Abstract
Human diseases have always been a significant turf of concern since the origin of mankind. It is cardinal to know the cause, treatment, and cure for every disease condition. With the advent and advancement in technology, the molecular arena at the microscopic level to study the mechanism, progression, and therapy is more rational and authentic pave than a macroscopic approach. Non-coding RNAs (ncRNAs) have now emerged as indispensable players in the diagnosis, development, and therapeutics of every abnormality concerning physiology, pathology, genetics, epigenetics, oncology, and developmental diseases. This is a comprehensive attempt to collate all the existing and proven strategies, techniques, mechanisms of genetic disorders including Silver Russell Syndrome, Fascio- scapula humeral muscular dystrophy, cardiovascular diseases (atherosclerosis, cardiac fibrosis, hypertension, etc.), neurodegenerative diseases (Spino-cerebral ataxia type 7, Spino-cerebral ataxia type 8, Spinal muscular atrophy, Opitz-Kaveggia syndrome, etc.) cancers (cervix, breast, lung cancer, etc.), and infectious diseases (viral) studied so far. This article encompasses discovery, biogenesis, classification, and evolutionary prospects of the existence of this junk RNA along with the integrated networks involving chromatin remodelling, dosage compensation, genome imprinting, splicing regulation, post-translational regulation and proteomics. In conclusion, all the major human diseases are discussed with a facilitated technology transfer, advancements, loopholes, and tentative future research prospects have also been proposed.
Keywords: Non‐coding RNA, lncRNA, miRNA, Cancer, Genetic diseases, Neurodegenerative and cardiovascular diseases.
Introduction
RNA-the wonder of nature
The chief milestone in molecular biology was discovering the sequence of events in the flow of genetic information packed in DNA to the working cosmos of biological processes through proteins called the central dogma of molecular biology in 1958 by Francis Crick. However, with the advent of new technologies and robust next-generation sequencing, the enormous international consortiums such as the Functional Annotation of the Mammalian Genome (FANTOM) and the Encyclopaedia of DNA Elements (ENCODE) have described pervasive transcription (that 80 % of the DNA is transcribed into RNA but only a meager 1.5 % of that RNA actually translates into protein) (Carninci et al. 2005; Hangauer et al. 2013; Katayama et al. 2005; Okazaki et al. 2002). Thus the domain of RNA in the cellular world is actually separated into two diverse forms; the first one is non-coding RNA (ncRNA) which was once considered the evolutionary junk and lacking the potential to code protein and the other one is a much smaller chunk of protein-coding RNA. An RNA world hypothesis states that first life started with RNA, and later, its role was taken up by DNA being more stable and RNA was left with the job of a messenger. But eventually, it was unraveled that the RNA is just not endowed with the capacity to store genetic information alone but had been laden with a multitude of diverse, though latent catalytic functions that it makes it the most valuable contender in the field of epigenetics, disease, and undiscovered regulatory features despite of its dethronement. RNA is believed to evolve along with proteins and DNA during evolution (Robertson and Joyce 2012). The need of the hour is to understand their intricate role in diverse biological functions like development and homeostasis (Amaral et al. 2013). Figure 1 demonstrated the molecular events relating to noncoding RNA. Currently, the discrepancy between functional ncRNAs and junk RNA appears to be quite ambiguous. Also, it is an established fact that in prokaryotes, 80–95 % of the genome is protein-coding with only a fraction of ncRNA (differing even among closely associated strains) while eukaryotes have a relatively smaller fraction of protein-coding genes. With the increasing complexity in physiology, characteristics, and development of these organisms from lower non-chordates to humans, this proteome’s amount further decreases due to increases in introns and intergenic sequences that are transcribed but translationally tailored indifferently majorly via alternate splicing mechanisms (Frith et al. 2005; Mattick 2001). The RNA processing system, trans induction and transvection, DNA methylation, imprinting, RNA interference (RNAi), post-transcriptional gene silencing, chromatin modification, gene editing, splicing, dosage compensation, gene regulation mechanisms, and transcriptional gene silencing are also more sophisticated and complex in eukaryotes (Mattick 2003; Mattick and Gagen 2001; Mattick and Makunin 2005). Certain chromatin signatures binding transcription factors, histone modifications H3K4me3, H3K36me3, H3K9acand DNase1 hypersensitivity prove the existence of transcription of ncRNA in the intergenic region. ncRNA acts as conveyers of regulatory signals for the stimuli received at sensory genetic elements (Guttman et al. 2009, 2011).
Fig. 1.
Timeline of molecular discoveries relating noncoding RNA
Prokaryotic evolutionary history buttresses the fact that they still rely on protein-based regulatory architecture while eukaryotes have eventually adopted a digital regulatory remedy by developing new mechanisms and infrastructure to regulate penetrance and expressivity of disease, phenotypic trait expression, developmental programming via a repertoire of ncRNAs. Thus, the study of ncRNA concerning these integrated mechanisms is imperative to understand their role in health and diseases (Claverie 2005; Gagen and Mattick 2005; Mattick 2004).
Discovery, types, distribution of various ncRNAs
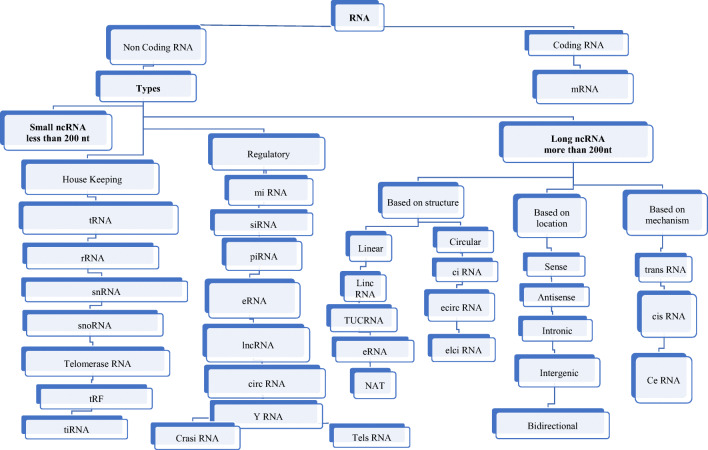
Figure 2 shows the different types of RNA existing in living cells. The ncRNAs are categorized broadly in two domains on the basis of transcript size, small ncRNA (< 200 nucleotides) and long ncRNA (> 200 nucleotides). The significant dimensions where ncRNA are instrumental include RNA maturation, RNA processing, signaling, gene expression, and protein synthesis (Kung et al. 2013; Morris and Mattick 2014; Palazzo and Lee 2015). There is a fascinating correlation between the amount of ncRNA and the level of conservation of species. It is estimated that the total RNA molecules per cell is around 107 ncRNA comprise of majorly snRNA, snoRNA, miRNA, rRNA, lncRNA etc. Human lncRNA are abundant and diverse, nearly 53,000 different lncRNA are known but only roughly 1000 are present in sufficiently high copy number to authentically justify their functional importance (Djebali et al. 2012). But estimates are unreliable as some of these code for short functional peptides and others may actually be misinterpreted as untranslated mRNA. Short ncRNA comprises another group that is believed to be enhancers (eRNA) regulating the structural aspects of a chromosome near the transcription sites. These are present in very small amounts (smaller than even lncRNA) (Fu 2014; Yan et al. 2019; Panni et al. 2020).
Fig. 2.
