cases, as compared with controls, had higher PoAF PRS values (Figure 2). After adjusting for sex, age, type and year of cardiac surgery, clinical and procedural predictors of PoAF, and 4 principle components, the PoAF PRS was significantly associated with PoAF (odds ratio, 1.92 [95% CI, 1.63–2.29]; P=6.8×10−14). Adding the AF PRS to the standard model with clinical predictors also significantly increased the C index from 0.742 to 0.782 (difference, 0.040 [95% CI, 0.019–0.060]; Table 2). A comparison of the two models using the likelihood ratio test demonstrated a significant improvement in model fit (χ2 test, 62.4; P=2.83×10−15) while maintaining good calibration (Figures I and II in the Data Supplement). In sum, the addition of the AF PRS improved discrimination, as compared with a model comprising clinical predictors.
Figure 2.
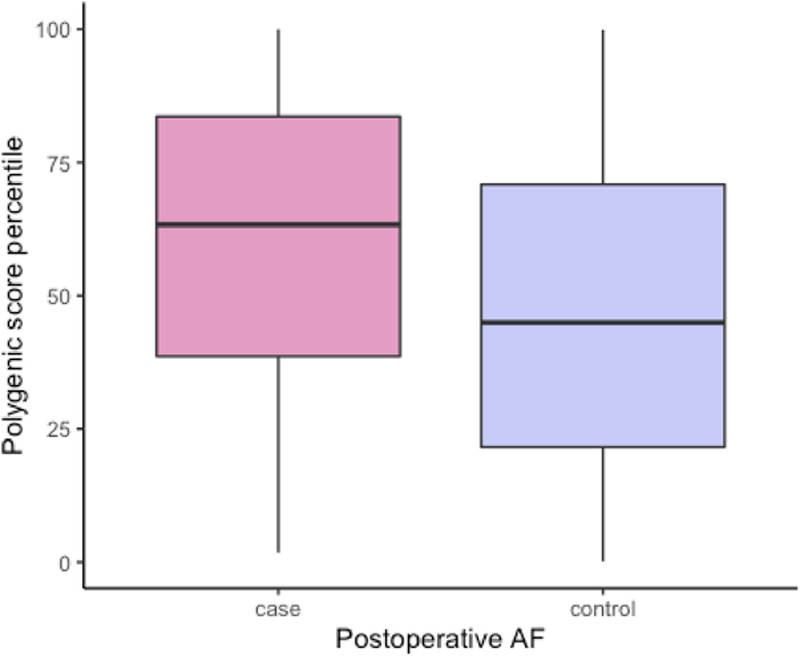
Box plots summarizing the distributions of the postoperative atrial fibrillation (AF) polygenic risk score (PRS) among cases and controls (median and interquartile range for PRS: cases, 0.34 [−0.32 to 1.02]; controls, −0.18 [−0.77 to 0.56]).
Table 2.
C Statistics Evaluating the Performance of the PRS in the Study Population
Net Reclassification Improvement
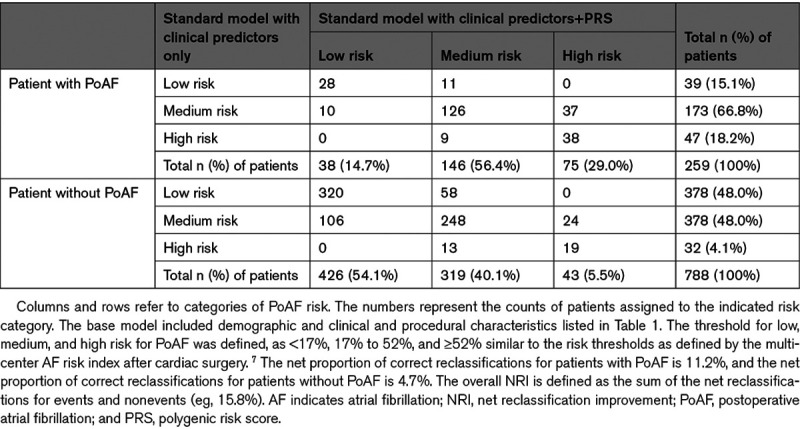
The Integrated Discrimination Improvement—a continuous measure of reclassification enhancement—was significant (Integrated Discrimination Improvement, 0.06 [95% CI, 0.04–0.07]) for a model that included the PRS, as compared with a model comprising clinical and procedural characteristics. The net reclassification improvement was used to assess reclassification among low-risk (<17%), intermediate-risk (17%–52%), and high-risk (≥52%) categories. Among individuals who developed PoAF, addition of the PRS to the clinical model increased the proportion of subjects categorized as high risk from 18% to 29.0% (Table 3). Among controls, the proportion categorized as low risk increased from 48% to 54%. Thus, addition of the PRS improved reclassification primarily by increasing risk estimates among cases who fell in the intermediate-risk categories and decreasing risk estimates for the intermediate-risk patients among controls. The net reclassification improvement estimates for cases and controls were 0.112 and 0.047, respectively, and the overall net reclassification improvement estimate was significant (net reclassification improvement, 0.159 [95% CI, 0.066–0.274]).
