Abstract
The oxysterol sulfate, 25-hydroxycholesterol 3-sulfate (25HC3S), has been shown to play an important role in lipid metabolism, inflammatory response, and cell survival. However, the mechanism(s) of its function in global regulation is unknown. The current study investigates the molecular mechanism by which 25HC3S functions as an endogenous epigenetic regulator. To study the effects of oxysterols/sterol sulfates on epigenetic modulators, 12 recombinant epigenetic enzymes were used to determine whether 25HC3S acts as their endogenous ligand. The enzyme kinetic study demonstrated that 25HC3S specifically inhibited DNA methyltransferases (DNMTs), DNMT1, DNMT3a, and DNMT3b with IC50 of 4.04, 3.03, and 9.05 × 10−6 M, respectively. In human hepatocytes, high glucose induces lipid accumulation by increasing promoter CpG methylation of key genes involved in development of nonalcoholic fatty liver diseases. Using this model, whole genome bisulfate sequencing analysis demonstrated that 25HC3S converts the 5mCpG to CpG in the promoter regions of 1,074 genes. In addition, we observed increased expression of the demethylated genes, which are involved in the master signaling pathways, including MAPK-ERK, calcium-AMP-activated protein kinase, and type II diabetes mellitus pathways. mRNA array analysis showed that the upregulated genes encoded for key elements of cell survival; conversely, downregulated genes encoded for key enzymes that decrease lipid biosynthesis. Taken together, our results indicate that the expression of these key elements and enzymes are regulated by the demethylated signaling pathways. We summarized that 25HC3S DNA demethylation of 5mCpG in promoter regions is a potent regulatory mechanism.
Supplementary key words: cholesterol metabolites, oxysterol sulfation, oxysterol, oxysterol sulfate, high glucose, MAPK-ERK signaling, calcium-AMPK signaling, DNMT, CpG methylation, metabolic regulation
Abbreviations: AMPK, AMP-activated protein kinase; CREB, cAMP-response element binding protein; DMR, differentially methylated region; DNMT, DNA methyltransferase; DUSP, dual-specificity phosphatase; 25HC, 25-hydroxycholesterol; 27HC, 27-hydroxycholesterol; 25HC3S, 25-hydroxycholesterol 3-sulfate; 27HC3S, 27-hydroxycholesterol 3-sulfate; HDAC, histone deacetylase; HG, high glucose; HMGR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; KEGG, Kyoto Encyclopedia of Genes and Genomes; LINE-1, long interspersed nucleotide element 1; 5mC, 5-methylcytosine; NAFLD, nonalcoholic fatty liver disease; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Xol, cholesterol; Xol3S, cholesterol 3-sulfate
Epigenetic modification plays a major role in the regulation and coordination of gene expression. Methylation at position 5 of cytosine [5-methylcytosine (5mC)] in DNA is an important epigenetic modification that regulates gene expression among other functions of the genome (1). Cytosine methylation of CpG in the promoter region is inversely correlated with transcriptional activity of associated genes as it causes chromatin condensation and thus gene silencing (2). Recently published literature has demonstrated that dysregulation of CpG methylation and gene expression is important in metabolism and can affect tissue function and therefore the metabolic state (3). It has been shown that cytosine methylation is catalyzed by DNA methyltransferases [DNMTs (DNMT1, DNMT3a, and DNMT3b)], which have been reported to play an important role in the regulation of DNA methylation (4, 5). However, the specific molecular-molecular interactions, the regulators of these enzyme activities, and their authentic endogenous ligands, agonist(s), and antagonist(s) are unknown.
The potent regulatory oxysterol sulfate, 25-hydroxycholesterol 3-sulfate (25HC3S; 5-cholesten-3β,25-diol 3-sulfate), has been reported to play important roles in lipid metabolism, inflammatory responses, and cell survival since its discovery in hepatocyte nuclei (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18). Although some other sterol sulfates have been discovered and shown to be important cell regulatory metabolites [cholesterol sulfate, 27-hydroxycholesterol 3-sulfate (27HC3S), 27HCDS, and 25HCDS] (19, 20, 21, 22), 25HC3S appears to be the most potent regulator and has been used as a biomedicine for therapy of acute and chronic diseases, which are at phase II clinical trials (DUR928 represents 25HC3S; www.durect.com). 25HC3S decreases SREBP-1c/SREBP-2 activities and nuclear liver oxysterol receptor levels, subsequently decreasing cholesterol (Xol) and triglyceride accumulation in vitro in hepatocytes and in vivo in liver tissues in nonalcoholic fatty liver disease (NAFLD) mouse models (11, 13). Furthermore, 25HC3S has been shown to increase nuclear PPARγ and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) mRNA levels and downregulate NF-κB, which consequently decreases proinflammatory cytokine expression and secretion, suppresses inflammatory responses, inhibits apoptosis, and promotes cell survival in human macrophages and in vivo lipopolysaccharide-induced acute multiple organ injury in mouse models (7). Although the detailed molecular and biochemical mechanism is not fully understood, its role in global regulation indicates that 25HC3S most likely acts as an endogenous epigenetic regulator. The aim of the current study was to elucidate the molecular mechanism of 25HC3S as an epigenetic ligand by enzyme kinetic analysis and whole genomic analysis. The results showed that the expression of key genes via DNA 5mCpG demethylation in promoter regions is involved in master signaling pathways, which regulate cell stress responses.
Materials and methods
Materials
Cell culture reagents and supplies were purchased from GIBCO BRL (Grand Island, NY); Huh-7 cells were obtained from American Type Culture Collection (Rockville, MD). The reagents for real-time RT-PCR were from AB Applied Biosystems (Warrington, UK). The chemicals used in this research were obtained from Sigma Chemical Co (St. Louis, MO) or Bio-Rad Laboratories (Hercules, CA). All solvents were obtained from Fisher (Fair Lawn, NJ) otherwise indicated.
Cell culture
Huh-7 and HepG-2 cells were cultured in DMEM supplemented with 10% heat-inactivated FBS, high glucose (HG; 4.5 g/l) at 37°C in a humidified atmosphere of 5% CO2.
Extraction and determination of DNA and mRNA levels
After culturing Huh-7 cells in DMEM with HG for 72 h followed by treating with 25 μM 25HC3S for 4 h, genomic DNA from 5,000 cells were extracted using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Each sample, 2 μg, was sent to EpigenDx, Inc (Hopkinton, MA) for analysis of global methylation bisulfite sequencing. The same samples, 6 μg, were sent to Novogene Co, Ltd (Tianjin, China) for analysis of whole genome bisulfite sequencing (WGBS). Total RNA was isolated using the Promega SV total RNA isolation system (Madison, WI) with DNase treatment. Each sample, 2 μg, was used for the first-stranded cDNA synthesis as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Real-time RT-PCR was performed using SYBR Green as the indicator on ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplifications of β-actin or GAPDH were used as internal controls. Relative mRNA expression was quantified with the comparative cycle threshold (Ct) method using the primer set shown in supplemental Table S1 and was expressed as 2−ΔΔCt as described previously (23).
Chemical synthesis and characterization of sterol sulfates, 25HC3S, cholesterol 3-sulfate, and 27HC3S
5-Cholesten-3β,25-diol 3-sulfate (25HC3S); 5-cholesten-3β-ol, 3-sulfate [cholesterol 3-sulfate (Xol3S)]; and 5-cholesten-3β,27-diol 3-sulfate (27HC3S) were synthesized as previously described with mild modification (6). Briefly, a mixture of 25-hydroxycholesterol, Xol, or 27-hydroxycholesterol (6.5 mg, 0.016 mmol) and triethylamine-sulfur trioxide (3.5 mg, 0.019 mmol) was dissolved in dry pyridine (300 μl) and was stirred at room temperature for 2 h. The solvents were evaporated at 40°C under nitrogen gas stream, and the syrup was added into 2 ml of 50% acetonitrile (loading buffer). The products were applied to a 6 cc Oasis cartridge (Waters), which had been primed by methanol (15 ml) and water (15 ml). The cartridge was successively washed with the loading buffer (15 ml), water (15 ml), methanol (15 ml), 50% methanol (15 ml), 5% ammonia hydroxide in 10% methanol (15 ml), and 5% ammonia hydroxide in 50% methanol (15 ml). The retained sulphated sterol was eluted with 5% ammonia hydroxide in 80% methanol (10 ml), respectively. After dilution with 10 times volume of acetonitrile, the solvents were evaporated to dryness under nitrogen gas stream, and the sterol sulphates were obtained in white powder form.
