Abstract
BACKGROUND
Colon cancer cell lines are widely used for research and for the screening of drugs that specifically target the stem cell compartment of colon cancers. It was reported that colon cancer carcinoma specimens contain a subset of leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)-expressing stem cells, these so-called “tumour-initiating” cells, reminiscent in their properties of the normal intestinal stem cells (ISCs), may explain the apparent heterogeneity of colon cancer cell lines. Also, colon cancer is initiated by aberrant Wnt signaling in ISCs known to express high levels of LGR5. Furthermore, in vivo reports demonstrate the clonal expansion of intestinal adenomas from a single LGR5-expressing cell.
AIM
To investigate whether colon cancer cell lines contain cancer stem cells and to characterize these putative cancer stem cells.
METHODS
A portable fluorescent reporter construct based on a conserved fragment of the LGR5 promoter was used to isolate the cell compartments expressing different levels of LGR5 in two widely used colon cancer cell lines (Caco-2 and LoVo). These cells were then characterized according to their proliferation capacity, gene expression signatures of ISC markers, and their tumorigenic properties in vivo and in vitro.
RESULTS
The data revealed that the LGR5 reporter can be used to identify and isolate a classical intestinal crypt stem cell-like population from the Caco-2, but not from the LoVo, cell lines, in which the cancer stem cell population is more akin to B lymphoma Moloney murine leukemia virus insertion region 1 homolog (+4 crypt) stem cells. This sub-population within Caco-2 cells exhibits an intestinal cancer stem cell gene expression signature and can both self-renew and generate differentiated LGR5 negative progeny. Our data also show that cells expressing high levels of LGR5/enhanced yellow fluorescent protein (EYFP) from this cell line exhibit tumorigenic-like properties in vivo and in vitro. In contrast, cell compartments of LoVo that are expressing high levels of LGR5/EYFP did not show these stem cell-like properties. Thus, cells that exhibit high levels of LGR5/EYFP expression represent the cancer stem cell compartment of Caco-2 colon cancer cells, but not LoVo cells.
CONCLUSION
Our findings highlight the presence of a spectrum of different ISC-like compartments in different colon cancer cell lines. Their existence is an important consideration for their screening applications and should be taken into account when interpreting drug screening data. We have generated a portable LGR5-reporter that serves as a valuable tool for the identification and isolation of different colon cancer stem cell populations in colon cancer lines.
Keywords: Colorectal cancer, Colon cancer cell lines, Intestinal stem cell, Cancer stem cell, Leucine-rich repeat-containing G protein-coupled receptor 5, Heterogenicity
Core Tip: The intestinal epithelium harbors two distinct pools of putative stem cells, the leucine-rich repeat-containing G protein-coupled receptor 5+ (LGR5) stem cell population and the B lymphoma Moloney murine leukemia virus insertion region 1 homolog+ stem cell population. Colon cancer cell lines such as Caco-2 and LoVo are extensively used in colon cancer research, and express high levels of LGR5. Here, we aimed to investigate whether colon cancer cell lines contain cancer stem cells and characterized these cells. Using an LGR5 reporter, we characterized LGR5+ cells and revealed that Caco-2 cell line contains a classical intestinal stem cell-like population (LGR5+). However, in LoVo cell lines, stem cell-like population is more akin to the B lymphoma Moloney murine leukemia virus insertion region 1 homolog+ stem cells.
INTRODUCTION
Colorectal cancer (CRC) remains a leading cause of morbidity and mortality worldwide[1], highlighting the need for more effective therapeutics. It is now well established that both blood and solid cancers are initiated and propagated from a subset of rare cells, called cancer stem cells (CSCs), which are resistant to chemotherapy or radiation[2-4]. Current evidence indicates that CRC is indeed a disease of colon stem cells[2,5,6] that are resistant to current therapeutics and can rapidly proliferate to re-establish the tumor[7]. The intestinal crypt-villous structure harbors two distinct pools of putative stem cells[8]. One pool is located at the crypt base and is characterized by the expression of leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), and the other pool resides at +4 position and consists of B lymphoma Moloney murine leukemia virus (Mo-MLV) insertion region 1 homolog (Bmi-1) and telomerase reverse transcriptase (TERT) expressing cells[9-11]. The hierarchy of these stem cell pools in the normal crypt and their respective contributions to colon cancer and relapse following therapy is under debate. Some suggest both pools contribute equally to maintenance of the crypt, following a pattern of neutral drift[12], while others propose that Lgr5+ stem cells (SCs) comprise the active population of the crypt and that Bmi-1+ or TERT+ cells are quiescent SCs that represent a reserve pool of SCs with the ability to replace Lgr-5+ cells in case of loss or injury[8,13]. The existence of CSCs within intestinal tumors was most elegantly demonstrated by Schepers et al[14] using lineage re-tracing with a multicolor Cre-reporter R26R-confetti targeted to a single adenomatous polyposis coli allele in mouse intestinal stem (LGR5+) cells, demonstrating that these intestinal adenomas were maintained by CSCs that exist in a cellular hierarchy. LGR5 is a receptor for R-spondins and activates potent Wnt signal enhancers such as Rnf43 and Znrf3[15]. LGR5 is present in various stem cell compartments throughout the body[16], including the intestine, and is an exquisite marker of intestinal stem cells (ISCs) capable of forming the entire intestinal mucosa[9]. LGR5+ ISCs persist in adenoma and can initiate CRC through the activation of the Wnt/β-catenin signaling pathway[17].
Human colon cancer cell lines such as Caco-2 and LoVo are extensively used in drug screening and colon cancer research and express high levels of LGR5[18-20]. To establish criteria for the selection of suitable cell lines for drug discovery, several studies have investigated variability between the colon cell lines, identifying characteristic differences in gene expression signature, epigenetic and genetic make-up, migratory abilities, and proliferative capacities between different intestinal cancer and cell lines, including Caco-2 and LoVo[21,22]. However, the CSC population(s) within these cell lines remains to be fully characterized. This is important since anticancer drugs often attempt to target these CSCs[23,24].
To isolate the CSC compartment from colon cancer cell lines we constructed a reporter based on a conserved promoter fragment of the LGR5 gene. After validating the fidelity of this LGR5 reporter, we showed that it can be used as a genetic tool to isolate LGR5-expressing CSCs from Caco-2 and LoVo human colon cancer cell lines. Surprisingly, our data revealed that these two widely used colon cancer lines possess different stem cell compartments. We conclude that this portable LGR5 reporter constitutes a valuable tool for the development of colon cancer therapeutics specifically designed to ablate these CSCs.