Table 3.
Reclassification of the PoAF Risk
Discussion
To our knowledge, this study examined for the first time whether a polygenic predictor for AF risk enhanced risk stratification for incident AF in a real-world clinical population of patients undergoing cardiac surgery. The PoAF PRS was strongly associated with incident AF risk, independent of conventional clinical predictors for PoAF. Furthermore, the PoAF PRS classifier enhanced the discrimination and reclassification of a predictive risk model. Collectively, our findings support the concept that incorporating PoAF PRS along with conventional clinical predictors of PoAF may enhance risk prediction for PoAF in patients undergoing CABG or valve surgery.
Our findings support and extend prior observations that AF genetic risk is associated with PoAF after cardiac surgery. Several potential genetic factors have been studied and implicated including noncoding polymorphisms within the chromosome 4q25 region that have been associated with the development of AF in both ambulatory15,16 and cardiac surgery cohorts.11,13 We previously observed in a candidate gene study an association between variants of GRK5 gene polymorphisms and PoAF in patients who exclusively received perioperative β-blocker therapy and underwent CABG surgery.12 We also observed in genome-wide association studies an association between a variant in LY96 with relevance to activation and modulation of innate immune responses and a decreased risk for PoAF after CABG surgery.11
In parallel to these studies that either identified a single genetic variation or validated a previously identified single genetic marker in the cardiac surgery setting, large-scale population-based studies observed that when several significant SNPs associated with AF were combined into a genetic risk score, such scores showed a more profound association with AF independent from traditional clinical risk factors. Indeed, a 12-SNP AF genetic risk score that was based on 9 loci was associated with 4-fold to 5-fold increased risk between those in the highest versus the lowest tails of the AF genetic risk score in case-referent and cohort studies.17 Similarly, the Women’s Genome Health Study reported an association between an AF genetic risk score based on 12 SNPs and a higher risk for incident AF.18 Recently, Lubitz et al19 examined associations between AF genetic risk scores and incident AF in 5 prospective studies of 18 919 population-based individuals of the European ancestry. They found that predictive models with AF PRSs with 25 to 129 SNPs were significantly associated with new-onset AF beyond associations for conventional clinical AF risk factors. Thus, by using our well-characterized cohort of cardiac surgery patients, our present findings extend these prior reports and demonstrate that AF polygenic risk is also associated with incident PoAF after cardiac surgery.
