Abstract
STUDY QUESTION
Is the microRNA (miRNA) expression pattern of cumulus oophorus cells (COCs) in women undergoing medically assisted reproduction (MAR) procedures differentially modulated according to patient age and gonadotropin treatment strategy?
SUMMARY ANSWER
Maternal age is an independent factor impacting miRNA expression in COCs while gonadotropin treatment may affect follicular miRNA expression and IVF efficacy.
WHAT IS KNOWN ALREADY
Epigenetic mechanisms in female infertility are complex and poorly studied. DNA methylation, histone modifications, miRNAs and nucleosome positioning influence cellular machinery through positive and negative feedback mechanisms either alone or interactively. miRNAs are important regulators during oogenesis, spermatogenesis and early embryogenesis, and are reported to play a role in regulating crosstalk between the oocyte and COCs. Although miRNome analysis has been performed in female human reproductive tissues (endometrium, myometrium, cervix and ovaries), epigenetic modifications in women with infertility have not been explored in detail. In addition, the impact of gonadotropin treatments during MAR on miRNA expression in COCs has not been fully investigated.
STUDY DESIGN, SIZE, DURATION
This study was carried out in 53 COC samples obtained from mature metaphase II (MII) oocytes in 53 women undergoing MAR treatment. A total of 38 samples for assay development were pooled by maternal age and gonadotropin treatment into four predetermined subgroups: ≥36 years and recombinant human FSH (r-hFSH), n = 10; ≥36 years and r-hFSH+ recombinant human-luteinizing hormone (r-hLH), n = 10; ≤35 years and r-hFSH, n = 9; ≤35 years and r-hFSH+r-hLH, n = 9. miRNome profiles were determined and compared between subgroups. Expression of defined miRNAs was validated in the remaining fifteen samples, representative of each subgroup, by quantitative polymerase chain reaction (PCR).
PARTICIPANTS/MATERIALS, SETTING, METHODS
COCs were processed for miRNA-enriched total RNA extraction and pooled in homogeneous subgroups to obtain a sufficient amount and quality of starting material to perform the analysis. Each pooled sample underwent miRNA profiling using PCR assay system to examine expression of 752 human miRNAs without pre-amplification. Data were analyzed using the delta-delta Ct method for relative quantitation and prediction of target genes (with at least four algorithms predicting the same miRNA-gene interaction pair (HIT)>4). The miRSystem database provided functional annotation enrichment (raw P-value <0.05) of co-expressed miRNAs.
MAIN RESULTS AND THE ROLE OF CHANCE
We found distinctive miRNA expression profiles in each subgroup correlating with age and MAR stimulation. In addition, a number of selective and co-expressed miRNAs were revealed by comparative analysis. A cluster of 37 miRNAs were commonly but differentially expressed in all four pools. Significant differences were observed in expression regulation of 37 miRNAs between age groups (≤35 or ≥36) in women receiving r-hFSH+r-hLH compared to those receiving r-hFSH alone. Higher concentrations and increased numbers of miRNAs were recorded in younger than in older patients, regardless of treatment. Functional and expression studies performed to retrieve common miRNome profiles revealed an enrichment of biological functions in oocyte growth and maturation, embryo development, steroidogenesis, ovarian hyperstimulation, apoptosis and cell survival, glucagon and lipid metabolism, and cell trafficking. The highest scored pathways of target genes of the 37 common miRNAs were associated with mitogen-activated protein kinase (MAPK) signaling pathways, G alpha signaling, transcription regulation, tight junctions, RNA polymerase I and III, and mitochondrial transcription. We identified a potential age- and MAR stimulation-dependent signature in the miRNA landscape of COCs.
LIMITATIONS, REASONS FOR CAUTION
We cannot rule out the possibility that other unknown individual genetic or clinical factors may have interfered with the reported results. Since miRNA profiling was conducted with a predefined array of target probes, other miRNA molecules, potentially modulated by age and hormonal stimulation, may have been missed in this study.
WIDER IMPLICATIONS OF THE FINDINGS
miRNA expression in COCs is modulated by gonadotropin treatment and correlates strongly with age. A better understanding of the expression patterns and functions of miRNAs may lead to the development of novel therapeutics to treat ovarian dysfunction and improve fertility in older women.
STUDY FUNDING/COMPETING INTEREST
This study was funded by Merck KGaA, Darmstadt, Germany. All authors declared no competing interest, except SL and TD who are fully employed by Merck KGaA.
TRIAL REGISTRATION NUMBER
N/A
Keywords: miRNA, epigenetics, gonadotropin stimulation, aging, cumulus oophorus cells, medically assisted reproduction
Introduction
From birth, human ovaries have a determinate number of follicles holding immature primary oocytes. Follicular development is a constant process: at any time, follicles can be in different phases of development. Hormones, particularly FSH, have a crucial role in the release of the mature oocyte and the block of folliculogenesis (Scarica et al., 2019). Most follicles expire without completing development, though a few are able to develop fully and produce a secondary oocyte, released by rupture of the follicle during ovulation (Holesh et al., 2019). The maturing human oocyte is influenced by follicular fluid and cumulus oophorus cells (COCs) (Mendoza et al., 2002; Poulsen et al., 2019). COCs are granulosa-derived cells arranged in close contact with the oocyte, supporting its maturation and cooperating in the control of access of spermatozoa to the oocyte (Tanghe et al., 2002; de los Santos et al., 2012). The critical functional role of COCs suggests that their dysfunction might impact on female fertility. However, while IVF strategies evaluate COC structural properties and oocyte morphological features, both are subject to interpretation by the operators and may therefore be variable (Lin et al., 2003). In recent years, although advanced IVF techniques have been designed and different conditions of infertility have become treatable, implantation and pregnancy rates after embryo transfer are still low. Substantially, reproductive function is dependent on a woman’s age; women aged 35 years and over begin to experience a decline in the quality and quantity of oocytes as well as irregular menstruation (Igarashi et al., 2015). Age has an impact on the reduction of ovarian reserve, which leads to a decrease in competence of the oocyte/embryo. Aging insults can lead to a reduction in the success of medically assisted reproduction (MAR), an increase in aneuploidies, reduced mitochondrial activity and a reduction in overall reproduction (Cimadomo et al., 2018). Therefore, identifying additional biomarkers able to define the ‘oocytic phenotype’ and to predict the efficacy of IVF treatment and successful fertilization is crucial.
The advent of ‘omics’ technologies disclosed hundreds of genes with a functional role in oocyte growth and maturation, endometrial receptivity, embryo development and embryo-endometrial signaling (Egea et al., 2014). Nevertheless, the molecular mechanisms downstream the expression of these genes are poorly understood. Recently, epigenetics has emerged as a ‘hot’ new field of study for prognosis, diagnosis and therapy in reproductive medicine (Pisarska et al., 2019). Crucial epigenetic changes occur from fertilization to embryo formation up to the birth. Transcriptional and proteomic rearrangement occurs through post-transcriptional gene regulation and/or intercellular communication with surrounding COCs (Biase and Kimble, 2018). However, epigenetic mechanisms in female infertility are complex and poorly explored. DNA methylation, histone modifications, microRNAs (miRNAs) and nucleosome positioning influence cellular machinery through positive and negative feedback mechanisms either alone or interactively (Pinborg et al., 2016; Giacone et al., 2019). It was recently demonstrated that miRNAs are important regulators during oogenesis (Imbar and Eisenberg 2014; Maalouf et al., 2016; Tesfaye et al., 2018), spermatogenesis (Yadav and Kotaja, 2014; Hilz et al., 2016, Luo et al., 2016) and early embryogenesis (Liu et al., 2016; Paul et al., 2019). miRNomes were performed in different female human reproductive tissues including uterus (endometrium, myometrium and cervix) and ovaries (Battaglia et al., 2016; Yerushalmi et al., 2018). Similarly, some evidence reports a role for miRNAs in regulating oocyte and COC crosstalk (Tong et al., 2014).
Our work aimed to understand the expression patterns and functions of miRNAs, and may lead to the development of novel treatment for ovarian dysfunction, improve fertility and, potentially, design minimally invasive approaches. We identified a potential miRNA signature in COCs from different groups of patients, enrolled according to specific inclusion and exclusion criteria, associated with oocyte quality and prognostic response to fertilization therapy. By further exploring the link between oocyte aging and epigenetic modifications, miRNAs may prove to be instrumental in unraveling the molecular mechanisms underlying oocyte ontogeny and reproductive competence.
Materials and methods
COC collection
COCs retrieved during pickup were isolated from the follicular fluid and incubated at 37°C and 6% CO2 for 2 h in Sydney IVF Fertilization Medium (Cook Medical, Limerick, Ireland). Using 18 G needles, a portion of roughly two-thirds of the cumulus oophorus mass was mechanically dissociated and exposed to enzymatic action of 80 IU/ml hyaluronidase (BioCare Europe S.r.l.—Irvine Scientific, Milan, Italy). Cumulus cells’ collection was performed individually from each oocyte. After denudation, oocytes were examined to assess maturational stage and turned away for sperm injection, while the cumulus cells were aspirated from the hyaluronidase, diluted 1:2 with HEPES-Human Tubaric Fluid (HTF) (Irvine Scientific) and pelleted. The samples are washed with PBS and stored at −80°C until use for experimental analysis. Only cumulus cells collected from mature metaphase II stage (MII) oocytes were included in the study. COC samples were collected from 53 women undergoing MAR treatment with ART. Patients were recruited for the study based on the following inclusion criteria: nonsmokers, Caucasian women undergoing controlled ovarian stimulation cycles, 18≤ age ≤43,18≤ BMI ≤27. A total of 38 patients eligible for the study were classified by maternal age and gonadotropin treatment into four predetermined subgroups according to age and type of gonadotropin used for ovarian stimulation: POOL I ≥ 36 years and recombinant human FSH (r-hFSH), n = 10; POOL II ≥36 and r-hFSH+ recombinant human-luteinizing hormone (r-hLH), n = 10; POOL III ≤35 and r-hFSH, n = 9; POOL IV ≤35 and r-hFSH+r-hLH, n = 9. Average age was 39 and 32 years in the older and younger patient subgroups respectively. In addition to these 38 patients, samples from a further 15 women were used for validation, based on age and MAR treatment as representative of each pool.