MATERIALS AND METHODS
Cell culture
Human colon cancer cell lines (LoVo, Caco-2, and SW480), a human fibroblast cell line (CRL-2429), a mouse fibroblast cell line (3T3), and a mouse motor neuron cell line (NSC-34) were obtained from ATCC and maintained in Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA, United States) supplemented with fetal bovine serum (FBS, Bovogen Biologicals, Keilor East, Australia) to a final concentration of 10% and passaged every 3-4 d at 80% confluency. The human neuroblastoma cell line (SH-SY5Y from ATCC) was maintained in DMEM supplemented with 20% FBS and passaged every 2-3 d. All cell lines used for experiments were at early (a maximum of 20-25) passage number.
Construction of LGR5 promoter-based reporter constructs
Conservation of the LGR5 gene and its promoter was assessed using evolutionary conserved region browser and multiple alignments of several vertebrate genomes. The human LGR5 gene is located on chromosome 12 and has a conserved region upstream of the main promoter. This gene segment [LGR5 promoter element (983 bp)] was amplified from RP11-59F15 bacterial artificial chromosome clones (obtained from the Australian Genome Research Facility, Ltd., Melbourne, Australia) by polymerase chain reaction (PCR) (Supplementary Figure 1A) using a set of primers (Table 1). Sequences for specific restriction sites (Nde I, Nhe I, Sac I, and Xho I) were added to allow the insertion of the amplicon into the pEYFP-N1 vector (Clontech, Mountain View, CA, United States). Subsequently, this purified deoxyribonucleic acid (DNA) fragment was A-tailed and ligated into pGEM®-T Easy Vector System I (Madison, WI, Promega). White colonies were selected and plasmid DNA isolated, purified, and sequenced. Verified plasmids containing the DNA of interest, which is also free of PCR-introduced mutations, were then digested and ligated into the pEYFP-N1 vector. A negative control clone was constructed by the deletion of the promoter insert from the promoter-pEYFP-N1 clone. The original pEYFP-N1 vector (Clontech) with the CMV promoter was used as a positive control.
Table 1.
Primer sequences
|
Gene
|
Forward primer 5’-3’
|
Reverse primer 5’-3’
|
| GAPDH | TGAAGCTGGAGAAGGAGAAG | ATCGGCCTGTGTATATCCC |
| LGR5 | CCTCTGCTGGCTTTTAGGTG | AGCAGTTTTCAGGCCTTTCA |
| OLFM4 | ACTGTCCGAATTGACATCATGG | TTCTGAGCTTCCACCAAAACTC |
| ASCL2 | ACCTGCGTACCTTGCTTTGG | GAAATCTGCGAGTTCCCGGT |
| Amplification/cloning of the LGR5 promoter element | CATATGCTAGCTCGAGCTCACTTCGACTTCCTCACCCCGC | AAGCTTGGTGCCCGAAGTAGGGGGCCA |
Transient transfection
Cell lines at 50%-60% confluence was transfected with LGR5 promoter-pEYFP-N1 DNA using FuGENE® HD Transfection Reagent (Roche, Basel, Switzerland) following the manufacturer instructions. LoVo, Caco-2, SW480, human fibroblast CRL2923, 3T3, 4T1, and EMT6 cell lines were transfected in 3:1 transfection reagent to DNA ratio in serum-free medium. For the SY5Y cell line, the ratio was 3:2. Eight to ten hours post-transfection, serum was added to the culture medium. All cultures were kept at 37 °C in a 5% CO2 atmosphere until microscopic analyses were performed using an inverted fluorescence microscope (IX51, Nikon, Tokyo, Japan) 24-48 h post-transfection to examine enhanced yellow fluorescent protein (EYFP) expression levels.
Flow cytometry
For flow cytometry and fluorescence-activated cell sorting (FACS), cultured cells were harvested by incubation in a non-enzymatic cell dissociation buffer (Gibco® Life Technologies, Waltham, MA, United States) for 7-10 min. Next, cells were washed in phosphate buffer saline (PBS) and resuspended in 0.5 mL PBS for analysis. Flow cytometric analysis of EYFP expression of live cells was conducted using an Accuri flow cytometer (BD Biosciences, San Jose, CA, United States). FACS sorting was performed using an Influxcell sorter (BD Biosciences). Negative controls were used in every analysis to set the background fluorescence. Propidium Iodide Staining Solution (Invitrogen) was added to discriminate dead cells. Data analysis was performed on at least 10000 cells per sample as assessed by CFlow software (BD Biosciences). Fluorescence gates for positive cells were set to attain the false-positive rates of < 1%.
Colony PCR
A single colony of bacterial cells was suspended in a reaction volume of 50 µL, containing 1 µL of PCR primers (10 µmol/L each), 5 µL of 10 × PCR Buffer Minus Mg2+, 1 µL of 10 mmol/L dNTP mixture, 1.5 µL of 50 mmol/L MgCl2, and Milli-Q water. The cycle parameters of the reaction were as follows: Initial denaturation 95 °C for 3 min; 35 cycles of 30 s for denaturation at 95 °C, 30 s for annealing at 58 °C, and 1 min for the extension at 72 °C then final elongation at 72 °C for 2 min.
Quantitative real-time PCR
RNA extraction and complementary DNA (cDNA) synthesis: RNA was extracted from approximately 5 × 106 cells using RNeasy mini kit (Qiagen, Valencia, CA, United States) following the manufacturer's instructions. DNAse I treatment (on column) was performed during the extraction procedure as recommended by the kit manual. RNA quantification was performed using Nanodrop 1000. One microgram of RNA was used for cDNA synthesis using iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA, United States) following the manufacturer's instructions. As a control for genomic DNA contamination, a 'no reverse transcriptase reaction' was carried out for every batch of cDNA synthesized.
Quantitative real-time PCR analysis: SsoFast™ EvaGreen® Supermix was used as the Master-mix and quantitative real-time PCR (qPCR) was carried out with a C1000™ Thermal Cycler (Bio-Rad Laboratories, Inc.). Each gene was amplified in triplicate in a reaction volume of 10 µL, containing 10 µL qPCR primers (Table 1), 5 µL of 2 × SsoFast EvaGreen Supermix, 0.2 µL cDNA, and Milli-Q water. The cycling parameters of the reaction were as follows: 95 °C for 3 min for enzyme (Sso7-fusion polymerase) activation; 40 cycles of 10 s for denaturation at 95 °C, and 30 s for annealing and extension at 60 °C followed by a Melt Curve analysis to ensure reaction specificity. Glyceraldehyde-3-phosphate dehydrogenase was amplified in parallel and used for normalization. The amplification product was then analyzed using the 2-ΔCt method. ‘No reverse transcriptase’ and water controls were included in each qPCR run to exclude genomic DNA or sample cross-contamination.