Enzyme kinetic study of 5-cholesten-3β,27-diol 3-sulfate
For the DNMT1 activity assay, the substrate solution, 0.001 mg/ml poly(dI-dC):poly(dI-dC) in 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 5 mM DTT, 1 mM PMSF, 5% glycerol, 0.01% Brij35, and 1% DMSO were used. For the DNMT3a/DNMT3b activity assay, 0.0075 mg/ml lambda DNA in 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 5 mM DTT, 1 mM PMSF, 5% glycerol, and 1% DMSO were used. The indicated DNMT1, DNMT3a, and DNMT3b were added to the appropriate substrate solution and gently mixed. Amounts of Xol, 25-hydroxycholesterol (25HC), 27-hydroxycholesterol (27HC), Xol3S, 25HC3S, or 27HC3S ranging from 5.08E-09 to 0.0001 M in DMSO were added to the reaction mixture by using Acoustic Technology (Echo 550; LabCyte Inc, Sunnyvale, CA). The mixtures were first incubated for 15 min, then adenosyl-l-methionine, S-[methyl-3H] was added to the reaction mixture to initiate the reaction, and the mixture was incubated for 60 min at 30°C. Following incubation, the reaction mixture was finally transferred to filter paper for detection of radioactivity counts.
Analysis of global methylation, long interspersed nucleotide element 1 assay
For global DNA methylation analysis, 500 ng of extracted genomic DNA was treated with bisulfite using the EZ DNA Methylation kit (Zymo Research, Inc, CA). PCR and product purification were performed as per the manufacturer's protocol (GE Healthcare Life Sciences). The PCR products, 10 μl, were sequenced by pyrosequencing on the PSQ96 HS System following the manufacturer's instructions (Pyrosequencing, Qiagen). The methylation status of each CpG site was determined individually as an artificial C/T SNP using QCpG software (Pyrosequencing, Qiagen). The methylation level at each CpG site was calculated as the percentage of the methylated alleles divided by the sum of all methylated and unmethylated alleles. The mean methylation level was calculated using methylation levels of all measured CpG sites within the targeted region of each gene. Each experiment included non-CpG cytosines as internal controls to detect incomplete bisulfite conversion of the input DNA. In addition, a series of unmethylated and methylated DNA were included as controls in each PCR assay. Furthermore, PCR bias testing was performed by mixing unmethylated control DNA with in vitro methylated DNA at different ratios (0, 5, 10, 25, 50, 75, and 100%), followed by bisulfite modification, PCR, and pyrosequencing analysis.
Analysis of human WGBS
Each sample, 5.2 μg of genomic DNA spiked with 26 ng lambda DNA, was fragmented by sonication to 200–300 bp with Covaris S220, followed by end repair and adenylation. Cytosine-methylated barcodes were ligated to sonicated DNA per the manufacturer's instructions. These DNA fragments were treated twice with bisulfite using EZ DNA Methylation-Gold TM Kit (Zymo Research) before the resulting single-stranded DNA fragments were PCR amplified using KAPA HiFi Hot Start Uracil and Ready Mix (2X). Library concentration was quantified by Qubit® 2.0 Flurometer (Life Technologies, CA) and quantitative PCR, and the insert size was assayed on an Agilent Bioanalyzer 2100 system (Agilent Technologies).
The library preparations were sequenced on an Illumina Hiseq 2500/4000 or NovaSeq platform, and 125/150 bp paired-end reads were generated. Image analysis and base calling were performed with Illumina CASAVA pipeline. Trimmomatic (Trimmomatic-0.36, Bjorn Usadel Lab) software was used for quality control. Bismark software (version 0.16.3; Krueger F., 2011) was used to perform alignments of bisulfite-treated reads to a reference genome (-X 700 --dovetail). DSS software (Bioconductor) (24) was used to identify differentially methylated regions (DMRs). KOBAS software (Center for Bioinformatics, Peking University) (25) was used to test the statistical enrichment of DMR-related genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Transcriptional profiling and data analysis
Total RNA was extracted and purified from HepG-2 cells using SV Total RNA Isolation System (Promega, Madison, WI). cDNAs were prepared and analyzed using GeneChip® Human Genome U133 Plus 2.0 Array, Affymetrix (Santa Clara, CA) as previously described with technical support from Shanghai Biotechnology Corporation. Direct target genes in the present study were selected based on more than 2-fold of reduction together with array detect signal more than 5-fold in both samples. Genes showing fold changes greater than 2 and array-detected signals greater than 7 in at least one sample were selected as differently expressed genes. DAVID software (https://david.ncifcrf.gov/conversion.jsp) was used to analyze gene ontological enrichment of differently expressed genes.
Results
25HC3S specifically inactivates DNMT activities
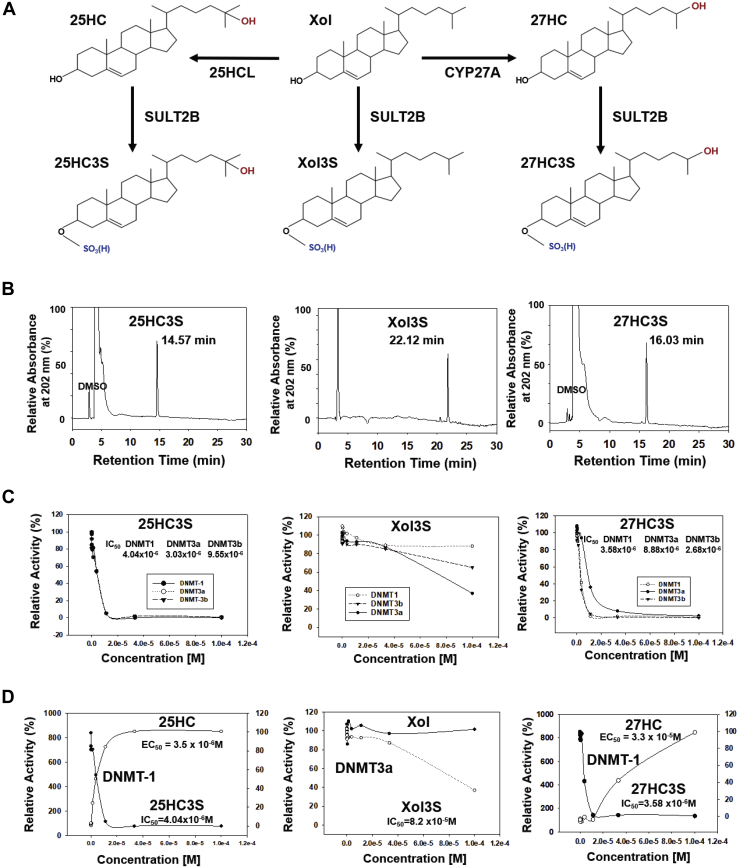
In order to study the effects of sterol sulfates on the epigenetic regulating targets, 25HS3S, Xol3S, and 27HC3S (Fig. 1A) were synthesized and purified to more than 95% purity using triethylamine sulphate complex methods as shown in Fig. 1B. The major epigenetic regulation includes DNA and histone methylation, demethylation, acetylation, and deacetylation (26). The enzymes involved in the process are DNA and histone methyltransferases/demethylases, and acetyltransferases/deacetylases, although their detailed mechanisms of regulation are not completely understood (27). To study the effects of nuclear oxysterols/sterol sulfates on epigenetic modulators, 12 recombinant enzymes, DNMT1, DNMT3a, DNMT3b, giant congenital nevi 3, p300 (histone acetyltransferase), PCAF (KAT2B lysine acetyltransferase 2B), histone deacetylase 1 (HDAC1), HDAC2, HDAC3, HDAC6, HDAC10, and KDM6B-JMJD3 (lysine demethylase 6B), were used to determine whether 25HC and 25HC3S, 27HC and 27HC3S, or Xol and Xol3S act as their endogenous ligand(s). Results show that 25HC3S significantly inhibits only DNMT-1, DNMT-3a, and DNMT-3b activities with IC50 = 4.04, 3.03, and 9.05 × 10−6 M, respectively (Fig. 1C, left), while its precursor 25HC activates DNMT-1 activity by 8-fold with EC50 = 3.5 × 10−6 M (Fig. 1D, left). As controls, Xol as well as Xol3S did not significantly affect enzymatic activities although Xol3S slightly inhibits DNMT3a with IC50 = 8.2 × 10−5 M, which is most likely not physiologically significant (Fig. 1C, middle). Compared with 25HC3S, 27HC3S did inhibit DNMTs with similar IC50 = 3.58 × 10−6 M for DNMT1, 8.88 × 10−6 M for DNMT3a, and 2.68 × 10−6 M for DNMT3b as shown in Fig. 1C (right). In contrast, its precursor 27HC was much less potent in activation of DNMT-1 with EC50 = 3.3 ×10−5 M and had no effect on other enzymes (Fig. 1D, right). In contrast to the three DNMTs, the nine other epigenetic enzymes are not affected by these oxysterols or sterol sulfates (data not shown). As a positive control, S-adenosyl homocysteine inhibited DNMT1 activity by 95% at 1 μM (data not shown), as previously reported (28). The results demonstrated that both 25HC3S and 27HC3S are potent inhibitors of DNMTs. However, only 25HC3S has been discovered in vivo in human hepatocyte nuclei: first found in concentration of 20 μg/g (∼40 μM) following overexpression of mitochondrial Xol delivery protein, StarD1 (17). The kinetic study shows that the IC50s are between 1 and 10 μM. Recently, 25HC3S (DUR928) has been used in clinical trial and shown to have physiological and pharmaceutical significance at an oral dose (30–300 mg/person). In addition, 27HC3S has been detected in sera of patients with sterol sulfatase deficiency (29). Its concentration within cells has never been determined. Furthermore, its true physiological function is unknown.
