Abstract
Coronavirus disease 2019 (COVID-19) is a devastating worldwide pandemic infection caused by a severe acute respiratory syndrome namely coronavirus 2 (SARS-CoV-2) that is associated with a high spreading and mortality rate. On the date this review was written, SARS-CoV-2 infected about 96 million people and killed about 2 million people. Several arguments disclosed the high mortality of COVID-19 due to acute respiratory distress syndrome or change in the amount of angiotensin-converting enzyme 2 (ACE2) receptor expression or cytokine storm strength production. In a similar pattern, hepatic impairment patients co-infected with SARS-CoV-2 exhibited overexpression of ACE2 receptors and cytokine storm overwhelming, which worsens the hepatic impairment and increases the mortality rate. In this review, the impact of SARS-CoV-2 on hepatic impairment conditions we overviewed. Besides, we focused on the recent studies that indicated cytokine storm as well as ACE2 as the main factors for high COVID-19 spreading and mortality while hinting at the potential therapeutic strategies.
Keywords: COVID-19, Hepatic impairment, SARS-CoV-2, Cytokine storm, Immuno-modulators receptors
Core Tip: Implications of fast coronavirus disease 2019 (COVID-19) outbreak are huge annoying problems that affected countries' health and economies around the world. Patients associated with hepatic impairment are considered at a high-risk target for severe acute respiratory syndrome namely coronavirus 2 severity and mortality. A good understanding of virus machinery provides excellent ideas about how to combat this monster. We provide detailed information about the recent mechanisms of COVID-19 pathogenesis, implications on several hepatic disorders, and potential therapeutic strategies.
INTRODUCTION
Coronaviruses are classified into four categories, namely α, β, γ, and δ type. The α- and β-coronaviruses could infect mammals, while γ- and δ- coronaviruses appear to infect birds[1]. Six coronaviruses reported as human-affected viruses include α-coronaviruses NL63 and 229E, low-pathogenic β-coronaviruses OC43 and HKU1, these viruses cause mild respiratory symptoms as common cold[2]. The remaining two types were identified as β-coronavirus, which unusually induced fatal respiratory tract infections[3]. Between 2002 and 2003, severe acute respiratory syndrome (SARS) resulted in the outbreak of SARS0[4]. In 2012 the Middle East respiratory syndrome (MERS) originated and circulated in camels[5].
In 2019, a newly discovered coronavirus is a SARS coronavirus 2 (SARS-CoV-2), which induces a contagious disease coronavirus disease 2019 (COVID-19) and is currently spreading nationwide[6]. Consequently, the third identified zoonotic coronavirus detected after SARS and MERS cause potentially fatal respiratory tract infections[7,8]. The early cases of COVID-19 were linked to the Huanan seafood market, Wuhan, China[9,10].
On January 12, 2020, the World Health Organization (WHO) temporarily named this new virus the 2019 novel coronavirus (2019-nCoV)[11]. On January 30, 2020, the WHO declared the 2019-nCoV epidemic a public health emergency of international burden[11]. After that February 11, 2020, the WHO formally named the disease caused by 2019-nCoV as COVID-19[12]. On the same day, the international committee of taxonomy named this virus as SARS-CoV-2 and classified it as coronavirus under the family Coronaviridae, subfamily Orthocoronavirinae, based on the genotypic and serological characterization[11,13,14].
SARS-CoV-2 is a new generation of β-coronavirus which shares 79% and 50% genome sequencing with SARS and MERS, respectively[15]. This information confirmed some of the available evidence that SARS-CoV-2 was generated from bats and pangolins[14]. Other studies suggest that SARS-CoV-2 was a chimeric virus obtained from the fusion of a bat's coronavirus and a coronavirus of unknown origin[16]. In the same context, the study conducted by Sun et al[17] showed that the structure of SARS-CoV-2 was similar to the coronavirus isolated from Chinese chrysanthemum-headed bats in 2015. So far, no confirmation has been made of the zoonotic source of SARS-CoV-2 yet[18].
SARS-CoV-2 has a 29891-nucleotide genome (encoding 9860 amino acids)[19] with a diameter of about 50-200 nm[20]. Two-thirds of viral RNA, primarily in the first open reading frames (ORFs) (ORF1a/b), converts into two non-structural polyproteins pp1a, pp1ab, and encodes 16 non-structural proteins (NSP). In contrast, the remaining ORFs encode structural and accessory proteins[21,22]. Six ORFs of functions are arranged between 5′ to 3′ (ORF1a/ORF1b), spike glycoprotein (S), a small envelope protein (E), matrix protein (M), and nucleocapsid protein (N)[21]. Moreover, seven putative ORFs encoding accessory proteins are disseminated between the structural genes[23,24]. The glycoprotein S is responsible for the receptor-binding site and membrane fusion[25]. The envelope protein E has essential roles in releasing and accumulating viruses[26], besides protein M[27]. Additionally, a NSP plays many functions during the viral cycle[28].
SARS-CoV-2 AND ANGIOTENSIN-CONVERTING ENZYME 2
Only specific and permissive cells inside the host may be infected. Specifically, cells that generate proteins and viral factors which are required for virus replication. As soon as the virus enters the cell, the virus recruits cells to produce essential viral encoded proteins involved in replicating the virus's genetic material[29]. The viral replication process generally known as cytopathic effects, could induce biochemical changes resulting in cellular damage. Interestingly, SARS-CoV-2 relies on specific cellular receptors to invade the cells that carry these factors like other types of coronaviruses[30].
The surface of SARS-CoV-2 is covered by several types of glycosylated S proteins that facilitate the virus's attachment to the host angiotensin-converting enzyme 2 (ACE2), hence mediating the entry of the virus into the host cells[31]. This action could activate S protein by a specific type of serine protease located on the host cell membrane called TM protease serine 2. After the virus recruited the host cell, it releases its RNA and uses the host cells to replicate and synthesize its functional proteins[32].
SARS-CoV-2 is predicted to have organ tropism beyond the respiratory tract, also including the kidney, heart, skeletal muscle, central nervous system, liver, and gastrointestinal tract. The spread of infection through the body is reasonable for developing COVID-19 systemic symptoms and exacerbating pre-existing disorders[33,34]. Therefore, patients with severe COVID-19 illness show multiple organ damage, such as acute kidney injury, acute pulmonary injury, heart injury, and liver injury[35].
SARS-CoV-2 may induce liver injury via a similar mechanism. Furthermore, we cannot neglect angiotensin II's action; once SARS-CoV-2 binds to ACE2 receptors it stimulates angiotensin II to mediate the tissue injury[36]. These findings confirm that angiotensin II levels are increased in healthy patients with COVID-19 and are directly correlated with viral load[37].
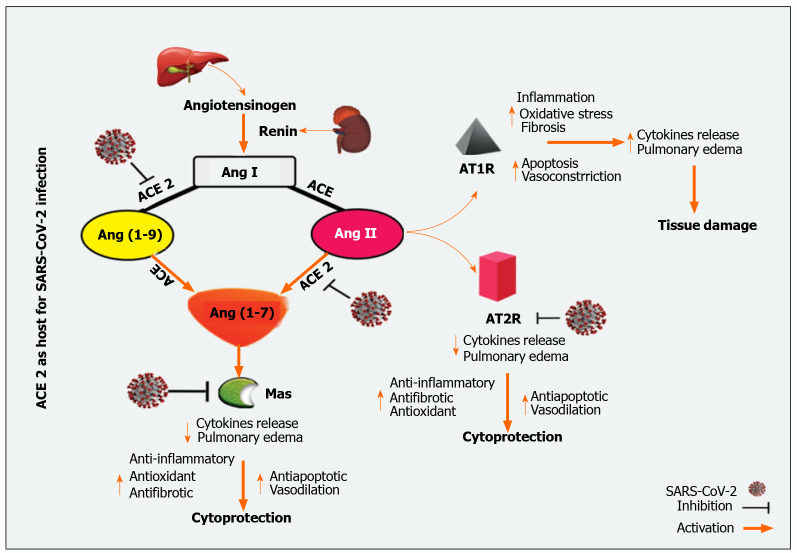
Hepatic sharing of ACE2 receptors is unreasonable; it is widely distributed in the endothelial layer but not in the sinusoidal endothelium[38]. On the surface of cholangiocytes, ACE2 receptors have more significant expressions than hepatocytes[39]. Cholangiocytes ACE2 receptors expression was close to that of type II alveoli cells in the lungs, indicating that the liver may be a strong candidate for SARS-CoV-2[40]. In Contrast, Immunohistochemical stains for Kupffer cells and lymphocytes T and B for ACE2 are negative[41]. In fact, in patients with COVID-19, SARS-CoV-2 has been detected in their liver and can induce liver injury through different pathways[42]. The general mechanism of ACE2 mediated SARS-CoV-2 infection is outlined in Figure 1.
Figure 1.
Angiotensin-converting enzyme 2 as a host for severe acute respiratory syndrome coronavirus 2 infection. SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; ACE: Angiotensin-converting enzyme; Ang: angiotensin; ATR: Angiotensin receptors type.
SARS-CoV-2 and devastating cytokine storm
Cytokine storm is an augmented cytokine output attributable to an irregular immune response to various stimuli like viral infections, rheumatic disorders, cancer, sepsis, multiple organ failure, and drugs that contribute to unnecessary immune cell activation[43]. Consequently, the rising aggressive systemic hyperinflammatory reaction is associated with the release of many pro-inflammatory cytokines responsible for critical illness[43,44]. Cytokine storm has been found in various viral infections, including influenza H5N1, H1N1 viruses, and two SARS-CoV and MERS-CoV coro-naviruses[45-48]. Recent data reported that patients infected by SARS-CoV-2 show massive levels of cytokines and chemokines and are implicated with COVID-19 severity and high mortality rate[49].
The growing cytokine storm results in the continuous activation and expansion of innate and adaptive immune cells to produce a harmonized cascade of inflammatory responses, resulting in a devastating cytokine storm[50]. Indeed, the result of a mixture of several immune active molecules is the cytokine storm's product. The main components involved in forming the cytokine storm are interferons (IFNs) and interleukins (ILs)[51]. However, other inflammatory biomarkers such as c-reactive protein (CRP), fibroblast growth factor, macrophage-colony stimulating factor (M-CSF), interferon gamma-induced protein 10, monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory proteins-1α and 1β, platelet-derived growth factor, tumor necrosis factor-alpha (TNF-α), and vascular endothelial growth factor (VEGF) are markedly detected in the COVID-19 sever case-patients[52-54]. Hence, the uncontrolled response in severe COVID-19 patients is correlated with unfavorable clinicopathological consequences.