MAR protocol
This study was conducted at the Outpatient Fertility Unit of the University of Campania ‘Luigi Vanvitelli’. Patients included in the study underwent ovarian stimulation according to our standard protocol. Briefly, all the patients underwent a standard downregulation with GnRH analogue hormone at a dose of 0.1 mg/day (triptrolin, Decapeptyl, Ipsen, Milan, Italy) until serum or estradiol levels were ≤40 ng/mL and no follicle >7 mm was observed; POOL I patients received r-hFSH (Gonal-F; Serono, Rome, Italy) at a daily dose of 300 IU for 4 days; POOL II patients received r-hFSH (Gonal-F) and r-hLH (Pergoveris; Serono, Rome, Italy) at a daily dose of 300 IU r-hFHS + 150 IU r-hLH for 4 days; POOL III patients received r-hFSH (Gonal-F) at a daily dose of 150 IU for 4 days; and POOL IV patients received r-hFSH (Gonal- F) and r-hLH (Pergoveris) at a daily dose of 150 IU r-hFSH + 75 IU r-hLH for 4 days. By the fifth day therapy was personalized according to monitoring of ovarian response by hormonal and ultrasonographic assessment every second–third day. When at least three follicles had reached a diameter of 18 mm, a single s.c. bolus of 10,000 IU hCG (Gonasi HP 10000; IBSA, Rome, Italy) was administered. Trans-vaginal follicular aspiration was performed 34–36 h after hCG administration. All patients signed informed consent forms (as part of their initial intake) to be part of the study. The experimental study was conducted in accordance with the principles of the Helsinki Declaration of 1975, using clinical practice procedures routinely performed during IVF cycles. These procedures did not pose any additional risks to the patients, and all medical decisions concerning individual patients were not affected by the study. The study was approved by the Institutional Review Board (ref. no. 498 12/09/2017).
RNA isolation
Total RNA was isolated from COCs using the following protocol. COCs were collected by centrifugation and resuspended in 1 ml of QIAzol Lysis Reagent (Qiagen, Milan, Italy), vigorously shaken and stored at −80°C overnight. The next day, samples were added with 100-μL 2-bromo-3-chloropropane (Sigma-Aldrich, Milan, Italy), gently shaken and incubated for 15 min at room temperature. After 15-minute centrifugation at 12000 rpm at 4°C, the supernatants were placed in a fresh tube and supplemented with 500-μL cold isopropyl alcohol. The RNA precipitation reaction was carried out for 30 min at −80°C followed by 30-min centrifugation at 12000 rpm at 4°C. The pellets were subsequently resuspended in 1-mL cold 70% ethanol, and the samples were centrifuged again for 10 min at 7500 rpm at 4°C. The pellets were then dried at 42°C for a few minutes and resuspended in diethyl pyrocarbonate (DEPC)-treated H2O.
miRCURY LNA™ universal RT microRNA PCR
cDNA synthesis and real-time quantitative polymerase chain reaction (PCR) were performed using the miRCURY LNA™ Universal RT microRNA PCR System (Qiagen) according to the manufacturer’s instructions. Real-time PCRs were run on a 7900HT thermocycler (Applied Biosystems) using the thermal cycling parameters suggested by the manufacturer’s protocol. Raw Ct values were calculated using RQ manager software v.1.2.1 (Applied Biosystems) with manual settings for threshold and baseline. All miRCURY assays were analyzed using a ΔRn threshold of 60 and baseline subtraction using cycles 1–10. miRNA profiles were determined using the delta-delta Ct method and compared between subgroups.
miRNA real-time PCR
Following RNA extraction, the miRNA fraction was converted into cDNA using miScript II RT Kit (Qiagen): 10 ng RNA was used with 1X HiSpec buffer, 1X miScriptRT, 1X miScript Nucleics Mix and DEPC-H2O for 60 min at 37°C and then 5 min at 95°C. Subsequently, miRNA pre-amplification was performed with 1/10 of cDNA volume using miScriptPreAMP Kit (Qiagen) according to the manufacturer’s protocol. Real-time PCR was performed with QuantiTect SYBR Green PCR Kit (Qiagen), requiring the use of 1/25 of cDNA pre-amplified in presence of 1X QuantiTect SYBR Green PCR Master Mix, miScript Universal Primer and primer specific for target miRNA (Qiagen). The thermal protocol was as follows: 95°C for 15 min plus 40 cycles at 94°C for 30 s, 58°C for 34 s and 70°C for 34 s.
Statistical analysis
Data were evaluated using Student’s t-test. Statistical differences in miRNA expression were considered significant when they exceeded a 95% confidence interval (false discovery rate (FDR) ≤ 0.05) and fold change (FC) ≥2.
Target prediction analysis
The prediction of target genes was performed using the miRSystem database accomplished by a value HIT ≥ 4, included validated genes, and observed/expected (O/E) ratio ≥ 2. In particular, the HIT value represents the number of algorithms predicting the same miRNA–gene interaction pair, while O/E parameter indicates the ratio between observed identification probability for a given gene set as miRNA predicted targets, and the expected probability of the proportion of all miRNAs in the miRSystem database predicted to target that gene, pair. By the miRSystem, we provided functional and pathway annotations, of predicted target genes (raw P-value <0.05) of co-expressed miRNAs.
Results
miRNome profiling of COCs isolated from MAR-treated patients
Cumulus expansion and oocyte maturation are key processes in ovulation. To elucidate the role of miRNAs in human folliculogenesis and ovulation, COCs obtained from 53 patients undergoing MAR treatment were analyzed. By real-time PCR, miRNA profiles of COCs from mature MII oocytes were obtained and compared, after categorizing 38 patients into four subgroups according to age and MAR treatment: POOL I (≥36 years and r-hFSH, n = 10); POOL II (≥36 years and r-hFSH+r-hLH, n = 10); POOL III (≤35 years and r-hFSH, n = 9); POOL IV (≤35 years and r-hFSH+r-hLH, n = 9). Clinical and biological features of patients are reported in Table I.
Table I.
Clinical and biological features of patients enrolled in the study, pooled according to medically assisted reproduction (MAR) stimulation and age.
|
Pool 1
(n = 10) |
Pool 2
(n = 10) |
Pool 3
(n = 9) |
Pool 4
(n = 9) |
|
|---|---|---|---|---|
| Age (years) | ≥36 (39.5 ± 2.63) | ≥36 (39 ± 2.67) | ≤35 (31.11 ± 2.37) | ≤35 (32.88 ± 2.62) |
| Stimulation protocol | r-hFSH | r-hFSH + r-hLH | r-hFSH | r-hFSH + r-hLH |
| BMI | 25.89 ± 1.10 | 20.03 ± 2.30 | 21.10 ± 1.50 | 22.20 ± 2.30 |
| LH (mIU/mL) | 3.35 ± 1.75 | 6.27 ± 4.38 | 3.80 ± 1.29 | 7.38 ± 1.68 |
| FSH (mIU/mL) | 7.32 ± 1.09 | 12.37 ± 5.41 | 6.67 ± 1.04 | 6.23 ± 1.42 |
| AMH | 2.79 ± 2.26 | 0.65 ± 0.28 | 4.42 ± 3.47 | 1.73 ± 1.18 |
| E2 a | 1167.75 ± 443.42 | 817.5 ± 437.98 | 884.25 ± 338.99 | 1881.25 ± 556.76 |
| P a | 0.78 ± 0.41 | 0. 77 ± 0.22 | 1.86 ± 0.25 | 0.71 ± 0.14 |
| Endometrial thickness (mm) | 8.00 ± 1.50 | 7.05 ± 2.00 | 10.75 ± 1.63 | 9.50 ± 0.75 |
| Follicles> 16 mm (n) | 7.25 ± 2.75 | 3.00 ± 1.82 | 7.25 ± 2.50 | 8.50 ± 4.35 |
| Oocyte retrieval (n) | 7.25 ± 0.41 | 2.50 ± 1.29 | 8.25 ± 1.25 | 8.50 ± 4.80 |
| Oocyte MII (n) | 6,30 ± 3.65 | 3,20 ± 1.47 | 8,60 ± 5.57 | 4.67 ± 2.28 |
| Fertilization rate (%) | 83% | 70% | 84% | 81% |
| β-HCG + (%) | 30% | 10% | 33% | 33% |
| AFC (n) | 5.3 ± 2.21 | 4.2 ± 1.3 | 10.83 ± 3.06 | 8 ± 2.78 |
r-hLH, recombinant human luteinizing hormone; r-hFSH, recombinant human follicle stimulation hormone; AMH, anti-Müllerian hormone; E2, estradiol; P: progesterone; MII, metaphase II; AFC, antral follicle count.