Fig. 1.
Synthesis and enzyme kinetic studies of Xol3S, 25HC3S, and 27HC3S. A: The biosynthesis of Xol3S, 25HC3S, and 27HC3S in the cells. B: HPLC profile of purified 25HC3S, Xol3S, and 27HC3S. C: Effects of 25HC3S, 27HC3S, Xol3S, and their precursors, 25HC, 27HC, and cholesterol, on DNMT1/DNMT3a/DNMT3b activities. The concentration dependent, 0–0.001 M (10 points), effects of 25HC3S, Xol3S, and 27HC3S on the enzyme activities. D: Comparison of 25HC with 25HC3S, cholesterol with Xol3S, and 27HC with 27HC3S are shown. The results in (B) represent one of the five experiments; (C) and (D), one of three experiments. 25HC, EC50 = 2.33 ± 1.38 × 10−6 M (DNMT1; mean ± SD; n = 3); 25HC3S, IC50 = 3.35 ± 2.68 × 10−6 M (DNMT1; mean ± SD; n = 3).
25HC3S decreases 5mCpG levels in global promoter regions
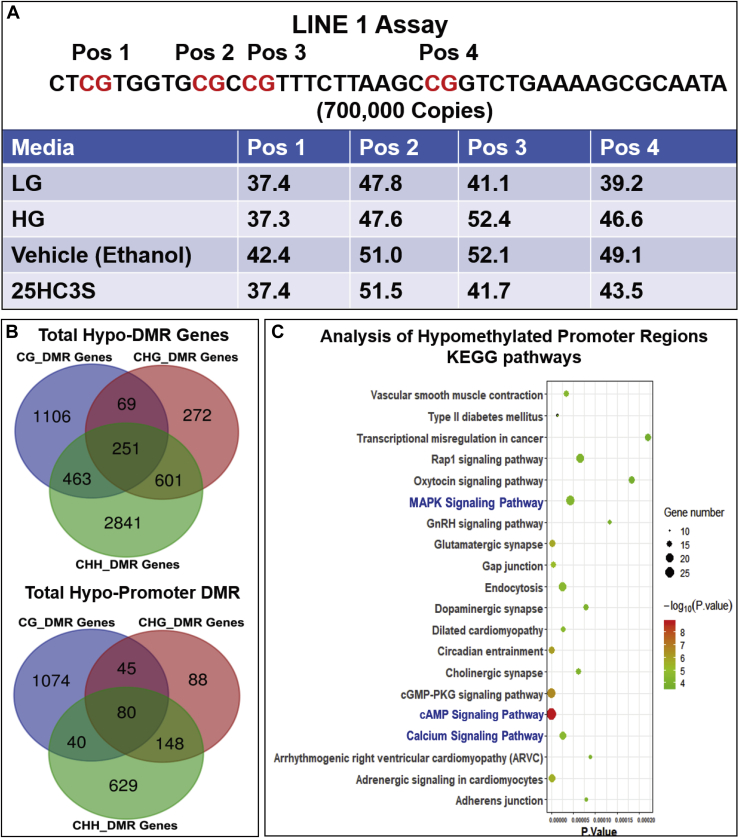
Previous studies have shown that global DNA methylation and the methylation of specific genes are involved in adipogenesis (30), lipid metabolism (31, 32), and inflammation in visceral adipose tissues (33), which, in turn, are related to the specific etiology of metabolic syndrome. To study the effects of 25HC3S on methylation status of 5mCpG in global promoter regions, long interspersed nucleotide element 1 (LINE-1) analysis was first performed to estimate global demethylation. Methylation usually occurs in repetitive elements, such as LINE elements. There are ∼500,000 LINE elements and 750 million copies in total human genome. Human LINE-1 is a retrotransposable region (promoter region) and has only 700,000 copies, which correlates to ∼17% of the human genome (34). The specific sequence includes four CpG dinucleotides (Pos 1, 2, 3, and 4), which serve as methylation/demethylation targets in LINE-1. As shown in Fig. 2A, culturing Huh-7 cells in HG media, Pos 3 and Pos 4 had higher methylation, whereas all four Pos increased methylation after culturing cells in ethanol control. Reduction of methylation (demethylation) at Pos1 (−5%), Pos3 (−10%), and Pos 4 (−5.6%) occurred after incubating cells with 25HC3S for 4 h. The results indicate that 25HC3S significantly reduce 5mCpG methylation in promoter regions induced by HG or ethanol.
Fig. 2.
Effects of 25HC3S on DNA methylation in hepatocytes by global methylation sequencing analysis. Huh-7 cells were cultured in HG media for 72 h and then treated with 25 mM 25HC3S for 4 h. The levels of global methylation were estimated by LINE-1 assay. A: Four CpG sites in promoter regions of LINE-1 element were chosen as the target positions as shown. Detailed global methylation was measured by WGBS. B: Venn diagrams of hypomethylated DMR-associated genes (DMGs) in 25HC3S and vehicle libraries under CG, CHG, and CHH contexts of whole genome (up) and promoter regions (low). C: KOBAS software was used to test the statistical enrichment of DMR-related genes in the KEGG pathways. High enrichment of hypomethylated DMRs in promoter regions in KEGG pathways. The detailed KEGG pathways are shown in supplemental Table S3.
Profiles of whole genome-wide DNA methylation in 25HC3S-treated human hepatocytes
To understand the possible cellular functions of 5mCpG demethylation in 25HC3S-treated Huh-7 cells, the cells were harvested for the construction of bisulfite-treated genomic DNA libraries. In these two libraries, more than 80% of cytosine residues were covered by at least 10 reads in “human reference genome (hg38)”. The depth and density of the sequencing were enough for a high-quality genome-wide methylation analysis. Meanwhile, the efficiencies of bisulfite conversion, represented by the lambda DNA to the libraries, were over 99%, providing reliable and accurate results for the WGBS (supplemental Table S2).
CpG methylation and demethylation are well documented to relate with gene expression (35). A total of about 7,136 DMRs under CG context were identified as hypomethylated regions located in 1,106 genes (differential methylated genes). In 97% (1,074) of the differential methylated genes, the hypomethylated regions were identified in their promoters (Fig. 2B). The hypomethylated genes were highly enriched in 75 KEGG pathways (P < 0.05) (supplemental Table S3). The top 20 pathways (from the most significant, P < 10−9) were shown in Fig. 2C. Among these pathways, MAPK-ERK and calcium-cAMP signaling are believed as the master pathways regulating cell survival, antioxidants, antiapoptosis (36, 37), energy metabolism, and lipid homeostasis (38, 39). Previous report has showed that HG incubation, an in vitro model for study of NAFLD, induces lipid accumulation via increasing DNA promoter methylation signaling (28). It was noted that the hypermethylated 5mCpG in the promoter regions induced by HG were demethylated by 25HC3S. 25HC3S demethylated 5mCpG in promoter regions of 23 genes in MAPK signaling pathway (Table 1), 19 genes in calcium pathway (Table 2), and 28 genes in cAMP pathway (Table 3). Interestingly, none of the hypermethylated DMRs was significantly enriched in any KEGG signaling pathway. The chromosome and sequence location of the hypermethylated 5mCpG by HG and the hypomethylated CpG by 25HC3S in promoter regions are compared in the tables. It is observed that these genes are also involved in many other KEGG pathways including insulin, type II diabetes mellitus, and cyclic guanosine monophosphate-protein kinase G signaling pathways. The results indicate that the global regulatory mechanisms of 25HC3S are through demethylation of 5mCpG in promoter regions of the key genes, such as the dual-specificity phosphatase (DUSP) and calcium channel families, involved in MAPK-ERK and calcium-cAMP master signaling pathways.