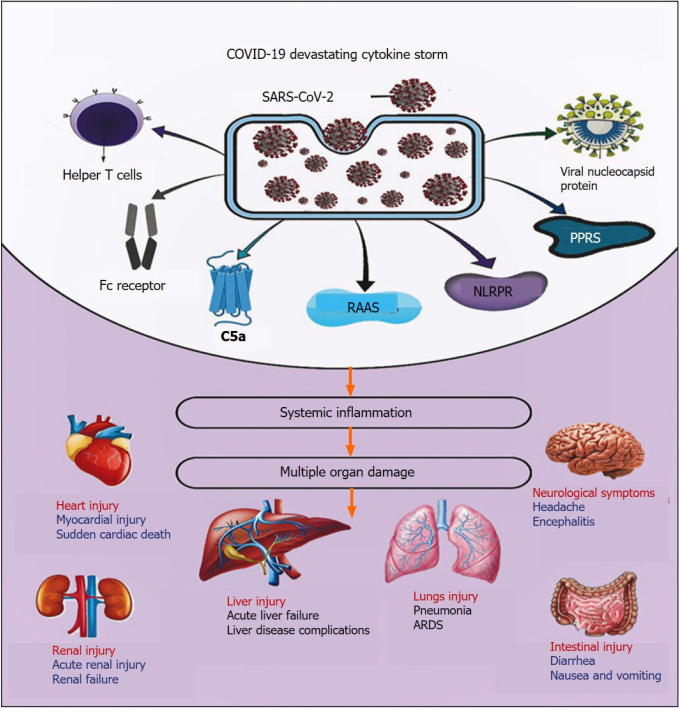
During COVID-19 illness, several pathological factors have been reported to participate in the initiation and propagation of SARS-CoV-2-induced cytokine storm (Figure 2).
Figure 2.
Coronavirus disease 2019 and devastating cytokine storm. COVID-19: Coronavirus disease 2019; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; C5a: Complement component 5a; RAAS: Renin-angiotensin-aldosterone system; NLRPR: Nucleotide-binding oligomerization domain, Leucine-rich repeat and pyrin domain-containing receptor; PPRs: Pattern recognition receptors; ARDS: Acute respiratory distress syndrome.
Renin angiotensin-aldosterone system
The Renin-angiotensin-aldosterone system (RAAS) is a significant regulator of several physiology and pathophysiology conditions such as maintenance of fluid and electrolyte homeostasis with the maintenance of vascular tone[55,56]. Recent evidence implies that RAAS has autocrine and paracrine effects in addition to the classical circulating endocrine effects. Hence, RAAS plays pivotal roles in cellular growth, migration, differentiation, apoptosis, inflammation, thrombosis, and fibrosis[11,57-59].
When renin is secreted directly into the bloodstream by juxtaglomerular cells, this secreted renin cleaves the liver-released substrate, angiotensinogen, to generate the inactive peptide, angiotensin (Ang) I, which is then converted by endothelial ACE to Ang II[56,60]. On the other hand, ACE2 cleaves Ang I to produce Ang (1-9) peptide. Besides, Ang II and Ang (1-9) can metabolize by ACE or other peptidases to form Ang 1-7[61,62].
The RAAS is a double-edged sword that acts through two distinct opposing arms: the ACE/Ang II/AT1 receptor axis, which is responsible for main actions, and the ACE-2/Ang-(1-7) /Mas receptor axis, the counter-regulatory arm[63,64]. The ACE catalytic activity contributes to an increase of Ang II levels and an increase in Ang-(1-7) catabolism, while ACE2 catalytic activity is primarily based on Ang I and Ang II and leads to the formation of Ang (1-7)[64,65]. Ang II plays a central role in the RAAS via angiotensin II type 1 and 2 receptors (AT1R and AT2R); thus, stimulating the ACE/Ang II/AT1 receptor axis which results in regulating vasoconstriction, fibrotic remodeling, inflammatory response, and production of reactive oxygen species (ROS)[66]. Ang II itself will directly activate the nuclear factor-kappaB (NF-κB) pathway through the phosphorylation of the p65 subunit of NF-κB; therefore, increases the output of IL-6, IL-1β, TNF-α, and IL-10[67,68]. Ang II also affects mitogen-activated protein kinases (MAPKs; extracellular signal-regulated kinases 1/2, c-Jun N-terminal kinases, p38-MAPK), which have a high impact on pro-inflammatory cytokines release[68,69]. Besides, a spontaneous association of Ang II with host immune cells, e.g., neutrophils, T and B lymphocytes, and tissue-resident cells results in the release of pro-inflammatory cytokines, including prostaglandins, IL-6, IL-1β, TNF-α, VEGF, and IFN-γ, as well as the activation of kinase plethora [e.g., Janus kinase (JAK) and p38 MAPK][70,71].
As soon as the SARS-CoV-2 spike (S)-protein binds to ACE2[72], the serum level of Ang II was significantly increased, which in turn mediated trans-signaling of the IL-6/soluble IL-6 receptor-a complex in to finally activate of signal transducer and activator of transcription 3 (STAT3)[73]. STAT-3 has been reported to up-regulated several inflammatory genes like NF-κB. Besides activation of NF-κB by SARS-CoV-2 mediated STAT3 activation, SARS-CoV-2 itself can activate NF-κB by binding with pattern recognition receptors (PRRs). The net results trigger a cytokine storm accompanied by acute respiratory distress syndrome (ARDS) and multiple organ damage[68,74]. During COVID-19, ACE2 depletion and ACE/Ang II/AT1R axis activity are highly augmented[75]. Therefore, a therapeutic modality by administering exogenous ACE2 to patients with COVID-19 can potentially impact COVID-19 severity[76]. On the other hand, targeting the ACE/Ang-II/AT1R axis downstream, such as the IL-6/STAT3 axis, should be considered to avoid ARDS inflammation and end-organ damage induced by cytokine storm[68,76].
In contrast to the ACE/Ang II/AT1R pathway, the Ang (1-7) interacts with the Mas receptor to provide vasodilation and antiproliferative effects[77]. Indeed, activating Mas receptors induce activation of phospholipase A, phosphoinositide 3 kinases/protein kinase B axis, endothelial nitric oxide synthase, and intracellular calcium accumulation[78]. Additionally, Ang (1-7) down-regulates p38 MAPK and NF-κB expression harmony with the suppression of inflammatory markers such as IL-6, IL-8, and TNF-α[68]. Furthermore, Ang (1-7) via the Mas receptor activation attenuated Ang II-induced intercellular adhesion molecule-1, vascular cell adhesion molecule-1, and MCP-1[68,79]. Together, activation of these pathways suggests that Ang (1-7) has antiproliferative, anti-thrombotic, and anti-inflammatory functions and ameliorates tissue damages[70,78].
Interestingly, down-regulation of the ACE-2/Ang (1-7)/Mas receptors axis was proposed to be increased during COVID-19; since the virus uses the enzyme's peptidase domain to enter into the cells, there is a down-regulation of ACE-2 with ACE up-regulation[63]. Thus, it contributes to a massive release of cytokines and inflammatory responses. Since Ang (1-7) plays a critical role in counteracting the pro-inflammatory activity of RAAS, it has been proposed that Ang (1-7) or one of its associated agents could be administered to patients with COVID-19 since it protects against activation of inflammatory mediators in a cytokine outbreak[68]. Additionally, Ang (1-7) was previously reported to inhibit liver fibrogenesis[80]. Thus, it may be helpful to attenuate liver injury in patients with liver impairments infected with SARS-CoV-2.
Fc receptor
SARS-CoV-2 is a specific antibody with the Fc receptor. Indeed, SARS-CoV-2 recruits several immune cells, e.g., granulocytes, monocytes, and macrophages, and activates the complement cascade, resulting in high virus reproduction and violent infection[81,82]. Since the liver includes several cells related to the immune response, so, antibody-dependent, ACE2-independent pathway was activated[83]. As a result, SARS-CoV-2 infection overactivated and secreted excessive amounts of cytokines and chemokines such as TNF-α, IFN-γ, IL-6, IL-8[82].
Viral nucleocapsid proteins
Viral nucleocapsid proteins fused with the infected cells' host cell membrane, which permits the viral nucleocapsid protein to stay on the surface of the cell membrane[84]. This fusion makes the proteins easily recognizable by antigen-presenting cells that activate the immune response[83]. The normal antigen-presenting cells are dendritic cells, macrophage cells, monocytes, plasma cells, etc., introduced to viral particles CD8+ cytotoxic and CD4+ regulatory T lymphocytes, the major histocompatibility complexes[85].
PRRs
Several PRRs are the first line receptors that detect pathogenic infection like SARS-CoV-2. After the virus enters the body, it produces pathogen-associated molecular patterns (PAMPs), which recognizes by PRRs and fires the standard innate immune system[86]. The membrane-bound toll-like receptor (TLR) family among PRRs, predominantly recognizes PAMPs in the extracellular system and to a lesser degree in the intracellular milieu[87]. Activation of this signal contributes to increasing the expression of transcription factors that induce pro-inflammatory cytokine production, such as NF-κB, and activate immune defense against viral infection via the IFN type I pathway[85,88].
Nucleotide-binding oligomerization domain, Leucine-rich Repeat and Pyrin domain-containing (nod-like receptor protein) receptor
Another family of pathogenic detecting receptors is nod-like receptor protein (NLRP) receptors (NLRP1, NLRP3, NLRP7, and NLRC4), which are the cytosol of endogenous danger-associated molecular patterns expressed inside the cell[89,90]. The activation of these receptors by SARS-CoV-2 has been reported to associate with the adhesion of molecules' activation, inflammatory response, and triggering innate immune cells[91]. This reaction contributes to the activation of pro-inflammatory mediators cytokines production such as TNF-α, IL-1, IL-6, IL-10, or type 1 interferons[92,93].
T-helper type 1 cell
The T-helper type 1 (Th1) cell response can be triggered by the elevation of inflammatory cytokines, which plays a significant role in providing a memory response against the virus and adaptive immunity. It is essential to coordinate humoral and cytotoxic T cell responses during viral infections[94]. Besides, elevated levels of Th2 cells secrete cytokines in patients with COVID-19, e.g., IL-4 and IL-10, which inhibit inflammatory responses[95]. Furthermore, there was a significant increase in cytokines secreted by Th1 and Th2 cells, such as TNF-α, IL-6, IL-18, IL-4, and IL-10. Additionally, IL-2 and IL-6 are significantly elevated in patients with COVID-19 and correlate with the disease's seriousness[96]. Interestingly, hepatocytes, Kupffer cell, and hepatic stellate cells after being infected with SARS-CoV-2 immune cells after overactivation and secretion of excessive cytokines, e.g., IL-6, IL-8, TNF-α, IFN-γ, which are involved in severe cytokine storm and causes tissue damage[85,97]. Compared to a reduced level of naive B cells, plasma B cells significantly increased in COVID-19 patients, several naive B-cell receptor isotypes [immunoglobulin heavy chain (IGHV) 3-15, IGHV3-30, and IGHV3-11] were previously used in the manufacture of other virus vaccines which were also reported and identified in patients[98]. Furthermore, the novel target genes in cytokine storm are IL-1β, IL-6, and the granulocyte-macrophage colony-stimulating factor (GM-CSF), while TNFSF13, IL-18, IL-2, and IL-4 seemed to help patients' recovery from COVID-19[99].
Complement activation
Viral infection has been reported to stimulate the complement cascade and consequently, initiate specific inflammatory responses[100]. Among these complements, complement factor 5a (C5a) is the most potent inflammatory peptide in the complement cascade, which increases the production of pro-inflammatory cytokines such as IL-6, IL-1, and TNF-α from macrophages under the effect of TLR-2, TLR-4, and TLR-9[101,102]. Terminal complement variable C5b-9 stimulates the release of IL-6 from vascular smooth muscle cells by stimulating the redox-sensitive transcription factor NF-κB, activator protein 1, and MCP-1[68,100]. Besides, C3a overexpression results in increased production of IL-1, IL-6, and TNF-α[103].