Different Types of existing RNAs in living cells. RNA is either coding or non-coding. Housekeeping RNAs are fundamental and expressed primarily while regulatory noncoding RNAs are synthesised at specific phase of development or in response to some external stimuli. Various types of ncRNA include (1) lnc RNA (long non coding RNA 200nt, 5`cap and poly A tail) (2) ciRNA: circular intronic RNA (introns of protein coding genes) 3). circ RNA (circular RNA) head to tail splicing of exons. 4). eRNA (enhancer associated RNA) 5) siRNA (small interfering RNA)21-22nt. 6) piRNA (piwi interacting RNA) major role in transposon defence, germ cell development.7). miRNA (micro-RNA) 22-23nt highly conserved and plays role in post transcriptional gene silencing. 8) tRF (tRNA-derived RNA fragment) and tiRNA (tRNA-derived stress-induced RNAs) 9) YRNA functions during DNA replication in vertebrates and RNA stability. 10) TUCRNA (transcribed ultra-conserved RNAs) 11) Natural antisense transcripts (NAT) RNA 12) Telomeres RNA (TEL) is localized in clusters at the nuclear envelope. 13) ElciRNA retained intron circRNAs
Other exotic RNAs that are immensely stable are circular RNA and are vital for cell survival. Intergenic RNA are significantly low in number, with only one copy number per cell. In total, around 21,000 human lncRNAs are synthesized from less than 1 % of the human genome (Volders et al. 2013). Surprisingly as few as 166 lncRNA are biologically valid in terms of their role in homeostasis, cell physiology, development, and diseases. It is likely that pervasive transcription occurs very frequently in the human genome leading to the formation of this huge and diverse population of ncRNA. Thus, inappropriate transcription in nucleosome-free DNA; lack of strictly regulated heterochromatin; inappropriate binding of transcription factors often leads to cryptic transcription of DNA into ncRNA. It is also true that improper RNA created through non -specific transcription would be checked and degraded by various quality control mechanisms working at the molecular level in the cell until there is flawed RNA degradation machinery in a cell (Palazzo et al. 2013). Then the question arises if the transcription process is so unpredictable and faulty, then why evolution has supported ncRNA theory. A convincing answer was fetched through the work of Kimura et al. on population genetics, which shows that nature`s ability to remove slightly harmful mutations depends on the size of the population of that species. More clearly, natural selection will not prevent formation of these ncRNA until these are detrimental to the existence of the organism (Abdel-Haleem 2007).
Biogenesis of ncRNA
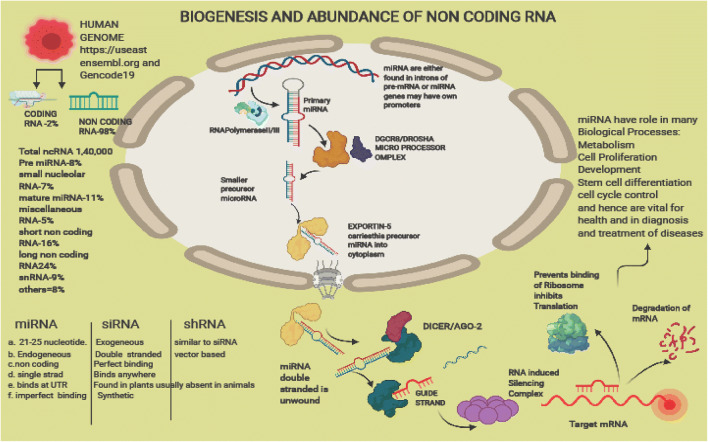
Throughout the human genome, a large number of genes are responsible for the formation of ncRNAs, and also, there may be separate transcriptional units (working autonomously) for the production of ncRNA. This biogenesis process includes transcription, maturation in the nucleus, export to the cytoplasm for processing and generation of functional RNA. Figure 3 shows the biogenesis of ncRNA and their role in health and disease. Transcription of polycistrons takes place by RNA polymerase II/III giving rise to large precursors (pri-miRNA: hairpin loop; 5’capping; 3’polyadenylation). This then undergoes two processing steps:
Fig. 3.
Biogenesis of noncoding RNA. Double stranded RNA are the major precursors which may be produced by inverted repeats in DNA, or two different RNA having complementary sequences may be simultaneously transcribed, or replication of double stranded RNA virus. Steps in biogenesis include (1) These may be formed in introns of pre-mRNA or miRNA genes having own promoters (transcribed by RNA polymerase II as large pri-miRNA hair pin loop like structure. This is recognised by DGCR8 (DiGeorge syndrome chromosomal region 8) and in association with Drosha forms Micro Processor Complex which cuts the RNA into smaller precursor microRNA. (2) This is then exported out of the nucleus into the cytoplasm with EXPORTIN-5 molecule. (3) In cytoplasm it is recognised by DICER helicase with RNase motif which cleaves the stem loop and releases microRNA molecule.4. Argonaute protein AGO-2 associates with miRNA/DICER, unwinds the double stranded RNA and releases one of the starnds. 5. It retains one of the strands known as Guide strand and other proteins now associate with it forming RISC (RNA Induced Silencing Complex) which guides this complex to the target complementary mRNA either by degrading mRNA or by inhibiting translation by preventing ribosomes from binding
The microprocessor (DGCR8) recognizes and governs the cleavage of pri-miRNA by Drosha (RNAse III endonuclease), leading to pre-miRNA formation that is translocated from nucleus to cytoplasm by RAN-GTP and Exportin-5.
In the second step Dicer (RNase III endonuclease) cuts the precursor near terminal end and hence RNA duplex is released, which interacts with Argonaute proteins in association with RISC (miRNA-induced silencing complex) (Beermann et al. 2016). Detailed mechanisms are discussed below:
Drosha/DGCR8-dependent pathway
miRNAs in animals are synthesised through a series of cleavages actions (over the hairpin precursors) brought about by 2 RNase III enzymes. DGCR8 protein (DiGeorge syndrome chromosomal region 8) recognises pri-miRNA hairpin and recruits Drosha RNase III enzyme (class II). Thus a ~ 55–70 pre-miRNA hairpin is cleaved near the terminal loop end by the Dicer (RNase III enzyme) after transporting it to cytoplasm. Effector complexes are formed by association of miRNAs with Argonaute (AGO) proteins. Its basic function is to cleave target miRNA. Argonaute effectors carry miRNA/miRNA as duplex and thus give rise to TNA induced silencing complex (Okamura et al. 2007).
Drosha/DGCR8-independent pathways
Mirtrons are microRNAs hairpins present in the spliced-out introns that are linearized by lariat debranching. Pol III termination and RNaseZ process the MHV68 tRNA-shRNA fusions into pre-miRNA-like hairpins. Endogeneous short hairpin RNA (Endo-shRNAs) lack attachment point for Drosha/DGCR8 but are loaded onto Ago1-4. Some Transposable elements, hpRNA etc., having additional dsRNA character, become prey to Dicer protein to produce siRNA (Yang and Lai 2011).
Integrated networks of ncRNA: Major disease mechanisms
ncRNA: a regulator of chromatin (chromatin modulation)
Many ncRNA work by recruiting multisubunit chromatin modifying complexes on the DNA for regulation of transcription; nucleosome positioning; chromatin marking or histone modifications (Affymetrix and Cold Spring Harbor Laboratory 2009). All these either enhance or repress the expression of target specific genes. Examples of such interactions include, lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) and NEAT (nuclear enriched abundant transcript 1) that are co-located but define different patterns of gene activation. Similarly, some miRNAs interact with promoter region of the gene (miR-24-1 induces eRNA and promoter binding, thus resulting in stronger RNA polymerase II action). Another example is hY1.RNA,that represses cell proliferation and non-coding human YRNA serve as biomarkers of cancer (Christov et al. 2008).
ncRNA: interaction with proteins (splicing regulation)
Some ncRNA alter the disease progression and fundamental cellular processes by binding to specific protein complexes that critically effect gene expression. Spliceosomes are formed by snRNA and other proteins leading to splicing mechanism (Lafontaine 2015). RNP (ribonucleoproteins) complexes also aids in post-transcriptional modification of pre-snRNA, pre-tRNA and pre-rRNA. LncRNA and circRNAs help recruit proteins (McHugh et al. 2015). For example, expression of circRNA-Foxo3 increases during cancer cell apoptosis and so is a potential treatment for suppressing tumour growth. Similarly, SAF-A and LBR proteins interact with lncRNA Xist for female development (Du et al. 2017).
