Abstract
Mycovirus diversity is generally analyzed from isolates of fungal culture isolates at a single point in time as a snapshot. The stability of mycovirus composition within the same geographical location over time remains unclear. Not knowing how the population fluctuates in the field can be a source of unpredictability in the successful application of virocontrol. To better understand the changes over time, we monitored the interannual dynamics and abundance of mycoviruses infecting Sclerotinia sclerotiorum at a rapeseed-growing field for three years. We found that the virome in S. sclerotiorum harbors unique mycovirus compositions each year. In total, sixty-eight mycoviruses were identified, among which twenty-four were detected in all three successive years. These twenty-four mycoviruses can be classified as the members of the core virome in this S. sclerotiorum population, which show persistence and relatively high transmissibility under field conditions. Nearly two-thirds of the mycoviruses have positive-sense, single-stranded RNA genomes and were found consistently across all three years. Moreover, twenty-eight mycoviruses are newly described, including four novel, multi-segmented narnaviruses, and four unique bunyaviruses. Overall, the newly discovered mycoviruses in this study belong to as many as twenty families, into which eight were first identified in S. sclerotiorum, demonstrating evolutionarily diverse viromes. Our findings not only shed light on the annual variation of mycovirus diversity but also provide important virus evolutionary clues.
Keywords: Sclerotinia sclerotiorum, core mycoviruses, interannual dynamics, diversity, evolution
1. Introduction
Many fungi are pathogenic and cause severe disease in a variety of plants and animals, including humans (Sharon and Shlezinger 2013). Phytopathogenic fungi are responsible for approximately 70-80% of plant diseases (Oerke 2006). When resistant cultivars are not available, the application of chemical fungicides is the primary method for controlling fungal diseases in economically important crops. However, environmentally friendly biological controls, including virocontrol with hypovirulence-associated mycoviruses, have been considered a viable alternative approach (García-Pedrajas et al. 2019).
Viruses are the most abundant and the most diverse organisms that exist almost everywhere on Earth (Güemes et al. 2016). They infect all types of life forms, from animals and plants to microorganisms, including bacteria and fungi (Koonin et al. 2006). Metagenomics or metatranscriptomics have revealed a remarkable diversity of viruses sampled from various cellular organisms in the past few decades (Zhang et al. 2019). The uncovering of the virosphere diversity has rapidly transformed our understanding of virus diversity and evolution (Zhang et al. 2018). A typical virus has a coat protein that encapsidate the viral genome to form a virion (Krupovic and Koonin 2017). However, many positive-sense single-stranded RNA viruses are capsidless, including Narnaviridae, Mitoviridae, Hypoviridae, Endornaviridae, Umbravirus, among several others (Dolja et al. 2012).
Mycoviruses replicate in all lineages within kingdom Fungi and have been described in phytopathogenic, entomopathogenic, medical, and edible fungi (Pearson et al. 2009; Xie and Jiang 2014; Ghabrial et al. 2015; Kotta-Loizou and Coutts 2017). Most mycovirus infections cause few or negligible symptoms detectable in the host (Ghabrial and Suzuki 2009). However, a portion of the mycoviruses can reduce fungal growth, sporulation, virulence, and mycotoxin production and cause fruiting bodies to deform (Xie and Jiang 2014). Mycoviruses have also been reported to enhance abiotic stress tolerance, pathogenicity, or the fitness of their fungal host (Ahn and Lee 2001; Marquez et al. 2007; Ozkan and Coutts 2015). In practical use, some of the mycoviruses known to induce hypovirulence have been developed as virocontrol agents against plant fungal diseases (Nuss 2005; Ghabrial et al. 2015).
Sclerotinia sclerotiorum is a devastating phytopathogenic fungus with a worldwide distribution and can infect more than 700 species of plants, including important crops and numerous weeds (Farr and Rossman 2020). Sclerotinia stem rot caused by S. sclerotiorum is one of the most serious diseases of annual rapeseed, potentially resulting in yield losses as great as 80% and significantly reducing oil content and quality (Sharma et al. 2015). Sclerotinia sclerotiorum harbors a great diversity with eighty-nine mycoviruses documented in the GenBank database (update date: 26 September 2020), some of which have great potential to be developed as virocontrol agents. For instance, a circular single-stranded DNA virus that confers hypovirulence has been shown to manage sclerotinia disease effectively under field conditions (Yu et al. 2010; Yu et al. 2013; Zhang et al. 2020).
In this study, to investigate the spread and prevalence of mycoviruses in S. sclerotiorum, we focused on mycovirus biodiversity and the annual variation within a single field for three years at the end of growing seasons. The results suggest that even within a small geographical area, the S. sclerotiorum virome consists of a highly variable mycovirus community that not only includes a stably maintained core population under field conditions but also harbors a dynamic portion of the mycovirus compositions. We aimed to define novel mycoviruses in exploring the diversity and evolution of viruses detected from a field population of S. sclerotiorum, as well as initiating a platform for continuously monitoring fungus–mycovirus dynamics in natural fungal communities.
2. Materials and methods
2.1 Fungus isolation and purification
From the pith of rapeseed stems in the late stage of the disease, sclerotia were collected annually for three years from 2016 to 2018. More than 100 sclerotia were collected each year from diseased stems (one sclerotium from each diseased stem) that were randomly sampled from a single field (approximately 667 m2) located in Wuxue City (N29°59′3.36″, E115°27′58.84″), Hubei Province, China. All sclerotia were isolated and purified to obtain 100 isolates of S. sclerotiorum. A total of 300 isolates were purified for three years. All isolates were cultured on potato dextrose agar at 20 °C and stored at 4 °C.
2.2 Total RNA extraction and RNA sequencing
Total RNA of each isolate was extracted from mycelia using an RNAiso Plus kit (TaKaRa, China) according to the manufacturer’s instructions and then treated with DNase I. The concentration of each RNA extraction was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). Total RNA was stored at −80 °C until being used for RNA-Seq analysis. Equal amounts of total RNA from 100 isolates collected annually were pooled in a single library according to the collection year. Three RNA libraries were generated and used for sequence analysis. All library preparations and sequencing steps were performed by GENEWIZ Inc. (China). rRNA was depleted from total RNA using the Illumina Ribo Zero rRNA removal Kit (Plant Leaf). Three libraries were constructed according to the manufacturer’s protocol (NEBNext Ultra Directional RNA Library Prep Kit for Illumina). Paired-end (150 bp) sequencing of the three cDNA libraries was performed on the HiSeq 2500 platform (Illumina, USA). To obtain the 5' and 3' terminal sequences of the viral genome, the previously described method was used (Potgieter et al. 2009). Primers used in this study are listed in Supplementary Table S1.