Measured at HGC administration.
The data were expressed as mean ± SD.
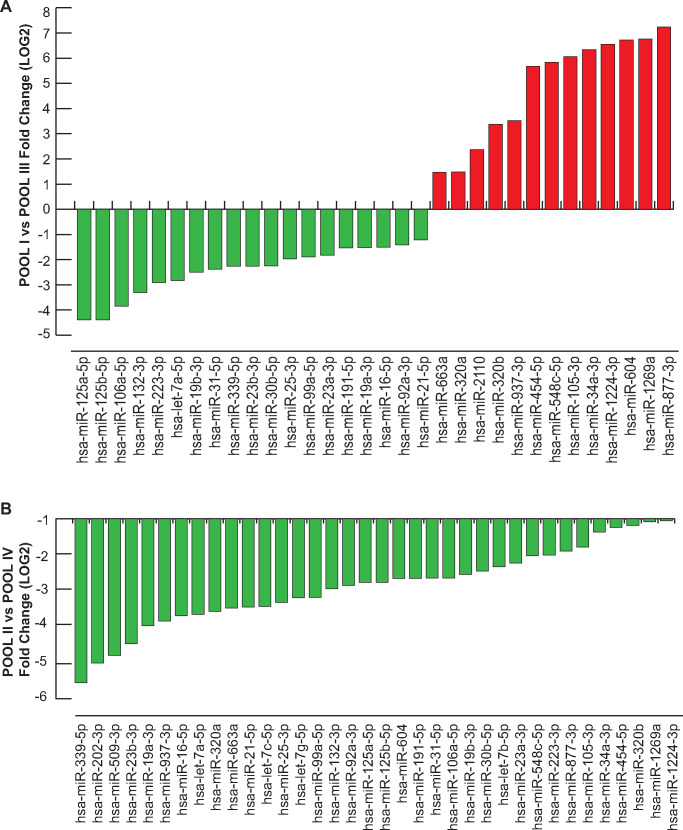
By exploring miRNome profiles, we found specific miRNA expression patterns in each patient pool (Fig. 1A). A total of 77 miRNAs were detected in POOL I, 143 in POOL II, 222 in POOL III and 169 in POOL IV. Venn diagrams showing the comparative analysis of different groups are depicted in Fig. 1B–E, illustrating the number of selective and co-expressed miRNAs in two conditions at a time. Additionally, distinct miRNAs were found expressed exclusively in different patient subgroups (Supplementary Table SI).
Figure 1.
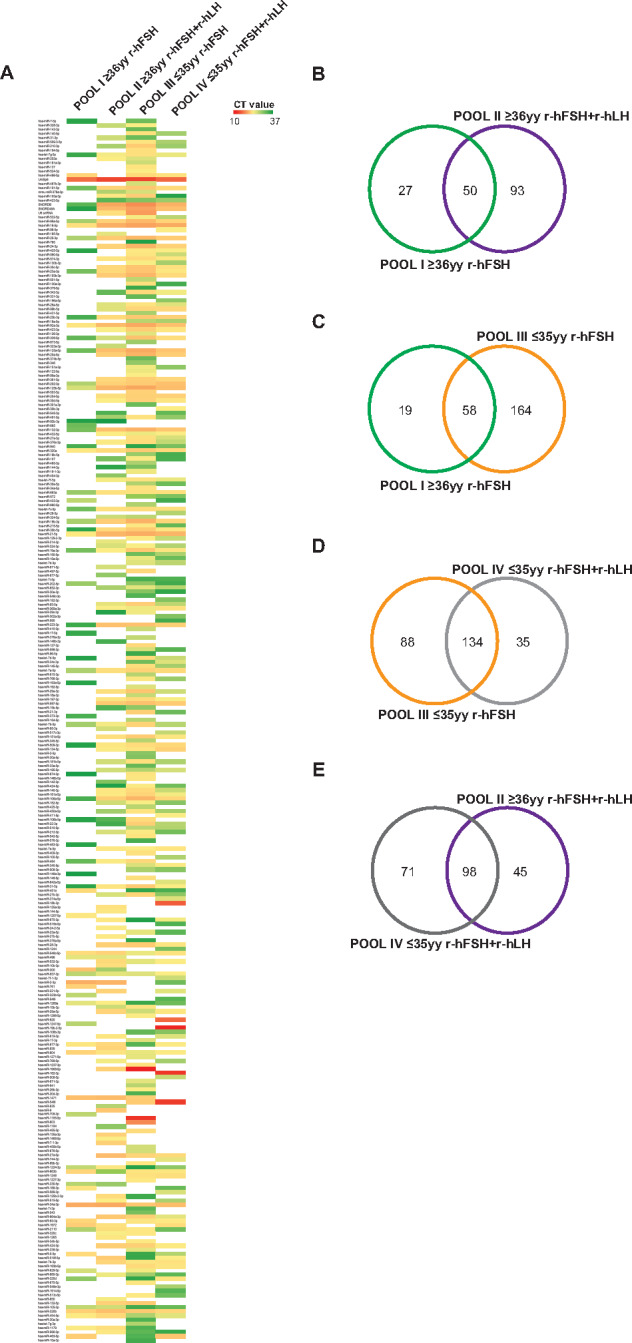
miRNA profiling of cumulus oophorus cells (COCs). (A) Heat map of miRNAs expressed in four different pools (POOL I ≥ 36 years and recombinant human follicle stimulating hormone (r-hFSH), n = 10; POOL II ≥36 years and r-hFSH recombinant human-luteinizing hormone (r-hLH), n = 10; POOL III ≤35 years and r-hFSH, n = 9; POOL IV ≤35 years and r-hFSH+r-hLH, n = 9). (B–E) Venn diagrams illustrating the number of selective and co-expressed miRNAs in the two indicated pools. The color code in the heat maps is linear with green as the lowest and red as the highest expression according to 37≤ Cycle threshold (Ct) ≥ 10. miRNAs not included in the analysis are shown in white. yy, years.
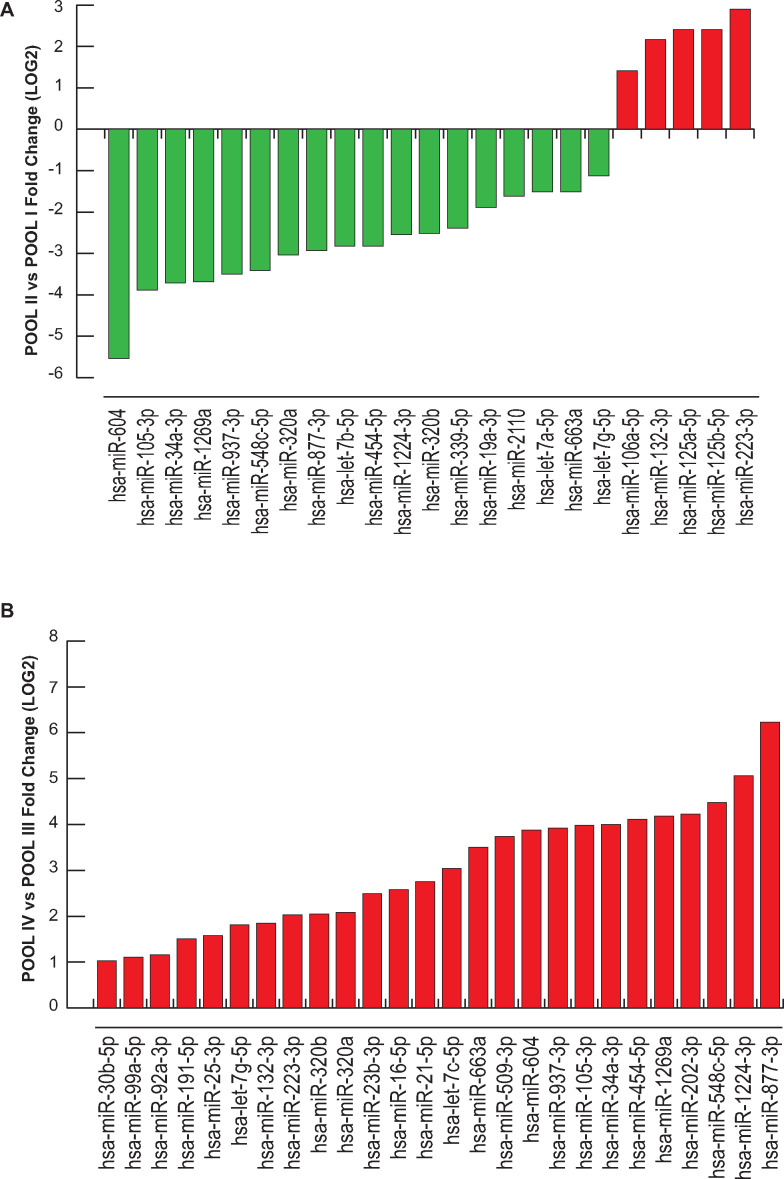
Differential expression analysis was performed using the delta-delta Ct method, and a statistically significant threshold was applied (cutoff Ct ≥37 and ≤10; FC (2^−ΔΔCt) = ±2). A cluster of 37 miRNAs resulted commonly expressed but differentially regulated in the four pools (excluding UniSp6, SNORD38B and SNORD49A; Table II). A hierarchical clustering of miRNAs differentially expressed in COCs in the different subgroups is illustrated in Figs. 2 and 3. We found distinctive miRNAs in aged COCs obtained from women undergoing the same MAR treatment: 19 downregulated and 13 upregulated miRNAs in POOL I vs POOL III (Fig. 2A); 36 downregulated miRNAs in POOL II vs POOL IV (Fig. 2B). In contrast, by comparing the pools based on MAR therapy, we found that r-hFSH+r-hLH treatment regulated distinct clusters of miRNAs: 18 downregulated and 5 upregulated miRNAs in POOL II vs POOL I (Fig. 3A); 26 upregulated miRNAs in POOL IV vs POOL III (Fig. 3B). In addition, 13 miRNAs were commonly expressed but differentially regulated upon r-hLH addition in both the older and younger women (miR-877-3p, miR-34a-3p, miR-105-3p, miR-454-5p, miR-663a, miR-937-3p, miR-320a, miR-1269a, miR-604, miR-548c-5p, miR-320b, miR-132-3p, let-7g-5p), while miR-1224-3p and miR-223-3p exhibited a similar upregulated expression trend independently of age. Indeed, a comparative analysis between differentially expressed miRNAs in POOL I vs POOL III and in POOL II vs POOL IV revealed: 31 common miRNAs (miR-877-3p, let-7a-5p, miR-1224-3p, miR-21-5p, miR-34a-3p, miR-25-3p, miR-105-3p, miR-454-5p, miR-19b-3p, miR-663a, miR-106a-5p, miR-92a-3p, miR-937-3p, miR-23a-3p, miR-30b-5p, miR-320a, miR-125a-5p, miR-1269a, miR-16-5p, miR-339-5p, miR-604, miR-548c-5p, miR-31-5p, miR-19a-3p, miR-320b, miR-99a-5p, miR-23b-3p, miR-191-5p, miR-132-3p, miR-125b-5p, miR-223-3p); one exclusive miRNA from differential analysis in POOL I vs POOL III (miR-2110, upregulated in older compared to younger women, both groups stimulated with r-hFSH alone, FC (log2) = 2.369881333); five miRNAs distinctively downregulated in the older POOL II compared to the younger POOL IV, both groups stimulated with r-hFSH+r-hLH (miR-202-3p, FC (log2) = −4.9869; let-7b-5p, FC (log2) = −2.366236125; let-7c-5p, FC (log2) = −3.4895; miR-509-3p, FC (log2) = −4.7617; let-7g-5p, FC (log2) = −3.244).
