Abstract
Glycogen storage disease type Ia (GSD-Ia), deficient in glucose-6-phosphatase-α (G6PC), is characterized by impaired glucose homeostasis and a hallmark of fasting hypoglycemia. We have developed a recombinant adeno-associated virus (rAAV) vector-mediated gene therapy for GSD-Ia that is currently in a phase I/II clinical trial. While therapeutic expression of the episomal rAAV-G6PC clinical vector is stable in mice, the long-term durability of expression in humans is currently being established. Here we evaluated CRISPR/Cas9-based in vivo genome editing technology to correct a prevalent pathogenic human variant, G6PC-p.R83C. We have generated a homozygous G6pc-R83C mouse strain and shown that the G6pc-R83C mice manifest impaired glucose homeostasis and frequent hypoglycemic seizures, mimicking the pathophysiology of GSD-Ia patients. We then used a CRISPR/Cas9-based gene editing system to treat newborn G6pc-R83C mice and showed that the treated mice grew normally to age 16 weeks without hypoglycemia seizures. The treated G6pc-R83C mice, expressing ≥ 3% of normal hepatic G6Pase-α activity, maintained glucose homeostasis, displayed normalized blood metabolites, and could sustain 24 h of fasting. Taken together, we have developed a second-generation therapy in which in vivo correction of a pathogenic G6PC-p.R83C variant in its native genetic locus could lead to potentially permanent, durable, long-term correction of the GSD-Ia phenotype.
Graphical abstract

Chou and colleagues generated a new glycogen storage disease type Ia mouse model, homozygous for a pathogenic G6PC variant, and provided proof of concept that a CRISPR/Cas9 genome editing approach enables the treated mice to maintain glucose homeostasis and survive long term, which may offer permanent, durable correction for the disease.
Introduction
Glycogen storage disease type Ia (GSD-Ia; MIM: 232200) is an autosomal recessive disorder caused by pathogenic variants in the G6PC gene encoding glucose-6-phosphatase-α (G6Pase-α or G6PC), a key enzyme in glucose homeostasis.1, 2, 3 G6Pase-α catalyzes the hydrolysis of glucose-6-phosphate (G6P) to glucose and phosphate in the terminal step of gluconeogenesis and glycogenolysis. GSD-Ia patients manifest a phenotype of impaired glucose homeostasis characterized by fasting hypoglycemia, hepatomegaly, nephromegaly, hypercholesterolemia, hypertriglyceridemia, hyperuricemia, lactic acidemia, and growth retardation.1, 2, 3 There is no known cure for GSD-Ia. The current dietary therapies4,5 can help patients to attain near-normal growth and pubertal development, if adhered to strictly. However, the underlying pathological process remains uncorrected, and long-term complications, including hepatocellular adenoma/carcinoma and renal disease, still persist in GSD-Ia patients.1, 2, 3
To develop new therapies for GSD-Ia, we developed a recombinant adeno-associated virus (rAAV) vector-mediated gene augmentation therapy for GSD-Ia,6, 7, 8 which was licensed to Ultragenyx Pharmaceutical (Novato, CA, USA) that is currently in a phase I/II clinical trial (NCT03517085). However, the rAAV-G6PC clinical vector is episomally expressed,9 and the clinical durability of in vivo rAAV gene augmentation strategies remains unclear. Long-term durability is essential for a pediatric lethal disease, which must last over many decades of life. While the current clinical trial will provide improved understanding, we have sought to explore alternative genetic technologies for GSD-Ia therapies that may be more durable than episomal expression. To do this, we evaluated the efficiency of the CRISPR/Cas9-based gene editing technology10, 11, 12, 13, 14 to correct a high-prevalence pathogenic G6PC-p.R83C variant using a mouse model of GSD-Ia.
The most prevalent pathogenic variant identified in Caucasian GSD-Ia patients is G6PC-p.R83C, representing 32% of diseased alleles.15 It is particularly common in the Ashkenazi Jewish population, with a carrier frequency of 1.4%. Based on mutational and active site labeling studies, the current paradigm for the G6Pase-α reaction mechanism is that G6PC-p.H176 initiates a nucleophilic attack on the phosphate of G6P to form a phosphohistidine-enzyme transition state, which is stabilized by hydrogen bonding to G6PC-p.R83.16 Transient expression assays have shown that the G6PC-p.R83C variant is devoid of G6Pase catalytic activity.17 The vital role of G6PC-p.R83 residue in G6Pase-α catalysis is further verified by studies showing that substituting G6PC-p.R83 with any other amino acid residue completely abolishes G6Pase-α catalytic activity.17
In this study, we generated a homozygous G6pc-R83C mouse strain that exhibits the same symptoms of the G6pc-knockout (G6pc−/−) mouse strain,18 which mimics the symptoms of impaired glucose homeostasis characteristic of human GSD-Ia.1, 2, 3 We then treated newborn G6pc-R83C mice with a CRISPR/Cas9-based dual AAV system and showed that the strategy restored hepatic G6Pase-α activity in the G6pc-R83C mice and increased their 16-week survival rate from 0% to 100%. At age 8 and 16 weeks, the treated G6pc-R83C mice expressing ≥ 3% of normal hepatic G6Pase-α activity displayed a normalized metabolic phenotype and could sustain 24 h of fasting. Taken together, we have developed a second-generation genetic therapy for in vivo correction of a pathogenic variant in its native genetic locus, which may offer a durable therapeutic for GSD-Ia.