2.3 Virus-related sequence assembly and annotation
Adapter sequences and low-quality reads were removed from the raw reads of each library using the Trimmomatic program (version 0.36) (Bolger et al. 2014) with default parameter settings. The remaining reads were mapped to the S. sclerotiorum genome (http://fungi.ensembl.org/Sclerotinia_sclerotiorum_1980_uf_70_gca_001857865/Info/Index) using HISAT2 (version 2.1.0) (Kim et al. 2015). Unmapped reads were extracted from the BAM file using SAMtools (version: 1.9) (Li et al. 2009) and were then assembled de novo using Trinity (version 2.5.0) (Haas et al. 2013) and SPAdes (version 3.11.1) (Bankevich et al. 2012). After the removal of all contigs shorter than 1,000 bp, the remaining contigs were filtered for redundancy at 90% nucleotide identity using CD-HIT (Fu et al. 2012). To identify virus-related contigs, the assembled contigs were compared against the NCBI nonredundant protein database using DIAMOND BLASTX (version 0.9.10) (Buchfink et al. 2015). We set the e-value below 1 × 10−5 to maintain high sensitivity and a low false-positive rate. Contigs matched to the same reference sequence were merged into longer contigs using DNAMAN software (version 8.0; Lynnon Biosoft, Quebec, Canada). Nucleotide sequences, potential ORFs, and deduced amino acid sequences were analyzed using DNAMAN.
2.4 Quantification of relative transcript abundances
We used the assembled contigs and S. sclerotiorum transcripts as templates for mapping with the program Bowtie2 (Langmead and Salzberg 2012). Next, we measured the abundance of these sequences as the number of transcripts per million (TPM) within each sequencing library. The relative abundance of each viral transcript was calculated as the TPM using RSEM (Li et al. 2011) and visualized in a heat map by TBtools (Chen et al. 2020).
2.5 Phylogenetic analysis of mycoviruses
To determine the evolutionary relationships among the newly identified viruses, the RdRp encoded by the putative mycoviruses was subjected to BLASTp analysis with the NCBI-nr database. The core conserved domain of RdRp was aligned with MAFFT (version 7.427) using the E-INS-i settings (Nakamura et al. 2018). All alignments were trimmed with trimAl (v1.4) (Capella-Gutierrez et al. 2009) to remove low-quality regions through heuristic selection under the automatic method (-automated1) based on similarity statistics. The maximum likelihood (ML) phylogenetic trees were constructed using the IQ-TREE (version 1.6.11) (Nguyen et al. 2015) with 1,000 bootstrap replicates and the best-fit amino acid substitution models were identified using ModelFinder (Kalyaanamoorthy et al. 2017). Nodes supported by a bootstrap value below 70% were collapsed in TreeGraph 2 (Stöver and Müller 2010). Phylogenetic trees were visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). The percent identity matrix among protein-coding sequences was generated using Clustal Omega for multiple sequence alignment (Li et al. 2015).
2.6 Availability of data and materials
The raw sequence reads from the metagenomic libraries are available at the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA598316. The sequences reported in this research have been deposited in GenBank databases, and the assembled contig sequences are available from GenBank databases under the accession numbers indicated in Supplementary Table S2. All virus nucleotide sequences (Fasta format), the unaligned and aligned datasets used in the phylogenetic analyses (Fasta format), and the phylogenetic trees (Newick format) that were generated in this work are available at the Figshare website (DOI: 10.6084/m9.figshare.11474709).
3. Results
3.1 Metatranscriptomic identification of mycoviruses from S. sclerotiorum in a single rapeseed field
We initially analyzed the growth rate, colony morphology, and pathogenicity for the 100 isolates of S. sclerotiorum collected in May 2017, and the results showed that these isolates were significantly different in these three biological characteristics (Supplementary Fig. S1). Nearly 50% of the isolates grew fast and had strong pathogenicity, while some isolates grew slowly with reduced pathogenicity (Supplementary Fig. S1C and D). These phenotypic differences indicate that there may be abundant mycoviruses in the selected field site. To understand the interannual dynamics in diversity and abundance of mycoviruses in this field, a total of 300 isolates were collected and used to construct three metatranscriptomic sequencing libraries according to the collection year. Total coverage of more than 100 million reads passing the filtering criteria was obtained for each library. For simplicity, we named the three libraries 2016SS, 2017SS, and 2018SS based on the collection year. Based on the analysis of the assembled contigs, a total of sixty-eight putative mycoviruses with nearly complete genomes (containing complete ORFs) or partial genomes were identified (Supplementary Table S2).
3.2 Novel positive-sense single-stranded RNA viruses
In this study, the forty-three putative viruses encoded by fifty-two contigs as diverse as previously characterized +ssRNA viruses (Supplementary Table S2), signifying that S. sclerotiorum does harbor abundant +ssRNA mycoviruses with great diversity regardless of the geographical origins.
Four novel multi-segmented narnaviruses were identified, which were temporarily named as Sclerotinia sclerotiorum narnavirus 1 (SsNV1), SsNV2, SsNV3, and SsNV4, representing the first discovery of narnaviruses in S. sclerotiorum. RT-PCR analysis of the cDNA of each isolate included in the NGS sample 2017SS showed that all segments of the four segmented narna-like viruses coexisted in the corresponding fungal isolate (Supplementary Fig. S2). The 5' and 3' terminal sequences of the four multi-segmented narnaviruses were obtained. The 5' and 3'-terminal ∼40 nt were highly conserved in the multi-segments of each narnavirus (Supplementary Fig. S3A). SsNV1 and SsNV2, respectively, have quad- and tri-segmented genomes (Fig. 1A). Interestingly, complete RdRps of SsNV1 and SsNV2 were encoded by two different RNA segments. RNA1 of SsNV1 and SsNV2 harbored the G, F, A, and B motifs of the RdRp at the C-terminus, whereas RNA2 of these two viruses contained the C, D, and E motifs at its N-terminus (Fig. 1A and Supplementary Fig. S3B). RNA1 of SsNV1 and SsNV2 encoded proteins share less than 62% identity to other narnaviruses, while proteins encoded by RNA2, RNA3, and RNA4 of these two viruses share no significant similarity with known sequences in either the NCBI nr database or in the Pfam domain database. In addition, the other seventeen narnaviruses with multi-segmented genomes were identified from public data (NCBI SRA and TSA database) (Fig. 1B and Supplementary Table S3). The phylogenetic tree suggested that these divided RdRp narnaviruses formed an independent phylogenetic cluster, which was significantly distant from other non-segmented narnaviruses (Fig. 1B). Therefore, we propose a new family, Polynarnaviridae, to accommodate this virus clade.
Figure 1.
Genome organizations of narna-like viruses and phylogenetic relationships of the members in the ‘Narna-Mito-Botourmia’ clade. (A) Genome structure models for the narna-like viruses identified in this study. The colored boxes indicate hypothetical ORFs. (B) The ML tree was constructed based on the multiple amino acid sequence alignment of the core conserved domain of RdRps using IQ-TREE with the best-fit model ‘Blosum62+F+R7’. The first identified viruses are marked by red colors, and the identified polynarnaviruses from NCBI SRA or TSA database are marked by blue colors. Sequence information of all selected viruses were supplied in Supplementary Table S4. The numbers next to each branch represent the bootstrap support based on 1,000 replicates. All branch lengths are drawn to a scale of amino acid substitutions per site. Host taxa are shown by circles delineated in different colors. The abbreviation ‘WX’ represents Wuxue (WX) city, which was used to distinguish the identified viruses in this study from the previously reported mycoviruses with more than 90% amino acid identity.