ncRNA crosstalk with ncRNA in proteonomics
These interactions result in translation, transcription, epigenetics, pathological and physiological processes involving competitive endogenous RNA (ceRNA). For example, lncRNA-lncRNA plays a vital role in oncogenesis and tumour suppressor pathways (Grossi et al. 2016). Similarly, a cooperative association of vmiR-124, miR-375 and let-7b suppress erbB2/erbB3 to treat breast cancer. Hence these could be used in therapies and diagnostic approach (Li et al. 2015).
ncRNA interaction with mRNA (antisense transcription)
A process where the target gene is repressed by the expression of lncRNA from the opposite template can prove miraculous for treating certain genetic abnormalities like spinocerebellar ataxia type 7, Angleman syndrome etc. Also, miRNA and mRNA interactions may silent mRNA Expression. Increased amounts of miRNA may act as oncogenes by silencing mRNA. For example, cell adhesion molecule 1 (CADM1) gene in osteosarcoma acts as a niRNA sponge and thus have opened doors for developing new therapies (Chugh 2010; Gao et al. 2020; Liang et al. 2013; Palmini et al. 2017).
ncRNA as potential diagnostic and therapeutic biomarkers
Hereditary diseases
Alpha thalassemia
It is a hereditary disease caused by mutation in chromosome 16 (shorter arm`s telomeric region: HBA1 and HBA2 genes) causing haemoglobinopathy (formation of haemoglobin alpha chains in RBC); anaemia, jaundice and splenomegaly. The deletion leads to formation of an abridged LUC7L transcript lnc RNA with loss of protein-coding capacity (Taher et al. 2018; Tufarelli et al. 2003).
Alzheimer`s disease
It is a neurodegenerative, age affected disease mainly caused due to the accumulation of amyloid-beta and tau protein owing to cellular stress. In this disease, there is impairment of speech, memory and cognition. Cellular stresses leads to an increase in expression of BACE1 AS (Beta Secretase − 1 antisense) lnc RNA which further upregulates the levels of beta-secretase and amyloid plaque (Faghihi et al. 2008). Also, LRP1 (Low density lipoprotein receptor related protein 1) helps in removal of amyloid-beta and cholesterol. Antisense lnc RNA LPR1-AS negatively regulate LRP1 level by binding with HMGB2 High-mobility group protein B2 (chromatin associated protein). In Alzheimer`s patients’ level of LRP1 significantly falls (Andersen et al. 2005; Deane et al. 2008; Holtzman et al. 2012). Another protein known to fall during this disease is SORL1 (Sortilin related receptor 1) and the absence of this protein leads to the accumulation of amyloid plaque and that further culminates into Alzheimer`s disease. This protein is malformed due to the alteration in the splicing of SORL1 pre mRNA leading to the formation of an antisense lnc RNA 51 A decreasing the level of protein isoform A. Thus an increase in the amount of 51 A leads to a rise in amyloid-beta accumulation (Andersen et al. 2006; Massone et al. 2011). Also, GABBR2 gene shows an altered pre mRNA splicing, transcription ending in the formation of 17 A lnc RNA (gamma amino butyric acid type B receptor) thus leading to reduced GABAergic signalling. CDR1-AS (cerebellar degeneration related antigen 1 antisense) shows elevated expression in the normal brain and suppressed in the hippocampus and cortex in Alzheimer`s patients. This is also known as circular RNA sponge for miR-7. Thus when the level of ciRS-7 falls, it is unable to cause degradation of BACE1 and APP hence level of amyloid beta abnormally increases (Hansen et al. 2011, 2013; Shi et al. 2017; Zhao et al. 2016).
Angleman syndrome
Patients suffering from this disease shows severe and complex mental disorder have abnormally happy and smiling excitable personality, seizures, speech impairment, intellectual disability and imprinting disorder pertaining to abnormal expression of UBE3A (Ubiquitin protein ligase E3A) gene caused by either deletion of 15q11-13 on maternal chromosome 15 or uniparental disomy of paternal chromosome 15 or mutation in UBE3A gene or imprinting defect in q11-13 region of chromosome 15 (Runte et al. 2001; Yamasaki et al. 2003). Usually in normal healthy people the UBEA3 maternally inherited gene is active and paternal allele is silenced leading to imprinting phenomenon. Thus, such patients lack E3ubiquitine ligase which causes neurogenetic disorder. Experimental studies have figured out a UBE3A-ATS antisense lnc RNA transcribed from paternal chromosome (upstream to the imprinting centre transcription start site is located). In a study an antisense oligonucleotide was developed against this UBE3A-ATS which prevented the suppression of UBE3A from the paternal allele. This therapeutic treatment with antisense oligonucleotide helps reduce symptoms (Meng et al. 2015).
Beckwith-Wiedemann syndrome
It is a genetic disorder characterised by over grown body parts, asymmetric growth pattern (hemi hyperplasia), larger infants than normal (macrosomia).The abnormality results from differential expression and regulation of genes on chromosome 11 (genome imprinting H19,IGF2, CDKN1C genes) .There are two imprinting centres involved IC1 and IC2 that comprise of genes that code for lncRNA. This disease results from hypomethylation of IC2 and binding of KCNQ1OT1 with PRC2 complex and DNA methyltransferase. Normally this gene shows only paternal allele expression but biallelic suppression of cdkn1c and kcnq1 results in the manifestation of clinical signs of this disease (Mohammad et al. 2010; Ounap 2016).
Cartilage hair‐hypoplasia (CHH) / metaphyseal chondrodysplasia
It is an autosomal recessive disorder of RMRP (RNA Component of Mitochondrial RNA Processing Endoribonuclease) gene on chromosome 9p13 characterised by the deformed and shortened limbs; anaemia; metaphyseal chondrodysplasia (malformed cartilage at the end of long bones); (hypopigmentation), dental abnormalities and malformed nails. Mutations in the non-coding region of RMRP causes this disease (Chang and Clayton 1989). This RMRP gene codes for lncRNA (this RNA is transcribed by RNA polymerase III in nucleus and then transported to mitochondria). RMRP lnc RNA acts as the precursor for miRNA synthesis, namely RMRP-S1 and RMRP-S2 as evident by Norther blotting technique and Photo Activatable Ribonucleoside enhanced crosslinking and immunoprecipitation (PAR-CLIP). Patients suffering from this disease are known to have decreased levels of RMRP-S1 and RMRP-S2 up to 70 % than normal healthy cells. These two miRNAs are known to regulate the balance between bone mineralization and chondrocyte growth by downregulating the expression of other genes (Rogler et al. 2014). Experimental shreds of evidence on HEK293 cell line showed that RMRP-S1 decreased the expression of 35 concerned genes whereas RMRP-S2 downregulated 902 other genes (Sparber et al. 2019).
Facioscapulohumeral muscular dystrophy (FSHD)
It is a genetic disease associated with deletion of D4Z4 microsatellite repeat on chromosome 4 at locus 4q35 (sub telomeric region) leading to gradual deterioration of muscular strength, atrophy (wasting of muscles) in higher limbs and facio muscles. In normal individuals, the D4Z4 array exhibits more than 11 repeats (mostly 11–150). These remain associated with Polycomb group proteins (PcG). This binding suppresses the expression of FSHD locus. In individuals where this D4Z4 copy number falls below 11, PcG binding also reduces to a great extent leading to transcription of the lncRNA DBE-T (D4Z4binding element), which activates the FSHD gene by recruiting TrxG protein (Trithorax-group proteins) ASHL1(Achaete-scute homolog 1) that regulates transcription. This causes large scale transcription of DUX4 ((Double Homeobox 4) gene that is majorly responsible for myopathy, cell death and muscular dystrophy. Experimental evidence show direct association between Ash1L protein and DBE-T (Bodega et al. 2009; Cabianca et al. 2012; Gabellini et al. 2002).