Results
Generation of the G6pc-R83C mouse model
The wild-type G6pc-R83, heterozygous G6pc-R83/R83C, and homozygous G6pc-R83C mice in a mixed C57BL/129 background designated as G6pc-R83, G6pc-R83/R83C, and G6pc-R83C mice, respectively, were used in this study. The G6pc-R83C mice could survive to age 7 days in the absence of any forms of therapy. For longer-term survival of the G6pc-R83C mice, glucose therapy was administered, starting at age 8 days. Biochemical and phenotypic studies were conducted in 3-week-old G6pc-R83C mice using age-matched G6pc-R83 and G6pc-R83/R83C littermates as controls. The 3-week-old G6pc−/− mice18 were used as an additional control. GSD-Ia is an autosomal recessive disorder,1, 2, 3 and the G6pc-R83 and G6pc-R83/R83C mice displayed a similar phenotype and were collectively named as the control mice. This was consistent with previous studies using the G6pc−/− mouse model.18
Liver and kidney microsomal G6Pase-α enzyme activities in 3-week-old G6pc-R83 mice were 364.3 ± 32.1 and 620.0 ± 21.1 units, respectively, and in 3-week-old G6pc-R83/R83C mice they were 221.2 ± 20.8 and 374.1 ± 26.7 units, respectively (Figure 1A). Liver and kidney microsomal G6Pase-α enzyme activities in 3-week-old G6pc+/+ mice were 355.3 ± 21.1 and 425.2.0 ± 31.4 units, respectively, and in 3-week-old G6pc+/− mice they were 212.3 ± 28.1 and 233.0 ± 21.1 units, respectively (Figure 1A). The liver and kidney microsomal G6Pase-α enzyme activities in 3-week-old G6pc−/− mice were 1.0 ± 0.3 and 4.0 ± 0.6 units, respectively (Figure 1A). The limitation of the lower level of quantitation for the microsomal G6Pase-α assay is 2 units, confirming the null phenotype of the liver and near null activity of the kidney. The slightly higher background of G6P hydrolytic activity in the G6pc−/− kidney may reflect a very low level of other kidney phosphatases measured by this assay. The liver and kidney microsomal G6Pase-α enzyme activities in 3-week-old G6pc-R83C mice were 1.6 ± 0.2 and 6.5 ± 0.6 units, respectively (Figure 1A), indicating that the G6pc-R83C mice were similar to the G6pc−/− mice with a null enzymatic activity for the G6pc locus. Studies have shown that G6Pase-α is expressed heterogeneously throughout the entire liver, but its expression is restricted to the cortex of the kidney.1 Transient expression assays in COS-1 cells have shown that the G6PC-p.R83C construct directed the synthesis of wild-type levels of an immunoreactive G6Pase-α variant lacking enzymatic activity.17 Enzyme histochemical analysis confirmed that G6Pase-α activity in the control mice was distributed heterogeneously throughout the liver and in the cortex of the kidney (Figure 1B). As expected, G6Pase-α activity was not detectable in the liver or kidney sections of the G6pc-R83C mice (Figure 1B). Using a monoclonal antibody against human G6Pase-α,19 we showed that liver microsomal preparations from G6pc-R83, G6pc-R83C, and G6pc+/+ mice expressed an immunoreactive G6Pase-α protein, in contrast to its absence in liver microsome preparations of the G6pc−/− mice (Figure 1C).
Figure 1.
The G6pc-R83C mice manifest impaired glucose homeostasis
The G6pc-R83, G6pc-R83/R83C, and G6pc-R83C mice were designated as R83, R83/R83C, and R83C, respectively. The R83 and R83/R83C mice displaying a similar phenotype were collectively named as the control mice. (A) Liver and kidney microsomal G6Pase-α activity in 3-week-old R83 (n = 5), R83/R83C (n = 7), R83C (n = 12), G6pc+/+ (n = 6), G6pc+/− (n = 6), and G6pc−/− (n = 12) mice. (B) Histochemical analysis of G6Pase-α activity. Each image represents an individual mouse. The arrow indicates kidney cortex. Scale bars, 100 μm. (C) Western blot analysis of liver microsomal G6Pase-α using a monoclonal antibody against G6Pase-α.19 (D) Postnatal growth of control (○) and R83C (●) mice. Each point represents 10 animals. (E) Size comparison of R83 and R83C mice at age 3 weeks, showing growth retardation of the G6pc-R83C mice. (F) Liver and kidney weight of 3-week-old control (n = 10) and R83C (n = 10) mice. Data represent the mean ± SEM. ∗∗p < 0.005.
The G6pc-R83C mice weighed significantly less than their control littermates throughout their postnatal development (Figure 1D), suggesting that the growth of G6pc-R83C mice is retarded, a characteristic of human GSD-Ia.1, 2, 3 Indeed, at age 3 weeks, the G6pc-R83C mice were about half the size of the control littermates (Figure 1E) and displayed marked hepatomegaly and nephromegaly (Figure 1F), characteristics of GSD-Ia.1, 2, 3
Phenotype of the G6pc-R83C mice
The G6pc-R83C mice displayed hypoglycemia, hypertriglyceridemia, hypercholesterolemia, and hyperuricemia (Figure 2A). In contrast to human GSD-1a patients, serum lactate concentrations were not elevated in the G6pc-R83C mice (Figure 2A), as also previously observed in the G6pc−/− mice.18 In gluconeogenic organs, G6P participates in multiple metabolic pathways, including glycogen synthesis, glycolysis, and glucose production in the endoplasmic reticulum. A deficiency in G6Pase-α reprograms G6P metabolism, leading to reduced glucose production, increased glycogen synthesis, and enhanced glycolysis. Compared to the controls, hepatic glucose levels in the G6pc-R83C mice were markedly reduced, along with increased hepatic levels of triglyceride, glycogen, and G6P (Figure 2B). While serum levels of lactate were unchanged, hepatic lactate levels were significantly increased in the G6pc-R83C mice (Figure 2B).
Figure 2.
Phenotype analysis of the G6pc-R83C mice
The G6pc-R83, G6pc-R83/R83C, and G6pc-R83C mice were designated as R83, R83/R83C, and R83C, respectively. The R83 and R83/R83C mice displaying a similar phenotype were collectively named as the control mice. (A) Serum glucose, triglyceride, cholesterol, lactate, and uric acid levels in 3-week-old control (n = 12) and R83C (n = 12) mice. (B) Liver glucose, triglyceride, glycogen, lactate, and G6P levels in 3-week-old control (n = 12) and R83C (n = 12) mice. (C) H&E-stained liver and kidney sections in 3-week-old R83 and R83C mice. Each plate represents an individual mouse. Scale bar, 20 μm. (D) Oil red O staining of liver and kidney from 3-week-old R83 and R83C mice. Scale bar, 20 μm. Data represent the mean ± SEM. , ∗∗p < 0.005.