Table II.
List of microRNAs (miRNAs) commonly expressed in patient pools.
| miRNAs commonly expressed | FC POOL I vs POOL III | FC POOL II vs POOL IV | FC POOL II vs POOL I | FC POOL IV vs POOL III |
|---|---|---|---|---|
| hsa-let-7a-5p | 0.14 | 0.08 | 0.35 | 0.65 |
| hsa-let-7b-5p | 1.35 | 0.19 | 0.14 | 0.98 |
| hsa-let-7c-5p | 1.28 | 0.09 | 0.57 | 8.20 |
| hsa-let-7g-5p | 0.81 | 0.11 | 0.46 | 3.51 |
| hsa-miR-105-3p | 66.98 | 0.29 | 0.07 | 15.75 |
| hsa-miR-106a-5p | 0.07 | 0.16 | 2.67 | 1.19 |
| hsa-miR-1224-3p | 93.35 | 0.48 | 0.17 | 33.38 |
| hsa-miR-125a-5p | 0.05 | 0.14 | 5.29 | 1.77 |
| hsa-miR-125b-5p | 0.05 | 0.14 | 5.29 | 1.77 |
| hsa-miR-1269a | 109.00 | 0.47 | 0.08 | 18.08 |
| hsa-miR-132-3p | 0.10 | 0.13 | 4.50 | 3.60 |
| hsa-miR-16-5p | 0.35 | 0.07 | 1.25 | 5.97 |
| hsa-miR-191-5p | 0.35 | 0.15 | 1.27 | 2.85 |
| hsa-miR-19a-3p | 0.35 | 0.06 | 0.27 | 1.55 |
| hsa-miR-19b-3p | 0.18 | 0.17 | 0.97 | 1.03 |
| hsa-miR-202-3p | 0.82 | 0.03 | 0.72 | 18.79 |
| hsa-miR-2110 | 5.17 | 1.11 | 0.33 | 1.52 |
| hsa-miR-21-5p | 0.43 | 0.09 | 1.36 | 6.70 |
| hsa-miR-223-3p | 0.13 | 0.24 | 7.48 | 4.07 |
| hsa-miR-23a-3p | 0.28 | 0.21 | 0.83 | 1.13 |
| hsa-miR-23b-3p | 0.21 | 0.05 | 1.25 | 5.62 |
| hsa-miR-25-3p | 0.25 | 0.10 | 1.13 | 2.98 |
| hsa-miR-30b-5p | 0.21 | 0.18 | 1.72 | 2.04 |
| hsa-miR-31-5p | 0.19 | 0.15 | 0.99 | 1.23 |
| hsa-miR-320a | 2.79 | 0.08 | 0.12 | 4.23 |
| hsa-miR-320b | 10.34 | 0.44 | 0.18 | 4.15 |
| hsa-miR-339-5p | 0.21 | 0.02 | 0.19 | 1.85 |
| hsa-miR-34a-3p | 80.63 | 0.38 | 0.08 | 15.96 |
| hsa-miR-454-5p | 51.38 | 0.42 | 0.14 | 17.25 |
| hsa-miR-509-3p | 0.53 | 0.04 | 0.92 | 13.32 |
| hsa-miR-548c-5p | 57.38 | 0.24 | 0.09 | 22.32 |
| hsa-miR-604 | 105.19 | 0.15 | 0.02 | 14.71 |
| hsa-miR-663a | 2.76 | 0.09 | 0.35 | 11.31 |
| hsa-miR-877-3p | 151.22 | 0.26 | 0.13 | 75.00 |
| hsa-miR-92a-3p | 0.38 | 0.13 | 0.80 | 2.24 |
| hsa-miR-937-3p | 11.43 | 0.07 | 0.09 | 15.12 |
| hsa-miR-99a-5p | 0.27 | 0.11 | 0.85 | 2.16 |
Figure 2.
Differential miRNA expression analysis according to age. (A) miRNAs regulated in COCs of the two r-FSH treatment groups, POOL I (≥36 years) vs POOL III (≤35 years). (B) miRNAs regulated in COCs of the two r-FSH+r-LH treatment groups, POOL II (≥36 years) vs POOL IV (≤35 years). Fold change is shown in log2 scale.
Figure 3.
Differential miRNA expression analysis according to ovarian stimulation for treatment with assisted reproduction technology. (A) miRNAs regulated in COCs of the two older groups (≥36 years) under different ovarian stimulation protocols, POOL II (r-hFSH+r-hLH) vs POOL I (r-hFSH). (B) miRNAs regulated in COCs of the two younger groups (≤35 years) under different medically assisted reproduction stimulation, POOL IV (r-hFSH+r-hLH) vs POOL III (r-hFSH). Fold change is shown in log2 scale.
Identification of differentially expressed miRNA target genes—in silico analysis
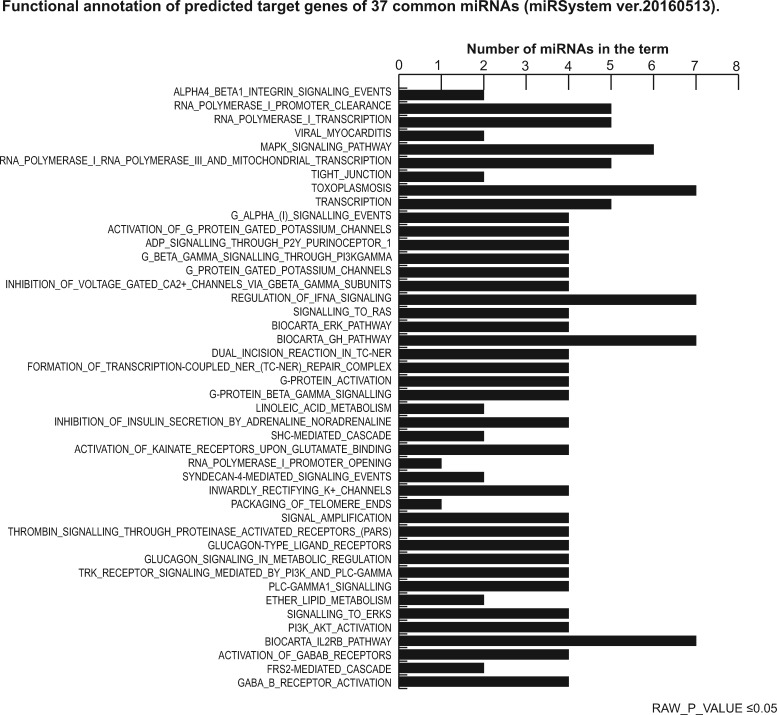
Precise target prediction is crucial to reveal miRNA functions. Target prediction and functional analysis for the 37 common miRNAs were performed using the miRSystem database and applying a stringent miRNA target filter tool: HIT value ≥4; O/E ratio ≥2; functional annotation raw P-value ≤0.05; Kyoto Encyclopedia of Genes and Genomes (KEGG) ranking score ≥1. We identified 3781 putative miRNA target genes, which were experimentally validated and displayed highly confident prediction (Supplementary Table SII). The target gene list of the 37 miRNAs commonly expressed in each pool was used to interpret the biological functions affected by these miRNAs. Functional annotation and relative KEGG pathway enrichment were performed. The top scored processes affected by the 37 common miRNAs are reported in Figs. 4 and 5, showing significantly enriched pathways: mitogen-activated protein kinase (MAPK) signaling pathways, G-protein signaling, transcription regulation, tight junction, RNA polymerase I and III, hormone response signaling and mitochondrial transcription. Putative classification according to biological functions suggested a role for these miRNAs in regulating gene activity related to oocyte growth and maturation, embryo development, steroidogenesis, ovarian hyperstimulation, polycystic ovary syndrome (PCOS), apoptosis and cell survival, glucagon and lipid metabolism and cell trafficking (Table III).