Hirschsprung disease (HSCR)
It is a condition where a new born`s large intestine (colon) does not have proper nerve supply to the muscles of colons (abnormal cell migration of neural cells to the colon), leading to problems with passing stool, severe constipation, gas problems. A genetic mutation, malfunctioning of miRNA involved in nerve cell formation in intestine, leading to aganglionosis of neural plexus of the colon are the major causes of this disease. Recently it has been established that a circular lncRNA cir-ZNF609 plays a pivotal role in this disease (Peng et al. 2017, 2019). It was experimentally determined that in HSCR patients, miR-150-5p level increases while level of cir-ZNF609 dramatically reduces. Thus cir-ZNF609 is a potential biomarker in detecting HSCR (de Lorijn et al. 2006). Actually, miR-150-5p regulates the level of AKT3, assessed by qRT-PCR (which is responsible for cell migration and differentiation) (Peng et al. 2017). Thus, any fall in or knockdown of cir-ZNF609 would disrupt the regulatory loop of AKT3 (Serine/Threonine Kinase) and cause unstructured cell migration.
Huntington diseases
Huntington’s disease usually occurs after 30 years of age in adults and causes cognitive, movement and psychiatric abnormalities and is related as a neurodegenerative and autosomal dominant disorder (Duyao et al. 1993). In normal individuals, the HTT gene has a maximum of 36 repeats of CAG. In contrast, any further increase in the repeats leads to exponential production of a mutant protein leading to cytotoxicity. At least 7 lncRNA are known to dysregulate in such patients and many of which hold site for the attachment of transcriptional repressor REST (RE1-Silencing Transcription factor). TUG1 (Taurine Up Regulated 1) and NEAT 1 (Nuclear Paraspeckle Assembly Transcript 1) are known to increase while MEG3 and HTTAS (Huntingtin antisense) are known to decrease by forming epigenetic ribonucleoprotein complexes (Johnson 2012).
Opitz-Kaveggia syndrome/FG syndrome
It is an X-linked recessive disease due to a mutation in the MED12 gene at Xq13. The patients are due to disruption, usually mentally retarded, have down slanted palpebral fissures, hypertelorism, behaviour problems, and broad thumbs. A multiprotein complex serves to assist interaction between transcription factors and RNA polymerase II. MED 12 (mediator complex subunit 12) is one of the subunits of this complex that acts as both activator and repressor (Ding et al. 2008). When the level of MED12 decreases, its binding capacity with ncRNA-a1 and ncRNA-a3 also decreases (Lai et al. 2013).
Prader‐willi syndrome
This disease results from the deletion of a 108 kb region of paternal chromosome 15 (15q11-13) leading to hypogonadism, obesity, mental retardation. This region of the chromosome codes for (Small Nucleolar RNA, C/D Box 116 Cluster) SNORD116 (Duker et al. 2010). Thus, such patients lack sno-lncRNA. In normal individuals, these sno- lncRNA bind to FOX2. But in the effected patients, these FOX2 (alternate splicing factors) are excessively and freely available, leading to unorganized development due to altered pre-mRNA splicing patterns (Yin et al. 2012).
Pseudohypoparathyroidism type 1b (PHP1b)
It is an autosomal dominant imprinting disorder where GNAS (Guanine Nucleotide binding protein, Alpha Stimulating activity polypeptide) region of maternal chromosome 20 is involved in proximal tubules of the kidney (Chen et al. 2009; Plagge et al. 2008). This GNAS complex actually comprises of two non-coding lncRNA genes (A/S-1 and A/B) and three protein-coding genes (NESP55, GNAS, GNAS-XL). This GNAS complex produces alpha subunit of G protein responsible for signal transduction in kidney proximal tubule) in context of parathyroid hormone. When this GNAS complex is irresponsive, then Gα is absent and causes an increase in the level of phosphorous and decrease in the level of Ca in the blood leading to this disease. It has been experimentally determined that when a region of A/S1 lncRNA (exon 3 and 4) is deleted, then A/S1 on maternal chromosome gets transcribed thus inhibiting NESP55 expression and in turn expression of GNAS gene (Chillambhi et al. 2010).
Silver Russell syndrome
This disease results from hypomethylation of imprinted genomic region IC1 on 11p15.5 chromosome locus that leads to an increase in H19 lncRNA gene and inhibits protein-coding gene IGF2. The characteristic features include retarded and stunted growth and dwarfism.
Spinocerebral ataxia type 7 SCA7
It is a CAG repeat expansion, autosomal dominant neurodegenerative disorder of ATXN7 gene (Ataxin 7 protein-coding gene) present on short arm of chromosome 3. In normal conditions, this gene codes for Ataxin 7 protein (having a polyglutamine tract of 4–35 amino acids), while in a diseased state, it is composed of 37–400 amino acids (Paulson et al. 2000). Ataxin 7 actually is a part of STAGA (Spt-Ada-Gcn5-Acetyltransferase is a transcription coactivation complex). SCAANT1 (spinocerebellar ataxia-7 antisense non-coding transcript1) is an lncRNA that inhibits the ATXN7. But due to abnormally high CAG repeats, SCAANT1 is not formed efficiently and thus the expression of ATXN7 turns on and cause SCA7disease (Tan et al. 2014). Another lncRNA (lnc-SCA7) is involved in this disease manifestation. When CAG repeats increase, then a mediator microRNA (having binding site for Atxn7) miRNA-124 decreases that leads to increase in lncRNA SCA-7 and Atxn7. This increased expression in retina and cerebellum causes ataxia, dysarthria, dysphagia, blindness, and loss of motor control (Tay et al. 2014).
Spinocerebral ataxia type 8
It is also a neurological disease caused due to CTG expansion in lncRNA gene ATXN8OS (71- 1300 repeats) along with CAG expansion in ATXN8 gene (as discussed above) in which the patient show rapid involuntary movement of eyes, uncoordinated walk, slurred speech and is autosomal dominantly inherited (Todd and Paulson 2010). It involves both RNA and Protein level interference. ATXN8OS gene is collocated with a very important alternative splicing protein factor MBLN1 which is responsible for the normal working of GABA transporters and GABAergic signaling (confirmed by RNA-FISH). In SCA8 patients, this GABT4 amount rises to an enormous amount due to delocalization of ATXN8OS and MBLN1, leading to a changed slicing pattern (Daughters et al. 2009).
Spinal muscular atrophy
Another autosomal recessive neurodegenerative disease is SMA. Here duplication and alteration of SMN1 (Survival Motor Neuron 1) gene occur resulting in SMN2 gene (Lefebvre et al. 1995) Thus, the protein translated from such abnormal gene leads to muscle weakening, death of neurons in anterior part of spinal cord. Also the number of SMN1 gene decides the severity of the disease phenotype. Greater the number of SMN2 copies, the milder is the disease. Thus the best therapy is to increase the SMN2 level endogenously in the patient (Feldkötter et al. 2002). The concern was to discover lncRNA that could mimic SMN pre mRNA, thus SMN AS-1(SMN Antisense 1) was transcribed from SMN locus that negatively regulates SMN mRNA level and interacts with PRC2 complex. Also, this treatment lacked any cytotoxicity and thus proved a very effective therapy (Zhao et al. 2010).
Heart diseases
World Health Organization counts Coronary Artery Disease as the leading cause of death worldwide (http://www.who.int/cardiovascular_ diseases). Though numerous prognostic markers are available including N-terminal pro b-type natriuretic peptide (NT-pro BNP) and highly sensitive troponins, however the accurate determination of heart attack is still unpredicted. The ncRNA have been known as crucial components of pathological processes relating coronary artery disease. Since these are readily stable and detected in both blood and urine due to their remarkable stability studying their role can prove as an effective therapy. Table 1 show the involvement of ncRNAs in pathophysiology of heart diseases.