Hematoxylin and eosin (H&E) staining showed that the G6pc-R83C mice displayed no histological abnormalities except marked glycogen storage in liver hepatocytes, which created a uniform mosaic architecture with compression of the sinusoids (Figure 2C), similar to that seen in GSD-Ia patients. There was also marked glycogen accumulation in the tubular epithelial cells of the kidney, resulting in enlargement and compression of the glomeruli (Figure 2C). Hepatomegaly in GSD-Ia is caused by elevated accumulation of both glycogen and neutral fat.1, 2, 3 Oil red O staining confirmed the increases in neutral fat accumulation in the liver of the G6pc-R83C mice (Figure 2D). Interestingly, little or no fat accumulation occurred in the kidney of the G6pc-R83C mice (Figure 2D). In conclusion, the G6pc-R83C mice manifest growth retardation, hypoglycemia, hepatomegaly, nephromegaly, hyperlipidemia, and hyperuricemia, with elevated hepatic lactate mimicking human GSD-Ia patients1, 2, 3 and the previously characterized G6pc−/− mice.18
Correcting the pathological manifestations of the G6pc-R83C mice by gene editing
We examined the efficacy of CRISPR/Cas9 to correct metabolic abnormalities in G6pc-R83C mice using a dual AAV strategy that targets primarily the liver.11 Two AAV8 vectors, AAV8-mG6pc-guide RNA (gRNA) and AAV8-SaCas9-gRNA, were used (Figure 3A). Newborn G6pc-R83C mice were infused via the temporal vein using two different viral doses, maintaining the AAV8-mG6pc-gRNA:AAV8-SaCas9-gRNA vector ratio at 10:1. Dose AAV-H (AAV-high) (n = 20) consisted of AAV8-mG6pc-gRNA (1 × 1014 virus particle [vp]/kg) + AAV8-SaCas9-gRNA (1 × 1013 vp/kg), and dose AAV-L (AAV-low) (n = 21) consisted of AAV8-mG6pc-gRNA (1 × 1013 vp/kg) + AAV8-SaCas9-gRNA (1 × 1012 vp/kg). Glucose therapy was not administered to either group. Phenotypic correction was assessed in the treated mice at age 8 weeks, a time point at which there is no further liver mitosis.20
Figure 3.
In vivo correction of the human G6PC-p.R83C variant in the G6pc-R83C mice by a dual AAV-CRISPR-Cas9 strategy
(A) Schematic diagram of the two AAV vectors, AAV8-mG6pc-gRNA and AAV8-SaCas9-gRNA. (B) HDR and indels in AAV-H liver cells analyzed at age 8 weeks.
We examined the targeted region of the G6pc gene by next-generation sequencing (NGS) of PCR amplicons in the liver tissues of 8-week-old AAV-H (n = 16) and AAV-L (n = 7) mice. The insertions or deletions (indels) in the G6pc locus of the AAV-H mice were detected in 18.4% ± 1.5% of G6pc alleles, and the homology-directed repair (HDR)-based correction of T to C mutation was observed in 0.7% ± 0.06% of G6pc alleles (Figure 3B). The indels in the AAV-L mice were detected only in 1% of G6pc alleles, and the HDR-based correction of the T to C mutation was nondetectable in the AAV-L mice.
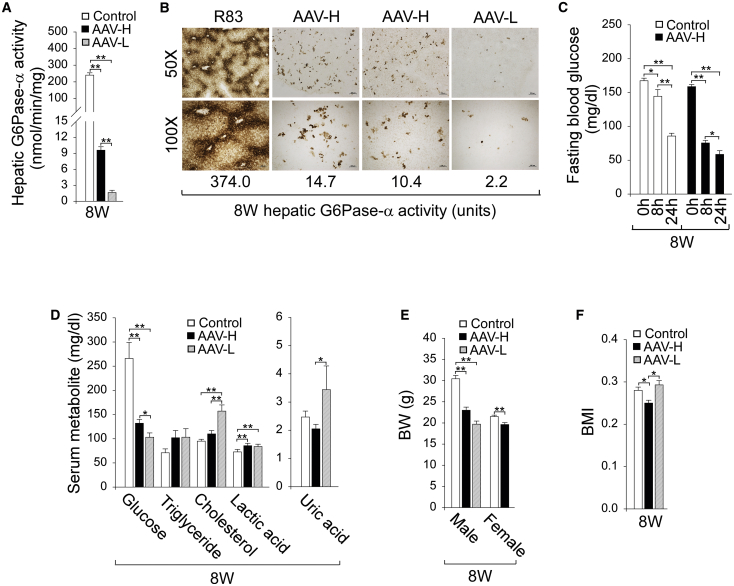
In the absence of glucose therapy, the untreated G6pc-R83C mice had a 4-week survival rate of 17% and a 6-week survival rate of 0%. In contrast, the AAV-H and AAV-L mice had an 8-week survival rate of 100% and 33% (7 out of 21 mice), respectively. Liver microsomal G6Pase-α enzyme activities in 8-week-old control mice averaged 240.0 ± 13.9 units, representing 100% of normal hepatic G6Pase-α activity (Figure 4A). Liver microsomal G6Pase-α enzyme activity in 8-week-old AAV-H (n = 20) mice was 9.6 ± 0.6 units, representing 4% of normal hepatic G6Pase-α activity, while for the low dose AAV-L (n = 7) mice, G6Pase activity was 1.6 ± 0.2 units, just 0.7% of normal hepatic G6Pase-α activity (Figure 4A).
Figure 4.