The complement cascade plays aberrant pivotal roles in the pathogenesis of SARS-CoV-2. It promotes viral nucleocapsid protein-mediated auto-activation of mannan-binding lectin serine peptidase-2 (MASP-2)[104]. Indeed, MASP-2 is considered the main serine protease in the lectin pathway. It binds to the mannose-binding lectin pathway to induce the downstream complement cascade, which in turn accelerates inflammatory responses. In two patients with COVID-19 who received an anti-C5a antibody, a positive clinical response was improved as evidence by enhancing lung oxygenation and alleviating systemic inflammation[105,106]. Hence anti-C5a antibodies could be a new therapeutic option for combating COVID-19 severity.
COVID-19 and hepatic dysfunction
Patients with digestive issues and co-infected with SARS-CoV-2 show a higher risk of mortality than patients without digestive problems. In severe cases of COVID-19, liver dysfunction was observed and associated with extensive activation of coagulative and fibrinolytic pathways as well as alteration of platelets, neutrophils, and lymphocytes profiles[8]. In parallel, patients with chronic liver disease (CLD), hepatitis viruses [hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus, and hepatitis E virus], hepatotropic viruses infection, non-alcoholic fatty liver disease, and non-alcoholic steatohepatitis are more susceptible to COVID-19 and may present worse outcomes from ARDS compared with the other critically ill patients[82,107]. Alqahtani et al[108] performed a meta-analysis that showed that patients with a previous history of liver disease have a 57.33% chance of severing COVID-19 infection and a 17.65 % higher mortality rate than other patients[108]. A recent multicenter cohort study showed that hepatic decompensation was positively associated with COVID-19 disease, which increasing the risk of death from 26.2% to 63.2%[109]. This rate could be related to low platelets and lymphocytes in those patients or due to cirrhosis-related immune dysfunction[97]. Besides, COVID-19 is markedly distinguished by an increase in cytokine secretion, which induces hepatocytes injury, contributing to the loss of hepatic regeneration and worse clinical outcome, especially in patients with CLD[97,110]. In contrast, Lippi et al[111] indicated that CLD plays a minor role in affecting patients' progression, severity, or mortality[111].
The pattern of liver damage in COVID-19 is essentially considered during COVID-19. The liver injury can be associated with the virus's immediate cytopathic effect, unregulated immune response, sepsis, hypoxia, and drug-inducing liver injury[112,113]. Also, COVID-19 causes underlying chronic hepatic disorders to exacerbate and contributes to liver decompensation with higher mortality[114]. Recent general studies show that about 2%-11% of COVID-19 patients have underlying CLD, and 14%-53% developed hepatic dysfunction with COVID-19[8,14,115,116]. In patients with severe COVID-19, the ratio of hepatic injury is highly set in contrast to mild patients' rate. Hence, hepatic injury frequency can represent 58.06%, 51%, and 78% in patients' death from COVID-19[14]. Cai et al[117] conducted a study on 417 patients with COVID-19 which showed that about 76.3% of patients had abnormal liver test results and 21.5% exhibited liver injury during hospitalization[117]. Li et al[118] indicated that patients with abnormal liver activity (58.8% and 66.7%) could be more likely to have moderate to severe COVID-19[118].
It is understood that cholangiocytes play an essential role in liver regeneration and immune response, and cholangiocytes ACE2 expression is greater than that of hepatocytes[38,116,119]. Thus, it was proposed that the liver injury that resulted in SARS-CoV-2 infection could be due to the damage caused by virus infection to bile duct cells and not liver cells[113]. The expression of ACE2 in cholangiocytes was also observed, it is about 20 times higher than in hepatocytes[13]. Additionally, gamma-glutamyl transferase, a diagnostic biomarker for cholangiocyte injury, has also been identified to be highly up-regulated in severe cases of COVID-19[120,121]. However, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) are elevated predominantly in COVID-19. Generally, it is 1-2 times more than the normal range, which averages to modestly elevated total bilirubin (TBIL) levels early in the disease process[112,115,116]. On the contrary, the study conducted by Chen et al[122] showed an elevation of ALT and AST serum levels to 7590 U/L and 1445 U/L, respectively, in patients with severe COVID-19[122].
Several studies have found that AST, ALT, and TBIL were substantially higher in the intensive care unit (ICU) patients than in non-ICU patients[123,124]. A retrospective cohort study performed by Hundt et al[125] reported abnormal liver tests [AST, ALT, Alkaline phosphatase (ALP), and TBIL] in the patients with COVID-19. Most patients with abnormal liver tests had an elevation limit of 1-2 times more than the normal limits. It has also been reported that abnormal liver tests were directly proportionated with poorer clinical outcomes[125].
Hypothesizes of hepatic injury during COVID-19 illness
Hepatic injury in COVID-19 patients could result from a cytokine storm rather than direct cytopathic effects of the virus itself[112,113,126]. The direct cytopathic effects of SARS-CoV-2 can induce stress on endoplasmic reticulum and mitochondrial dysfunction[127]. Moreover, the human body can initiate immune-mediated inflammation, such as cytokine storm resulting in liver damage or hepatic failure in critical COVID-19 patients[128-130]. Hypoxia is also a common characteristic of extreme COVID-19 illness that promotes ROS production and initiates the release of multiple pro-inflammatory factors to induce more liver damage[82]. Electron microscope examination of liver obtained from patients infected with SARS-CoV-2 revealed that SARS-CoV-2 causes endoplasmic reticulum tension that induces de novo lipogenesis. Lipogenesis could also contribute to the production of non-specific inflammatory changes, including hepatocyte swelling and steatosis, mild hepatic sinus cell proliferation, and Kupffer cell hyperplasia[82].
Effect of SARS-CoV-2 on several types of liver disorders
As we mentioned before, SARS-CoV-2 is remarkably impactful on patients with chronic diseases, including chronic hepatic impairments. The impact of SARS-CoV-2 on several types of liver disorders is outlined in Table 1.
Table 1.
Implication of severe acute respiratory syndrome coronavirus 2 on patients with hepatic disorders
|
Hepatic disorders
|
Main findings of the study
|
Ref.
|
| NAFLD | NAFLD is associated with a higher risk of symptomatic, severe, and progressive COVID-19 | Hashemi et al[137], 2020 |
| SARS-CoV-2 infection in patients with NAFLD required ICU admission and mechanical ventilation concomitant with increased NAFLD progression to NASH | Sachdeva et al[212], 2020 | |
| Liver cirrhosis | Patients with liver cirrhosis and COVID-19 are related to worse clinical outcomes and a high mortality rate than patients with COVID-19 alone | Kushner and Cafardi[139], 2020 |
| SARS-CoV-2 co-infection augmented liver injury as evidenced by worsens decompensated clinical status | Sarin et al[143], 2020 | |
| Cirrhotic patients with COVID-19 had a higher risk of mortality than COVID-19 patients alone, but they are equally mortality rate with cirrhosis patients without COVID-19 | Bajaj et al[144], 2021 | |
| HCC | Patients with HCC consider a risk group, and HCC is positively related to deterioration symptoms and bad outcomes in COVID-19 | Zhang et al[213], 2020 |
| Patients with cancer are more susceptible to infection and poorer prognosis of COVID-19 | Liang et al[147], 2020 | |
| Hepatitis B | Patients infected with HBV tend to have a more severe form of COVID-19 | Chen et al[150], 2020 |
| SARS-CoV-2 and HBV co-infection showed monocytopenia, lymphopenia, and thrombocytopenia, as well as metabolic disorders | Liu et al[151], 2021 | |
| COVID-19 may induce HBV reactivation, but it rarely occurs | Aldhaleei et al[152], 2020 | |
| Hepatitis C | Patients infected with HBV or HCV showed a high risk of mortality and morbidity if co-morbid with COVID-19 | Mirzaie et al[155], 2020 |
| Patient with hepatic c and COVID-19 has an undesirable clinical outcome | Mostardeiro et al[156], 2020 | |
| HCV pre-existing was associated with high mortality | Mangia et al[157], 2020 |
NAFLD: Non-alcoholic fatty liver disease; HCC: Hepatocellular carcinoma; NASH: Non-alcoholic steatohepatitis; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; COVID-19: Coronavirus disease 2019; HBV: Hepatitis B virus; HCV: Hepatitis C virus; ICU: Intensive care units.
Non-alcoholic fatty liver disease and COVID-19
Several studies have documented that obesity is a significant mortality factor in COVID-19 patients. The need for overall survival and mechanical ventilation correlated with obesity[131]. Typically the ACE2 expression level in adipose tissue is greater than that of lung tissue[97]. These clarify adipose tissue's susceptibility to invasion by SARS-CoV-2, and then the virus can spread to other organs[132]. Obese patients are at high risk for non-alcoholic fatty liver disease (NAFLD), which in turn has a higher chance of developing severe COVID-19 with a higher probability of abnormal liver function and a more extended viral shedding period[133]. NAFLD is associated with the development and severity of COVID-19[97,133]. NAFLD patients also elevate cytokine levels, rendering them more vulnerable to COVID-19 related to excessive cytokine production[134]. Experimentally, it has been shown that ACE2 expression increases in chronic liver damage and NAFLD[135]. Patients with COVID-19 exhibit increased serum levels of MCP-1, which exacerbate steatohepatitis; thus, the virus can increase NAFLD progression to Non-alcoholic steatohepatitis (NASH)[136]. Multicenter retrospective by Hashemi et al[137] study the clinical outcome of COVID-19 and CLD. Patients with high NAFLD require ICU admission and mechanical ventilation. There is no overall mortality among CLD patients mainly due to NAFLD[137]. According to the evidence available, NAFLD is an independent risk factor for extreme COVID-19, but most studies did not separate NAFLD from its more extreme NASH[138].
Liver cirrhosis and COVID-19
Patients with liver cirrhosis have a potentially higher risk of SARS-CoV-2 infection, a higher risk of a severe condition, and a higher risk of liver decompensation[139]. Liver cirrhosis in SARS-CoV-2 infected patients is described as a predictor of mortality[140,141]. A decrease in ACE2 via SARS-CoV-2 induced internalization is predicted to exacerbate liver fibrosis and augment the disease's severity, especially in the long term. Therefore, the impact of COVID-19 on long-term liver-related outcomes is also worth considering in patients with cirrhosis[142]. A multicenter matched cohort study by Kushner and Cafardi[139] patients with cirrhosis plus COVID-19 were observed to have mortality equal to cirrhosis patients alone higher than those with COVID-19 alone[139]. Sarin et al[143] The APCOLIS study of pre-existing liver disease indicated that in CLD patients, SARS-CoV-2 infection causes severe liver damage, decompensating one-fifth of the cirrhosis and worsening the already decompensated clinical status[143]. Bajaj et al[144] multicenter North American contemporaneously enrolled study, compared to patients with cirrhosis alone or cirrhosis and COVID-19 patients had similar mortality rate, but higher than patients with COVID-19 alone[144].