Table 1.
Molecular mechanisms involving ncRNAs in heart diseases
| Types of cardiac cells | Noncoding RNA involved | Mechanism | Effect |
|---|---|---|---|
| Smooth muscle cell | ANRIL |
Inhibits PRC-1/CBX7 PRC-2/EZH2 |
Increases proliferation |
| SENCR | Reduces cell migration | ||
| H19 |
Inhibits HIF1α miR675 |
Causes cell death | |
| Endothelial Cells | STEEL | Regulation of PARP1 |
Increased expression of eNOS KLF2 |
| MEG3 |
Regulate p53 leading to VEGF expression and Inhibits miR-9 miR-26 miR-138 |
Inhibits angiogenesis | |
| PUNISHER | Leads to vascular differentiation | ||
| MIAT |
Innervate miR-1246, miR200a, miR-29 |
Leading to vascular leakage | |
| MALAT1 | Control cell cycle regulatory genes | Increases proliferation | |
| MANTIS |
Regulate ICAM1, SMAD6, COUP-TFII, SOX18 BRG1 |
Increases angiogenesis and reduces inflammation | |
| Inflammatory cells | lncRNA CCL2 |
Regulates IGF2BP2 hnRNPU |
Increases the expression of CCL2 |
| MALAT1 | Regulates miR503 | Reduces inflammation | |
| Blood serum | GAS5 | Regulates EZH2 | Increases lipid accumulation |
| CHROME |
Inhibits miR-128, miR-27b, miR-33a/b |
Leads to cholesterol metabolism | |
| LeXis | Regulation via RALY | Reduces cholesterol | |
| MeXis | ABCA1 through DDX17 | Decreases cholesterol level |
Abbreviations: CHROME cholesterol homeostasis regulator of miRNA expression eNOS endothelial NO synthase,EZH2 enhancer of zeste homolog 2,GAS5 growth arrest-specific transcript 5,ICAM-1 intercellular adhesion molecule 1,LeXis liver-expressed LXR-induced sequence, lncRNA long noncoding RNA,MALAT1 metastasis-associated lung adenocarcinoma transcript 1,MEG3 maternally expressed gene 3,MeXis macrophage-expressed LXR induced Sequence, MIAT myocardial infarction associated Transcript, miR miRNA,PRC2 polycomb repressive complex 2,SENCR smooth muscle and endothelial cell–enriched migration/differentiation associated lncRNA, STEEL spliced-transcript endothelial enriched lncRNA, HOXD-AS1, or HAGLR, TNF tumor necrosis factor, VEGF vascular endothelial growth factor and VEGFR2 vascular endothelial growth factor receptor 2
Atherosclerosis
It is called hardening of major arteries due to plaque (fats/cholesterol rich diet) build-up that may lead to vascular disease, stroke and heart attack. Several genes and lncRNA regulates the vascular endothelial injury, fatty streaks, cell adhesion, apoptosis and angiogenesis. For instance, lncRNA H19, p38 and p65 have been expressed significantly in vascular myocytes and prevent their apoptosis via MAPK and NF- κB signal transduction (Pan 2017). Similarly, another lncRNA SMILR (Smooth Muscle Enriched Long Non coding RNA) can be best suited for diagnosis as well as treatment of atherosclerosis since this controls apoptosis and proliferation of cardiac muscle cells (Ballantyne et al. 2016). Also, miR 148a, miR-122 and miR-33 control fatty acid metabolism leading to lesion of atherosclerosis (Jaé and Dimmeler 2020).
Myocardial infarction
When necrosis of heart muscle occurs, accompanied by inflammatory reaction and apoptosis due to persistent lack of oxygen, it leads to heart failure. Inhibiting the NF-κB signal transduction pathway can prevent the heart from this ischemic injury. It has been observed that lncRNA MIAT (myocardial infarction associated transcript) gets upregulated during myocardial infarction. If somehow, the amount of MIAT is reduced, it even inhibits NF-κB activation, and thus can prevent a myocardial injury (Nishikido et al. 2016). Another lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) is known to prevent apoptosis of myocardial cells by suppressing PTEN (phosphatase and tensin homolog) gene expression through miR-320 (Li et al. 2017).
Hypertension
When the arterial blood pressure persistently remains above 140/90 the individual is said to be a patient of hypertension. It is observed in numerous studies that lncRNA AK098656 is excessively secreted in vascular muscles cells of hypertension patients as this lncRNA is known to degrade various proteins (extracellular matrix fibrin 1 and myosin heavy chains-11) and narrow the arteries (Jin et al. 2018).
Cardiac developmental disorder
For the proper functioning of heart, the lineage commitment of cardiac cells is imperative. The development of progenitor cells of cardiovascular tissue is regulated by a lncRNA Bvht (Braveheart), which activates MesP1 (mesoderm posterior 1). Another lncRNA Fendrr bind to PRC2 polycomb repressive complex 2 and Trithorax-group proteins/ mixed-lineage leukemia TrxG/MLL (chromatin remodelling complex) for smooth development of lateral mesoderm (Grote et al. 2013; Zhang et al. 2019).
Cardiac fibrosis
It refers to unusual hardening of valves of heart due to the deposition of extracellular matrix (ECM) in muscles of heart owing to abnormal proliferation of cardiac fibroblasts. Cardiac fibroblasts are flat or spindle shaped; more than 50 % of cells in connective tissue; lack basement membrane and functions include maintaining 3D structure, biochemical and electrical conductivity of heart along with production of ECM. During the cardiomyopathy, the proliferation of ECM is triggered, resulting in scaring of the site of necrosis in cardiac muscle cells leading to infarction. Many ncRNAs are now studied and evaluated for their impact on cardiac fibrosis. For instance, the levels of miR21 increase drastically during myocardial infarction which in turn stimulates the ERK-MAP signal transduction pathway leading to cardiac hypertrophy (Ghosh et al. 2012). miR 21 also regulates TGF-β receptor III and MMP2 (Matrix Metallopeptidase 2 ) pathway. Another miR- 29 falls during myocardial infarction, which depresses ECM components like elastin and fibrillin (Liang et al. 2012). miR101 acts as transcription silencer of c-fos, lnc RNA–NR024118 and cdkn1c expression is downregulated by angiotensin II via AT1 dependent signalling pathway (Jiang et al. 2015).
Cancer
It is now well evident that ncRNA are of fundamental worth in various types of cancers. Some of these ncRNA are exaggeratedly expressed in some cancers causing abnormal activation of CSC (Cancer Stem Cells) and deregulation of various signal transduction pathways conveying cell cycle. This usually ends up in causing a relay renewal and differentiation of other cells. Thus they play a pivotal role in tumour initiation and progression. Also in many cases these ncRNA are known to get suppressed, thus hindering normal cell division. Because of these vital capacities and diverse roles these are suitable candidates in diagnosis as well as treatment of cancer. Many other therapies are effective in reducing the progression or size of carcinomas but a therapy serving the curative purpose is the need of the hour.
Major candidates and their perspective strategies: lncRNA and miRNA
LncRNA
These lncRNA are highly specific (60 %) to a particular type of cancer; produced in only small amounts; variable in sequence but quite unique in secondary structures and exhibit both stimulatory as well as repressive role in various types of cancers. These may adopt any of the following strategies: (a) Translational inhibition (b) Shear splicing (c) Degradation of target mRNA (d) Formation of ribonucleoprotein complex (e) Association with transcription factors (Diederichs 2014; Srikantan et al. 2000). Few other strategies are elaborated as below:
Chromatin remodelling
LncRNA Evf2 suppresses the SWI/SNF and in turn downregulate Dlx6/Dlx5. Also a remarkable example is lncRNA HOTAIR which controls chromatin remodelling and inhibit the expression of HOXD genes by associating PRC2. Thus lncRNA play vital role in causing various types of cancer simply by modulation of the transcription capacity of various genes by altering their exposure to RNA polymerase enzyme, transcription factors and enhancers/modulators (Bond et al. 2009).