Table 1.
Demethylation of 5mCpG in promoter regions of MAPK signaling genes
| Gene Name | DMR Location in Promoter Region |
DMR (Methylation %) |
|||
|---|---|---|---|---|---|
| Chromosome | Start | End | HG-LG | 25HC3S-Vehicle | |
| RAC3 | Chr17 | 82031108 | 82031736 | −26.18 | |
| MAPK8 | Chr10 | 48306510 | 48306561 | −29.24 | |
| DUSP8 | Chr11 | 1572973 | 1573032 | −30.51 | |
| 1573583 | 1573855 | +41.72 | |||
| MAX | Chr14 | 65102273 | 65102358 | −31.75 | |
| DUSP7 | Chr3 | 52056416 | 52056485 | −40.47 | |
| 52056538 | 52056778 | +31.75 | |||
| NTF3 | Chr12 | 5432796 | 5432882 | −33.48 | |
| CACNA1D | Chr3 | 53494583 | 53494705 | −20.46 | |
| 53493469 | 53494019 | +49.03 | |||
| CACNA1H | Chr16 | 1194928 | 1195015 | −26.48 | |
| 1194670 | 1195215 | +41.26 | |||
| CACNA1A | Chr19 | 13226635 | 13226711 | −39.91 | |
| 13238959 | 13239237 | +9.51 | |||
| MAPK1 | Chr22 | 21867450 | 21867512 | −7.66 | |
| 21867333 | 21867621 | +20.15 | |||
| HRAS | Chr11 | 535071 | 535127 | −18.85 | |
| 536242 | 537214 | +42.8 | |||
| PDGFB | Chr22 | 39243967 | 39244084 | −30.51 | |
| 39242292 | 39242477 | +29.29 | |||
| CACNA1C-AS1 | Chr12 | 2691335 | 2691438 | −20.97 | |
| CACNB2 | Chr10 | 18140401 | 18140547 | −21.91 | |
| 18340356 | 18341053 | +47.13 | |||
| MAP4K1 | Chr19 | 38596260 | 38596495 | −34.3 | |
| RAPGEF2 | Chr4 | 1.59E+08 | 1.59E+08 | −36.05 | |
| CTB-186G2.1 | Chr19 | 38596260 | 38596495 | −34.3 | |
| RELA | Chr11 | 65661914 | 65662035 | −35.1 | |
| GADD45B | Chr19 | 2474806 | 2474873 | −28.6 | |
| AL671762.1 | Chr6 | 31828019 | 31828149 | −27.17 | |
| CACNG8 | Chr19 | 53961377 | 53961479 | −31.18 | |
| MAP4K4 | Chr2 | 1.02E+08 | 1.02E+08 | −13.63 | |
| 1.02E+08 | 1.02E+08 | +17.93 | |||
| TGFB3 | Chr14 | 75982395 | 75982557 | −23.74 | |
LG, low glucose.
After culturing Huh-7 cells in DMEM with HG for 72 h followed by treating with ethanol (vehicle) and 25 μM 25HC3S for 4 h, genomic DNA from 5,000 cells were extracted using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Each sample (6 μg) was used for analysis of the WGBS. The KEGG analysis shows that the demethylated genes are involved in MAPK signaling pathway (P = 0.00087). Of the 257 total genes in the MAPK signaling pathway, 23 were demethylated by the 25HC3S treatment. Of these 23 genes, 10 were found to be methylated by an HG environment (shown in bold). The first column represents the gene name, the second column (DMR location in promoter region) shows the location of differential methylation region in the chromosome, and the third column [DMR (methylation %)] shows the methylation rates by HG and demethylation rates induced by 25HC3S.
Table 2.
Demethylation of 5mCpG in promoter regions of calcium signaling genes
| Gene Name | DMR Location in Promoter Region |
DMR (Methylation %) |
|||
|---|---|---|---|---|---|
| Chromosome | Start | End | HG-LG | 25HC3S-Vehicle | |
| CACNA1A | Chr19 | 13226635 | 13226711 | −39.91 | |
| 13238959 | 13239237 | +9.51 | |||
| DRD5P2 | Chr1 | 1.43E+08 | 1.43E+08 | −34.73 | |
| EDNRB | Chr13 | 77919496 | 77919743 | −34.09 | |
| ADRB1 | Chr10 | 1.14E+08 | 1.14E+08 | −33.1 | |
| GRIN2A | Chr16 | 10183302 | 10183414 | −19.63 | |
| 10084383 | 10084724 | +8.62 | |||
| GNA11 | Chr19 | 3094383 | 3094442 | −32.66 | |
| 3092574 | 3092876 | +6.13 | |||
| SPHK1 | Chr17 | 76383180 | 76383291 | −24.74 | |
| CACNA1C-AS1 | Chr12 | 2691335 | 2691438 | −20.97 | |
| HRH1 | Chr3 | 11154292 | 11154354 | −25.48 | |
| ITPKB | Chr1 | 2.27E+08 | 2.27E+08 | −25.12 | |
| 2.27E+08 | 2.27E+08 | +23.84 | |||
| PLCD1 | Chr3 | 38029863 | 38030016 | −24.31 | |
| 38029901 | 38030042 | +26.02 | |||
| GNAS | Chr20 | 58888598 | 58888893 | −25.68 | |
| 58888560 | 58888756 | +23.2 | |||
| AC005264.2 | Chr19 | 3156392 | 3156457 | −15.76 | |
| CACNA1D | Chr3 | 53494583 | 53494705 | −20.46 | |
| 53493469 | 53494019 | +49.03 | |||
| GNAQ | Chr9 | 78032198 | 78032440 | −11.92 | |
| GRIN2C | Chr17 | 74861739 | 74861853 | −37.6 | |
| 74854901 | 74855100 | +36.66 | |||
| SLC8A3 | Chr14 | 70188747 | 70189089 | −28.51 | |
| 70046033 | 70046320 | +65.66 | |||
| CACNA1H | Chr16 | 1194928 | 1195015 | −26.48 | |
LG, low glucose.
Preparation of cells and DNA methylation are as described in Table 1. The KEGG analysis shows that the demethylated genes are involved in calcium signaling pathway (P = 0.00066). Of the 180 total genes in the calcium signaling pathway, 19 were demethylated by the 25HC3S treatment. Of these 19 genes, 10 were found to be methylated by an HG environment (shown in bold). The first column represents the gene name, the second column (DMR location in promoter region) shows the location of differential methylation region in the chromosome, and the third column [DMR (methylation %)] shows the methylation rates by HG and demethylation rates induced by 25HC3S.
Table 3.