Hepatocellular carcinoma and COVID-19
In general, cancer patients, especially those who have recently received cancer treatment, have a greater chance of infection and a worse outcome. Besides, epidemiological data suggest that patients with cancer are more vulnerable to SARS-CoV-2 infection[145]. Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer globally, accounting for about 6% of all cancer incidences[146]. HCC patients are more susceptible than other cancers to the consequences of the COVID-19 pandemic since the liver damage caused by SARS-CoV-2 could complicate current hepatitis in most HCC patients[146]. A retrospective cohort study by Yang et al[96] confirmed that cancer patients had deteriorating symptoms and bad results from SARS-CoV-2 infection[96]. Besides, these patients have poor nutritional status-related anemia and hypo-proteinemia that impair their immunity, rendering them more vulnerable to severe disease[96].
A prospective nationwide cohort study in China on cancer patients, including HCC patents and COVID-19, was more vulnerable to severe disease. They had an increased chance of mortality and ICU admission. Recent chemotherapy within a month also increased the risk of severe disease[147]. Most HCC and CLD patients fall into a high-risk group and are expected to have worse outcomes[114]. Vigorous screening for COVID-19 disease should be prescribed in cancer patients undergoing antitumor therapy, and medications that induce suppression of immunity should be stopped, or their dosages decreased in the event of COVID-19[148].
Hepatitis B and COVID-19
For patients with chronic hepatitis B and are in the immune tolerance phase, further tests are required to confirm whether these patients have active viral replication and repeated liver damage after co-infection with SARS-CoV-2[82]. Several Chinese clinical trials have found patients with hepatitis B infection are also present among patients with severe COVID-19 cases. That suggests that patients infected with HBV tend to have a more severe form of SARS-CoV-2[85,149]. Chen et al[150] found that 47% of patients with HBV are observed with severe COVID-19 cases[150]. However, other studies have shown that chronic viral hepatitis does not seem to be proportional to the severity of COVID-19[115]. Liu et al[151] retrospective study reported that COVID-19 was not substantially affected by co-infection with HBV. However, at the onset of COVID-19, patients co-infected with SARS-CoV-2 and HBV showed more severe monocytopenia, thrombocytopenia, hypoalbuminemia, and hepatic deficiency in lipid metabolism[151]. SARS-CoV-2 and HBV co-infection produces lymphopenia that can cause HBV reactivation. However, it has been reported just in one case of a patient with potential HBV reactivation, which seems to be an unusual event[152]. Chinese medical association and Chinese society of hepatology indicated that patients with hepatitis B administrated antiviral therapy, discontinuation of anti-HBV drugs, or failure to receive anti-HBV treatment can lead to the reactivation of HBV, especially during SRSA-CoV-2 infection following high-dose hormone therapy[82].
Hepatitis C and COVID-19
HCV infection is a dominant factor for liver diseases, including liver cirrhosis, hepatocellular carcinoma, and a common liver transplantation cause[153]. All over the world, HCV infection is the primary cause of liver-related mortality and morbidity[154]. Similarly, the symptoms of hepatic impairments are widespread among patients with COVID-19 and HCV[155]. Therefore, the American associations of liver disease evaluated the underline liver diseases' status, e.g., hepatitis A, hepatitis B, and hepatitis C, in COVID-19 patients and found that co-infection between HCV and SARS Cov-2 increased liver enzyme levels, especially in pediatric[112]. Besides, a systematic preprint review by Mirzaie et al[155] suggested that patients with hepatitis B and/or hepatitis C present a high risk of morbidity to COVID-19 and require further investigation to overcome a significant marker of mortality due to co-infection[155]. Mostardeiro et al[156] reported that in a case study patient with hepatitis C and COVID-19 co-morbidity has an undesirable clinical outcome[156]. Correspondingly, a cohort study by Mangia et al[157] has been elucidated that HCV pre-existing associated with high mortality; meanwhile, HCV antibodies suggestive as a protective agent against COVID-19[157].
On the other hand, countries co-operated and adapted to develop a screening model to detect HCV as a hepatitis elimination program[158]. Nowadays, due to the spreading of the COVID-19 pandemic, it extends beyond the direct morbidity and mortality associated with coinfection COVID-19 may act as a barrier to reduce hepatitis C care, result in a decrease in HCV serological testing and identification[159]. Hence, Blach et al[160] based on their mathematical models, predicted scenario of the 1-year delay which resulted in an additional 44800 liver cancers and 72300 deaths from HCV globally by 2030[160]. Thus, rapid HCV testing in the context of SARS-CoV-2 screening programs may be the only solution for achieving the WHO’s 2030 HCV elimination target[161].
Therapeutic approaches for aborting COVID-19 cytokine storm
As mentioned before, SARS-CoV-2 infections may cause cytokine storm to result in hyperactivation of T lymphocytes and release massive amounts of cytokines such as IL-6, IL-1β, TNF-α, and others[162]. Neutralization of these cytokines offer an excellent therapeutic avenue in combating the COVID-19 outbreak. Here, several therapeutic options can be used for terminating SARS-CoV-2-induced cytokine storm.
IL-6 inhibitors
IL-6 acts as a critical catalyst during cytokine storm, specifically during COVID-19[163]. Hence, IL-6 may be the main accused acute phase response, including fever, the elevation of CRP and ferritinemia, ARDS, multiorgan damage in cases of severe COVID-19[90,164]. Anti-IL-6 biologics targeted the IL-6 receptor or IL-6 itself with anti-inflammatory properties[44]. So, anti-IL-6 agents like tocilizumab, siltuximab, and sarilumab are all humanized monoclonal antibodies that are generated to the IL-6 receptor[165,166]. Currently, tocilizumab (TCZ) is used in active rheumatoid arthritis, juvenile idiopathic arthritis, autoimmune rheumatic diseases[165,167], temporal arteritis, and giant-cell arteritis[168]. Recently, TCZ is used to attenuate the adverse effects of the cytokine storm-induced by chimeric antigen receptor T cell treatment[169,170]. Giving TCZ a potential therapeutic option for critically ill patients with COVID-19 who have a significant elevation in the level of IL-6[171]. However, the treatment of COVID-19 with TCZ is an off-label use[172]. However, based on existing evidence, it could be a right and safe choice to compete with cytokine storm during COVID-19. A retrospective study by Nasonov and Samsonov[172] indicated that tocilizumab directly enhances clinical outcomes in severe and critical COVID-19 patients, no noticeable adverse reactions were observed, and it is effectively reducing mortality[172]. Another retrospective study by Luo et al[173] reported that in COVID-19 patients with a risk of cytokine storm, TCZ is an excellent treatment choice. They are also recommended a repeat dose of TCZ for severe COVID-19 patients with elevated IL-6[173]. Because of its common uses in rheumatoid arthritis and other auto-inflammatory disorders, TCZ hepatic side effect is well known. Typically mild elevations of serum aminotransferase are observed[174]. In contrast, TCZ may be correlated with HBV reactivation. Hence, HBV serological test could be a part of routine pretreatment work-up[175].
IL-1 inhibitors
IL-1 is an active pro-inflammatory cytokine secreted during cytokine storm[49]. It can lower pain thresholds, on the other hand it is one of the cytokines playing a dominant role in tissue damage[176,177]. The IL-1 receptor antagonist, anakinra, canakinumab, and Rilonacept are already indicated in conditions characterized by sustained fevers and systemic inflammatory response such as rheumatoid arthritis, familial Mediterranean fever, and cryopyrin-associated periodic syndrome[169]. Anakinra is the first biologic recombinant IL-1R antagonist. Anakinra inhibits both IL-1α and IL-1β via competitively IL-1R binding[178]. As previously reported, anakinra is effective in attenuating cytokine storm[169,178]. These promising results suggest the role of anakinra in combating CS in patients with COVID-19[179]. A retrospective cohort study by Huet et al[180] showed that in patients with COVID-19, anakinra decreased both the need for invasive mechanical ventilation in the ICU and mortality rate without significant side effects[180]. Anakinra is not a hepatotoxic agent but is more likely to indirectly trigger acute liver damage through activity against IL-1 or the immune system. Moreover, it has not been related to hepatitis B reactivation[181].
TNF-α inhibitors
The viral spike protein of SARS-Cov2 induced a TNF-α converting enzyme-dependent shedding of the ACE2 ectodomain, facilitating viral penetration into cells and promoting tissue injury via increased TNF-α production[90,182]. TNF-α is a crucial cytokine produced in all inflammatory conditions, and autoimmune diseases stimulate inflammation and oxidative stress production. TNF-α is mainly produced by monocytes, macrophages, B cells, and other tissues[93,183]. Activation of TNF-α increases IL-1 and IL-6 release[49]. TNF-α inhibitors such as adalimumab, etanercept, and infliximab are used to manage several conditions like rheumatoid arthritis, ankylosing spondylitis, psoriatic arthritis, and inflammatory bowel disease[88,169]. Consequently, attenuate IL-6, IL-1, and VEGF levels as well as adhesion molecules and angiogenic factors. In patients with COVID-19, both plasma and tissues show an excessive amount of TNF-α[184]. Therefore, the anti-TNF-α antibody markedly reduces TNF-α in the blood, indicating that anti-TNF-α antibody could have potential anti-inflammatory benefits during COVID-19[185]. Additionally, it may cause downregulation of ACE2 expression and shedding[186,187]. So far, in treating patients with COVID-19, TNF-α blockers have not been proposed, but TNF-α blockers' effectiveness in treating patients with COVID-19 needs more priority[188,189].
JAK/STAT inhibitors
The JAK family and the adaptor-associated protein kinase 1 (AAK1) is a member of the numb-associated kinase family that plays a role in viral particles endocytosis, which act as a regulator of clathrin-mediated endocytosis[190,191]. Inhibitors of AAK1 can prevent viral particles entry into the cell[40,50]. Phosphorylated cytokine receptors recruit STAT transcription factors that modulate gene transcription. Furthermore, inhibiting activated cytokine receptors' phosphorylation through Janus kinase inhibitors, which could suppress cytokine signaling pathways[165]. Thus, AAK1 inhibitors have been suggested as possible candidates for the treatment of COVID-19. By inhibiting the JAK-STAT pathway, and cellular viral entry in COVID-19[192,193]. Janus kinase inhibitors such as baricitinib, filgotinib, fostamatinib, peficitinib, tofacitinib, and upadacitinib are previously indicated in managing the treatment of rheumatoid arthritis and autoimmune diseases[194,195]. Baricitinib is a JAK inhibitor with an extreme affinity to AAK1-binding[190]. Therefore, it inhibits the JAK-STAT pathway, that is used to suppress pro-inflammatory cytokine and attenuate the risk of cytokine storm[191,196]. Baricitinib is supposed to have dual effects in COVID 19 patients via reducing viral cell entry and inflammation[190]. One consideration related to the use of Baricitinib could be superior to other JAK-STAT signaling inhibitors, as it is relatively safe[190,197]. However, baricitinib is in harmony with increased thromboembolic events, which affect the risk of developing these events in patients with COVID-19[198]. However, it is also probable that the JAK inhibitors might affect the activity of numerous cytokines, e.g., INF-α, a potent mediator of antiviral response, may be impaired by JAK inhibitors[199]. Data by Bronte et al[200] suggests that baricitinib prevented the progression of COVID-19 by regulating patients' immune response, accordingly improving clinical outcomes[200].