SsNV3 and SsNV4 have bi-segmented genomes (Fig. 1A). The RNA1 segment contains an ORF encoding an essential RdRp, and the RNA2 fragment contains an ORF encoding no similarity with the known proteins in the NCBI nr database. BLASTp analysis showed that the protein with the top hit for ORF1 of SsNV3 was RdRp from Plasmopara viticola lesion-associated orfanplasmovirus 4 (PvLaOrfPlV4) (33.75% identity), whereas that for ORF1 of SsNV4 was RdRp from PvLaOrfPlV5 (72.16% identity) (Supplementary Table S2). However, different from the genomes of PvLaOrfPlV (1–5) that were reported to contain a single RNA segment, SsNV3 and SsNV4 have two RNA segments. Interestingly, the proteins encoded by RNA2 of SsNV3 are most similar to coat proteins of the known viruses, based on the prediction by I-TASSER and Phyre2 (Supplementary Fig. S4). Therefore, we speculate that the genomes of SsNV3 and SsNV4 may not be naked, but encapsulated in virions. Phylogenetic analysis demonstrated that SsNV3 and SsNV4 grouped as a cluster with those unsegmented narnaviruses, but clustered into a separate branch with PvLaOrfPlV (1–5) (Fig. 1B).
Twelve novel mycoviruses share RdRp identity with members of the Botourmiaviridae family, ranging from 23% to 82% (Supplementary Fig. S5A). The obtained genomes ranged from 1.99 to 3.85 kb in length, and each genome contained a single complete ORF (Supplementary Fig. S5B). Phylogenetic analysis suggested that twelve botourmiaviruses clustered into three well-supported clades, among which five botourmiaviruses belong to the genus Penoulivirus, four belong to the genus Botoulivirus, and another three belong to the genus Scleroulivirus (Fig. 1B).
We identified a novel virus belonging to the family Fusariviridae, tentatively named Sclerotinia sclerotiorum fusarivirus 2 (SsFV2). The assembled genome of SsFV2 is 6,281 nt in length and encodes two nonoverlapping ORFs (Supplementary Fig. S6A). ORF1 encodes a polyprotein with conserved RdRp and helicase (Hel) domains showing 51.29% identity with previously described Rutstroemia firma fusarivirus 1. ORF2 encodes a hypothetical protein of unknown function. Phylogenetic analysis based on the conserved RdRps showed that SsFV2 and other members of the Fusariviridae family are divided into three branches with good support (Supplementary Fig. S6B). For this reason, we propose the division of this family into three genera: Alpha-, Beta-, and Gammafusarivirus, among which SsFV2 belongs to the Alphafusarivirus genus.
One mycovirus, Sclerotinia sclerotiorum virga-like virus 1 (SsVV1), was identified. The SsVV1 genome consists of two RNA segments, designated as RNA1 (1,800 nt) and RNA2 (1,878 nt), which encode RdRp and methyltransferase (Met), respectively (Supplementary Fig. S7A). The predicted amino acid sequence of RNA1 showed 44% identity with the RdRp of Agaricus bisporus virus 16 (AbV16; KY357502). The Met encoded by RNA2 of SsVV1 had an identity of 29% with AbV16. AbV16 is a proposed member of the Mycovirgaviridae family (Nerva et al. 2019), whose members exhibit four RNA segments, containing a single ORF: segment 1 encoding RdRp, segment 2 encoding Met, and segments 3 and 4 encoding proteins of unknown function (Deakin et al. 2017). Although both RNA1 and RNA2 of SsVV1 were similar to those of AbV16, we failed to detect the other two segments corresponding to those of AbV16 from our data. Phylogenetic analysis revealed that SsVV1 clusters in a clade with AbV16 along with several other viruses recently discovered in soil samples and fungal transcriptome data (Supplementary Fig. S7B).
We also identified three mycoviruses belonging to the Deltaflexiviridae family, tentatively named Sclerotinia sclerotiorum deltaflexivirus 1-WX (SsDFV1-WX), SsDFV2-WX, and SsDFV3. The replicases of SsDFV1-WX and SsDFV2-WX shared 99% and 98% identity with SsDFV1 (NC_038977) and SsDFV2 (NC_040649), respectively (Supplementary Fig. S8). The assembled genome of SsDFV3 consisted of 7,375 nt with two predicted ORFs. ORF1 encodes a putative replication-associated polyprotein and contains three conserved domains: Met, Hel, and RdRp (Supplementary Fig. S7A). A BLASTp search using the deduced amino acid sequence showed that ORF1 has a low identity (<40%) with the replication-associated polyprotein of members in the Deltaflexiviridae family (Supplementary Fig. S8). Phylogenetic analysis revealed that SsDFV3 was in a well-supported clade with other deltaflexiviruses (Supplementary Fig. S7B).
A 4470 nt contig was similar to barnavirus, tentatively named Sclerotinia sclerotiorum barnavirus 1 (SsBNV1). SsBNV1 was a monosegmented +ssRNA virus with four ORFs (Supplementary Fig. S9A). The four ORFs encoded a protein of unknown function, a putative serine protease domain, RdRp, and a capsid protein. The predicted RdRp shared 39% identity with the Mushroom bacilliform virus belonging to the family Barnaviridae. SsBNV1 within a well-supported clade of the Barnaviridae family is closely related to members of Solemoviridae and Luteoviridae families (Supplementary Fig. S9B).
In addition to the mycoviruses mentioned above, other detected +ssRNA viruses and dsRNA viruses included members of Mitoviridae, Gammaflexiviridae, Tymoviridae, Hypoviridae, Solemoviridae, Ambiguiviridae, Endornaviridae, Botybirnavirus, and one unassigned virus species. Due to space limitations, these mycoviruses are described in Supplementary Text S1.