Demethylation of 5mCpG in Promoter Regions of cAMP Signaling Genes
| Gene Name | DMR Location in Promoter Region |
DMR (Methylation %) |
|||
|---|---|---|---|---|---|
| Chromosome | Start | End | HG-LG | 25HC3S-Vehicle | |
| RAC3 | Chr17 | 82031108 | 82031736 | −26.18 | |
| MAPK8 | Chr10 | 48306510 | 48306561 | −29.24 | |
| PTCH1 | Chr9 | 95508446 | 95508561 | −12.09 | |
| ADCY5 | Chr3 | 1.23E+08 | 1.23E+08 | −30.44 | |
| 1.23E+08 | 1.23E+08 | +15.8 | |||
| GLI3 | Chr7 | 42228171 | 42228223 | −19.62 | |
| 42184839 | 42185064 | +8.27 | |||
| PPP1CB | Chr2 | 28751716 | 28751798 | −10.93 | |
| 28793684 | 28794075 | +7.5 | |||
| GNAS | Chr20 | 58888598 | 58888893 | −25.68 | |
| 58888560 | 58888756 | +23.2 | |||
| CACNA1D | Chr3 | 53494583 | 53494705 | −20.46 | |
| 53493469 | 53494019 | +49.03 | |||
| GRIN2A | Chr16 | 10183302 | 10183414 | −19.63 | |
| MAPK1 | Chr22 | 21867450 | 21867512 | −7.66 | |
| 21867333 | 21867621 | +20.15 | |||
| GABBR2 | Chr9 | 98708549 | 98708610 | −21.84 | |
| CREB5 | Chr7 | 28489448 | 28489601 | −29.31 | |
| 28776194 | 28776325 | +27.34 | |||
| GRIN3B | Chr19 | 1000331 | 1000405 | −34.67 | |
| CACNA1C-AS1 | Chr12 | 2691335 | 2691438 | −20.97 | |
| VAV2 | Chr9 | 1.34E+08 | 1.34E+08 | −33.11 | |
| 1.34E+08 | 1.34E+08 | +44.31 | |||
| ABCC4 | Chr13 | 95301634 | 95301736 | −33.06 | |
| PDE4D | Chr5 | 59893498 | 59893718 | −27.84 | |
| 59215742 | 59215941 | +39.66 | |||
| ROCK2 | Chr2 | 11344628 | 11344733 | −23.14 | |
| ADCY1 | Chr7 | 45574500 | 45574591 | −14.76 | |
| 45574683 | 45574877 | +25.79 | |||
| ADCY4 | Chr14 | 24334570 | 24334719 | −18.63 | |
| 24334404 | 24334719 | +32.6 | |||
| DRD5P2 | Chr1 | 1.43E+08 | 1.43E+08 | −34.73 | |
| ADRB1 | Chr10 | 1.14E+08 | 1.14E+08 | −33.1 | |
| GIPR | Chr19 | 45668710 | 45668767 | −10.04 | |
| 45669075 | 45669744 | +43.62 | |||
| RELA | Chr11 | 65661914 | 65662035 | −35.1 | |
| AFDN | Chr6 | 1.68E+08 | 1.68E+08 | −30.68 | |
| BAD | Chr11 | 64286088 | 64286179 | −42.33 | |
| ATP1A3 | Chr19 | 41999134 | 41999316 | −15.75 | |
| GRIN2C | Chr17 | 74861739 | 74861853 | −37.64 | |
| 74854901 | 74855100 | +36.66 | |||
LG, low glucose.
Preparation of cells and DNA methylation are as described in Table 1. The KEGG analysis shows that the demethylated genes are significantly involved in cAMP signaling pathway (P = 3.69E-07). Of the 200 total genes in the cAMP signaling pathway, 28 were demethylated by the 25HC3S treatment. Of these 28 genes, 13 were found to be methylated by an HG environment (shown in bold). The first column represents the gene name, the second column (DMR location in promoter region) shows the location of differential methylation region in the chromosome, and the third column [DMR (methylation %)] shows the methylation rates by HG and demethylation rates induced by 25HC3S.
Relationship between 5mCpG demethylation in promoter regions and gene expression: 25HC3S decreases HG-induced 5mCpG levels in promoter regions
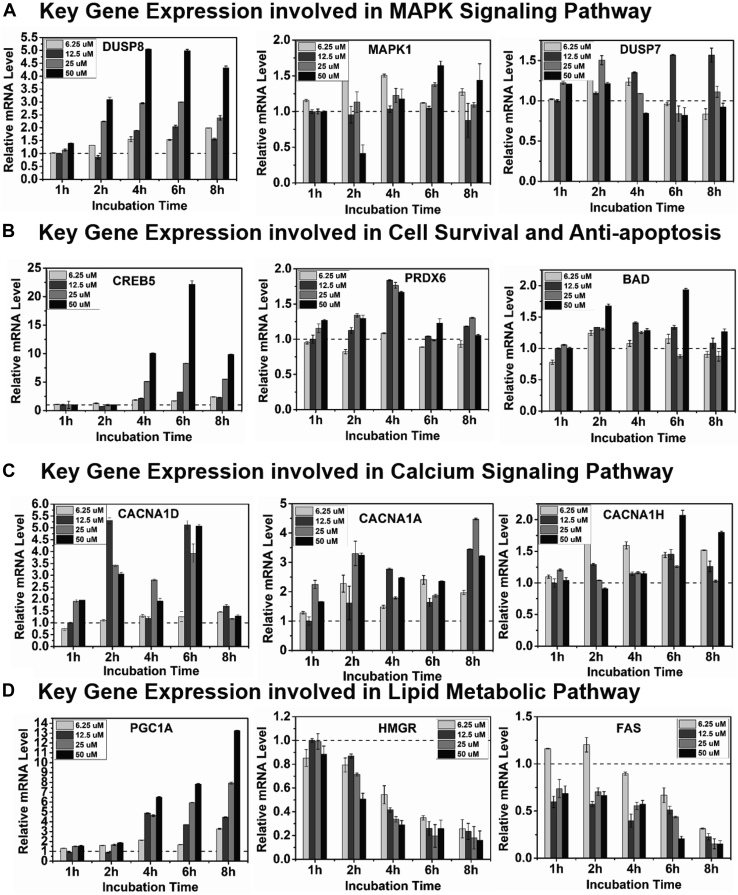
To explore the relationship of promoter 5mCpG demethylation and gene expression from the results of KEGG pathway analysis, the expression of key genes (DUSP7, DUSP8, and MAPK1) and their target genes cAMP response element-binding protein 5 (CREB5), peroxiredoxin 6, and BCL2-associated agonist of cell death in the MAPK pathway, as well as the key genes CACNA1D (calcium voltage-gated channel subunit alpha1 D,), CACNA1A (calcium voltage-gated channel subunit alpha1 A), and CACNA1H (calcium voltage-gated channel subunit alpha1 H) (encoding for calcium voltage-gated channel subunits) and their targeting genes [PGC1A, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR), and FAS] in the calcium-AMP-activated protein kinase (AMPK) pathway were determined by RT-PCR analysis. The DUSP-MAPK signaling pathway is the major pathway involved in cell survival/death and antioxidization (37, 40), and the calcium signaling pathway controls lipid and energy metabolism (38, 39). As expected, 25HC3S increased expression of DUSP8 by 5-fold and its targeting gene, CREB5, by up to 20-fold, which is the key element involved in cell survival and death (Fig. 3A, B). Meanwhile, 25HC3S treatment significantly increased expression of key genes involved in the calcium signaling pathway, and its downstream element, PGC1A, by 12-fold, whereas it decreased expression of HMGR and FAS genes by ∼90%, which encode the key enzymes controlling energy metabolism in mitochondria, Xol biosynthesis, and fatty acid biosynthesis, as shown in Fig. 3C, D.
Fig. 3.
A: Expression of key genes related to signaling pathways. Huh-7 cells were cultured in HG media for 72 h and treated with 25HC3S at 6.25, 12.5, 25, and 50 μM for 1, 2, 4, 6, and 8 h. Key genes and their targeting gene expression were determined by RT-PCR analysis. The expressions of DUSP8, DUSP7, and MAPK1 in MAPK signaling pathway are shown. B: Their target genes, CREB5, PRDX6, BAD. C: Key genes, CACNA family, in calcium-AMPK pathway. D: Their targeting genes PGC1A (PPARG coactivator 1 alpha), HMGR, and FAS. The graphs represent one of three experiments. Each value represents the mean of triplicate determination ± SD.