Corticosteroids
Corticosteroids have a broad spectrum of cellular immune responses, different pharmacological activity, and wide therapeutic applications[201,202]. Glucocorticoids can inhibit innate and adaptive immune responses which are significantly used in inflammatory conditions and autoimmune disorders[203]. These anti-inflammatory and immunosuppressive effects occur in different mechanisms through direct actions on gene expression, transcription factors, and glucocorticoids' receptors' second messenger cascades[203]. Corticosteroids receptor complex induces gene transcription of many anti-inflammatory genes, involving IκB, which have immunosuppressive effects by inhibiting the activation of NF-κB signaling resulting in downregulation of IL-1β, IL-4, IL-10, IL-13, GM-CSF, TNF-α, and transforming growth factor-β[165,204,205].
Additionally, Corticosteroids reduce T cells and macrophages proliferation, activation, differentiation, and survival. Corticosteroids diminishes Th1 and macrophage pro-inflammatory cytokines IL-1β, IL-2, IL-6, TNF-α, and IL-17[206,207]. The uses of corticosteroids suppress the immune response, reduce viral clearance, and provoke viral replication[202]. Corticosteroids are widely used in critically ill patients with SARS and MERS infections[44]. Evidence has shown that corticosteroids' delayed their viral clearance[201]. The controversy on the use of corticosteroids remains far from definitive in COVID-19 patients. Corticosteroids decrease the pro-inflammatory cytokine's transcription, thus avoiding prolonged cytokine reaction and cytokine storm[208]. It allows corticosteroids in low doses and short duration and specific conditions with COVID-19 and requiring respiratory support the wat to survive[209]. In patients with COVID-19, trials indicated that dexamethasone in low dose, i.e., 6 mg once daily orally for ten days reduced the death rate by one-third in patients who undergo ventilation and one-fifth in patients with oxygen supplementation[210]. Today, dexamethasone is widely used in severe COVID-19, including in patients with pre-existing CLD[211].
CONCLUSION
The outbreak of COVID-19 disease is the most annoying problem all over the world. This situation was developed due to a mutant version of the SARS virus called the SARS-CoV-2 virus. Studying the virus structure, mode of infection, and pathogenesis mechanism provided excellent hints about how we can fight it. In this review, we concluded that the most destructive power of the virus is the generation of violent cytokine storm, which is probably the cause of high mortality rates in hepatic and non-hepatic patients. Moreover, this review concluded that the possible ways to combat aggressive cytokine chaos might be a potential therapy for the COVID-19 patient co-morbid with liver disorders. Finally, this review encourages the scientists to study the SARS-CoV-2 virus cytokine storm in deep to find more effective and promising therapy for COVID-19 illness.
Footnotes
Conflict-of-interest statement: No potential conflicts of interest and financial support for this work.
Manuscript source: Invited manuscript
Peer-review started: January 27, 2021
First decision: February 10, 2021
Article in press: March 29, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Egypt
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pawlowska M S-Editor: Fan JR L-Editor: A P-Editor: Liu JH
Contributor Information
Fares E M Ali, Department of Pharmacology and Toxicology, Faculty of Pharmacy, Al-Azhar University, Assiut Branch, Assiut 71524, Egypt. faresali@azhar.edu.eg.
Zuhair M Mohammedsaleh, Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, University of Tabuk, Tabuk 71491, Saudi Arabia.
Mahmoud M Ali, Pre-graduated students, Faculty of Pharmacy, Al-Azhar University, Assiut Branch, Assiut 71524, Egypt.
Osama M Ghogar, Pre-graduated students, Faculty of Pharmacy, Al-Azhar University, Assiut Branch, Assiut 71524, Egypt.
References
- 1.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J. SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat. Int J Biol Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Páramo JA. [Coagulopathy and thrombosis: similarities and differences among pathogenic coronaviruses] An Sist Sanit Navar. 2020;43:245–249. doi: 10.23938/ASSN.0885. [DOI] [PubMed] [Google Scholar]
- 4.Zhong NS, Zheng BJ, Li YM, Poon , Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris , Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Morens DM, Fauci AS. Emerging Pandemic Diseases: How We Got to COVID-19. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singhal T. A Review of Coronavirus Disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang X, Du R, Wang R, Cao T, Guan L, Shi H. Comparison of hospitalized patients with acute respiratory distress syndrome caused by covid-19 and H1N1. Chest. 2020:158. doi: 10.1016/j.chest.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn JY, Sohn Y, Lee SH, Cho Y, Hyun JH, Baek YJ, Jeong SJ, Kim JH, Ku NS, Yeom JS, Roh J, Ahn MY, Chin BS, Kim YS, Lee H, Yong D, Kim HO, Kim S, Choi JY. Use of Convalescent Plasma Therapy in Two COVID-19 Patients with Acute Respiratory Distress Syndrome in Korea. J Korean Med Sci . 2020;35:e149. doi: 10.3346/jkms.2020.35.e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Q, Huang D, Ou P, Yu H, Zhu Z, Xia Z, Su Y, Ma Z, Zhang Y, Li Z, He Q, Liu L, Fu Y, Chen J. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 15.Fani M, Teimoori A, Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol . 2020:15. [Google Scholar]
- 16.Segreto R, Deigin Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: SARS-CoV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. Bioessays. 2021;43:e2000240. doi: 10.1002/bies.202000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun P, Lu X, Xu C, Sun W, Pan B. Understanding of COVID-19 based on current evidence. J Med Virol. 2020;92:548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan S, Siddique R, Shereen MA, Ali A, Liu J, Bai Q, Bashir N, Xue M. Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: Biology and Therapeutic Options. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Hoeven B, Oudshoorn D, Koster AJ, Snijder EJ, Kikkert M, Bárcena M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen FS. How Viruses Invade Cells. Biophys J. 2016;110:1028–1032. doi: 10.1016/j.bpj.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, Li T, Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serafim RB, Póvoa P, Souza-Dantas V, Kalil AC, Salluh JIF. Clinical course and outcomes of critically ill patients with COVID-19 infection: a systematic review. Clin Microbiol Infect. 2021;27:47–54. doi: 10.1016/j.cmi.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparks MA, South A, Welling P, Luther JM, Cohen J, Byrd JB, Burrell LM, Batlle D, Tomlinson L, Bhalla V, Rheault MN, Soler MJ, Swaminathan S, Hiremath S. Sound Science before Quick Judgement Regarding RAS Blockade in COVID-19. Clin J Am Soc Nephrol. 2020;15:714–716. doi: 10.2215/CJN.03530320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Feng C, Zhang Z, Wang L, Peng L, Chen L, Qin Y, Zhao D, Tan S, Yin L, Xu J, Zhou C, Jiang C, Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chai X, Hu L, Zhang Y, Han W, Lu Z, Ke A, Zhou J, Shi G, Fang N, Fan J, Cai J, Lan F. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. 2020 Preprint. Available from: bioRxiv:2020.2002.2003.931766.
- 39.Kumar P, Sharma M, Kulkarni A, Rao PN. Pathogenesis of Liver Injury in Coronavirus Disease 2019. J Clin Exp Hepatol. 2020;10:641–642. doi: 10.1016/j.jceh.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salamanna F, Maglio M, Landini MP, Fini M. Body Localization of ACE-2: On the Trail of the Keyhole of SARS-CoV-2. Front Med (Lausanne) 2020;7:594495. doi: 10.3389/fmed.2020.594495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, Lou Y, Gao D, Yang L, He D, Wang MH. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrido I, Liberal R, Macedo G. Review article: COVID-19 and liver disease-what we know on 1st May 2020. Aliment Pharmacol Ther. 2020;52:267–275. doi: 10.1111/apt.15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalaiyarasu S, Kumar M, Senthil Kumar D, Bhatia S, Dash SK, Bhat S, Khetan RK, Nagarajan S. Highly pathogenic avian influenza H5N1 virus induces cytokine dysregulation with suppressed maturation of chicken monocyte-derived dendritic cells. Microbiol Immunol. 2016;60:687–693. doi: 10.1111/1348-0421.12443. [DOI] [PubMed] [Google Scholar]
- 46.Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, Chan JFW, Woo PCY, Yuen KY. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 47.Woo PC, Tung ET, Chan KH, Lau CC, Lau SK, Yuen KY. Cytokine profiles induced by the novel swine-origin influenza A/H1N1 virus: implications for treatment strategies. J Infect Dis. 2010;201:346–353. doi: 10.1086/649785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herold T, Jurinovic V, Arnreich C, Hellmuth JC, von Bergwelt-Baildon M, Klein M, Weinberger T. Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. 2020 Preprint. Available from: medRxiv:2020.2004.2001.20047381.