Figure 4.
Functional annotation of putative target genes of 37 common miRNAs (using miRSystem ver.20160513). Raw P-value ≤0.05. MAPK, mitogen-activated protein kinase; SHC, Src Homology 2 Domain Containing; PLC-GAMMA1, Phospholipase C, gamma 1; PI3K, Phosphatidylinositol 3-Kinase; AKT, Serine/Threonine Kinase; FRS-2, fibroblast growth factor receptor substrate 2; GABA_B, receptor: gamma-aminobutyric acid receptor.
Table III.
List of the 44 processes of associated miRNA target genes.
| CATEGORY | TERM | TERM_ID | MIRS_IN_ THE_TERM | RAW_P_ VALUE |
|---|---|---|---|---|
| PATHWAY_INTERACTION_ DATABASE | ALPHA4_BETA1_INTEGRIN_SIGNALING_EVENTS | 200222 | 2 | 0.000857429 |
| REACTOME | RNA_POLYMERASE_I_PROMOTER_CLEARANCE | REACT_1974 | 5 | 0.00231918 |
| REACTOME | RNA_POLYMERASE_I_TRANSCRIPTION | REACT_1309 | 5 | 0.00249289 |
| KEGG | VIRAL_MYOCARDITIS | 5416 | 2 | 0.0039772 |
| KEGG | MAPK_SIGNALING_PATHWAY | 4010 | 6 | 0.00543667 |
| REACTOME | RNA_POLYMERASE_I_RNA_POLYMERASE_III_AND_MITOCHONDRIAL_TRANSCRIPTION | REACT_21352 | 5 | 0.00656541 |
| KEGG | TIGHT_JUNCTION | 4530 | 2 | 0.0131535 |
| KEGG | TOXOPLASMOSIS | 5145 | 7 | 0.0131535 |
| REACTOME | TRANSCRIPTION | REACT_1788 | 5 | 0.0223681 |
| REACTOME | G_ALPHA_(I)_SIGNALLING_EVENTS | REACT_19231 | 4 | 0.0277461 |
| REACTOME | ACTIVATION_OF_G_PROTEIN_GATED_POTASSIUM_CHANNELS | REACT_75831 | 4 | 0.0330182 |
| REACTOME | ADP_SIGNALLING_THROUGH_P2Y_PURINOCEPTOR_1 | REACT_19140 | 4 | 0.0330182 |
| REACTOME | G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | REACT_19290 | 4 | 0.0330182 |
| REACTOME | G_PROTEIN_GATED_POTASSIUM_CHANNELS | REACT_75780 | 4 | 0.0330182 |
| REACTOME | INHIBITION_OF_VOLTAGE_GATED_CA2+_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | REACT_25004 | 4 | 0.0330182 |
| REACTOME | REGULATION_OF_IFNA_SIGNALING | REACT_25216 | 7 | 0.0330182 |
| REACTOME | SIGNALLING_TO_RAS | REACT_12033 | 4 | 0.0355656 |
| BIOCARTA | BIOCARTA_ERK_PATHWAY | 4 | 0.0368342 | |
| BIOCARTA | BIOCARTA_GH_PATHWAY | 7 | 0.0368342 | |
| REACTOME | DUAL_INCISION_REACTION_IN_TC-NER | REACT_2222 | 4 | 0.0368342 |
| REACTOME | FORMATION_OF_TRANSCRIPTION-COUPLED_NER_(TC-NER) REPAIR_COMPLEX | REACT_1941 | 4 | 0.0368342 |
| REACTOME | G-PROTEIN_ACTIVATION | REACT_15457 | 4 | 0.0368342 |
| REACTOME | G-PROTEIN_BETA_GAMMA_SIGNALLING | REACT_19388 | 4 | 0.0368342 |
| KEGG | LINOLEIC_ACID_METABOLISM | 591 | 2 | 0.0380994 |
| REACTOME | INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | REACT_18339 | 4 | 0.0380994 |
| REACTOME | SHC-MEDIATED_CASCADE | REACT_21374 | 2 | 0.0380994 |
| REACTOME | ACTIVATION_OF_KAINATE_RECEPTORS_UPON_GLUTAMATE_BINDING | REACT_21312 | 4 | 0.0393612 |
| REACTOME | RNA_POLYMERASE_I_PROMOTER_OPENING | REACT_2232 | 1 | 0.0393612 |
| PATHWAY_INTERACTION_ DATABASE | SYNDECAN-4-MEDIATED_SIGNALING_EVENTS | 200135 | 2 | 0.0406196 |
| REACTOME | INWARDLY_RECTIFYING_K+_CHANNELS | REACT_75918 | 4 | 0.0406196 |
| REACTOME | PACKAGING_OF_TELOMERE_ENDS | REACT_7963 | 1 | 0.0406196 |
| REACTOME | SIGNAL_AMPLIFICATION | REACT_20524 | 4 | 0.0406196 |
| REACTOME | THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_(PARS) | REACT_21384 | 4 | 0.0418745 |
| REACTOME | GLUCAGON-TYPE_LIGAND_RECEPTORS | REACT_18377 | 4 | 0.0431261 |
| REACTOME | GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION | REACT_1665 | 4 | 0.0431261 |
| PATHWAY_INTERACTION_ DATABASE | TRK_RECEPTOR_SIGNALING_MEDIATED_BY_PI3K_AND_PLC-GAMMA | 200215 | 4 | 0.0443744 |
| REACTOME | PLC-GAMMA1_SIGNALLING | REACT_12079 | 4 | 0.0443744 |
| KEGG | ETHER_LIPID_METABOLISM | 565 | 2 | 0.0456192 |
| REACTOME | SIGNALLING_TO_ERKS | REACT_12058 | 4 | 0.0456192 |
| REACTOME | PI3K_AKT_ACTIVATION | REACT_12464 | 4 | 0.0480988 |
| BIOCARTA | BIOCARTA_IL2RB_PATHWAY | 7 | 0.0493336 | |
| REACTOME | ACTIVATION_OF_GABAB_RECEPTORS | REACT_25330 | 4 | 0.0493336 |
| REACTOME | FRS2-MEDIATED_CASCADE | REACT_21247 | 2 | 0.0493336 |
| REACTOME | GABA_B_RECEPTOR_ACTIVATION | REACT_25031 | 4 | 0.0493336 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; SHC, Src homology 2 domain containing; PLC-GAMMA1, phospholipase C, gamma 1; PI3K, phosphatidylinositol 3-kinase; AKT, serine/threonine kinase; FRS-2, fibroblast growth factor receptor substrate 2; TRK, tropomyosin receptor kinase; GABA_B: receptor, gamma-aminobutyric acid receptor.
Validation of selected miRNA expression levels
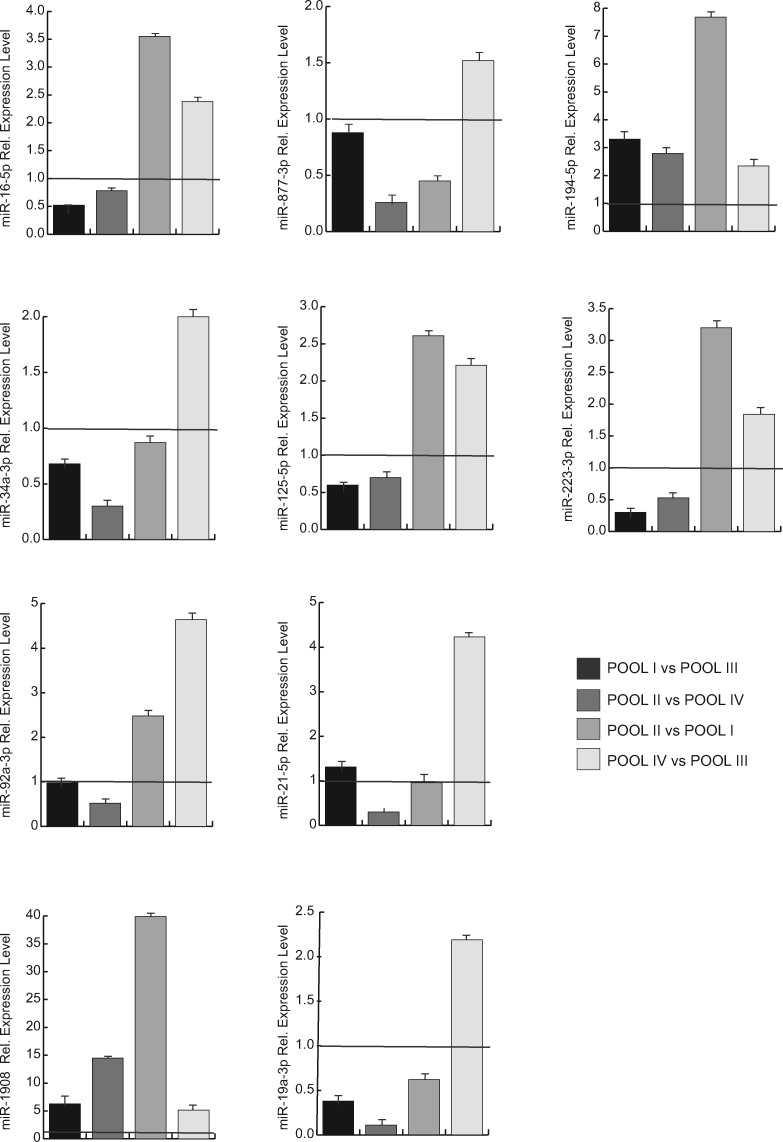
To further validate the modulation of miRNA expression in different pools, we tested the results obtained by real-time PCR. We selected 10 miRNAs (miR-19a-3p, miR-1908, miR-223-3p, miR-125a-5p, miR-34a-3p, miR-21-5p, miR-92a-3p, miR-194-5p, miR-877-3p, miR-16-5p) that were estimated to be differentially expressed, common or exclusive to each pool. Real-time PCR was performed in samples from 15 women undergoing MAR treatment representative of different pools. Clinical and biological features of the 15 patients included in the validation analysis are reported in Table IV. Data confirmed the results obtained by miRNome profiling (Fig. 6).