Transcriptional array analysis in hepatocytes
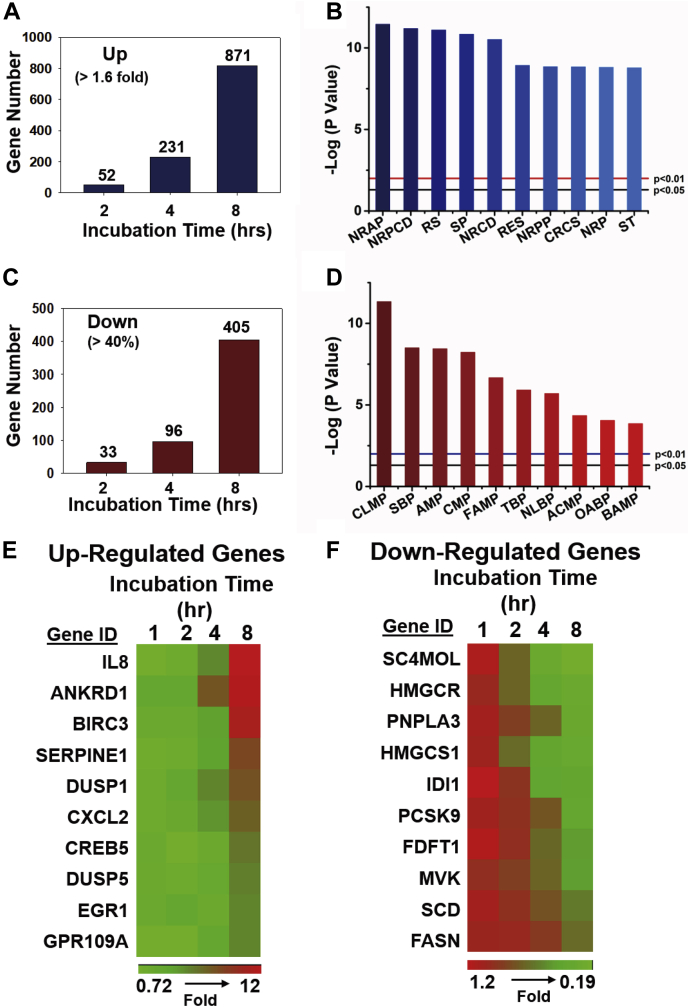
To examine the effect of 25HC3S on whole gene expression in human hepatocytes, GeneChip Human Genome U133A Plus 2.0 Array analysis of 38,500 full-length genes and expressed sequence tag clusters showed that treatment with 25HC3S in HpG-2 cells significantly modulated many clusters of gene expressions. The major clusters affected are genes involved in Xol and triglyceride metabolism, cell survival, and inflammation. Genes associated with Xol and triglyceride biosynthesis were significantly downregulated, whereas genes associated with cell survival, proliferation, and antioxidization were significantly upregulated as shown in Fig. 4. Altogether, 25HC3S modulated the transcription of 1,276 genes (>1.6-fold) in a time-dependent manner. Genetic analysis of different gene ontological processes, a collection of genes associated with a specific biological functional process, revealed that at 8 h, the majority of upregulated pathways are involved in cell survival (Fig. 4A, B); in contrast, majority of downregulated genes are involved in lipid metabolism (Fig. 4C, D). The upregulated genes related with antiapoptosis (increased by 3- to 12-fold at 8 h) are listed in Fig. 4E; and the downregulated genes related with lipid metabolism (decreased by 50–95%) are listed in Fig. 4F. The detailed individual upregulated genes are listed in supplemental Table S4; the downregulated genes are listed in supplemental Table S5. Many studies have shown that epigenetic modification could globally regulate gene expression involved in vital cellular functions, including metabolism, inflammation, and cell death/proliferation (41, 42). Our data demonstrate that 25HC3S epigenetically regulates gene expressions via DNA 5mCpG demethylation in promoter regions.
Fig. 4.
Effect of 25HC3S on transcription levels in hepatocytes. HepG-2 cells were cultured in HG media and treated with 25 μM of 25HC3S for 2, 4, and 8 h. A: The upregulated genes (>1.6-fold) are shown. B: Enrichment of upregulated genes (8 h) to gene ontological groups are shown. C: Downregulated genes (reduction >40%). D: Enrichment of downregulated genes (8 h) to gene ontological groups. E: Heat map of upregulated genes related to this study as shown. F: Heat map of downregulated genes related to this study are shown. ACMP, acyl-CoA metabolic process; AMP, alcohol metabolic process; BAMP, bile acid metabolic process; CLMP, cellular lipid metabolic process; CMP, cholesterol metabolic process; CRCS, cellular response to chemical stimulus; FAMP, fatty acid metabolic process; NLBP, neutral lipid biosynthetic process; NRAP, negative regulation of apoptotic process; NRCD, negative regulation of cell death; NRP, negative regulation of phosphorylation; NRPCD, negative regulation of programmed cell death; NRPP, negative regulation of protein phosphorylation; OABP, organic acid biosynthetic process; RES, response to stress; RS, regulation of signaling; SBP, steroid biosynthetic process; SP, regulation of phosphorylation; ST, signal transduction; TBP, triglyceride biosynthetic process.
Discussion
This study uncovered the mechanisms of 25HC3S, an endogenous and novel intracellular regulatory molecule, and demonstrated that 25HC3S epigenetically regulates lipid metabolism, cell survival, and inflammatory responses via DNA 5mCpG demethylation by inhibiting DNMTs. Unlike oxysterols that are known for activating DNMT-1, the sulfated oxysterol 25HC3S specifically inhibited DNMT-1, DNMT-3a, and DNMT-3b, which demethylated 5mCpG in promoter regions, increased gene expression, and upregulated master signaling pathways, MAPK, calcium, AMPK, and CREB signaling pathways. The results from this study suggested an important regulatory mechanism by which intracellular oxysterols and oxysterol sulfates cooperatively regulate critical cell signaling pathways at transcriptional levels in the nuclei. The results are consistent with all our previous findings that 25HC3S plays in the global regulation (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17). Oxysterol sulfation appears to play equally important physiological and pathophysiological roles as protein phosphorylation, inositol phosphorylation, and sphingosine phosphorylation in regulating cellular functions. However, the signaling activities of the pathways that are induced at the transcript level by 25HC3S have not been directly measured. The concept represents a new complex field, and we are far away from fully understanding how exactly the epigenetic modification affects gene expression.
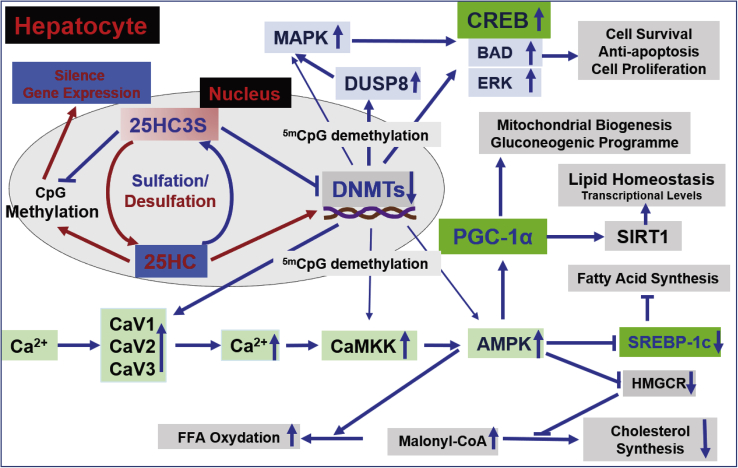
25HC3S decreases lipid accumulation and suppresses inflammatory responses in vitro in hepatocytes and in vivo NAFLD models (6, 7, 8, 9, 10, 13). Incubation of hepatocytes in HG has been widely used as an in vitro model for study of NAFLD (43, 44). A recent report shows that HG induces lipid accumulation via increasing nuclear 25HC levels and DNA CpG methylation in promoter regions of key genes involved in insulin, type II diabetes, and NAFLD signaling pathways in human hepatocytes (28). The present study shows that the addition of 25HC3S to human hepatocytes reversed the methylation induced by HG, increased hypomethylated CpG in promoter regions of the key genes, and increased their targeting gene expression. The results indicate that the CpG demethylation by 25HC3S is the mechanism for its function of global regulation: decreasing lipid accumulation, anti-inflammatory responses, antioxidants, and anticell death, which benefit recovery from NAFLD.
The DUSP family is a subset of protein tyrosine phosphatases, many of which dephosphorylate MAPKs and hence are referred to as MAPK phosphatases (45). DUSP8, a unique member of DUSP family, plays an important role in signal transduction of the phosphorylation-mediated MAPK pathway, which regulates responses to oxidative stress and cell death signals in various human diseases (37). The results from this study showed that 25HC3S greatly demethylated 5mCpG in promoter regions of DUSP genes, including DUSP8, DUSP1, and DUSP7, and their downstream genes, CREB5, peroxiredoxin 6, BCL2-associated agonist of cell death, and ERK, and increased their expression. The transcribed proteins from these genes are response for cell survival and proliferation (46, 47). Therefore, the effects of 25HC3S on promoting cell survival/proliferation and alleviating oxidative stress occur through inhibiting DNMTs and increasing expression of the DUSP family, especially DUSP8 and their downstream elements, as shown in Fig. 5. The results are consistent with our previous reports that 25HC3S promoted hepatic regeneration, minimized multiple organ injury, and decreased mortality in mouse disease models (8).
Fig. 5.
Sulfation of 25HC as an epigenetic regulatory pathway. 25HC is an endogenous agonist of DNMT-1 that methylates CpG in promoter regions and subsequently silences gene expression, resulting in cell death and lipogenesis. 25HC can be sulfated to 25HC3S, which acts as an endogenous ligand and inhibits activities of DNMTs. 25HC3S demethylates 5mCpG in promoter regions and successively increases gene expression. The eminent pathways regulated by the sulfation of oxysterol are involved in energy and lipid metabolisms, MAPK-ERK, and calcium-AMPK. 25HC3S significantly increases DUSPs and CREB expressions, which activate MAPK-ERK pathway, including CREB, BAD, and ERK, and subsequently regulate cell survival and death. 25HC3S decreases lipid biosynthesis and reduces lipid accumulation by demethylating 5mCpG in promoter regions, increasing expression of key genes involved in calcium channels and AMPK, and activating corresponding signaling pathways, which result in increased oxidation of FFA, and decreased biosynthesis of cholesterol and FFA. The global regulation by sulfation of oxysterol suggests the physiological and pathophysiological significance of this regulatory mechanism.