- 54.Miao Y, Fan L, Li JY. Potential Treatments for COVID-19 Related Cytokine Storm - Beyond Corticosteroids. Front Immunol. 2020;11:1445. doi: 10.3389/fimmu.2020.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, Penninger JM, Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Border WA, Noble NA. Interactions of transforming growth factor-beta and angiotensin II in renal fibrosis. Hypertension . 1998;31:181–188. doi: 10.1161/01.hyp.31.1.181. [DOI] [PubMed] [Google Scholar]
- 58.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos RA, Simoes e Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, Heringer-Walther S, Pinheiro SV, Lopes MT, Bader M, Mendes EP, Lemos VS, Campagnole-Santos MJ, Schultheiss HP, Speth R, Walther T. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci USA. 2003;100:8258–8263. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui CC, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 62.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 63.Dalan R, Bornstein SR, El-Armouche A, Rodionov RN, Markov A, Wielockx B, Beuschlein F, Boehm BO. The ACE-2 in COVID-19: Foe or Friend? Horm Metab Res. 2020;52:257–263. doi: 10.1055/a-1155-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simões e Silva AC, Silveira KD, Ferreira AJ, Teixeira MM. ACE2, angiotensin-(1-7) and Mas receptor axis in inflammation and fibrosis. Br J Pharmacol. 2013;169:477–492. doi: 10.1111/bph.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tikellis C, Thomas MC. Angiotensin-Converting Enzyme 2 (ACE2) Is a Key Modulator of the Renin Angiotensin System in Health and Disease. Int J Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fyhrquist F, Saijonmaa O. Renin-angiotensin system revisited. J Intern Med. 2008;264:224–236. doi: 10.1111/j.1365-2796.2008.01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mancini L, Quinzi V, Mummolo S, Marzo G, Marchetti EJAS. Angiotensin-Converting Enzyme 2 as a Possible Correlation between COVID-19 and Periodontal Disease. Appl Sci . 2020;10:6224. [Google Scholar]
- 68.Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I. COVID-19 cytokine storm: The anger of inflammation. Cytokine. 2020;133:155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol. 2007;292:C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 70.Iwasaki M, Saito J, Zhao H, Sakamoto A, Hirota K, Ma D. Inflammation Triggered by SARS-CoV-2 and ACE2 Augment Drives Multiple Organ Failure of Severe COVID-19: Molecular Mechanisms and Implications. Inflammation. 2021;44:13–34. doi: 10.1007/s10753-020-01337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ji Y, Liu J, Wang Z, Liu N. Angiotensin II induces inflammatory response partly via toll-like receptor 4-dependent signaling pathway in vascular smooth muscle cells. Cell Physiol Biochem. 2009;23:265–276. doi: 10.1159/000218173. [DOI] [PubMed] [Google Scholar]
- 72.Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egyp J Med Hum Genet. 2020;21:54. doi: 10.1186/s43042-020-00099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mustafa MI, Abdelmoneim AH, Mahmoud EM, Makhawi AM. Cytokine Storm in COVID-19 Patients, Its Impact on Organs and Potential Treatment by QTY Code-Designed Detergent-Free Chemokine Receptors. Mediators Inflamm. 2020;2020:8198963. doi: 10.1155/2020/8198963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saba L, Gerosa C, Fanni D, Marongiu F, La Nasa G, Caocci G, Barcellona D, Balestrieri A, Coghe F, Orru G, Coni P, Piras M, Ledda F, Suri JS, Ronchi A, D'Andrea F, Cau R, Castagnola M, Faa G. Molecular pathways triggered by COVID-19 in different organs: ACE2 receptor-expressing cells under attack? Eur Rev Med Pharmacol Sci . 2020;24:12609–12622. doi: 10.26355/eurrev_202012_24058. [DOI] [PubMed] [Google Scholar]
- 76.Hirano T, Murakami M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020;18:1561–1566. doi: 10.1016/j.cgh.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amraei R, Rahimi N. COVID-19, Renin-Angiotensin System and Endothelial Dysfunction. Cells. 2020;9 doi: 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Medina-Enríquez MM, Lopez-León S, Carlos-Escalante JA, Aponte-Torres Z, Cuapio A, Wegman-Ostrosky T. ACE2: the molecular doorway to SARS-CoV-2. Cell Biosci. 2020;10:148. doi: 10.1186/s13578-020-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sansoè G, Aragno M, Wong F. Pathways of hepatic and renal damage through non-classical activation of the renin-angiotensin system in chronic liver disease. Liver Int. 2020;40:18–31. doi: 10.1111/liv.14272. [DOI] [PubMed] [Google Scholar]
- 81.Liu C, Jiang ZC, Shao CX, Zhang HG, Yue HM, Chen ZH, Ma BY, Liu WY, Huang HH, Yang J, Wang Y, Liu HY, Xu D, Wang JT, Yang JY, Pan HQ, Zou SQ, Li FJ, Lei JQ, Li X, He Q, Gu Y, Qi XL. [Preliminary study of the relationship between novel coronavirus pneumonia and liver function damage: a multicenter study] Zhonghua Gan Zang Bing Za Zhi. 2020;28:107–111. doi: 10.3760/cma.j.issn.1007-3418.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian D, Ye Q. Hepatic complications of COVID-19 and its treatment. J Med Virol. 2020;92:1818–1824. doi: 10.1002/jmv.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sahin TT, Akbulut S, Yilmaz S. COVID-19 pandemic: Its impact on liver disease and liver transplantation. World J Gastroenterol. 2020;26:2987–2999. doi: 10.3748/wjg.v26.i22.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273, Table of Contents. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iqbal MS, Sardar N, Akmal W, Sultan R, Abdullah H, Qindeel M, Dhama K, Bila M. Role of toll-like receptors in coronavirus infection and immune response. J Exp Biol Agric Sci . 2020;8:S66–S78. [Google Scholar]
- 88.Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jamilloux Y, Henry T, Belot A, Viel S, Fauter M, El Jammal T, Walzer T, François B, Sève P. Should we stimulate or suppress immune responses in COVID-19? Autoimmun Rev. 2020;19:102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iannaccone G, Scacciavillani R, Del Buono MG, Camilli M, Ronco C, Lavie CJ, Abbate A, Crea F, Massetti M, Aspromonte N. Weathering the Cytokine Storm in COVID-19: Therapeutic Implications. Cardiorenal Med. 2020;10:277–287. doi: 10.1159/000509483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franchi L, Warner N, Viani K, Nuñez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 93.Kany S, Vollrath JT, Relja B. Cytokines in Inflammatory Disease. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marchingo JM, Sinclair LV, Howden AJ, Cantrell DA. Quantitative analysis of how Myc controls T cell proteomes and metabolic pathways during T cell activation. Elife. 2020;9 doi: 10.7554/eLife.53725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. J Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Metawea MI, Yousif WI, Moheb I. COVID 19 and liver: An A-Z literature review. Dig Liver Dis. 2021;53:146–152. doi: 10.1016/j.dld.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Dong L, Cui X, Miao Y, Wang D, Dong J, Xiao C, Chen W, Wang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parvez MK. COVID-19 and coronaviral hepatitis: evidence of collateral damage. Future Virol . 2020;15:325–329. [Google Scholar]
- 100.Mellors J, Tipton T, Longet S, Carroll M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front Immunol. 2020;11:1450. doi: 10.3389/fimmu.2020.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hawlisch H, Belkaid Y, Baelder R, Hildeman D, Gerard C, Köhl J. C5a negatively regulates toll-like receptor 4-induced immune responses. Immunity. 2005;22:415–426. doi: 10.1016/j.immuni.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 102.Zhang X, Kimura Y, Fang C, Zhou L, Sfyroera G, Lambris JD, Wetsel RA, Miwa T, Song WC. Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood. 2007;110:228–236. doi: 10.1182/blood-2006-12-063636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laudisi F, Spreafico R, Evrard M, Hughes TR, Mandriani B, Kandasamy M, Morgan BP, Sivasankar B, Mortellaro A. Cutting edge: the NLRP3 inflammasome links complement-mediated inflammation and IL-1β release. J Immunol. 2013;191:1006–1010. doi: 10.4049/jimmunol.1300489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Noris M, Benigni A, Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98:314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vlaar APJ, de Bruin S, Busch M, Timmermans SAMEG, van Zeggeren IE, Koning R, Ter Horst L, Bulle EB, van Baarle FEHP, van de Poll MCG, Kemper EM, van der Horst ICC, Schultz MJ, Horn J, Paulus F, Bos LD, Wiersinga WJ, Witzenrath M, Rueckinger S, Pilz K, Brouwer MC, Guo RF, Heunks L, van Paassen P, Riedemann NC, van de Beek D. Anti-C5a antibody IFX-1 (vilobelimab) treatment vs best supportive care for patients with severe COVID-19 (PANAMO): an exploratory, open-label, phase 2 randomised controlled trial. Lancet Rheumatol. 2020;2:e764–e773. doi: 10.1016/S2665-9913(20)30341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Annane D, Heming N, Grimaldi-Bensouda L, Frémeaux-Bacchi V, Vigan M, Roux AL, Marchal A, Michelon H, Rottman M, Moine P Garches COVID 19 Collaborative Group. Eculizumab as an emergency treatment for adult patients with severe COVID-19 in the intensive care unit: A proof-of-concept study. EClinicalMedicine. 2020;28:100590. doi: 10.1016/j.eclinm.2020.100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kudaravalli P, Saleem SA, Ibeche B, John S. Case series and review of liver dysfunction in covid-19 patients. Eur J Gastroenterol Hepat. 2020 doi: 10.1097/MEG.0000000000001806. [DOI] [PubMed] [Google Scholar]
- 108.Alqahtani JS, Oyelade T, Aldhahir AM, Alghamdi SM, Almehmadi M, Alqahtani AS, Quaderi S, Mandal S, Hurst JR. Prevalence, Severity and Mortality associated with COPD and Smoking in patients with COVID-19: A Rapid Systematic Review and Meta-Analysis. PLoS One. 2020;15:e0233147. doi: 10.1371/journal.pone.0233147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moon AM, Webb GJ, Aloman C, Armstrong MJ, Cargill T, Dhanasekaran R, Genescà J, Gill US, James TW, Jones PD, Marshall A, Mells G, Perumalswami PV, Qi X, Su F, Ufere NN, Barnes E, Barritt AS, Marjot T. High mortality rates for SARS-CoV-2 infection in patients with pre-existing chronic liver disease and cirrhosis: Preliminary results from an international registry. J Hepatol. 2020;73:705–708. doi: 10.1016/j.jhep.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bonnel AR, Bunchorntavakul C, Reddy KR. Immune dysfunction and infections in patients with cirrhosis. Clin Gastroenterol Hepatol. 2011;9:727–738. doi: 10.1016/j.cgh.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 111.Lippi G, de Oliveira MHS, Henry BM. Chronic liver disease is not associated with severity or mortality in Coronavirus disease 2019 (COVID-19): a pooled analysis. Eur J Gastroenterol Hepatol. 2021;33:114–115. doi: 10.1097/MEG.0000000000001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fix OK, Hameed B, Fontana RJ, Kwok RM, McGuire BM, Mulligan DC, Pratt DS, Russo MW, Schilsky ML, Verna EC, Loomba R, Cohen DE, Bezerra JA, Reddy KR, Chung RT. Clinical Best Practice Advice for Hepatology and Liver Transplant Providers During the COVID-19 Pandemic: AASLD Expert Panel Consensus Statement. Hepatology. 2020;72:287–304. doi: 10.1002/hep.31281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D, Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231–1240. doi: 10.1016/j.jhep.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.APASL Covid-19 Task Force, Lau G, Sharma M. Clinical practice guidance for hepatology and liver transplant providers during the COVID-19 pandemic: APASL expert panel consensus recommendations. Hepatol Int. 2020;14:415–428. doi: 10.1007/s12072-020-10054-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai Q, Huang D, Yu H, Zhu Z, Xia Z, Su Y, Li Z, Zhou G, Gou J, Qu J, Sun Y, Liu Y, He Q, Chen J, Liu L, Xu L. COVID-19: Abnormal liver function tests. J Hepatol. 2020;73:566–574. doi: 10.1016/j.jhep.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li L, Li S, Xu M, Yu P, Zheng S, Duan Z, Liu J, Chen Y, Li J. Risk factors related to hepatic injury in patients with corona virus disease 2019. 2020 Preprint. Available from: medRxiv: 2020.2002.2028.20028514.