3.3 Novel negative-sense single-stranded RNA viruses
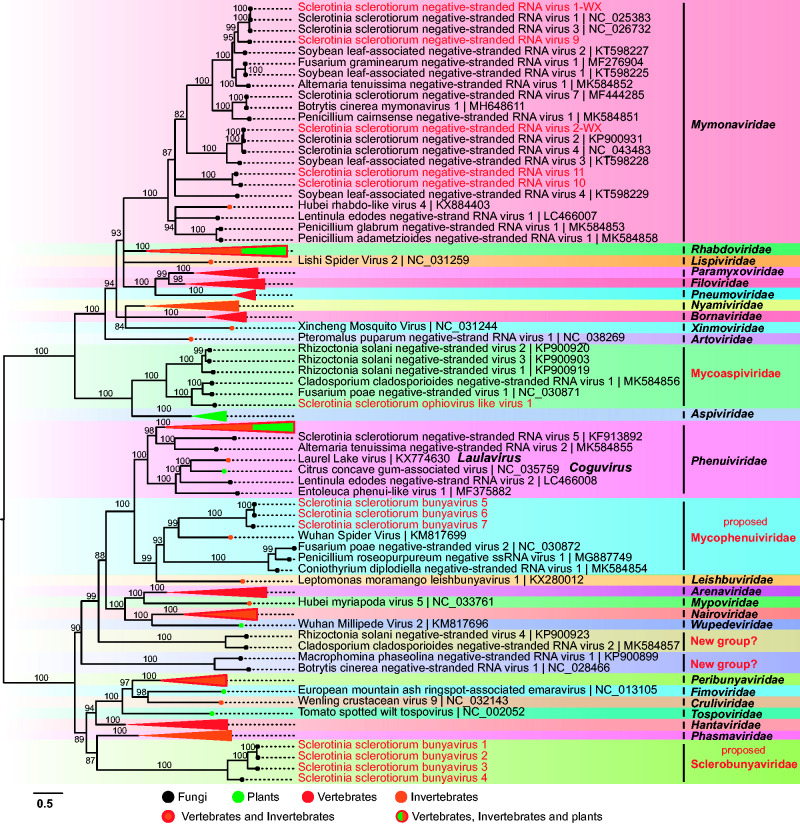
We identified five -ssRNA mycoviruses belonging to the family Mymonaviridae. The sequences of these five mycoviruses ranged from 9,623 to 9,982 nt, suggesting that the complete or nearly complete genomes of all these viruses were obtained based on the deep-sequencing data. The L proteins of Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV1-WX) and SsNSRV2-WX shared 100% and 99% identity with SsNSRV1 (KJ186782.1) and SsNSRV2 (KP900931.1), respectively (Supplementary Fig. S10A). Similar to that of other members of Mymonaviridae, the genome of SsNSRV9 contained five nonoverlapping ORFs (ORF I–V) arranged linearly in the viral genome (Supplementary Fig. S10B). The protein encoded by ORF IV in SsNSRV9 shared 56% identity with the L protein in SsNSRV1 and SsNSRV3 (Supplementary Fig. S10A). In addition, a conserved gene-junction sequence (3′-UAUU(U/A)AAUAAAACUUAGGAA-5′) was found downstream of each ORF (Supplementary Fig. S10D), which is a common feature in mononegaviruses (Whelan et al. 2004). Notably, SsNSRV10 and SsNSRV11 contained four ORFs (ORF I–IV) (Supplementary Fig. S10B), but there was only a transcription start signal with no stop signal upstream of ORF I and only a transcription stop signal with no start signal downstream of ORF IV (Supplementary Fig. S10D); therefore, we believe that these two mycoviruses may have only four ORFs. However, SsNSRV9 is similar to the previously reported SsNSRV1 in that there is no transcription initiation signal in their 3'-UTR (Liu et al. 2014). Phylogenetic analysis of the L protein confirmed that SsNSRV10 and SsNSRV11 formed an independent evolutionary branch, while SsNSRV9 was clustered with SsNSRV1 and related viruses (Fig. 2).
Figure 2.
ML phylogenetic analysis depicting the evolutionary relationships of the negative-stranded RNA viruses. The best-fit model for constructing the phylogenetic tree is ‘LG+F+R8’. The name of the virus family or genus is shown on the right. This is same as that of Fig. 1.
Sclerotinia sclerotiorum ophiovirus-like virus 1 (SsOpLV1) with 7,817 nt in length shows the highest similarity to Fusarium poae negative-stranded virus 1 within the recently proposed Mycoaspiviridae family (Chiapello et al. 2020). SsOpLV1 contained two linearly arranged nonoverlapping ORFs (Supplementary Fig. S10C). The ORF1 encoded a protein (143 aa) of unknown function, and ORF2 encoded a polyprotein with the RdRp domain. Phylogenetic analysis based on RdRp indicated that mycoophioviruses are closely related to the viruses of the Aspiviridae family (Fig. 2).
Seven novel mycoviruses related to the members of Bunyavirales were identified. The genomes for the seven bunyaviruses (Sclerotinia sclerotiorum bunyavirus 1-7 (SsBYV1-7)) ranged from 6.4 to 8.0 kb, and contain a single ORF (Supplementary Fig. S11A). The L proteins encoded by the seven bunyaviruses shared less than 26% identity with those of known bunyaviruses (Supplementary Table S2). We failed to find any additional segments related to bunyavirus M or S proteins in the assembled contig database. The alignment of the conserved motifs of the L proteins encoded by the seven bunyaviruses and those in the other reported bunyaviruses indicates that these mycoviruses contain eight conserved motifs (Supplementary Fig. S12), which represent highly conserved central regions of RdRps from members of the Bunyavirales order (Amroun et al. 2017). The conserved endonuclease domain and motif G (Supplementary Fig. S11B and C) were also identified in the N-terminal of the L protein encoded by three bunyaviruses (SsBYV5-7). However, the endonuclease domain and motif G were not detected in the L proteins encoded by the four other bunyaviruses. A phylogenetic tree was constructed using the core regions of the L protein. Seven sclerotinia bunyaviruses were assigned into two clades. Four bunyaviruses (SsNYV1-4) were well supported in one phylogenetic cluster, which was significantly distant from the other bunyaviruses, suggesting that these four bunyaviruses represent a new virus lineage. Therefore, we propose the establishment of a new family, Sclerobunyaviridae, to accommodate these similar bunyaviruses. The other three bunyaviruses (SsNYV5-7) formed well-supported clades with an invertebrate virus (Wuhan spider virus) and several fungal bunyaviruses but were significantly phylogenetically distant from members of the Phenuiviridae family. We therefore propose the establishment of a new family, Mycophenuiviridae, to accommodate these similar bunyaviruses that infect fungi and invertebrates (Fig. 2).
3.4 Diversity of the S. sclerotiorum virome in a single rapeseed field
We examined the proportion of mycovirus-associated reads in each library, which we found to be 4.07% for 2016SS, 9.51% for 2017SS, and 2.89% for 2018SS (Fig. 3A). The sixty-eight mycoviruses we identified from these three libraries represent four genome types: positive-sense single-stranded RNA (+ssRNA), negative-sense single-stranded RNA (-ssRNA), double-stranded RNA (dsRNA), and single-stranded DNA (ssDNA) (Fig. 3B). Among these sixty-eight mycoviruses, the amino acid sequences of twenty-two mycoviruses showed high identity (≥90%) to previously reported viruses, twenty-eight mycoviruses represented novel mycoviruses sharing low amino acid sequence identity (<70%) with previously recognized viruses, and eighteen mycoviruses had ≥70% and <90% amino acid identity with previously recognized viruses (Supplementary Table S2). The characteristics of all identified mycoviruses, including their contig length and best hit in the NCBI-nr database, are summarized in Supplementary Table S2. Based on the amino acid sequences of the replicase encoded by the assembled contigs, all identified mycoviruses were classified. Nineteen of sixty-eight (27.9%) mycoviruses had a lower identity (<45%) than previously characterized viruses, and eleven of these nineteen could not be placed into any established family. At the genus level, thirty-eight mycoviruses could be assigned to eleven established genera, while the remaining thirty mycoviruses could be assigned to thirteen proposed genera and one unassigned genus.