Soft agar assay
To test anchorage-independent growth in soft agar, a layer of 0.6% agar (Agar Noble; Difco Laboratories, Detroit, MI, United States) in 2 × DMEM containing 20% FBS was plated in 35-mm dishes and placed in the fridge overnight. The next day, Geneticin resistant (400-500 µg/mL of Geneticin) cells were sorted for EYFP expression into high, low, and no-EYFP expressing cells using the Influxcell sorter (BD Biosciences). Cells were next washed with PBS and pelleted by centrifugation at 300 g for 3 min. Cells (2 × 104) were suspended in 2 mL of equal volumes of 0.6% agar (Agar Noble; Difco Laboratories) and 2 × DMEM containing 20% FBS and overlaid as a second layer. This was overlaid over a layer of 3 mL of 0.3% agar in the same medium, in 35 mm dishes. Once a week, 300 µL of medium containing 0.3% agar was added. After 21 d, plates were stained with 0.5 mL of 0.005% Crystal Violet for an hour. Colonies larger than 0.25 mm in diameter were then counted using ImageJ software.
In vivo tumorigenicity assay
Six- to eight-week-old non-obese diabetic (NOD)/severe combined immunodeficient (SCID) mice were obtained from Animal Resources Centre (Perth, Australia) and were housed in the animal house facility of the Australian Institute for Bioengineering and Nanotechnology, The University of Queensland. All procedures were approved by the University of Queensland Animal Ethics Committee (UQCCR/115/13/NHMRC) and conformed to the Animal Care and Protection Act Qld (2002) and the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (8th edition, 2013). Sorted cells were suspended in a 1:1 mixture of medium and Matrigel (BD Biosciences) and injected subcutaneously into NOD/SCID mice (2 × 105 cells per animal in a total volume of 50 µL) into the ventral side of the flank (n = 3 animals per group). Tumor growth was monitored and measured with calipers daily. Three weeks after the injection, mice were anesthetized by CO2 inhalation, and tumors were excised, measured, weighed, and fixed in 4% paraformaldehyde for paraffin embedding. Tissue sections were stained with hematoxylin-eosin.
RESULTS
A conserved LGR5 enhancer-based promoter fragment recapitulates endogenous LGR5 expression
Due to the lack of a reliable anti-LGR5 antibody, isolation of LGR5 expressing cells via FACS or magnetic-ACS was not possible in our hands. We, therefore, constructed an EYFP reporter driven by a conserved LGR5 promoter fragment (Supplementary Figure 1A). To test whether the LGR5 promoter-based construct correctly reported on LGR5 expression, we transiently transfected the LGR5-EYFP reporter as well as a negative control vector (promoter-less vector or empty-EYFP vector) and a positive control vector (CMV-EYFP) into colon cancer cell lines Caco-2, LoVo, and SW480 that display a high endogenous expression of LGR5 and into CRL2429, 3T3, NSC-34 and SH-SY5Y cells that do not express LGR5 messenger RNA (mRNA). Transient transfection of the LGR5 reporter construct in these LGR5-expressing and LGR5-negative cell lines revealed that EYFP expression correlated well with endogenous LGR5 mRNA expression level (Supplementary Figure 1B). As expected, transfection of the promoter-less construct (empty-EYFP vector) did not result in YFP expression, whereas the CMV-driven EYFP vector showed widespread (30%-60% of cells) expression (Supplementary Figure 1B), indicating that the absence of fluorescence in LGR5 negative lines was not due to ineffective delivery of plasmid DNA.
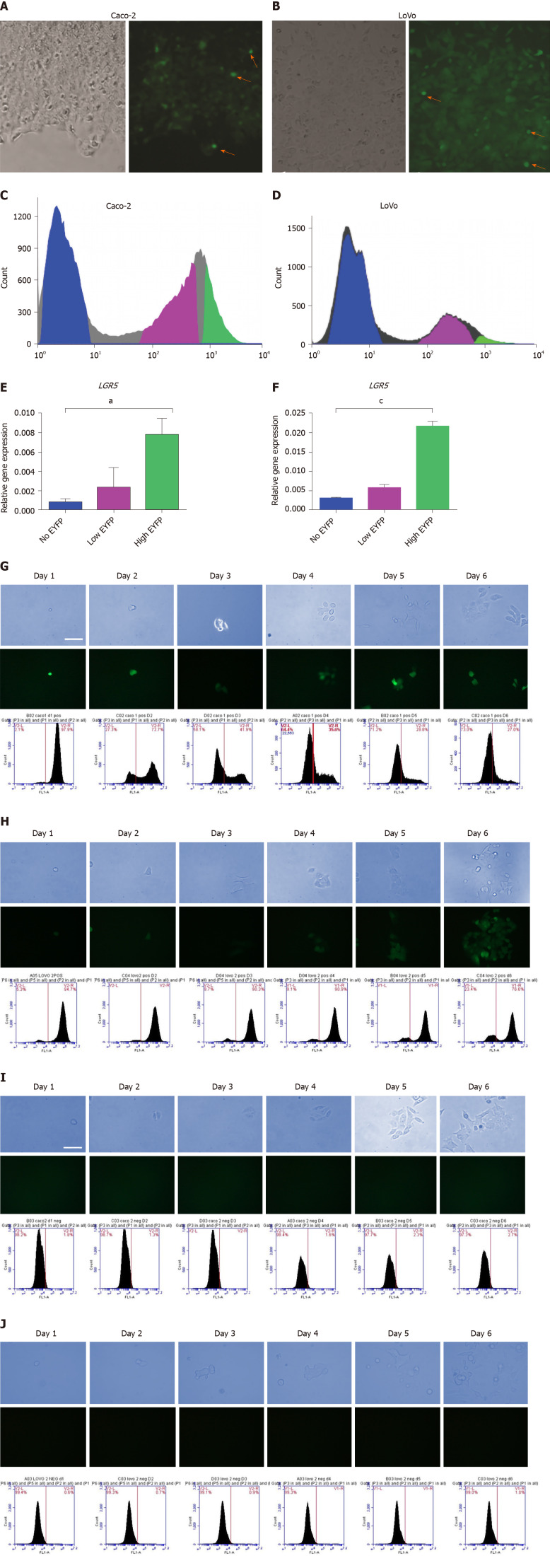
To generate stably expressing reporter lines, Caco-2 and LoVo cell lines were transfected with a linearized LGR5 reporter vector, single-cell seeded, and maintained under Geneticin selection (400 µg/mL for LoVo cell line and 500 µg/mL for Caco-2 cell line) for 1 mo. Two single cell-derived clones from each cell line were picked, expanded, and cryopreserved. Fluorescence microscopy analysis revealed that these clonal LGR5/EYFP Caco-2 and LGR5/EYFP LoVo cultures contained a subset of cells with a range of EYFP fluorescence, including small EYFP fluorescence bright cells (Figure 1A and B; orange arrows), which was validated using FACS (Figure 1C and D). Cells transfected with promoter-less plasmid showed no EYFP expression in any of the surviving clones, whereas the majority of CMV-promoter transfected clones homogeneously expressed EYFP, as expected. We hypothesized that cells expressing high levels of LGR5/EYFP in the colon cancer cell lines would constitute the stem cell compartment of Caco-2 and LoVo cell lines. To test this hypothesis, we sorted the cells into three populations according to their EYFP expression (EYFPnegative, EYFPlow, and EYFPhigh cells). As anticipated, the qPCR analysis showed that LGR5 mRNA expression was 8-fold higher in EYFP high cells than in the EYFP negative fraction in both LGR5/EYFP Caco-2 and LGR5/EYFP LoVo clonal lines (Figure 1E and F, respectively).