- 119.Banales JM, Huebert RC, Karlsen T, Strazzabosco M, LaRusso NF, Gores GJ. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol. 2019;16:269–281. doi: 10.1038/s41575-019-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020;40:998–1004. doi: 10.1111/liv.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Testino G, Pellicano R. Alcohol consumption in the COVID-19 era. Minerva Gastroenterol Dietol. 2020;66:90–92. doi: 10.23736/S1121-421X.20.02698-7. [DOI] [PubMed] [Google Scholar]
- 122.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garnier-Crussard A, Forestier E, Gilbert T, Krolak-Salmon P. Novel Coronavirus (COVID-19) Epidemic: What Are the Risks for Older Patients? J Am Geriatr Soc. 2020;68:939–940. doi: 10.1111/jgs.16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal Liver Tests in COVID-19: A Retrospective Observational Cohort Study of 1,827 Patients in a Major U.S. Hospital Network. Hepatology. 2020;72:1169–1176. doi: 10.1002/hep.31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gu J, Han B, Wang J. COVID-19: Gastrointestinal Manifestations and Potential Fecal-Oral Transmission. Gastroenterology. 2020;158:1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nardo AD, Schneeweiss-Gleixner M, Bakail M, Dixon ED, Lax SF, Trauner M. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2021;41:20–32. doi: 10.1111/liv.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alqahtani SA, Schattenberg JM. Liver injury in COVID-19: The current evidence. United European Gastroenterol J. 2020;8:509–519. doi: 10.1177/2050640620924157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghoda A, Ghoda M. Liver Injury in COVID-19 Infection: A Systematic Review. Cureus. 2020;12:e9487. doi: 10.7759/cureus.9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chand N, Sanyal AJ. Sepsis-induced cholestasis. Hepatology. 2007;45:230–241. doi: 10.1002/hep.21480. [DOI] [PubMed] [Google Scholar]
- 131.Kassir R. Risk of COVID-19 for patients with obesity. Obes Rev. 2020;21:e13034. doi: 10.1111/obr.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bourgeois C, Gorwood J, Barrail-Tran A, Lagathu C, Capeau J, Desjardins D, Le Grand R, Damouche A, Béréziat V, Lambotte O. Specific Biological Features of Adipose Tissue, and Their Impact on HIV Persistence. Front Microbiol. 2019;10:2837. doi: 10.3389/fmicb.2019.02837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ji D, Qin E, Xu J, Zhang D, Cheng G, Wang Y, Lau G. Non-alcoholic fatty liver diseases in patients with COVID-19: A retrospective study. J Hepatol. 2020;73:451–453. doi: 10.1016/j.jhep.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Prins GH, Olinga P. Potential implications of COVID-19 in non-alcoholic fatty liver disease. Liver Int. 2020;40:2568. doi: 10.1111/liv.14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM, Angus PW. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54:1790–1796. doi: 10.1136/gut.2004.062398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boeckmans J, Rodrigues RM, Demuyser T, Piérard D, Vanhaecke T, Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol. 2020;94:1367–1369. doi: 10.1007/s00204-020-02734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hashemi N, Viveiros K, Redd WD, Zhou JC, McCarty TR, Bazarbashi AN, Hathorn KE, Wong D, Njie C, Shen L, Chan WW. Impact of chronic liver disease on outcomes of hospitalized patients with COVID-19: A multicentre United States experience. Liver Int. 2020;40:2515–2521. doi: 10.1111/liv.14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saviano A, Wrensch F, Ghany MG, Baumert TF. Liver disease and COVID-19: from Pathogenesis to Clinical Care. Hepatology. 2020 doi: 10.1002/hep.31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kushner T, Cafardi J. Chronic Liver Disease and COVID-19: Alcohol Use Disorder/Alcohol-Associated Liver Disease, Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis, Autoimmune Liver Disease, and Compensated Cirrhosis. Clin Liver Dis (Hoboken) 2020;15:195–199. doi: 10.1002/cld.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lei HY, Ding YH, Nie K, Dong YM, Xu JH, Yang ML, Liu MQ, Wei L, Nasser MI, Xu LY, Zhu P, Zhao MY. Potential effects of SARS-CoV-2 on the gastrointestinal tract and liver. Biomed Pharmacother. 2021;133:111064. doi: 10.1016/j.biopha.2020.111064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marjot T, Moon AM, Cook JA, Abd-Elsalam S, Aloman C, Armstrong MJ, Pose E, Brenner EJ, Cargill T, Catana MA, Dhanasekaran R, Eshraghian A, García-Juárez I, Gill US, Jones PD, Kennedy J, Marshall A, Matthews C, Mells G, Mercer C, Perumalswami PV, Avitabile E, Qi X, Su F, Ufere NN, Wong YJ, Zheng MH, Barnes E, Barritt AS 4th, Webb GJ. Outcomes following SARS-CoV-2 infection in patients with chronic liver disease: An international registry study. J Hepatol . 2021;74:567–577. doi: 10.1016/j.jhep.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao F, Huang ZM. The impact of COVID-19 on the clinical outcome of patients with cirrhosis deserves more attention and research. J Hepatol. 2020;73:1568–1569. doi: 10.1016/j.jhep.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarin SK, Choudhury A, Lau GK, Zheng MH, Ji D, Abd-Elsalam S, Hwang J, Qi X, Cua IH, Suh JI, Park JG, Putcharoen O, Kaewdech A, Piratvisuth T, Treeprasertsuk S, Park S, Wejnaruemarn S, Payawal DA, Baatarkhuu O, Ahn SH, Yeo CD, Alonzo UR, Chinbayar T, Loho IM, Yokosuka O, Jafri W, Tan S, Soo LI, Tanwandee T, Gani R, Anand L, Esmail ES, Khalaf M, Alam S, Lin CY, Chuang WL, Soin AS, Garg HK, Kalista K, Batsukh B, Purnomo HD, Dara VP, Rathi P, Al Mahtab M, Shukla A, Sharma MK, Omata M APASL COVID Task Force. APASL COVID Liver Injury Spectrum Study (APCOLIS Study- NCT 04345640) Pre-existing liver disease is associated with poor outcome in patients with SARS CoV2 infection; The APCOLIS Study (APASL COVID-19 Liver Injury Spectrum Study) Hepatol Int. 2020;14:690–700. doi: 10.1007/s12072-020-10072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bajaj JS, Garcia-Tsao G, Biggins SW, Kamath PS, Wong F, McGeorge S, Shaw J, Pearson M, Chew M, Fagan A, de la Rosa Rodriguez R, Worthington J, Olofson A, Weir V, Trisolini C, Dwyer S, Reddy KR. Comparison of mortality risk in patients with cirrhosis and COVID-19 compared with patients with cirrhosis alone and COVID-19 alone: multicentre matched cohort. Gut. 2021;70:531–536. doi: 10.1136/gutjnl-2020-322118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Derosa L, Melenotte C, Griscelli F, Gachot B, Marabelle A, Kroemer G, Zitvogel L. The immuno-oncological challenge of COVID-19. Nat Cancer. 2020;1:946–964. doi: 10.1038/s43018-020-00122-3. [DOI] [PubMed] [Google Scholar]
- 146.Chan SL, Kudo M. Impacts of COVID-19 on Liver Cancers: During and after the Pandemic. Liver Cancer. 2020;9:491–502. doi: 10.1159/000510765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, Li C, Ai Q, Lu W, Liang H, Li S, He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jyotsana N, King MR. The Impact of COVID-19 on Cancer Risk and Treatment. Cell Mol Bioeng. 2020:1–7. doi: 10.1007/s12195-020-00630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen X, Jiang Q, Ma Z, Ling J, Hu W, Cao Q, Mo P, Yao L, Yang R, Gao S, Gui X, Hou W, Xiong Y, Li J, Zhang Y. Clinical Characteristics of Hospitalized Patients with SARS-CoV-2 and Hepatitis B Virus Co-infection. Virol Sin. 2020;35:842–845. doi: 10.1007/s12250-020-00276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu R, Zhao L, Cheng X, Han H, Li C, Li D, Liu A, Gao G, Zhou F, Liu F, Jiang Y, Zhu C, Xia Y. Clinical characteristics of COVID-19 patients with hepatitis B virus infection - a retrospective study. Liver Int . 2021;41:720–730. doi: 10.1111/liv.14774. [DOI] [PubMed] [Google Scholar]
- 152.Aldhaleei WA, Alnuaimi A, Bhagavathula AS. COVID-19 Induced Hepatitis B Virus Reactivation: A Novel Case From the United Arab Emirates. Cureus. 2020;12:e8645. doi: 10.7759/cureus.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pokorska-Śpiewak M, Śpiewak M. Management of hepatitis C in children and adolescents during COVID-19 pandemic. World J Hepatol. 2020;12:485–492. doi: 10.4254/wjh.v12.i8.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Indolfi G, Easterbrook P, Dusheiko G, El-Sayed MH, Jonas MM, Thorne C, Bulterys M, Siberry G, Walsh N, Chang MH, Meyers T, Giaquinto C, Wirth S, Chan PL, Penazzato M. Hepatitis C virus infection in children and adolescents. Lancet Gastroenterol Hepatol. 2019;4:477–487. doi: 10.1016/S2468-1253(19)30046-9. [DOI] [PubMed] [Google Scholar]
- 155.Mirzaie H, Vahidi M, Shokoohi M, Darvishian M, Sharifi H, Sharafi H, Karamouzian M. COVID-19 among patients with hepatitis B or hepatitis C: A systematic review. 2020 Preprint. Available from: medRxiv: 2020.2010.2022.20216317.