The calcium, AMPK, and PPAR signaling pathways are the master ones involved in regulation of energy, lipids, and carbohydrate metabolisms (38, 48). The Ca2+/calmodulin-dependent protein kinase and AMPK signaling pathway increases expression and decreases acetylation of PGC-1α, which regulates mitochondrial biogenesis and lipid metabolism (49). The present data from analysis of whole genome-wide DNA methylation (genomic level) and transcriptional analysis of GeneChip Human Genome U133Aplus2.0 Array (mRNA level) showed that 25HC3S treatment significantly demethylated 5mCpG in the promoter regions of key genes including calcium channels, as well as genes of Ca2+/calmodulin-dependent protein kinase and AMPK, increased their expression, and modulated downstream elements. The results provided convincing evidence that 25HC3S globally regulated metabolic pathways mainly via the calcium-AMPK signaling pathway as shown in Fig. 5. Interestingly, 25HC and 25HC3S are potent modulators in regulating DNA methylation. 25HC methylates CpG, and 25HC3S demethylates 5mCpG, while also downregulating and upregulating expression of the key genes. PGC-1α is a key regulator of mitochondrial biogenesis, oxidative phosphorylation, and mitochondrial antioxidant defense, and it is also responsible for maintaining metabolic homeostasis (50). PGC-1α expression is upregulated by the CREB protein and the AMPK signaling pathway. The present finding shows that 25HC3S upregulates expression of CREB and AMPK via demethylating 5mCpG in their promoter regions and subsequently increases intracellular PGC-1α levels (Fig. 3), which provides a detailed mechanism for how 25HC3S functions as proposed in Fig. 5. 25HC3S suppresses DNMT activities and demethylates 5mCpG in the key promoter regions. The demethylation upregulates gene expression and increases MAPK-CREB signaling, which blocks cell apoptosis and induces cell proliferation. The demethylation also upregulates calcium-AMPK signaling, resulting in inhibition of SREBP-1 activity, which inhibits fatty acid and triglyceride biosynthesis. The AMPK signaling pathway also inhibits HMGR expression. Through this process, decreases in Xol biosynthesis, increases in the levels of malonyl-CoA, and the oxidation of fatty acids are shown in Fig. 5.
The results from 11 previous clinical trials, including phase 1 studies in healthy subjects and phases 1b/2a studies in patients, have demonstrated the safety and pharmacological significance of 25HC3S (DUR928; www.durect.com). 25HC3S is currently undergoing phase 2 of clinical development for treating patients with NAFLD. Treatment with 25HC3S significantly decreases lipid accumulation in liver tissues and alleviate lipid-injured liver functions in the patients with nonalcoholic steatohepatitis (Safety and signals of 4 weeks oral DUR-928 in NASH subjects; at The Liver Meeting Digital Experience™, American Association for the Study of Liver Diseases, November 13–16, 2020).
Data availability
All data described are contained within the article (Virginia Commonwealth University/McGuire VA Medical Centre; Yaping.wang@va.gov).
Conflict of interest
S. R. and Virginia Commonwealth University obtained license-related payments from DURECT Corporation, Cupertino, CA. All other authors declare that they have no conflicts of interest with the contents of this article.
Supplemental data
This article contains supplemental data.
Acknowledgments
This work was supported by VA Merit Review grant and DURECT Research Agreement to Virginia Commonwealth University.
Author contributions
Y. W. designed and conducted the experiments and wrote the article; S. R. designed experiments, wrote the article, and is the principle investigator of the project; W. L., J. E. B., L. C., W. M. P., and P. B. H. gave a suggestion and edited the article.
Supplemental data
Tables S1 to S5
References
- 1.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu W., McIntosh C., Lister R., Zhu I., Han Y., Ren J., Landsman D., Lee E., Briones V., Terashima M., Leighty R., Ecker J.R., Muegge K. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24:1613–1623. doi: 10.1101/gr.172015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellano-Castillo D., Moreno-Indias I., Sanchez-Alcoholado L., Ramos-Molina B., Alcaide-Torres J., Morcillo S., Ocaña-Wilhelmi L., Tinahones F., Queipo-Ortuño M.I., Cardona F. Altered adipose tissue dna methylation status in metabolic syndrome: relationships between global DNA methylation and specific methylation at adipogenic, lipid metabolism and inflammatory candidate genes and metabolic variables. J. Clin. Med. 2019;8 doi: 10.3390/jcm8010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gowher H., Jeltsch A. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans. 2018;46:1191–1202. doi: 10.1042/BST20170574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren W., Gao L., Song J. Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation. Genes. 2018;9 doi: 10.3390/genes9120620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren S., Li X., Rodriguez-Agudo D., Gil G., Hylemon P., Pandak W.M. Sulfated oxysterol, 25HC3S, is a potent regulator of lipid metabolism in human hepatocytes. Biochem. Biophys. Res. Commun. 2007;360:802–808. doi: 10.1016/j.bbrc.2007.06.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L., Shen S., Ma Y., Kim J.K., Rodriguez-Agudo D., Heuman D.M., Hylemon P.B., Pandak W.M., Ren S. 25-Hydroxycholesterol-3-sulfate attenuates inflammatory response via PPARγ signaling in human THP-1 macrophages. Am. J. Physiol. Endocrinol. Metab. 2012;302:E788–E799. doi: 10.1152/ajpendo.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren S., Ning Y. Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 2014;306:E123–E130. doi: 10.1152/ajpendo.00552.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L., Kim J.K., Bai Q., Zhang X., Kakiyama G., Min H.K., Sanyal A.J., Pandak W.M., Ren. 5-cho S. 5-cholesten-3β,25-diol 3-sulfate decreases lipid accumulation in diet-induced nonalcoholic fatty liver disease mouse model. Mol. Pharmacol. 2013;83:648–658. doi: 10.1124/mol.112.081505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X., Bai Q., Kakiyama G., Xu L., Kim J.K., Pandak W.M., Jr., Ren S. Cholesterol metabolite, 5-cholesten-3β-25-diol-3-sulfate, promotes hepatic proliferation in mice. J. Steroid Biochem. Mol. Biol. 2012;132:262–270. doi: 10.1016/j.jsbmb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y., Xu L., Rodriguez-Agudo D., Li X., Heuman D.M., Hylemon P.B., Pandak W.M., Ren S. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1369–E1379. doi: 10.1152/ajpendo.90555.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning Y., Kim J.K., Min H.K., Ren S. Cholesterol metabolites alleviate injured liver function and decrease mortality in an LPS-induced mouse model. Metab. Clin. Exp. 2017;71:83–93. doi: 10.1016/j.metabol.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Xu L., Bai Q., Rodriguez-Agudo D., Hylemon P.B., Heuman D.M., Pandak W.M., Ren S. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45:821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- 14.Li X., Pandak W.M., Erickson S.K., Ma Y., Yin L., Hylemon P., Ren S. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta,25-diol 3-sulfate, in hepatocytes. J. lipid Res. 2007;48:2587–2596. doi: 10.1194/jlr.M700301-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Bai Q., Xu L., Kakiyama G., Runge-Morris M.A., Hylemon P.B., Yin L., Pandak W.M., Ren S. Sulfation of 25-hydroxycholesterol by SULT2B1b decreases cellular lipids via the LXR/SREBP-1c signaling pathway in human aortic endothelial cells. Atherosclerosis. 2011;214:350–356. doi: 10.1016/j.atherosclerosis.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Q., Zhang X., Xu L., Kakiyama G., Heuman D., Sanyal A., Pandak W.M., Yin L., Xie X., Ren S. Oxysterol sulfation by cytosolic sulfotransferase suppresses liver X receptor/sterol regulatory element binding protein-1c signaling pathway and reduces serum and hepatic lipids in mouse models of nonalcoholic fatty liver disease. Metab. Clin. Exp. 2012;61:836–845. doi: 10.1016/j.metabol.