Our study also further highlights the ability of common genetic variations associated with AF genetic risk to predict PoAF and to capture complimentary information beyond the effects observed for well-characterized clinical and procedure-related risk factors for PoAF.19 Similar to established risk factors, the improvements associated with the PoAF PRS are incremental. Thus, our findings are similar to the observations made by the study of Lubitz et al19 and underscore the challenges of improving clinical prediction models even when significantly associated predictors such as genetic predictors are included.20
Our observation that the PoAF PRS, which was constructed and selected from a pool of previously identified SNPs associated with AF, was associated with PoAF after cardiac surgery highlights the polygenic nature of postcardiac surgery AF. These findings also indicate and reinforce previous observations that true PoAF PRS susceptibility variants are present among SNPs that did not achieve genome-wide significance in studies of patients undergoing cardiac surgery.11–13 Given the relatively small number and sample size of the genetic studies for PoAF after cardiac surgery, there is a need for future type of studies with merging contemporary cardiac surgery datasets with available genetic information to improve power and to confirm previously identified genetic variations or discover additional susceptibility genes for PoAF after cardiac surgery. Until then, it remains to be determined whether assessment of PoAF PRS genetic risk using polygenic predictors derived from larger sample sizes will further increase the predictive accuracy and discriminative ability of clinical risk prediction models.
Limitations
This study has several limitations. First, in our retrospective single-center study, we identified potential cases of PoAF to review using the International Classification of Diseases, Ninth and Tenth Revision, codes, and thus, there is a potential possibility that cases with PoAF may have been underestimated. However, the frequency of PoAF observed in our study was similar to those reported from recent studies.11–13 Second, in our study, we used a previously validated methodology for developing our PoAF PRS, but the application of this PoAF PRS to predict PoAF will have to be externally validated to test for its performance and predictive accuracy in an independent cohort of cardiac surgery patients. The underlying etiology for PoAF, compared with AF, in the ambulatory setting could be multifactorial.11,12 Therefore, applying a PRS for AF that was originally developed in ambulatory subjects to patients undergoing cardiac surgery could not fully capture and characterize the genetic contribution to postcardiac surgery AF. Thus, future studies are needed to develop and validate a PRS specifically for postcardiac surgery AF and characterize how such a PRS would compare to the one described in our study. Finally, patients enrolled in our study were Whites with European descent, and, therefore, our findings may not be generalizable to subjects of other ancestral groups.
Conclusions
As highlighted by the current Society of Cardiovascular Anesthesiologists/European Association of Cardiothoracic Anaesthetists Practice Advisory for the Management of Perioperative AF in Patients undergoing Cardiac Surgery, improved risk stratification through risk score models allows stratification patients into risk groups and facilitates adherence to the evidence-based recommendations for the prevention of PoAF.21 The findings of our study that the addition of PRS incrementally improved the ability to predict PoAF risk, above standard clinical predictors, after CABG or valve surgery further reinforces the recommendations of this practice advisory, and potentially highlights the opportunity to apply preventative measures, such a combination of prophylactic perioperative β-blocker and amiodarone administration, selectively to patients at high risk for PoAF.
The use of PRS to predict postoperative complications including PoAF after cardiac surgery will likely become increasingly feasible in the future with the advancement and increasing frequency of genetic testing in clinical settings. Therefore, when genetic information is available, a PRS could be readily incorporated to enhance risk prediction for PoAF.
Sources of Funding
Publication of this article was supported, in part, by the National Library of Medicine (R01-LM010685-11 to L. Bastarache), National Institute of General Medical Sciences (R01-GM130791 to Dr Mosley), the American Heart Association (16FTF30130005 and 18SFRN34230089 to Dr Mosley), and the Vanderbilt Institute for Clinical and Translation Research Grant (VR52365 to Dr Kertai).
Disclosures
None.
Supplemental Materials
Expanded Methods
Online Figures I and II
Nonstandard Abbreviations and Acronyms
- AF
- atrial fibrillation
- CABG
- coronary artery bypass grafting
- PoAF
- postoperative atrial fibrillation
- PRS
- polygenic risk score
- SNP
- single-nucleotide polymorphism
For Sources of Funding and Disclosures, see page 157.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.120.003269.
Contributor Information
Jonathan D. Mosley, Email: jonathan.d.mosley@vumc.org.
Jing He, Email: jing.he@vumc.org.
Abinaya Ramakrishnan, Email: abinaya.ramakrishnan@vanderbilt.edu.
Mark J. Abdelmalak, Email: mark.j.abdelmalak@vanderbilt.edu.
Yurim Hong, Email: yurim.hong@vanderbilt.edu.