- 156.Mostardeiro LR, Antoniolli ECA, Xavier JW. Coronavirus in a patient with hepatitis C: case report. J Bras Patolo Med Lab. 2020:56. [Google Scholar]
- 157.Mangia A, Cenderello G, Verucchi G, Ciancio A, Fontana A, Piazzolla V, Minerva N, Squillante MM, Copetti M. Is positivity for hepatitis C virus antibody predictive of lower risk of death in COVID-19 patients with cirrhosis? World J Clin Cases. 2020;8:5831–5834. doi: 10.12998/wjcc.v8.i22.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Waked I, Esmat G, Elsharkawy A, El-Serafy M, Abdel-Razek W, Ghalab R, Elshishiney G, Salah A, Abdel Megid S, Kabil K, El-Sayed MH, Dabbous H, El Shazly Y, Abo Sliman M, Abou Hashem K, Abdel Gawad S, El Nahas N, El Sobky A, El Sonbaty S, El Tabakh H, Emad E, Gemeah H, Hashem A, Hassany M, Hefnawy N, Hemida AN, Khadary A, Labib K, Mahmoud F, Mamoun S, Marei T, Mekky S, Meshref A, Othman A, Ragab O, Ramadan E, Rehan A, Saad T, Saeed R, Sharshar M, Shawky H, Shawky M, Shehata W, Soror H, Taha M, Talha M, Tealaab A, Zein M, Hashish A, Cordie A, Omar Y, Kamal E, Ammar I, AbdAlla M, El Akel W, Doss W, Zaid H. Screening and Treatment Program to Eliminate Hepatitis C in Egypt. N Engl J Med. 2020;382:1166–1174. doi: 10.1056/NEJMsr1912628. [DOI] [PubMed] [Google Scholar]
- 159.Sperring H, Ruiz-Mercado G, Schechter-Perkins EM. Impact of the 2020 COVID-19 Pandemic on Ambulatory Hepatitis C Testing. J Prim Care Community Health. 2020;11:2150132720969554. doi: 10.1177/2150132720969554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Blach S, Kondili LA, Aghemo A, Cai Z, Dugan E, Estes C, Gamkrelidze I, Ma S, Pawlotsky JM, Razavi-Shearer D, Razavi H, Waked I, Zeuzem S, Craxi A. Impact of COVID-19 on global HCV elimination efforts. J Hepatol. 2021;74:31–36. doi: 10.1016/j.jhep.2020.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Giacomelli A, Pagani G, Conti F, Bassoli C, Galli M. Detecting HCV infection by means of mass population SARS-CoV-2 screening: a pilot experience in Northern Italy. J Hepatol. 2021 doi: 10.1016/j.jhep.2020.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Misra DP, Agarwal V, Gasparyan AY, Zimba O. Rheumatologists' perspective on coronavirus disease 19 (COVID-19) and potential therapeutic targets. Clin Rheumatol . 2020;39:2055–2062. doi: 10.1007/s10067-020-05073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alijotas-Reig J, Esteve-Valverde E, Belizna C, Selva-O'Callaghan A, Pardos-Gea J, Quintana A, Mekinian A, Anunciacion-Llunell A, Miró-Mur F. Immunomodulatory therapy for the management of severe COVID-19. Beyond the anti-viral therapy: A comprehensive review. Autoimmun Rev. 2020;19:102569. doi: 10.1016/j.autrev.2020.102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rossi-Semerano L, Hermeziu B, Fabre M, Koné-Paut I. Macrophage activation syndrome revealing familial Mediterranean fever. Arthritis Care Res (Hoboken) 2011;63:780–783. doi: 10.1002/acr.20418. [DOI] [PubMed] [Google Scholar]
- 165.Burrage DR, Koushesh S, Sofat N. Immunomodulatory Drugs in the Management of SARS-CoV-2. Front Immunol. 2020;11:1844. doi: 10.3389/fimmu.2020.01844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lotfi M, Hamblin MR, Rezaei N. COVID-19: Transmission, prevention, and potential therapeutic opportunities. Clin Chim Acta. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ding C, Jones G. Anti-interleukin-6 receptor antibody treatment in inflammatory autoimmune diseases. Rev Recent Clin Trials. 2006;1:193–200. doi: 10.2174/157488706778250168. [DOI] [PubMed] [Google Scholar]
- 168.Rouse BT, Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol. 2010;10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rizk JG, Kalantar-Zadeh K, Mehra MR, Lavie CJ, Rizk Y, Forthal DN. Pharmaco-Immunomodulatory Therapy in COVID-19. Drugs . 2020;80:1267–1292. doi: 10.1007/s40265-020-01367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dholaria BR, Bachmeier CA, Locke F. Mechanisms and Management of Chimeric Antigen Receptor T-Cell Therapy-Related Toxicities. BioDrugs. 2019;33:45–60. doi: 10.1007/s40259-018-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Atal S, Fatima Z. IL-6 Inhibitors in the Treatment of Serious COVID-19: A Promising Therapy? Pharmaceut Med. 2020;34:223–231. doi: 10.1007/s40290-020-00342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nasonov E, Samsonov M. The role of Interleukin 6 inhibitors in therapy of severe COVID-19. Biomed Pharmacother. 2020;131:110698. doi: 10.1016/j.biopha.2020.110698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Luo P, Liu Y, Qiu L, Liu X, Liu D, Li J. Tocilizumab treatment in COVID-19: A single center experience. J Med Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Boettler T, Marjot T, Newsome PN, Mondelli MU, Maticic M, Cordero E, Jalan R, Moreau R, Cornberg M, Berg T. Impact of COVID-19 on the care of patients with liver disease: EASL-ESCMID position paper after 6 mo of the pandemic. JHEP Rep. 2020;2:100169. doi: 10.1016/j.jhepr.2020.100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chen LF, Mo YQ, Jing J, Ma JD, Zheng DH, Dai L. Short-course tocilizumab increases risk of hepatitis B virus reactivation in patients with rheumatoid arthritis: a prospective clinical observation. Int J Rheum Dis. 2017;20:859–869. doi: 10.1111/1756-185X.13010. [DOI] [PubMed] [Google Scholar]
- 176.Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, Antinori S, Galli M. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin Exp Rheumatol. 2020;38:337–342. [PubMed] [Google Scholar]
- 177.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, Yan KW, Chan KH, Tsang NC, Guan Y, Yuen KY, Peiris JS. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Monteagudo LA, Boothby A, Gertner E. Continuous Intravenous Anakinra Infusion to Calm the Cytokine Storm in Macrophage Activation Syndrome. ACR Open Rheumatol. 2020;2:276–282. doi: 10.1002/acr2.11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Langer-Gould A, Smith JB, Gonzales EG, Castillo RD, Figueroa JG, Ramanathan A, Li BH, Gould MK. Early identification of COVID-19 cytokine storm and treatment with anakinra or tocilizumab. Int J Infect Dis. 2020;99:291–297. doi: 10.1016/j.ijid.2020.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huet T, Beaussier H, Voisin O, Jouveshomme S, Dauriat G, Lazareth I, Sacco E, Naccache JM, Bézie Y, Laplanche S, Le Berre A, Le Pavec J, Salmeron S, Emmerich J, Mourad JJ, Chatellier G, Hayem G. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet] Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases, 2012. [Google Scholar]
- 182.Brojakowska A, Narula J, Shimony R, Bander J. Clinical Implications of SARS-CoV-2 Interaction With Renin Angiotensin System: JACC Review Topic of the Week. J Am Coll Cardiol. 2020;75:3085–3095. doi: 10.1016/j.jacc.2020.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lefebvre AL, McAuliffe L. Targeted Immunomodulatory Therapy: An Overview. R I Med J (2013) 2016;99:19–22. [PubMed] [Google Scholar]
- 184.Fara A, Mitrev Z, Rosalia RA, Assas BM. Cytokine storm and COVID-19: a chronicle of pro-inflammatory cytokines. Open Biol. 2020;10:200160. doi: 10.1098/rsob.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mendoza VMM. Interleukin-17: A potential therapeutic target in COVID-19. J Infect. 2020;81:e136–e138. doi: 10.1016/j.jinf.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Sasazuki T, Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natl Acad Sci USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mahase E. Covid-19: what treatments are being investigated? BMJ. 2020;368:m1252. doi: 10.1136/bmj.m1252. [DOI] [PubMed] [Google Scholar]
- 188.Feldmann M, Maini RN, Woody JN, Holgate ST, Winter G, Rowland M, Richards D, Hussell T. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Robinson PC, Richards D, Tanner HL, Feldmann M. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2:e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, Phelan A, Rawling M, Savory E, Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stebbing J, Phelan A, Griffin I, Tucker C, Oechsle O, Smith D, Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Richardson PJ, Corbellino M, Stebbing J. Baricitinib for COVID-19: a suitable treatment? Lancet Infect Dis. 2020;20:1013–1014. doi: 10.1016/S1473-3099(20)30270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15:23. doi: 10.1186/s12964-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sepriano A, Kerschbaumer A, Smolen JS, van der Heijde D, Dougados M, van Vollenhoven R, McInnes IB, Bijlsma JW, Burmester GR, de Wit M, Falzon L, Landewé R. Safety of synthetic and biological DMARDs: a systematic literature review informing the 2019 update of the EULAR recommendations for the management of rheumatoid arthritis. Ann Rheum Dis. 2020;79:760–770. doi: 10.1136/annrheumdis-2019-216653. [DOI] [PubMed] [Google Scholar]
- 195.Zhang J, Xie B, Hashimoto K. Current status of potential therapeutic candidates for the COVID-19 crisis. Brain Behav Immun. 2020;87:59–73. doi: 10.1016/j.bbi.2020.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jamilloux Y, El Jammal T, Vuitton L, Gerfaud-Valentin M, Kerever S, Sève P. JAK inhibitors for the treatment of autoimmune and inflammatory diseases. Autoimmun Rev. 2019;18:102390. doi: 10.1016/j.autrev.2019.102390. [DOI] [PubMed] [Google Scholar]
- 197.Tanner T, Wahezi DM. Hyperinflammation and the utility of immunomodulatory medications in children with COVID-19. Paediatr Respir Rev. 2020;35:81–87. doi: 10.1016/j.prrv.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jorgensen SCJ, Tse CLY, Burry L, Dresser LD. Baricitinib: A Review of Pharmacology, Safety, and Emerging Clinical Experience in COVID-19. Pharmacotherapy. 2020;40:843–856. doi: 10.1002/phar.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Favalli EG, Biggioggero M, Maioli G, Caporali R. Baricitinib for COVID-19: a suitable treatment? Lancet Infect Dis. 2020;20:1012–1013. doi: 10.1016/S1473-3099(20)30262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bronte V, Ugel S, Tinazzi E, Vella A, De Sanctis F, Canè S, Batani V, Trovato R, Fiore A, Petrova V, Hofer F, Barouni RM, Musiu C, Caligola S, Pinton L, Torroni L, Polati E, Donadello K, Friso S, Pizzolo F, Iezzi M, Facciotti F, Pelicci PG, Righetti D, Bazzoni P, Rampudda M, Comel A, Mosaner W, Lunardi C, Olivieri O. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J Clin Invest. 2020;130:6409–6416. doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tufan A, Avanoğlu Güler A, Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci. 2020;50:620–632. doi: 10.3906/sag-2004-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vandewalle J, Luypaert A, De Bosscher K, Libert C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol Metab. 2018;29:42–54. doi: 10.1016/j.tem.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 204.Ristimäki A, Narko K, Hla T. Down-regulation of cytokine-induced cyclo-oxygenase-2 transcript isoforms by dexamethasone: evidence for post-transcriptional regulation. Biochem J. 1996;318 (Pt 1):325–331. doi: 10.1042/bj3180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Almawi WY, Melemedjian OK. Negative regulation of nuclear factor-kappaB activation and function by glucocorticoids. J Mol Endocrinol. 2002;28:69–78. doi: 10.1677/jme.0.0280069. [DOI] [PubMed] [Google Scholar]
- 206.Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol Cell Endocrinol. 2011;335:2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Franchimont D. Overview of the actions of glucocorticoids on the immune response: a good model to characterize new pathways of immunosuppression for new treatment strategies. Ann N Y Acad Sci. 2004;1024:124–137. doi: 10.1196/annals.1321.009. [DOI] [PubMed] [Google Scholar]
- 208.Montón C, Ewig S, Torres A, El-Ebiary M, Filella X, Rañó A, Xaubet A. Role of glucocorticoids on inflammatory response in nonimmunosuppressed patients with pneumonia: a pilot study. Eur Respir J. 1999;14:218–220. doi: 10.1034/j.1399-3003.1999.14a37.x. [DOI] [PubMed] [Google Scholar]
- 209.Sharun K, Tiwari R, Dhama J, Dhama K. Dexamethasone to combat cytokine storm in COVID-19: Clinical trials and preliminary evidence. Int J Surg. 2020;82:179–181. doi: 10.1016/j.ijsu.2020.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mahase E. Covid-19: Low dose steroid cuts death in ventilated patients by one third, trial finds. BMJ. 2020;369:m2422. doi: 10.1136/bmj.m2422. [DOI] [PubMed] [Google Scholar]
- 211.Boettler T, Newsome PN, Mondelli MU, Maticic M, Cordero E, Cornberg M, Berg T. Care of patients with liver disease during the COVID-19 pandemic: EASL-ESCMID position paper. JHEP Rep. 2020;2:100113. doi: 10.1016/j.jhepr.2020.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sachdeva S, Khandait H, Kopel J, Aloysius MM, Desai R, Goyal H. NAFLD and COVID-19: a Pooled Analysis. SN Compr Clin Med. : 2020: 1–4. doi: 10.1007/s42399-020-00631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhang L, Zhu F, Xie L, Wang C, Wang J, Chen R, Jia P, Guan HQ, Peng L, Chen Y, Peng P, Zhang P, Chu Q, Shen Q, Wang Y, Xu SY, Zhao JP, Zhou M. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann Oncol. 2020;31:894–901. doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed] [Google Scholar]