Phenotypic analysis of the AAV-H and AAV-L mice at age 8 weeks
The G6pc-R83 and G6pc-R83/R83C mice displaying a similar phenotype were collectively named as the control mice. Biochemical analysis was conducted in 8-week-old control (n = 28), AAV-H (n = 20), and AAV-L (n = 7) mice. (A) Liver microsomal G6Pase-α activity. (B) Histochemical analysis of hepatic G6Pase-α activity. Each image represents an individual mouse. Scale bars, 200 μm (50×) and 100 μm (100×). The numbers represent hepatic G6Pase-α activity expressed in the mice. (C) Fasting blood glucose levels in control (n = 16) and AAV-H (n = 12) mice. (D) Serum glucose, triglyceride, cholesterol, lactate, and uric acid levels in mice not subjected to fasting. (E) BW values. (F) BMI values. Data represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.005.
Enzyme histochemical analysis showed that G6Pase-α in 8-week-old control mice was distributed throughout the liver with significantly higher levels in proximity to blood vessels (Figure 4B). G6Pase-α in AAV-H and AAV-L mice was also distributed throughout the liver but with foci containing markedly higher levels of enzymatic activity and with a substantial proportion of hepatocytes harboring little or no G6Pase-α activity (Figure 4B). Despite the low enzyme activity in the AAV-L mice, enzymatic active G6Pase-α protein was detectable in liver sections. Our study also suggests that uniform hepatic G6Pase-α expression is not required for rescuing the GSD-Ia phenotype, as was shown previously in the rAAV-treated G6pc−/− mice.7
The hallmark of GSD-Ia is fasting hypoglycemia.1, 2, 3 The mean blood glucose level of the control mice (n = 23) before commencing fasting was 167.4 ± 3.8 mg/dL, which decreased 49% to 85.7 ± 4.1 mg/dL after 24 h of fasting (Figure 4C), while the AAV-H mouse blood glucose levels decreased 63% from 158.7 ± 3.2 to 58.7 ± 5.5 mg/dL (Figure 4C). Because of the low survival rate of the AAV-L mice, we did not examine their fasting glucose profile.
Compared to control mice, serum glucose levels were at the low end of the normal range for AAV-H (132 ± 7.7 mg/dL) and AAV-L (103 ± 9 mg/dL) mice (Figure 4D) but were significantly higher than serum glucose levels of 82.1 ± 5.9 mg/dL in 3-week-old untreated G6pc-R83C mice (Figure 2A). Importantly, the AAV-H mice displayed normal serum levels of triglyceride, cholesterol, and uric acid, although they manifested a mild lactic acidemia (Figure 4D). Compared to 3-week-old untreated G6pc-R83C mice (Figure 2A), the AAV-L mice displayed improved hyperlipidemia (Figure 4D).
The average body weight (BW) of male and female control mice differed greatly at age 8 weeks (Figure 4E). The BW of 8-week-old male and female AAV-H mice were 76% and 91%, respectively, of their age- and sex-matched control mice (Figure 4E). The body mass index (BMI) values21 of the AAV-H mice were significantly lower than those of their control littermates (Figure 4F), suggesting that the AAV-H mice were considerably leaner. Six of the 7 AAV-L mice that survived to age 8 weeks were male. The BW of male AAV-L mice was 65% of that of the male control mice (Figure 4E). The BMI values of AAV-L and control mice were statistically similar (Figure 4F).
The AAV-H and AAV-L mice continued manifesting hepatomegaly (Figure 5A). Again, the LW/BW values of the 8-week-old AAV-H and AAV-L mice were inversely correlated to their hepatic G6Pase-α activity restored. H&E and oil red O staining showed that the 8-week-old AAV-H and AAV-L mice exhibited no hepatic histological abnormalities except increased glycogen and neutral fat storage (Figure 5B). The extents of these increases were inversely correlated to hepatic G6Pase-α activity restored. As expected, hepatic glucose levels were markedly reduced in both groups of mice (Figure 5C). Moreover, both AAV-H and AAV-L mice displayed increased hepatic levels of glycogen, triglyceride, lactate, and G6P, and the increases were again inversely correlated to hepatic G6Pase-α activity restored (Figure 5C).
Figure 5.
Biochemical analysis of AAV-H and AAV-L mice at age 8 weeks
The G6pc-R83 and G6pc-R83/R83C mice displaying a similar phenotype were collectively named as the control mice. The analysis was conducted in 8-week-old control (n = 28), AAV-H (n = 20), and AAV-L (n = 7) mice. (A) Liver weight (LW)/BW values. (B) H&E and Oil Red O-stained liver sections in 8-week-old control, AAV-H, and AAV-L mice. The numbers represent hepatic G6Pase-α activity expressed in the mice. Each plate represents an individual mouse. Scale bar, 20 μm. (C) Hepatic levels of glucose, triglyceride, glycogen, lactate, and G6P. (D) Hepatic levels of G6pt transcript. (E) Serum insulin levels. Data represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.005.
Glucose homeostasis is maintained by the coupled action of the G6Pase-α/G6P transporter (G6PT) complex,1, 2, 3 and the reduced G6Pase-α activity can be offset by an increase in G6PT expression.7 In AAV-H and AAV-L mice, hepatic G6pt mRNA levels were increased 1.8- and 1.9-fold, respectively, over that of the controls (Figure 5D). Compared to control mice, serum insulin levels were significantly lower in both AAV-H and AAV-L mice (Figure 5E), as previously shown in AAV-treated G6pc−/− mice.7
The AAV-H mice maintain glucose homeostasis at age 16 weeks
The normalization of glucose homeostasis seen in 8-week-old AAV-H mice suggests that the CRISPR/Cas9 strategy could lead to longer-term duration of in vivo correction in the G6pc-R83C mice. Indeed, the 16-week survival rate of the AAV-H mice was also 100%. Liver microsomal G6Pase-α activities in 16-week-old control mice (n = 10) averaged 236.2 ± 17.3 units, similar to their activity at age 8 weeks (Figure 6A). Liver microsomal G6Pase-α activities in 16-week-old AAV-H mice (n = 10) averaged 7.5 ± 1.2 units (Figure 6A), representing 3.2% of normal hepatic G6Pase-α activity. Importantly, the 16-week-old AAV-H mice could sustain 24 h of fasting (Figure 6B).