Table IV.
Clinical and biological features of 15 women enrolled in the study for cumulus oophorus cell (COC) miRNA validation by real-time PCR.
| N° of validation samples | Pool 1 | Pool 2 | Pool 3 | Pool 4 |
|---|---|---|---|---|
| (n = 8) | (n = 1) | (n = 4) | (n = 2) | |
| Age (years) | ≥36 (38.63 ± 1.3) | ≥36 (41) | ≤ 35 (30 ± 0.82) | ≤ 35 (32 ± 2.83) |
| Stimulation Protocol | r-hFSH | r-hFSH + r-hLH | r-hFSH | r-hFSH + r-hLH |
| BMI | 22.43 ± 2.51 | 22.11 | 21.73 ± 3.77 | 22.02 ± 0.14 |
| LH (mIU/mL) | 7.74 ± 2.05 | 4 | 6.86 ± 2.66 | 6.97 ± 3.32 |
| FSH (mIU/mL) | 8.7 ± 2.12 | 5.6 | 6.35 ± 0.80 | 8.05 ± 0.77 |
| AMH | 2.09 ± 0.92 | 0.92 | 1.56 ± 0.33 | 2.55 ± 0.78 |
| E2 a | 2106.375 ± 1091.39 | 1112 | 2194.5 ± 874.63 | 1434 ± 790.54 |
| P a | 1.09 ± 0.27 | 0.98 | 1.1 ± 0.29 | 0.97 ± 0.18 |
| Endometrial thickness (mm) | 10.5 ± 2 | 11 | 9.87 ± 1.54 | 9.5 ± 0.14 |
| Follicles> 16 mm (n) | 5 ± 2.39 | 5 | 7.5 ± 1.73 | 5.5 ± 0.7 |
| Oocyte retrieval (n) | 10.5 ± 5.09 | 8 | 7.75 ± 2.5 | 9 ± 6.65 |
| Oocyte MII (n) | 5.80 ± 1.64 | 8 | 4.75 ± 1.25 | 5 ± 2.28 |
| Fertilization rate (%) | 57.5% | 80 | 60% | 92.3% |
| β-HCG + (%) | 60% | 0 | 50% | 0% |
| AFC (n) | 10.25 ± 5.41 | 5 | 9.75 ± 4.19 | 12.5 ± 3.53 |
Measured at HGC administration.
The data were expressed as mean ± SD.
Figure 6.
Real-time PCR analysis of expression levels of selected differentially expressed miRNAs in COCs pools. The relative expression of miRNA normalized for RNU6B is shown and plotted as a fold change between different pools: POOL IV vs POOL III; POOL II vs POOL I; POOL II vs POOL IV; POOL I vs POOL III. Results represent the mean ± SD.
Discussion
Folliculogenesis is a complex process that involves intra-follicular and oocyte-derived paracrine signals to create the most appropriate microenvironment for oocyte development. A proper environment is achieved through the fine-tuned transcriptional and post-transcriptional expression of a plethora of genes. The essential equilibrium in signal production defines sophisticated molecular networks that govern successful fertilization and embryo development.
Communication between the oocyte and its companion COCs is crucial in the acquisition of developmental competency of the oocyte (Liu et al., 2015; Andrei et al., 2018). Although several studies recently suggested the role of post-translational regulation by miRNAs in ovarian follicular development and ovulation (Imbar and Eisenberg, 2014; Maalouf et al., 2016; Tesfaye et al., 2018; Yerushalmi, 2018), no conclusive evidence has yet been presented.
This study investigated the molecular and regulatory mechanisms mediated by miRNAs in COCs affecting the regulation of oocyte competence in women subjected to MAR. Patients undergoing MAR treatment were divided into four subgroups based on age and gonadotropin treatment strategy, and a number of selective (Supplementary Table SI) and co-expressed (Table III) miRNAs were identified.
Taken together, our findings suggest that aging and MAR stimulation modulate the miRNA landscape as well as the functional properties of COCs. A correlation between increasing maternal age and decreasing births is known to exist (Nelson and Lawlor, 2011). This is due to a reduction in ovarian reserve and oocyte quality, which may contribute to failure of MAR. We investigated the impact of aging on miRNA expression in COCs by performing a differential analysis between pools of women receiving the same treatment but with different age ranges. Differential analysis of miRNome between POOL I and POOL III showed 32 hits regulated in COCs from older compared to younger women (Fig. 2A). In contrast, a cluster of 36 downregulated miRNAs was identified in COCs differentially regulated in older women stimulated with r-hFSH+r-hLH (POOL II vs POOL IV; Fig. 2B). We identified a set of 13 miRNAs (hsa-miR-663a, hsa-miR-320a, hsa-miR-211, hsa-miR-320b, hsa-miR-937-3p, hsa-miR-454-5p, hsa-miR-548c-5p, hsa-miR-105-3p, hsa-miR-34a-3p, hsa-miR-1224-3p, hsa-miR-604, hsa-miR-1269a, hsa-miR-877-3p) upregulate in younger group but after combined r-hFSH+r-hLH treatment are downregulated. In addition, our data clarified the influence of combined r-hFSH+r-hLH therapy on miRNA expression. We found a total of 18 downregulated and 5 upregulated miRNAs in the older groups (POOL II vs POOL I, ≥36 years). Addition of r-hLH to MAR therapy in COCs from younger women (POOL IV vs POOL III, ≤35 years) upregulated 26 miRNAs (miR-877 3p, miR-1224 3p, miR-21-5p, miR-34a-3p, miR-202-3p, miR-25-3p, miR-105-3p, miR-454-5p, miR-663a, miR-92a-3p, let-7c-5p, miR-937-3p, miR-509-3p, miR-30b-5p, miR-320a, miR-1269a, miR-16-5p, miR-604, miR-548c-5p, miR-320b, miR-99a-5p, miR-23b-3p, miR-191-5p, miR132-3p, let-7g-5p, miR-223-3p), which were lower expressed in the older POOL II than in POOL IV. Only miR-223-3p was upregulated by additional stimulation of r-hLH in both age ranges. Interestingly, all comparative analyses showed a prominent r-hLH action on miRNA profiling, mainly downregulating their expression. Functional annotation indicated that almost all these miRNAs target genes in the MAPK signaling pathway. To elucidate the functions of these miRNAs in relation to folliculogenesis and their impact on oocyte quality, we deeply examined our results and found that the miRNOME profile of aged COCs, regardless of hormone stimulation, exhibits a diminished number of miRNAs with a lower basal expression level. The observed trend of miRNA downregulation, albeit to a lesser extent, was also confirmed by comparing different gonadotropin treatments (r-hFSH+r-hLH vs r-hFSH) in aged patient groups. A massive upregulation of shared miRNAs was found in younger patients treated with r-hFSH+r-hLH compared to the corresponding r-hFSH patient group. This finding supports the view that ovarian stimulation with a combination of two different gonadotropins, namely r-hFSH+r-hLH, drives a significant modulation of miRNA expression, particularly exacerbated in younger women.
It is tempting to speculate that miRNA composition and arrangement are associated with the aging of COCs and, consequently, determine oocyte quality and function. Comparative analysis also revealed 37 common miRNAs that were differentially expressed in all four pools. Output data revealed a significant enrichment of critical miRNAs in oocyte function. The let-7 family members (let-7b, let-7a-5p, let-7c-5p, let-7g-5p) we re-expressed and modulated in all clusters were investigated. Unlike r-hFSH patient groups, the majority of let-7 miRNAs were all significantly downregulated in the older r-hFSH+r-hLH group compared to the corresponding younger pool. The let-7 miRNA family members have been implicated in several events during folliculogenesis and embryo implantation in various species: let-7 family members are the most highly expressed miRNAs in mouse ovary (Ahn et al., 2010); high levels of let-7a were observed in quiescent mouse embryos and seem to prevent embryo implantation as well as inhibiting expression of Dicer (Cheong et al., 2014); expression of let-7a, -7 b, -7c, -5p, -7d, -7 h and -7i was found consistently increased in activated follicles (Wong et al., 2018); and low expression of let7-b correlates with the probability of obtaining a blastocyst (Timofeeva et al., 2019).