Figure 3.
The diversity of mycoviruses detected in S. sclerotiorum. (A) Proportion of each taxonomic category in each library based on read counts. (B) The percentage of each virus genome type. (C) The percentage of each positive, single-stranded RNA virus at the family level. (D) Mycoviruses belonging to different genome types presented in each library. (E) Sankey diagram showing the compositions of mycovirome from different years of S. sclerotiorum field isolates.
+ssRNA mycoviruses were the most diverse class within the three libraries, compared to other types (Fig. 3B). Forty-three mycoviruses were +ssRNA viruses, accounting for up to 63.2% of the identified viruses, suggesting that S. sclerotiorum harbors abundant +ssRNA mycoviruses with high diversity (Fig. 3B). Among the +ssRNA mycoviruses, members of the Botourmiaviridae and Mitoviridae families were most prevalent, accounting for up to 44.2% (Fig. 3C). The eleven identified dsRNA mycoviruses included seven botybirnaviruses and four fusagraviruses (Supplementary Fig. S13). We also identified an ssDNA mycovirus that shared 100% identity to Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1) belonging to the Genomoviridae family. Finally, the mycoviruses with -ssRNA genomes were classified into three orders: Bunyavirales (seven viruses), Mononegavirales (five viruses), and Serpentovirales (one virus). These results suggest that mycoviruses present in S. sclerotiorum are diverse, even though all isolates originated from a single rapeseed field.
3.5 Interannual dynamics of members in the S. sclerotiorum virome
The composition of virome was different among the three libraries. The list of putative mycoviruses was summarized for each library, and we found that 47, 52, and 35 different mycoviruses were detected in the 2016SS, 2017SS, and 2018SS libraries, respectively (Fig. 3D). In all three libraries, the number of +ssRNA mycoviruses was greater than that of -ssRNA mycoviruses. The number of dsRNA mycoviruses was less than that of -ssRNA mycoviruses, and the number of ssDNA mycoviruses was the lowest, with only one of these viruses found in all libraries (Fig. 3D). The libraries and the taxonomy of mycoviruses were summarized in a Sankey diagram (Fig. 3E).
Mycovirus occurrence changed within sampling times as follows: 35.3% of mycoviruses were detected in all three years, 25.8% were detected twice, and 39.4% were detected once. More than 60% of the identified dsRNA mycoviruses and -ssRNA mycoviruses were detected once, suggesting that these mycoviruses may have difficulty spreading widely in S. sclerotiorum or that they may be easily eliminated. In total, twenty-four mycoviruses were found each year, accounting for 35.3% of all identified viruses (Fig. 4A), suggesting that these mycoviruses commonly exist in the population of S. sclerotiorum and are potential core mycoviruses in the virome of S. sclerotiorum. Among these twenty-four mycoviruses, nineteen were +ssRNA mycoviruses, including five mitoviruses, five botourmiaviruses, three hypoviruses, two deltaflexiviruses, a narnaviruses, a gammaflexivirus, a tymovirus, and a betaendornavirus. Additionally, there are five other mycoviruses, including three -ssRNA mycoviruses in the Sclerotimonavirus genus, one dsRNA mycovirus in the Botybirnavirus genus, and one ssDNA mycovirus in the Gemycircularvirus genus (Supplementary Table S2).
Figure 4.
The abundances of mycoviruses in S. sclerotiorum from different years. (A) The number of shared mycoviruses in the virome of S. sclerotiorum in three years. (B) The relative abundance of mycoviruses at the family level in the same year, normalized by column. (C) Heat map displaying the relative abundance of the mycoviruses in different years according to the TPM values, normalized by row. The red columns show high abundance, and grey columns indicate not detected. Mycoviruses detected in all three libraries are marked with green dots.
The relative abundances of the identified viral families showed differences among the years. Mycoviruses within the Mitoviridae family were the most abundant, and their TPM values accounted for more than 63.4% of all mycovirus TPM values (Fig. 4B). Botourmiaviruses were the second most common after mitoviruses in the 2016SS library, accounting for 23.2% of the mycovirus TPM values, while the family accounted for only 2.0% in the 2018SS library. In the 2017SS library, the abundance levels of mycoviruses within the Botybirnavirus genus and Botourmiaviridae family were lower than the abundance of mycoviruses belonging to the Mitoviridae family, accounting for 12.4% and 6.8% of the TPM values, respectively. In the 2018SS library, the abundance of mycoviruses within the Mymonaviridae family accounted for more than 17.3% of the total abundance, while mycoviruses in this family accounted for less than 2.0% in the other two libraries. A heat map of the relative abundances of all mycoviruses identified in the different libraries is presented in Fig. 4C. As shown in the heat map, almost all the mycoviruses had changing abundances between sampling years.
4. Discussion
Metatranscriptomics sequencing has dramatically expanded our understanding of virus diversity and allowed novel mycoviruses to be increasingly discovered in phytopathogenic fungi (Marzano et al. 2016; Kotta-Loizou and Coutts 2017; Mu et al. 2017; Arjona-Lopez et al. 2018; Nerva et al. 2019; Chiapello et al. 2020). However, the understanding on the interannual dynamics of mycovirus under natural conditions is limited at a population level. In the present research, we performed viral metatranscriptomics sequencing to investigate the interannual dynamics and characterize unbiased viral communities in S. sclerotiorum collected from a single rapeseed-growing field for three consecutive years. As expected, the species and number of the identified mycoviruses showed variation in an individual year, but it is noteworthy that twenty-four mycoviruses detected in all three years are considered to be core mycoviruses in S. sclerotiorum virome. Additionally, some of the mycoviruses discovered in this study were classified into families of Narnaviridae, Polynarnaviridae, Gammaflexiviridae, Mycovirgaviridae, Barnaviridae, Mycoaspiviridae, Mycophenuiviridae, Sclerobunyaviridae with S. sclerotiorum as the host for the first time. The findings enrich our understanding on the diversity of mycoviruses and provided important evolutionary clues.