Figure 6.
Biochemical analysis of AAV-H mice at age 16 weeks
The G6pc-R83 and G6pc-R83/R83C mice displaying a similar phenotype were collectively named as the control mice. The analysis was conducted in 16-week-old control (n = 10) and AAV-H (n = 10) mice. (A) Liver microsomal G6Pase-α activity. (B) Fasting blood glucose levels in 16-week-old control (n = 5) and AAV-H (n = 5) mice. (C) Serum levels of glucose, triglyceride, cholesterol, lactate, and uric acid in mice not subjected to fasting. (D) BW and LW/BW values. (E) BMI values. (F) Serum insulin levels. Data represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.005.
Compared to control mice, serum glucose levels remained low in the 16-week-old AAV-H mice (Figure 6C). Significantly, the AAV-H mice displayed normal serum levels of triglyceride, cholesterol, lactic acid, and uric acid (Figure 6C). At age 16 weeks, the average BW of the AAV-H mice was 79% of their age-matched control mice (Figure 6D), and the AAV-H mice continued manifesting hepatomegaly (Figure 6D). However, the average BMI value of the AAV-H mice was significantly lower than that of the controls (Figure 6E), again suggesting that the AAV-H mice were leaner. At age 16 weeks, serum insulin levels in the AAV-H mice were significantly lower (Figure 6F), similar to those observed in the AAV-H mice at age 8 weeks.
Discussion
Our current work explores the use of the CRISPR/Cas9 technology10, 11, 12, 13, 14 to correct a pathogenic GSD-Ia variant in its native genetic locus. We first used CRISPR/Cas9-based technology to generate a G6pc-R83C mouse line homozygous for G6PC-p.R83C, the most prevalent pathogenic variant identified in Caucasian GSD-Ia patients.15 We showed that the G6pc-R83C mice were devoid of hepatic and renal G6Pase-α activity and died prematurely, with a 4-week survival rate of 17%. Moreover, the G6pc-R83C mice manifest impaired glucose homeostasis and frequent hypoglycemic seizures, like the symptoms of the global G6pc−/− mice.18 Indeed, the G6pc-R83C mice manifested essentially the same pathophysiology as human GSD-Ia patients, characterized by hypoglycemia, growth retardation, hepatomegaly, nephromegaly, hypertriglyceridemia, hypercholesterolemia, and hyperuricemia.1, 2, 3 The glycogen storages in the mice are markedly increased in liver hepatocytes and renal tubular epithelial cells, like that seen in human GSD-Ia patients. Moreover, hepatic levels of glucose were reduced and hepatic levels of triglyceride, glycogen, G6P, and lactate were increased in the G6pc-R83C mice. Our study establishes that the phenotype of the G6pc-R83C mice mimics that of human GSD-Ia patients1, 2, 3 and G6pc−/− mice.18
Using the G6pc-R83C mice, we explore the efficacy of CRISPR/Cas9 to correct metabolic abnormalities in these mice using a dual AAV strategy that targets primarily the liver.11 We used two different virally delivered doses of the editing reagents: AAV-H and AAV-L. The indels in AAV-H and AAV-L mice were low, at 18% and 1% of the G6pc alleles, respectively. The HDR could be detected only in the AAV-H mice at 0.7% of the G6pc alleles. However, the CRISPR/Cas9 technology markedly increased the survival of the G6pc-R83C mice. In contrast to the untreated G6pc-R83C mice with a 0% survival rate to age 6 weeks, the AAV-H and AAV-L mice had a 100% and 33% survival rate, respectively, to age 8 weeks. Indeed, the AAV-H mice had a 100% survival rate to age 16 weeks. Direct enzyme activity measurements showed that hepatic G6Pase-α activity in AAV-H mice was restored to ≥ 3% of normal hepatic G6Pase-α activity at both age 8 and 16 weeks. The AAV-H mice grew normally, displayed no hepatic histological abnormalities, and exhibited normalized blood metabolite profiles. In contrast to GSD-Ia patients and untreated G6pc-R83C mice, which cannot tolerate a short fast, the AAV-H mice could sustain 24 h of fasting. It is important to emphasize that metabolic abnormalities, including hyperlipidemia, hyperuricemia, lactic acidemia, hepatomegaly, and nephromegaly, are secondary to recurrent hypoglycemia because the loss of glucose homeostasis reprograms cellular G6P metabolism, leading to the clinical manifestations seen in GSD-Ia patients.1, 2, 3 We have previously shown that rAAV-G6PC-treated G6pc−/− mice expressing ≥ 3% of normal hepatic G6Pase-α activity maintain glucose homeostasis, survive long term, and have no hepatic tumors.7,8 The viability and glucose control observed for the AAV-H mice suggest that they may survive long term and lack hepatocellular adenoma (HCA)/hepatocellular carcinoma (HCC). Our study indicates that CRISPR/Cas9 can correct a G6PC pathogenic variant in its native genetic locus, leading to a permanent therapeutic correction.
Studies have shown that AAV8 targets primarily the liver,11,22 and the rAAV8 vector does not transduce the kidney efficiently.22 As expected, the dual AAV8 strategy failed to transduce the kidney, and kidney microsomal G6Pase-α activity in 8-week-old AAV-H mice was 6.4 ± 0.7 units, indistinguishable from kidney microsomal G6Pase-α activity of 6.5 ± 0.6 units in 3-week-old untreated G6pc-R83C mice (Figure 1A). Rocca et al.23 have shown that retrograde renal vein injection of a rAAV9 vector efficiently targets the kidney cortex and medulla. To correct renal disease in GSD-Ia, we will examine whether retrograde renal vein injection of CRISPR/Cas9-based dual AAV9 vectors can normalize kidney function in the G6pc-R83C mice.