M. Benjamin Shoemaker, Email: moore.b.shoemaker@vumc.org.
Dan M. Roden, Email: Dan.Roden@VUMC.org.
Lisa Bastarache, Email: lisa.bastarache@vumc.org.
References
- 1.Kaireviciute D, Aidietis A, Lip GY. Atrial fibrillation following cardiac surgery: clinical features and preventative strategies. Eur Heart J. 2009; 30:410–425. doi: 10.1093/eurheartj/ehn609 [DOI] [PubMed] [Google Scholar]
- 2.Helgadottir S, Sigurdsson MI, Ingvarsdottir IL, Arnar DO, Gudbjartsson T. Atrial fibrillation following cardiac surgery: risk analysis and long-term survival. J Cardiothorac Surg. 2012; 7:87. doi: 10.1186/1749-8090-7-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigurdsson MI, Longford NT, Heydarpour M, Saddic L, Chang TW, Fox AA, Collard CD, Aranki S, Shekar P, Shernan SK, et al. Duration of postoperative atrial fibrillation after cardiac surgery is associated with worsened long-term survival. Ann Thorac Surg. 2016; 102:2018–2026. doi: 10.1016/j.athoracsur.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobrev D, Aguilar M, Heijman J, Guichard JB, Nattel S. Postoperative atrial fibrillation: mechanisms, manifestations and management. Nat Rev Cardiol. 2019; 16:417–436. doi: 10.1038/s41569-019-0166-5 [DOI] [PubMed] [Google Scholar]
- 5.Dunham WC, Weinger MB, Slagle J, Pretorius M, Shah AS, Absi TS, Shotwell MS, Beller M, Thomas E, Vnencak-Jones CL, et al. CYP2D6 genotype-guided metoprolol therapy in cardiac surgery patients: rationale and design of the pharmacogenetic-guided metoprolol management for postoperative atrial fibrillation in cardiac surgery (PREEMPTIVE) pilot study. J Cardiothorac Vasc Anesth. 2020; 34:20–28. doi: 10.1053/j.jvca.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal N, Kertai MD. Perioperative precision medicine: where are we in 2020? Curr Opin Anaesthesiol. 2020; 33:463–474. doi: 10.1097/ACO.0000000000000858 [DOI] [PubMed] [Google Scholar]
- 7.Mathew JP, Fontes ML, Tudor IC, Ramsay J, Duke P, Mazer CD, Barash PG, Hsu PH, Mangano DT; Investigators of the Ischemia Research and Education Foundation; Multicenter Study of Perioperative Ischemia Research Group. A multicenter risk index for atrial fibrillation after cardiac surgery. JAMA. 2004; 291:1720–1729. doi: 10.1001/jama.291.14.1720 [DOI] [PubMed] [Google Scholar]
- 8.Shen J, Lall S, Zheng V, Buckley P, Damiano RJ, Jr, Schuessler RB. The persistent problem of new-onset postoperative atrial fibrillation: a single-institution experience over two decades. J Thorac Cardiovasc Surg. 2011; 141:559–570. doi: 10.1016/j.jtcvs.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariscalco G, Biancari F, Zanobini M, Cottini M, Piffaretti G, Saccocci M, Banach M, Beghi C, Angelini GD. Bedside tool for predicting the risk of postoperative atrial fibrillation after cardiac surgery: the POAF score. J Am Heart Assoc. 2014; 3:e000752. doi: 10.1161/JAHA.113.000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldron NH, Cooter M, Piccini JP, Anstrom KJ, Klinger RY, Kertai MD, Podgoreanu MV, Stafford-Smith M, Newman MF, Mathew JP. Predictive ability of perioperative atrial fibrillation risk indices in cardiac surgery patients: a retrospective cohort study. Can J Anaesth. 2018; 65:786–796. doi: 10.1007/s12630-018-1119-x [DOI] [PubMed] [Google Scholar]
- 11.Kertai MD, Li YJ, Ji Y, Qi W, Lombard FW, Shah SH, Kraus WE, Stafford-Smith M, Newman MF, Milano CA, et al. ; Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team. Genome-wide association study of new-onset atrial fibrillation after coronary artery bypass grafting surgery. Am Heart J. 2015; 170:580–90.e28. doi: 10.1016/j.ahj.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kertai MD, Li YW, Li YJ, Shah SH, Kraus WE, Fontes ML, Stafford-Smith M, Newman MF, Podgoreanu MV, Mathew JP; Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team. G protein-coupled receptor kinase 5 gene polymorphisms are associated with postoperative atrial fibrillation after coronary artery bypass grafting in patients receiving β-blockers. Circ Cardiovasc Genet. 