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren S., Hylemon P., Zhang Z.P., Rodriguez-Agudo D., Marques D., Li X., Zhou H., Gil G., Pandak W.M. Identification of a novel sulfonated oxysterol, 5-cholesten-3beta,25-diol 3-sulfonate, in hepatocyte nuclei and mitochondria. J. Lipid Res. 2006;47:1081–1090. doi: 10.1194/jlr.M600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Li X., Ren S. Cholesterol metabolites 25-hydroxycholesterol and 25-hydroxycholesterol 3-sulfate are potent paired regulators: from discovery to clinical usage. Metabolites. 2020;11 doi: 10.3390/metabo11010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prah J., Winters A., Chaudhari K., Hersh J., Liu R., Yang S.H. Cholesterol sulfate alters astrocyte metabolism and provides protection against oxidative stress. Brain Res. 2019;1723:146378. doi: 10.1016/j.brainres.2019.146378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X., Cheng Q., Xu L., Yan J., Jiang M., He J., Xu M., Stefanovic-Racic M., Sipula I., O'Doherty R.M., Ren S., Xie X. Cholesterol sulfate and cholesterol sulfotransferase inhibit gluconeogenesis by targeting hepatocyte nuclear factor 4α. Mol. Cell. Biol. 2014;34:485–497. doi: 10.1128/MCB.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi Y., Shi X., Zhu J., Guan X., Garbacz W.G., Huang Y., Gao L., Yan J., Xu M., Ren S., Ren S., Liu Y., Ma X., Li S., Xie W. Regulation of Cholesterol Sulfotransferase SULT2B1b by Hepatocyte Nuclear Factor 4α Constitutes a Negative Feedback Control of Hepatic Gluconeogenesis. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng L.J., Griffiths W.J., Nazer H., Yang Y., Sjövall J. High levels of (24S)-24-hydroxycholesterol 3-sulfate, 24-glucuronide in the serum and urine of children with severe cholestatic liver disease. J. Lipid Res. 1997;38:926–934. [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Park Y., Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics (Oxford, England) 2016;32:1446–1453. doi: 10.1093/bioinformatics/btw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics (Oxford, England) 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 26.Nowacka-Zawisza M., Wiśnik E. DNA methylation and histone modifications as epigenetic regulation in prostate cancer (Review) Oncol. Rep. 2017;38:2587–2596. doi: 10.3892/or.2017.5972. [DOI] [PubMed] [Google Scholar]
- 27.Handy D.E., Castro R., Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Chen L., Pandak W.M., Heuman D., Hylemon P.B., Ren S. High glucose induces lipid accumulation via 25-hydroxycholesterol DNA-CpG methylation. iScience. 2020;23:101102. doi: 10.1016/j.isci.2020.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Guijo A., Oji V., Hartmann M.F., Traupe H., Wudy S.A. Simultaneous quantification of cholesterol sulfate, androgen sulfates, and progestagen sulfates in human serum by LC-MS/MS. J. Lipid Res. 2015;56:1843–1851. doi: 10.1194/jlr.D061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malodobra-Mazur M., Cierzniak A., Dobosz T. Oleic acid influences the adipogenesis of 3T3-L1 cells via DNA Methylation and may predispose to obesity and obesity-related disorders. Lipids Health Dis. 2019;18:230. doi: 10.1186/s12944-019-1173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Wang J., Wang L., Feng G., Li G., Yu M., Li Y., Liu C., Yuan X., Zang G., Li Z., Zhao L., Ouyang H., Quan Q., Wang G. Impaired lipid metabolism by age-dependent DNA methylation alterations accelerates aging. Proc. Natl. Acad. Sci. U.S.A. 2020;117:4328–4336. doi: 10.1073/pnas.1919403117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Chen L., Shi Y., Liu N., Wang Z., Yang R., Yan B., Liu X., Lai W., Liu Y., Xiao D., Zhou H., Cheng Y., Cao Y., Liu S., Xia Z. DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism. Epigenetics Chromatin. 2019;12:59. doi: 10.1186/s13072-019-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellano-Castillo D., Morcillo S., Clemente-Postigo M., Crujeiras A.B., Fernandez-Garcia J.C., Torres E., Tinahones F.J., Macias-Gonzalez M. Adipose tissue inflammation and VDR expression and methylation in colorectal cancer. Clin. Epigenetics. 2018;10:60. doi: 10.1186/s13148-018-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander E.S. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.Y., Kim T.R., Jeong H.H., Sohn K.A. Integrative pathway-based survival prediction utilizing the interaction between gene expression and DNA methylation in breast cancer. BMC Med. genomics. 2018;11(Suppl 3):68. doi: 10.1186/s12920-018-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemos F.O., Ehrlich B.E. Polycystin and calcium signaling in cell death and survival. Cell calcium. 2018;69:37–45. doi: 10.1016/j.ceca.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding T., Zhou Y., Long R., Chen C., Zhao J., Cui P., Guo M., Liang G., Xu L. DUSP8 phosphatase: structure, functions, expression regulation and the role in human diseases. Cell Biosci. 2019;9:70. doi: 10.1186/s13578-019-0329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kouba S., Ouldamer L., Garcia C., Fontaine D., Chantome A., Vandier C., Goupille C., Potier-Cartereau M. Lipid metabolism and calcium signaling in epithelial ovarian cancer. Cell calcium. 2019;81:38–50. doi: 10.1016/j.ceca.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Greineisen W.E., Speck M., Shimoda L.M., Sung C., Phan N., Maaetoft-Udsen K., Stokes A.J., Turner H. Lipid body accumulation alters calcium signaling dynamics in immune cells. Cell calcium. 2014;56:169–180. doi: 10.1016/j.ceca.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou X.M., Zhang T., Da Z., Wu X.A. CHPF promotes lung adenocarcinoma proliferation and anti-apoptosis via the MAPK pathway. Pathol. Res. Pract. 2019;215:988–994. doi: 10.1016/j.prp.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Smith Z., Ryerson D., Kemper J.K. Epigenomic regulation of bile acid metabolism: emerging role of transcriptional cofactors. Mol. Cell. Endocrinol. 2013;368:59–70. doi: 10.1016/j.mce.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R., Deivendran S., Santhoshkumar T.R., Pillai M.R. Signaling coupled epigenomic regulation of gene expression. Oncogene. 2017;36:5917–5926. doi: 10.1038/onc.2017.201. [DOI] [PubMed] [Google Scholar]
- 43.Swaminathan K., Kumar S.M., Clemens D.L., Dey A. Inhibition of CYP2E1 leads to decreased advanced glycated end product formation in high glucose treated ADH and CYP2E1 over-expressing VL-17A cells. Biochim. Biophys. Acta. 2013;1830:4407–4416. doi: 10.1016/j.bbagen.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Fan R., Cui J., Ren F., Wang Q., Huang Y., Zhao B., Wei L., Qian X., Xiong X. Overexpression of NRK1 ameliorates diet- and age-induced hepatic steatosis and insulin resistance. Biochem. biophysical Res. Commun. 2018;500:476–483. doi: 10.1016/j.bbrc.2018.04.107. [DOI] [PubMed] [Google Scholar]
- 45.Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 46.Wu J., Wang S.T., Zhang Z.J., Zhou Q., Peng B.G. CREB5 promotes cell proliferation and correlates with poor prognosis in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2018;11:4908–4916. [PMC free article] [PubMed] [Google Scholar]
- 47.Bui N.L., Pandey V., Zhu T., Ma L., Basappa, Lobie P.E. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018;415:177–186. doi: 10.1016/j.canlet.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Zang Y., Fan L., Chen J., Huang R., Qin H. Improvement of lipid and glucose metabolism by capsiate in palmitic acid-treated HepG2 cells via activation of the AMPK/SIRT1 signaling pathway. J. Agric. Food Chem. 2018;66:6772–6781. doi: 10.1021/acs.jafc.8b01831. [DOI] [PubMed] [Google Scholar]
- 49.Iwabu M., Yamauchi T., Okada-Iwabu M., Sato K., Nakagawa T., Funata M., Yamaguchi M., Namiki S., Nakayama R., Tabata M., Ogata H., Kubota N., Takamoto I., Hayashi Y.K., Yamauchi N. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 50.Rius-Pérez S., Torres-Cuevas I., Millán I., Ortega Á L. PGC-1α, inflammation, and oxidative stress: an integrative view in metabolism. Oxid. Med. Cell Longev. 2020;2020:1452696. doi: 10.1155/2020/1452696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S5
Data Availability Statement
All data described are contained within the article (Virginia Commonwealth University/McGuire VA Medical Centre; Yaping.wang@va.gov).