Blood glucose homeostasis is maintained by the G6PT/G6Pase-α complex.1, 2, 3 G6PT transports G6P from the cytoplasm into the endoplasmic reticulum lumen, and G6Pase-α, with its active site inside the lumen, hydrolyzes intraluminal G6P to glucose. We have shown that in rAAV-G6PC-treated G6pc−/− mice, the decreased G6Pase-α expression is accompanied by increased G6PT expression.7 As expected, in AAV-H mice, the reduced G6Pase-α activity is associated with elevated G6pt expression. Moreover, the AAV-H mice also displayed reduced levels of blood insulin. Collectively, our results suggest that elevated G6pt expression along with reduced blood insulin has enabled the AAV-H mice expressing ≥ 3% of normal hepatic G6Pase-α activity to maintain a functional glucose homeostasis.
The current study benefited by treating newborn mice when the liver is undergoing rapid mitotic expansion.20 In any human therapy, a gene-editing therapeutic reagent would be delivered to a more mature liver, with little if any mitotic activity. The efficiency of the CRISPR/Cas9 technology we used depends significantly on homology-directed recombination.10, 11, 12, 13, 14 Given the current proof of concept, we are now developing an alternative CRISPR/Cas9-based strategy to correct the G6PC-p.R83C variant in the G6pc-R83C mice using the non-homologous end joining repair mechanism and short double-stranded DNA oligonucleotides (dsODNs).24 This strategy uses lipid nanoparticles to deliver CRISPR reagents that consist of Cas9 mRNA, a G6pc exon-II-specific gRNA, and a dsODN when transcribed to RNA, consists of a 3′ splice site and G6pc exon-II region of the G6pc-R83C mice.24 The dsODN insertional technique bypasses the need for the HDR machinery and functions in both dividing and non-dividing cells, which can potentially correct genetic mutations in tissues with low rates of homologous recombination. In addition, the lipid delivery of reagents of a defined half-life reduces the risk of constitutively expressed viral-encoded reagents that remain active after gene correction and may lead to increased risks, such as a longitudinal increase in off-target cleavage.
In summary, we have generated a murine model of GSD-Ia harboring the pathogenic human G6PC-p.R83C variant and shown that the G6pc-R83C mice manifest a phenotype mimicking human GSD-Ia. Using these mice, we have shown that the CRISPR-Case9-based gene editing, when administered to newborn mice, can effectively correct the G6PC variant in the liver and normalize metabolic abnormalities in the G6pc-R83C mice. Our study provides a proof of concept for CRISPR-based correction of a prevalent pathogenic G6PC variant in a mouse model.
Materials and methods
Generation of the G6pc-R83C mouse
The heterozygous G6pc-R83/R83C mice in the C57BL/6 background were generated by microinjecting Cas9 mRNA, lead gRNA (spacer sequence: GTTTGGACAACGCCCG TATTG), and a donor template of 2 kb that substitutes CGC (R) with TGC (C) at codon 83 in exon 2 of the mouse G6pc gene into the C57BL/6 embryos. The resulting animals were screened by Sanger sequencing of topoisomerase (TOPO)-cloned PCR products to identify mice harboring the mutant allele. The G6pc-R83/R83C mice carrying one R83 (wild type) and one R83C allele in the C57BL/6 background were used to start the breeding colony. We have previously found that the global G6pc−/− mice18 in either the C57BL/6 or the 129S4/SvJaeJ background die within 24 h after birth, while in a mixed C57BL/129 background they survived to age 7 days without any forms of therapy and longer if the mice were supported by a glucose therapy (J.Y.C., unpublished data). As expected, the homozygous G6pc-R83C mice in the C57BL/6 background died within 24 h after birth. We therefore generated wild-type, heterozygous G6pc-R83/R83C and homozygous G6pc-R83C mice in the mixed C57BL/129 background, which were designated as G6pc-R83, G6pc-R83/R83C, and G6pc-R83C mice, respectively.
Dual AAV editing strategy and phenotype analysis
All animal studies were conducted under an animal protocol approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Animal Care and Use Committee. Mice were maintained on a standard NIH-31 Open formula mouse/rat sterilizable diet from Envigo (Madison, WI, USA). To sustain the survival of the G6pc-R83C mice beyond age 1 week, a glucose therapy consisting of daily subcutaneous injection of 100–150 μL of 15% glucose per mouse was started at age 8 days.
To correct the p.R83C mutation in the G6pc-R83C mice, two AAV8 vectors (Figure 3) were generated, both expressing the same single-strand gRNA (spacer sequence: GTTTGGACAATGCCCATACTG). One vector, AAV8-mG6pc-gRNA, harbors a 503 bp G6pc donor sequence to be used as the template for HDR, a single gRNA driven by a U6 promoter, and the TurboGFP marker directed by the cytomegalovirus (CMV) promoter/enhancer. The second vector, AAV8-SaCas9-gRNA, expresses Staphylococcus aureus (Sa) Cas9 directed by a liver-specific human thyroxine binding globulin (TBG) promoter and the single gRNA directed by the U6 promoter. The rAAV vectors were infused into newborn G6pc-R83C mice via the temporal vein as described.25 Glucose therapy was not administered to the AAV-treated G6pc-R83C mice. For all studies, age-matched wild-type and G6pc-R83/R83C littermates were used as controls. Fasting blood glucose analysis of mice consisted of blood sampling via the tail vein at 0, 8, and 24 h after food deprivation. Blood glucose levels were measured using the HemoCue Glucose 201 System (HemoCue America, Brea, CA, USA).
H&E staining was performed on liver sections preserved in 10% neutral buffered formalin, and Oil Red O staining was performed on cryopreserved optimal cutting temperature compound (OCT) embedded liver sections following the standard procedures. The stained sections were visualized using the Imager A2m microscope with Axiocam 506 camera and the ZEN 2.6 software (Carl Zeiss, White Plains, NY, USA).