Epigenetic modifications
For example the linc RNA-HOTAIR (HOX antisense intergenic RNA) expressed on chromosome 12q13.13, homeobox C (HOXC) locus gets highly upregulated in breast cancers. This intergenic RNA alters the H3K27 methylation pattern and hence progression of cancer (Young et al. 2012). Thus epigenetic modifications like methylation and acetylation is one of the favourite strategies concerning cancer studies.
Developmental abnormalities: Genome imprinting
During genome imprinting methylation and acetylation of only either of the two alleles (either from mother or father) occurs and hence only one of them express to give the character. Recently association of genome imprinting with tumours have been established. LncRNA IRAIN (IGF1R Antisense Imprinted Non-Protein Coding RNA) works as a tumour suppressing agent and hence gene imprinting conversion leads to suppression of this gene and initiate tumour formation (Zhang et al. 2019).
MicroRNAs
Micro RNA can exhibit several roles either as tumour suppressor gene or oncogene. It mostly deregulate most involved genes responsible for apoptosis, cell division, cell migration, invasion, metastasis, proliferation of tumour cells etc. Thus virtually in every cancer they do have their involvement either as diagnostic marker, or cell cycle disruption or even in therapeutics. For instance miR 21 is involved in almost all different types of cancer: leukaemia, colorectal cancer, pancreatic cancer, brain cancer. There are recorded evidences that during cancer of glial cells of nervous system, miR 21 interfere with the PTEN (phosphatase and tensin homolog) and EGFR pathways by activating the epidermal growth factor receptor, thus supress the activation of CYCLIN D, and AKT (Chan et al. 2005; Si et al. 2007). Similarly it has been observed in some studies that miR34 family. MiR34a is useful for normal cell growth and proliferation of healthy cells however, any deletion in this gene induces tumour formation in prostrate and pancreas. Whenever DNA damage occurs this miR34 is triggered by tumour suppressor gene TP53 (Liu et al. 2011).
piRNA
The piRNA are the RNA produced independently with the aid of Dicer protein and are about 30 nucleotides long to maintain the germline. piRNA − 651 was recently reported to increase abruptly during cancer while piRNA − 823 decreased. Hence these are potential biomarkers of gastric cancer specifically (Cheng et al. 2011, 2012).
Breast cancer
It is the most prevalent type of cancer among women worldwide and second highest cause of death. Various environmental factors, stress, hormonal imbalance triggers this disease. However role of lncRNA seems to be the core concern. BANCR (BRAF-activated non-coding RNA) is a 693 bp lncRNA which gets upregulated during breast cancer. If Epithelial Mesenchymal transition is repressed, then BANCR usually gets supressed and prevents the uncontrolled cell proliferation. Also NORAD (Non-Coding RNA Activated by DNA Damage) is another important lncRNA known to increase during breast cancer and causes malignant progression of cancer by innervating TGF-β signalling (involving RUNX2- Runt-related transcription factor-2) (Flockhart et al. 2012; Zhou et al. 2019).
Cervical cancer
Certain viruses induce uncontrolled proliferation in cells of cervix causing cancer. Many lncRNA are known to play vital role in cell cycle regulation and apoptosis. It was worked out in several studies on cervical cancer that lncRNA MEG3 (Maternally Expressed 3) expression is down regulated and determines the size and metastasis of cancer (Zhang et al. 2017). Another antisense, RNA, homeobox transcriptional antisense RNA gets upregulated during the cervical cancer. Thus, this is a remarkable biomarker (Eoh et al. 2017).
Lung cancer
Lung cancer is a deadly and one of the most bothering diseases whose impact is rising with increasing pollution. The ncRNAs are emerging as valuable and potential diagnostic indicators of cancer. GACAT3 (gastric cancer associated transcript 3) is an lncRNA whose expression significantly rises during lung cancer while GAS5 gets downregulated (Ding et al. 2015; Yang et al. 2018). The molecular networks pertaining to lung cancer are highly intricate, thus more research is required in this area.
Viral infection
Viruses have been in the past and are now evidently emerging as the major threat to global public health including hepatitis C virus (HCV), dengue virus (DENV), Zika virus (ZIKV) yellow fever virus, West Nile virus, Ebola virus and severe acute respiratory syndrome coronavirus (SARS-CoV). Many research advancements have pointed the role of non-coding RNAs in regulating the critical interplay between viral-host interactions (Holmes et al. 2016; Metsky et al. 2017). Thus, elucidation of the complex pathogenic and physiological mechanism during infection is necessary for the efficacious control of these deadly viruses. Novel sequencing methodologies have revealed that a large number of regulatory non-coding RNAs are expressed in human cells in reaction to viral infection. Few of these are utilised by viruses for their survival in host cells while others are crucial for regulation of the immune system. In the evolving field of RNA diagnostics and therapeutics, such dealings in roles of ncRNAs can be seen as a promising antiviral targets. Only a little work has been carried on virus-host ncRNA interactions, while a gamut of interactions await discovery.
Dengue virus
Dengue fever caused by mosquito borne single stranded RNA Dengue virus (DENV), a flavivirus. During this viral infection miRNA − 378 level drastically reduces in blood mononuclear cells inhibiting Granzyme B serine protease that causes apoptosis in cells (Liu et al. 2016).
Hepatitis C virus
This is another flavivirus having envelope and single-stranded RNA as the genetic material that is associated with the development of various liver diseases including cirrhosis, fibrosis and hepatocellular carcinoma. miRNA-122 normally helps in managing circadian rhythm and lipid metabolism. But during viral infection it provides attachment site for HCV RNA that further protects viral RNA from degradation and also stimulates its protein synthesis and viral replication (Luna et al. 2015). This miRNA is a tumour suppressor but it attenuates the usual host targets.
Japanese encephalitis virus
Swelling of brain accompanied with high fever is a result of flavivirus infection. miR-15b is upregulated during JEV infection of glial cells and mouse brains. This miRNA acts on Ring Finger Protein (RNF125) and inhibits it leading to increased RIG-I, IL-6, CCL2, IFN-type I. Thus, this microRNA may prove a promising player to treat inflammation in neurons (Zhu et al. 2015).
Enterovirus 71 (EV71)
This single stranded RNA virus causes hand, foot, and mouth disease (HFMD) and neurological complications in children. It has been experimentally determined that miR-296-5p binds to the EV71 RNA and supress viral replication (Zheng et al. 2013).
Human immunodeficiency virus
This virus attacks monocytes and cluster of differentiation 4+ (CD4+) T-lymphocytes and exhibit complicated progression through acute, chronic, and AIDS stages triggering cell death pathways. Two very valuable microRNA: miRNA-34a and miRNA − 217 are known to be involved that suppress the protein synthesis of SIRT1 (Silent Mating Type Information Regulation 2 Homolog 1, responsible for deacetylation of nuclear factor kappa B (NF-kB) and suppresses transactivation of HIV-1 long terminal repeat (LTR) (Jopling et al. 2005; Scheel et al. 2016). Also it has been p21 (cyclin dependant kinase inhibitor protein) also inhibit HIV infection, however the HIV replication increased if p21 was inhibited by miRNA Let-7c, miR-34a-5p (Balasubramaniam et al. 2018; Farberov et al. 2015). These evidences establish that one of the best strategies to cure this diseases is to target host.