Many of the common miRNAs identified drive specific granulosa cell functions such as apoptosis, follicular development, ovarian hyperstimulation, cumulus versus mural modulation, and fertility in PCOS. Of these, miR-223-3p was found to be increased after r-hFSH+r-hLH stimulation in both the older and younger pools. Interestingly, miR-223-3p is reported upregulated in cumulus versus mural granulosa cells (Velthut-Meikas et al., 2013) and in human follicular fluid versus oocytes (Machtinger et al., 2017), and seems to affect embryo implantation by suppressing leukemia inhibitory factor expression in endometrium of pregnant mice (Dong et al., 2016). In addition, high levels of miR-15a-5p repress granulosa cell proliferation inducing apoptosis through B-cell lymphoma 2 (BCL2) and BCL2-associated agonist off cell death (BAD), and also by regulating PI3K/AKT/mTOR signaling pathway in young women with poor ovarian response (Zhang et al., 2017). Similarly, miR-202-3p mediates FSH action on oogenesis (Bouchareb et al., 2017). miR-34a was also identified as a promoter of ovarian apoptosis by inhibiting BCL2 (Hou et al., 2019). miR-21 and miR-23a affect cell survival (Yan et al., 2012), while miR-378, miR-21-5p and miR-509-3p regulate oocyte maturation, embryo development and estradiol secretion (Tesfaye et al., 2018).
Interestingly, upregulation of miR-320a and miR-320b was observed in the group of older women receiving r-hFSH treatment. miR-320 is reported to regulate proliferation and steroid production by targeting E2F1 and SF-1 in follicular development (Yin et al., 2014).
Although the idea of differential miRNA expression according to the type of gonadotropin treatment in COCs has long been mooted, very few studies have as yet explored this hypothesis. Scalici et al. (2016) provided clear evidence that r-FSH and highly purified human menopausal gonadotropin stimulation elicit a different expression profile of miR-29a and miR-140 in follicular fluid, and a dramatic upregulation of target miRNAs was observed by increasing the total dose of administered gonadotropins. Since miR-29a levels predicted pregnancy outcome with higher sensitivity in this study, a counteracting synergistic effect driven by gonadotropin treatment(s) has the potential to negatively affect oocyte quality and embryo competence.
Interestingly, our data provide also new and prominent evidence about the role of miRNA mediated regulation on MAPK signaling, in concert to other crucial processes in oocyte ontogeny. Performing target prediction analysis and relative functional annotation using the miRSystem database for the 37 common miRNAs, we identified 36 top-scored pathways associated with MAPK signaling pathways, G-alpha signaling, transcription regulation, tight junctions, RNA polymerase I and III, mitochondrial transcription, RAS signaling, extracellular signal-regulated protein kinase (ERK) signaling and signaling by Class III histone deacetylases (Fig. 5). We recognize the role of MAPK pathway in the regulation of gene expression, cellular growth and survival. Furthermore, we documented that many proteins directly associated to phosphorylation/dephosphorylation processes of MAPK/ERK signaling are fine-regulated by miRNAs and regulate oocyte meiotic cell cycle and maturation. Our findings were supported by characterization of the miRNA regulators of the human ovulatory cascade, properly in COCs, and about the related pathway by Yerushalmi and Moreno groups (Moreno et al., 2015; Yerushalmi et al., 2018). Specifically, we would like to highlight the known role of some predicted target genes in MAPK signaling, such as Nik-related kinase (NRK), in development of trophoblast lineage cells (Morioka et al., 2017); neurofibromatosis-type 1 (NFI), in initiating nucleosome remodeling in oocytes (Belikov et al., 2004); tropomyosin receptor kinases (Trk) receptors, in follicular growth and oocyte survival in the mammalian ovary (Paredes et al., 2004; Harel et al., 2006; Kerr et al., 2009); neurotrophins and neurotrophic tyrosine kinase (NTRK) receptors, in ovarian follicle development (Nilsson et al., 2009) (Seifer et al., 2006); brain-derived neurotrophic factor (BDNF), in nuclear and cytoplasmic maturation of the oocyte (Kawamura et al., 2005).
Figure 5.
Top enriched pathways of associated 37 common miRNAs (using miRSystem ver.20160513). Rank score ≥1. KEGG, Kyoto Encyclopedia of Genes and Genomes; WNT signaling, wingless-type MMTV integration site signaling; ERBB signaling, Erb-B Receptor Tyrosine Kinase signaling.
The bi-directional and dynamic communication between the oocyte and its companion COCs seems to be mediated by miRNA activity mainly on epidermal growth factor (EGF)-EGFR-RAS-c-Jun N-terminal kinases (JNK), IL1-IL1R-p38, RAC/CDC42-serine/threonine p21-activating kinases (PAK)-ERK and neurotrophic factor-mediated Trk receptor networks (by NDEx and KEGG databases analysis). Moreover, putative classification according to biological functions suggests a role for the identified miRNAs in regulating gene activity associated with oocyte growth and maturation, embryo development, steroidogenesis, ovarian hyperstimulation, PCOS, apoptosis and cell survival, glucagon and lipid metabolism, and cell trafficking. We validated selected miRNA expression by real-time PCR in a separate group of 15 patients representative of the four pools.
Taken together, our findings confirm that maternal age is an independent factor impacting miRNA expression in COCs and that gonadotropin treatment may affect follicular miRNA expression and post-transcriptional regulation in women undergoing IVF. The co-treatment of r-hFSH and r-hLH, particularly in older patients, seems to exert the most effective epigenetic remodeling in COCs both in miRNA content and in expression profiling. Whether and to what extent the observed epigenetic impact of r-hLH may account for the increased rate of euploidy, and positive pregnancy outcome previously reported in older patients and poor responders represents an intriguing research challenge that warrants dedicated clinical and experimental efforts. Finally, we also would like to speculate about the identification of a potential miRNA signature in COCs in which MAPK-associated pattern appears to have a crucial role on oocyte development, maturation, selection and function.
Conclusions
Here, we describe the results of what is to our knowledge the largest comprehensive study performed to date addressing the impact of gonadotropin treatment and age on miRNome expression profiles in an IVF setting. Our findings clearly show that miRNA expression in COCs is modulated by gonadotropin treatment and is age-dependent. The identification of miRNA signatures associated with age and/or MAR stimulation in COCs may be useful to discriminate the epigenetic impact of age and/or gonadotropin treatment (r-hFSH vs r-hFSH+r-hLH) on the cellular machinery of COCs and their relevance to oocyte quality and embryo competence.
Supplementary data
Supplementary data are available at Human Reproduction online.
Data availability
The data underlying this article are incorporated and available in the article and in its online supplementary material.
Supplementary Material
Acknowledgments
Special thanks to Catherine Fisher for language editing.
Authors’ roles
C.D. performed the majority of experiments and wrote the manuscript. F.C. performed validation experiments. F.G., S.L. and T.D. were involved in the study design and data interpretation. D.V. contributed to study design, data collection and interpretation. F.C., N.C., S.B., V.P. and E.G. collected COC samples and clinical data. L.A. conceived the study and wrote the manuscript.
Funding
This study was funded by Merck KGaA, Darmstadt, Germany.
Conflict of interest
The authors declare that they have no conflict of interest except S.L. and T.D. who are fully employed by Merck KGaA.