2014; 7:625–633. doi: 10.1161/CIRCGENETICS.113.000451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, Perry TE, Muehlschlegel JD, Aranki S, Donahue BS, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009; 2:499–506. doi: 10.1161/CIRCGENETICS.109.849075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SH, Jurgens SJ, Weng LC, Pirruccello JP, Roselli C, Chaffin M, Lee CJ, Hall AW, Khera AV, Lunetta KL, et al. Monogenic and polygenic contributions to atrial fibrillation risk: results from a National Biobank. Circ Res. 2020; 126:200–209. doi: 10.1161/CIRCRESAHA.119.315686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010; 42:240–244. doi: 10.1038/ng.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darbar D, Roden DM. Genetic mechanisms of atrial fibrillation: impact on response to treatment. Nat Rev Cardiol. 2013; 10:317–329. doi: 10.1038/nrcardio.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME, et al. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol. 2014; 63:1200–1210. doi: 10.1016/j.jacc.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J. 2013; 34:2243–2251. doi: 10.1093/eurheartj/eht033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubitz SA, Yin X, Lin HJ, Kolek M, Smith JG, Trompet S, Rienstra M, Rost NS, Teixeira PL, Almgren P, et al. ; AFGen Consortium. Genetic Risk Prediction of Atrial Fibrillation. Circulation. 2017; 135:1311–1320. doi: 10.1161/CIRCULATIONAHA.116.024143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007; 115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402 [DOI] [PubMed] [Google Scholar]
- 21.Muehlschlegel JD, Burrage PS, Ngai JY, Prutkin JM, Huang CC, Xu X, Chae SH, Bollen BA, Piccini JP, Schwann NM, et al. Society of Cardiovascular Anesthesiologists/European Association of Cardiothoracic Anaesthetists Practice Advisory for the management of perioperative atrial fibrillation in patients undergoing cardiac surgery. Anesth Analg. 2019; 128:33–42. doi: 10.1213/ANE.0000000000003865 [DOI] [PubMed] [Google Scholar]
- 22.Zhong X, Yin Z, Jia G, Zhou D, Wei Q, Faucon A, Evans P, Gamazon ER, Li B, Tao R, et al. Electronic health record phenotypes associated with genetically regulated expression of CFTR and application to cystic fibrosis. Genet Med. 2020; 22:1191–1200. doi: 10.1038/s41436-020-0786-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007; 316:1491–1493. doi: 10.1126/science.1142842 [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38:904–909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 25.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. ; Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016; 48:1279–1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, Lin H, Arking DE, Smith AV, Albert CM, et al. ; METASTROKE Consortium of the ISGC; Neurology Working Group of the CHARGE Consortium; AFGen Consortium. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017; 49:946–952. doi: 10.1038/ng.3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982; 247:2543–2546 [PubMed] [Google Scholar]
- 28.Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P, Tzoulaki I. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020; 323:636–645. doi: 10.1001/jama.2019.22241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ, O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012; 5:113–121. doi: 10.1161/CIRCGENETICS.111.961342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeileis A, Hothorn T. Diagnostic checking in regression relationship. R News. 2002; 2:7–10 [Google Scholar]
- 31.Kundu S, Aulchenko YS, van Duijn CM, Janssens AC. PredictABEL: an R package for the assessment of risk prediction models. Eur J Epidemiol. 2011; 26:261–264. doi: 10.1007/s10654-011-9567-4 [DOI] [PMC free article] [PubMed] [Google Scholar]