Phosphohydrolase assays
Liver microsome isolation and microsomal phosphohydrolase assays were performed as described previously.18 In phosphohydrolase assays, reaction mixtures (50 μL) containing 50 mM sodium cacodylate buffer (pH 6.5), 2 mM EDTA, 10 mM G6P, and appropriate amounts of microsomal preparations were incubated at 30°C for 10 min. Disrupted microsomal membranes were prepared by incubating intact membranes in 0.2% deoxycholate for 20 min at 4°C. Non-specific phosphatase activity was estimated by pre-incubating disrupted microsomal preparations at pH 5 for 10 min at 37°C to inactivate the acid-labile G6Pase-α. One unit of G6Pase-α activity represents one nmol G6P hydrolysis per minute per mg microsomal protein. The lower level of quantitation for the microsomal G6Pase-α assay is 2 units.
Enzyme histochemical analysis of G6Pase-α was performed as described previously.7 Briefly, 10-μm-thick liver or kidney tissue sections were incubated for 10 min at room temperature in a solution containing 40 mM Tris-maleate (pH 6.5), 10 mM G6P, 300 mM sucrose, and 3.6 mM lead nitrate. After rinsing, liver sections were incubated for 2 min at room temperature in 0.09% ammonium sulfide solution, and the trapped lead phosphate was visualized following conversion to the brown-colored lead sulfide.
Serum and liver metabolite measurement
To measure serum metabolites, blood was collected by intracardiac bleeding from non-fasted, anesthetized mice at sacrifice. Serum glucose, total cholesterol, and uric acid were analyzed using Glucose Liquid Stable Reagent, Cholesterol Liquid Stable Reagent, and Uric Acid Liquid Stable Reagent, respectively, from Thermo Fisher Scientific (Waltham, MA, USA). Serum triglyceride was analyzed using the Serum Triglyceride Determination Kit from Sigma Diagnostics (St. Louis, MO, USA), serum lactate was analyzed using the EnzyFluo L-Lactate Assay Kit from BioAssay Systems (Hayward, CA, USA), and serum insulin was analyzed using the Ultra-Sensitive Mouse Insulin ELISA Kit from Crystal Chem (Downers Grove, IL, USA).
Liver tissues were homogenized in 5% NP-40, incubated for 5 min at 99°C, and centrifuged to remove insoluble material. Hepatic glycogen, G6P, glucose, and lactate in the deproteinized supernatants were analyzed using Glycogen Colorimetric Assay Kit II, Glucose-6-Phosphate Colorimetric Assay Kit, Glucose Colorimetric Assay Kit II, and Lactate Colorimetric/Fluorometric Assay Kit, respectively, from BioVision (Milpitas, CA, USA). Hepatic triglycerides were isolated by homogenizing 10–20 mg liver tissues in 300 μL of 5% NP-40, incubating for 5 min at 85°C, then cooling down to room temperature. After repeating the heating/cooling step once, tissue samples were centrifuged for 15 min at 17,000 × g, and triglycerides in the supernatant solutions were measured using the Triglyceride Quantification Colorimetric/Fluorometric Kit from BioVision (Milpitas, CA, USA).
NGS
NGS was used to determine the percentage of cells in the liver that contained indels or had undergone HDR. In brief, frozen mouse livers were ground to a powder form in Geno/Grinder 2010 (Ops Diagnostics, Lebanon, NJ, USA), and genomic DNA was isolated from the liver powder using the PrepGem Universal Kit (MicroGEM US, Charlottesville, VA, USA). Next, genomic DNA representing 5 × 105 cells was subjected to PCR to produce a 965-nucleotide fragment harboring mouse G6pc exon 2 using the primer pair: forward, ACCCGATGTCAAAAGAGACAGGTG and reverse, ATCTGGATCAGGCTGCTAGGAAGG. To add specificity for mouse G6pc, a second PCR was performed using a nested primer pair: forward, GAAACTGTGGGCTTCCGATA and reverse, ACAGGTGACAGGGAACTGCT. Then, the NGS adapters were added, and the resulting amplicons were sequenced using the Nextseq 550 System (Illumina, San Diego, CA, USA). The sequences were compiled, and the percentage of each editing outcome determined. The reads consisted of p.R83C mutant alleles (unedited), alleles with indels near the site of the Cas9-mediated double-stranded DNA break, and alleles that demonstrate HDR by converting the p.R83C mutation back to wild-type.
Quantitative real-time RT-PCR
Total RNA was extracted from liver tissues using the TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), purified further using the RNeasy Mini Kit (QIAGEN, Germantown, MD, USA), and cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The mRNA expression was quantified by real-time RT-PCR using the TaqMan Gene Expression Assay and Applied Biosystems QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). Data were normalized to Rpl19 mRNA.
Statistical analysis
The unpaired t test was performed using the GraphPad Prism Program, version 8 (GraphPad Software, CA, USA). Values were considered statistically significant at p <0.05.
Acknowledgments
We thank CRISPR Therapeutics, Inc., for co-development of the G6pc-R83C mouse model, design and production of the AAV vectors, and technical support for NGS analysis. This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, United States, and The Children's Fund for Glycogen Storage Disease Research, United States.
Author contributions
I.A. designed the research, performed the experiments, analyzed the data, and wrote the paper. L.Z. and H.-D.C. performed the experiments and edited the manuscript. B.C.M. analyzed the data and edited the manuscript. J.Y.C. designed the research, acquired the funding, analyzed the data, and wrote the paper.