Figure 1.
Proliferation and differentiation analysis of the stem cell compartment in stably transfected clones. A and B: Phase contrast and fluorescence images show colonies of stably transfected Caco-2 (A) and LoVo (B) cells expressing the enhanced yellow fluorescent protein (EYFP) reporter (red arrows show small cells expressing high level of enhanced yellow fluorescent protein, EYFP); C and E: Fluorescence-activated cell sorting (FACS) sorting of leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)/EYFP expression in Caco-2 clone (C) and LoVo clones (E) into EYFPhigh, EYFPlow, and EYFPnegative populations; D and F: Quantitative real-time polymerase chain reaction analysis of LGR5 messenger ribonucleic acid expression in EYFPhigh, EYFPlow, and EYFPnegative cells relative to glyceraldehyde-3-phosphate dehydrogenase gene in Caco-2 clones (D) and LoVo clones (F); G: Phase contrast images show proliferation of EYFPhigh cells from LGR5/EYFP Caco-2 clone; fluorescence images show self-renewal of LGR5/EYFP stem cells and down-regulation of LGR5/EYFP in differentiated progeny in LGR5/EYFP Caco-2. Sequential FACS analysis shows EYFP expression in LGR5/EYFP Caco-2 clone over 6 d in culture; H: Phase contrast images show proliferation of EYFPnegative and cells from LGR5/EYFP Caco-2 clone; fluorescence images show self-renewal of LGR5 stem cells and down-regulation of LGR5 in differentiated progeny in LGR5/EYFP Caco-2. Sequential FACS analysis shows EYFP expression in LGR5/EYFP Caco-2 clone over 6 d in culture; I and J: Phase contrast images show proliferation of EYFPhigh (I) and EYFPnegative (J) cells from LGR5/EYFP LoVo clone; fluorescent pictures showing self-renewal of LGR5 stem cells and down-regulation of LGR5/EYFP in differentiated progeny in LGR5/EYFP LoVo; sequential FACS analysis of EYFP expression in LGR5/EYFP LoVo clone over 6 d in culture. LGR5: Leucine-rich repeat-containing G protein-coupled receptor 5; EYFP: Enhanced yellow fluorescent protein.
In agreement with the idea that LGR5 marks a stem cell compartment, flow cytometric analysis of sorted EYFPnegative, EYFPlow, and EYFPhigh cells from each of the lines revealed that over the course of several days, EYFPhigh cells gave rise to EYFP negative “daughter” cells (Figure 1G and H). Time-lapse immunofluorescence and phase-contrast images of cultured EYFPhigh sorted Caco-2 cells confirmed that EYFP negative cells emerged from the small EYFPhigh cells over time (Figure 1G). No cells acquired EYFP expression amongst derivatives of FACS-sorted EYFP-negative cells (Figure 1I). LGR5-EYFP LoVo clones exhibited identical behaviors (Figure 1H and J). We interpreted these data to mean that LGR5high (EYFPhigh) cells possess stem cell-like properties, given they are capable of self-renewal and generate a distinct EYFP-negative cell population, incapable of re-acquiring expression of the LGR5 reporter.
EYFPhigh-expressing cells of Caco-2 and LoVo CRC cell lines show an ISC gene expression signature
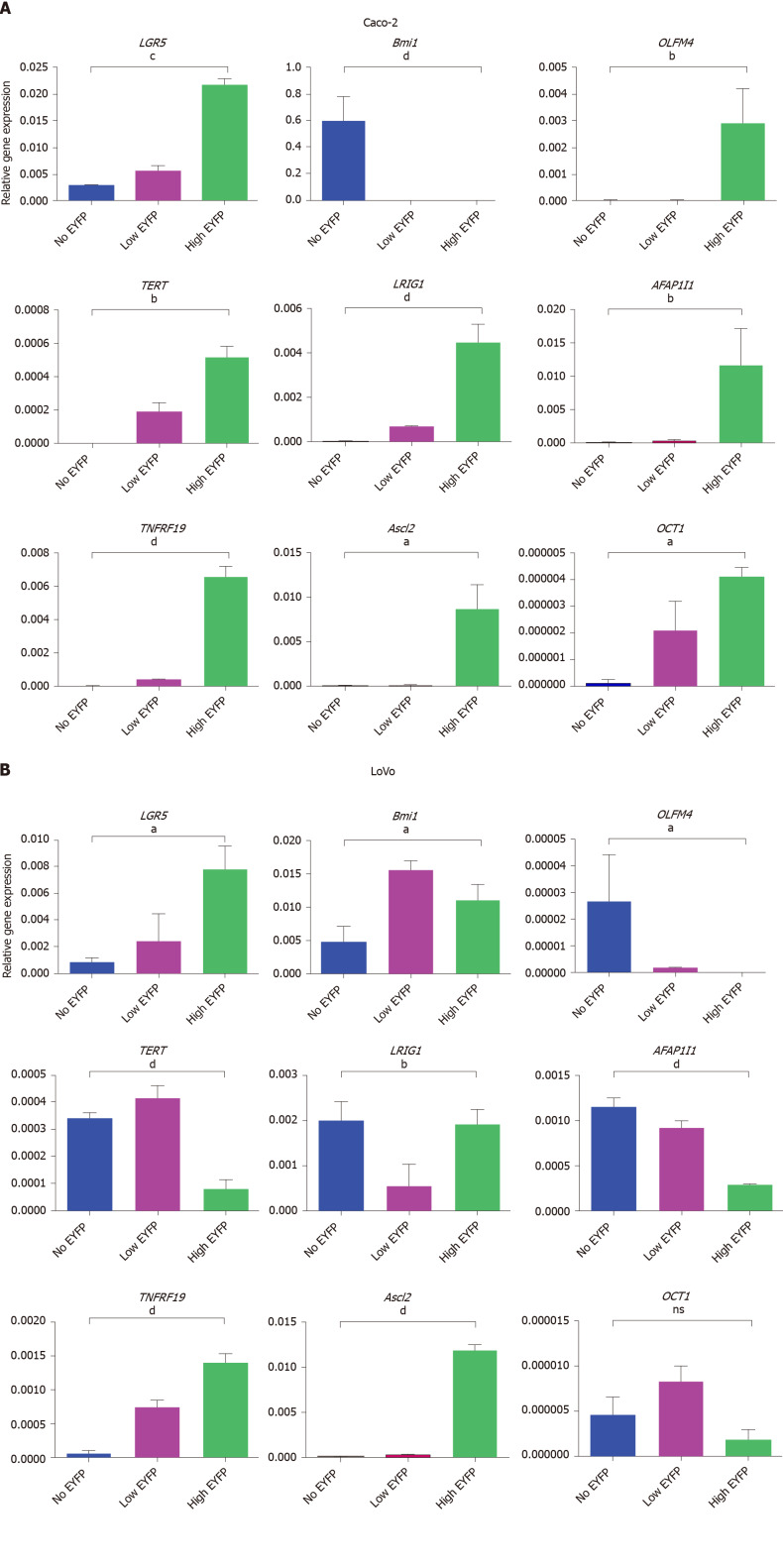