References
- Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, Milosavljevic A, Marra MA, Rajkovic A.. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod 2010;16:463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei D, Nagy RA, van Montfoort A, Tietge U, Terpstra M, Kok K, van den Berg A, Hoek A, Kluiver J, Donker R.. Differential miRNA expression profiles in cumulus and mural granulosa cells from human pre-ovulatory follicles. MicroRNA 2018;8:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia R, Vento ME, Ragusa M, Barbagallo D, La Ferlita A, Di Emidio G, Borzi P, Artini PG, Scollo P, Tatone C. et al. MicroRNAs are stored in human MII oocyte and their expression profile changes in reproductive aging. Biol Reprod 2016;95:131. [DOI] [PubMed] [Google Scholar]
- Belikov S, , Holmqvist P-H, , Åstrand C, , Wrange Ö. Nuclear factor 1 and octamer transcription factor 1 binding preset the chromatin structure of the mouse mammary tumor virus promoter for hormone induction. J Biol Chem 2004;279:49857–49867. [DOI] [PubMed] [Google Scholar]
- Biase FH, Kimble KM.. Functional signaling and gene regulatory networks between the oocyte and the surrounding cumulus cells. BMC Genomics 2018;19:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchareb A, Le Cam A, Montfort J, Gay S, Nguyen T, Bobe J, Thermes V.. Genome-wide identification of novel ovarian-predominant miRNAs: new insights from the medaka (Oryzias latipes). Sci Rep 2017;7:40241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong AW, Pang RT, Liu WM, Kottawatta KS, Lee KF, Yeung WS.. MicroRNA Let-7a and dicer are important in the activation and implantation of delayed implanting mouse embryos. Hum Reprod 2014;29:750–762. [DOI] [PubMed] [Google Scholar]
- Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L.. Impact of maternal age on oocyte and embryo competence. Front Endocrinol (Lausanne) 2018;9:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos MJ, Garcia-Laez V, Beltran-Torregrosa D, Horcajadas JA, Martinez-Conejero JA, Esteban FJ, Pellicer A, Labarta E.. Hormonal and molecular characterization of follicular fluid, cumulus cells and oocytes from pre-ovulatory follicles in stimulated and unstimulated cycles. Hum Reprod 2012;27:1596–1605. [DOI] [PubMed] [Google Scholar]
- Dong X, Sui C, Huang K, Wang L, Hu D, Xiong T, Wang R, Zhang H.. MicroRNA-223-3p suppresses leukemia inhibitory factor expression and pinopodes formation during embryo implantation in mice. Am J Transl Res 2016;8:1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Egea RR, Puchalt NG, Escriva MM, Varghese AC.. OMICS: current and future perspectives in reproductive medicine and technology. J Hum Reprod Sci 2014;7:73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacone F, Cannarella R, Mongioi LM, Alamo A, Condorelli RA, Calogero AE, La Vignera S.. Epigenetics of male fertility: effects on assisted reproductive techniques. World J Mens Health 2019;37:148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel S, Jin S, Fisch B, Feldberg D, Krissi H, Felz C, Freimann S, Tan SL, Ao A, Abir R. Tyrosine kinase B receptor and its activated neurotrophins in ovaries from human fetuses and adults. Mol Hum Reprod2006;12:357-365. [DOI] [PubMed] [Google Scholar]
- Hilz S, Modzelewski AJ, Cohen PE, Grimson A.. The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development 2016;143:3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holesh JE, Bass ANLord M.. Physiology, Ovulation. Treasure Island, FL: StatPearls Publishing, 2020. [PubMed] [Google Scholar]
- Hou Y, Wang Y, Xu S, Qi G, Wu X.. Bioinformatics identification of microRNAs involved in polycystic ovary syndrome based on microarray data. Mol Med Rep 2019;20:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Takahashi T, Nagase S.. Oocyte aging underlies female reproductive aging: biological mechanisms and therapeutic strategies. Reprod Med Biol 2015;14:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbar T, Eisenberg I.. Regulatory role of microRNAs in ovarian function. Fertil Steril 2014;101:1524–1530. [DOI] [PubMed] [Google Scholar]
- Kawamura K, , Kawamura N, , Mulders S M, , Sollewijn Gelpke MD, , Hsueh AJW. Ovarian brain-derived neurotrophic factor (BDNF) promotes the development of oocytes into preimplantation embryos. Proc Natl Acad Sci USA 2005;102:9206–9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, , Garcia-Rudaz C, , Dorfman M, , Paredes A, , Ojeda SR. NTRK1 and NTRK2 receptors facilitate follicle assembly and early follicular development in the mouse ovary. Reproduction 2009; 138:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang SY, Lan KC, Huang HW, Chang CY, Tsai MY, Kung FT, Huang FJ.. Human oocyte maturity in vivo determines the outcome of blastocyst development in vitro. J Assist Reprod Genet 2003;20:506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang X, Shi C, Lin J, Chen G, Wu B, Wu L, Shi H, Yuan Y, Zhou W. et al. Altered microRNAs expression profiling in cumulus cells from patients with polycystic ovary syndrome. J Transl Med 2015;13:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Niu Z, Li Q, Pang RT, Chiu PC, Yeung WS.. MicroRNA and embryo implantation. Am J Reprod Immunol 2016;75:263–271. [DOI] [PubMed] [Google Scholar]
- Luo LF, Hou CC, Yang WX.. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 2016;578:141–157. [DOI] [PubMed] [Google Scholar]
- Maalouf SW, Liu WS, Pate JL.. MicroRNA in ovarian function. Cell Tissue Res 2016;363:7–18. [DOI] [PubMed] [Google Scholar]
- Machtinger R, Rodosthenous RS, Adir M, Mansour A, Racowsky C, Baccarelli AA, Hauser R.. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: an exploratory study. J Assist Reprod Genet 2017;34:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza C, Ruiz-Requena E, Ortega E, Cremades N, Martinez F, Bernabeu R, Greco E, Tesarik J.. Follicular fluid markers of oocyte developmental potential. Hum Reprod 2002;17:1017–1022. [DOI] [PubMed] [Google Scholar]
- Morioka Y, , Nam JM, , Ohashi T. Nik-related kinase regulates trophoblast proliferation and placental development by modulating AKT phosphorylation. PLoS ONE 2017;12:e0171503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JM, Núñez MJ, Quiñonero A, Martínez S, de la Orden M, Simón C, Pellicer A, Díaz-García C, Domínguez F. Follicular fluid and mural granulosa cells microRNA profiles vary in in vitro fertilization patients depending on their age and oocyte maturation stage. FertilSteril 2015;104:1037–1046. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Lawlor DA.. Predicting live birth, preterm delivery, and low birth weight in infants born from in vitro fertilisation: a prospective study of 144,018 treatment cycles. PLoS Med 2011;8:e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Dole G, Skinner MK.. Neurotrophin NT3 promotes ovarian primordial to primary follicle transition. Reproduction 2011;138:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul ABM, Sadek ST, Mahesan AM.. The role of microRNAs in human embryo implantation: a review. J Assist Reprod Genet 2019;36:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A, , Romero C, , Dissen G A, , Dechiara T M, , Reichardt L, , Cornea A, , Ojeda S R, , Xu B. TrkB receptors are required for follicular growth and oocyte survival in the mammalian ovary. Dev Biol 2004;267:430–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinborg A, Loft A, Romundstad LB, Wennerholm UB, Soderstrom-Anttila V, Bergh C, Aittomaki K.. Epigenetics and assisted reproductive technologies. Acta Obstet Gynecol Scand 2016;95:10–15. [DOI] [PubMed] [Google Scholar]
- Pisarska MD, Chan JL, Lawrenson K, Gonzalez TL, Wang ET.. Genetics and epigenetics of infertility and treatments on outcomes. J Clin Endocrinol Metab 2019;104:1871–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LC, Pla I, Sanchez A, Grondahl ML, Marko-Varga G, Yding Andersen C, Englund ALM, Malm J.. Progressive changes in human follicular fluid composition over the course of ovulation: quantitative proteomic analyses. Mol Cell Endocrinol 2019;495:110522. [DOI] [PubMed] [Google Scholar]
- Scalici E, Traver S, Mullet T, Molinari N, Ferrieres A, Brunet C, Belloc S, Hamamah S.. Circulating microRNAs in follicular fluid, powerful tools to explore in vitro fertilization process. Sci Rep 2016;6:24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarica C, Cimadomo D, Dovere L, Giancani A, Stoppa M, Capalbo A, Ubaldi FM, Rienzi L, Canipari R.. An integrated investigation of oocyte developmental competence: expression of key genes in human cumulus cells, morphokinetics of early divisions, blastulation, and euploidy. J Assist Reprod Genet 2019;36:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifer DB, , Feng B, , Shelden RM. Immunocytochemical evidence for the presence and location of the neurotrophin–Trk receptor family in adult human preovulatory ovarian follicles. Am J Obstet Gynecol 2006;194:1129–1134. [DOI] [PubMed] [Google Scholar]
- Tanghe S, Van Soom A, Nauwynck H, Coryn M, de Kruif A.. Minireview: functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol Reprod Dev 2002;61:414–424. [DOI] [PubMed] [Google Scholar]
- Tesfaye D, Gebremedhn S, Salilew-Wondim D, Hailay T, Hoelker M, Grosse-Brinkhaus C, Schellander K.. MicroRNAs: tiny molecules with a significant role in mammalian follicular and oocyte development. Reproduction 2018;155:R121–R135. [DOI] [PubMed] [Google Scholar]
- Timofeeva AV, Chagovets VV, Drapkina YS, Makarova NP, Kalinina EA, Sukhikh GT.. Cell-free, embryo-specific sncRNA as a molecular biological bridge between patient fertility and IVF efficiency. Int J Mol Sci 2019;20:2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong XH, Xu B, Zhang YW, Liu YS, Ma CH.. Research resources: comparative microRNA profiles in human corona radiata cells and cumulus oophorus cells detected by next-generation small RNA sequencing. PLoS One 2014;9:e106706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velthut-Meikas A, Simm J, Tuuri T, Tapanainen JS, Metsis M, Salumets A.. Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Mol Endocrinol 2013;27:1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong QW, Sun MA, Lau SW, Parsania C, Zhou S, Zhong S, Ge W.. Identification and characterization of a specific 13-miRNA expression signature during follicle activation in the zebrafish ovary. Biol Reprod 2018;98:42–53. [DOI] [PubMed] [Google Scholar]
- Yadav RP, Kotaja N.. Small RNAs in spermatogenesis. Mol Cell Endocrinol 2014;382:498–508. [DOI] [PubMed] [Google Scholar]
- Yan G, Zhang L, Fang T, Zhang Q, Wu S, Jiang Y, Sun H, Hu Y.. MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Lett 2012;586:3263–3270. [DOI] [PubMed] [Google Scholar]
- Yerushalmi GM, Salmon-Divon M, Ophir L, Yung Y, Baum M, Coticchio G, Fadini R, Mignini-Renzini M, Dal Canto M, Machtinger R. et al. Characterization of the miRNA regulators of the human ovulatory cascade. Sci Rep 2018;8:15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wang X, Yao G, Lu M, Liang M, Sun Y, Sun F.. Transactivation of micrornA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. J Biol Chem 2014;289:18239–18257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhong W, Li WP, Chen ZJ, Zhang C.. miR-15a-5p levels correlate with poor ovarian response in human follicular fluid. Reproduction 2017;154:483–496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are incorporated and available in the article and in its online supplementary material.