SARC-COV
It includes seven types: HCoV-NL63, HCoV-OC43, SARS-CoV1, HCoV-229E, Middle East respiratory syndrome coronavirus (MERS-CoV), HCoV-HKU1 and SARS-CoV2. It has ′- and 3′ UTRs (untranslated regions) with 5′ cap, a 3′ poly-A tail on single-stranded RNA genome. It leads to Cytokine storm culminating into acute respiratory distress syndrome (Huang et al. 2020). Relative studies on SARC-CoV have verified that it leads to differential triggering of host as well as viral miRNAs. It has been observed that expression of miR-9 and NFKB1 elevates during infection by HCoV-OC43 virus. RNA binding protein (N protein) binds to viral genome and hence helps in viral replication. Also N-protein binds to miR-9 and thus triggers inflammatory reaction and is considered as N protein-miRNA–mediated interaction (Cui et al. 2015; Lai et al. 2014). Another important strategy involved is apurinic/apyrimidinic endonuclease 1 which has unique RNA cleaving property (Kim et al. 2010). SARS-CoV generally targets bronchoalveolar stem cells (BASCs) and upregulates miR-214, miR-17 and miR-574-5p (deal with evading host immune response) while miR-98 and miR-223 gets down regulated leading to cytokines inflammatory exaggerated response (Leon-Icaza et al. 2019). The most infectious current pandemic SARS-CoV2 also sees its therapeutic taking non coding RNA as the base. A great deal of strategies have been worked on to cure this disease, but little is yet known with ultimate success. Bioinformatics and computer based predictions concerning miRNA are opening new doors of hope. These in-silico comparison studies have been able to successfully predict few microRNA targeting genes [Polymerase acidic protein (PA), major capsid protein VP1, polymerase basic protein 2 (PB2), ATP synthase F1 subunit β (ATP5B), interferon β (IFN-β)and Erb-B2 receptor tyrosine kinase 2 (ERBB2)] namely: hsa-miR-98, hsa-miR-126, hsa-miR-125a-5p, hsa-miR-101, hsa-miR-380-5, hsa-miR-222, hsa-miR-378, hsa-let-7a and hsa-miR-23b (Arisan et al. 2020; Chen and Zhong 2020; Fulzele et al. 2020; Saçar Demirci and Adan 2020).
Conclusions and future perspectives
The amazing and complex molecular processes that orchestrate the fundamental process of life are now known to be governed by an intricate and complicated ncRNA world. Rapid advancements in bioinformatics, sequencing technologies, proteomics and microarrays have led to the discovery of a colossal array of ncRNA which comprises the major chunk of regulators of cellular mechanisms primarily associated with eukaryotic complexity. With these diverse ncRNAs with integrated, complex networks and biological pathways, it seems further confusing to understand the individual role of these non-coding RNAs. Another baffling concern is that most of the ncRNAs seem to be falsely annotated. For this, new technological approaches need to be adopted tools for network analysis, in silico predictors and especially a deep understanding of mitochondrial miRNA that may give a boost to this field and may also help in unfolding certain latent molecular mechanisms of these ncRNA. During evolution, nature has been selecting them from times immemorial; thus, we believe that mitochondrial RNA studies would be pathfinding. Mitochondrial DNA shows maternal inheritance and is a vital organelle for the perpetuation of multicellular life. Therefore, switching the research towards mitochondrial RNA would evade some perplexing molecular pathways since it is free from recombination and seems to be carrying highly preserved informative centers.
Acknowledgements
The research presented in this article was supported by DST-SERB grant (EEQ/2020/000188) (JSB); National Institutes of Health (NIH) grants AG042178, AG047812, NS105473, AG060767, AG069333, AG066347 and R41 AG060836 (PHR).
Author contributions
Each author has made substantial contributions to the conception (JSB, GKB, NK, PHR) or design of the work (JSB, NK, IPS, USN); acquisition (NK, GKB, USN, IPS), or interpretation of data (GKB, NK, IPS, APR, USN); or have drafted the work (JSB, PHR, GKB, NK) and substantively revised it (GKB, NK, IPS, USN, APR, PHR); and has approved the submitted version (All Authors).
Data availability
Not Applicable.
Declarations
Conflict of interest
None declared.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not Applicable.
Conflict of interest
None declared.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gurjit Kaur Bhatti and Naina Khullar contributed equally to this work.
Contributor Information
Gurjit Kaur Bhatti, Email: bhattigk@yahoo.com.
Naina Khullar, Email: naina306@gmail.com.
Inderpal Singh Sidhu, Email: inderpalsidhu76@yahoo.co.in.
Uma Shanker Navik, Email: usnavik@gmail.com.
Arubala P. Reddy, Email: arubala.reddy@ttu.edu
P. Hemachandra Reddy, Email: hemachandra.reddy@ttuhsc.edu.
Jasvinder Singh Bhatti, Email: jasvinder.bhatti@cup.edu.in.
References
- Abdel-Haleem H. The origins of genome architecture. J Hered. 2007;98:633–634. doi: 10.1093/jhered/esm073. [DOI] [Google Scholar]
- Affymetrix ETP, Cold Spring Harbor Laboratory ETP Post-transcriptional processing generates a diversity of 5’-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12:254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- Andersen OM, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OM, et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR. Biochemistry. 2006;45(11):2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- Arisan ED, Dart A, Grant GH, Arisan S, Cuhadaroglu S, Lange S, Uysal-Onganer P (2020) The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses 12. 10.3390/v12060614 [DOI] [PMC free article] [PubMed]
- Balasubramaniam M, Pandhare J, Dash C (2018) Are microRNAs important players in HIV-1 infection? An update. Viruses 10. 10.3390/v10030110 [DOI] [PMC free article] [PubMed]
- Ballantyne MD, et al. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133:2050–2065. doi: 10.1161/circulationaha.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- Bodega B, et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.Can-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Chen M, et al. Central nervous system imprinting of the G protein G(s)alpha and its role in metabolic regulation. Cell Metab. 2009;9:548–555. doi: 10.1016/j.cmet.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhong L. Genomics functional analysis and drug screening of SARS-CoV-2. Genes Dis. 2020;7:542–550. doi: 10.1016/j.gendis.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Chillambhi S, Turan S, Hwang DY, Chen HC, Jüppner H, Bastepe M. Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab. 2010;95:3993–4002. doi: 10.1210/jc.2009-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Trivier E, Krude T. Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br J Cancer. 2008;98:981–988. doi: 10.1038/sj.bjc.6604254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh R. Experimental therapies and clinical trials in bone sarcoma. J Natl Compr Canc Netw. 2010;8:715–725. doi: 10.6004/jnccn.2010.0052. [DOI] [PubMed] [Google Scholar]
- Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian. Cells J Virol. 2015;89:9029–9043. doi: 10.1128/jvi.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorijn F, Kremer LC, Reitsma JB, Benninga MA. Diagnostic tests in Hirschsprung disease: a systematic review. J Pediatr Gastroenterol Nutr. 2006;42:496–505. doi: 10.1097/01.mpg.0000214164.90939.92. [DOI] [PubMed] [Google Scholar]
- Deane R, et al. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9:955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duker AL, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Eoh KJ, Kim HJ, Lee JY, Nam EJ, Kim S, Kim SW, Kim YT. Upregulation of homeobox gene is correlated with poor survival outcomes in cervical cancer. Oncotarget. 2017;8:84396–84402. doi: 10.18632/oncotarget.21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. J Cell Sci. 2015;128:1607–1616. doi: 10.1242/jcs.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- Fu X-D. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulzele S, Sahay B, Yusufu I, Lee TJ, Sharma A, Kolhe R, Isales CM. COVID-19 virulence in aged patients might be impacted by the host cellular MicroRNAs abundance/profile. Aging Dis. 2020;11:509–522. doi: 10.14336/ad.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Gagen MJ, Mattick JS. Inherent size constraints on prokaryote gene networks due to “accelerating" growth. Theory Biosci. 2005;123:381–411. doi: 10.1016/j.thbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Gao SS, Wang YJ, Zhang GX, Zhang WT. Potential diagnostic value of miRNAs in peripheral blood for osteosarcoma: A meta-analysis. J Bone Oncol. 2020;23:100307. doi: 10.1016/j.jbo.2020.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Nagpal V, Covington JW, Michaels MA, Vaughan DE. Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): differential expression of microRNAs during EndMT. Cell Signal. 2012;24:1031–1036. doi: 10.1016/j.cellsig.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi E, Sanchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta. 2016;1859:200–208. doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Dudas G, Rambaut A, Andersen KG. The evolution of Ebola virus: Insights from the 2013–2016 epidemic. Nature. 2016;538:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaé N, Dimmeler S. Noncoding RNAs vascular diseases. Circ Res. 2020;126:1127–1145. doi: 10.1161/circresaha.119.315938. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhang F, Ning Q. Losartan reverses the down-expression of long noncoding RNA-NR024118 and Cdkn1c induced by angiotensin II in adult rat cardiac fibroblasts. Pathol Biol (Paris) 2015;63:122–125. doi: 10.1016/j.patbio.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Jin L, et al. AK098656, a novel vascular smooth muscle cell-dominant long noncoding RNA promotes hypertension . Hypertension. 2018;71:262–272. doi: 10.1161/hypertensionaha.117.09651. [DOI] [PubMed] [Google Scholar]
- Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Katayama S, et al. Antisense transcription in the mammalian transcriptome . Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim WC, King D, Lee CH. RNA-cleaving properties of human apurinic/apyrimidinic endonuclease 1 (APE1. Int J Biochem Mol Biol. 2010;1:12–25. [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present and future . Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol. 2015;22:11–19. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with mediator to enhance chromatin architecture and transcription . Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai FW, Stephenson KB, Mahony J, Lichty BD. Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-κB activation . J Virol. 2014;88:54–65. doi: 10.1128/jvi.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Leon-Icaza SA, Zeng M, Rosas-Taraco AG (2019) microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA 1:1–7 [DOI] [PMC free article] [PubMed]
- Li Y, et al. Construction and analysis of lncRNA-lncRNA synergistic networks to reveal clinically relevant lncRNAs in cancer . Oncotarget. 2015;6:25003–25016. doi: 10.18632/oncotarget.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou J, Huang K. Inhibition of the lncRNA Mirt1 attenuates acute myocardial infarction by suppressing NF-κB activation . Cell Physiol Biochem. 2017;42:1153–1164. doi: 10.1159/000478870. [DOI] [PubMed] [Google Scholar]
- Liang H, et al. A novel reciprocal loop between microRNA-21 and TGFβRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. 2012;44:2152–2160. doi: 10.1016/j.biocel.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Liang W, Gao B, Fu P, Xu S, Qian Y, Fu Q. The miRNAs in the pathgenesis of osteosarcoma. Front Biosci (Landmark Ed) 2013;18:788–794. doi: 10.2741/4142. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. Suppressed expression of miR-378 targeting gzmb in NK cells is required to control dengue virus infection. Cell Mol Immunol. 2016;13:700–708. doi: 10.1038/cmi.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JM, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massone S, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2005) Small regulatory RNAs in mammals. Hum Mol Genet 14 Spec No 1:R121-132. 10.1093/hmg/ddi101 [DOI] [PubMed]
- McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsky HC, et al. Zika virus evolution and spread in the Americas. Nature. 2017;546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt 1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikido T, Oyama J, Shiraki A, Komoda H, Node K. Deletion of Apoptosis Inhibitor of Macrophage (AIM)/CD5L attenuates the inflammatory response and infarct size in acute myocardial infarction. J Am Heart Assoc. 2016;5:e002863. doi: 10.1161/jaha.115.002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Ounap K. Silver-Russell Syndrome and Beckwith-Wiedemann Syndrome: Opposite phenotypes with heterogeneous molecular etiology. Mol Syndromol. 2016;7:110–121. doi: 10.1159/000447413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Mahadevan K, Tarnawsky SP. ALREX-elements and introns: two identity elements that promote mRNA nuclear export . Wiley Interdiscip Rev RNA. 2013;4:523–533. doi: 10.1002/wrna.1176. [DOI] [PubMed] [Google Scholar]
- Palmini G, Marini F, Brandi ML (2017) What is new in the miRNA world regarding osteosarcoma and chondrosarcoma? Molecules 22. 10.3390/molecules22030417 [DOI] [PMC free article] [PubMed]
- Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:322–328. [PubMed] [Google Scholar]
- Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks . Biochim Biophys Acta Gene Regul Mech. 2020;1863:194417. doi: 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Bonini NM, Roth KA. Polyglutamine disease and neuronal cell death. Proc Natl Acad Sci U S A. 2000;97:12957–12958. doi: 10.1073/pnas.210395797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, et al. Correction: Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2019;10:3313–3314. doi: 10.18632/oncotarget.26963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge A, Kelsey G, Germain-Lee EL. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J Endocrinol. 2008;196:193–214. doi: 10.1677/joe-07-0544. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF (2012) The origins of the RNA world. Cold Spring Harb Perspect Biol 4. 10.1101/cshperspect.a003608 [DOI] [PMC free article] [PubMed]
- Rogler LE, et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet. 2014;23:368–382. doi: 10.1093/hmg/ddt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A . Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Saçar Demirci MD, Adan A (2020) Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 8:e9369. 10.7717/peerj.9369 [DOI] [PMC free article] [PubMed]
- Scheel TK, et al. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. 2016;19:409–423. doi: 10.1016/j.chom.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-kappaB-dependent manner. FEBS J. 2017;284:1096–1109. doi: 10.1111/febs.14045. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Sparber P, Filatova A, Khantemirova M, Skoblov M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med Genomics. 2019;12:42. doi: 10.1186/s12920-019-0487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan V, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher AT, Weatherall DJ, Cappellini MD. Thalassaemia. Lancet. 2018;391:155–167. doi: 10.1016/S0140-6736(17)31822-6. [DOI] [PubMed] [Google Scholar]
- Tan JY, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21:955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Volders PJ, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yan Q, Zhu C, Guang S, Feng X (2019) The functions of non-coding RNAs in rRNA regulation. Front Genetics 10. 10.3389/fgene.2019.00290 [DOI] [PMC free article] [PubMed]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang W, Cheng SQ, Yang RL. High expression of lncRNA GACAT3 inhibits invasion and metastasis of non-small cell lung cancer to enhance the effect of radiotherapy. Eur Rev Med Pharmacol Sci. 2018;22:1315–1322. doi: 10.26355/eurrev_201803_14473. [DOI] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 2012;4:427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lin Z, Gao Y, Yao T. Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer. J Exp Clin Cancer Res. 2017;36:5. doi: 10.1186/s13046-016-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem. 2019;92:103214. doi: 10.1016/j.bioorg.2019.103214. [DOI] [PubMed] [Google Scholar]
- Zhao J et al (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40:939–953. 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed]
- Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ (2016) Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel) 7. 10.3390/genes7120116 [DOI] [PMC free article] [PubMed]
- Zheng Z, et al. Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J Virol. 2013;87:5645–5656. doi: 10.1128/jvi.02655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Ou Q, Wang G, Zhang W, Hao Y, Li W. High long non-coding RNA NORAD expression predicts poor prognosis and promotes breast cancer progression by regulating TGF-β pathway. Cancer Cell Int. 2019;19:63. doi: 10.1186/s12935-019-0781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. MicroRNA-15b modulates Japanese encephalitis virus-mediated inflammation via targeting RNF 125. J Immunol. 2015;195:2251–2262. doi: 10.4049/jimmunol.1500370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not Applicable.