If LGR5/EYFPhigh cell populations in Caco-2 and LoVo clones represent an ISC compartment, then these cells should show higher mRNA expression of typical ISC marker genes compared to LGR5/EYFPlow and LGR5/EYFPnegative fractions. Our data (Figure 2A) show that EYFPhigh cells in the Caco-2 cell line indeed display significantly increased expression of the stem cell markers LGR5, olfactomedin 4 (OLFM4), TERT, leucine-rich repeated and immunoglobulin-like domains 1 (LRIG1), AFAP1I1, TNFRF19, organic cation transporter 1 (OCT1), and achaete-scute complex homolog 2 as compared to cells with low or no EYFP expression, but conspicuously do not express B lymphoma Moloney murine leukemia virus insertion region 1 homolog (Bmi1). Surprisingly, in the LoVo cell line (Figure 2B) EYFPhigh cells expressing high LGR5 reporter activity possessed low levels of OLFM4, AFAP1I1, and LRIG1 compared to EYFPlowand EYFPnegative fractions. EYFPhigh cells in the LoVo cell line instead robustly express Bmi1 as compared to EYFPnegative cells but exhibit lower hTERT as compared to EYFPnegative and EYFPlow fractions. These data suggest that in the LoVo cell lines LGR5 marks a different cancer cell compartment than in Caco-2 cells. Collectively, our data indicate that the LGR5/EYFP reporter faithfully marks a population of LGR5 expressing CSCs in the Caco-2 cell line but that this stem cell population is distinct from the EYFPhigh fraction in the LoVo cell line that appears to mark a Bmi1high/LGR5 TERTlow expressing a subset of cells.
Figure 2.
Quantitative polymerase chain reaction analysis. A and B: Quantitative polymerase chain reaction analysis of intestinal stem cell markers and the cancer stem cell marker organic cation transporter 1 in leucine-rich repeat-containing G protein-coupled receptor 5/enhanced yellow fluorescent protein (LGR5/EYFP)high, LGR5/EYFPlow, and LGR5/EYFPnegative fractions of the Caco-2 cell line (A) and the LoVo cell line (B). The expression value of each gene was normalized to glyceraldehyde-3-phosphate dehydrogenase gene. Data were then analyzed by one-way analysis of variance. aP < 0.05, bP < 0.01, cP < 0.001, dP < 0.0001. LGR5: Leucine-rich repeat-containing G protein-coupled receptor 5; EYFP: Enhanced yellow fluorescent protein; Bmi1: B lymphoma Moloney murine leukemia virus insertion region 1 homolog; OCT1: organic cation transporter 1; TERT: Telomerase reverse transcriptase; ASCL2: Achaete-scute complex homolog 2.
LGR5/EYFPhigh cells in Caco-2 colon cancer cell line exhibit increased colony-forming ability
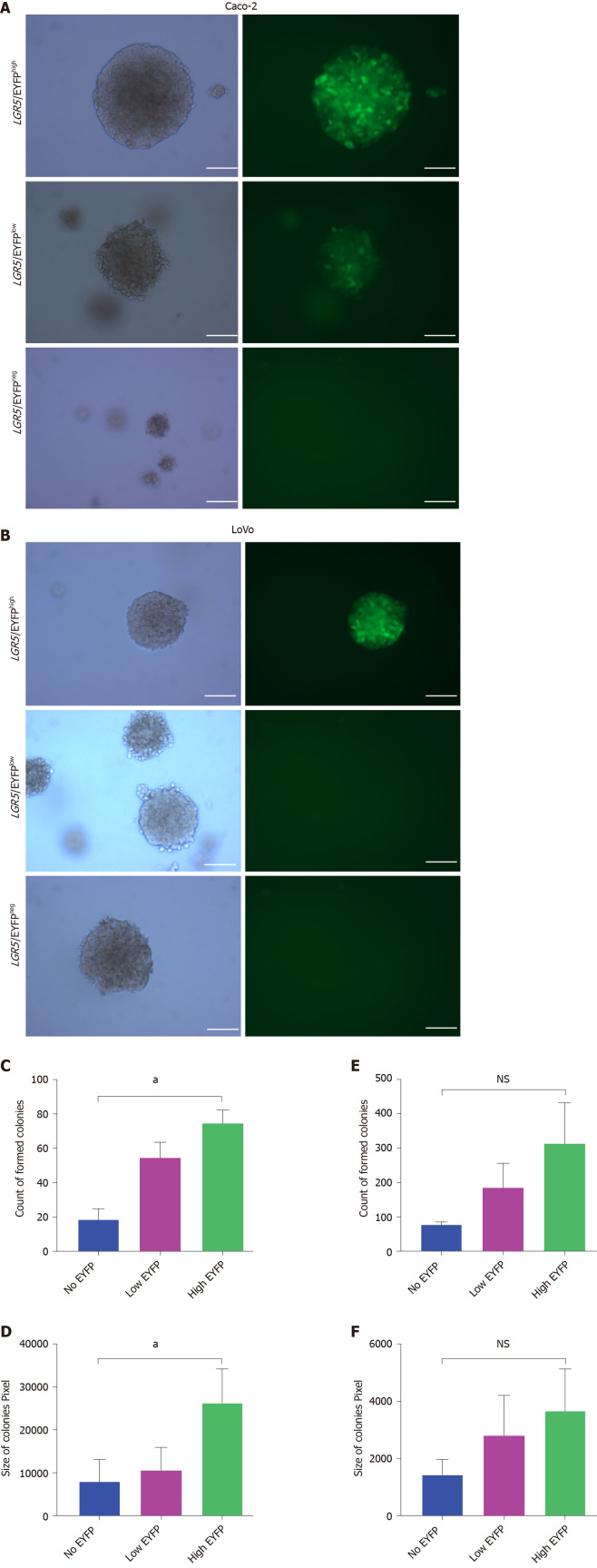
To assess whether LGR5 expressing subsets in the Caco-2 and LoVo lines exhibit functional differences we employed the anchorage-independent tumor stem cell colony-forming assay[25]. Using this assay, we assessed the in vitro anchorage-independent growth of LGR5/EYFPhigh, LGR5/EYFPlow, and LGR5/EYFPnegative cell populations of the LGR5/EYFP Caco-2 and LGR5-EYFP LoVo lines (Figure 3A and B). Our data showed that after 3-wk culture in soft agar, the number of colonies formed by the three cell populations of Caco-2 and LoVo cell lines was comparable (Figure 3C and E) but that the colony sizes of the LGR5/EYFPhigh cell populations in Caco-2 line were considerably larger (> 0.25 mm in diameter) than those in LGR5/EYFPlow and LGR5/EYFPnegative cell populations (Figure 3D and F), suggesting LGR5high cells in Caco-2 cell line exhibit increased proliferation and/or reduced cell death rates.