References
- 1.Chou J.Y., Matern D., Mansfield B.C., Chen Y.T. Type I glycogen storage diseases: disorders of the glucose-6-phosphatase complex. Curr. Mol. Med. 2002;2:121–143. doi: 10.2174/1566524024605798. [DOI] [PubMed] [Google Scholar]
- 2.Chou J.Y., Jun H.S., Mansfield B.C. Glycogen storage disease type I and G6Pase-β deficiency: etiology and therapy. Nat. Rev. Endocrinol. 2010;6:676–688. doi: 10.1038/nrendo.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou J.Y., Jun H.S., Mansfield B.C. Type I glycogen storage diseases: disorders of the glucose-6-phosphatase/glucose-6-phosphate transporter complexes. J. Inherit. Metab. Dis. 2015;38:511–519. doi: 10.1007/s10545-014-9772-x. [DOI] [PubMed] [Google Scholar]
- 4.Greene H.L., Slonim A.E., O’Neill J.A., Jr., Burr I.M. Continuous nocturnal intragastric feeding for management of type 1 glycogen-storage disease. N. Engl. J. Med. 1976;294:423–425. doi: 10.1056/NEJM197602192940805. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y.T., Cornblath M., Sidbury J.B. Cornstarch therapy in type I glycogen-storage disease. N. Engl. J. Med. 1984;310:171–175. doi: 10.1056/NEJM198401193100306. [DOI] [PubMed] [Google Scholar]
- 6.Yiu W.H., Lee Y.M., Peng W.T., Pan C.-J., Mead P.A., Mansfield B.C., Chou J.Y. Complete normalization of hepatic G6PC deficiency in murine glycogen storage disease type Ia using gene therapy. Mol. Ther. 2010;18:1076–1084. doi: 10.1038/mt.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y.M., Jun H.S., Pan C.-J., Lin S.R., Wilson L.H., Mansfield B.C., Chou J.Y. Prevention of hepatocellular adenoma and correction of metabolic abnormalities in murine glycogen storage disease type Ia by gene therapy. Hepatology. 2012;56:1719–1729. doi: 10.1002/hep.25717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim G.Y., Lee Y.M., Kwon J.H., Cho J.H., Pan C.-J., Starost M.F., Mansfield B.C., Chou J.Y. Glycogen storage disease type Ia mice with less than 2% of normal hepatic glucose-6-phosphatase-α activity restored are at risk of developing hepatic tumors. Mol. Genet. Metab. 2017;120:229–234. doi: 10.1016/j.ymgme.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty D.M., Young S.M., Jr., Samulski R.J. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- 10.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankowicz F.P., Jarrett K.E., Lagor W.R., Bissig K.D. CRISPR/Cas9: at the cutting edge of hepatology. Gut. 2017;66:1329–1340. doi: 10.1136/gutjnl-2016-313565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneller J.L., Lee C.M., Bao G., Venditti C.P. Genome editing for inborn errors of metabolism: advancing towards the clinic. BMC Med. 2017;15:43. doi: 10.1186/s12916-017-0798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lino C.A., Harper J.C., Carney J.P., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou J.Y., Mansfield B.C. Mutations in the glucose-6-phosphatase-α (G6PC) gene that cause type Ia glycogen storage disease. Hum. Mutat. 2008;29:921–930. doi: 10.1002/humu.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh A., Shieh J.-J., Pan C.-J., Sun M.-S., Chou J.Y. The catalytic center of glucose-6-phosphatase. HIS176 is the nucleophile forming the phosphohistidine-enzyme intermediate during catalysis. J. Biol. Chem. 2002;277:32837–32842. doi: 10.1074/jbc.M201853200. [DOI] [PubMed] [Google Scholar]
- 17.Lei K.-J., Pan C.J., Liu J.L., Shelly L.L., Chou J.Y. Structure-function analysis of human glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1a. J. Biol. Chem. 1995;270:11882–11886. doi: 10.1074/jbc.270.20.11882. [DOI] [PubMed] [Google Scholar]
- 18.Lei K.-J., Chen H., Pan C.-J., Ward J.M., Mosinger B., Jr., Lee E.J., Westphal H., Mansfield B.C., Chou J.Y. Glucose-6-phosphatase dependent substrate transport in the glycogen storage disease type-1a mouse. Nat. Genet. 1996;13:203–209. doi: 10.1038/ng0696-203. [DOI] [PubMed] [Google Scholar]
- 19.Cho J.-H., Kim G.Y., Pan C.-J., Anduaga J., Choi E.-J., Mansfield B.C., Chou J.Y. Downregulation of SIRT1 signaling underlies hepatic autophagy impairment in glycogen storage disease type Ia. PLoS Genet. 2017;13:e1006819. doi: 10.1371/journal.pgen.1006819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham S.C., Alexander I.E. AAV-mediated gene delivery to the mouse liver. Methods Mol. Biol. 2019;1937:213–219. doi: 10.1007/978-1-4939-9065-8_12. [DOI] [PubMed] [Google Scholar]
- 21.Bahary N., Leibel R.L., Joseph L., Friedman J.M. Molecular mapping of the mouse db mutation. Proc. Natl. Acad. Sci. USA. 1990;87:8642–8646. doi: 10.1073/pnas.87.21.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y.M., Pan C.J., Koeberl D.D., Mansfield B.C., Chou J.Y. The upstream enhancer elements of the G6PC promoter are critical for optimal G6PC expression in murine glycogen storage disease type Ia. Mol. Genet. Metab. 2013;110:275–280. doi: 10.1016/j.ymgme.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocca C.J., Ur S.N., Harrison F., Cherqui S. rAAV9 combined with renal vein injection is optimal for kidney-targeted gene delivery: conclusion of a comparative study. Gene Ther. 2014;21:618–628. doi: 10.1038/gt.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart C., Beaudry K., Bayles T., Spencer S.J., Woodward N., Jayaraman M., Leonard J.P., Ho T.W., Carlo T. American Society of Gene & Cell Therapy Abstract; 2020. Insertion of short double-stranded oligonucleotides using CRISPR/Cas9 as a therapeutic approach for glycogen storage disease type 1a. [Google Scholar]
- 25.Gombash Lampe S.E., Kaspar B.K., Foust K.D. Intravenous injections in neonatal mice. J. Vis. Exp. 2014;93:e52037. doi: 10.3791/52037. [DOI] [PMC free article] [PubMed] [Google Scholar]