In this study, sixty-eight mycoviruses were detected in S. sclerotiorum, and besides one unclassified mycovirus, the other sixty-seven mycoviruses were assigned into twenty families, suggesting that S. sclerotiorum harbors a highly complex mycovirus composition, even in a single crop field. Moreover, twenty-eight mycoviruses were previously uncharacterized, indicating mycoviral communities in S. sclerotiorum are still poorly covered by current databases. Two virome analyses were previously conducted in S. sclerotinia. Twenty-eight mycoviruses were detected from 138 isolates of S. sclerotiorum in the USA, belonging to eight families (nine genera) (Marzano et al. 2016), of which members from seven families (eight genera) were found in the present research. Fifty-seven mycoviruses were detected from eighty-four isolates in Australia, belonging to thirteen families (fourteen genera) (Mu et al. 2017), of which members within ten families (eleven genera) were detected in our samples. Mitoviruses are the most prevalent in S. sclerotiorum samples from Australia and the USA, and the proportion of them in those two viromes of S. sclerotiorum are 43.9% (25/57) and 60.7% (17/28), respectively. Unlike samples from the United States and Australia, botourmiaviruses were most prevalent in this virome survey of S. sclerotiorum from China, accounting for 17.6% (12/68), followed by mitoviruses (7/68) and botybirnaviruses (7/68). Based on the percentage of genome types, our findings were similar to those of previously reported results (Marzano et al. 2016; Mu et al. 2017), with +ssRNA viruses as the dominant population accounting for more than 60% of the identified mycoviruses. S. sclerotiorum virome showed a similar mycovirus composition despite location as S. sclerotiorum isolates from Australia, USA, and China are from different continents separated by oceans. Therefore, we boldly speculated that the interaction system between S. sclerotiorum and its core virome had been initiated before continental plate separation.
The S. sclerotiorum isolates of the USA and Australia were collected from different geographical regions and plant hosts, while strains of the Chinese group were isolated from a smaller annual rapeseed-growing field. However, as mentioned above, the mycovirus composition in a single field was found similar to the previously reported virome, although the numbers of species were more diverse. Moreover, most of these mycoviruses did not exhibit any specific association to geography or plant host. This result also revealed that the virome of S. sclerotiorum from a single crop field in the same region might not differ significantly from the overall virome of S. sclerotiorum sampled from other plant hosts and geographical origins. Whether viromes in other species of fungal populations have similar results is still unclear and yet to be explored for the dynamics among the plant disease severity, fungal hosts, and mycoviruses at population levels.
Previous reports brought insights into the important roles viruses play in natural ecosystems (Suttle 2007; Dell’Anno et al. 2015). Understanding the temporal dynamics of viromes could help us determine the drivers of community stability and ecosystem function generally in nature (Rodriguez-Brito et al. 2010; Arkhipova et al. 2018; Ignacio-Espinoza et al. 2019) and specifically in agricultural fields. For instance, the virus population can co-vary with prokaryotic abundance by preying on the microbes and change the microbial community composition, and thereby modulating ecosystem functions such as carbon and nitrogen cycling in the natural environment (Suttle 2007). The temporal dynamics of several environmental viromes have been characterized in recent decades, including bioaerosol (Prussin et al. 2019), ocean (Chow and Fuhrman 2012), sphagnum peatlands (Ballaud et al. 2016), and sediment ecosystems (Helton et al. 2012). In this study, we found that the mycovirus communities exhibited fluctuating temporal abundance, with the viral populations in 2017 being larger than those in the other two years. In contrast to mycoviruses, another study analyzed plant viruses from six native plant species over four years in the Nature Conservancy’s Tallgrass Prairie Preserve, Osage County, Oklahoma, USA. The taxonomic composition of plant viruses was found to have a close relationship with the plant host species but no clear relationship with the sampling site or year (Thapa et al. 2015). Although most mycoviruses cause latent infections, the mycoviruses associated with hypovirulence/hypervirulence or other biological functions have been widely reported in various phytopathogenic or nonpathogenic fungi (Marquez et al. 2007; Ghabrial et al. 2015; Kotta-Loizou and Coutts 2017; Nerva et al. 2019). Mycoviruses also can influence the plant–endophyte relationship by changing the virulence or fitness of the fungal host under altered environmental conditions (Bao and Roossinck 2013). Bio-priming treatment of rapeseeds with the hypovirulent strain DT-8 infected with a mycovirus, SsHADV-1, could influence the structure and composition of overall fungal communities in Sclerotinia stem rot (Qu et al. 2020). Therefore, we believe that mycoviruses may play an essential role in fungal communities in the agricultural ecosystem via complex fungi–mycovirus interactions. However, the temporal dynamics of virome have not been explored at a fungal community level. Therefore, our study represents the first effort in quantitatively and qualitatively describing the temporal dynamics of the virome in fungi. Previous research confirmed that the occurrence of sclerotinia disease was affected by many factors including temperature, humidity, and host resistance (Sharma et al. 2015). However, beyond abiotic and host factors, we speculated that the virulence of the S. sclerotiorum population taken as a whole was also greatly affected by mycoviruses for the following reasons. First, the isolates of S. sclerotiorum collected from a single crop field showed significant virulence variation (Attanayake et al. 2013; Abán et al. 2018), and a wide variety of mycoviruses have been discovered in S. sclerotiorum (Jiang et al. 2013; Marzano et al. 2016; Mu et al. 2017). Secondly, some mycoviruses could not only confer hypovirulence on S. sclerotiorum but also have the potential to convert S. sclerotiorum into an endophyte and enhance the resistance of rapeseed against the phytopathogens, for example Botrytis cinerea and S. sclerotiorum (Zhang et al. 2020). Third, biopriming with hypovirulent strain DT-8 could reduce the total abundance of the potential phytopathogenic fungi (Qu et al. 2020). Therefore, we speculate that the interannual dynamics of virome in S. sclerotiorum may be the hidden driver in shaping the fungal community structure that then may reduce the occurrence of diseases caused by other fungal pathogens.
It is generally thought that the RdRp of all RNA viruses is encoded by a single segment of gene and the phylogeny of RNA viruses is fully characterized based on the viral RdRp (te Velthuis 2014; Wolf et al. 2018). We found the RdRps of two novel multi-segmented polynarnaviruses encoded by two different segments and this similar result was recently reported in Aspergillus fumigatus, Magnaporthe oryzae, and Oidiodendron maius (Chiba et al. 2020; Sutela et al. 2020). By analyzing NCBI SRA and TSA data, we found that seventeen polynarnaviruses containing multi-segmented genomes with their RdRp conserved domains located in different segments. This finding suggests that multi-segmented polynarnaviruses are widely distributed in fungi, and their genomes possibly contain up to seven segments. Based on the understanding of the lifestyle of multipartite viruses and multi-segmented viruses (Lowen 2018), the genome segmentation of polynarnaviruses may be advantageous for viral replication and recombination. We also found two narnaviruses, SsNV3 and SsNV4, with bi-segmented genomes. Based on Phyre2 results, the protein encoded by RNA 2 of SsNV3 showed the similarity to a -ssRNA virus, but I-TASSER results indicated the similarity to dsRNA viruses. We speculated that the difference between the two software results could be due to the poor conservation of viral coat proteins, and the 3D structure of most viral coat proteins has not been resolved, and there is no available adequate data related to the narnavirus coat proteins in the database. This may also be the reason why SsNV4 did not predict the information associated with the viral coat protein. We believed that more narnaviruses that could have coat proteins would be identified with the release of these two viral sequences in the future. We speculated that these two narnaviruses may have obtained genes related to coat proteins from other viruses or hosts, or that capsidless narnaviruses have lost their coat protein genes during the evolutionary process. These two viruses revealed an unanticipated evolutionary link between encapsidated and capsidless RNA viruses. In summary, these newly characterized mycoviruses with unique genome features have a distant phylogenetic relationship from all known viruses and formed an independent evolutionary lineage that supported the establishment of a new genus or family.