Figure 3.
Invitro anchor independent assay of Caco-2 and LoVo colon cancer cell line. Sorted cells were cultured for 2 wk in soft agar before analysis. A-F: Representative images of colonies formed by cells with different leucine-rich repeat-containing G protein-coupled receptor 5/enhanced yellow fluorescent protein levels in Caco-2 cell line (A) and in LoVo cell line (B). Scale bar, 50 µm; the (C) and (E) number, and (D) and (F) size of colonies formed by Caco-2 and LoVo clones respectively were assessed using crystal violet staining. Colonies larger than 0.25 mm in diameter were then counted. Columns and error bars represent means ± SD of two independent experiments using duplicate measurements in each experiment. aP < 0.05. LGR5: Leucine-rich repeat-containing G protein-coupled receptor 5; YFP: Yellow fluorescent protein; EYFP: Enhanced yellow fluorescent protein.
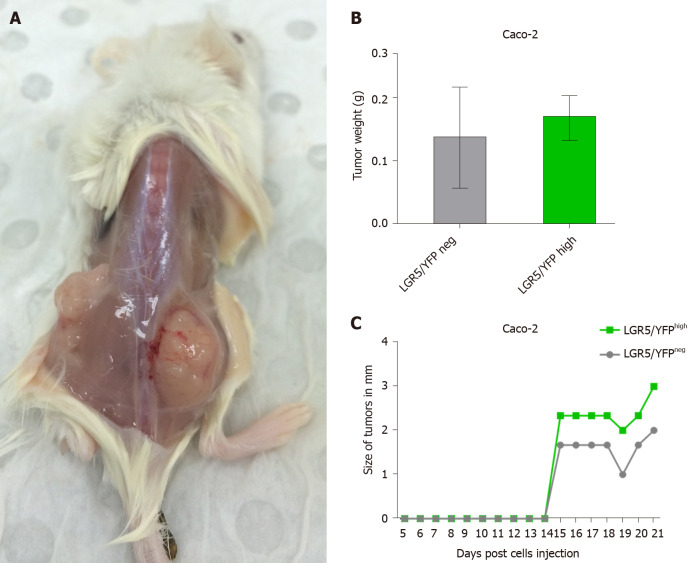
To assess further the tumorigenicity of the LGR5/EYFPhigh cell compartments of Caco-2 and LoVo lines, LGR5/EYFPhigh and EYFPnegative sorted cells were injected subcutaneously into the right and left flanks, respectively, of NOD-SCID mice (n = 3), and tumor growth was monitored daily before excision of tumors after 3 wk. As shown in Figure 4A, LGR5/EYFPhigh Caco-2 cells were able to form tumors when injected into NOD-SCID mice. LGR5/EYFPhigh formed larger tumors than the LGR5/EYFPnegative cells (Figure 4B and C).
Figure 4.
Stable leucine-rich repeat-containing G protein-coupled receptor 5/enhanced yellow fluorescent proteinhigh expressing Caco-2 cells line possesses stem cell-like properties in vivo. Sorted leucine-rich repeat-containing G protein-coupled receptor 5/enhanced yellow fluorescent protein (LGR5/EYFP)negative and LGR5/EYFPhigh cells were injected subcutaneously in left and right flanks (respectively) of non-obese diabetic- severe combined immunodeficient mice and monitored for 3 wk. A: Representative images of tumors formed by LGR5/EYFPhigh (right flank) and LGR5/EYFPnegative (left flank) cells; B and C: Tumor weights (B) and size (C) resulting from LGR5/EYFPnegative and LGR5/EYFPhigh sorted cells. Columns and error bars represent means ± SD of two independent experiments using duplicate measurements for each experiment. neg: Negative; LGR5: Leucine-rich repeat-containing G protein-coupled receptor 5; YFP: Yellow fluorescent protein.
In the LoVo cell line, on the other hand, both LGR5/EYFPhigh cells and LGR5/ EYFPnegative sorted cells had almost comparable tumorigenic properties (Supplementary Figure 2).
DISCUSSION
The heterogeneity of cancer cell lines has been recognized, but it was not until the 1980s that it was shown that the human DLD-1 colon cancer cell line, originally established from a single human colon carcinoma[26], contains subpopulations of cells with different morphology, karyotype, and cloning efficiency in soft agarose that produce histologically distinct tumors when injected into athymic mice. Recent studies have explained this apparent heterogeneity of colon cancer cell lines when it was discovered that colon cancer carcinoma contains a subset of stem cells expressing LGR5, termed "tumor-initiating” cells specimens[27]. This was strongly supported by lineage tracing studies[28] showing that colon cancer is initiated by aberrant Wnt signaling in ISCs, known to express high levels of LGR5. Furthermore, in vivo lineage tracing experiments on intestinal adenomas demonstrated the clonal expansion of these adenomas from a single LGR5-expression cell[14].
In light of this evidence, we decided to investigate whether colon cancer cell lines widely used for drug-screening[21] contain CSCs and developed a reporter that would allow their isolation and characterization. Here, we showed that a 983 bp fragment of the LGR5 promoter faithfully reports on endogenous LGR5 expression in the colon cancer cell lines LoVo and Caco-2. Interestingly, we revealed that the ISC populations in these two cell lines exhibit different gene expression and functional properties. Our data show that the Caco-2 cell line contains a population of highly proliferative cells that exhibit a classic intestinal CSC gene expression signature that is marked by a high expression of LGR5, in agreement with previous findings[28,29]. These cells can both self-renew and generate more differentiated LGR5 negative progeny and also exhibit an in vitro tumorigenic potential. In support of the notion that LGR5/EYFPhigh cells within Caco-2 colon cancer cell line represent the CSC compartment, our limited gene expression analysis revealed that LGR5/EYFPhigh cells display a transcriptional signature very similar to that of the ISC, including LGR5, Olfm4, and Ascl2[29-31]. Our data are also in agreement with Sato et al[9] who showed that only very high LGR5-expressing cells are the ISCs in normal intestinal tissue. In agreement with a study by Maddox et al[32], our data further show that the LGR5/EYFPhigh fraction of the Caco-2 cell line exhibits a high expression of OCT1, a known ISC marker. The LGR5/EYFPhigh fraction of the Caco-2 colon cancer cells also showed the highest telomerase expression (TERT), which is a characteristic of most CSCs, associated with the ability to proliferate indefinitely[33,34]. Conspicuously, the LGR5/EYFPhigh fraction of the Caco-2 acquires Bmi1 expression upon loss of LGR5 expression, in agreement with the proposed cellular differentiation hierarchy of the intestinal crypt[10]. Collectively, these data suggest that the LGR5/EYFPhigh fraction of the Caco-2 colon cancer cell line indeed represents a CSC-like population. These data are in close agreement with a similar study in SW480 and HT-29 human colon cancer cell lines in which LGR5 was also found to mark a CSC population[35].
In contrast to the Caco-2 cell line, in the LoVo colon cancer cell line, LGR5/EYFPhigh cells showed the lowest expression of the ISC markers hTERT, LRIG1, AFAP1I1, and , and low expression of the CSC marker OCT1, suggesting that in LoVo cells LGR5 marks a different cell population, i.e. not a stem cell-like population. Indeed, our data show that in the LoVo cell line, cells with high LGR5/EYFP expression possess a gene expression signature more akin to Bmi-expressing stem cells of the crypt. In LoVo cells it is rather the LGR5negative fraction that highly expresses ISC marker OCT1[32], OLFM4[36,37], and Bmi1[10], suggesting that this cell line originated from a CSC population other than LGR5+cells. Supporting this suggestion, in a recent review on the intestinal hemostasis and plasticity in humans, the writer argued that there is a wide range of cell types in the intestinal epithelium that can revert to stem cell fate and regenerate the epithelium post-injury[38]. Moreover, Bmi1 marks a distinct pool of ISCs that mainly reside above the crypt base[39]. Lineage tracing has shown that this Bmi1+ stem cell pool, the so-called reserve stem cell pool, is able to regenerate intestinal epithelial tissue in the absence of LGR5+ ISCs[10,39] and also give rise to LGR5+ cells in normal and injured intestinal epithelium[40]. In support of this idea, a number of other human colon cancer cell lines (such as LIM1899, LIM1215, LIM2537, and LIM1863)[41] have previously been shown to exhibit increased anchorage-independent growth and enhanced tumorigenicity in xenograft experiments upon ablation of LGR5.
CONCLUSION
We conclude that we have generated a portable LGR5-reporter that should prove a valuable tool for the identification and isolation of different colon CSC populations in colon cancer lines. The characterization of such populations is important for drug screening and design. Since LGR5 marks adult stem cell populations in a number of tissues throughout the body[42], it will be interesting to determine whether our LGR5 promoter reporter can also be used to isolate such LGR5 expressing CSCs from other human cancer cell lines such as glioblastoma[43], esophageal adenocarcinomas[44], ovarian primary tumors[45], gastric tumors[46], and hepatomas[47-50] that are also marked by high LGR5 expression.
Our data further indicate that LGR5/EYFPhigh expression marks two different intestinal crypt stem cell population in the Caco-2 and LoVo cell lines. Since Caco-2 and LoVo cell lines are widely used for screening of colon cancer drugs[51], our findings of different stem cell populations in these lines likely has important implications for such drug screening programs.
ARTICLE HIGHLIGHTS
Research background
The intestinal crypt-villus structure harbors two distinct pools of putative stem cells, the active [leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)+ cells] and the quiescent stem cell populations (B lymphoma Moloney murine leukemia virus insertion region 1 homolog+ or telomerase reverse transcriptase+ cells). Current evidence indicates that CRC is indeed a disease of colon stem cells that are resistant to current therapeutics and can rapidly proliferate to re-establish the tumor. Furthermore, it was reported that colon cancer carcinoma specimens contain a subset of LGR5-expressing stem cells. Human colon cancer cell lines, such as Caco-2 and LoVo, are extensively used in drug screening and colon cancer research and express high levels of LGR5.
Research motivation
To establish criteria for the selection of suitable cell lines for drug discovery and reveal potential variability between colon cell lines, we here identify the cancer stem cell (CSC) population(s) within colon cancer cell lines, quantify differences in gene expression signatures in these cells, and assess their proliferative and tumorgenicity capacities.
Research objectives
The present study aimed to isolate cells expressing LGR5 in Caco-2 and LoVo colon cancer cell lines and investigate whether these cells constitute the CSC compartments in these cell lines. This was achieved through the creation of a transgenic proliferating stem cell-specific reporter construct based on the proximal promoter of LGR5 gene.
Research methods
Using a portable fluorescent reporter construct based on a conserved fragment of the LGR5 promoter, subpopulations of cells from the colon cancer cell lines (Caco-2 and LoVo) were sorted into three cell compartments expressing different levels of LGR5 (high-, low-, and no-expression of LGR5). Next these cell compartments were characterized based on their gene expression signatures, proliferation, and tumorgenicity properties.
Research results
Cells expressing high levels of LGR5 with Caco-2 colon cancer cell line appear to represent a CSC-like population. In contrast, in the LoVo cell line, the LGR5 negative fraction possessed some features of the intestinal stem cells, e.g., a specific gene expression signature, suggesting that this cell line was likely derived from a CSC population other than LGR5+ cells.
Research conclusions
LGR5 marks the stem cell compartments of the Caco-2, but not LoVo, cell lines. Thus, it would appear that different colon cancer lines possess properties of different stem cell compartments. The portable LGR5 reporter outlined in this study constitutes a valuable tool for the identification and purification of LGR5-expressing cells from mixed cell populations.
Research perspectives
The portable LGR5 reporter described in this study constitutes a valuable tool for the identification and purification of LGR5-expressing cells from mixed cell populations.
ACKNOWLEDGEMENTS
We thank Professor McGuckin M (MMRI, Brisbane) for providing human colon cancer cell lines (Caco-2, LoVo, and SW480) and Dr. Rolfe B (AIBN, Brisbane) for providing mouse NSC-34 cells.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Institutional Review Board at The University of Queensland (approval No. 2019000159).
Institutional animal care and use committee statement: All animal experiments conformed to the internationally accepted principles for the care and use of laboratory animals, The Australian Institute of Bioengineering and Nanotechnology, Brisbane; Australia (approval No. AIBN/065/12/SCA/LEJEUNE/KACST).
Conflict-of-interest statement: All authors have nothing to disclose.
ARRIVE guidelines statement: The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
Manuscript source: Unsolicited manuscript
Peer-review started: December 1, 2020
First decision: December 21, 2020
Article in press: February 24, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bordonaro M S-Editor: Gao CC L-Editor: Filipodia P-Editor: Ma YJ
Contributor Information
Samah Abdulaali Alharbi, Physiology Department, College of Medicine, Umm Al-Qura University, Makkah 24231, Saudi Arabia; Department of Stem Cell Engineering Group, Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Brisbane 4072, QLD, Australia. saaharbi@uqu.edu.sa.