Supplementary Material
Acknowledgments
We would like to thank Mr. Huang Huang in Huazhong Agricultural University and Mr. Zaiwu Zheng in the Plant Protection Station of Wuxue City of Hubei Province for his help in collecting samples, Dr. Kenichi Tsuda from Huazhong Agricultural University for constructive suggestions, and Amanda Gutek for proof-reading the manuscript. This work was financially supported by the National Science Foundation of China (31722046), the National Key Research and Development Program of China (2017YFD0201100), and the earmarked fund for China Agriculture Research System (CARS-12).
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Abán C. L. et al. (2018) ‘Molecular, Morphological and Pathogenic Diversity of Sclerotinia sclerotiorum Isolates from Common Bean (Phaseolus vulgaris) Fields in Argentina’, Plant Pathology, 67: 1740–8. [Google Scholar]
- Ahn I.-P., Lee Y.-H. (2001) ‘A Viral Double-Stranded RNA up Regulates the Fungal Virulence of Nectria Radicicola’, Molecular Plant-Microbe Interactions: Mpmi, 14: 496–507. [DOI] [PubMed] [Google Scholar]
- Amroun A. et al. (2017) ‘Bunyaviridae RdRps: Structure, Motifs, and RNA Synthesis Machinery’, Critical Reviews in Microbiology, 43: 753–78. [DOI] [PubMed] [Google Scholar]
- Arjona-Lopez J. M. et al. (2018) ‘Novel, Diverse RNA Viruses from Mediterranean Isolates of the Phytopathogenic Fungus, Rosellinia Necatrix: Insights into Evolutionary Biology of Fungal Viruses’, Environmental Microbiology, 20: 1464–83. [DOI] [PubMed] [Google Scholar]
- Arkhipova K. et al. (2018) ‘Temporal Dynamics of Uncultured Viruses: A New Dimension in Viral Diversity’, The ISME Journal, 12: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanayake R. N. et al. (2013) ‘Sclerotinia sclerotiorum Populations Infecting Canola from China and the United States Are Genetically and Phenotypically Distinct’, Phytopathology, 103: 750–61. [DOI] [PubMed] [Google Scholar]
- Ballaud F. et al. (2015) ‘Dynamics of Viral Abundance and Diversity in a Sphagnum-Dominated Peatland: Temporal Fluctuations Prevail over Habitat’, Frontiers in Microbiology, 6: 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A. et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’, Journal of Computational Biology: A Journal of Computational Molecular Cell Biology, 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Roossinck M. J. (2013). Multiplexed Interactions: Viruses of Endophytic Fungi. Advances in Virus Research, 86: 37–58. [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014) ‘Trimmomatic: A Flexible Trimmer for Illumina Sequence Data’, Bioinformatics (Oxford, England), 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al. (2020) ‘TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data’, Molecular Plant, 13: 1194–202. [DOI] [PubMed] [Google Scholar]
- Chiapello M. et al. (2020) ‘Analysis of the Virome Associated to Grapevine Downy Mildew Lesions Reveals New Mycovirus Lineages’, Virus Evolution, 6: veaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y., Oiki S., Yaguchi T., Urayama S.-I., Hagiwara D. (2020). ‘Discovery of divided RdRp sequences and a hitherto unknown genomic complexity in fungal viruses.’ Virus Evolution, 7: veaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. E. T., Fuhrman J. A. (2012) ‘Seasonality and Monthly Dynamics of Marine Myovirus Communities’, Environmental Microbiology, 14: 2171–83. [DOI] [PubMed] [Google Scholar]
- Deakin G. et al. (2017) ‘Multiple Viral Infections in Agaricus bisporus - Characterisation of 18 Unique RNA Viruses and 8 ORFans Identified by Deep Sequencing’, Scientific Reports, 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Anno A., Corinaldesi C., Danovaro R. (2015) ‘Virus Decomposition Provides an Important Contribution to Benthic Deep-Sea Ecosystem Functioning’, Proceedings of the National Academy of Sciences, 112: E2014–E2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V. V., Koonin E. V. (2012) ‘Capsid-Less RNA Viruses’, in eLS. J. Wiley and L. Sons, Ltd: Chichester. [Google Scholar]
- Farr D. F., Rossman A. Y. (2020) ‘Fungal Databases’, U.S. National Fungus Collections, ARS, USDA.
- Fu L. et al. (2012) ‘CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data’, Bioinformatics (Oxford, England), 28: 3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pedrajas M. et al. (2019) ‘Mycoviruses in Biological Control: From Basic Research to Field Implementation’, Phytopathology, 109: 1828–39. [DOI] [PubMed] [Google Scholar]
- Ghabrial S. A. et al. (2015) ‘50-plus Years of Fungal Viruses’, Virology, 479–480: 356–68. [DOI] [PubMed] [Google Scholar]
- Ghabrial S. A., Suzuki N. (2009) ‘Viruses of Plant Pathogenic Fungi’, Annual Review of Phytopathology, 47: 353–84. [DOI] [PubMed] [Google Scholar]
- Güemes A. G. C. et al. (2016) ‘Viruses as Winners in the Game of Life’, Annual Review of Virology, 3: 197–214. [DOI] [PubMed] [Google Scholar]
- Haas B. J. et al. (2013) ‘De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis’, Nature Protocols, 8: 1494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helton R. R. et al. (2012) ‘Interannual Dynamics of Viriobenthos Abundance and Morphological Diversity in Chesapeake Bay Sediments’, FEMS Microbiology Ecology, 79: 474–86. [DOI] [PubMed] [Google Scholar]
- Ignacio-Espinoza J. C., Ahlgren N. A., Fuhrman J. A. (2020) ‘Long-Term Stability and Red Queen-like Strain Dynamics in Marine Viruses’, Nature Microbiology, 5: 265–71. [DOI] [PubMed] [Google Scholar]
- Jiang D. et al. (2013) ‘Viruses of the Plant Pathogenic Fungus Sclerotinia sclerotiorum’, Advances in Virus Research, 86: 215–48. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S. et al. (2017) ‘ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates’, Nature Methods, 14: 587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015) ‘HISAT: A Fast Spliced Aligner with Low Memory Requirements’, Nature Methods, 12: 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Senkevich T. G., Dolja V. V. (2006) ‘The Ancient Virus World and Evolution of Cells’, Biology Direct, 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotta-Loizou I., Coutts R. H. A. (2017) ‘Studies on the Virome of the Entomopathogenic Fungus Beauveria bassiana Reveal Novel dsRNA Elements and Mild Hypervirulence’, PLoS Pathogens, 13: e1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E. V. (2017) ‘Multiple Origins of Viral Capsid Proteins from Cellular Ancestors’, Proceedings of the National Academy of Sciences of the United States of America, 114: E2401–E2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011) ‘RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome’, BMC Bioinformatics, 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 1000 Genome Project Data Processing Subgroup. et al. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics (Oxford, England), 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2015) ‘The EMBL-EBI Bioinformatics Web and Programmatic Tools Framework’, Nucleic Acids Research, 43: W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. et al. (2014) ‘Fungal Negative-Stranded RNA Virus That is Related to Bornaviruses and Nyaviruses’, Proceedings of the National Academy of Sciences of the United States of America, 111: 12205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen A. C. (2018) ‘It’s in the Mix: Reassortment of Segmented Viral Genomes’, PLOS Pathogens, 14: e1007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez L. M. et al. (2007) ‘A Virus in a Fungus in a Plant: Three-Way Symbiosis Required for Thermal Tolerance’, Science, 315: 513–5. [DOI] [PubMed] [Google Scholar]