Dmitry A Ovchinnikov, Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Brisbane 4072, QLD, Australia.
Ernst Wolvetang, Department of Stem Cell Engineering Group, Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Brisbane 4072, QLD, Australia.
Data sharing statement
No additional data are available.
References
- 1.Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi RE, Corcione F. Worldwide burden of colorectal cancer: a review. Updates Surg. 2016;68:7–11. doi: 10.1007/s13304-016-0359-y. [DOI] [PubMed] [Google Scholar]
- 2.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odoux C, Fohrer H, Hoppo T, Guzik L, Stolz DB, Lewis DW, Gollin SM, Gamblin TC, Geller DA, Lagasse E. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res. 2008;68:6932–6941. doi: 10.1158/0008-5472.CAN-07-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garza-Treviño EN, Said-Fernández SL, Martínez-Rodríguez HG. Understanding the colon cancer stem cells and perspectives on treatment. Cancer Cell Int. 2015;15:2. doi: 10.1186/s12935-015-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YS, Hsu HC, Tseng KC, Chen HC, Chen SJ. Lgr5 promotes cancer stemness and confers chemoresistance through ABCB1 in colorectal cancer. Biomed Pharmacother. 2013;67:791–799. doi: 10.1016/j.biopha.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 10.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363–371. doi: 10.1002/stem.1031. [DOI] [PubMed] [Google Scholar]
- 12.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 15.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leushacke M, Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene. 2012;31:3009–3022. doi: 10.1038/onc.2011.479. [DOI] [PubMed] [Google Scholar]
- 17.Uchida H, Yamazaki K, Fukuma M, Yamada T, Hayashida T, Hasegawa H, Kitajima M, Kitagawa Y, Sakamoto M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci. 2010;101:1731–1737. doi: 10.1111/j.1349-7006.2010.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li AP. Screening for human ADME/Tox drug properties in drug discovery. Drug Discov Today. 2001;6:357–366. doi: 10.1016/s1359-6446(01)01712-3. [DOI] [PubMed] [Google Scholar]
- 19.Lieberman MM, Patterson GM, Moore RE. In vitro bioassays for anticancer drug screening: effects of cell concentration and other assay parameters on growth inhibitory activity. Cancer Lett. 2001;173:21–29. doi: 10.1016/s0304-3835(01)00681-4. [DOI] [PubMed] [Google Scholar]
- 20.Balimane PV, Patel K, Marino A, Chong S. Utility of 96 well Caco-2 cell system for increased throughput of P-gp screening in drug discovery. Eur J Pharm Biopharm. 2004;58:99–105. doi: 10.1016/j.ejpb.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Christensen J, El-Gebali S, Natoli M, Sengstag T, Delorenzi M, Bentz S, Bouzourene H, Rumbo M, Felsani A, Siissalo S, Hirvonen J, Vila MR, Saletti P, Aguet M, Anderle P. Defining new criteria for selection of cell-based intestinal models using publicly available databases. BMC Genomics. 2012;13:274. doi: 10.1186/1471-2164-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang C, Ang BT, Pervaiz S. Cancer stem cell: target for anti-cancer therapy. FASEB J. 2007;21:3777–3785. doi: 10.1096/fj.07-8560rev. [DOI] [PubMed] [Google Scholar]
- 24.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34:732–740. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamburger AW, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197:461–463. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- 26.Dexter DL, Spremulli EN, Fligiel Z, Barbosa JA, Vogel R, VanVoorhees A, Calabresi P. Heterogeneity of cancer cells from a single human colon carcinoma. Am J Med. 1981;71:949–956. doi: 10.1016/0002-9343(81)90312-0. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 29.Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P, Clevers H, Sancho E, Mangues R, Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 31.Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, Clevers H. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 32.Maddox J, Shakya A, South S, Shelton D, Andersen JN, Chidester S, Kang J, Gligorich KM, Jones DA, Spangrude GJ, Welm BE, Tantin D. Transcription factor Oct1 is a somatic and cancer stem cell determinant. PLoS Genet. 2012;8:e1003048. doi: 10.1371/journal.pgen.1003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Hum Mol Genet. 2001;10:677–685. doi: 10.1093/hmg/10.7.677. [DOI] [PubMed] [Google Scholar]
- 34.Hiyama E, Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96:1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirsch D, Barker N, McNeil N, Hu Y, Camps J, McKinnon K, Clevers H, Ried T, Gaiser T. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis. 2014;35:849–858. doi: 10.1093/carcin/bgt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 37.Schuijers J, van der Flier LG, van Es J, Clevers H. Robust cre-mediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem Cell Reports. 2014;3:234–241. doi: 10.1016/j.stemcr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buczacki S. Fate plasticity in the intestine: The devil is in the detail. World J Gastroenterol. 2019;25:3116–3122. doi: 10.3748/wjg.v25.i25.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 41.Walker F, Zhang HH, Odorizzi A, Burgess AW. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One. 2011;6:e22733. doi: 10.1371/journal.pone.0022733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakata S, Campos B, Bageritz J, Bermejo JL, Becker N, Engel F, Acker T, Momma S, Herold-Mende C, Lichter P, Radlwimmer B, Goidts V. LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol. 2013;23:60–72. doi: 10.1111/j.1750-3639.2012.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Rahden BH, Kircher S, Lazariotou M, Reiber C, Stuermer L, Otto C, Germer CT, Grimm M. LgR5 expression and cancer stem cell hypothesis: clue to define the true origin of esophageal adenocarcinomas with and without Barrett's esophagus? J Exp Clin Cancer Res. 2011;30:23. doi: 10.1186/1756-9966-30-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, Kirschmeier P, Samatar AA. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 46.Wu C, Xie Y, Gao F, Wang Y, Guo Y, Tian H, Li Y, Fan W. Lgr5 expression as stem cell marker in human gastric gland and its relatedness with other putative cancer stem cell markers. Gene. 2013;525:18–25. doi: 10.1016/j.gene.2013.04.067. [DOI] [PubMed] [Google Scholar]
- 47.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Vries RG, Huch M, Clevers H. Stem cells and cancer of the stomach and intestine. Mol Oncol. 2010;4:373–384. doi: 10.1016/j.molonc.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 50.Fujii Y, Yoshihashi K, Suzuki H, Tsutsumi S, Mutoh H, Maeda S, Yamagata Y, Seto Y, Aburatani H, Hatakeyama M. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci USA. 2012;109:20584–20589. doi: 10.1073/pnas.1208651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bu XD, Li N, Tian XQ, Huang PL. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell. 2011;43:201–206. doi: 10.1016/j.tice.2011.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.