- Marzano S.-Y. L. et al. (2016) ‘Identification of Diverse Mycoviruses through Metatranscriptomics Characterization of the Viromes of Five Major Fungal Plant Pathogens’, Journal of Virology, 90: 6846–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu F. et al. (2017) ‘Virome Characterization of a Collection of Sclerotinia sclerotiorum from Australia’, Frontiers in Microbiology, 8: 2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. et al. (2018) ‘Parallelization of MAFFT for Large-Scale Multiple Sequence Alignments’, Bioinformatics (Oxford, England), 34: 2490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerva L. et al. (2019) ‘Mycoviruses Mediate Mycotoxin Regulation in Aspergillus Ochraceus’, Environmental Microbiology, 21: 1957–68. [DOI] [PubMed] [Google Scholar]
- Nerva L. et al. (2019) ‘Isolation, Molecular Characterization and Virome Analysis of Culturable Wood Fungal Endophytes in Esca Symptomatic and Asymptomatic Grapevine Plants’, Environmental Microbiology, 21: 2886–904. [DOI] [PubMed] [Google Scholar]
- Nguyen L.-T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss D. L. (2005) ‘Hypovirulence: Mycoviruses at the Fungal-Plant Interface’, Nature Reviews. Microbiology, 3: 632–42. [DOI] [PubMed] [Google Scholar]
- Oerke E. (2006) ‘Crop Losses to Pests’, The Journal of Agricultural Science, 144: 31–43. [Google Scholar]
- Ozkan S., Coutts R. H. A. (2015) ‘Aspergillus fumigatus Mycovirus Causes Mild Hypervirulent Effect on Pathogenicity When Tested on Galleria mellonella’, Fungal Genetics and Biology: FG & B, 76: 20–6. [DOI] [PubMed] [Google Scholar]
- Pearson M. N. et al. (2009) ‘Mycoviruses of Filamentous Fungi and Their Relevance to Plant Pathology’, Molecular Plant Pathology, 10: 115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potgieter A. C. et al. (2009) ‘Improved Strategies for Sequence-Independent Amplification and Sequencing of Viral Double-Stranded RNA Genomes’, The Journal of General Virology, 90: 1423–32. [DOI] [PubMed] [Google Scholar]
- Prussin A. J. et al. (2019) ‘Seasonal Dynamics of DNA and RNA Viral Bioaerosol Communities in a Daycare Center’, Microbiome, 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z. et al. (2020) ‘Bio-Priming with a Hypovirulent Phytopathogenic Fungus Enhances the Connection and Strength of Microbial Interaction Network in Rapeseed’, Npj Biofilms and Microbiomes, 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B. et al. (2010) ‘Viral and Microbial Community Dynamics in Four Aquatic Environments’, The ISME Journal, 4: 739–51. [DOI] [PubMed] [Google Scholar]
- Sharma P. et al. (2015) ‘Sclerotinia sclerotiorum (Lib) de Bary Causing Sclerotinia Rot in Oilseed Brassicas: A Review’, Journal of Oilseed Brassica, 6(Special Issue): 1–44. [Google Scholar]
- Sharon A., Shlezinger N. (2013) ‘Fungi Infecting Plants and Animals: Killers, Non-Killers, and Cell Death’, PLoS Pathogens, 9: e1003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver B. C., Müller K. F. (2010) ‘TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses’, BMC Bioinformatics, 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutela S. et al. (2020) ‘The Virome from a Collection of Endomycorrhizal Fungi Reveals New Viral Taxa with Unprecedented Genome Organization’, Virus Evolution, 6: veaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. (2007) ‘Marine Viruses — Major Players in the Global Ecosystem’, Nature Reviews. Microbiology, 5: 801–12. [DOI] [PubMed] [Google Scholar]
- te Velthuis A. J. (2014) ‘Common and Unique Features of Viral RNA-Dependent Polymerases’, Cellular and Molecular Life Sciences: CMLS, 71: 4403–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa V. et al. (2015) ‘Determinants of Taxonomic Composition of Plant Viruses at the Nature Conservancy’s Tallgrass Prairie Preserve’, Virus Evolution, 1: vev007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S. P. J., Barr J. N., Wertz G. W. (2004) ‘Transcription and Replication of Nonsegmented Negative-Strand RNA Viruses’. Curr Top Microbiol Immunol, 283: 61–119. [DOI] [PubMed] [Google Scholar]
- Wolf Y. I. et al. (2018) ‘Origins And Evolution of the Global RNA Virome’, mBio, 9: e02329–02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Jiang D. (2014) ‘New Insights into Mycoviruses and Exploration for the Biological Control of Crop Fungal Diseases’, Annual Review of Phytopathology, 52: 45–68. [DOI] [PubMed] [Google Scholar]
- Yu X. et al. (2010) ‘A Geminivirus-Related DNA Mycovirus That Confers Hypovirulence to a Plant Pathogenic Fungus’, Proceedings of the National Academy of Sciences of the United States of America, 107: 8387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. et al. (2013) ‘Extracellular Transmission of a DNA Mycovirus and Its Use as a Natural Fungicide’, Proceedings of the National Academy of Sciences of the United States of America, 110: 1452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. (2020) ‘A 2-kb Mycovirus Converts a Pathogenic Fungus into a Beneficial Endophyte for Brassica Protection and Yield Enhancement’, Molecular Plant, 13: 1420–33. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-Z. et al. (2019) ‘Expanding the RNA Virosphere by Unbiased Metagenomics’, Annual Review of Virology, 6: 119–39. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-Z., Shi M., Holmes E. C. (2018) ‘Using Metagenomics to Characterize an Expanding Virosphere’, Cell, 172: 1168–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads from the metagenomic libraries are available at the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA598316. The sequences reported in this research have been deposited in GenBank databases, and the assembled contig sequences are available from GenBank databases under the accession numbers indicated in Supplementary Table S2. All virus nucleotide sequences (Fasta format), the unaligned and aligned datasets used in the phylogenetic analyses (Fasta format), and the phylogenetic trees (Newick format) that were generated in this work are available at the Figshare website (DOI: 10.6084/m9.figshare.11474709).